Summary
Transposable elements are parasitic genomic elements that can be deleterious for host gene function and genome integrity. Heterochromatic histone modifications are involved in the repression of transposons. However, it remains unknown how these histone modifications mark different types of transposons during embryonic development. Here we document the variety of heterochromatic epigenetic signatures at parasitic elements during development in Xenopus tropicalis, using genome-wide ChIP-sequencing data and ChIP-qPCR analysis. We show that specific subsets of transposons in various families and subfamilies are marked by different combinations of the heterochromatic histone modifications H4K20me3, H3K9me2/3 and H3K27me3. Many DNA transposons are marked at the blastula stage already, whereas at retrotransposons the histone modifications generally accumulate at the gastrula stage or later. Furthermore, transposons marked by H3K9me3 and H4K20me3 are more prominent in gene deserts. Using intra-subfamily divergence as a proxy for age, we show that relatively young DNA transposons are preferentially marked by early embryonic H4K20me3 and H3K27me3. In contrast, relatively young retrotransposons are marked by increasing H3K9me3 and H4K20me3 during development, and are also linked to piRNA-sized small non-coding RNAs. Our results implicate distinct repression mechanisms that operate in a transposon-selective and developmental stage-specific fashion.
Keywords: Transposons, H4K20me3, H3K9me2/3, H3K27me3, piRNA, evolution
Introduction
Large parts of the human, mouse and frog genomes (resp. 46%, 37% and 35%) consist of repetitive and transposable elements (TEs) (Lander et al. 2001; Waterston et al. 2002; Hellsten et al. 2010). Different TEs can be distinguished based on their evolutionary origin and functional properties (Koonin et al. 2015; Muñoz-López & García-Pérez 2010). DNA transposons and retrotransposons represent two main classes with different replication strategies. DNA transposons move through the genome via cut and paste or rolling cycle mechanisms, while retrotransposons require RNA intermediates for a copy-paste mode of amplification. Depending on the transcriptional regulatory elements that they are equipped with, retrotransposons are divided further into subclasses: long terminal repeats (LTR), long interspersed nuclear elements (LINE) and short interspersed nuclear elements (SINE). Finally, within a subclass TEs are grouped into unique families and subfamilies based on their evolutionary origin (Bao et al. 2015). Xenopus does contain all main repeat families that are found in mammals. However, whereas most TEs in mammals fall in the retrotransposon class, 70% of all TEs in Xenopus are DNA transposons (Hellsten et al. 2010).
It has been shown in human, mouse and frogs that TEs are actively and dynamically transcribed during early embryogenesis (Grau et al. 2014; Levin & Moran 2011). TEs contain functional elements, such as enhancers, promoters, polyadenylation signals, insulators and transcription factor binding sites. As a consequence TE spreading has had a major influence on host evolution (Friedli & Trono 2015). However, TEs also form a threat for the integrity of the host genome, since insertion can cause loss-of-function mutations. Insertion sites of retro- and DNA transposons in the Xenopus genome negatively correlate with the location of exons (Shen et al. 2013). This implies that TEs are more often found outside than inside coding regions, since gene disruption has a negative influence of survival of the zygote. Besides the loss-of-function mutations, also the introduction of cis-regulatory elements upon TE transposition can perturb host gene regulation (Friedli & Trono 2015). Therefore hosts have developed repressive defense mechanisms that restrain TE proliferation to some extent.
Various epigenetic modifications are involved in transcriptional repression. Besides DNA methylation (mainly on CpG dinucleotides, meCG), multiple histone modifications are involved in epigenetic silencing, such as histone H3 di- and trimethylation of lysine K9 (H3K9me2/3), histone H3 trimethylation of lysine K27 (H3K27me3) and histone H4 trimethylation of lysine K20 (H4K20me3) (Lu et al. 2008; Fischle et al. 2003; Schultz et al. 2002; Jacobs & Khorasanizadeh 2002). Whereas chromatin decorated with H3K9me2/3 and H4K20me3 is often referred to as constitutive heterochromatin, chromatin with H3K27me3 is known as facultative heterochromatin, because it is found on genic regions in a cell type-specific manner (Trojer & Reinberg 2007). The repressive histone modifications can mediate transcriptional repression by chromosomal condensation via the recruitment of effector proteins such as Heterochromatin protein 1 (HP1) (Jacobs & Khorasanizadeh 2002; Lu et al. 2008; Schultz et al. 2002; van Kruijsbergen et al. 2015).
DNA methylation (meCG) is necessary for repressing distinct TEs in committed cells. For example, in mouse fibroblasts repression of LINE and ERV retrotransposons is dependent on meCG (Bulut-Karslioglu et al. 2014), but repression of Alu SINE retrotransposons rather depends on H3K9me3 (Varshney et al. 2015). While needed in differentiated cells, meCG is dispensable for repression of LINE and ERV in mouse embryonic stem (ES) cells (Bulut-Karslioglu et al. 2014; Matsui et al. 2010; Karimi et al. 2011; Hutnick et al. 2010; Martens et al. 2005).
A substantial part of TEs in pre-gastrula E6.25 mouse embryos is marked with H3K9me2 and/or H3K27me3 (Zylicz et al. 2015). However, less than 1% of the marked TEs showed increased expression in mouse embryos and ES cells lacking the G9a and EZH2 methyltransferases responsible for these modifications (Dong et al. 2008; Leeb et al. 2010; Maksakova et al. 2013; Zylicz et al. 2015). In contrast, the transcription of TEs marked with H3K9me3 increased in mouse ES cells deficient for the H3K9me3 methyltransferases SETDB1 or SUV39H1,2 (Wolf & Goff 2009; Matsui et al. 2010; Karimi et al. 2011; Bulut-Karslioglu et al. 2014). H4K20me3 was found to function downstream of H3K9me3 in silencing retrotransposons (Matsui et al. 2010). However, H4K20me3 can also function independently of H3K9me3, as occurs at DNA transposon family Charlie in mouse ES cells and at the retrotransposon family IAP in quiescent cells (Bierhoff et al. 2014; Martens et al. 2005). All together these studies have demonstrated that heterochromatic histone modifications are needed to restrain TEs.
Targeting of histone modifiers to TEs can be achieved via small non-coding RNAs such as piRNAs. piRNAs are derived from the transcribed RNA intermediates of retrotransposons, which are bound by Argonaute proteins (Vagin et al. 2006; Kalmykova et al. 2005). These Argonaute protein-containing complexes interact with H3K9 methyltransferases (SUV39 and SETDB) and are targeted to genomic DNA by the piRNAs (Klenov et al. 2011; Wang & Elgin 2011; Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013; Sienski et al. 2015). A second mechanism to recruit histone modifiers involves DNA-binding zinc-finger proteins. The zinc-finger protein KRAB-ZFP interacts with TRIM28/KAP1 and the H3K9 methyltransferase SETDB1/ESET (Wolf & Goff 2009; Frietze et al. 2010; Rowe et al. 2010). A third way to recruit histone modifiers involves transcription factors. H3K9me2 catalyzing enzymes G9a/GLP are guided to their genomic targets via interaction with DNA-binding proteins, such as REST and SNAIL1 (Dong et al. 2012; Roopra et al. 2004).
piRNA and KAP1 driven recruitment mechanisms change over evolutionary time (Castro-Diaz et al. 2014; Pezic et al. 2014). piRNA dependent H3K9me3 deposition occurs at full length LINEs, but not at degraded LINEs in germ cells (Pezic et al. 2014). Zinc-finger proteins bind to specific DNA motifs and co-evolve with TEs (Jacobs et al. 2014). KAP1 also binds more often at relatively young subfamilies of LINE L1 in human and mouse ES cells (Castro-Diaz et al. 2014). The youngest LINE L1s, however, are not bound by KAP1 either and are silenced via a meCG dependent mechanism (Castro-Diaz et al. 2014).
It has been shown that H3K9me3 is required for repression of retrotransposons in gastrulating Xenopus embryos (Herberg et al. 2015). Furthermore, also in frogs small RNAs are involved in repression of TEs by mediating deposition of H3K9me3 and H4K20me3 (Faunes et al. 2012; Harding et al. 2014). However, it is not known which TEs are repressed by histone modifications and what the dynamics are during development. Here we report on the histone modifications present on all TEs of different classes and families using genome-wide ChIP-sequencing data and ChIP-qPCR analysis. We show that specific subsets of TEs in different families and subfamilies are marked by different combinations of heterochromatic histone modifications and that these patterns have different developmental dynamics and are more prominent in gene deserts. We show that early embryonic marking of retrotransposons is linked to small RNAs. Using intra-subfamily divergence as a proxy for age, we show that relatively young DNA transposon subfamilies are marked by early embryonic H4K20me3 and H3K27me3, while at relatively young retrotransposon subfamilies H3K9me3 and H4K20me3 accumulate during development. Our study implicates dynamic repression mechanisms that operate in a developmental stage-specific and a TE-selective fashion.
Materials and methods
Animal procedures
Xenopus tropicalis embryos were obtained by in vitro fertilization. Embryos grew at 23°C in 10% Marc’s Modified Ringer’s solution (MMR) (88 mM NaCl; 2 mM KCl; 2 mM CaCl2; 1 mM MgCl2; 5 mM HEPES, pH 7.4) and were dejellied in 10% MMR + 3% cysteine (pH 7.8-8). Animal use was conducted under the DEC permission (Dutch Animal Experimentation Committee) RU-DEC 2012-116 and 2014-122 to G.J.C.V.
ChIP-qPCR
Embryos were fixed in 1% formaldehyde, methanol-free (Thermo Scientific #28906) for 30 minutes. Formaldehyde was quenched with 125 mM glycine in 25% MMR. Embryos were homogenized in a low-salt buffer (20 mM Tris, pH 8; 70 mM KCl; 1 mM EDTA; 10% glycerol; 5 mM DTT; 0.125% Igepal; cOmplete Protease Inhibitor Cocktail (Roche #04693132001)) (300 embryos/2 mL) and sonicated until DNA fragments had a size of 0.2-2 kb. Yolk was removed by spinning it down. For each ChIP, chromatin extract from 15 embryo equivalents was two-fold diluted with IP buffer (50 mM Tris, pH 8; 100 mM NaCl; 2 mM EDTA; 1 mM DTT; 1% Igepal; Complete Protease Inhibitor Cocktail) for overnight incubation with the antibody: anti-H3K9me2 (Diagenode C15410060, 1 μg); anti-H3K9me3 (Abcam ab8898, 2 μg); anti-H4K20me3 (Abcam ab9053, 2 μg). DNA bound by antibody was captured using 1/10 volume of Dynabeads Protein G during a 1 hour incubation. The beads were washed with ChIP1 buffer (IP buffer + 0.1% deoxycholate), ChIP2 buffer (ChIP1 buffer + 400 mM NaCl), ChIP3 buffer (ChIP1 buffer + 250 mM LiCl), ChIP1 buffer and TE buffer (10 mM Tris-Cl, pH 7.5; 1 mM EDTA). Chromatin was eluted from the beads in 0.1 M NaHCO3, pH 8.8 + 1% SDS. NaCl (final 25 mM) and 5 μg proteinase K were added for reversal at 65°C. DNA was purified by phenol:chloroform:isoamyl alcohol (25:24:1) extraction. DNA was precipitated by adding 1/10 volume NaOAc (3M, pH 5.2), 2.5 volumes ethanol and glycogen (2 μg/μL) at −20°C. DNA was washed in 70% ethanol and dissolved in TE. qPCR was carried out with iQ Custom SYBR Green Super mix (BioRad) on a CFX96 Real-Time PCR Detection System (BioRad) using an annealing temperature of 60°C. Primers for qPCR (Supplementary table 5) were designed in Primer3 (v.0.4.0) and purchased from Biolegio (Nijmegen, The Netherlands).
Data analysis
Mapping
We used RepeatMasker (version open-4.0.3) to identify repeats in Xenopus tropicalis genome JGI7.1 using all frog repeats in the included RepBase repeat library (release 20130422). ChIP data (Hontelez et al. 2015) (read length = 42 bp, Supplementary table 6) was mapped to the reference Xenopus tropicalis genome JGI7.1 using BWA version 0.6.1-r104 (Li & Durbin 2009). Duplicates were marked using bamUtil version 1.0.2 (http://genome.sph.umich.edu/wiki/BamUtil). RNA-seq reads (Collart et al. 2014) (Ribozero, 4 hpf & 7 hpf, read length = 60 bp) were mapped with GSNAP (version 2012-07-20) (Wu & Nacu 2010). After adapter clipping with fastx_clipper (part of FASTX Toolkit 0.0.13.2) (http://hannonlab.cshl.edu/fastx_toolkit/), small RNA reads (Harding et al. 2014) were mapped with BWA using standard settings (read length in Fig. 4A). MethylC-seq reads (read length > 105 bp) (Bogdanović et al. 2016) were mapped to in silico bisulfite-converted reference genome JGI7.1 with Bowtie alignment algorithm as described previously (Bogdanović et al. 2016), but with allowing two mismatches in the seed. For this mapping we excluded multi-mappers (reads mapping to multiple locations), after which >90% of all genomic C was covered.
Quantification of reads
Duplicate reads were removed from the ChIP-seq data (samtools view -F 1024). After eliminating duplicate reads (but not multimapped reads) Reads Per Kilobase of transcript per Million mapped reads (RPKM) was calculated for all annotated transposons using peakstats.py (version 2.1) (http://dx.doi.org/10.5281/zenodo.50023). Enrichment for the histone modifications was calculated by dividing the RPKM of ChIP-seq data by the RPKM of an input control track (non-ChIPed, stage 9, all reads excluding duplicates). This division over the input control track corrects for inaccuracies in the genome assembly, as repetitive DNA is hard to assemble into larger contigs. The calculation of enrichment values relative to stage 30 input DNA gave similar results, which is expected because embryos of different stages contain the same genomic DNA (Supplementary Fig. 1).
Peakstats.py (using –remove_dup option to eliminate duplicated reads) was also used to calculate RPKM for RNA and small RNA at each annotated transposon location.
We calculated the fraction meCG ((sum of all Cs)/(sum of all C+Ts)) for each annotated transposon for which C+T coverage > 4 using bedtools map (version 2.20.1), output sum (Quinlan & Hall 2010). Next, subfamily annotation according to Repbase was used to calculate median RPKM (for small RNA), average RPKM (RNA), or median enrichment (over input for ChIP or fraction for meCG) for each transposon subfamily (Supplementary table 1). Subfamily sizes are included in Supplementary table 4: ‘number of sequences in subfamily library’.
Blast alignment
After aligning the Repbase derived Xenopus repeat library to the Xenopus tropicalis genome (assembly JGI7.1) we generated subfamily specific sequence libraries (with a maximum of 1000, evenly distributed along the chromosomes). For each sequence forming part of each of these libraries, we performed a BLAST search against its whole subfamily and summed the result of the obtained bitscore divided by its length . This quantity that scales linearly with the number of homologs of a sequence and its level of similarity serves as an approximation of the recent activity of the gene family.
Generation of plots
Plots were generated in R. PAM clustering of subfamily median enrichment for H4K20me3, H3K9me2/3 and H3K27me3 was done using the package “cluster”, version 2.0.3. Heatmaps of the clustering were visualized with “heatmap.2()” using the “gplots” package, version 2.17.0 (http://cran.r-project.org/web/packages/gplots/index.html). All other plots were generated with the package “ggplot2”, version 2.1.0 (http://ggplot2.org).
Results
Majority of transposons not decorated by repressive histone modifications
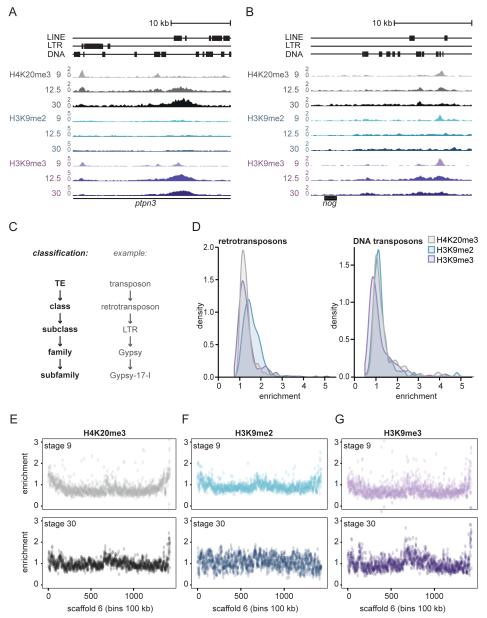
We have generated genome-wide epigenome reference maps, including the constitutive heterochromatin modifications H4K20me3, H3K9me2 and H3K9me3 (Hontelez et al. 2015). These marks were profiled for Xenopus tropicalis embryos in developmental stages 9, 10.5, 12.5, 16 and 30, which represent blastula, early gastrula, late gastrula, neurula and organogenesis. We observed that most enrichment for these heterochromatin modifications is found in the non-unique portion of the genome and that they show different dynamics during development (Fig. 1A, B). We validated peaks sets of the ChIP-sequencing data using ChIP-qPCR, and also checked whether quantitative differences in ChIP-sequencing signals across developmental stages could be reproduced by ChIP-qPCR (Supplementary Fig. 2). The heterochromatin-marked repeats can be found at intronic regions (Fig. 1A), as well as in relatively close proximity to genes, for example downstream of noggin (Fig. 1B). Given that 35% of the X.tropicalis genome consists of TEs and that some TEs obviously are not enriched for heterochromatic histone modifications (Fig. 1A, B), we wondered what the distribution of histone modifications over distinct TEs is.
Fig. 1. A subset of transposons acquires H4K20me3, H3K9me2 or H3K9me3.
A, B) Genome browser views of the first intron of ptpn3 (panel A) and downstream of nog (panel B), showing H4K20me3 (grey), H3K9me2 (blue) and H3K9me3 (purple) signal (RPKM) at TEs in developmental stages 9, 12.5 and 30 (top to bottom). C) TEs can be classified in DNA and retrotransposons, with subclasses, families and subfamilies. D) Distribution of histone modification enrichment at retrotransposon (left) and DNA transposon (right) subfamilies. Median H4K20me3, H3K9me2 and H3K9me3 enrichment over input was calculated for all TE subfamilies. For each subfamily we used the stage (9, 10.5, 12.5, 16 or 30) with maximal enrichment. E, F, G) Chromosome scale enrichment of histone modifications at Scaffold 6. Histone modification RPKM enrichment over input DNA of E) H4K20me3, F) H3K9me2 and G) H3K9me3 was calculated in bins of 100kb. Top: stage 9. Bottom: stage 30.
Transposable elements can be classified into DNA transposons and retrotransposons, with subclasses, families and subfamilies for each (Fig. 1C, Supplementary table 1). In our analysis we included ChIP-sequencing reads that can map to multiple locations to identify the genomic locations of repeats enriched for heterochromatic histone modifications. To identify the TE subfamilies that are enriched for these histone modifications, we calculated the median Reads Per Kilobase per Million mapped reads (RPKM) for each TE subfamily. The enrichment was calculated by dividing the subfamily median RPKM by the median RPKM of the input track (Supplementary table 1, see Methods). Most retro- and DNA transposon subfamilies were not or barely enriched for H4K20me3, H3K9me2 or H3K9me3 (Fig. 1D).
Heterochromatic modifications are expected to be enriched at subtelomeric and pericentric chromatin. This is also observed for most Xenopus chromosome-sized scaffolds, showing moderate enrichment (~1.5-2.5-fold) of H4K20me3 and H3K9me3 throughout development and of H3K9me2 at the blastula stage, in large 100 kb bins of genomic sequence (Fig. 1E, F, G, Supplementary Fig. 3).
Dynamics and co-occurrence
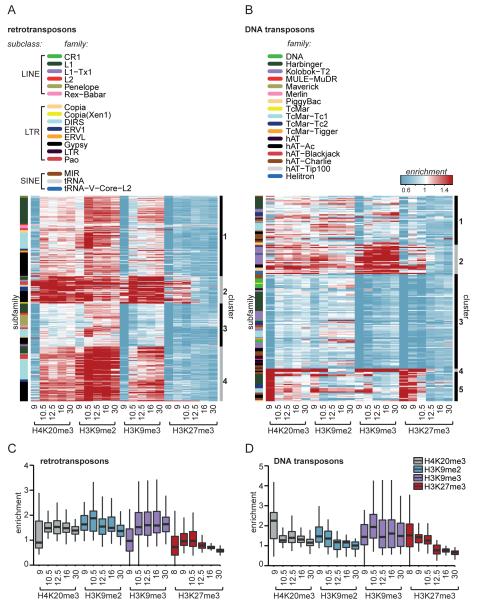
To gain insight into the global dynamics and co-occurrence of histone modifications at TEs, a clustering analysis of retro- and DNA transposon subfamilies was performed, using the subfamily median enrichments for H4K20me3, H3K9me2/3 and H3K27me3 (Fig. 2A, B, Supplementary table 2, 3). Furthermore, we also quantified the behavior of the histone modifications for the most strongly enriched retro- and DNA transposons (>2-fold compared to background) (Fig. 2C, D).
Fig. 2. Variation in repressive histone modification dynamics at retro- and DNA transposons.
A) Retro- and B) DNA transposon subfamilies were clustered (PAM, cluster-bar right of heatmap) based on median enrichment over input for H4K20me3, H3K9me2/3 and H3K27me3 during stages 9, 10.5, 12.5, 16 and 30. The family to which a subfamily belongs is depicted in the left side-bar. C, D) The general dynamics of median enrichment of repressive histone marks were plotted for only enriched (>2-fold over input) C) retro- and D) DNA transposon subfamilies. The upper and lower hinges correspond to the 25th and 75th percentiles and the horizontal line in between represents the median.
The majority of enriched retrotransposons subfamilies gained H3K9me3 and H4K20me3 between blastula (stage 9) and early gastrula (stage 10.5) (Fig. 2A, clusters 1, 2, 4). In contrast, cluster 5 of the DNA transposon subfamilies was strongly enriched mainly for H4K20me3 and H3K27me3 at the blastula stage, but subsequently lost these histone modifications later in development (Fig. 2B). These differences were also reflected in the enrichment of all retrotransposons and DNA transposons that were at least two-fold enriched over input for one of the marks in one stage of development (Fig. 2C, D). Despite the divergent heterochromatic signatures of retro- and DNA transposons, for both TE classes the most strongly enriched clusters also had the lowest transcript levels (Supplementary Fig. 4) (Collart et al. 2014). Their distinct dynamic patterns, which can also be observed at individual loci (Fig. 1A), suggest that retro- and DNA transposons interact differently with the host during the course of embryonic development.
At both retrotransposons and DNA transposons, the levels of H4K20me3 and H3K9me3 were more similar to each other than to H3K9me2 (Fig. 2A, B and Supplementary Fig. 5 and 6). Even though subfamilies decorated with one of the three constitutive heterochromatin marks were often also marked by the other two marks, the strength of the enrichment for the three marks was different. For example, in cluster 1 of retrotransposons H3K9me2 was relatively abundant compared to H4K20me3 and H3K9me3 (Fig. 2A), whereas for the DNA transposons in cluster 2 H3K9me3 marking was relatively strong compared to H4K20me3 and H3K9me2 (Fig. 2B).
Interestingly, the facultative heterochromatin mark H3K27me3 co-occurred with constitutive heterochromatin marks during late blastula and early gastrula (Fig. 2A, B). The correlation of H3K27me3 was highest with H4K20me3 on DNA transposons, but only during these early stages (DNA transposons Spearman rho 0.9 and 0.1, retrotransposons 0.8 and 0.1 at resp. stages 9 and 30). A substantial number of DNA transposon subfamilies was already marked with H3K27me3 at stage 8, before the onset of zygotic genome activation (Fig. 2B), which is different from the H3K27me3 dynamics observed on genic regions, where H3K27me3 starts to accumulate between blastula and gastrula stages (Hontelez et al. 2015; van Heeringen et al. 2014). To examine the occurrence of the other modifications at early blastula stages, we performed ChIP-qPCR for H3K9me2/3 and H4K20me3. At one gypsy and two helitron elements the H3K9me2 modification was found at stage 8, and for one harbinger element H3K9me2/3 and H4K20me3 were already enriched before stage 9; for the most part however, these histone modifications strongly increased between stages 8 and 9 (Supplementary Fig. 7).
Whereas retro- and DNA transposons had different dynamic patterns, the various TE subclasses, families and subfamilies could not be distinguished by heterochromatic histone modification signatures (Fig. 2A, B; subfamily indicated by color code in side-bar). For example, subfamilies belonging to two of the largest retrotransposon families L1 (subclass LINE) and Gypsy (subclass LTR) were assigned to all four clusters (Fig. 2A). Similarly, subfamilies belonging to the same family of DNA transposons were spread over all clusters (Fig. 2B). These data document the variation in heterochromatic histone modifications within TE classes and families.
Gene density and repressive histone modifications
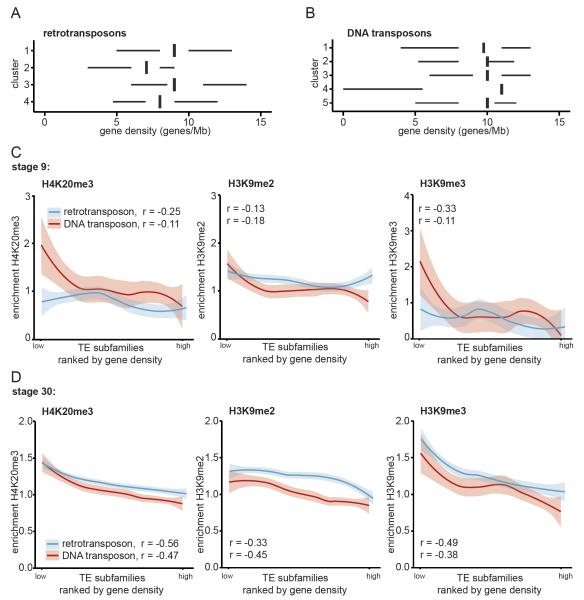
Given the variability in histone marking between TE subfamilies, we wondered whether the genomic environment could be involved in the observed differences. Cis-regulatory elements within TEs can potentially perturb transcription regulation when located near or within the genes (Casa & Gabellini 2012). Hence, regulation of TEs in a gene-dense context might be different from TEs located in gene-poor loci.
Gene densities (genes/Mbp) were calculated for annotated TEs by counting the number of genes within 1 megabase of the centre of the TE. Median gene density was calculated for all TE subfamilies (Supplementary table 1) and subfamilies were grouped according to the clusters of Fig. 2. Strikingly, subfamilies from the retrotransposon clusters (clusters 2 and 4) with the highest enrichment for H4K20me3 and H3K9me3 were located at relatively gene-poor regions (Fig. 3A). The gene densities of subfamilies from the DNA transposon clusters, however, were more similar to each other (Fig. 3B).
Fig. 3. Strongest enrichment for H4K20me3 and H3K9me3 at transposons in gene desert.
For each TE subfamily the median gene density was calculated 500.000 bp up- and downstream of the center of each TE. A) Retro- and B) DNA transposon subfamilies were grouped according to clusters determined in Fig. 2A, B. Median gene densities for the subfamilies were plotted for each cluster. The left and right hinges correspond to the 25th and 75th percentiles and the vertical line in between represents the median. C, D) All TE subfamilies were ranked by gene density and median H4K20me3 (left), H3K9me2 (middle) and H3K9me3 (right) enrichments during stage C) 9 and D) 30 were plotted for retro- (blue) and DNA transposon (red) subfamilies. Loess was used as smoothing method, with a 0.95 confidence interval. Spearman correlation between histone mark and gene density was determined (left corner).
To probe these relationships further, we analysed the correlation between gene density and repressive histone modifications. TE subfamilies were ranked by gene density and the subfamily median enrichment for the histone modifications in stage 9 and 30 were plotted (Fig. 3C, D respectively). At stage 30 gene density and constitutive heterochromatin marks were inversely correlated for both retro- and DNA transposons, which was more pronounced for H4K20me3 and H3K9me3 compared to H3K9me2 (Fig. 3D). Furthermore, these inverse patterns are less evident earlier in development (Fig. 3C). No inverse patterns with gene density could be observed for the facultative heterochromatin mark H3K27me3, the enhancer-binding protein p300, or the active promoter mark H3K4me3 (Supplementary Fig. 8).
Together these results imply that TEs with heterochromatic marks are more likely to be located in in gene deserts, most likely reflecting a TE integration bias (Shen et al. 2013).
H4K20me3, H3K9me3 and small RNAs co-occur at retrotransposons
Upon zygotic genome activation, Polycomb Repressive Complex 2 (PRC2) catalyzes H3K27me3 deposition on genic regions lacking DNA methylation (Hontelez et al. 2015). On the other hand, DNA methylation can stimulate H3K9me3 deposition in genic regions by recruiting histone H3K9 methyltransferase SETDB1 (Matsumura et al. 2015). Small RNAs can also direct deposition of heterochromatin modifications, since it guides protein complexes that recruit H3K9 methyltransferases (Klenov et al. 2011; Wang & Elgin 2011; Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013). We examined to which extent DNA methylation and small RNAs could be involved in depositing heterochromatin modifications at TEs.
We used recently published bisulfite sequencing profiles (Bogdanović et al. 2016) to analyze the DNA methylation status of TEs (Supplementary table 1). Median DNA methylation at stage 9 was calculated for all TE subfamilies, and subfamilies were grouped according to the clusters described in Fig. 2 (Supplementary Fig. 9). High DNA methylation levels were observed for all subfamily clusters (methylation fraction > 0.9). This indicates that DNA methylation status does not distinguish between TEs that do or do not obtain repressive histone modifications. Furthermore, it indicates that H3K27me3 is not deposited according to DNA methylation logic at TEs, in contrast to what has been observed for genic regions (Hontelez et al. 2015).
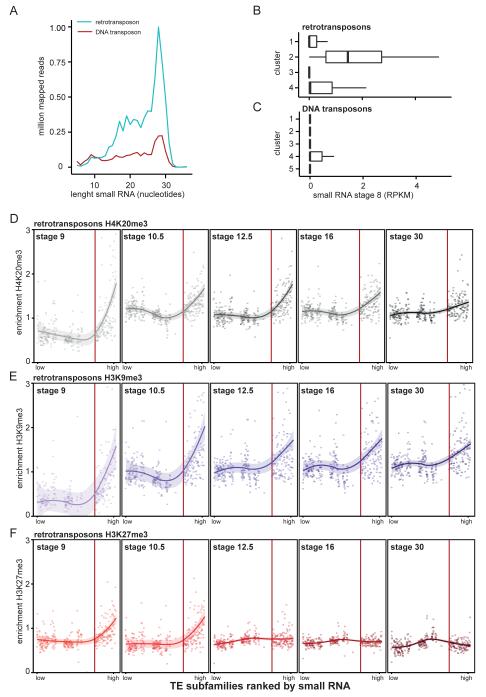
To assess the relationship between small RNAs and histone modifications we used a small RNA data set of stage 8, 10 and 18 embryos, size-selected for a range of 18-30 nucleotides (Harding et al. 2014). Small RNAs with a length of 28-29 nucleotides, most likely piRNA (Lim & Kai 2015), were most abundant among small RNA reads mapping to TEs (Fig. 4A, Supplementary Fig. 10). Approximately five-fold more small RNA reads mapped to retrotransposons than to DNA transposons (Fig. 4A), despite the fact that the sequence coverage of DNA transposons in the collection of repetitive elements exceeds that of retrotransposons by a factor of 2.5. Small RNA reads mainly mapped to approximately a quarter of all TE subfamilies (Supplementary Fig. 11, Supplementary table 1). Although small RNA coverage of TE subfamilies changed to some extent during development, their ranking for small RNA coverage was largely stable (Supplementary Fig. 11, Supplementary table 1).
Fig. 4. Small RNA aligns to retrotransposon subfamilies that obtain H4K20me3, H3K9me3 and H3K27me3 first.
A) Stage 8 small RNA reads that aligned to retro- (blue) and DNA transposons (red) were split based on nucleotide length and counted. B, C) Median small RNA (RPKM stage 8) was calculated for each TE subfamily and plotted according to B) retro- and C) DNA transposon clusters determined in Fig. 2A, B. The left and right hinges correspond to the 25th and 75th percentiles and the vertical line in between represents the median. D, E, F) All TE subfamilies were ranked by amount of small RNA (stage 8) and median D) H4K20me3 E) H3K9me3 and F) H3K27me3 enrichments were visualized for retrotransposons. Histone marks were plotted for stage 9, 10.5, 12.5, 16 and 30 (left to right). The line was plotted using Loess smoothing method, with a 0.95 confidence interval (line shade). All subfamilies left of the red line have a median small RNA RPKM of zero.
Next, we compared the small RNA data with our repressive histone modification profiles. Median small RNA coverage (RPKM in stage 8) was analyzed for the different clusters of TE subfamilies (cf. Fig. 2). Interestingly, small RNAs were most abundant for the retrotransposon cluster with the strongest enrichment for H4K20me3 and H3K9me3 (Fig. 4B, C). Moreover, the clusters with most small RNA transcripts corresponded with the clusters lowest in gene density (Fig. 3A, B, Fig. 4B, C). To probe these relationships further, we analysed the correlation between small RNA and repressive histone modifications. TE subfamilies were sorted based on abundance of small RNA during stage 8. Subsequently subfamily median enrichments for the histone modifications were plotted. At stage 9 H4K20me3 and H3K9me3 were predominantly present at retrotransposon subfamilies that exhibited relatively strong small RNA coverage (Fig. 4D, E, to the right of red line, median RPKM >0). Retrotransposons with less small RNA did obtain these repressive histone modifications later in development (Fig. 4D, E, left of red vertical line). By contrast, the correlations between H4K20me3 or H3K9me3 and small RNA were less pronounced for DNA transposons (Supplementary Fig. 12). Furthermore, the pattern of H3K9me2 enrichment was different from the other modifications as it was not correlated with small RNA (Supplementary Fig. 12, 13).
We found similar trends when comparing small RNA occurrence with H3K27me3 enrichment at TEs in early developmental stages (Fig. 4F, Supplementary Fig. 12). At stage 9, H3K27me3 is predominantly present at retrotransposon subfamilies that exhibit relatively strong small RNA coverage. However, TE subfamilies earliest enriched for H4K20me3 and H3K9me3 remained marked with these modifications, while H3K27me3 diminished during development.
These results show that the presence of small RNA in the embryo is linked to heterochromatic histone modifications at retrotransposons.
Epigenetic variation within TE families derived from age of TE
TE subfamilies that belong to the same TE family do have variable epigenetic signatures (sidebar Fig. 2A, B). It has been reported that both small RNA-dependent and KAP1-dependent recruitment of histone modifying enzymes occur at relatively young TEs (Pezic et al. 2014; Castro-Diaz et al. 2014). We therefore asked if TE age provides an explanation for the variation in epigenetic regulation within TEs families.
For each subfamily we performed a sequence alignment for all individual TEs within the subfamily. Parasitic elements erode by mutations over evolutionary time. Therefore, we expected young TE subfamilies to have higher alignment scores than old subfamilies. We used the alignment bitscore as a proxy for age (Supplementary table 4). Bitscores were corrected for TE fragment length (Supplementary Fig. 14).
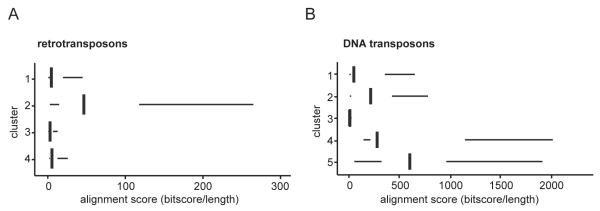
Alignment scores were analyzed for the clusters of TE subfamilies (cf. Fig. 2A, B) (Fig. 5A, B). Among the retrotransposon clusters, the cluster most strongly enriched for H4K20me3 and H3K9me3 (cluster 2) had the highest alignment score (Fig. 5A). This indicates that this cluster contained the youngest retrotransposon subfamilies. Among the DNA transposons, the cluster that increased in H3K9me3 binding (cluster 2) also had a higher alignment score than the clusters that lacked heterochromatic histone modifications (cluster 1, 3) (Fig. 5B). DNA transposon subfamilies with the highest alignment scores were in cluster 5. This indicates that the DNA transposons that lost repressive marks from blastula to tailbud (cluster 5) were younger than the DNA transposons that acquired H3K9me3 during these developmental stages (cluster 2).
Fig. 5. Transposon clusters enriched with H3K9me3, H4K20me3 and H3K27me3 contain relatively young TEs.
A, B) TEs belonging to the same TE subfamily were pairwise aligned to each other using BLAST. The median alignment score was determined for each TE subfamily by dividing the bitscores by the length of the aligned sequences. A) Retro- and B) DNA transposons were grouped according to clusters determined in Fig. 2A, B. The median alignment scores of the subfamilies were plotted for each cluster. The left and right hinges correspond to the 25th and 75th percentiles and the vertical line in between represents the median. The alignment scores of retro- and DNA transposons cannot be directly compared to each other, because of differences in library sizes (Supplementary Fig. 14A, B).
These results indicate that recruitment of histone modifying enzymes occurs at relatively young retro- and DNA transposons. Furthermore, our study implies that different repression mechanisms operate at young relative to older DNA transposons.
Discussion
TEs can form a threat for the integrity of the host genome (Friedli & Trono 2015). To protect host genome integrity, inserted TEs should be tightly regulated during embryogenesis. In most cases the repression of TEs is independent of genes, but in some cases regulatory sequences of protein-coding genes are regulated by heterochromatic histone modifications. For example, H4K20me3 and the Kmt5b and -5c (Suv4-20h1 and -h2) methyl transferases play a role in the repression of the mouse Oct4 (Pou5f1) and Xenopus oct25 (pou5f3.2) gene and the regulated exit from pluripotency during lineage commitment (Nicetto et al. 2013). In some cases TEs and their active repression may have been co-opted to stably repress genes during development. Our study indicates that TE subfamilies acquiring heterochromatic histone modifications are relatively young. Our data documents the variation in heterochromatic histone modifications within TE classes and families. Not only genomic location and evolutionary age are linked to these differences, but also small RNA abundance.
Epigenetic dynamics
Retrotransposons that gained H3K9me3 during Xenopus development also gained H4K20me3. This similar behavior of modifications at retrotransposons is different from what has been observed in mouse embryos, where H3K9me3 appears a few cleavage cycles before H4K20me3 (Burton & Torres-Padilla 2014). Furthermore, H3K9me3 is never completely removed during mouse early development, since de novo H3K9me3 deposition already occurs before parentally inherited H3K9me3 is fully diluted out (Burton & Torres-Padilla 2014). We, however, could barely detect H3K9me3 before zygotic genome activation, which suggests that the mark is being removed more drastically than what has been described for mice. This seems to be true for many histone modifications, which appear to be largely absent before the mid-blastula transition, to be re-established in the blastula or at subsequent stages (Akkers et al. 2009; Hontelez et al. 2015; van Heeringen et al. 2014).
The histone modification differences between frog and mice may originate from the difference in the duration of the embryonic cell division cycles. Before gastrulation, cell cycles in Xenopus are completed in 30 minutes, while mouse embryos reach the two cell stage 24 hours after fertilization (O’Farrell et al. 2004). From work with Drosophila embryos it is known that histone modifications are re-established on newly replicated chromatin with a delay depending on the histone modification (Petruk et al. 2013; Petruk et al. 2012), so it is possible that the developmental acquisition of these modifications depends on cell cycle lengthening.
The dynamics of heterochromatic histone modifications at retro- and DNA transposons are different. While most retrotransposons in our study gained heterochromatic marks, a substantial subset of DNA transposon subfamilies lost them during development. Interestingly, this inversed dynamics at the two TE classes might be linked to the disparate risk of TE amplification. Retrotransposons use RNA intermediates; therefore amplification of retrotransposons is more likely to take place after zygotic genome activation. DNA transposons, however, can only increase in copy number if their transposition occurs during S phase of the cell cycle. This can occur if a transposon cut from one of the newly synthesised daughter strands is pasted into a region that has not replicated yet, resulting in an additional copy in one of the two daughter cells. Therefore, DNA transposons may form a bigger threat for the integrity of the host genome when genome replication occurs more frequently, which is before zygotic genome activation. The repressive histone modification dynamics (Fig. 2A, B) may therefore partly reflect amplification risk at different times of development as well as the activity of the mechanisms that generate the defensive histone modification response.
Recruiting histone modifiers
piRNAs bind to Argonaute complexes and can mediate silencing of TEs by recruiting H3K9 methyltransferases SUV3-9 and SETDB1 (Klenov et al. 2011; Wang & Elgin 2011; Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013). Once H3K9me3 is deposited, HP1 can bind to this mark and recruit SUV4-20, which catalyzes H4K20me3 (Jacobs & Khorasanizadeh 2002). In our study 28 bp long small RNAs mostly aligned to retrotransposon clusters (cluster 2 and 4) which were also marked by H3K9me3 and H4K20me3. Therefore, it is likely that during Xenopus embryogenesis piRNAs are involved in guiding H3K9 and H4K20 methyltransferases to retrotransposons.
In contrast to the H3K9me3-enriched retrotransposons, the DNA transposon cluster (cluster 2) that gained substantial H3K9me3 marking during development was not enriched for small RNAs. This suggests that DNA transposons recruit H3K9 methyltransferases in a piRNA independent way, for example via KRAB domain-containing zinc-finger proteins, which bind TRIM28/KAP1 and SETDB1 (Frietze et al. 2010).
We found another intriguing set of DNA transposon subfamilies (cluster 5) that was heavily marked with H4K20me3 independent of H3K9me3 deposition. We could not identify a link between small RNAs and these TEs. At ERVs in mouse ES cells, H3K9me3 binding by HP1 protein was important for spreading of H4K20me3, but was dispensable for initial H4K20me3 deposition (Maksakova et al. 2011). So, piRNA and H3K9me3-HP1 independent recruitment mechanisms can guide H4K20 methyltransferases to TEs, for example by long non-coding RNA. Recently, it was shown that SUV4-20H2 is recruited to IAP retrotransposons by long non-coding RNA in quiescent and terminally differentiated cells (Bierhoff et al. 2014). This process was also independent from H3K9me3 and HP1.
Retrotransposons minimally enriched for H3K9me3 often did obtain H3K9me2. SUV3-9 (KMT1A) can catalyze both methylation forms, whereas methyltransferase G9a (EHMT2) can catalyze dimethylation, but not trimethylation of H3K9 (Rice et al. 2003). Therefore, recruitment of G9a rather than Suv3-9 could result in selective dimethylation of H3K9. Furthermore, the low correlation between piRNA and H3K9me2 indicates that it is unlikely that piRNA-dependent mechanisms (involving Suv3-9 or Setdb1) are involved in the recruitment of H3K9 di-methyltransferases. Additionally, H3K9me2-mediated TE silencing can also be achieved independent of KAP1 at MERVLs in mouse ES cells (Maksakova et al. 2013). Instead of piRNA or KAP1-driven mechanisms, DNA-binding proteins might be involved in guiding H3K9 dimethyltransferases towards TEs. It has been shown, for example, that REST, SNAIL1 and JARID2 recruit G9a/GLP (Shirato et al. 2009; Roopra et al. 2004; Dong et al. 2012).
Concluding remarks
Overall, our study shows that epigenetic regulation is variable between TE subfamilies. The epigenetic variation we described can very well rely on different strategies to recruit histone modifiers. Therefore it will be interesting to decipher the interplay of the factors involved in different recruitment mechanisms like piRNA, KAP1 and DNA-binding proteins.
Supplementary Material
Highlights.
H4K20me3 and H3K27me3 mark specific DNA transposons during early development
Retrotransposon subfamilies acquire H3K9me3 and H4K20me3 during development
Retrotransposon subfamilies decorated with H3K9me3 and H4K20me3 generate small RNA
Relatively young retro- and DNA transposon obtain heterochromatic histone marks
Acknowledgements
We thank Ozren Bogdanović for his help with the bisulfite-seq analysis. We thank Georgios Georgiou for bioinformatics support. This work has been supported by the US National Institutes of Health (NICHD, grant R01HD069344). S.J.v.H. was supported by the Netherlands Organization for Scientific Research (NWO-ALW) [863.12.002].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akkers RC, et al. A hierarchy of H3K4me3 and H3K2me3 acquisition in spatial gene regulation in Xenopus embryos. Developmental Cell. 2009;17(3):425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. [Accessed March 5, 2016];Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 2015 6(1):11. doi: 10.1186/s13100-015-0041-9. Available at: http://mobilednajournal.biomedcentral.com/articles/10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H, et al. [Accessed February 24, 2016];Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Molecular cell. 2014 54(4):675–82. doi: 10.1016/j.molcel.2014.03.032. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24768537. [DOI] [PubMed] [Google Scholar]
- Bogdanović O, et al. [Accessed March 2, 2016];Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nature genetics. 2016 doi: 10.1038/ng.3522. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26928226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, et al. [Accessed January 26, 2016];Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Molecular cell. 2014 55(2):277–90. doi: 10.1016/j.molcel.2014.05.029. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24981170. [DOI] [PubMed] [Google Scholar]
- Burton A, Torres-Padilla M-E. [Accessed February 24, 2016];Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nature reviews. Molecular cell biology. 2014 15(11):723–34. doi: 10.1038/nrm3885. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25303116. [DOI] [PubMed] [Google Scholar]
- Casa V, Gabellini D. [Accessed October 28, 2015];A repetitive elements perspective in Polycomb epigenetics. Frontiers in genetics. 2012 3:199. doi: 10.3389/fgene.2012.00199. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3465993&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Diaz N, et al. [Accessed February 24, 2016];Evolutionally dynamic L1 regulation in embryonic stem cells. Genes & development. 2014 28(13):1397–409. doi: 10.1101/gad.241661.114. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4083085&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, et al. [Accessed April 12, 2016];High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development (Cambridge, England) 2014 141(9):1927–39. doi: 10.1242/dev.102012. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3994770&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, et al. [Accessed February 24, 2016];G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. The Journal of clinical investigation. 2012 122(4):1469–86. doi: 10.1172/JCI57349. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22406531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong KB, et al. [Accessed February 24, 2016];DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. The EMBO journal. 2008 27(20):2691–701. doi: 10.1038/emboj.2008.193. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2572176&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faunes F, et al. [Accessed February 24, 2016];Characterization of small RNAs in Xenopus tropicalis gastrulae. Genesis (New York, N.Y. : 2000) 2012 50(3):260–70. doi: 10.1002/dvg.22012. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22253037. [DOI] [PubMed] [Google Scholar]
- Fischle W, et al. [Accessed January 3, 2015];Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes & development. 2003 17(15):1870–81. doi: 10.1101/gad.1110503. Available at: http://genesdev.cshlp.org/content/17/15/1870.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedli M, Trono D. [Accessed September 30, 2015];The Developmental Control of Transposable Elements and the Evolution of Higher Species. Annual review of cell and developmental biology. 2015 doi: 10.1146/annurev-cellbio-100814-125514. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26393776. [DOI] [PubMed] [Google Scholar]
- Frietze S, et al. [Accessed February 22, 2016];ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PloS one. 2010 5(12):e15082. doi: 10.1371/journal.pone.0015082. Available at: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau JH, et al. [Accessed February 24, 2016];LTR retroelements are intrinsic components of transcriptional networks in frogs. BMC genomics. 2014 15:626. doi: 10.1186/1471-2164-15-626. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4131045&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding JL, et al. [Accessed February 24, 2016];Small RNA profiling of Xenopus embryos reveals novel miRNAs and a new class of small RNAs derived from intronic transposable elements. Genome research. 2014 24(1):96–106. doi: 10.1101/gr.144469.112. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3875865&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeringen SJ, et al. [Accessed December 22, 2014];Principles of nucleation of H3K27 methylation during embryonic development. Genome research. 2014 24(3):401–10. doi: 10.1101/gr.159608.113. Available at: http://genome.cshlp.org/content/24/3/401.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, et al. [Accessed February 23, 2016];The genome of the Western clawed frog Xenopus tropicalis. Science (New York, N.Y.) 2010 328(5978):633–6. doi: 10.1126/science.1183670. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2994648&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberg S, et al. [Accessed September 30, 2015];Histone H3 lysine 9 trimethylation is required for suppressing the expression of an embryonically activated retrotransposon in Xenopus laevis. Scientific Reports. 2015 5:14236. doi: 10.1038/srep14236. Available at: http://www.nature.com/srep/2015/150921/srep14236/full/srep14236.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontelez S, et al. [Accessed December 18, 2015];Embryonic transcription is controlled by maternally defined chromatin state. Nature Communications. 2015 6:10148. doi: 10.1038/ncomms10148. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4703837&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutnick LK, et al. [Accessed February 24, 2016];Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. The Journal of biological chemistry. 2010 285(27):21082–91. doi: 10.1074/jbc.M110.125674. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2898347&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FMJ, et al. [Accessed November 16, 2015];An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014 516(7530):242–5. doi: 10.1038/nature13760. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4268317&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. [Accessed February 21, 2016];Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science (New York, N.Y.) 2002 295(5562):2080–3. doi: 10.1126/science.1069473. Available at: http://science.sciencemag.org.ru.idm.oclc.org/content/295/5562/2080.abstract. [DOI] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA. [Accessed February 24, 2016];Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic acids research. 2005 33(6):2052–9. doi: 10.1093/nar/gki323. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1074743&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, et al. [Accessed January 29, 2016];DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell stem cell. 2011 8(6):676–87. doi: 10.1016/j.stem.2011.04.004. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3857791&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, et al. [Accessed February 24, 2016];Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proceedings of the National Academy of Sciences of the United States of America. 2011 108(46):18760–5. doi: 10.1073/pnas.1106676108. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3219103&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Krupovic M, Yutin N. [Accessed February 24, 2016];Evolution of double-stranded DNA viruses of eukaryotes: from bacteriophages to transposons to giant viruses. Annals of the New York Academy of Sciences. 2015 1341:10–24. doi: 10.1111/nyas.12728. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4405056&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kruijsbergen I, Hontelez S, Veenstra GJC. [Accessed May 30, 2015];Recruiting polycomb to chromatin. The international journal of biochemistry & cell biology. 2015 67:177–87. doi: 10.1016/j.biocel.2015.05.006. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. [Accessed July 10, 2014];Initial sequencing and analysis of the human genome. Nature. 2001 409(6822):860–921. doi: 10.1038/35057062. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11237011. [DOI] [PubMed] [Google Scholar]
- Leeb M, et al. [Accessed February 24, 2016];Polycomb complexes act redundantly to repress genomic repeats and genes. Genes & development. 2010 24(3):265–76. doi: 10.1101/gad.544410. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2811828&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL, Moran JV. [Accessed March 12, 2015];Dynamic interactions between transposable elements and their hosts. Nature Reviews Genetics. 2011 12(9):615–627. doi: 10.1038/nrg3030. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3192332&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. [Accessed July 9, 2014];Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009 25(14):1754–60. doi: 10.1093/bioinformatics/btp324. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2705234&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RSM, Kai T. [Accessed November 17, 2015];A piece of the pi(e): the diverse roles of animal piRNAs and their PIWI partners. Seminars in Cell & Developmental Biology. 2015 47-48:17–31. doi: 10.1016/j.semcdb.2015.10.025. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26582251. [DOI] [PubMed] [Google Scholar]
- Lu X, et al. [Accessed February 20, 2016];The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nature structural & molecular biology. 2008 15(10):1122–4. doi: 10.1038/nsmb.1489. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2648974&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, et al. [Accessed February 24, 2016];Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics & chromatin. 2013 6(1):15. doi: 10.1186/1756-8935-6-15. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3682905&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, et al. [Accessed February 24, 2016];H3K9me3-binding proteins are dispensable for SETDB1/H3K9me3-dependent retroviral silencing. Epigenetics & chromatin. 2011 4(1):12. doi: 10.1186/1756-8935-4-12. Available at: http://epigeneticsandchromatin.biomedcentral.com/articles/10.1186/1756-8935-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JHA, et al. [Accessed February 24, 2016];The profile of repeat-associated histone lysine methylation states in the mouse epigenome. The EMBO journal. 2005 24(4):800–12. doi: 10.1038/sj.emboj.7600545. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=549616&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, et al. [Accessed November 20, 2015];Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010 464(7290):927–31. doi: 10.1038/nature08858. Available at: http://dx.doi.org/10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Matsumura Y, et al. [Accessed March 27, 2016];H3K4/H3K9me3 Bivalent Chromatin Domains Targeted by Lineage-Specific DNA Methylation Pauses Adipocyte Differentiation. Molecular cell. 2015 60(4):584–96. doi: 10.1016/j.molcel.2015.10.025. Available at: http://www.cell.com/article/S1097276515008163/fulltext. [DOI] [PubMed] [Google Scholar]
- Muñoz-López M, García-Pérez JL. [Accessed November 9, 2015];DNA transposons: nature and applications in genomics. Current genomics. 2010 11(2):115–28. doi: 10.2174/138920210790886871. Available at: /pmc/articles/PMC2874221/?report=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicetto D, et al. [Accessed April 14, 2016];Suv4-20h histone methyltransferases promote neuroectodermal differentiation by silencing the pluripotency-associated Oct-25 gene. PLoS genetics. 2013 9(1):e1003188. doi: 10.1371/journal.pgen.1003188. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3561085&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell PH, Stumpff J, Su TT. [Accessed March 25, 2016];Embryonic cleavage cycles: how is a mouse like a fly? Current biology : CB. 2004 14(1):R35–45. doi: 10.1016/j.cub.2003.12.022. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2712630&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, et al. [Accessed February 6, 2015];Stepwise histone modifications are mediated by multiple enzymes that rapidly associate with nascent DNA during replication. Nature communications. 2013 4:2841. doi: 10.1038/ncomms3841. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3874871&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, et al. [Accessed July 12, 2014];TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012 150(5):922–33. doi: 10.1016/j.cell.2012.06.046. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3432699&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezic D, et al. [Accessed February 24, 2016];piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes & development. 2014 28(13):1410–28. doi: 10.1101/gad.240895.114. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4083086&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. [Accessed July 9, 2014];BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England) 2010 26(6):841–2. doi: 10.1093/bioinformatics/btq033. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2832824&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, et al. [Accessed March 25, 2016];Histone Methyltransferases Direct Different Degrees of Methylation to Define Distinct Chromatin Domains. Molecular Cell. 2003 12(6):1591–1598. doi: 10.1016/s1097-2765(03)00479-9. Available at: http://www.sciencedirect.com/science/article/pii/S1097276503004799. [DOI] [PubMed] [Google Scholar]
- Roopra A, et al. [Accessed February 24, 2016];Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Molecular cell. 2004 14(6):727–38. doi: 10.1016/j.molcel.2004.05.026. Available at: http://www.ncbi.nlm.nih.gov/pubmed/15200951. [DOI] [PubMed] [Google Scholar]
- Rowe HM, et al. [Accessed January 27, 2016];KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010 463(7278):237–40. doi: 10.1038/nature08674. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20075919. [DOI] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ. [Accessed February 24, 2016];Multiple roles for Piwi in silencing Drosophila transposons. Genes & development. 2013 27(4):400–12. doi: 10.1101/gad.209767.112. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3589557&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, et al. [Accessed February 19, 2016];SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & development. 2002 16(8):919–32. doi: 10.1101/gad.973302. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=152359&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JJ, et al. [Accessed March 27, 2016];Genomic dynamics of transposable elements in the western clawed frog (Silurana tropicalis) Genome biology and evolution. 2013 5(5):998–1009. doi: 10.1093/gbe/evt065. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3673623&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato H, et al. [Accessed February 24, 2016];A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. The Journal of biological chemistry. 2009 284(2):733–9. doi: 10.1074/jbc.M804994200. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19010785. [DOI] [PubMed] [Google Scholar]
- Sienski G, et al. [Accessed February 24, 2016];Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes & development. 2015 29(21):2258–71. doi: 10.1101/gad.271908.115. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26494711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. [Accessed February 24, 2016];Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012 151(5):964–80. doi: 10.1016/j.cell.2012.10.040. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3504300&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, et al. [Accessed February 24, 2016];Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes & development. 2013 27(4):390–9. doi: 10.1101/gad.209841.112. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3589556&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. [Accessed March 10, 2016];Facultative heterochromatin: is there a distinctive molecular signature? Molecular cell. 2007 28(1):1–13. doi: 10.1016/j.molcel.2007.09.011. Available at: http://www.cell.com/article/S1097276507006235/fulltext. [DOI] [PubMed] [Google Scholar]
- Vagin VV, et al. [Accessed February 24, 2016];A distinct small RNA pathway silences selfish genetic elements in the germline. Science (New York, N.Y.) 2006 313(5785):320–4. doi: 10.1126/science.1129333. Available at: http://www.ncbi.nlm.nih.gov/pubmed/16809489. [DOI] [PubMed] [Google Scholar]
- Varshney D, et al. [Accessed February 24, 2016];SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nature communications. 2015 6:6569. doi: 10.1038/ncomms7569. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4382998&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Elgin SCR. [Accessed February 24, 2016];Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proceedings of the National Academy of Sciences of the United States of America. 2011 108(52):21164–9. doi: 10.1073/pnas.1107892109. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3248523&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, et al. [Accessed January 2, 2015];Initial sequencing and comparative analysis of the mouse genome. Nature. 2002 420(6915):520–62. doi: 10.1038/nature01262. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12466850. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. [Accessed February 24, 2016];Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009 458(7242):1201–4. doi: 10.1038/nature07844. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2676211&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. [Accessed November 8, 2014];Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics (Oxford, England) 2010 26(7):873–81. doi: 10.1093/bioinformatics/btq057. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2844994&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylicz JJ, et al. [Accessed November 12, 2015];Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. eLife. 2015 4:e09571. doi: 10.7554/eLife.09571. Available at: http://elifesciences.org/content/4/e09571.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.