Abstract
To complete the infection cycle efficiently, the virus must hijack the host systems in order to benefit for all the steps and has to face all the defense mechanisms from the host. This review involves a discussion of how these positive and negative factors regulate the viral RNA accumulation identified for the Bamboo mosaic virus (BaMV), a single-stranded RNA virus. The genome of BaMV is approximately 6.4 kb in length, encoding five functional polypeptides. To reveal the host factors involved in the infection cycle of BaMV, a few different approaches were taken to screen the candidates. One of the approaches is isolating the viral replicase-associated proteins by co-immunoprecipitation with the transiently expressed tagged viral replicase in plants. Another approach is using the cDNA-amplified fragment length polymorphism technique to screen the differentially expressed genes derived from N. benthamiana plants after infection. The candidates are examined by knocking down the expression in plants using the Tobacco rattle virus-based virus-induced gene silencing technique following BaMV inoculation. The positive or negative regulators could be described as reducing or enhancing the accumulation of BaMV in plants when the expression levels of these proteins are knocked down. The possible roles of these host factors acting on the accumulation of BaMV will be discussed.
Keywords: Bamboo mosaic virus, host factors, virus replication, virus movement, defense proteins
Introduction
When a positive-sense RNA virus infects a host cell, it needs to produce its progeny and move it to the neighboring cells efficiently. In general, the entire infection cycle starts at the viral RNA entry, using the host translation system to produce the viral-specific replicase, transition the viral template from translation status to replication status, targeting the specific organelle for replication, rearrangement of the cellular membrane, recruitment of ancillary proteins to the replication site, viral RNA replication to synthesize the minus- and plus-strand RNAs, subgenomic RNA synthesis in some species, and finally, the viral-encoded movement proteins (MPs) and coat proteins accumulated for cell-to-cell movement and encapsidation, respectively. Some of the viral-encoded proteins that evolved not only fulfilled a specific role apart from amplification, but also performed counter-defense functions such as silencing suppressors against the virus-induced gene silencing system (Qu and Morris, 2005) or preventing the spread of the gene silencing signal (Voinnet et al., 2016).
Bamboo mosaic virus (BaMV) is a positive-sense, single-stranded RNA virus, belonging to the genus Potexvirus of the family Alphaflexiviridae. The genome of BaMV is approximately 6.4 kb in length with a 5′ m7GpppG structure and a 3′-end poly(A) tail and contains five open reading frames (ORFs) (Lin et al., 1994). The 3′ untranslated region (UTR) was demonstrated to form a complexed structure (including a cloverleaf-like structure, a major stem-loop, and a pseudoknot) and acted as a cis-acting element for minus-strand RNA synthesis, polyadenylation, an intracellular trafficking signal, and long distance movement (Cheng and Tsai, 1999; Tsai et al., 1999; Chen et al., 2005; Lin et al., 2005; Cheng et al., 2013). ORF 1 encodes replicase, including a capping enzyme domain (Li et al., 2001a; Huang et al., 2004), a helicase-like domain (Li et al., 2001b), and an RNA-dependent RNA polymerase domain (Li et al., 1998). ORFs 2-4 overlapping genes termed “triple-gene-block” (TGB) encode the MPs (TGBp1, TGBp2, and TGBp3) involved in virus movement (Lin et al., 1994). ORF 5 encodes the capsid protein (CP) for virus encapsidation, movement, and symptom development (Lin et al., 1994; Lan et al., 2010; Lee et al., 2011; Hung et al., 2014). Furthermore, a satellite RNA (satBaMV) was identified to associate with BaMV and could be amplified by BaMV replicase (Lin and Hsu, 1994; Lin et al., 2006). The tertiary structures of the 5′ and 3′ UTRs of satBaMV were revealed to be similar to those of BaMV (Huang et al., 2009; Chen et al., 2010).
As mentioned previously, the positive-sense RNA virus has to establish an efficient replication after entry into a host cell; the host factors are usually required to join and form a multi-functional replication complex (Ahlquist et al., 2003; Nagy and Pogany, 2008; Huang et al., 2012a). The replicase complex isolated from Qβ-infected cells is composed of bacterial proteins, elongation factors EF-Tu and -Ts and ribosomal protein S1, and Qβ RdRp for plus-strand RNA synthesis (Blumenthal and Carmichael, 1979; Blumenthal, 1980). Additional bacteria protein HF1, a ribosome-associated protein, is required for the complex to synthesize the minus-strand RNA (Barrera et al., 1993). The eukaryotic translational elongation factor 1a (eEF1a) was revealed to be part of the replicase complex in tobamoviruses, tymoviruses, potyviruses, and tobusviruses (Joshi et al., 1986; Mans et al., 1991; Dreher et al., 1999; Nishikiori et al., 2006; Yamaji et al., 2006; Thivierge et al., 2008; Li et al., 2010; Luan et al., 2016).
A few strategies were used to identify the host factors involved in virus infection cycles. By screening the host cDNA library constructed in yeast with the two-hybrid technique, one can discover the specific host factor that interacted with the viral-encoded target protein, such as the replicase, MPs, or CP (Ren et al., 2000; Nagy, 2008; Schoelz et al., 2011). The virus-encoded proteins can also be used as a ligand to co-immunoprecipitation the possible candidates for interaction with the host (DeBlasio et al., 2015, 2016). In the UV cross-linking competition technique, host proteins could be identified as interacting with the viral RNA, such as the 5′ or 3′ UTRs (Lin et al., 2007; Huang et al., 2012b; Hyodo et al., 2014). The identities of the interacted candidates derived from co-immunoprecipitation or UV cross-linking techniques could be revealed by LC/MS/MS. The cDNA-amplified length polymorphism (AFLP), a highly sensitive and efficient technique used for studying gene expression (Money et al., 1996) and demonstrated to deliver reproducible results (Bachem et al., 1996; Ditt et al., 2001), was used to screen the host’s differentially expressed genes in a post-virus infection (Cheng et al., 2010). The up- and downregulated cDNA fragments could be easily visualized and compared when run in parallel on the gel. These differentially expressed cDNA fragments could be straightforwardly isolated, amplified, cloned, and sequenced.
To reveal the relationship of the interacting host proteins with viral proteins or RNAs and the differentially expressed proteins in a post-virus infection, the Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) technique (Ruiz et al., 1998; Ratcliff et al., 2001) could be used to knock down the expressions and examine their effect on virus accumulation (Cheng et al., 2010). The results derived from the specific gene knockdown experiment (i.e., a loss of function) can be further confirmed by the complementary results derived from a transient expression of the same gene (i.e., a gain of function). These results would reveal whether the specific gene is playing an assistant or defense role in the virus’ life cycle. Furthermore, the results of viral accumulation in the specific gene knockdown plants and protoplasts could specify that the host factor is acting in the replication or movement step of the infection cycle.
The following sections provide discussions of how the host factors identified by the techniques described above participate in BaMV replication and movement. The study of virus infection mechanisms and hosts’ responses to them will provide a better understanding of the relationship between pathogens and hosts. This learning would lead to designing the better strategies for pathogen control on plants.
The Factors Involved in Assisting BaMV RNA Replication
The entire replication processes of a positive-sense RNA virus could be divided into a few different steps. First, once BaMV enters a host cell, with bamboo as a natural host and N. benthamiana as an experimental host, the RNA genome is used as a template for translation to synthesize the replication enzyme, replicase (Figure 1). At this stage, the 3′ UTR is playing a critical role in trapping a few different factors that would lead to the next step of the replication process, targeting the replication site. At least four host proteins, glutathione transferase U4 (NbGSTU4), the eEF1a, chloroplasts PGK (chlPGK), and heat shock protein 90 (Hsp90), were discovered to interact with the 3′ UTR of BaMV. In particular, the eEF1a was shown to play a negative role in BaMV RNA replication (Lin et al., 2007). Since the binding site of the eEF1a in the 3′ UTR of BaMV is overlapped with that of the RdRp, the eEF1a might play a role in the template switch that blocks viral replication during translation. The eEF1a has commonly been demonstrated to bind the tRNA-like structure of Brome mosaic virus (BMV) (Bastin and Hall, 1976) and Turnip yellow mosaic virus (TYMV) (Joshi et al., 1986). This interaction was claimed to function in the negative-regulation of TYMV minus-strand RNA synthesis (Matsuda et al., 2004); however, a similar interaction in West Nile virus was revealed to facilitate minus-strand RNA synthesis (Davis et al., 2007).
FIGURE 1.
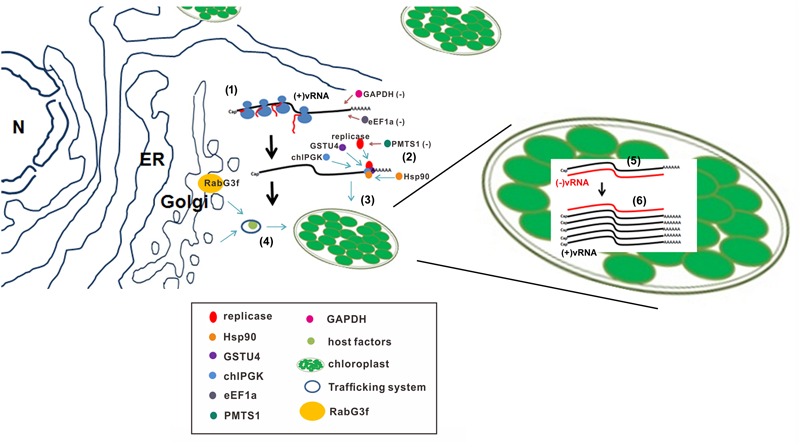
A schematic representation of the model for BaMV RNA-replicase-host factors interaction in the replication steps. (1) Once BaMV RNA entering the cell, the viral replicase is translated using the host translation system. (2) The 3′ untranslated region of BaMV genomic RNA shown as (+)vRNA is interacted with several host factors which regulate BaMV replication positively including chloroplast phosphoglycerate kinase (chlPGK), heat shock protein 90 (Hsp90), thioredoxin transferase GSTU4, and negatively indicated with red arrows including elongation factor 1a (eEF1a) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Another host factor a putative methyltransferase (PMTS1) is also shown as a negative regulator for BaMV replication through the interaction with viral replicase. (3) The (+)vRNA is transported to the chloroplasts through the interaction with chlPGK for replication. (4) Some of host factors required for BaMV replication can be transported into chloroplasts by the endomembrane trafficking system using one of the Rabs, NbRabG3f. (5) The minus-strand RNA shown as (-)vRNA is synthesized inside the chloroplasts. (6) The plus-strand (+)vRNA is then synthesized following with (-)vRNA synthesis.
The nucleus-encoded chlPGK interacting with the 3′ UTR (Lin et al., 2007) was demonstrated to play a role in ushering the viral RNA and its associated proteins, including replicase, into chloroplasts for replication (Figure 1) (Cheng et al., 2013). The genomic RNA was visualized in the chloroplast by confocal microscopy after being labeled with a green fluorescent protein fusion MS2 coat protein construct (NLS-MS2-GFP) that could recognize the BaMV RNA containing the MS2 hairpins (Cheng et al., 2013). The advantage of BaMV targeting a chloroplast for replication is that it might be a way to hide from the host scavenging system, including the RNA silencing pathway. To get into the chloroplast for replication, the entire replication complex—including the viral RNA [approximately 6.4 kb plus the poly(A) tail], replicase, and other associated host factors—must be transporting into the chloroplasts through the chloroplast transporting complex. This gigantic viral RNA-protein complex can enter into the chloroplasts because Hsp90 interacting with the 3′ UTR of BaMV was demonstrated to play a positive role in the very early event of BaMV replication (Huang et al., 2012b). Heat shock proteins acting as chaperones on protein complex folding, protein degradation, and protein translocation across membranes (Mayer and Bukau, 2005; Taipale et al., 2010) could help transport the viral RNP complex into the chloroplasts (Figure 1). HSPs were shown to be assisting the viral RNA recruitment and viral replication complexes (VRCs) assembly (Pogany et al., 2008; Wang et al., 2009a,b; Huang et al., 2012b). Accordingly, Hsp90 involved in the early event of BaMV replication could be implied to assist the viral RNP complex entry into the chloroplasts and stimulate the replication complex assembly on the right location in order to initiate the minus-strand RNA synthesis.
Another 3′ UTR-associated protein, NbGSTU4, was demonstrated to be upregulated post BaMV infection and involved in assisting the replication of BaMV in vitro and in vivo (Chen et al., 2013). In general, GSTs are involved in the antioxidation process; the oxidative stress triggered by a pathogen infection could be attenuated via the enzymatic reaction of GSTs. A chloroplast is one of the major reactive oxygen species (ROS) producer in which the relative concentration of ROS should be higher than that of other organelles. Once BaMV enters into the chloroplasts for replication, the viral RNAs face the obstacle of the higher levels of ROS produced either from the photosynthesis process or the virus infection. NbGSTU4 moving with BaMV RNA in the presence of Glutathione (GSH) could play a role in eliminating the effects of ROS. We have demonstrated that NbGSTU4 could interact with the 3′ UTR in the physiological concentration of GSH, which is approximately 10 mM (Chen et al., 2013). These results imply that NbGSTU4 could also be one of the host proteins associated with viral RNA and was transported together into the chloroplasts.
A 5′ to 3′ exonuclease (XRN4), a protein component enriched in BaMV RdRp preparation, was demonstrated to enhance the accumulation of BaMV (Lee et al., 2015). The members of the XRN_N family are divided into two groups, the cytoplasmic (XRN1/PACMAN or XRN4) and the nuclear enzymes (XRN2/RAT1 and XRN3), and display the RNase activities for mRNA degradation (Kastenmayer and Green, 2000; Souret et al., 2004). Although XRN4 was revealed to conduct antiviral activity against Tomato bushy stunt virus (TBSV) and Tobacco mosaic virus (TMV) (Cheng et al., 2007; Jaag and Nagy, 2009; Peng et al., 2011), the presence of XRN4 could elevate the accumulation of BaMV. Because XRN4 was demonstrated to have a role in reducing the activities of siRNA- and miRNA-mediated RNA decay in Arabidopsis (Souret et al., 2004), downregulated the silencing pathway might result in a positive regulation of BaMV RNA accumulation.
Some host factors identified by cDNA-AFLP could influence the replication of BaMV in an indirect manner so that these proteins could not be revealed by using the strategy of detecting the direct interaction with viral products. NbRabG3f was demonstrated to be a positive regulator in BaMV replication by loss- and gain-of-function assays (Huang et al., 2016). Rabs are a group of small GTPases involved in vesicles transport, uncoating, tethering, and fusion (Seabra et al., 2002). A deletion mutant of NbRabG3f failure in membrane-anchoring lost the ability to assist the accumulation of BaMV. A mutant with the fixed GDP-bound RabG3f (T22N) was trapped at the Golgi and could not assist the accumulation of BaMV. Overall, these results suggest that NbRabG3f is involved in a vesicle budding from the Golgi and transports the cargos containing the unidentified host factors to the destination site for BaMV replication (Figure 1).
Based on the host factors identified so far for BaMV replication, BaMV RNA entry into the host cell would require chlPGK to usher the viral RNA into the chloroplasts. The transport of the viral RNP complex needs the chaperon Hsp90 to cross the membrane and assemble the functional replication complex. During this process of trafficking from the cytoplasm to the chloroplasts, NbXRN4 might be involved in reducing the activities of siRNA-mediated silencing. Once the viral RNP is transported into the chloroplasts, the VRC needs the anti-oxidant enzyme NbGSTU4 to neutralize the oxidative stress inside the chloroplasts for an efficient minus-strand RNA synthesis (Figure 1).
The Factors Involved in Viral RNA Movement
Through a biochemical analysis of the BaMV movement complex isolated by co-immunoprecipitation using an anti-TGBp3 antibody, the movement trafficking complex was revealed to harbor not only the MPs TGBp1, TGBp2, and TGBp3, but also the coat protein and replicase (Chou et al., 2013). The TGBps-mediated cell-to-cell trafficking was proposed to be in two possible paths: TGBps-associated virion complex traffics alongside the endoplasmic reticulum (ER) network, or the virions and the MPs would associate with the TGBp2-induced vesicles (Chou et al., 2013; Liou et al., 2015).
A few host factors were identified to participate in the process of BaMV movement. A RabGTPase-activating protein (GAP) designated as NbRabGAP1 was demonstrated to participate in BaMV cell-to-cell and systemic movements (Huang et al., 2013). Rabs, a family of small GTPases, are known to be involved in all aspects of intracellular vesicle budding, targeting, docking, and fusion (Johansen et al., 2009; Mizuno-Yamasaki et al., 2012; Cherfils and Zeghouf, 2013). Two Rabs regulators, guanine nucleotide exchange factors (GFFs), and GAPs play roles in recycling Rabs for vesicles trafficking, in which GEFs exchange GDP for GTP and GAPs accelerate GTP hydrolysis (Bos et al., 2007). The results of the mutational analysis of NbRabGAP1in BaMV accumulation suggest that the fully GAP function of NbRabGAP1 is essential to support the efficient movement of BaMV. The proposed role of NbRabGAP1 in BaMV movement is that NbRabGAP1is to trigger one of the RabGTPases (not yet identified) to release the vesicles containing the viral movement complex trafficking to the plasmodesmata (PD) (Figure 2), similar to those revealed in Chinese wheat mosaic virus (Andika et al., 2013), or to shuttle TGB proteins from the PD via the endocytotic pathway back to the ER, like those found in Potato mop top virus (Haupt et al., 2005).
FIGURE 2.
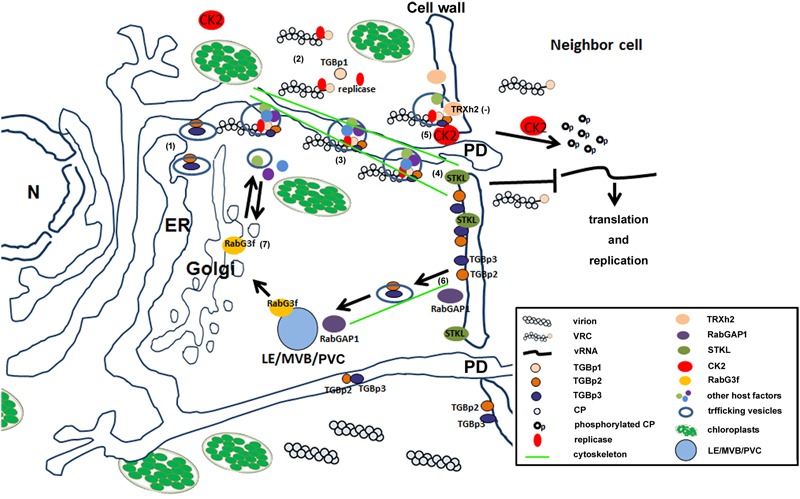
A schematic view of the hypothetical model for BaMV movement. The intracellular trafficking of BaMV movement complex is proposed to form the vesicle and trafficking through the cytoskeleton toward the plasmodesmata (PD). The steps of the BaMV movement are illustrated as (1) the movement proteins of BaMV, TGBp2 and TGBp3, are synthesized on the endoplasmic reticulum (ER) and transported via vesicles possibly regulated by one of the Rab-GTPases; (2) the newly synthesized viral RNA is assembled with the capsid protein (CP), movement protein TGBp1, and the viral replicase to form a competent viral replication complex (VRC); (3) the VRC and possibly some other host factors are recruited to TGBp2/TGBp3-containing vesicle to form a movement complex and trafficking toward the PD; (4) the host factor STKL localized on the plasma membrane might control the gate of the PD; (5) the host factor CK2α targeting the CP of VRC and release the vRNA from the VRC to the neighbor cell for further translation or replication; (6) TGBp2 and TGBp3 are released from the movement complex after disassembly on the PD and might shuttle back from the plasma membrane to late endosome/multivesicle bodies/prevacuolar compartments (LE/MVB/PVC) with the help of RabGAP1 to activate the Rab (unidentified yet); (7) RabG3f is possibly involved in shuttling the viral movement proteins back to Golgi and ER. One of the host factors, TRXh2, is shown to play a negative role indicated as (-) in hindering the movement through the interaction with viral movement protein TGBp2.
A serine/threonine kinase-like protein from N. benthamiana (NbSTKL), an upregulated gene that is post BaMV inoculation, was demonstrated to be critical in the movement step of the BaMV infection cycle (Cheng et al., 2013). The results from the sequence analysis and the intracellular localization indicated that NbSTKL is plasma membrane-associated through myristoylation at glycine, the second amino acid from the N-terminus. The mutant that lost the kinase activity (NbSTKL/D224A) or failed to associate with the plasma membrane (NbSTKL/G2A) also failed to enhance the movement of BaMV. These results suggest that NbSTKL might target a specific factor on the membrane, regulating the gating of the PD for the passage of BaMV (Figure 2). Furthermore, another kinase, casein kinase 2α (CK2α), which interacts with BaMV CP in PD, might assist the release of viral RNA from the RNP movement complex during the virial RNP complex passage through the PD (Figure 2) (Hung et al., 2014).
Taken together, the identification of NbRabGAP1 involved in the movement of BaMV supports the idea that the movement could be made through the vesicle trafficking path (Chou et al., 2013; Liou et al., 2015). To reach an efficient movement of BaMV requires at least two kinases, NbSTKL and CK2α, gating the PD and releasing the viral RNA from the RNP complex (Figure 2). Obviously, some other factors are also required for this process such as the target of NbSTKL.
The Factors Involved in Defense Against Viral RNA Replication
As mentioned previously, once a virus enters a host cell, it needs not only to seek the host factors for assistance, but also to face the challenges from the host itself. In plants, there already exist a few defense mechanisms such as the RNA silencing pathway and the elector-induced hypersensitive reaction. In addition, some novel proteins in host cells could display anti-viral activities beyond their already known functions. A putative methyltransferase (PMTS1) once interacted with BaMV RdRp, screened by a yeast two-hybrid technique, and displayed an inhibitory effect on the RdRp activity with a dosage-dependent fashion (Figure 1) (Cheng et al., 2009). PMTS1 comprises an N-terminal signal peptide predicted to target mitochondria or chloroplasts and two putative AdoMet-binding motifs in the middle region. Removing the signal peptide or abolishing the AdoMet-binding activity of PMTS1 would cause the loss of its inhibitory effect. Because BaMV has been demonstrated to replicate in chloroplasts, the signal peptide of PMTS1 targeting the chloroplasts is highly recommended.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) found in the purified RdRp complexes could bind the stem-loop C/poly(A) in the 3′ UTRs of satBaMV and the pseudoknot/poly(A) in the 3′ UTR of BaMV (Prasanth et al., 2011). Further analysis indicates that the cytosolic, GAPDH, could inhibit the replication of BaMV/satBaMV (Figure 1). The purified recombinant, GAPDH, could specifically inhibit the synthesis of minus-strand RNA of BaMV/satBaMV in an in vitro replication assay. GAPDH is a multifunctional enzyme involved in quite diverse activities in cells, including glycolysis, cellular dysfunction, cell death, apoptosis, association with cytoskeleton and vesicles transport, exportation of nuclear RNA, and DNA repair (Tristan et al., 2011). In Arabidopsis, cytosolic GAPDH was found to be a prominent target of H2O2-dependent oxidation (Hancock et al., 2005), but could be reversible back in the presence of reductant GSH (Bedhomme et al., 2012). Although the functions of GAPDH involved in BaMV replication are not clear, the simple interaction of GAPDH with the 3′ UTRs of BaMV and satBaMV could block the accessibility of RdRp for initiating the minus-strand RNA synthesis.
The Factors Involved in Defense Against Viral RNA Movement
Regarding the movement of potexviruses, both MP and CP are vital for efficient cell-to-cell movement and vascular transport (Verchot-Lubicz, 2005; Verchot-Lubicz et al., 2010). Post-translational modification, including ubiquitination, sumoylation, glycosylation, and phosphorylation of viral proteins, were issued as parts of an important process in modulating the structures and functions of viral proteins (Barajas and Nagy, 2010; Alcaide-Loridan and Jupin, 2012; Mathur et al., 2012; Perez Jde et al., 2013; Samuilova et al., 2013; Xiong and Wang, 2013). Maintaining the protein structural integrity, including those modifications of MP and CP, is critical for virus movement.
One of the thioredoxin proteins, NbTRXh2, an upregulated gene post BaMV inoculation, was demonstrated to restrict the movement of BaMV (Chen et al., 2017). NbTRXh2 was localized at the plasma membrane through myristoylation at the Glycine of the second amino acid from the N-terminus. NbTRXh2 was revealed to target the MP TGBp2 to reduce its disulfide bond (Figure 2). Also, the two conserved cysteins forming the disulfide bond were demonstrated to play a key role in BaMV movement (Tseng et al., 2009). Therefore, NbTRXh2 targets TGBp2, which could result in the loss of the structural integrity of TGBp2 and their failure to interact with other movement-associated proteins, including TGBps1 and 3.
Summary and Future Prospective
Taking all the available results into account, it can be concluded that some of the host factors are unique to BaMV, while some of them could be applied to other viruses. Some host factors could assist virus replication and movement, but some of them are involved in resisting virus infection. This review summarized a few host factors identified with different strategies and their possible roles in BaMV infection. However, to complete an accurate understanding of BaMV infection, more host factors need to be identified. Based on our current understandings, a few processes in BaMV infection cycle are still unclear. One of the most challenges is to uncover the process of how the newly synthesized RNAs are transported out of the chloroplasts where BaMV replicates. Hopefully, a much clearer picture of the infection cycle of BaMV can be obtained in the near future by knowing how of these factors involved.
Author Contributions
Y-PH wrote the viral replication part of the manuscript. I-HC wrote the viral movement part of the manuscript. C-HT wrote the rest part and edits the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by grant MOST 105-2311-B-005-002 from the Ministry of Science and Technology, Taiwan.
References
- Ahlquist P., Noueiry A. O., Lee W. M., Kushner D. B., Dye B. T. (2003). Host factors in positive-strand RNA virus genome replication. J. Virol. 77 8181–8186. 10.1128/JVI.77.15.8181-8186.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaide-Loridan C., Jupin I. (2012). Ubiquitin and plant viruses, let’s play together! Plant Physiol. 160 72–82. 10.1104/pp.112.201905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andika I. B., Zheng S., Tan Z., Sun L., Kondo H., Zhou X., et al. (2013). Endoplasmic reticulum export and vesicle formation of the movement protein of Chinese wheat mosaic virus are regulated by two transmembrane domains and depend on the secretory pathway. Virology 435 493–503. 10.1016/j.virol.2012.10.024 [DOI] [PubMed] [Google Scholar]
- Bachem C. W., Van Der Hoeven R. S., De Bruijn S. M., Vreugdenhil D., Zabeau M., Visser R. G. (1996). Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 9 745–753. 10.1046/j.1365-313X.1996.9050745.x [DOI] [PubMed] [Google Scholar]
- Barajas D., Nagy P. D. (2010). Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein. Virology 397 358–368. 10.1016/j.virol.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera I., Schuppli D., Sogo J. M., Weber H. (1993). Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J. Mol. Biol. 232 512–521. 10.1006/jmbi.1993.1407 [DOI] [PubMed] [Google Scholar]
- Bastin M., Hall T. C. (1976). Interaction of elongation factor 1 with aminoacylated Brome mosaic virus and tRNA’s. J. Virol. 20 117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedhomme M., Adamo M., Marchand C. H., Couturier J., Rouhier N., Lemaire S. D., et al. (2012). Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem. J. 445 337–347. 10.1042/BJ20120505 [DOI] [PubMed] [Google Scholar]
- Blumenthal T. (1980). Interaction of host-coded and virus-coded polypeptides in RNA phage replication. Proc. R. Soc. Lond. B Biol. Sci. 210 321–335. 10.1098/rspb.1980.0137 [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Carmichael G. G. (1979). RNA replication: function and structure of Qbeta-replicase. Annu. Rev. Biochem. 48 525–548. 10.1146/annurev.bi.48.070179.002521 [DOI] [PubMed] [Google Scholar]
- Bos J. L., Rehmann H., Wittinghofer A. (2007). GEFs and GAPs: critical elements in the control of small G proteins. Cell 129 865–877. 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Chen I. H., Chen H. T., Huang Y. P., Huang H. J., Shenkwen L. L., Hsu Y. H., et al. (2017). A thioredoxin NbTRXh2 from Nicotiana benthamiana negatively regulates the movement of Bamboo mosaic virus. Mol. Plant Pathol. 10.1111/mpp.12532 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. H., Chiu M. H., Cheng S. F., Hsu Y. H., Tsai C. H. (2013). The glutathione transferase of Nicotiana benthamiana NbGSTU4 plays a role in regulating the early replication of Bamboo mosaic virus. New Phytol. 199 749–757. 10.1111/nph.12304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. H., Chou W. J., Lee P. Y., Hsu Y. H., Tsai C. H. (2005). The AAUAAA motif of Bamboo mosaic virus RNA is involved in minus-strand RNA synthesis and plus-strand RNA polyadenylation. J. Virol. 79 14555–14561. 10.1128/JVI.79.23.14555-14561.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. C., Desprez A., Olsthoorn R. C. L. (2010). Structural homology between Bamboo mosaic virus and its satellite RNAs in the 5’ untranslated region. J. Gen. Virol. 91 782–787. 10.1099/vir.0.015941-0 [DOI] [PubMed] [Google Scholar]
- Cheng C. P., Jaag H. M., Jonczyk M., Serviene E., Nagy P. D. (2007). Expression of the Arabidopsis Xrn4p 5’-3’ exoribonuclease facilitates degradation of tombusvirus RNA and promotes rapid emergence of viral variants in plants. Virology 368 238–248. 10.1016/j.virol.2007.07.001 [DOI] [PubMed] [Google Scholar]
- Cheng C. P., Tsai C. H. (1999). Structural and functional analysis of the 3’ untranslated region of Bamboo mosaic potexvirus genomic RNA. J. Mol. Biol. 288 555–565. 10.1006/jmbi.1999.2716 [DOI] [PubMed] [Google Scholar]
- Cheng C. W., Hsiao Y. Y., Wu H. C., Chuang C. M., Chen J. S., Tsai C. H., et al. (2009). Suppression of Bamboo mosaic virus accumulation by a putative methyltransferase in Nicotiana benthamiana. J. Virol. 83 5796–5805. 10.1128/JVI.02471-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. F., Huang Y. P., Chen L. H., Hsu Y. H., Tsai C. H. (2013). Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiol. 163 1598–1608. 10.1104/pp.113.229666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. F., Huang Y. P., Wu Z. R., Hu C. C., Hsu Y. H., Tsai C. H. (2010). Identification of differentially expressed genes induced by Bamboo mosaic virus infection in Nicotiana benthamiana by cDNA-amplified fragment length polymorphism. BMC Plant Biol. 10:286 10.1186/1471-2229-10-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J., Zeghouf M. (2013). Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93 269–309. 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- Chou Y. L., Hung Y. J., Tseng Y. H., Hsu H. T., Yang J. Y., Wung C. H., et al. (2013). The stable association of virion with the triple-gene-block protein 3-based complex of Bamboo mosaic virus. PLoS Pathog. 9:e1003405 10.1371/journal.ppat.1003405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis W. G., Blackwell J. L., Shi P. Y., Brinton M. A. (2007). Interaction between the cellular protein eEF1A and the 3’-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 81 10172–10187. 10.1128/JVI.00531-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBlasio S. L., Johnson R., Mahoney J., Karasev A., Gray S. M., Maccoss M. J., et al. (2015). Insights into the polerovirus-plant interactome revealed by coimmunoprecipitation and mass spectrometry. Mol. Plant Microbe Interact. 28 467–481. 10.1094/MPMI-11-14-0363-R [DOI] [PubMed] [Google Scholar]
- DeBlasio S. L., Johnson R. S., Maccoss M. J., Gray S. M., Cilia M. (2016). Model system-guided protein interaction mapping for virus isolated from phloem tissue. J. Proteome Res. 15 4601–4611. 10.1021/acs.jproteome.6b00715 [DOI] [PubMed] [Google Scholar]
- Ditt R. F., Nester E. W., Comai L. (2001). Plant gene expression response to Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 98 10954–10959. 10.1073/pnas.191383498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T. W., Uhlenbeck O. C., Browning K. S. (1999). Quantitative assessment of EF-1alpha. GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J. Biol. Chem. 274 666–672. 10.1074/jbc.274.2.666 [DOI] [PubMed] [Google Scholar]
- Hancock J. T., Henson D., Nyirenda M., Desikan R., Harrison J., Lewis M., et al. (2005). Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol. Biochem. 43 828–835. 10.1016/j.plaphy.2005.07.012 [DOI] [PubMed] [Google Scholar]
- Haupt S., Cowan G. H., Ziegler A., Roberts A. G., Oparka K. J., Torrance L. (2005). Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell 17 164–181. 10.1105/tpc.104.027821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. L., Han Y. T., Chang Y. T., Hsu Y. H., Meng M. S. (2004). Critical residues for GTP methylation and formation of the covalent m(7)GMP-enzyme intermediate in the capping enzyme domain of Bamboo mosaic virus. J. Virol. 78 1271–1280. 10.1128/JVI.78.3.1271-1280.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. P., Chen J. S., Hsu Y. H., Tsai C. H. (2013). A putative Rab-GTPase activation protein from Nicotiana benthamiana is important for Bamboo mosaic virus intercellular movement. Virology 447 292–299. 10.1016/j.virol.2013.09.021 [DOI] [PubMed] [Google Scholar]
- Huang Y. P., Jhuo J. H., Tsai M. S., Tsai C. H., Chen H. C., Lin N. S., et al. (2016). NbRABG3f, a member of Rab GTPase, is involved in Bamboo mosaic virus infection in Nicotiana benthamiana. Mol. Plant Pathol. 17 714–726. 10.1111/mpp.12325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. W., Hu C. C., Lin C. A., Liu Y. P., Tsai C. H., Lin N. S., et al. (2009). Structural and functional analyses of the 3′ untranslated region of Bamboo mosaic virus satellite RNA. Virology 386 139–153. 10.1016/j.virol.2009.01.019 [DOI] [PubMed] [Google Scholar]
- Huang Y. W., Hu C. C., Lin N. S., Hsu Y. H. (2012a). Unusual roles of host metabolic enzymes and housekeeping proteins in plant virus replication. Curr. Opin. Virol. 2 676–682. 10.1016/j.coviro.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Huang Y. W., Hu C. C., Liou M. R., Chang B. Y., Tsai C. H., Meng M. H., et al. (2012b). Hsp90 interacts specifically with viral RNA and differentially regulates replication initiation of Bamboo mosaic virus and associated satellite RNA. PLoS Pathog. 8:e1002726 10.1371/journal.ppat.1002726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. J., Huang Y. W., Liou M. R., Lee Y. C., Lin N. S., Meng M. H., et al. (2014). Phosphorylation of coat protein by protein kinase CK2 regulates cell-to-cell movement of Bamboo mosaic virus through modulating RNA binding. Mol. Plant Microbe Interact. 27 1211–1225. 10.1094/MPMI-04-14-0112-R [DOI] [PubMed] [Google Scholar]
- Hyodo K., Kaido M., Okuno T. (2014). Host and viral RNA-binding proteins involved in membrane targeting, replication and intercellular movement of plant RNA virus genomes. Front. Plant Sci. 5:321 10.3389/fpls.2014.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag H. M., Nagy P. D. (2009). Silencing of Nicotiana benthamiana Xrn4p exoribonuclease promotes tombusvirus RNA accumulation and recombination. Virology 386 344–352. 10.1016/j.virol.2009.01.015 [DOI] [PubMed] [Google Scholar]
- Johansen J. N., Chow C. M., Moore I., Hawes C. (2009). AtRAB-H1b and AtRAB-H1c GTPases, homologues of the yeast Ypt6, target reporter proteins to the Golgi when expressed in Nicotiana tabacum and Arabidopsis thaliana. J. Exp. Bot. 60 3179–3193. 10.1093/jxb/erp153 [DOI] [PubMed] [Google Scholar]
- Joshi R. L., Ravel J. M., Haenni A. L. (1986). Interaction of Turnip yellow mosaic virus Val-RNA with eukaryotic elongation factor EF-1 [alpha]. Search for a function. EMBO J. 5 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmayer J. P., Green P. J. (2000). Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 97 13985–13990. 10.1073/pnas.97.25.13985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P., Yeh W. B., Tsai C. W., Lin N. S. (2010). A unique glycine-rich motif at the N-terminal region of Bamboo mosaic virus coat protein is required for symptom expression. Mol. Plant Microbe Interact. 23 903–914. 10.1094/MPMI-23-7-0903 [DOI] [PubMed] [Google Scholar]
- Lee C. C., Ho Y. N., Hu R. H., Yen Y. T., Wang Z. C., Lee Y. C., et al. (2011). The interaction between Bamboo mosaic virus replication protein and coat protein is critical for virus movement in plant hosts. J. Virol. 85 12022–12031. 10.1128/JVI.05595-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. C., Lin T. L., Lin J. W., Han Y. T., Huang Y. T., Hsu Y. H., et al. (2015). Promotion of Bamboo mosaic virus accumulation in Nicotiana benthamiana by 5′– > 3′ exonuclease NbXRN4. Front. Microbiol. 6:1508 10.3389/fmicb.2015.01508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. I., Chen Y. J., Hsu Y. H., Meng M. H. (2001a). Characterization of the AdoMet-dependent guanylyltransferase activity that is associated with the N terminus of Bamboo mosaic virus replicase. J. Virol. 75 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. I., Cheng Y. M., Huang Y. L., Tsai C. H., Hsu Y. H., Meng M. (1998). Identification and characterization of the Escherichia coli-expressed RNA-dependent RNA polymerase of Bamboo mosaic virus. J. Virol. 72 10093–10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. I., Shih T. W., Hsu Y. H., Han Y. T., Huang Y. L., Meng M. S. (2001b). The helicase-like domain of plant potexvirus replicase participates in formation of RNA 5’ cap structure by exhibiting RNA 5’-triphosphatase activity. J. Virol. 75 12114–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pogany J., Tupman S., Esposito A. M., Kinzy T. G., Nagy P. D. (2010). Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 6:e1001175 10.1371/journal.ppat.1001175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. W., Chiu H. N., Chen I. H., Chen T. C., Hsu Y. H., Tsai C. H. (2005). Structural and functional analysis of the cis-acting elements required for plus-strand RNA synthesis of Bamboo mosaic virus. J. Virol. 79 9046–9053. 10.1128/JVI.79.14.9046-9053.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. W., Ding M. P., Hsu Y. H., Tsai C. H. (2007). Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucleic Acids Res. 35 424–432. 10.1093/nar/gkl1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. K., Hu C. C., Lin N. S., Chang B. Y., Hsu Y. H. (2006). Movement of potexviruses requires species-specific interactions among the cognate triple gene block proteins, as revealed by a trans-complementation assay based on the Bamboo mosaic virus satellite RNA-mediated expression system. J. Gen. Virol. 87 1357–1367. 10.1099/vir.0.81625-0 [DOI] [PubMed] [Google Scholar]
- Lin N. S., Hsu Y. H. (1994). A satellite RNA associated with Bamboo mosaic potexvirus. Virology 202 707–714. 10.1006/viro.1994.1392 [DOI] [PubMed] [Google Scholar]
- Lin N. S., Lin B. Y., Lo N. W., Hu C. C., Chow T. Y., Hsu Y. H. (1994). Nucleotide sequence of the genomic RNA of Bamboo mosaic potexvirus. J. Gen. Virol. 75 2513–2518. 10.1099/0022-1317-75-9-2513 [DOI] [PubMed] [Google Scholar]
- Liou M. R., Hu C. C., Chou Y. L., Chang B. Y., Lin N. S., Hsu Y. H. (2015). Viral elements and host cellular proteins in intercellular movement of Bamboo mosaic virus. Curr. Opin. Virol. 12 99–108. 10.1016/j.coviro.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Luan H., Shine M. B., Cui X., Chen X., Ma N., Kachroo P., et al. (2016). The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 172 221–234. 10.1104/pp.16.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R. M., Pleij C. W., Bosch L. (1991). tRNA-like structures. Structure, function and evolutionary significance. Eur. J. Biochem. 201 303–324. 10.1111/j.1432-1033.1991.tb16288.x [DOI] [PubMed] [Google Scholar]
- Mathur C., Jimsheena V. K., Banerjee S., Makinen K., Gowda L. R., Savithri H. S. (2012). Functional regulation of PVBV nuclear inclusion protein-a protease activity upon interaction with viral protein genome-linked and phosphorylation. Virology 422 254–264. 10.1016/j.virol.2011.10.009 [DOI] [PubMed] [Google Scholar]
- Matsuda D., Yoshinari S., Dreher T. W. (2004). eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology 321 47–56. 10.1016/j.virol.2003.10.028 [DOI] [PubMed] [Google Scholar]
- Mayer M. P., Bukau B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 62 670–684. 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E., Rivera-Molina F., Novick P. (2012). GTPase networks in membrane traffic. Annu. Rev. Biochem. 81 637–659. 10.1146/annurev-biochem-052810-093700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Money T., Reader S., Qu L. J., Dunford R. P., Moore G. (1996). AFLP-based mRNA fingerprinting. Nucleic Acids Res. 24 2616–2617. 10.1093/nar/24.13.2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P. D. (2008). Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 46 217–242. 10.1146/annurev.phyto.121407.093958 [DOI] [PubMed] [Google Scholar]
- Nagy P. D., Pogany J. (2008). Multiple roles of viral replication proteins in plant RNA virus replication. Methods Mol. Biol. 451 55–68. 10.1007/978-1-59745-102-4_4 [DOI] [PubMed] [Google Scholar]
- Nishikiori M., Dohi K., Mori M., Meshi T., Naito S., Ishikawa M. (2006). Membrane-bound Tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 80 8459–8468. 10.1128/JVI.00545-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Yang J., Yan F., Lu Y., Jiang S., Lin L., et al. (2011). Silencing of NbXrn4 facilitates the systemic infection of Tobacco mosaic virus in Nicotiana benthamiana. Virus Res. 158 268–270. 10.1016/j.virusres.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Perez Jde J., Udeshi N. D., Shabanowitz J., Ciordia S., Juarez S., Scott C. L., et al. (2013). O-GlcNAc modification of the coat protein of the potyvirus Plum pox virus enhances viral infection. Virology 442 122–131. 10.1016/j.virol.2013.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., Stork J., Li Z., Nagy P. D. (2008). In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc. Natl. Acad. Sci. U.S.A. 105 19956–19961. 10.1073/pnas.0810851105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K. R., Huang Y. W., Liou M. R., Wang R. Y. L., Hu C. C., Tsai C. H., et al. (2011). Glyceraldehyde 3-phosphate dehydrogenase negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. J. Virol. 85 8829–8840. 10.1128/JVI.00556-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Morris T. J. (2005). Suppressors of RNA silencing encoded by plant viruses and their role in viral infections. FEBS Lett. 579 5958–5964. 10.1016/j.febslet.2005.08.041 [DOI] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez A. M., Baulcombe D. C. (2001). Technical advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25 237–245. 10.1046/j.0960-7412.2000.00942.x [DOI] [PubMed] [Google Scholar]
- Ren T., Qu F., Morris T. J. (2000). HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to Turnip crinkle virus. Plant Cell 12 1917–1926. 10.1105/tpc.12.10.1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M. T., Voinnet O., Baulcombe D. C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10 937–946. 10.1105/tpc.10.6.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuilova O., Santala J., Valkonen J. P. (2013). Tyrosine phosphorylation of the triple gene block protein 3 regulates cell-to-cell movement and protein interactions of Potato mop-top virus. J. Virol. 87 4313–4321. 10.1128/JVI.03388-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoelz J. E., Harries P. A., Nelson R. S. (2011). Intracellular transport of plant viruses: finding the door out of the cell. Mol. Plant 4 813–831. 10.1093/mp/ssr070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra M. C., Mules E. H., Hume A. N. (2002). Rab GTPases, intracellular traffic and disease. Trends Mol. Med. 8 23–30. 10.1016/S1471-4914(01)02227-4 [DOI] [PubMed] [Google Scholar]
- Souret F. F., Kastenmayer J. P., Green P. J. (2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15 173–183. 10.1016/j.molcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Taipale M., Jarosz D. F., Lindquist S. (2010). HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11 515–528. 10.1038/nrm2918 [DOI] [PubMed] [Google Scholar]
- Thivierge K., Cotton S., Dufresne P. J., Mathieu I., Beauchemin C., Ide C., et al. (2008). Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 377 216–225. 10.1016/j.virol.2008.04.015 [DOI] [PubMed] [Google Scholar]
- Tristan C., Shahani N., Sedlak T. W., Sawa A. (2011). The diverse functions of GAPDH: views from different subcellular compartments. Cell. Signal. 23 317–323. 10.1016/j.cellsig.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. H., Cheng C. P., Peng C. W., Lin B. Y., Lin N. S., Hsu Y. H. (1999). Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of Bamboo mosaic potexvirus RNA. J. Virol. 73 2703–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y. H., Hsu H. T., Chou Y. L., Hu C. C., Lin N. S., Hsu Y. H., et al. (2009). The two conserved cysteine residues of the triple gene block protein 2 are critical for both cell-to-cell and systemic movement of Bamboo mosaic virus. Mol. Plant Microbe Interact. 22 1379–1388. 10.1094/MPMI-22-11-1379 [DOI] [PubMed] [Google Scholar]
- Verchot-Lubicz J. (2005). A new cell-to-cell transport model for Potexviruses. Mol. Plant Microbe Interact. 18 283–290. 10.1094/MPMI-18-0283 [DOI] [PubMed] [Google Scholar]
- Verchot-Lubicz J., Torrance L., Solovyev A. G., Morozov S. Y., Jackson A. O., Gilmer D. (2010). Varied movement strategies employed by triple gene block-encoding viruses. Mol. Plant Microbe Interact. 23 1231–1247. 10.1094/MPMI-04-10-0086 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Lederer C., Baulcombe D. C. (2016). A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 166 780 10.1016/j.cell.2016.07.015 [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Stork J., Nagy P. D. (2009a). A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 83 3276–3287. 10.1128/JVI.02313-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. Y., Stork J., Pogany J., Nagy P. D. (2009b). A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology 394 28–38. 10.1016/j.virol.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong R., Wang A. (2013). SCE1, the SUMO-conjugating enzyme in plants that interacts with NIb, the RNA-dependent RNA polymerase of Turnip mosaic virus, is required for viral infection. J. Virol. 87 4704–4715. 10.1128/JVI.02828-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji Y., Kobayashi T., Hamada K., Sakurai K., Yoshii A., Suzuki M., et al. (2006). In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 347 100–108. 10.1016/j.virol.2005.11.031 [DOI] [PubMed] [Google Scholar]


