CLUH binds mRNAs implicated in intermediate metabolism and oxidative phosphorylation, but the physiological and molecular significance of these interactions is unclear. Schatton et al. use new constitutive and liver-specific Cluh knockouts to define the function of CLUH in catabolic and energy-converting pathways as a regulator of the translation and stability of target mRNAs.
Abstract
Mitochondria are essential organelles that host crucial metabolic pathways and produce adenosine triphosphate. The mitochondrial proteome is heterogeneous among tissues and can dynamically change in response to different metabolic conditions. Although the transcriptional programs that govern mitochondrial biogenesis and respiratory function are well known, posttranscriptional regulatory mechanisms remain unclear. In this study, we show that the cytosolic RNA-binding protein clustered mitochondria homologue (CLUH) regulates the expression of a mitochondrial protein network supporting key metabolic programs required under nutrient deprivation. CLUH exerts its function by controlling the stability and translation of target messenger RNAs. In the absence of Cluh, mitochondria are severely depleted of crucial enzymes involved in catabolic energy-converting pathways. CLUH preserves oxidative mitochondrial function and glucose homeostasis, thus preventing death at the fetal–neonatal transition. In the adult liver, CLUH ensures maximal respiration capacity and the metabolic response to starvation. Our results shed new light on the posttranscriptional mechanisms controlling the expression of mitochondrial proteins and suggest novel strategies to tailor mitochondrial function to physiological and pathological conditions.
Introduction
Mitochondria are complex organelles involved in ATP production and in several crucial biosynthetic and catabolic pathways. Their function is heterogeneous in different tissues, as reflected by qualitative and quantitative differences in their proteome (Mootha et al., 2003; Calvo and Mootha, 2010). The number of mitochondria and the expression of the mitochondrial proteome can dynamically change in response to cellular and tissue demands. Transcriptional networks enhancing mitochondrial biogenesis ensure long-term adaptation of mitochondrial function. For example, an increase in mitochondrial biogenesis has been observed in the skeletal muscle upon exercise and in the brown fat during adaptive thermogenesis. The transcriptional coactivator PGC1-α is activated in response to various cell signals and regulates nuclear DNA-binding transcription factors, which in turn control the expression of nuclear-encoded mitochondrial proteins, including respiratory chain subunits, and of proteins involved in replication, transcription, translation, and lipid metabolism (Scarpulla, 2002; Hock and Kralli, 2009). However, it is not clear whether transcriptional programs can provide specificity and flexibility to the mitochondrial adaptive responses required to reshape the mitochondrial proteome to boost specific metabolic functions, such as upon abrupt and transient changes in nutrient availability as it occurs at birth.
Clustered mitochondria homologue (CLUH) is an evolutionarily conserved protein that earned its name for the clustering of mitochondria next to the nucleus, which is observed in its absence in species as distant as yeast, amoeba, flies, plants, and mammals (Fields et al., 1998, 2002; Logan et al., 2003; Cox and Spradling, 2009; Gao et al., 2014). We recently discovered that CLUH is a cytosolic RNA-binding protein specific for a subset of mRNAs encoding mitochondrial proteins (Gao et al., 2014), making CLUH an excellent candidate for exerting a posttranscriptional regulatory role in their expression. CLUH is a 150-kD protein containing a highly conserved clustered mitochondria central domain followed by tetratricopeptide repeat domains in the C-terminal part of the protein. Recent data in the fly suggest that the tetratricopeptide repeat domains may favor RNA binding (Sen and Cox, 2016). In HeLa cells, CLUH binds several mRNAs implicated in intermediate metabolism and oxidative phosphorylation (OXPHOS; Gao et al., 2014), but the physiological and molecular significance of the interaction between CLUH and bound mRNAs is still unclear.
In this study, we develop constitutive and liver-specific Cluh knockouts (KOs) in the mouse and demonstrate that CLUH orchestrates the expression of mitochondrial proteins that function in crucial catabolic and energy-converting pathways by promoting the translation and ensuring the stability of its target mRNAs. In the absence of CLUH, mice fail to adapt their mitochondrial metabolism to conditions of nutrient deficiency, such as during the fetal–neonatal transition and starvation.
Results
Cluh−/− mice die shortly after birth
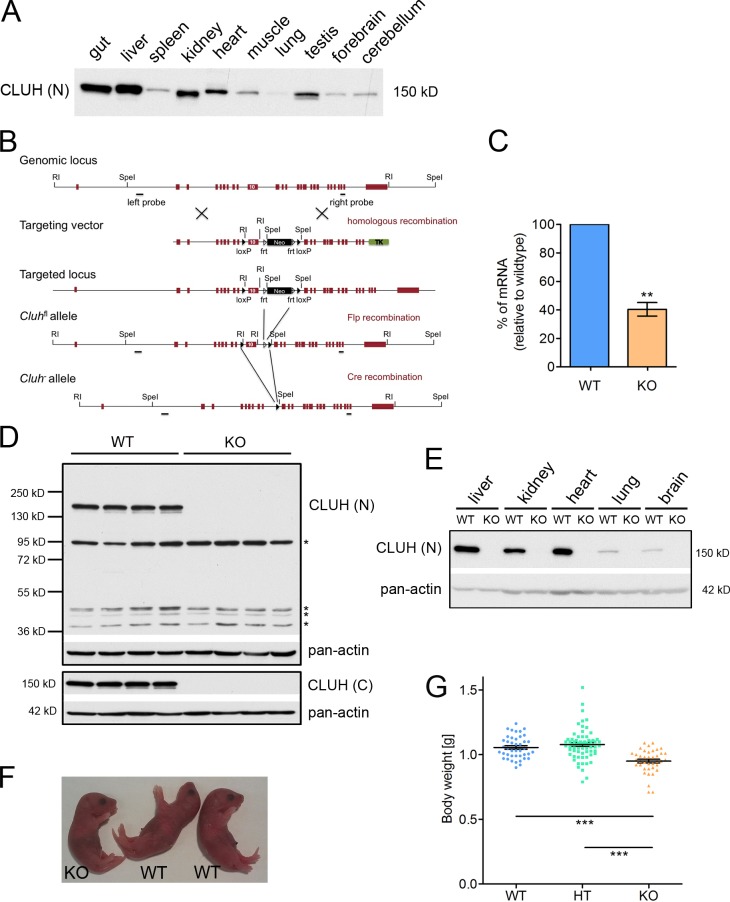
CLUH is ubiquitously expressed, but higher levels are found in the liver, gut, kidney, and heart (Fig. 1 A), raising the question of the physiological role of CLUH in different tissues. To this purpose, we generated a conditional Cluh mouse model (Cluhfl/fl), in which exon 10, encoding part of the conserved clustered mitochondria central domain, was flanked by loxP sites (Fig. 1 B). Upon Cre recombination, deletion of exon 10 created a premature stop codon in the transcript, which is predicted to undergo nonsense-mediated decay. Cluhfl/fl mice were first crossed with a Cre-deleter transgenic mouse line to obtain full-body heterozygous (HT) Cluh+/− mice (Fig. 1 B). When we intercrossed these HT mice and genotyped their progeny at weaning, no Cluh−/− KO mice were obtained (Table 1). However, Cluh−/− dead pups were found in the cages at postnatal day (P) 0–1 with a significant enrichment compared with other genotypes (Table 1), indicating that they died shortly after birth. Indeed, when pups were delivered by C-section at embryonic day (E) 18.5, Cluh−/− mice were present at a Mendelian ratio (Table 1) and survived and breathed normally, excluding respiratory insufficiency as the cause of death. To confirm the efficient deletion of Cluh in this model, we analyzed Cluh expression in tissues from E18.5 embryos. Levels of Cluh transcript were lower, indicating that the mRNA was degraded (Fig. 1 C). Most importantly, no full-length or truncated forms of CLUH were detected by Western blot using antibodies recognizing the N-terminal or C-terminal part of the protein (Fig. 1, D and E).
Figure 1.
Generation and characterization of Cluh−/− mice. (A) CLUH expression in murine tissues at 5 wk of age. (B) The genomic locus of the Cluh gene, the targeting vector, and the resulting floxed (fl) and Cluh− alleles are depicted. Exons are represented as red boxes. frt, Flp recognition target site; RI, EcoRI. (C) Percentage of Cluh mRNA in KO livers relative to WT determined by quantitative RT-PCR. n = 4. **, P ≤ 0.01 (two-tailed t test). (D) Immunoblot of whole-liver lysates of E18.5 neonates probed with anti-CLUH antibodies against N-terminal (N) or C-terminal (C) epitopes. Asterisks denote unspecific signals. (E) Immunoblot of WT and KO tissues at E18.5 probed for CLUH. (D and E) Pan-actin was used as a loading control. (F) Representative picture of WT and KO mice at E18.5. (G) Body weight of WT (n = 42), HT (n = 68), and KO (n = 39) mice at E18.5. ***, P ≤ 0.001 (one-way analysis of variance). Graphs show means ± SEM.
Table 1. Percent distribution of genotypes of progeny of Cluh+/− ×Cluh+/− mice.
| Age | Cluh+/+ | Cluh+/− | Cluh−/− | n | P χ2-test |
|---|---|---|---|---|---|
| 3 wk | 39.1 | 60.9 | 0.0 | 499 | 5.4 × 10−39 |
| E18.5 | 25.9 | 49.6 | 24.5 | 286 | 0.94 |
| P0–1 (total) | 23.7 | 55.7 | 20.6 | 97 | 0.48 |
| P0–1 (dead) | 5.9 | 11.8 | 82.3 | 17 | 3.3 × 10−7 |
At E18.5, KO mice looked indistinguishable from wild-type (WT) and HT littermates but weighed ∼10–15% less (Fig. 1, F and G). We did not observe any remarkable abnormalities in the morphology of mice livers, lungs, hearts, or kidneys in the absence of CLUH, suggesting that gross development proceeds normally in these tissues (Fig. S1, A and B). No fibrosis or hypertrophy was detected in the heart (Fig. S1, C and D). Thus, CLUH deficiency allows embryonic development but causes neonatal lethality.
Cluh deficiency remodels the mitochondrial proteome
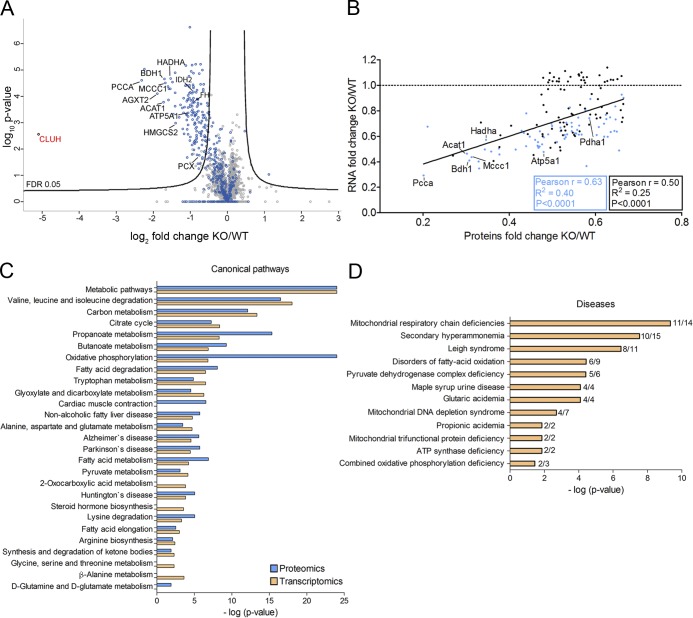
CLUH binds several mRNAs encoding mitochondrial proteins involved in different pathways (Gao et al., 2014). The availability of the mouse model gave us the opportunity to explore in an unbiased manner how the deletion of CLUH affects its mRNA targets and corresponding protein products. To this end, we compared proteomic and transcriptomic profiles of WT and KO livers at E18.5. Quantitative proteomic profiles were obtained from WT and KO total liver lysates using liquid chromatography–tandem mass spectrometry analysis in combination with a spike-in stable isotope labeling with amino acids in cell culture (SILAC) standard. We reliably detected and quantified ∼3,000 proteins, 525 of which were mapped to mitochondria (Fig. 2 A and Table S1). Remarkably, the abundance of ∼40% of these mitochondrial proteins was significantly reduced in the absence of CLUH (Fig. 2 A). Western blotting on liver and heart lysates of Cluh−/− mice confirmed the down-regulation of selected mitochondrial proteins, whereas other mitochondrial markers, such as VDAC1 and TOMM20, remained unchanged (Fig. S2, A–D).
Figure 2.
Protein and transcript levels of a subset of nuclear-encoded mitochondrial proteins are down-regulated in Cluh−/− livers. (A) Volcano plot of proteomics analyses of livers of E18.5 KO mice compared with WT. Names are indicated for all proteins relevant in the following experiments. Blue dots are mitochondrial proteins according to Mitocarta 2.0; gray dots indicate nonmitochondrial proteins. (B) Correlation of the abundance of significantly down-regulated proteins (FC ≥1.5) with corresponding transcript levels. Previously identified CLUH targets (Gao et al., 2014) are indicated in blue. The black line shows correlation regression of all genes. The black box shows statistics for all genes, and the blue box shows statistics only for identified CLUH targets. Names are indicated for genes for which mRNA stability is measured in the following experiments. The dashed line marks an RNA FC of 1. (C) Significantly enriched pathways highlighted by proteomics (blue) and transcriptomics (orange) data. (D) Disease implication analysis of proteomics data. In C and D, bars indicate log of p-values.
Transcriptome profiling using RNA sequencing revealed 460 genes differentially expressed in the liver in the absence of Cluh (at least 1.5-fold; P < 0.05; Table S2). Similar to the proteomic data, down-regulated genes encoded mitochondrial proteins that clustered in a highly interconnected network (Fig. S2 E). In fact, when we plotted the fold change (FC) of down-regulated proteins versus the FC of their transcripts, we found a rather good correlation (Pearson r = 0.50; P < 0.0001) that substantially improved when the analysis was restricted to CLUH targets previously identified by RNA immunoprecipitation in HeLa cells (Pearson r = 0.63; P < 0.0001; Fig. 2 B; Gao et al., 2014), leading to the hypothesis that CLUH targets are down-regulated both at the mRNA and protein level in the KO liver. mRNAs up-regulated in the absence of CLUH did not reveal any enriched functional pathways, nor did they cluster into one or a few networks (Fig. S2 F). Some of these up-regulated genes, however, pointed to delayed development (for instance, of Hba-x and Hbb-y) in the KO liver (Table S2). Many of these changes may be compensatory because they are not reflected at the protein level.
To shed light on the mitochondrial processes mostly affected by a lack of CLUH, we conducted pathway analysis of differentially expressed transcripts and proteins, which revealed a significant enrichment of metabolic pathways involved in amino acid degradation, tricarboxylic acid (TCA) cycle, fatty acid degradation, and OXPHOS (Fig. 2 C). Moreover, the mitochondrial proteins depleted in KO livers are implicated in human recessive disorders characterized by severe neonatal phenotypes, such as mitochondrial respiratory chain deficiencies, secondary hyperammonemia, Leigh syndrome, and mitochondrial DNA (mt-DNA) depletion syndromes (Fig. 2 D). We conclude that in the absence of CLUH, the steady-state levels of target mRNAs are decreased, leading to a reduction in the corresponding protein abundance. As a result, the mitochondrial proteome is depleted of crucial enzymes involved in energy-converting pathways and implicated in human metabolic diseases.
Lack of CLUH leads to mitochondrial clustering and respiratory deficiencies in vivo
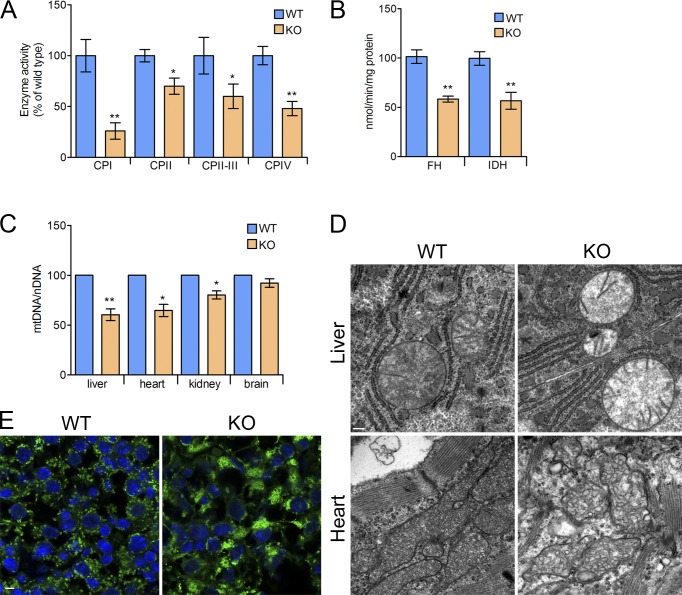
Liver proteomic data predicted that a lack of CLUH affects mitochondrial respiration. To this purpose, we measured the enzymatic activities of respiratory complexes I–IV in liver lysates from WT and KO mice at E18.5. All respiratory complexes were affected, but complex I activity was particularly decreased (Fig. 3 A). Because the TCA cycle represented one of the pathways most affected in the absence of CLUH, we assessed the activities of isocitrate dehydrogenase (IDH) and fumarate hydratase. These enzymatic activities were reduced by approximately half in the absence of CLUH, in agreement with the corresponding reduction of the protein level (Figs. 2 A and 3 B). Finally, we found that mt-DNA levels were decreased to a different extent in the liver, heart, and kidney of Cluh−/− mice (Fig. 3 C), as predicted by the disease enrichment analysis of proteomic data (Fig. 2 D). We then set out to assess whether the lack of CLUH affects the ultrastructure of mitochondria. Mitochondria still contained cristae in the liver and heart of KO mice; however, the matrix density was drastically reduced (Fig. 3 D). We also observed prominent mitochondrial clustering in the liver, reproducing the evolutionarily conserved hallmark of CLUH deficiency (Fig. 3 E). Thus, lack of CLUH causes the clustering of abnormal mitochondria and respiratory deficiencies in vivo.
Figure 3.
Cluh deficiency affects respiratory function, mt-DNA content, distribution, and ultrastructure of mitochondria. (A) Enzymatic activities of respiratory chain complexes in KO liver lysates relative to WT samples at E18.5. (B) Enzymatic activities of fumarate hydratase (FH) and IDH in whole-liver lysates at E18.5. (C) Quantitative PCR on DNA extracted from whole tissues. Graphs show mt-DNA amount of KO samples relative to WT levels, calculated by normalizing mt-Co1 expression (mitochondrial encoded) to Rmrp expression (nuclear encoded) and show mean ± SEM. n = 4. *, P ≤ 0.05; **, P ≤ 0.01 (two-tailed t test). (D) Representative electron micrographs of livers (n = 5) and hearts (n = 3) of WT and KO neonates at E18.5. Bar, 0.2 µm. (E) Immunofluorescence of livers of WT and KO neonates at E18.5 stained with anti-TOMM20 (green) and DAPI (blue). n = 3. Bar, 10 µm.
Cluh−/− mice develop hypoglycemia after birth
Neonatal mortality within the first 24–48 h after birth is often a feature of mice exhibiting homeostatic defects (Turgeon and Meloche, 2009). Birth is characterized by a shift from the glucose-rich placental nutrition to the high-fat, low-carbohydrate maternal milk diet and requires a metabolic switch from glycolysis to OXPHOS in several tissues (Cowet, 1992). Moreover, shortly after birth, before lactation starts, mice are challenged with a life-threatening starvation period, during which glucose levels are maintained by breaking down glycogen reserves and by inducing gluconeogenesis in the liver, a tissue that expresses high levels of CLUH (Kalhan and Bier, 2008). In liver mitochondria, catabolism of amino acids provides carbon skeletons as entry points for the TCA cycle, from which they can be completely oxidized or used for gluconeogenesis or ketogenesis (Kalhan and Bier, 2008). Because transcripts and proteins of crucial amino acid catabolic pathways were decreased in the absence of CLUH in the liver, we surmised that, in addition to respiratory deficiency, failure to convert amino acids into glucose contributes to the observed neonatal lethality in the KO mice.
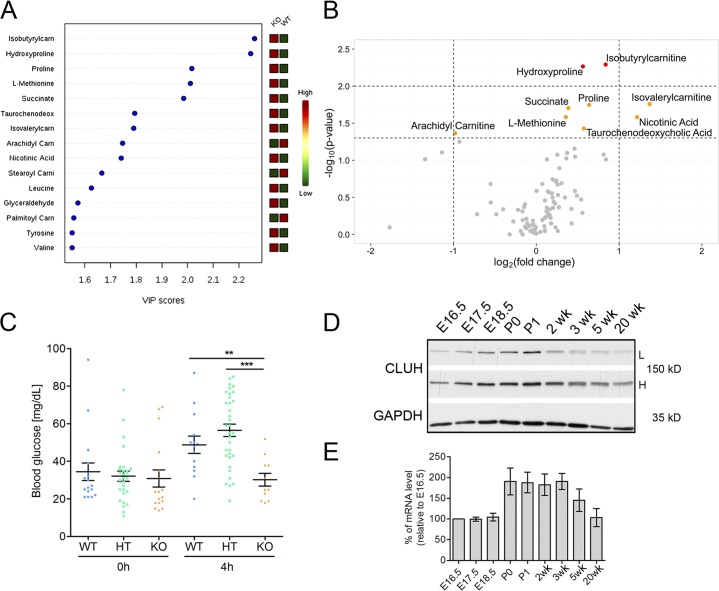
To test this hypothesis, we delivered mice at E18.5 by C-section and starved them for 4 h. Because the liver expresses high levels of CLUH and is a metabolic hub of crucial importance for responses to starvation, we subjected the livers from starved mice to targeted metabolomics analysis. The top-ranked metabolites that contributed to the separation of WT and KO samples included several amino acids and products of their catabolic degradation (Fig. 4 A). Proline and hydroxyproline, l-methionine, isobutyrylcarnitine, and isovalerylcarnitine, deriving from valine and leucine catabolism, respectively, accumulated at statistically significant levels in the absence of CLUH (Fig. 4 B). We also found increased levels of succinate, in agreement with a defect in the activity of succinate dehydrogenase, and of taurochenodeoxycholic acid, suggesting mild cholestasis (Fig. 4 B). Glycaemia was comparable among mice of different genotypes at birth; however, after 4 h of starvation, Cluh−/− mice had significantly lower glucose levels compared with WT and HT littermates (Fig. 4 C). Remarkably, CLUH expression increased in the liver during fetal development, peaked shortly after birth at P0–1, and decreased after weaning (Fig. 4, D and E). These data point to the critical role of Cluh in surviving postbirth starvation. In the absence of CLUH, catabolic pathways are impaired, leading to hypoglycemia.
Figure 4.
Metabolic alterations and hypoglycemia in Cluh−/− mice. (A) Targeted metabolomics of liver samples from WT and KO mice obtained by C-section at E18.5 and starved for 4 h. Partial least squares discriminant analysis variable importance in projection (PLSDA-VIP) plot depicting the top-ranked metabolites that contributed to the separation of WT and KO. n = 7. (B) Volcano plot of analyzed metabolites showing those reaching statistical significance. Dashed lines on the x axis indicate a log2 (KO/WT FC) of 1 and −1, respectively. Dashed lines on the y axis mark p-values corresponding to 0.05 and 0.01. (C) Glycaemia of E18.5 neonates immediately after C-section (WT, n = 17; HT, n = 28; KO, n = 17) or after 4 h of starvation (WT, n = 14; HT, n = 34; KO, n = 11). Graph shows means ± SEM. **, P ≤ 0.01; ***, P ≤ 0.001 (one-way analysis of variance). (D) Representative immunoblot of whole-liver lysates of WT mice at different ages. n = 3. GAPDH was used as a loading control. L, low exposure; H, high exposure. (E) Expression of Cluh in the liver was quantified at different ages using quantitative RT-PCR. Expression was normalized to GAPDH levels. The graph shows percentages of mRNA relative to E16.5 samples. Error bars are SEM. n = 3.
Cluh is required in the adult liver to increase respiratory capacity and mount a starvation response
An open question is whether CLUH supports general mitochondrial homeostasis or rather mediates the adaptation of mitochondrial function to specific metabolic situations, such as the fetal–birth transition. To address this point in the absence of a potential compensatory mechanism to survive the birth transition, we crossed Cluhfl/fl mice with a transgenic line that efficiently expresses the Cre recombinase only postnatally and specifically in hepatocytes (Alb-Cre). The resulting Li-CluhKO mice (genotype: Cluhfl/fl Alb-Crewt/tg) showed residual CLUH levels in the liver at P6, and a prominent depletion only after 4 wk (Fig. S3 A).
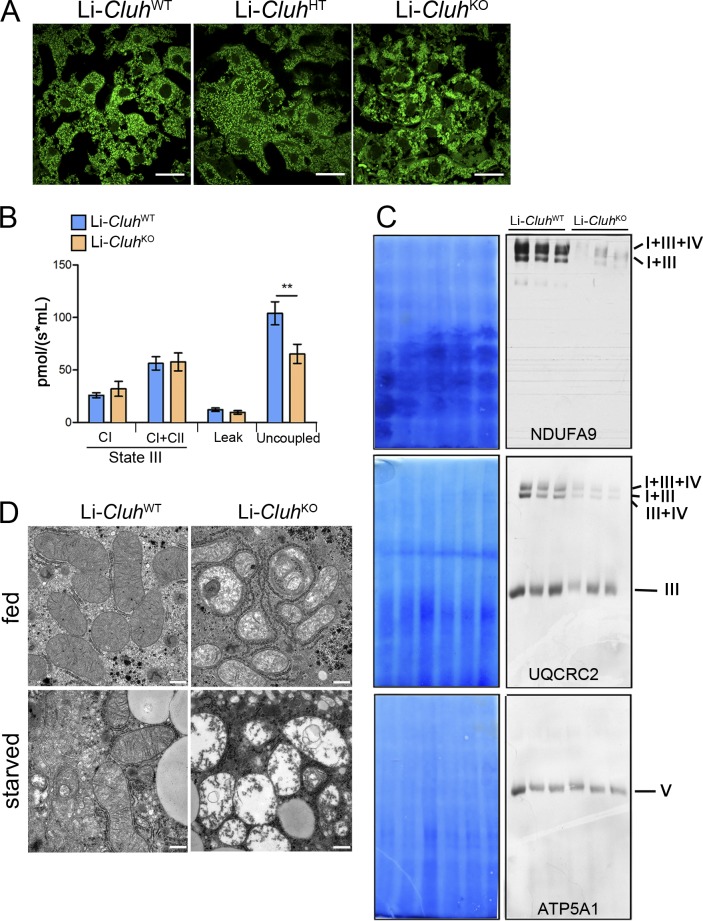
Analysis of the liver of Li-CluhKO mice at 8 wk of age showed an overall normal histological organization, with a tendency to increase in apoptotic cells and Ki-67–proliferating hepatocytes, but no signs of fibrosis or inflammation (Fig. S3, B and C). In line with these data, Li-CluhKO mice displayed a slight increase in serum alanine aminotransferase (ALT) levels, whereas AP activity was comparable to WT littermates (Fig. S3, D and E). As in neonates, mitochondria were clustered in livers lacking CLUH (Fig. 5 A).
Figure 5.
Liver-specific Cluh deletion affects mitochondrial distribution and structure, assembled respiratory supercomplexes, and respiratory capacity. (A) Representative confocal images of livers of 8-wk-old mice of the indicated genotypes. To analyze mitochondrial morphology, mice were crossed with a stop-mito-YFP reporter line activated by Cre recombination. n = 4. Bars, 5 µm. (B) Oxygen consumption of mitochondria isolated from livers of 8-wk-old mice. State III respiration was measured in the presence of pyruvate, malate, glutamate, and ADP (complex I [CI]), followed by addition of succinate (complex I + complex II [CI + CII]). The proton leak was measured after addition of oligomycin, whereas maximal respiration was assessed by CCCP titration. n = 5. Graph shows means ± SEM. **, P ≤ 0.01 (two-tailed t test). (C) Blue-native–PAGE of digitonin-solubilized mitochondria isolated from the liver of mice with the indicated genotypes at 8 wk of age. Corresponding Coomassie stainings are shown. Supercomplexes were detected using antibodies against NDUFA9 (complex I) and UQCRC2 (complex III). ATP5A1 was used to detect complex V. (D) Representative electron micrographs showing mitochondrial structures in the livers of 8-wk-old mice either fed ad libitum or starved for 24 h. n = 3. Bars, 0.5 µm.
To evaluate the respiratory capacity of liver mitochondria, we assessed oxygen consumption using high-resolution respirometry. When challenged with substrates for complex I or for complexes I and II, Cluh-deficient mitochondria revealed normal basal respiration, in agreement with the mild pathological abnormalities (Fig. 5 B). No signs of uncoupling were observed upon oligomycin addition. However, a reduction in the rate of maximal respiration was detected after dissipation of the membrane potential with carbonyl cyanide m-chlorophenyl hydrazone (CCCP), indicating that liver mitochondria were already working at their maximal capacity (Fig. 5 B). In addition, assembled respiratory chain supercomplexes were decreased (Fig. 5 C). Thus, loss of CLUH may become critical when the liver is challenged by more demanding energy needs.
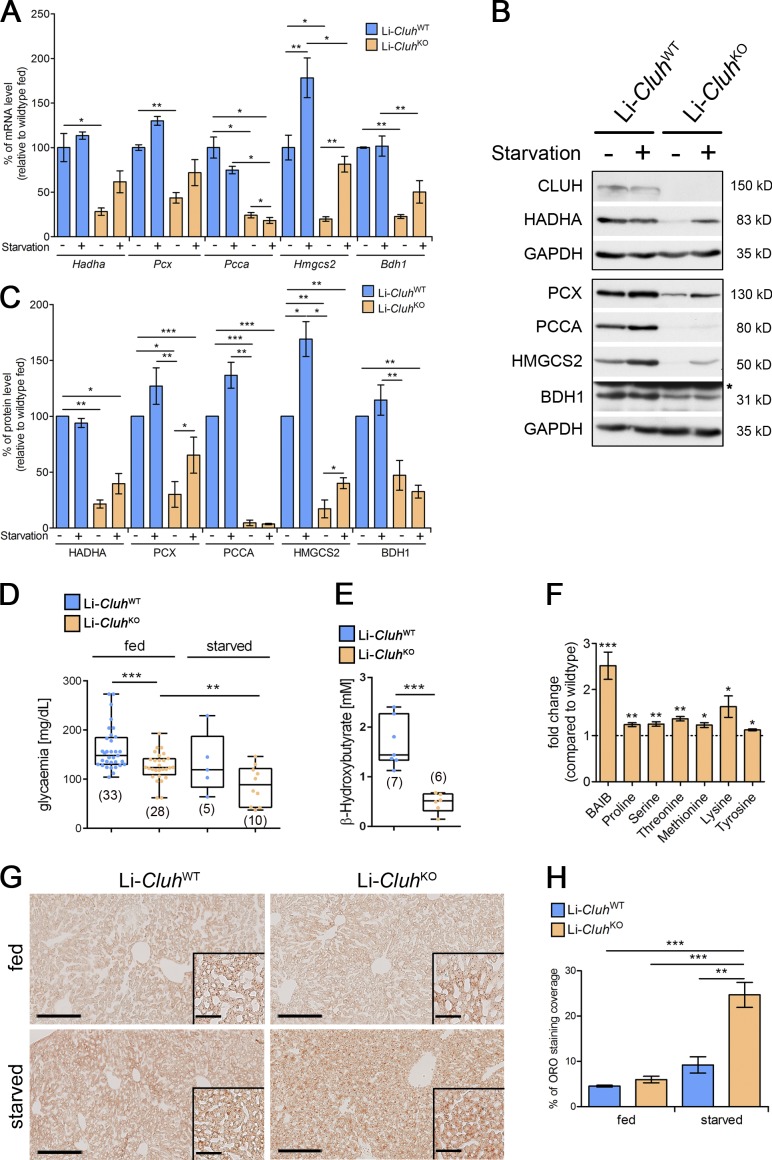
Similar to what happens at birth, during fasting, hepatocytes release glucose by breaking down glycogen and activating gluconeogenesis and convert fatty acids and amino acids to acetyl-CoA, which is then used to produce ketone bodies to sustain peripheral tissues. We noticed that upon starvation, ultrastructural abnormalities of mitochondria in the absence of CLUH became dramatic (Fig. 5 D). CLUH target genes that are especially down-regulated at both mRNA and protein levels in the full-body KO mice (Fig. 2 B), such as Pcca (valine, leucine, and isoleucine degradation), Hadha (β oxidation), Pcx (gluconeogenesis), and Bdh1 and Hmgcs2 (ketone body biosynthesis), play crucial roles in the liver during starvation. We therefore starved Li-CluhKO mice for 24 h and examined expression levels of these genes both at the mRNA and the protein level. Fasting induced a transcriptional response, increasing the expression of Hmgcs2 in both genotypes, but did not overtly affect the expression of the other genes in WT mice (Fig. 6 A). In contrast, a certain degree of up-regulation was observed in Li-CluhKO mice (Fig. 6 A). This notwithstanding, the mRNA and protein levels of all these genes were significantly decreased in CLUH-deficient livers compared with controls both under fed and starved conditions (Fig. 6, A–C). As a consequence, Li-CluhKO mice had lower blood glucose levels, and the hypoglycemia was enhanced upon starvation (Fig. 6 D). Moreover, production of the ketone body β-hydroxybutyrate was severely impaired (Fig. 6 E). In agreement with an impairment of amino acid catabolic pathways, we found increased levels of proline, serine, threonine, methionine, and lysine in the serum of Li-CluhKO upon starvation (Fig. 6 F). Moreover, β-aminoisobutyric acid (BAIB), a nonproteinogenic amino acid originating from the catabolism of thymine and valine, accumulated (Fig. 6 F). BAIB is one of the substrates of alanine-glyoxylate aminotransferase 2 (AGXT2), an enzyme that was profoundly depleted in the absence of CLUH in the proteomics analysis (Fig. 2 A). Finally, lipid droplets (LDs) were increased, a sign of inefficient β oxidation (Fig. 6, G and H).
Figure 6.
Cluh deletion in the adult liver impairs the metabolic response under starvation. (A) Quantitative RT-PCR of selected CLUH mRNA targets in the livers of 8-wk-old mice fed ad libitum or starved for 24 h. n = 3–4. Expression was normalized to GAPDH levels. Graph shows percentages of mRNA levels relative to WT fed samples. n = 4. (B) Representative immunoblots of selected CLUH mRNA targets in livers of 8-wk-old mice fed ad libitum or starved for 24 h. n = 3. GAPDH was used as a loading control. (C) Quantification of immunoblots shown in B. Graph shows percentages of protein levels relative to WT fed samples. n = 3. (D) Glycaemia of 8-wk-old mice fed ad libitum or starved for 24 h. (E) Serum levels of β-hydroxybutyrate after 24-h starvation of 8-wk-old mice. (D and E) Graphs show individual data points as boxplots. n values are indicated in parentheses. (F) Significantly changed amino acids and BAIB in serum of 8-wk-old mice starved for 24 h. Graph represents relative FC compared with WT samples (dashed line). n = 5. (G) Representative images of ORO staining in liver sections of 8-wk-old mice fed ad libitum or starved for 24 h. n = 4–6. Bars: (main images) 200 µm; (insets) 40 µm. (H) Quantification of ORO staining of 8-wk-old mice fed ad libitum or starved for 24 h. Graphs show means ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (two-tailed t test).
Collectively, these data demonstrate an important role of CLUH in the adult liver to adapt metabolism to conditions of high-energy demands. Moreover, CLUH promotes catabolic pathways upon starvation to provide energy intermediates such as glucose and ketone bodies to extrahepatic tissues.
Cluh controls the stability and translation of selected target mRNAs
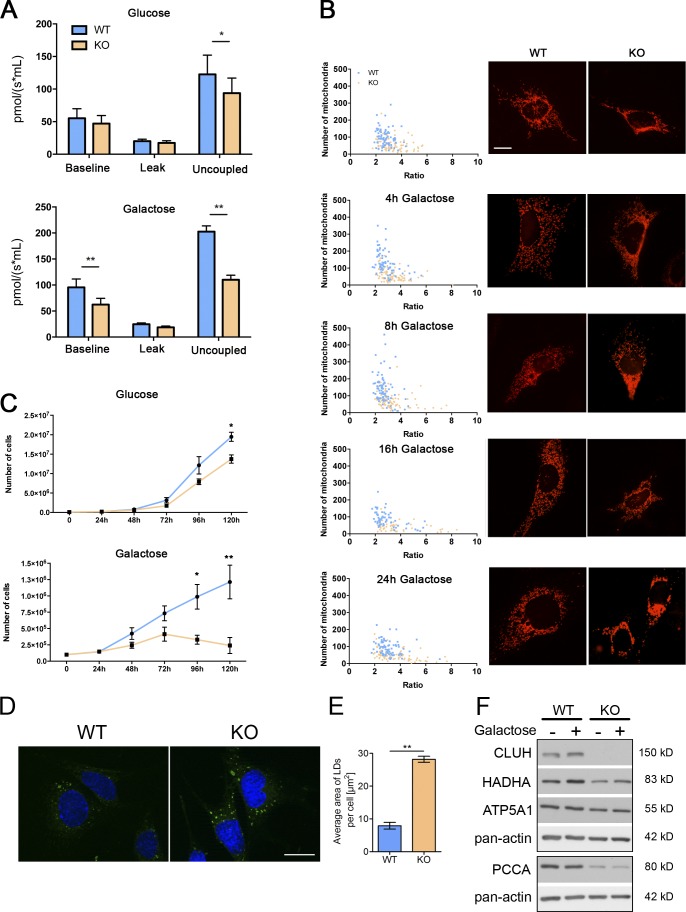
We explored whether Cluh-deficient mouse embryonic fibroblasts (MEFs) recapitulate our in vivo results. In agreement with results in the adult liver, basal respiration was normal in Cluh-deficient MEFs, but the maximal respiration capacity was reduced. This effect was more pronounced when the cells were shifted from glucose to galactose medium, forcing them to produce ATP by OXPHOS (Fig. 7 A). Under this condition, Cluh−/− MEFs displayed more pronounced growth defects and mitochondrial aggregation that increased with time (Fig. 7, B and C). LDs accumulated in Cluh-deficient MEFs as observed in the liver (Fig. 7, D and E). Furthermore, we confirmed depletion of ATP5A1, HADHA, and PCCA in Cluh−/− MEFs both in glucose and galactose media (Fig. 7 F). Thus, MEFs represent a good model system to examine the molecular mechanism of CLUH action on target mRNAs.
Figure 7.
Cluh-deficient MEFs mimic liver phenotypes. (A) Oxygen consumption of intact MEFs cultured in glucose or galactose medium. The proton leak was measured after the addition of oligomycin, whereas maximal respiration was assessed by CCCP titration. n ≥ 4. (B) Mitochondrial morphology in MEFs transfected with mito-mCherry. Graphs show the mean aspect ratio (area/perimeter) on the x axis and the number of mitochondria on the y axis for individual cells from three independent experiments. Right panels show representative images of mitochondrial morphology at the indicated time points. Bar, 12 µm. (C) Growth curves of MEFs cultured in glucose or galactose medium during five consecutive days. n = 3. (D) Representative images of LD staining in MEFs grown in glucose medium. Nuclei were stained with DAPI (blue), and LDs were stained with BODIPY 493/503 (green). Bar, 20 µm. (E) Quantification of LD staining shown in D. 50 cells were analyzed per genotype per experiment. Graph shows the mean area of LDs per cell. n = 3. (A, C, and E) Error bars are means ± SEM. *, P ≤ 0.05; ** P ≤ 0.01 (two-tailed t test). (F) Representative immunoblots of MEFs grown in glucose (−) or galactose (+) media for 20 h for selected CLUH mRNA targets. Pan-actin was used as a loading control. n = 3.
First, we used MEFs to inspect the distribution of Atp5a1 and Hadha mRNAs using single-molecule FISH. Consistent with observations in vivo, in the absence of CLUH, the number of detected Atp5a1 and Hadha mRNA molecules was decreased (Fig. S4, A and B). We also examined whether the distance between the detected mRNAs and mitochondria was affected in the absence of CLUH; however, no significant difference was apparent in either glucose or galactose media (Fig. S4 C).
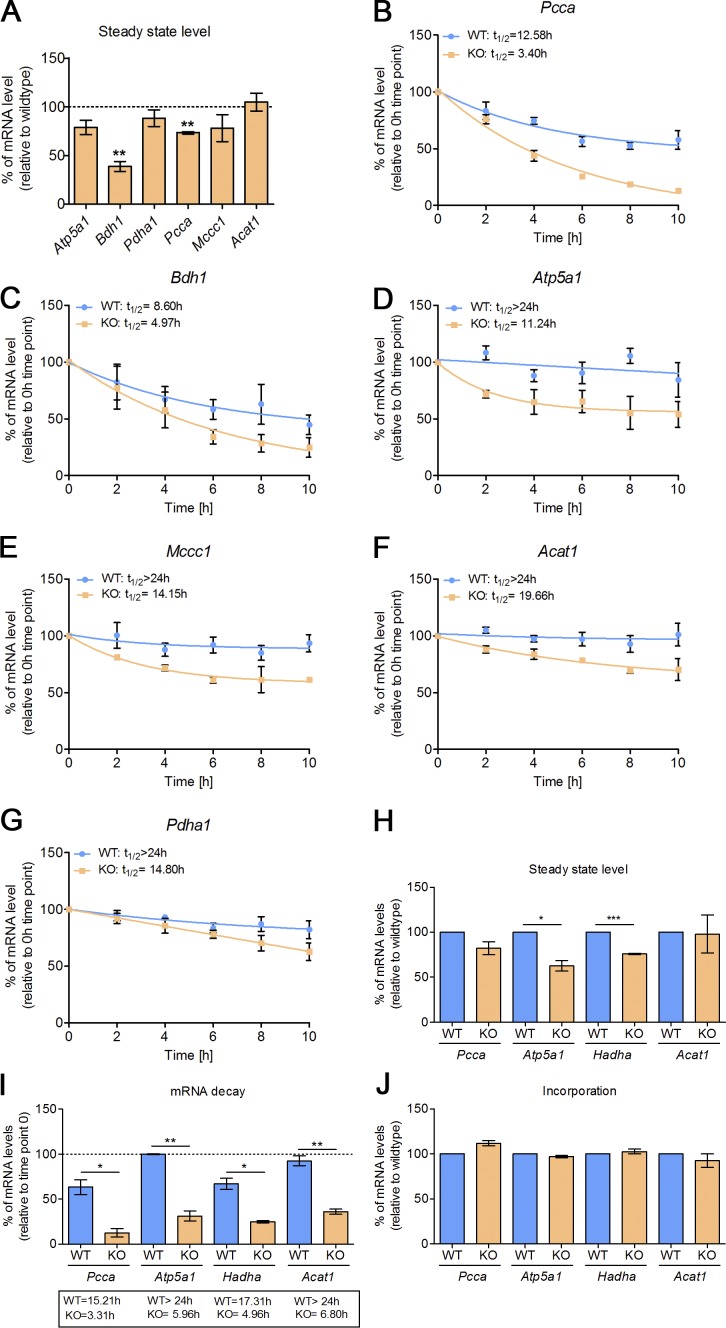
In the absence of CLUH, the steady-state levels of target mRNAs are reduced in tissues and MEFs (Figs. 2 B and 8 A), raising the question of the underlying mechanism. To test the hypothesis that CLUH controls the stability of its target mRNAs, we blocked transcription with actinomycin D for different time points, extracted RNA, and performed quantitative RT-PCR analysis to assess the decay rate of six CLUH targets (Pcca, Atp5a1, Pdha1, Bdh1, Acat1, and Mccc1), which were selected because of their low expression at both transcript and protein levels in vivo (Fig. 2 B). We found that the half-lives of these mRNAs were significantly shorter in the absence of CLUH (Fig. 8, B–G). To confirm these data in an independent way, we pulse-labeled RNA with 5-ethynyl uridine (EU) ribonucleotide homologues containing an alkyne reactive group for 24 h and chased with fresh medium for 10 h. We then used Click-iT chemistry to capture the newly synthesized pool of RNA. This method confirmed increased turnover of Pcca, Atp5a1, and Acat1 mRNAs. In addition, we determined a reduction in the half-life of Hadha (Fig. 8, H and I). Instead, the incorporation of EU was comparable at time point 0, strongly hinting at a higher transcription rate to compensate for the shorter half-life (Fig. 8 J). These data demonstrate a physiological requirement of CLUH to control the stability of at least a subset of mRNAs encoding mitochondrial proteins.
Figure 8.
CLUH controls the stability of target mRNAs. (A) Steady-state levels of mRNAs of KO cells relative to WT cells (dashed line) from experiments shown in B–G at time point 0. (B–G) Nonlinear regression decay trendlines of indicated mRNAs at different time points after actinomycin D treatment in WT and KO MEFs relative to time point 0. Calculated half-lives are indicated. Expression was normalized to GAPDH levels. (H) Steady-state levels of the indicated mRNAs in KO cells relative to WT levels from the experiment shown in I and J at time point 0 of whole-cell RNA. (I) Relative mRNA levels of pulled-down EU-incorporated mRNAs after 10-h chase relative to the corresponding time point 0 (dashed line). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (two-tailed t test). Calculated half-lives are indicated. (J) Relative mRNA levels of pulled down EU-incorporated mRNAs at time point 0 of KO cells relative to WT levels. Expression was normalized to GAPDH levels. Graphs represent means ± SEM. n = 3.
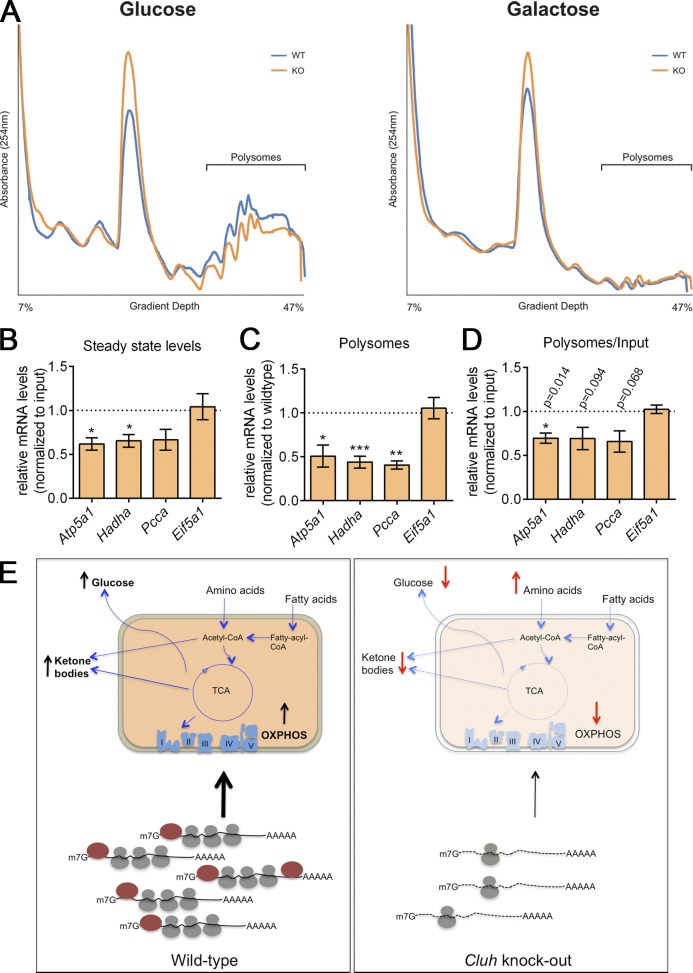
Because mRNA degradation and rate of translation are strictly coupled (Roy and Jacobson, 2013; Huch and Nissan, 2014), it is conceivable that increased decay of transcripts in the absence of CLUH may be caused at least in part by inefficient translation. To address this possibility, we fractionated polysomes by sucrose density gradient centrifugation from MEFs cultured in glucose or galactose for 20 h. We did not observe any overt differences in the polysome profiling of WT or KO MEFs; however, the absorbance of the monosome fraction was often higher in KO cells than in WT cells (Fig. 9 A). We then assessed the translation rate of three CLUH target mRNAs, Atp5a1, Hadha, and Pcca, along with one control mRNA, Eif5a, by measuring the amount of mRNA present in fractions corresponding to heavy polysomes containing three or more ribosomes using quantitative RT-PCR. We performed these experiments in MEFs cultured in galactose to maximize the rate of translation of these genes. The results demonstrated a reduction of Atp5a1, Hadha, and Pcca transcripts bound to heavy polysomes in Cluh KO cells that was still present after normalization to the input to account for differences at the steady-state level (Fig. 9, B–D).
Figure 9.
CLUH regulates translation of Atp5a1, Hadha, and Pcca. (A) Representative graphs of polysome profilings of WT and KO MEFs grown in glucose or galactose media for 20 h. n ≥ 4. (B) Steady-state levels of the indicated mRNAs in KO cells relative to WT cells (dotted line) in cell inputs from the experiments in C and D. (C) Relative mRNA levels of the indicated genes in polysome fractions of KO MEFs relative to WT (dotted line). (D) Relative mRNA levels of the indicated genes in polysome fractions of KO MEFs relative to WT (dotted line) after normalization to corresponding inputs. Graphs represent means ± SEM. n = 4. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (two-tailed paired t test), unless otherwise indicated. Firefly luciferase mRNA was used as a spike-in control in B–D and mRNA levels were normalized to luciferase levels. (E) Models of the physiological and molecular roles of CLUH. In the cytoplasm, CLUH (dark red) binds to mRNAs encoding specific mitochondrial proteins and promotes their stability and translation. As a result, mitochondria are enriched in proteins implicated in the conversion of fatty acids and amino acids in energy under nutrient deprivation. In the absence of CLUH, mRNAs become unstable and are less translated. As a consequence of the mitochondrial proteome reshaping, up-regulation of OXPHOS, ketone body production, and gluconeogenesis are impaired.
In conclusion, CLUH promotes translation and decreases the turnover of specific mRNAs coding for nuclear-encoded mitochondrial proteins, thus tuning their expression and affecting metabolic processes in mitochondria.
Discussion
We provide the first evidence that posttranscriptional regulatory mechanisms affecting the life cycle of mRNAs for nuclear-encoded mitochondrial proteins play a physiological role in shaping the mammalian mitochondrial proteome and adapting metabolism to changes in energy demand and nutrient availability. The RNA-binding protein CLUH is a key player controlling the expression of mitochondrial proteins involved in central catabolic pathways required for energy conversion. CLUH regulates the stability and translation of target mRNAs, thus expanding our view on regulatory mechanisms of mitochondrial function (Fig. 9 E).
We show that CLUH plays a central role in liver physiology under conditions of nutrient deprivation. The successful expression of a mitochondrial gene program required for the efficient breakdown of amino acids, for de novo synthesis of glucose, and for ketogenesis depends strongly on CLUH in vivo. Moreover, CLUH affects the expression of components of the respiratory chain and is required to enhance mitochondrial respiratory capacity. Notably, several of the genes that are most affected when CLUH is lacking are mutated in human diseases characterized by a multiorgan severe phenotype with neonatal lethality (Wanders et al., 2012; Lieber et al., 2013; Valle et al., 2016) or are associated with perinatal death when deleted in the mouse (Ibdah et al., 2001; Cotter et al., 2011). Consistently, CLUH is essential in the mouse for survival at birth when abrupt interruption of nutrients and a shift from glycolysis to β oxidation and OXPHOS occurs. In the adult liver, lack of CLUH impacts glucose homeostasis and ketone body production during starvation but does not lead to apoptotic cell death, as observed, for example, upon deletion of COX10, an essential assembly factor of complex IV (Diaz et al., 2008). This argues for the requirement of CLUH during specific metabolic conditions rather than to support general mitochondrial homeostasis. In agreement with this concept, CLUH deficiency does not affect mouse fetal or fly larval development (Sen et al., 2013), suggesting that its loss can be compensated in glycolytic conditions. Furthermore, deletion of Cluh orthologues in unicellular organisms or cell lines is compatible with survival and growth also in oxidative conditions (Zhu et al., 1997; Fields et al., 1998).
In the absence of CLUH, the expression of several previously identified target genes is reduced both at the RNA and protein levels in vivo. Our in vitro analysis performed on the most affected genes indicates that a combination of increased decay and reduced translation of corresponding mRNAs underlies this phenotype. We propose that CLUH acts with a similar mechanism for all of those mRNAs that are down-regulated in its absence. The mitochondrial proteins that were reduced in abundance in KO livers (whereas their transcripts were unaffected) were not previously identified in RNA immunoprecipitation using CLUH antibodies (Fig. 2 B; Gao et al., 2014). Because they were mainly components of respiratory chain complexes, we favor the hypothesis that they become secondarily unstable due to the lack of interacting partners.
The rate of mRNA turnover plays an important role in controlling gene expression (Garneau et al., 2007). CLUH may have a direct function to protect bound mRNAs from degradation. Other stabilizing RNA-binding proteins have been shown either to prevent mRNAs from localizing to sites of RNA decay or to compete for the binding of destabilizing factors (Lal et al., 2004; Bhattacharyya et al., 2006). CLUH may counteract the decay of bound transcripts until they are transported to the vicinity of mitochondria, where translation would occur. Although knowledge in mammalian systems is lagging behind, there is increasing evidence of targeting of mRNAs to the mitochondrial surface and cotranslational import of a subset of mitochondrial proteins in yeast, implicating the existence of posttranscriptional regulatory pathways (Marc et al., 2002; Margeot et al., 2002; Devaux et al., 2010; Williams et al., 2014). Drosophila melanogaster clueless has indeed been shown to bind ribosomes at the outer mitochondrial membrane (Sen and Cox, 2016). However, we did not observe obvious perturbations of the localization of Atp5a1 and Hadha mRNAs in respect to mitochondria in CLUH-deficient MEFs, but experiments in polarized cells are needed to rule out this possibility.
An alternative hypothesis is that CLUH is involved in the regulation of translation initiation. Degradation and translation of mRNAs are intricately coupled processes (Roy and Jacobson, 2013; Huch and Nissan, 2014), and it has been shown that stalled translation initiation, for instance after mutations in components of the eukaryotic initiation factor Eif3, activates mRNA decay pathways (Barnes, 1998; Schwartz and Parker, 1999). Interestingly, an interaction between Clu1 and the Eif3 complex has been previously observed in yeast (Vornlocher et al., 1999). Moreover, we found that the absorbance of the monosome fraction was increased in the absence of CLUH, suggesting a block in translation initiation. CLUH may allow the activation of translation of preexisting mRNAs upon nutrient deprivation, thus rapidly adjusting cell metabolism without the need for de novo transcription. Such a mechanism would explain previous observations of a rapid maturation of rat liver mitochondria in the first postnatal hour that depended on an increased rate of protein synthesis for mitochondrial proteins involved in bioenergetic and in metabolic functions (Valcarce et al., 1988). A role of CLUH in translational regulation is also supported by the finding that the Drosophila orthologue was found in polysome fractions (Sen and Cox, 2016).
Intriguingly, our results are somehow evocative of the function of the yeast PUF (Pumilio and FBF) family member Puf3p, an RNA-binding protein that specifically recognizes mRNAs encoding mitochondrial proteins. Puf3p targets these mRNAs to the outer mitochondrial membrane, thus promoting cotranslational import (Saint-Georges et al., 2008). Moreover, nonmitochondrial Puf3p represses the translation of target mRNAs by promoting their deadenylation and decay, whereas at the mitochondria, this repressing activity would be inhibited, resulting in translation (Quenault et al., 2011). CLUH, similar to Puf3p, exists in a cytosolic and mitochondria-targeted pool, binds mRNAs of mitochondrial proteins (Gao et al., 2014), and controls the turnover and translation of bound transcripts. Whether CLUH activity is regulated by posttranslational modifications that respond to the cell metabolic state is an attractive possibility that needs to be addressed in the future.
In conclusion, our data shed light on the mechanism whereby CLUH regulates its target mRNAs and reveal the physiological relevance of this posttranscriptional response to tailor mitochondrial function to conditions of nutrient deprivation.
Materials and methods
Generation of Cluh mouse models
All animal procedures were performed in accordance with European Union (EU directive 86/609/EEC), national (Tierschutzgesetz), and institutional guidelines and were approved by local authorities (Landesamt für Natur, Umwelt, und Verbraucherschutz Nordrhein-Westfalen, Germany). Mice with a conditional loxP-flanked exon 10 in the Cluh gene (Cluhfl/fl mice) were generated by gene targeting. The targeting vector for homologous recombination in mouse embryonic stem (ES) cells was constructed in the PL451 backbone containing a neo cassette for the positive selection of ES cells, flanked by two frt sites and one loxP site. The 4.8-kb left arm of homology was cloned in KpnI and EcoRI sites. An upstream loxP site was introduced with the reverse primer. Exon 10 of the Cluh gene was cloned into the EcoRI site. A 4.8-kb right arm of homology was then cloned using NotI and SacII, followed by the insertion of an MC1-HSV-TK cassette for negative selection of ES cells. The construct was linearized with ClaI and electroporated in C57BL/6N Bruce 4 ES cells (Köntgen et al., 1993). ES cells were picked and identified by Southern blots using probes situated outside the region of homology. Positive ES cell clones were injected into CB20 blastocysts and transferred into pseudopregnant females to obtain chimeric mice. Chimeras were crossed to C57BL/6N mice to obtain germline transmission. To remove the neomycin cassette in the offspring and generate Cluhfl/fl mice, mice were crossed with a transgenic line ubiquitously expressing the Flp recombinase (Rodríguez et al., 2000). Cluhfl/fl mice were further crossed with a CMV-Cre transgenic line (Schwenk et al., 1995) to generate a full-body KO and with an Alb-Cre transgenic line (Postic and Magnuson, 2000) to obtain a liver-specific KO (Li-CluhKO). WT littermates served as controls for Cluh−/− mice, whereas Cluhfl/fl mice without the Cre allele were used as controls for the conditional model. When specified, to visualize mitochondria in the liver, mice were crossed with ROSA26+/SmY mice (Sterky et al., 2011). In all cases, mouse lines were in a pure C57BL/6N background.
Mice were maintained in individually ventilated cages with specified pathogen–free hygiene levels, kept under a 12-h light cycle, and given a regular chow diet ad libitum. E18.5 pregnant females were euthanized, and fetuses were obtained by C-section. Reanimated neonates were kept in a humidified chamber at 30°C and fasted for 4 h. Adult animals were fasted for 24 h. For histological studies, adult mice were anesthetized intraperitoneally with xylazine/ketanest (10 mg/100 mg per kilogram body weight) and perfused intracardially with 4% PFA in PBS.
Blood and serum chemistry
Blood glucose was determined using a glucose meter with glucose test strips (Accu-Check; Aviva). ALT and AP levels were measured in the serum using a Cobas c111 analyzer (Roche). β-Hydroxybutyrate levels were measured with a Ketone Body Assay kit (Sigma-Aldrich).
Antibodies for Western blots
The following antibodies were used: polyclonal rabbit OPA1 (ab42364), monoclonal mouse PDHA1 (ab110330), monoclonal rabbit GOT2 (ab171739), monoclonal mouse ATP5A1 (ab14748), monoclonal rabbit CPT2 (ab181114), polyclonal rabbit HADHA (ab54477), and monoclonal rabbit PCX (ab128952) from Abcam; polyclonal rabbit CLUH from Aviva (ARP70642_P050) or from Novus Biologicals (NB100-93305); polyclonal rabbit SPG7 (Ferreirinha et al., 2004); monoclonal mouse GAPDH (MAB374), monoclonal mouse pan-actin (MAB1501R), and monoclonal mouse VDAC1 (529536) from EMD Millipore; polyclonal rabbit TOMM20 (sc-11415) and monoclonal mouse HMGCS2 (sc-376092) from Santa Cruz Biotechnology, Inc.; polyclonal rabbit UQCRC2 (HPA019146) from Sigma-Aldrich; monoclonal mouse NDUFA9 (459100), monoclonal mouse SDHA (459200), and monoclonal mouse COX5B (459110) and COX4I1 (A21348) from Invitrogen; and polyclonal rabbit OXCT1 (12175–1-AP), polyclonal rabbit OGDH (15212–1-AP), polyclonal rabbit BDH1 (15417–1-AP), and polyclonal rabbit PCCA (21988–1-AP) from ProteinTech.
Histology, immunohistochemistry, and immunofluorescence
Livers of perfused adult mice and freshly taken tissues of E18.5 neonates were fixed for 2 d in 4% PFA in PBS at 4°C. Tissues were dehydrated and embedded in paraffin, and then 7-µm sections were cut and deparaffinised. For hematoxylin/eosin staining, sections were incubated in Mayer’s hematoxylin solution for 5 min (livers), 3 min (hearts), or 4 min (lungs), washed and incubated for 1 min in eosin Y solution, and then were washed, dehydrated, and mounted with Eukitt (Fluka). For periodic acid–Schiff staining, sections were incubated in periodic acid for 5 min, washed, and incubated for 1.5 min (adult tissue) or 1 min (neonatal tissue) in Schiff’s reagent and then were washed again, counterstained with hematoxylin solution as described in the previous sentence, dehydrated, and mounted. For fibrosis staining, a Masson Trichrome staining kit (Sigma-Aldrich) was used according to the manufacturer’s instructions. For Oil red O (ORO) staining, after fixation tissues were dehydrated in sucrose and embedded in optimum cutting temperature mounting medium (Tissue-Tek), 10-µm cryostat sections were cut. Sections were incubated for 5 min in ORO solution, destained in water for 30 min, and then mounted in mounting solution (Aquatex; EMD Millipore). For image acquisition, the whole section was scanned with a brightfield slidescanner microscope (SCN400; Leica Biosystems). Five random images were taken at 15× magnification for each liver. Channels were split using ImageJ (National Institutes of Health), and the blue channel was used for quantification. Images were then thresholded to select the area covered by ORO staining and to obtain the percentage of area. The mean from five images was obtained to get the value for each animal. In total, five animals for each condition were analyzed.
Immunohistochemistry was performed as described previously (Kondylis et al., 2015). Paraffin liver sections were rehydrated, and antigen retrieval was performed by heating in 10 mM sodium citrate and 0.05% Tween 20, pH 6.2, or by proteinase treatment in the case of F4/80 staining. The following antibodies were used: anti-Ki67 (M724901; Dako), anti–active caspase-3 (9661; Cell Signaling Technology), anti–α-SMA (A2547; Sigma-Aldrich), and anti-F4/80 (AbD Serotec). Biotinylated secondary antibodies were purchased from Vector Laboratories. Stainings were visualized with ABC kit Vectastain Elite (Vector Laboratories) and DAB substrate (Dako). Quantification of stainings was performed as previously described (Kondylis et al., 2015). For immunofluorescence, sections were deparaffinized and rehydrated, followed by antigen retrieval by boiling sections in citrate buffer. Section were then washed, blocked in 10% goat serum in 0.4% Triton X-100 in PBS for 1 h, and then incubated with polyclonal rabbit TOMM20 antibody diluted in 5% goat serum in 0.4% Triton X-100 in PBS overnight at 4°C. Then, sections were washed and incubated in secondary antibody diluted in 5% goat serum in PBS for 2 h at RT, washed again, stained with DAPI (Sigma-Aldrich), and mounted with Fluorsave (EMD Millipore). For image acquisition, a gSTED superresolution and confocal microscope (TCS SP8; Leica Biosystems) with a HyD detector with a Plan-Apochromat 63×/1.2 NA water differential interference contrast (DIC) objective was used.
EM
Tissues were postfixed in 2% glutaraldehyde in 0.12 M phosphate buffer. After overnight fixation, tissues were treated with 1% osmium tetroxide in 0.12 M phosphate buffer and embedded in epon (Fluka). Ultrathin sections (70 nm) were cut and stained with uranyl acetate (Plano GmbH) and lead citrate (Electron Microscopy Sciences). Samples were analyzed with a transmission electron microscope (EM 902; ZEISS or CM10; Phillips) at an acceleration voltage of 80 kV, and pictures were acquired using a 2K-CCD (TRS Albert Tröndle) or SC200W (Orius) camera.
TCA cycle enzyme activities
Livers of E18.5 neonates were quickly homogenized in extraction buffer (20 mM Tris, pH 7.2, 250 mM sucrose, 2 mM EGTA, 40 mM KCl, and 1 mg/ml BSA) followed by spectrophotometric analysis using a spectrophotometer (Cary 50 Scan; Varian Inc.). Fumarate hydratase activity was determined at 250 nm (ε = 5,090 M/cm). Homogenate was added to buffer containing 10 mM KH2PO4, pH 7.8, 2 mM EDTA, 1 mg/ml BSA, and 0.1% Triton X-100, and activity was measured by the addition of 0.5 mM malate. NADP-IDH activity was determined at 340 nm (ε = 6,220 M/cm). Homogenate was added to buffer containing 2.5 mM Tris, pH 7.3, 1.5% Triton X-100, 1.5 mM MgCl2, and 1 mM NADP, and activity was measured by the addition of 125 mM isocitrate.
Respiratory chain enzyme activities
Livers of E18.5 neonates were quickly homogenized in phosphate buffer (50 mM KH2PO4, pH 7.4) followed by spectrophotometric analysis of isolated respiratory chain complex activities at 37°C using a spectrophotometer (UV-3600; Hitachi). NADH dehydrogenase activity was determined at 340 nm (ε = 6,220 M/cm) after the addition of 0.25 mM NADH, 0.25 mM decylubiquinone, and 1 mM KCN and controlling for rotenone sensitivity. Succinate dehydrogenase activity was measured at 600 nm (ε = 21,000 M/cm) after addition of 40 mM succinate, 35 µM dichlorophenolindophenol, and 1 mM KCN. The succinate cytochrome c reductase activity was followed at 540 nm (ε = 18,000 M/cm) in the presence of 40 mM succinate, 80 µg/ml cytochrome c, and 1 mM sodium azide. The specific II–III activity was defined as the flux difference before and after the addition of 1 µM antimycin A. The cytochrome c oxidase activity was assessed using a classical TMPD/ascorbate assay. In brief, oxygen consumption was assessed in the presence of 0.2 mM TMPD, 1 mM ascorbate, and 0.5 µM antimycin A. After a few minutes of stationary respiration, 2 mM KCN was injected into the chamber. The cytochrome c oxidase activity corresponds to the KCN-sensitive respiration. All chemicals were obtained from Sigma-Aldrich.
Oxygen consumption
100 µg of isolated mitochondria or 1 × 106 intact cells were used per experiment to measure oxygen consumption rates with Oxygraph-2k (Oroboros Instruments). Complex I activity was measured by the addition of 1 mM ADP, 5 mM pyruvate, 2 mM malate, and 10 mM glutamate. Complex I + complex II activity was obtained after addition of 10 mM succinate. Mitochondrial coupling was evaluated by the inhibition of ATP synthase by adding 1.5 µg/ml oligomycin and maximal respiration by a multiple-step CCCP titration. MEFs were cultured in galactose for 24 h before analysis.
Blue-native gel electrophoresis
Mitochondria were isolated from the liver as previously described (Edgar et al., 2009). Mitochondria were quantified using the Bradford method, and 25 µg were subjected to digitonin treatment (6 g digitonin per 1 g mitochondria) in lysis buffer (20 mM Tris, 0.1 mM EDTA, 50 mM NaCl, and 10% glycerol) during 15 min on ice after centrifugation at 4°C. Supernantant was combined with loading buffer (5% Coomassie Brilliant blue G-250, 100 mM Bis-Tris, and 500 mM 6-aminocaproic acid, pH 7) and loaded on 4–10% gradient gels. Gels were transferred to polyvinylidene fluoride membranes and blotted with the specified antibodies.
RNA sequencing
Total liver RNA was isolated using the ToTALLY RNA kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Libraries were prepared using the TruSeq RNA sample preparation kit (Illumina). Library preparation started with 1 µg of total RNA. After poly-A selection (using poly-T oligonucleotide-attached magnetic beads), mRNA was purified and fragmented using divalent cations under elevated temperature. The RNA fragments underwent reverse transcription using random primers, followed by second-strand cDNA synthesis with DNA Polymerase I and RNase H. After end repair and A-tailing, indexing adapters were ligated. The products were then purified and amplified (14 PCR cycles) to create the final cDNA libraries. After library validation and quantification (2200 TapeStation; Agilent Technologies), equimolar amounts of library were pooled. The pool was quantified by using the Peqlab KAPA Library Quantification kit (Roche) and the 7900HT Sequence Detection System (Applied Biosystems). The pool was sequenced by using a TruSeq PE Cluster kit (v3) and a TruSeq SBS kit (v3-HS) on a HiSeq 2000 sequencer with a paired-end (101 × 7 × 101 cycles) protocol (Illumina). Data were analyzed using QuickNGS, a highly scalable next-generation sequencing analysis pipeline developed in house (Wagle et al., 2015); basic read quality check was performed using FastQC (Babraham Bioinformatics), and read statistics were obtained with SAMtools. Reads were mapped to the human reference assembly, version GRCh38, using TopHat2 (Kim et al., 2013), and gene quantification was performed using a combination of Cufflinks (Trapnell et al., 2010) and the DEseq2 package (Anders and Huber, 2010) with genomic annotation from the Ensembl database (version 80). The results were uploaded into an in-house MySQL database and merged with annotations obtained with BiomaRt from Ensembl (version 80). The gene lists were filtered according to FC and p-values in comparison to the library-size normalized read counts between samples and controls. The sample and control means as well as FC and p-values were calculated with the DESeq2 package from the Bioconductor project. Wald’s test was used for significance testing. In contrast, gene expression for the individual samples was calculated by the Cufflinks package and returned as FPKM (fragments per kilobase of transcript per million mapped reads) values, which means that they have also been normalized by the molecule size. Enriched pathways were analyzed using the iPathwayGuide (Advaita Bio). Protein networks were assessed using STRING network analysis (STRING Consortium 2016). Only transcripts with an FC of >1.5 and a corresponding p-value of <0.05 were used for this analysis.
Proteomics
For proteomics analysis, livers of three WT and three KO mice and of one P1 SILAC mouse (fully labeled with [13C6]lysine; Krüger et al., 2008) were frozen in nitrogen immediately after dissection. Livers were powdered in nitrogen and lysed in buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM sodium chloride, 1 mM EDTA, 1% IGEPAL CA-630, 0.25% sodium deoxycholate, 0.1% SDS, and protease inhibitor cocktail (Sigma-Aldrich). 5 µg lysate of WT and Cluh−/− mice livers were mixed with 5 µg heavy liver lysate and precipitated with ice-cold acetone at −80°C. Dried pellet was resuspended in 8 M urea and 50 mM ammonium bicarbonate including a Complete Mini EDTA-free protease inhibitor cocktail (Roche). 5 mM DTT was added and incubated at 37°C for 1 h. 10 mM iodoacetamide was added and incubated for 30 min in the dark. Proteins were predigested with endoprotease Lys-C (Wako Pure Chemical Industries) with an enzyme/substrate ratio of 1:100 and incubated for 37°C for 4 h. After that, reactions were diluted with ammonium bicarbonate to a urea concentration of 2 M and digested with Lys-C with an enzyme/substrate ratio of 1:100 overnight at 37°C. The next day, digestion was stopped by the addition of 5% acetonitrile (ACN) and 1% trifluoroacetic acid, and StageTip purification was performed (Rappsilber et al., 2007). Samples were analyzed on a mass spectrometer (Q-Exactive Plus) coupled to an EASY nLC 1000 (Thermo Fisher Scientific). Peptides were separated on an in-house packed 50-cm analytical column (1.9-µm C18 beads; Dr. Maisch GmbH) using a binary buffer system consisting of (a) 0.1% formic acid (FA) in water and (b) 0.1% FA in ACN. Raw data were processed using MaxQuant 1.5.3.8 (Tyanova et al., 2016a) and the implemented Andromeda search engine (Cox et al., 2011). Tandem mass spectrometry spectra were correlated against the UniProt mouse database, including a list of common contaminants. We used 7- and 4.5-ppm tandem mass spectrometry tolerances for the first and main search, respectively. The false discovery rate at the peptide spectrum match and the protein level was controlled by the implemented decoy algorithm using the reverse database approach. Match-between runs and the Requantify option were enabled. N-terminal acetylation and oxidation at methionine residues were defined as variable modifications, whereas carbamidomethylation at cysteine residues was defined as a fixed modification. Data were further analyzed using the Perseus computational platform (Tyanova et al., 2016b). SILAC ratios were log2 transformed, and changes between KO and WT were determined as described previously (Zanivan et al., 2012). In brief, a one-sample two-tailed t test was used to calculate p-values (S0 was set to 0). The false discover rate was set to 0.05 (number of permutations, 500). Mitochondrial proteins were annotated based on the mitocarta database (Calvo et al., 2016). The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al., 2013) with the dataset identifier PXD005628. Significantly impacted pathways and diseases were analyzed using the iPathwayGuide (Advaita Bioinformatics).
Targeted metabolomics
Targeted metabolomics analyses of liver tissue were performed using ultra pressure liquid chromatography coupled to a triple quadrupole mass spectrometer. A detailed metabolite extraction protocol and instrument parameters are given elsewhere (Nikkanen et al., 2016). In brief, ∼20 mg of frozen liver tissues was homogenized by Precellys 24-bead homogenizer (1.4-mm beads), with 20 µl of the labeled internal standard mix and 500 µl of 100% ACN and 1% FA extraction solvent in the first step and 500 µl of 90/10% ACN/H2O + 1% FA in the second step extraction process. To obtain a robust statistical analysis, metabolomics data were normalized using the probabilistic quotient normalization method (Dieterle et al., 2006). After this procedure, using the median value of the WT group as reference, each metabolite value was rescaled to a common virtual concentration level. The data were further preprocessed with a log transformation. MetaboAnalyst 3.0 software (Xia et al., 2009) was used to conduct statistical analyses, and the unpaired two-sample t test was chosen to perform the comparisons. The Wilcoxon-Mann-Whitney test was preferred over the t test when a normal distribution did not show to be a good fit. Targeted metabolomics analyses of serum were performed as described previously (Mackay et al., 2015). In brief, 20 µl of serum was deproteinized by 1:20 dilution in a solution of 50% methanol, 30% ACN, and 20% water, vortexed for 5 min at 4°C, and then immediately centrifuged at 16,000 g for 15 min at 0°C. The supernatants were collected and analyzed by liquid chromatography–mass spectrometry. For the liquid chromatography separation, the Zic-pHilic column from EMD Millipore was used. Mobile phase A consisted of 20 mM ammonium carbonate plus 0.1% ammonia hydroxide in water. Mobile phase B consisted of ACN. The flow rate was kept at 100 ml/min, and the gradient was 0 min, 80% of B; 30 min, 20% of B; 31 min, 80% of B; and 45 min, 80% of B. The mass spectrometer (QExactive Orbitrap Plus; Thermo Fisher Scientific) was operated in a polarity switching mode. The data were analyzed using TraceFinder Software (Thermo Fisher Scientific). Statistical evaluation was performed using unpaired t tests of log-transformed data. Measured metabolites are listed in Table S3.
Cell culture
Immortalized MEFs obtained from WT and KO mice (Gao et al., 2014) were cultured in DMEM supplemented with 10% FetalClone III serum, 2% penicillin/streptomycin, and 2 mM l-glutamine (Thermo Fisher Scientific). To obtain the growth curve, 50,000 cells were seeded in different dishes and collected at the indicated time points. After cells were attached, medium was replaced with DMEM containing 4.5 g/liter glucose or 10 mM galactose supplemented with 10% dialysed serum (Invitrogen), 2% penicillin/streptomycin, and 2 mM l-glutamine. Cells were trypsinized at desired time points and counted with a hemocytometer to obtain the total amount of cells. The experiment was replicated four to five times per condition. Mitochondrial area/perimeter and the number of particles in MEFs were obtained using ImageJ and a macro developed by Dagda et al. (2009). Samples were analyzed using a spinning-disk confocal microscope (Ultra View VoX; PerkinElmer) with a Plan-Apochromat total internal reflection fluorescence 60×/1.49 NA oil DIC objective. Z stack images at 200-µm intervals were taken using an electron-multiplying charge-coupled device camera (C9100-50; CamLink). For LD staining, cells were fixed in 4% PFA in PBS for 30 min, incubated with 5 µM Bodipy 493/503 (Invitrogen) and DAPI in PBS for 20 min, washed in PBS, and mounted with Fluorsave. For image acquisition, an epifluorescence microscope (Imager.M2m) mounting an Apotome module and a 63×/1.4 NA Plan-Apochromat oil immersion objective lens (ZEISS) at RT was used. Images were acquired using an MRm camera (Axiocam) and AxioVision software (version 4.8 2.0; ZEISS). Images represent a single plane with a thickness of 1 µm.
RNAscope
Single-molecule RNA in situ hybridization was performed using the RNAscope 2.5 HD Fluorescent Reagent kit (Advanced Cell Diagnostics, Inc.). Target probes to detect murine Hadha (459331) and Atp5a1 (459311) were designed by the manufacturer. Cells were grown as indicated and fixed in 10% buffered formalin. Signal was amplified according to the manufacturer’s instructions and detected with an Amp4 Alt A-FL probe (Advanced Cell Diagnostics, Inc.). Mitochondria were stained using TOMM20 antibody combined with Alexa Fluor 546 secondary antibody (Thermo Fisher Scientific), and nuclei were stained with DAPI. Samples were analyzed using a spinning-disk confocal microscope with a Plan-Apochromat total internal reflection fluorescence 60×/1.49 oil DIC objective. Z stack images at 200-µm intervals were taken using an electron-multiplying charge-coupled device camera. To calculate the mean number of mRNAs per cell, a maximum projection was done, and particles were counted using an internal plugin of ImageJ. More than 80 cells were counted per treatment in three independent experiments. To obtain the frequency distribution of the distances, the images were deconvoluted with measured point-spread functions in Huygens Essential (Scientific Volume Imaging). The further processing was automated by a Fiji/ImageJ macro (Schindelin et al., 2012) by first reslicing the deconvoluted stacks to generate isometric voxels and then suppressing noise by a median filter (radius of 3 px). Mitochondria and RNA dots were segmented using the simple segmentation tool of the 3D ImageJ suite (Ollion et al., 2013), using a manual intensity threshold depending on the probe and the experiment and a minimum size limit of five voxels. Distances were calculated from the center of each RNA dot to the closest mitochondrial border. Negative distances represent colocalization between mitochondria and mRNA. Frequencies were obtained using Prism software (V6.0c; GraphPad Software), with a bin of 0.5 µm.
Measurement of mRNA stability
To assess mRNA stability, transcription of MEF cells was blocked by adding 5 µg/ml actinomycin D (Sigma-Aldrich). After 0, 2, 4, 6, 8, and 10 h, cells were harvested in TRIzol and stored at −80°C until RNA isolation was performed. Alternatively, the Click-iT Nascent RNA Capture kit (Thermo Fisher Scientific) was used according to the manufacturer’s instructions. In brief, 100,000 cells were seeded, and the next day, 0.2 mM EU was added for 24 h. After 24 h, cells were either immediately collected in TRIzol (total RNA at time point 0) or after 10-h incubation (total RNA at time point 10 h) in DMEM (4.5 g/liter glucose) without EU. RNA was isolated with TRIzol according to the manual, and 5 µg of total RNA was biotinylated with Click-iT chemistry. RNA was precipitated with ammonium acetate and EtOH, and 1 µg was used for pull-down of biotinylated RNAs with Dynabeads MyOne Streptavidin T1 beads (Thermo Fisher Scientific). EU-labeled RNA was retrotranscribed on the beads with the SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific). To calculate half-lives of mRNAs, the following equation was used: t1/2 = (t × log(2))/(log(No/Nf)); with t = time, No = percentage of mRNA at time point 0, and Nf = percentage of mRNA at time point 10 h.
Polysome profiling
MEFs were grown in glucose or for 20 h in galactose medium to 70–80% confluency. Before harvest, cells were incubated at 37°C for 15 min in medium containing 100 µg/ml cycloheximide (Sigma-Aldrich). Cells were washed and harvested in 4°C cold PBS supplemented with 100 µg/ml cycloheximide and pelleted at 21,000 g for 10 s at 4°C. Cells were lysed for 30 min on ice in buffer comprised of 20 mM Tris-HCl, pH 7.4, 30 mM KCl, 15 mM MgCl2, 0.5% Triton X-100 (vol/vol), 2 mM DTT, 1 mg/ml heparin, 100 µg/ml cycloheximide, 0.16 U/ml RNase inhibitor (RNasin Plus; Promega), and 1× EDTA-free protease cocktail. 3–4 mg lysate was applied on a continuous 7–47% sucrose gradient (mol weight/volume) and centrifuged at 97,658 g for 3 h at 4°C. Polysome fractions were obtained using the Foxy R1 Fraction Collector, and the polysome profile was detected in real-time using a UA-6 detector (Teledyne ISCO). Collected fractions were shock frozen in nitrogen and stored at −80°C. RNA extraction was performed with LS-TRIzol (Ambion) according to protocol, and monosome and polysome RNA fractions were pooled, followed by DNase treatment and 2.5 M LiCl precipitation at 4°C overnight. 0.5 ng Firefly luciferase mRNA (Promega) spike-in control was added before cDNA preparation.
Quantitative PCR
Total RNA was extracted using TRIzol reagent and retrotranscribed with the SuperScript First-Strand Synthesis system (Thermo Fisher Scientific) according to the manufacturer’s protocol. Quantitative PCR was performed using a thermocycler (Applied Biosystems) with SYBR green (Thermo Fisher Scientific). At least three biological replicates were performed per experiment, and GAPDH was used for normalization. The fold enrichment was calculated using the formula 2(−ΔΔCt). The following primers were used: Cluh forward, 5′-GGTAGCGGGCACGGTACA-3′, and reverse, 5′-GCATTGAGCACCCCAACAC-3′; Atp5a1 forward, 5′-TGGGCCGTGTGGTTGAC-3′, and reverse, 5′-CGTCTGCGGGTCTTGGAA-3′; Hadha forward, 5′-AAATCTGGGCCAACGACCAAA-3′, and reverse, 5′-CTTCCTGTGATATTCGTGTTGCT-3′; Pcx forward, 5′-AATGTCCGGCGTCTGGAGTA-3′, and reverse, 5′-ACGCACGAAACACTCGGAT-3′; Acat1 forward, 5′-TGTAAAAGACGGGCTAACTGATG-3′, and reverse, 5′-TGTTCCTGCCGTGAGATATTCAT-3′; Hmgcs2 forward, 5′-AGAGAGCGATGCAGGAAACTT-3′, and reverse, 5′-AAGGATGCCCACATCTTTTGG-3′; Nppb forward, 5′-GAGGTCACTCCTATCCTCTGG-3′, and reverse, 5′-GCCATTTCCTCCGACTTTTCTC-3′; Gapdh forward, 5′-AGGTCGGTGTGAACGGATTTG-3′, and reverse, 5′-TGTAGACCATGTAGTTGAGGTCA-3′; Bdh1 forward, 5′-TGCAACAGTGAAGAGGTGGAGAAG-3′, and reverse, 5′-CAAACGTTGAGATGCCTGCGTTGT-3′; Pdha1 forward, 5′-GAAATGTGACCTTCATCGGCT-3′, and reverse, 5′-TGATCCGCCTTTAGCTCCATC-3′; Pcca forward, 5′-GGTTTTAGGGGATAAACATGGCA-3′, and reverse, 5′-CCATTGCTTGGCGAGTTTCA-3′; Mccc1 forward, 5′-ACCATGAAGTATGGAACAACCC-3′, and reverse, 5′-TGCACACCCATCTTTTTGGCT-3′; Eif5a forward, 5′-TTAGTGTTCCCCAGCCTCATG-3′, and reverse, 5′-CAAATCTGGCTGGACGTCTCA-3′; and Firefly luciferase forward, 5′-AGTCAAGTAACAACCGCGAAAAA-3′, and reverse, 5′-TTCGGTACTTCGTCCACAAACA-3′. For mt-DNA quantification, whole-tissue DNA was extracted using standard techniques. The following primers were used: mt-Co1 forward, 5′-TGCTAGCCGCAGGCATTACT-3′, and reverse, 5′-CGGGATCAAAGAAAGTTGTGTTT-3′; and Rmrp forward, 5′-GCCTACACTGGAGTCGTGCTACT-3′, and reverse, 5′-CTGACCACACGAGCTGGTAGAA-3′.
Statistical analysis
For two-group comparison, two-tailed paired or unpaired nonparametric t tests were performed. For multiple group comparison, a one-way analysis of variance test was performed with a post-hoc Tukey’s test. χ2 tests were performed for the distribution of genotypes.
Online supplemental material
Fig. S1 shows histological analysis of the organs of Cluh−/− mice. Fig. S2 displays Western blots and relative quantification of different mitochondrial proteins in liver and heart lysates of WT and KO mice and shows the String network of down-regulated and up-regulated transcripts in the liver of KO mice. Fig. S3 shows additional phenotypic features of Li-CluhKO mice. Fig. S4 shows single-molecule FISH of Hadha and Atp5a1 mRNAs in WT and KO MEFs and a quantification of the number of mRNA molecules and of their distance to mitochondria. Table S1 is an analysis of proteomics data, and Table S2 reports transcripts up-regulated or down-regulated in the liver of Cluh KO mice in respect to WT mice (FC > 1.5, P < 0.05). Table S3 contains a list of measured metabolites.
Supplementary Material
Acknowledgments
We are grateful to Jie Gao for fruitful discussions, Anja Gruszczyk for assistance with C-sections, Hue-Tran Hornig-Do for support to measure TCA enzyme activities, Jatin Nandania for technical help in metabolomics sample analyses, Peter Frommolt for bioinformatics analysis of RNA sequencing data, and Christian Freese and Markus Krüger from the Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD)/Center for Molecular Medicine Cologne proteomics facility for help with proteomics analysis. Furthermore, we thank Christian Jüngst of the CECAD imaging facility, the CECAD proteomic facility, and Janine Altmüller and Christian Becker of the Cologne Center for Genomics.
This work was supported by Deutsche Forschungsgemeinschaft grants RU 1653/2-1 and SFB-1218, A05 to E.I. Rugarli.
The authors declare no competing financial interests.
Author contributions: E.I. Rugarli conceived the project and provided input for the experimental design and interpretations of results. D. Schatton designed and performed all experiments in Cluh−/− mice, measured ketone body production and gene expression of Cluhfl/fl:Alb-Cre mice, quantified LDs, and assessed RNA stability. D. Pla-Martin designed and performed experiments in Cluhfl/fl:Alb-Cre mice and characterized the MEF phenotype. M.-C. Marx performed polysome profiling and analyzed protein expression in MEFs. V. Kondylis contributed to the pathological analysis of Cluhfl/fl:Alb-Cre mice. H. Hansen and T. Corona designed the targeting strategy, and H. Hansen generated Cluh−/− and Cluhfl/fl mice. A. Mourier measured respiratory activities in Cluh−/− liver lysates. V. Velagapudi supervised the metabolomics experimentation and, together with A. Pessia, analyzed the data. I. Nemazanyy determined amino acids in the serum of Cluhfl/fl:Alb-Cre mice. P. Zentis quantified the distance of RNA molecules and mitochondria in RNAscope experiments. E. Barth provided technical assistance and performed EM. E.I. Rugarli and D. Schatton wrote the manuscript.
Footnotes
Abbreviations used:
- ACN
- acetonitrile
- ALT
- alanine aminotransferase
- BAIB
- β-aminoisobutyric acid
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- CLUH
- clustered mitochondria homologue
- DIC
- differential interference contrast
- EU
- 5-ethynyl uridine
- FA
- formic acid
- FC
- fold change
- HT
- heterozygous
- IDH
- isocitrate dehydrogenase
- KO
- knockout
- LD
- lipid droplet
- MEF
- mouse embryonic fibroblast
- mt-DNA
- mitochondrial DNA
- ORO
- Oil red O
- OXPHOS
- oxidative phosphorylation
- SILAC
- stable isotope labeling with amino acids in cell culture
- TCA
- tricarboxylic acid
- WT
- wild type
References
- Anders S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.A. 1998. Upf1 and Upf2 proteins mediate normal yeast mRNA degradation when translation initiation is limited. Nucleic Acids Res. 26:2433–2441. 10.1093/nar/26.10.2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., and Filipowicz W.. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 125:1111–1124. 10.1016/j.cell.2006.04.031 [DOI] [PubMed] [Google Scholar]
- Calvo S.E., and Mootha V.K.. 2010. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 11:25–44. 10.1146/annurev-genom-082509-141720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S.E., Clauser K.R., and Mootha V.K.. 2016. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44:1251–1257. 10.1093/nar/gkv1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter D.G., d’Avignon D.A., Wentz A.E., Weber M.L., and Crawford P.A.. 2011. Obligate role for ketone body oxidation in neonatal metabolic homeostasis. J. Biol. Chem. 286:6902–6910. 10.1074/jbc.M110.192369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowet R.M. 1992. Principles of Perinatal-Neonatal Metabolism. Springer-Verlag, New York. 1280 pp. [Google Scholar]
- Cox R.T., and Spradling A.C.. 2009. clueless, a conserved Drosophila gene required for mitochondrial subcellular localization, interacts genetically with parkin. Dis. Model. Mech. 2:490–499. 10.1242/dmm.002378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Dagda R.K., Cherra S.J. III, Kulich S.M., Tandon A., Park D., and Chu C.T.. 2009. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 284:13843–13855. 10.1074/jbc.M808515200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux F., Lelandais G., Garcia M., Goussard S., and Jacq C.. 2010. Posttranscriptional control of mitochondrial biogenesis: spatio-temporal regulation of the protein import process. FEBS Lett. 584:4273–4279. 10.1016/j.febslet.2010.09.030 [DOI] [PubMed] [Google Scholar]
- Diaz F., Garcia S., Hernandez D., Regev A., Rebelo A., Oca-Cossio J., and Moraes C.T.. 2008. Pathophysiology and fate of hepatocytes in a mouse model of mitochondrial hepatopathies. Gut. 57:232–242. 10.1136/gut.2006.119180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle F., Ross A., Schlotterbeck G., and Senn H.. 2006. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78:4281–4290. 10.1021/ac051632c [DOI] [PubMed] [Google Scholar]
- Edgar D., Shabalina I., Camara Y., Wredenberg A., Calvaruso M.A., Nijtmans L., Nedergaard J., Cannon B., Larsson N.G., and Trifunovic A.. 2009. Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab. 10:131–138. 10.1016/j.cmet.2009.06.010 [DOI] [PubMed] [Google Scholar]
- Ferreirinha F., Quattrini A., Pirozzi M., Valsecchi V., Dina G., Broccoli V., Auricchio A., Piemonte F., Tozzi G., Gaeta L., et al. . 2004. Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J. Clin. Invest. 113:231–242. 10.1172/JCI200420138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S.D., Conrad M.N., and Clarke M.. 1998. The S. cerevisiae CLU1 and D. discoideum cluA genes are functional homologues that influence mitochondrial morphology and distribution. J. Cell Sci. 111:1717–1727. [DOI] [PubMed] [Google Scholar]
- Fields S.D., Arana Q., Heuser J., and Clarke M.. 2002. Mitochondrial membrane dynamics are altered in cluA- mutants of Dictyostelium. J. Muscle Res. Cell Motil. 23:829–838. 10.1023/A:1024492031696 [DOI] [PubMed] [Google Scholar]
- Gao J., Schatton D., Martinelli P., Hansen H., Pla-Martin D., Barth E., Becker C., Altmueller J., Frommolt P., Sardiello M., and Rugarli E.I.. 2014. CLUH regulates mitochondrial biogenesis by binding mRNAs of nuclear-encoded mitochondrial proteins. J. Cell Biol. 207:213–223. 10.1083/jcb.201403129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N.L., Wilusz J., and Wilusz C.J.. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8:113–126. 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- Hock M.B., and Kralli A.. 2009. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 71:177–203. 10.1146/annurev.physiol.010908.163119 [DOI] [PubMed] [Google Scholar]
- Huch S., and Nissan T.. 2014. Interrelations between translation and general mRNA degradation in yeast. Wiley Interdiscip. Rev. RNA. 5:747–763. 10.1002/wrna.1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibdah J.A., Paul H., Zhao Y., Binford S., Salleng K., Cline M., Matern D., Bennett M.J., Rinaldo P., and Strauss A.W.. 2001. Lack of mitochondrial trifunctional protein in mice causes neonatal hypoglycemia and sudden death. J. Clin. Invest. 107:1403–1409. 10.1172/JCI12590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhan S.C., and Bier D.M.. 2008. Protein and amino acid metabolism in the human newborn. Annu. Rev. Nutr. 28:389–410. 10.1146/annurev.nutr.28.061807.155333 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., Polykratis A., Ehlken H., Ochoa-Callejero L., Straub B.K., Krishna-Subramanian S., Van T.M., Curth H.M., Heise N., Weih F., et al. . 2015. NEMO prevents steatohepatitis and hepatocellular carcinoma by inhibiting RIPK1 kinase activity-mediated hepatocyte apoptosis. Cancer Cell. 28:582–598. (published erratum appears in Cancer Cell 2015. 28:830) 10.1016/j.ccell.2015.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köntgen F., Süss G., Stewart C., Steinmetz M., and Bluethmann H.. 1993. Targeted disruption of the MHC class II Aa gene in C57BL/6 mice. Int. Immunol. 5:957–964. 10.1093/intimm/5.8.957 [DOI] [PubMed] [Google Scholar]
- Krüger M., Moser M., Ussar S., Thievessen I., Luber C.A., Forner F., Schmidt S., Zanivan S., Fässler R., and Mann M.. 2008. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 134:353–364. 10.1016/j.cell.2008.05.033 [DOI] [PubMed] [Google Scholar]
- Lal A., Mazan-Mamczarz K., Kawai T., Yang X., Martindale J.L., and Gorospe M.. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23:3092–3102. 10.1038/sj.emboj.7600305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber D.S., Calvo S.E., Shanahan K., Slate N.G., Liu S., Hershman S.G., Gold N.B., Chapman B.A., Thorburn D.R., Berry G.T., et al. . 2013. Targeted exome sequencing of suspected mitochondrial disorders. Neurology. 80:1762–1770. 10.1212/WNL.0b013e3182918c40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan D.C., Scott I., and Tobin A.K.. 2003. The genetic control of plant mitochondrial morphology and dynamics. Plant J. 36:500–509. 10.1046/j.1365-313X.2003.01894.x [DOI] [PubMed] [Google Scholar]
- Mackay G.M., Zheng L., van den Broek N.J.F., and Gottlieb E.. 2015. Analysis of cell metabolism using LC-MS and isotope tracers. In Methods in Enzymology. Vol. 561. Christian M.M., editor. Academic Press, Inc., Orlando, FL. 171–196. [DOI] [PubMed] [Google Scholar]
- Marc P., Margeot A., Devaux F., Blugeon C., Corral-Debrinski M., and Jacq C.. 2002. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 3:159–164. 10.1093/embo-reports/kvf025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeot A., Blugeon C., Sylvestre J., Vialette S., Jacq C., and Corral-Debrinski M.. 2002. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 21:6893–6904. 10.1093/emboj/cdf690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Bunkenborg J., Olsen J.V., Hjerrild M., Wisniewski J.R., Stahl E., Bolouri M.S., Ray H.N., Sihag S., Kamal M., et al. . 2003. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 115:629–640. 10.1016/S0092-8674(03)00926-7 [DOI] [PubMed] [Google Scholar]
- Nikkanen J., Forsström S., Euro L., Paetau I., Kohnz R.A., Wang L., Chilov D., Viinamäki J., Roivainen A., Marjamäki P., et al. . 2016. Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism. Cell Metab. 23:635–648. 10.1016/j.cmet.2016.01.019 [DOI] [PubMed] [Google Scholar]
- Ollion J., Cochennec J., Loll F., Escudé C., and Boudier T.. 2013. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 29:1840–1841. 10.1093/bioinformatics/btt276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C., and Magnuson M.A.. 2000. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 26:149–150. [DOI] [PubMed] [Google Scholar]
- Quenault T., Lithgow T., and Traven A.. 2011. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 21:104–112. 10.1016/j.tcb.2010.09.013 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., and Ishihama Y.. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., and Dymecki S.M.. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140. 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Roy B., and Jacobson A.. 2013. The intimate relationships of mRNA decay and translation. Trends Genet. 29:691–699. 10.1016/j.tig.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Georges Y., Garcia M., Delaveau T., Jourdren L., Le Crom S., Lemoine S., Tanty V., Devaux F., and Jacq C.. 2008. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One. 3 10.1371/journal.pone.0002293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R.C. 2002. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 286:81–89. 10.1016/S0378-1119(01)00809-5 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D.C., and Parker R.. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:5247–5256. 10.1128/MCB.19.8.5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F., Baron U., and Rajewsky K.. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080–5081. 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A., and Cox R.T.. 2016. Clueless is a conserved ribonucleoprotein that binds the ribosome at the mitochondrial outer membrane. Biol. Open. 5:195–203. 10.1242/bio.015313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A., Damm V.T., and Cox R.T.. 2013. Drosophila clueless is highly expressed in larval neuroblasts, affects mitochondrial localization and suppresses mitochondrial oxidative damage. PLoS One. 8 10.1371/journal.pone.0054283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F.H., Lee S., Wibom R., Olson L., and Larsson N.G.. 2011. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc. Natl. Acad. Sci. USA. 108:12937–12942. 10.1073/pnas.1103295108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., and Pachter L.. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon B., and Meloche S.. 2009. Interpreting neonatal lethal phenotypes in mouse mutants: insights into gene function and human diseases. Physiol. Rev. 89:1–26. 10.1152/physrev.00040.2007 [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., and Cox J.. 2016a The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11:2301–2319. 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., and Cox J.. 2016b The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13:731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- Valcarce C., Navarrete R.M., Encabo P., Loeches E., Satrústegui J., and Cuezva J.M.. 1988. Postnatal development of rat liver mitochondrial functions. The roles of protein synthesis and of adenine nucleotides. J. Biol. Chem. 263:7767–7775. [PubMed] [Google Scholar]
- Valle D., Beaudet A.L., Vogelstein B., Kinzler K.W., Antonarakis S.E., Ballabio A., Gibson K.M., and Mitchell G.. 2016. The Online Metabolic and Molecular Bases of Inherited Diseases. McGraw-Hill, Inc., New York. [Google Scholar]
- Vizcaíno J.A., Côté R.G., Csordas A., Dianes J.A., Fabregat A., Foster J.M., Griss J., Alpi E., Birim M., Contell J., et al. . 2013. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41:1063–1069. 10.1093/nar/gks1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vornlocher H.P., Hanachi P., Ribeiro S., and Hershey J.W.. 1999. A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiae TIF32 and TIF31 genes. J. Biol. Chem. 274:16802–16812. 10.1074/jbc.274.24.16802 [DOI] [PubMed] [Google Scholar]
- Wagle P., Nikolić M., and Frommolt P.. 2015. QuickNGS elevates Next-Generation Sequencing data analysis to a new level of automation. BMC Genomics. 16 10.1186/s12864-015-1695-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders R.J., Duran M., and Loupatty F.J.. 2012. Enzymology of the branched-chain amino acid oxidation disorders: the valine pathway. J. Inherit. Metab. Dis. 35:5–12. 10.1007/s10545-010-9236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.C., Jan C.H., and Weissman J.S.. 2014. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 346:748–751. 10.1126/science.1257522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Psychogios N., Young N., and Wishart D.S.. 2009. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37:652–660. 10.1093/nar/gkp356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanivan S., Krueger M., and Mann M.. 2012. In vivo quantitative proteomics: The SILAC mouse. In Integrin and Cell Adhesion Molecules: Methods and Protocols. Shimaoka M., editor. Humana Press, Totowa, NJ. 435–450. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Hulen D., Liu T., and Clarke M.. 1997. The cluA- mutant of Dictyostelium identifies a novel class of proteins required for dispersion of mitochondria. Proc. Natl. Acad. Sci. USA. 94:7308–7313. 10.1073/pnas.94.14.7308 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.