ABSTRACT
Most prokaryote-secreted proteins are transported to the cell surface using either the general secretion (Sec) or twin-arginine translocation (Tat) pathway. A majority of secreted proteins are anchored to the cell surface, while the remainder are released into the extracellular environment. The anchored surface proteins play a variety of important roles in cellular processes, ranging from facilitating interactions between cells to maintaining cell stability. The extensively studied S-layer glycoprotein (SLG) of Haloferax volcanii, previously thought to be anchored via C-terminal intercalation into the membrane, was recently shown to be lipidated and to have its C-terminal segment removed in processes dependent upon archaeosortase A (ArtA), a recently discovered enzyme. While SLG is a Sec substrate, in silico analyses presented here reveal that, of eight additional ArtA substrates predicted, two substrates also contain predicted Tat signal peptides, including Hvo_0405, which has a highly conserved tripartite structure that lies closer to the center of the protein than to its C terminus, unlike other predicted ArtA substrates identified to date. We demonstrate that, even given its atypical location, this tripartite structure, which likely resulted from the fusion of genes encoding an ArtA substrate and a cytoplasmic protein, is processed in an ArtA-dependent manner. Using an Hvo_0405 mutant lacking the conserved “twin” arginines of the predicted Tat signal peptide, we show that Hvo_0405 is indeed a Tat substrate and that ArtA substrates include both Sec and Tat substrates. Finally, we confirmed the Tat-dependent localization and signal peptidase I (SPase I) cleavage site of Hvo_0405 using mass spectrometry.
IMPORTANCE The specific mechanisms that facilitate protein anchoring to the archaeal cell surface remain poorly understood. Here, we have shown that the proteins bound to the cell surface of the model archaeon H. volcanii, through a recently discovered novel ArtA-dependent anchoring mechanism, are more structurally diverse than was previously known. Specifically, our results demonstrate that both Tat and Sec substrates, which contain the conserved tripartite structure of predicted ArtA substrates, can be processed in an ArtA-dependent manner and that the tripartite structure need not lie near the C terminus for this processing to occur. These data improve our understanding of archaeal cell biology and are invaluable for in silico subcellular localization predictions of archaeal and bacterial proteins.
KEYWORDS: archaea, archaeosortase, C-terminal processing, exosortase, Haloferax volcanii, Tat substrates, cell surface, protein secretion, secretion systems
INTRODUCTION
Prokaryotic cell surfaces are decorated with a diverse set of proteins that perform functions that are crucial to a variety of cellular processes, ranging from cell-cell communication to stress response and cell stability. Most surface proteins are transported across the cytoplasmic membrane via either the secretion (Sec) or the twin-arginine transport (Tat) pathway (1–3). The Sec pathway, which is found in all species of eukaryotes as well as those of both prokaryotic domains, transports proteins across the cytoplasmic membrane in an unfolded conformation. In most organisms, this pathway typically transports the major portion of secreted proteins across the cytoplasmic membrane (4). However, the haloarchaea, such as Haloferax volcanii, are predicted to use the Tat pathway, which transports proteins in a folded conformation, for a large portion of their secreted proteins (4, 5). The reason why many haloarchaeal proteins use the Tat pathway is perhaps because proteins transported in an unfolded conformation do not fold properly in highly saline extracytoplasmic environments lacking ATPase-driven chaperones (4, 6, 7). For example, nearly half of all H. volcanii-secreted proteins are predicted to be transported to the cell surface via the Tat pathway (5, 8). Once on the cell surface, a subset of secreted proteins are released into the extracellular environment. However, the vast majority are anchored to the cell surface, i.e., to the cytoplasmic membrane of archaea and Gram-positive bacteria, to the inner (cytoplasmic) membrane or the outer (surface) membrane of Gram-negative bacteria and a subset of archaea, or to the prokaryotic cell wall (7, 9–11). A diverse array of protein-anchoring pathways have evolved to facilitate retention of these secreted proteins at the cell surface, including a recently described mechanism that facilitates C-terminal lipid-mediated anchoring of prokaryotic proteins (12–14).
The H. volcanii S-layer glycoprotein (SLG), which is the sole component of the cell wall in this halophilic archaeon (15), contains a C-terminal tripartite structure consisting of a PGF motif, followed by a hydrophobic stretch and a positively charged region (14). C-terminal processing and lipid modification of this protein are dependent on a novel enzyme known as archaeosortase, which is homologous to the exosortase in Gram-negative bacteria (12–14). The predicted substrates of this enzyme contain a highly conserved C-terminal tripartite structure reminiscent of the substrates of a family of acyltransferases found in many Gram-positive bacterial species known as sortases (10, 14, 16, 17). The sortases process and subsequently covalently link their substrates to the bacterial cell wall. Although substrates of sortase and archaeosortase contain similar highly conserved tripartite structures and the enzymes each contain similar putative catalytic amino acid triads, no sequence or structural homology has been detected between the sortases and archaeosortases (14).
Recently, we determined that SLG processing and lipidation both depend on the presence of the PGF motif in this ArtA substrate (13). While only the highly abundant SLG in H. volcanii has been confirmed to be processed and anchored in an ArtA-dependent manner, in silico analyses predicted that many archaeal and bacterial genomes encode an archaeosortase and exosortase, respectively. These analyses have also shown that any genome that encodes either of these homologous enzymes also encodes secreted proteins that contain the highly conserved tripartite structure (14). In H. volcanii, in silico analyses have revealed that, in addition to the SLG, seven proteins carrying the signal peptide, as predicted by SignalP and/or the PRED-SIGNAL program, contain the highly conserved C-terminal tripartite structure of a predicted ArtA substrate (14, 18).
In addition to the previously confirmed ArtA substrate, SLG, the study described here revealed another ArtA substrate, Hvo_0405, a predicted Tat substrate containing the conserved tripartite structure of the ArtA substrates but that is closer to the center of the protein than to the C terminus. This provides an excellent opportunity to determine whether ArtA can recognize and process proteins in which the conserved tripartite structure is centrally located as opposed to C-terminally located, and whether a subset of Tat substrates can be processed and anchored to the cell surface in an ArtA-dependent manner.
Our in silico analyses have revealed that the more central location of the conserved tripartite structure in Hvo_0405 may be the result of a fusion of two genes, one encoding an ArtA substrate and the second encoding a protein containing a NifU-like domain that is commonly found in the Fe-S cluster scaffold protein, NifU of nitrogen-fixing bacteria and rhodobacterial species; the function(s) of these proteins has not yet been determined (19). While Hvo_0405 is not expressed under standard laboratory conditions, we have shown that when expressed in trans in H. volcanii, Hvo_0405 can be processed in an ArtA-dependent manner. Moreover, site-directed mutagenesis of the conserved “twin” arginines in the Tat signal peptide prevents processing of this protein. These data strongly suggest that this and other Tat substrates, in addition to a subset of Sec substrates that contain the conserved tripartite structure of ArtA substrates, can be processed in an ArtA-dependent manner.
RESULTS AND DISCUSSION
Sequence analysis of Hvo_0405.
In previous work, the H. volcanii genome was predicted to encode eight ArtA substrates (14). Subsequent analysis has revealed an additional H. volcanii polypeptide, Hvo_0405, containing a putative ArtA recognition motif that is not located at the C terminus, as it is in other predicted substrates. Instead, the PGF motif, followed by a hydrophobic stretch of amino acids and a positively charged region, is located 171 amino acids (aa) N-terminal to the stop codon of Hvo_0405 (Fig. 1A).
FIG 1.
Hvo_0405 is a fusion protein resulting from a single base deletion. (A) Predicted amino acid sequence alignment of Hvo_0405 with sequences of two proteins in other Haloferax species, which are encoded by genes adjacent to each other. Proteins homologous to the N-terminal region of Hvo_0405 contain a predicted Tat signal peptide, a signal peptidase I (SPase I)-processing site (arrowhead), conserved LVIVD repeat domains, and a PGF motif followed by a hydrophobic stretch and a positively charged region typical of ArtA substrates. Proteins homologous to the Hvo_0405 C-terminal region contain a NifU-like domain. An asterisk indicates positions which have a single, fully conserved residue, a colon indicates conservation between groups with strongly similar properties, and a period indicates conservation between groups with weakly similar properties. (B) Nucleotide alignment of the DNA sequences homologous to hvo_0405; the H. volcanii hvo_0405 gene appears to have lost a conserved cytosine 1,462 bp 3′ of the start site. This single-base-pair deletion (arrow) results in a frameshift, replacing the last five amino acids, eliminating the stop codon, and resulting in an in-frame intergenic region leading to the fusion that includes the N- and C-terminal regions of Hvo_0405. The protein and DNA sequence alignments shown include sequences from four Haloferax species, in addition to H. volcanii, which are representative of the sequences tested for all 11 Haloferax genomes available to date.
The Hvo_0405 region N-terminal to the PGF motif contains a predicted Tat signal peptide and several LVIVD repeat domains (Fig. 1A), which are predicted to form a four-β-stranded β-propeller domain that is often found in archaeal and bacterial proteins that localize to the cell surface (19, 20). The Hvo_0405 region C-terminal to the PGF motif contains a conserved NifU-like domain; originally discovered in proteins involved in nitrogen fixation, proteins containing a NifU-like domain are found in many species and typically bind a 4Fe-4S cluster (Fig. 1A) (19, 21, 22).
To determine whether the noncanonical ArtA substrate structure found in Hvo_0405 is conserved in other Haloferax spp., we performed BLAST searches for homologs and created a protein alignment using all Haloferax spp. sequenced to date. Surprisingly, H. volcanii Hvo_0405 demonstrated sequence homology to two polypeptides in other Haloferax species, including a protein containing LVIVD repeat domains and a NifU-like domain (Fig. 1A; also data not shown). The predicted protein containing the LVIVD repeat domains also contains a predicted N-terminal Tat signal peptide, in addition to the conserved ArtA recognition tripartite structure at the C terminus. Therefore, the most parsimonious explanation for the ArtA recognition motif not being localized to the C terminus in Hvo_0405 is that at some point during its evolutionary history, a mutation in the H. volcanii genome resulted in the fusion of two polypeptides that had previously been separately translated, with the polypeptide that became the N-terminal portion of the nascent fusion containing an ArtA recognition motif.
Since a fusion of two genes to form a gene encoding a single polypeptide requires a mutation or genome rearrangement, we sought to determine the precise genomic changes in H. volcanii that resulted in the fusion of Hvo_0405. We performed nucleotide alignments of the corresponding genomic regions for each of the 11 Haloferax species (Fig. 1B; also data not shown). We found that the nucleotide sequences are highly conserved between H. volcanii and the other Haloferax spp. but discovered a single-base-pair deletion in the H. volcanii hvo_0405 region encoding the charged domain of the tripartite structure. The deletion of this single cytosine base causes a frameshift that leaves the stop codon out of frame and an intergenic region that is translated without interruption, all while preserving the reading frame of the downstream sequence encoding the protein containing the NifU-like domain, thereby resulting in a single H. volcanii protein containing both the LVIVD repeat domains and the NifU-like domain. Interestingly, we also found that in Haloferax mediterranei, the otherwise conserved PGF motif is mutated to TGF. While we previously showed that a PGF motif is required for ArtA-dependent processing, this finding raises the question of whether a motif having a threonine rather than a proline at position +1 can be recognized by ArtA, potentially expanding the range of predicted ArtA substrates significantly (13, 14).
Construction and characterization of an H. volcanii Δhvo_0405 mutant strain.
To determine whether this fusion protein is functional and what role it might play in the cell, we created an H. volcanii Δhvo_0405 mutant strain and sought to characterize its defects. The Δhvo_0405 mutant strain (see Fig. S1 in the supplemental material) does not exhibit a discernible growth defect in comparison to the H53 background strain in a semidefined Casamino Acids (CA) medium (Fig. S2A). Additionally, when grown in CA liquid culture inoculated either with a colony or after liquid-to-liquid transfers, the cells of the Δhvo_0405 mutant strain display morphologies similar to those of the parental strain (Fig. S2B). Moreover, the deletion of hvo_0405, which is predicted to encode a cell surface protein containing low-complexity LVIVD repeat domains, does not result in any discernible surface adhesion phenotype under standard conditions (Fig. S2B). To date, we have been unable to reliably ascribe any phenotype to the loss of hvo_0405.
Characterization of ArtA-dependent processing of Hvo_0405.
Previous in silico studies have described only putative archaeosortase and exosortase substrates containing a tripartite structure located at the C terminus (14, 17). Since the Hvo_0405 tripartite structure is atypically located, we asked whether H. volcanii ArtA has a strict requirement that the tripartite structure of a substrate be located at the C terminus. To investigate this possibility, we constructed a plasmid containing the hvo_0405 coding sequence fused to a C-terminal polyhistidine tag under the control of a tryptophan-inducible promoter. Additionally, we created an antibody that detects only the N-terminal portion of Hvo_0405 (Hvo_0405-N-term), which contains the LVIVD repeat domains. When we induced expression of Hvo_0405 from a plasmid in cells containing ArtA, we observed both the full-length Hvo_0405 polypeptide, consisting of an N terminus containing LVIVD repeat domains and a C terminus containing a NifU-like domain, and processed forms of Hvo_0405 detected by immunoblotting using either anti-Hvo_0405-N-term or anti-penta-His antibody (Fig. 2). Strikingly, upon immunoblot analysis of proteins isolated from cells lacking ArtA, we observed only a band corresponding to the full-length form of Hvo_0405 (not yet processed by ArtA) (Fig. 2), strongly suggesting that Hvo_0405 is processed in an ArtA-dependent manner. To date, we have been unable to detect Hvo_0405 in nontransformed H53 parental strain cells grown under a variety of conditions (in semidefined versus complex medium or in biofilms) via immunoblotting with the anti-Hvo_0405-N-term antibody (Fig. 2; also data not shown). This result, which is consistent with mass spectrometric analyses that failed to identify Hvo_0405 peptides in H53 (data not shown) and microarray analysis not identifying significant hvo_0405 transcript (C. Daniels, unpublished data), may explain the absence of an observable phenotype for the Δhvo_0405 mutant strain.
FIG 2.
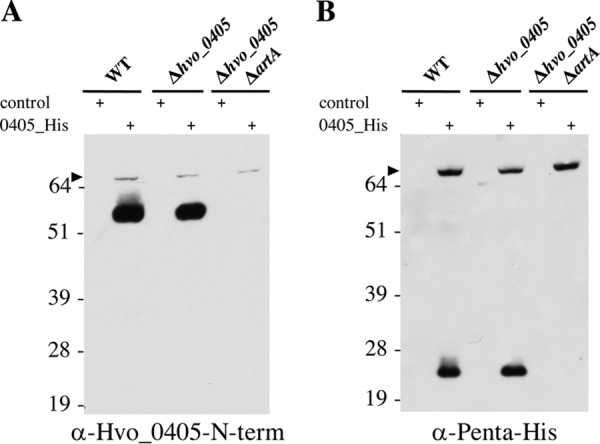
Hvo_0405 processing is dependent upon ArtA. Western blot analysis of proteins isolated from cell pellets generated from mid-log-phase liquid cultures for the wild type (WT), the Δhvo_0405 mutant, or the ΔartA Δhvo_0405 mutant transformed with pTA963 (control) or pTA963 expressing Hvo_0405 with a 6×His tag using anti-Hvo_0405-N-term antibodies recognizing the N terminus of Hvo_0405 (A) or anti-penta-His antibodies recognizing the C-terminal 6×His tag (B). Numbers indicate molecular masses in kilodaltons, and the arrowhead indicates the unprocessed full-length Hvo_0405. The images shown represent the results from at least three independent replicates tested.
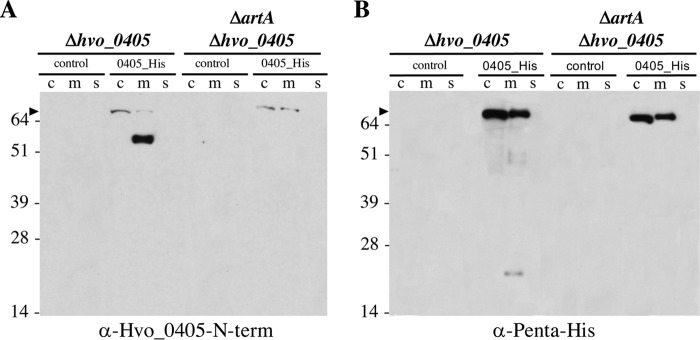
Canonical ArtA substrates, such as the SLG, are predicted to localize to the cell membrane. Since our data suggest that Hvo_0405 is an atypical ArtA substrate, we sought to determine the subcellular localization of the processed N- and C-terminal portions of Hvo_0405. To this end, we purified cytoplasmic, membrane, and supernatant fractions from both cells that express ArtA and cells that lack ArtA, each induced to express the Hvo_0405 construct. We found that although some full-length protein was observed in membrane fractions, full-length Hvo_0405 predominantly localizes to the cytoplasm, suggesting that this protein is posttranslationally inserted into the membrane, consistent with our in silico prediction of its being a Tat substrate (Fig. 1A and 3). Again, only full-length Hvo_0405 was observed in ArtA-deficient cells (Fig. 3), supporting our hypothesis that Hvo_0405 is processed in an ArtA-dependent manner. The processed N-terminal fragment containing the LVIVD repeat domains, which was detected with a signal intensity significantly greater than that of the full-length protein, localizes exclusively to the membrane fraction, suggesting that, like SLG, this ArtA-processed protein is membrane anchored (Fig. 3A). Unfortunately, attempts to detect lipid-modified Hvo_0405 were unsuccessful (data not shown). Using mass spectrometric analysis to resolve this was not possible due to the limited information available on the lipid modification site. While it is possible that Hvo_0405 is not lipid anchored, we cannot exclude the possibility that the Hvo_0405 protein concentration was too low to be detected using 14C-labeled mevalonic acid, which we had previously used to confirm the ArtA-dependent lipidation of the highly abundant SLG (13).
FIG 3.
Processed Hvo_0405 N- and C-terminal regions localized to the membrane. Western blot analysis of cytoplasmic (c), membrane (m), and supernatant (s) fractions for protein isolated from mid-log-phase liquid cultures of the Δhvo_0405 mutant or the ΔartA Δhvo_0405 mutant, transformed with pTA963 (control) or pTA963 expressing Hvo_0405 with a 6×His tag, using anti-Hvo_0405-N-term antibodies recognizing the N terminus of Hvo_0405 (A) or anti-penta-His antibodies recognizing the C-terminal 6×His tag (B). Numbers indicate molecular masses in kilodaltons, and the arrowhead indicates the unprocessed full-length Hvo_0405. The images shown represent the results from at least three independent replicates tested.
Interestingly, the processed C-terminal fragment containing the NifU-like domain is also exclusively localized to the membrane, but, consistent with results from whole-cell fractions, it is found in much lower abundance than the full-length form, possibly due to the degradation of this C-terminal region (Fig. 3B). While membrane association of the C-terminal region is likely due to the N-terminal transmembrane segment remaining after ArtA processing, in Haloferax species in which this NifU-like domain-containing protein is not fused to an ArtA substrate, it is likely a cytoplasmic protein. Therefore, this membrane-associated protein may be unstable because it is not adapted to function as a membrane protein.
Hvo_0405 is a Tat substrate.
As noted above, identification of full-length Hvo_0405 in the cytoplasm could be explained by the fact that, once translated, this protein, as predicted, is transported via the Tat pathway. We then sought to experimentally determine whether Hvo_0405 is indeed a Tat substrate. Since both Sec and Tat transport pathways are essential in H. volcanii, deleting the genes involved in these pathways to determine which transport pathway is used by Hvo_0405 is not feasible (5). Therefore, we generated a plasmid expressing a gene encoding an Hvo_0405 construct, in which the nucleotides encoding the highly conserved two arginine residues at the N terminus of Hvo_0405 (Fig. 1A), which are predicted to be crucial for Tat substrate recognition, were mutated to encode two lysine residues (Hvo_0405KK). We then expressed this tryptophan-inducible Hvo_0405KK mutant in trans in the Δhvo_0405 mutant strain to determine whether the mutated protein can still be transported properly. By immunoblot analysis of proteins isolated from cell membrane and cytoplasmic fractions, we found that the twin-arginine residues are required for Hvo_0405 processing, as the processed forms of Hvo_0405 in the membrane fractions were observed only in strains expressing Hvo_0405 that contain the twin-arginine residues, while the strain expressing the Hvo_0405KK mutant contained only full-length protein (Fig. 4).
FIG 4.

The Hvo_0405 Tat signal peptide is critical for ArtA-dependent C-terminal processing. Western blot analysis of proteins isolated from the cytoplasmic (c), membrane (m), and supernatant (s) fractions of the Δhvo_0405 mutant expressing the wild-type C-terminally His-tagged Hvo_0405 (Hvo_0405RR) or C-terminally His-tagged Hvo_0405 with a mutated twin lysine signal sequence (Hvo_0405KK), using either anti-Hvo_0405-N-term antibodies (A) or anti-penta-His antibodies recognizing the C-terminal 6×His tag (B). Numbers indicate molecular masses in kilodaltons, and the arrowhead indicates the unprocessed full-length Hvo_0405. The images shown represent the results from at least three independent replicates tested.
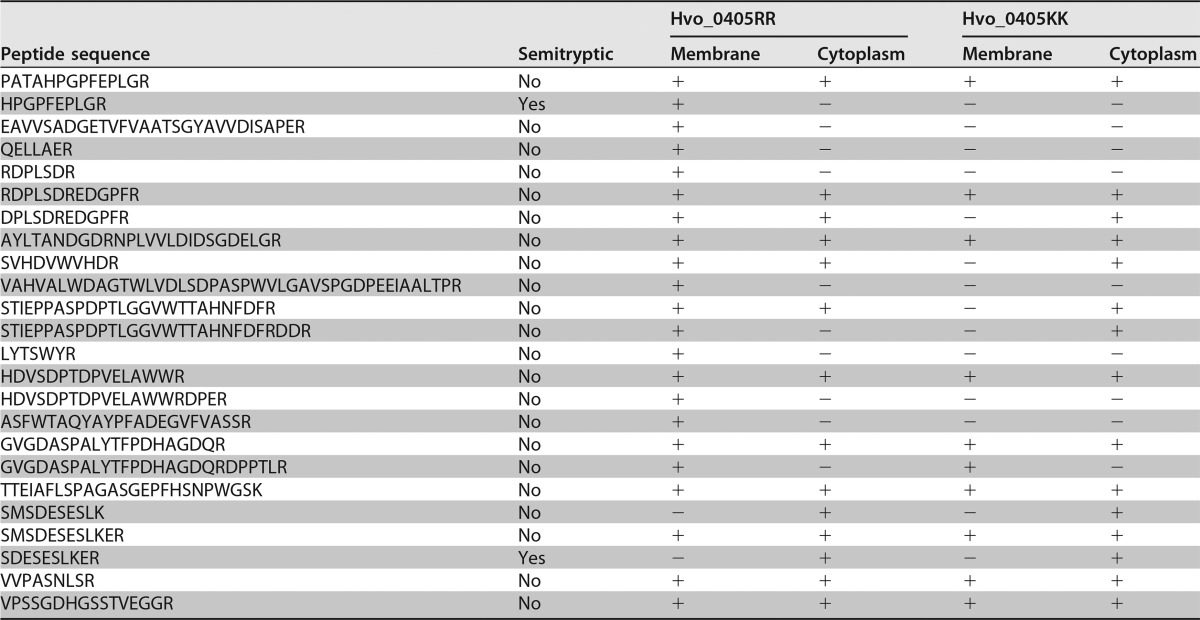
Interestingly, we found that a fraction of the unprocessed Hvo_0405KK continued to be localized to the membrane (Fig. 4). It is possible that the presence of this small fraction of unprocessed Hvo_0405KK is due to contamination from the cytoplasm, which would further support our hypothesis that Hvo_0405 is indeed a Tat substrate, as the Hvo_0405KK is no longer targeted to the membrane. However, we hypothesize that the likely reason for the Hvo_0405KK membrane localization is that the Sec machinery recognizes the hydrophobic region in the tripartite structure of the Hvo_0405KK mutant and targets the protein to the cytoplasmic membrane. Due to its at least partially folded state, the Hvo_0405KK mutant protein would be unable to properly translocate across the cytoplasmic membrane through the Sec pore, resulting in the full-length Hvo_0405KK mutant protein, which cannot be processed in an ArtA-dependent manner, localizing to the membrane. In this scenario, it would also be unlikely that the evolutionarily conserved signal peptidase I (SPase I), the peptidase that processes Sec and Tat substrate signal peptides, would be able to process Hvo_0405KK. To test this hypothesis, we performed mass spectrometric analysis of trypsin-digested proteins isolated from the membrane and cytoplasmic fractions of the Δhvo_0405 mutant strain expressing either the wild-type Hvo_0405RR or the mutated Hvo_0405KK. While multiple tryptic peptides were detected in the trypsin-treated proteins isolated from membrane fractions of strains expressing either Hvo_0405RR or Hvo_0405KK, a semitryptic peptide, HPGPFEPLGR, was found only in protein samples from the wild-type strain expressing Hvo_0405RR (Table 1). This semitryptic peptide likely represents the most N-terminal peptide of the mature Hvo_0405 protein, and its N-terminal sequence indicates the position of the SPase I cleavage site, which is consistent with the predicted signal peptide cleavage site identified by SignalP (23). In contrast, while we did not detect any semitryptic fragments in the membrane fractions from the mutated strain expressing Hvo_0405KK, a tryptic peptide including the SPase I-processing site, PATAHPGPFEPLGR, was identified in both replicates, suggesting that only the unprocessed signal peptide was present (Table 1). It should be noted that, despite the generally accepted “Keil rules” (24), tryptic cleavage before proline is possible (25). These results provide additional support for Hvo_0405 as a bona fide Tat substrate, as well as provide one of only a few in vivo-confirmed SPase I-processing sites for archaeal or bacterial Tat substrates.
TABLE 1.
Peptide identifications in strains expressing Hvo_0405RR or Hvo_0405KK
A second semitryptic peptide, SDESESLKER, was identified exclusively in protein samples isolated from the cytoplasmic fractions of strains expressing either Hvo_0405RR and Hvo_0405KK (Table 1). Interestingly, in all other Haloferax spp., this semitryptic fragment corresponds to the penultimate position of the region containing the start codon of the NifU-like domain-containing protein (Fig. 1). The removal of the N-terminal methionine is consistent with methionine aminopeptidase processing observed in many prokaryotic species (26). This observation supports our hypothesis that the DNA between the regions encoding the Hvo_0405 N-terminal region, which contains the LVIVD repeat domains, and the C-terminal region containing the NifU-like domain includes an internal promoter that continues to drive expression of the protein containing the NifU-like domain.
Using the TatFind and TatP programs, we also determined whether any of the other putative H. volcanii ArtA substrates are predicted to be Tat substrates (5, 8). Despite containing twin arginines in the charged region of the signal peptides of Hvo_2160 and Hvo_B0206, neither of these two predicted ArtA substrates were predicted by TatFind to be Tat substrates. Alternatively, sequence analysis with TatP predicted that Hvo_2160 is indeed carrying a Tat signal peptide. The discrepancies between these Tat substrate prediction programs indicate that it is imperative to carry out additional in vivo studies like the one presented here to obtain a larger training set for optimizing the subcellular localization prediction programs.
Concluding remarks.
In this work, we demonstrate that the predicted ArtA substrate Hvo_0405 is likely a fusion of an ArtA substrate and a cytoplasmic protein, resulting in a tripartite structure no longer located at the C terminus. Despite the atypical localization of this putative ArtA recognition motif, we demonstrate that Hvo_0405 is indeed processed in an ArtA-dependent manner. Interestingly, there are fewer positively charged residues following the PGF motif and hydrophobic stretch in Hvo_0405 than in homologs from other Haloferax species (one versus five to six) (Fig. 1). Since this paucity of positively charged residues apparently does not interfere with ArtA processing, we can conclude that either this is not a requirement for ArtA substrates, consistent with the putative ArtA substrate Hvo_1095 that also contains only a single positively charged residue at the C terminus, or that the role of these residues in anchoring the C terminus to the cytoplasmic membrane is subsumed by the large NifU-like domain of Hvo_0405. Together with our observation that the conserved PGF motif is TGF in the H. mediterranei homolog of LVIVD domain-containing N-terminal region of Hvo_0405, these findings suggest that ArtA has a broader substrate repertoire than previously thought (Fig. 1).
Our data also suggest additional mechanisms by which proteins can become associated with the cell membrane. The LVIVD repeat domain-containing N-terminal region of Hvo_0405 is likely membrane anchored via lipidation, similar to other ArtA substrates. It is intriguing that the NifU-like domain-containing C-terminal region is also membrane associated, likely due to the hydrophobic region that was originally part of the tripartite structure but now remains attached to the N terminus after ArtA-dependent processing. This is likely a novel cellular location for this NifU-like domain-containing protein, as it is almost certainly a cytoplasmic protein in other Haloferax spp., consistent with the identification of the second semitryptic peptide, exclusively in the cytoplasm (Table 1).
Using site-directed mutagenesis, we demonstrate that Hvo_0405 is a Tat substrate and that its processing is dependent upon the Tat pathway by both immunoblotting and mass spectrometry. Additionally, our mass spectrometry data confirm the SPase I-processing site in vivo. Prediction of SPase I-processing sites in Tat substrates has been challenging due to a paucity of confirmed SPase I-processing sites, evidenced by the failure of TatP to accurately predict the Hvo_0405-processing site. Studies like these will almost certainly provide better training sets for prediction programs, not only for accurate prediction of Sec and Tat substrates in halophilic archaea but also in the prediction of ArtA substrates in archaea and exosortase substrates in Gram-negative bacteria.
MATERIALS AND METHODS
Strains and growth conditions.
The plasmids and strains used in this study are listed in Table 2. H. volcanii strain H53 and its derivatives were grown at 45°C in liquid (orbital shaker at 250 rpm) or on solid semidefined Casamino Acids (CA) medium supplemented with tryptophan and uracil (both at 50 μg · ml−1 final concentration) (27). Solid medium plates contained 1.5% (wt/vol) agar. To ensure equal agar concentrations in all plates, agar was completely dissolved in the medium prior to autoclaving, and the autoclaved medium was stirred before plates were poured. Strains transformed with pTA963 were grown on CA medium supplemented with tryptophan (50 μg · ml−1 final concentration) (28). For selection of the deletion mutant, 5-fluoroorotic acid (5-FOA) (Toronto Research Biochemicals) (150 μg · ml−1 final concentration) and uracil (Sigma) (10 μg · ml−1 final concentration) were added to CA medium. Escherichia coli strains were grown at 37°C in NZCYM (Fisher Scientific) medium supplemented with ampicillin (Sigma) (100 μg · ml−1 final concentration) (29).
TABLE 2.
Plasmids and strains
| Plasmid or strain | Relevant characteristicsa | Reference or source |
|---|---|---|
| Plasmids | ||
| pET-22b(+) | Ampr; expression vector; T7 promoter, C-terminal 6×His tag | Novagen |
| pTA131 | Ampr; pyrE2+ marker | 32 |
| pTA963 | Ampr; pyrE2+ and hdrB+ markers; p.tnaA (Trp-inducible promoter) | 28 |
| pFH24 | pTA131 with insert: XhoI::700 bp 5′ of hvo_0405::700 bp 3′ of hvo_0405::XbaI | This study |
| pFH25 | pTA963 with insert: p.tnaA::NdeI::hvo_0405 cds::GGP linker::6×His::stop::EcoRI | This study |
| pJS143 | pET-22b(+) with insert: T7 promoter::lacO::ribosomal binding site::NdeI::hvo_0405 (bp 64–1344)::KL(HindIII)AAALE linker::6×His tag::stop | This study |
| pJS151 | pTA963 with insert: p.tnaA::Hvo_0405 cds with coding bases 7–12 mutated from CGCCGC to AAGAAG (changes RR to LL)::GGP linker::6×His tag::stop | This study |
| Strains | ||
| DH5α | E. coli F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| DL739 | E. coli Cmr; MC4100 recA dam-13::Tn9 | 31 |
| BL21(DE3) | E. coli Cmr; strain B F− ompT gal dcm lon hsdSB(rB− mB−) gal dcm (DE3 λ[lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λS)(pLysS[T7p20 orip15A]) | 33 |
| H53 | H. volcanii DS70 ΔpyrE2 ΔtrpA | 32 |
| AF103 | H53 ΔartA | 12 |
| FH24 | H53 Δhvo_0405 | This study |
| FH35 | H53 ΔartA Δhvo_0405 | This study |
| JS1 | H53 containing pTA963 | This study |
| JS13 | H53 containing pFH25 | This study |
| JS16 | FH24 containing pTA963 | This study |
| JS17 | FH24 containing pFH25 | This study |
| JS23 | FH35 containing pTA963 | This study |
| FH41 | FH35 containing pFH25 | This study |
| FH59 | FH24 containing pJS151 | This study |
Ampr, ampicillin resistance; cds, coding sequence; Cmr, chloramphenicol resistance.
Nucleotide and amino acid sequence analyses.
Protein and nucleotide alignments of Haloferax species Hvo_0405 homologs were performed with Clustal Omega using sequences from H. alexandrinus (gene locus tags RS08345 and RS08350), H. denitrificans (gene locus tags RS09235 and RS09240), H. elongans (gene locus tags RS04170 and RS04175), H. gibbonsii (gene locus tags ABY42_01850 and ABY42_01855), H. larsenii (gene locus tags RS15950 and RS15955), H. lucentense (gene locus tags RS15835 and RS15830), H. mediterranei (gene locus tags AFK18107 and AFK18108), H. mucosum (gene locus tags RS07215 and RS07210), H. prahovense (gene locus tags RS02190 and RS02195), and H. sulfurifontis (gene locus tags RS09845 and RS09840). Coding sequences were also determined by Geneious version 6.1.8 (Biomatters Ltd.). Signal sequence prediction was performed using the TatFind server (http://signalfind.org/tatfind.html) (6), SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) (23), and TatP 1.0 server (http://www.cbs.dtu.dk/services/TatP/) (30).
Plasmid construction.
The pFH24 plasmid, used in generating the Δhvo_0405 mutant strain, was constructed by first PCR amplifying 700 bp 5′ of hvo_0405 using primers FW_0405_XhoI (OP 1) and RV_0405_Up-DW as well as 700 bp 3′ of hvo_0405 using primers FW_0405_Up-DW and RV_0405_XbaI (OP 2). Subsequently, these two fragments were joined into a single amplicon by overlap extension PCR and cloned into pTA131 using XhoI and XbaI. Primers FW_0405_XhoI (OP 1) and RV_0405_XbaI (OP 2) were used to verify the insert by DNA sequencing.
The pFH25 plasmid used for complementation experiments was constructed by PCR amplifying the 1,971-bp hvo_0405 coding sequence with a 3′ GGP linker and 6×His tag using primers FW0405NdeI (IP 1) and RV0405EcoRIHis (IP 2). This amplicon was cloned into pTA963 using NdeI and EcoRI. Primers FW0405NdeI (IP 1) and RV0405EcoRIHis (IP 2) were used to verify the insert by DNA sequencing.
The pJS143 plasmid used for hvo_0405 expression in E. coli BL21(DE3) strain was constructed by PCR amplifying 1,281 bp of hvo_0405 (bp 64 to 1344) using primers Hvo-0405-1F and Hvo-0405-1R. This amplicon was cloned into pET-22b(+) using NdeI and HindIII. The insert was sequenced using primers T7 and T7term.
The pJS151 plasmid containing the hvo_0405 coding sequence with codons for the twin-arginine residues at the N terminus mutated to two lysine residues was constructed by PCR amplification using primers Hvo0405-3F and RV0405EcoRIHis (IP 2). This amplicon was cloned into pTA963 using NdeI and EcoRI. The insert was sequenced using primers pTA963-1F and pTA963-1R.
Polymerase, ligase, and restriction enzymes were purchased from New England BioLabs. Plasmids were initially transformed into E. coli DH5α cells and grown on NZCYM 1.5% agar plates or in liquid medium supplemented with ampicillin (100 μg · ml−1 final concentration). Plasmid preparations were performed using a QIAprep Miniprep (Qiagen) or PureLink plasmid prep (Invitrogen) kit. Prior to H. volcanii transformation, plasmids were passaged through the methylation-deficient E. coli strain DL739 (dam-13::Tn9) (31). All primers are listed in Table S1 in the supplemental material.
H. volcanii transformation and chromosomal substitution.
H. volcanii was transformed using the polyethylene glycol (PEG) method (27). Deletion of the hvo_0405 coding sequence in both the H53 and ΔartA mutant backgrounds was completed using the pFH24 plasmid, the homologous recombination (pop-in pop-out) method, and 5-FOA (32). To confirm the deletion of the hvo_0405 coding sequence in the resulting FH24 and FH35 strains, PCR amplification of the hvo_0405 region was performed using the FW_0405_XhoI (OP 1) and RV_0405_XbaI (OP 2) primers in both H53 and the ΔartA mutant, as well as the FH24 and FH35 deletion strains.
Subcellular fractionation of H. volcanii cultures.
H. volcanii transformants were cultured in CA liquid medium supplemented with tryptophan (50 μg · ml−1 final concentration) to an optical density at 600 nm (OD600) of approximately 0.5. Cells were separated from the supernatant by centrifugation at 6,000 × g for 5 min; subsequently, supernatants were spun a second time and frozen at −80°C along with the cell pellets. Cell pellets were fractionated further by first lysing the cells in phosphate-buffered saline (PBS) with 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) (Sigma), 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma) and 10 mM EDTA (Invitrogen) protease inhibitors. The cell lysate was centrifuged at 6,000 × g for 10 min to pellet unlysed cells, and the cleared cell lysate was then centrifuged at approximately 303,500 × g for 30 min in an Optima MAX-TL ultracentrifuge (Beckman). Subsequently, the membrane-rich pellet was resuspended in PBS. Both the membrane and cytoplasm fractions were centrifuged again at 303,500 × g for 30 min. Proteins in both the cytoplasm and culture supernatant fractions were precipitated using 10% trichloroacetic acid (Thermo Fisher Scientific). Protein pellets from membrane, cytoplasm, and culture supernatant fractions were resuspended in NuPAGE lithium dodecyl sulfate (LDS) sample buffer (Invitrogen) in volumes proportional to the original culture.
Anti-Hvo_0405-N-term antibody.
The anti-Hvo_0405-N-term antibody was generated by first amplifying a 1,281-bp fragment of hvo_0405 (bp 64 to 1344) that was 3′ of the predicted signal peptide and 5′ of predicted glycosylation sites and cloned into pET-22b(+) using NdeI and HindIII restriction sites to create pJS143. The pJS143 plasmid was transformed into the E. coli BL21(DE3) strain (33), and the hvo_0405 fragment, under the control of a T7 promoter, was expressed via genome-encoded T7 polymerase induction with isopropyl-β-d-1-thiogalactopyranoside (IPTG) (1 mM) (Sigma) in NZCYM supplemented with ampicillin (100 μg · ml−1 final concentration) at 37°C. After 4 to 5 h of induction, cells were chilled in ice water and pelleted at 4,000 × g for 15 min at 4°C. Cell pellets were frozen at −80°C.
Subsequently, the C-terminally 6×His-tagged N-terminal fragment of Hvo_0405 was purified by nickel affinity chromatography. Using Ni-nitrilotriacetic acid (Ni-NTA) Superflow beads (Qiagen) under denaturing conditions (8 M urea, 100 mM NaH2PO4, 10 mM Tris base), the purified N-terminal protein fragment of ArtA-processed Hvo_0405 was eluted according to the manufacturer's instructions. Purified protein was then concentrated using Amicon ultracentrifugal filters with a 3-kDa molecular mass cutoff (Millipore), and PBS was added twice in order to reduce the urea concentration to ∼0.4 M. The protein concentration was then determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Polyclonal antibodies were generated in two rabbits using Complete Freund's adjuvant over a 90-day production schedule (Cocalico Biologicals, Inc., Reamstown, PA). The two rabbits showed nearly identical immune responses. Specificity of the anti-Hvo_0405-N-term antibodies for Hvo_0405 was verified by immunoblotting against purified recombinant Hvo_0405.
Immunoblotting.
Liquid cultures were grown until mid-log phase (OD600, 0.4 to 0.5), and the cells were harvested by centrifugation at 4,300 × g for 10 min at 4°C. Cell pellets were resuspended and lysed in 1% (vol/vol) NuPAGE LDS supplemented with 100 mM dithiothreitol (DTT) (Sigma) and stored at −20°C. H. volcanii cell pellets or subcellular fractions were electrophoresed on either 10% or 12% Bis-Tris NuPAGE polyacrylamide gel (Invitrogen) with NuPAGE 3-(N-morpholino) propanesulfonic acid (MOPS)–sodium dodecyl sulfate (SDS) running buffer (Invitrogen). Proteins were then transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) using a semidry transfer apparatus at 15 V for 30 min (Bio-Rad). Next, the membrane was washed twice in PBS, blocked for 1 h in 3% bovine serum albumin (BSA) in PBS, and washed twice in PBS with 1% Tween 20 and once in PBS. For primary antibodies, either the mouse anti-penta-His antibody (catalog no. 34660, 200 μg · ml−1; Qiagen) was used at a 1:1,000 dilution in 3% BSA in PBS with sodium azide or the rabbit anti-Hvo_0405-N-term serum was used at a 1:10,000 dilution in 3% BSA in PBS with sodium azide. Subsequently, the membrane was washed twice in PBS with 1% Tween 20 and once in PBS. For secondary antibodies, either the horseradish peroxidase (HRP)-conjugated Amersham ECL anti-mouse IgG from sheep (GE) was used at a 1:20,000 dilution in 10% nonfat milk in PBS, or the HRP-conjugated Amersham ECL anti-rabbit IgG from donkey (GE) was used at a 1:60,000 dilution in 10% nonfat milk in PBS. The membrane was then washed two to four times in PBS with 1% Tween 20. HRP activity was assessed using the Amersham ECL Prime Western blotting detection reagent (GE) and autoradiography. At least three independent biological replicates were performed for each sample before one representative gel was selected for the immunoblotting result.
Growth curve.
H. volcanii liquid cultures were inoculated from colonies into 5 ml of liquid CA medium supplemented with tryptophan (50 μg · ml−1 final concentration), with continuous shaking at 45°C until mid-log phase (OD600, ∼0.4 to 0.5). Subsequently, approximately 6 μl of each culture (adjusted slightly for OD600 differences) was transferred into a well of a sterile 96-well plate containing 194 μl of fresh liquid medium (eight replicates of two biological replicates). Cultures were grown to stationary phase with continuous shaking, with OD600 recordings taken every 30 min using the BioTek PowerWave X2 microplate spectrophotometer, as described previously (12).
Light microscopy.
One milliliter of a mid-log-phase culture (OD600, 0.4 to 0.5) was concentrated by centrifugation at 4,911 × g for 1 min and pellets were resuspended in 10 μl of liquid CA medium. One microliter of the concentrated cells was transferred to a microscope slide and observed using the Eclipse TE2000-U inverted epifluorescence microscope (Nikon USA, Inc.), as described previously (12).
Adhesion assay.
H. volcanii surface adhesion assay was carried out based on the modified air-liquid interface (Ali) assay as described previously (34). Three milliliters of an H. volcanii mid-log-phase culture (OD600, 0.4 to 0.5) was transferred into each well of a sterile 12-well plate. Plastic coverslips (22 by 22 mm, 0.19 to 0.25 mm thick) were vertically inserted into each well, followed by overnight incubation at 45°C without shaking. The next day, the coverslips were gently retrieved from the well and the adhered cells were fixed by incubating the coverslips in 2% acetic acid for 3 min. Subsequently, the coverslips were stained in 0.1% (wt/vol) crystal violet solution for 10 min. After washing the excess stain with distilled water and air drying, the coverslips were examined by light microscopy.
LC-MS analysis.
Proteins from membrane fractions were solubilized with 2% SDS in 10 mM Tris-HCl (pH 7.6). For all samples, the protein concentration was determined by bicinchoninic acid assay (BCA protein assay kit; Thermo Scientific Pierce) before loading 50 μg of protein onto Amicon ultracentrifugal filters (0.5 ml; 30-kDa molecular mass cutoff; Millipore) for tryptic digestion using the filter-aided sample preparation method (35), as described previously (36). Peptides were dried in a vacuum centrifuge and stored at −20°C. Two biological replicates were processed and analyzed by liquid chromatography-mass spectrometry (LC-MS).
After reconstitution of the peptides in 2% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid in ultrapure water, chromatographic separation was performed on an UltiMate 3000 RSLCnano system (Thermo Scientific). The sample was desalted on a trap column (C18 PepMap 100, 300 μm by 5 mm, 5-mm particle size, 100-Å pore size; Thermo Scientific) for 5 min using 0.05% (vol/vol) trifluoroacetic acid (TFA) and 2% (vol/vol) acetonitrile in ultrapure water, at a flow rate of 10 μl · min−1. The trap column was then switched in line with the separation column (Acclaim PepMap100 C18, 75 mm by 15 cm, 2-mm particle size, 100-Å pore size; Thermo Scientific). For chromatographic separation, the mobile phases were composed of 0.1% (vol/vol) formic acid in ultrapure water and 80% acetonitrile (A) and 0.08% formic acid in ultrapure water (B), and the following gradient was employed using a flow rate of 300 nl · min−1: 5 min of 2.5% B, increase to 7.5% B over 4 min, increase to 40% B over 26 min, increase to 99% B over 1 min, and 10 min of 99% B. The column was reequilibrated with 2.5% B for 25 min.
The LC system was coupled via a nanospray source to a Q Exactive Plus mass spectrometer (Thermo Scientific) operating in positive-ion mode. MS data were acquired from 375 m/z to 2,000 m/z at a resolution of 70,000 for MS1. The automatic gain control (AGC) target was set to 106 and maximum injection time to 100 ms. The 12 most abundant precursor ions were triggered for fragmentation by higher-energy C-trap dissociation with a normalized collision energy of 27. MS2 spectra were acquired in a dynamic scan range with a fixed first mass of 150 m/z at a resolution of 17,500, with an AGC target of 105 and a maximum injection time of 120 ms. Precursor ions with an unassigned charge state as well as charge of 1 or greater than 5 were rejected. Fragmented ions were dynamically excluded for 15 s.
Peptide spectrum matches (PSMs) were identified with the Python framework Ursgal (version 0.4.0) (37) using the database search engines X! Tandem (38), MS-GF+ (39), and OMSSA (40). The database consisted of the UniProt H. volcanii proteome (proteome ID UP000008243) merged with sequences from the Common Repository of Adventitious Proteins (http://www.thegpm.org/crap/), resulting in a total database of 4,103 proteins. Decoy sequences were generated by peptide shuffling and included for all proteins. The Ursgal profile Q Exactive+ was used, including a precursor ion tolerance of 5 ppm and a fragment ion tolerance of 20 ppm. Additionally, carbamidomethylation of cysteine was set as a fixed modification and oxidation of methionine as well as N-terminal acetylation as potential modifications. Three missed cleavages and semienzymatic peptides were allowed. Statistical postprocessing of unified results was performed with Percolator (41). Since the error probability for each PSM should be controlled in order to validate the presence of certain peptides, filtering by posterior error probability (PEP) (≤0.01) was chosen instead of a global false-discovery rate. Results from all search engines for both replicates were combined.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Pohlschroder lab for helpful discussions. We thank Daniel Haft for helpful discussions and identifying the Hvo_0405 tripartite domain.
M.F.A.H., J.D.S., and M.P. were supported by the National Science Foundation grant MCB-1413158. M.P. is also supported by a Westfälische Wilhelms-Universität fellowship. M.F.A.H. also received funding from the University of Pennsylvania School of Arts and Sciences Dissertation Completion Fellowship. S.S. and M.H. were supported by a Deutsche Forschungsgemeinschaft grant.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00802-16.
REFERENCES
- 1.Denks K, Vogt A, Sachelaru I, Petriman NA, Kudva R, Koch HG. 2014. The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol Membr Biol 31:58–84. doi: 10.3109/09687688.2014.907455. [DOI] [PubMed] [Google Scholar]
- 2.Goosens VJ, van Dijl JM. 29 April 2016. Twin-arginine protein translocation. Curr Top Microbiol Immunol doi: 10.1007/82_2016_7. [DOI] [PubMed] [Google Scholar]
- 3.Berks BC. 2015. The twin-arginine protein translocation pathway. Annu Rev Biochem 84:843–864. doi: 10.1146/annurev-biochem-060614-034251. [DOI] [PubMed] [Google Scholar]
- 4.Dilks K, Rose RW, Hartmann E, Pohlschroder M. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol 185:1478–1483. doi: 10.1128/JB.185.4.1478-1483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dilks K, Giménez MI, Pohlschröder M. 2005. Genetic and biochemical analysis of the twin-arginine translocation pathway in halophilic archaea. J Bacteriol 187:8104–8113. doi: 10.1128/JB.187.23.8104-8113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rose RW, Brüser T, Kissinger JC, Pohlschröder M. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol 45:943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 7.Pohlschroder M, Dilks K, Hand NJ, Wesley Rose R. 2004. Translocation of proteins across archaeal cytoplasmic membranes. FEMS Microbiol Rev 28:3–24. doi: 10.1016/j.femsre.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Giménez MI, Dilks K, Pohlschröder M. 2007. Haloferax volcanii twin-arginine translocation substates include secreted soluble, C-terminally anchored and lipoproteins. Mol Microbiol 66:1597–1606. doi: 10.1111/j.1365-2958.2007.06034.x. [DOI] [PubMed] [Google Scholar]
- 9.Saier MH., Jr 2006. Protein secretion and membrane insertion systems in Gram-negative bacteria. J Membr Biol 214:75–90. doi: 10.1007/s00232-006-0049-7. [DOI] [PubMed] [Google Scholar]
- 10.Schneewind O, Missiakas D. 2014. Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochim Biophys Acta 1843:1687–1697. doi: 10.1016/j.bbamcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nass B, Poll U, Langer JD, Kreuter L, Kuper U, Flechsler J, Heimerl T, Rachel R, Huber H, Kletzin A. 2014. Three multihaem cytochromes c from the hyperthermophilic archaeon Ignicoccus hospitalis: purification, properties and localization. Microbiology 160:1278–1289. doi: 10.1099/mic.0.077792-0. [DOI] [PubMed] [Google Scholar]
- 12.Abdul Halim MF, Pfeiffer F, Zou J, Frisch A, Haft D, Wu S, Tolic N, Brewer H, Payne SH, Pasa-Tolic L, Pohlschroder M. 2013. Haloferax volcanii archaeosortase is required for motility, mating, and C-terminal processing of the S-layer glycoprotein. Mol Microbiol 88:1164–1175. doi: 10.1111/mmi.12248. [DOI] [PubMed] [Google Scholar]
- 13.Halim MFA, Karch KR, Zhou Y, Haft DH, Garcia BA, Pohlschroder M. 2015. Permuting the PGF-CTERM signature motif blocks both archaeosortase-dependent C-terminal cleavage and prenyl lipid attachment for the Haloferax volcanii S-layer glycoprotein. J Bacteriol 198:808–815. doi: 10.1128/JB.00849-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haft DH, Payne SH, Selengut JD. 2012. Archaeosortases and exosortases are widely distributed systems linking membrane transit with posttranslational modification. J Bacteriol 194:36–48. doi: 10.1128/JB.06026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sleytr UB, Schuster B, Egelseer E-M, Pum D. 2014. S-layers: principles and applications. FEMS Microbiol Rev 38:823–864. doi: 10.1111/1574-6976.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegel SD, Liu J, Ton-That H. 2016. Biogenesis of the Gram-positive bacterial cell envelope. Curr Opin Microbiol 34:31–37. doi: 10.1016/j.mib.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haft DH, Paulsen IT, Ward N, Selengut JD. 2006. Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol 4:29. doi: 10.1186/1741-7007-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagos PG, Tsirigos KD, Plessas SK, Liakopoulos TD, Hamodrakas SJ. 2009. Prediction of signal peptides in archaea. Protein Eng Des Sel 22:27–35. [DOI] [PubMed] [Google Scholar]
- 19.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adindla S, Inampudi KK, Guruprasad L. 2007. Cell surface proteins in archaeal and bacterial genomes comprising “LVIVD,” “RIVW” and “LGxL” tandem sequence repeats are predicted to fold as beta-propeller. Int J Biol Macromol 41:454–468. doi: 10.1016/j.ijbiomac.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Ouzounis C, Bork P, Sander C. 1994. The modular structure of NifU proteins. Trends Biochem Sci 19:199–200. doi: 10.1016/0968-0004(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 22.Py B, Gerez C, Angelini S, Planel R, Vinella D, Loiseau L, Talla E, Brochier-Armanet C, Garcia Serres R, Latour JM, Ollagnier-de Choudens S, Fontecave M, Barras F. 2012. Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Mol Microbiol 86:155–171. doi: 10.1111/j.1365-2958.2012.08181.x. [DOI] [PubMed] [Google Scholar]
- 23.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 24.Keil B. 1992. Specificity of proteolysis. Springer-Verlag, Berlin, Germany. doi: 10.1007/978-3-642-48380-6. [DOI] [Google Scholar]
- 25.Rodriguez J, Gupta N, Smith RD, Pevzner PA. 2008. Does trypsin cut before proline? J Proteome Res 7:300–305. doi: 10.1021/pr0705035. [DOI] [PubMed] [Google Scholar]
- 26.Falb M, Aivaliotis M, Garcia-Rizo C, Bisle B, Tebbe A, Klein C, Konstantinidis K, Siedler F, Pfeiffer F, Oesterhelt D. 2006. Archaeal N-terminal protein maturation commonly involves N-terminal acetylation: a large-scale proteomics survey. J Mol Biol 362:915–924. doi: 10.1016/j.jmb.2006.07.086. [DOI] [PubMed] [Google Scholar]
- 27.Dyall-Smith M. 2004. The Halohandbook: protocols for haloarchaeal genetics. University of Melbourne, Victoria, Australia. [Google Scholar]
- 28.Allers T, Barak S, Liddell S, Wardell K, Mevarech M. 2010. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl Environ Microbiol 76:1759–1769. doi: 10.1128/AEM.02670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blattner FR, Williams BG, Blechl AE, Denniston-Thompson K, Faber HE, Furlong L, Grunwald DJ, Kiefer DO, Moore DD, Schumm JW, Sheldon EL, Smithies O. 1977. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science 196:161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- 30.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blyn LB, Braaten BA, Low DA. 1990. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J 9:4045–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allers T, Ngo H-P, Mevarech M, Lloyd RG. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol 70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 34.Esquivel RN, Xu R, Pohlschroder M. 2013. Novel archaeal adhesion pilins with a conserved N terminus. J Bacteriol 195:3808–3818. doi: 10.1128/JB.00572-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 36.Esquivel RN, Schulze S, Xu R, Hippler M, Pohlschroder M. 2016. Identification of Haloferax volcanii pilin N-glycans with diverse roles in pilus biosynthesis, adhesion, and microcolony formation. J Biol Chem 291:10602–10614. doi: 10.1074/jbc.M115.693556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kremer LP, Leufken J, Oyunchimeg P, Schulze S, Fufezan C. 2016. Ursgal, universal python module combining common bottom-up proteomics tools for large-scale analysis. J Proteome Res 15:788–794. doi: 10.1021/acs.jproteome.5b00860. [DOI] [PubMed] [Google Scholar]
- 38.Craig R, Beavis RC. 2003. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom 17:2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 39.Kim S, Pevzner PA. 2014. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun 5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. 2004. Open mass spectrometry search algorithm. J Proteome Res 3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 41.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. 2007. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.