In this study, the authors use new reference models for 23 human centromeres and find that at all cell cycle phases centromeric CENP-A chromatin complexes are octameric nucleosomes with two molecules of CENP-A. This finding refutes previous models that have suggested that hemisomes may briefly transition to octameric nucleosomes.
Abstract
Chromatin assembled with centromere protein A (CENP-A) is the epigenetic mark of centromere identity. Using new reference models, we now identify sites of CENP-A and histone H3.1 binding within the megabase, α-satellite repeat–containing centromeres of 23 human chromosomes. The overwhelming majority (97%) of α-satellite DNA is found to be assembled with histone H3.1–containing nucleosomes with wrapped DNA termini. In both G1 and G2 cell cycle phases, the 2–4% of α-satellite assembled with CENP-A protects DNA lengths centered on 133 bp, consistent with octameric nucleosomes with DNA unwrapping at entry and exit. CENP-A chromatin is shown to contain equimolar amounts of CENP-A and histones H2A, H2B, and H4, with no H3. Solid-state nanopore analyses show it to be nucleosomal in size. Thus, in contrast to models for hemisomes that briefly transition to octameric nucleosomes at specific cell cycle points or heterotypic nucleosomes containing both CENP-A and histone H3, human CENP-A chromatin complexes are octameric nucleosomes with two molecules of CENP-A at all cell cycle phases.
Introduction
The ability of cells to properly segregate a complete set of chromosomes to each daughter cell during mitosis is dependent on a unique chromatin domain known as the centromere. Human centromeres are located on 1- to 4- Mb chromosomal regions that are composed of tandemly repeated arrays of a ∼171-bp element, termed α-satellite DNA repeats (Willard, 1985; Cleveland et al., 2003). In a proportion of centromeric DNA (Blower et al., 2002; Sullivan and Karpen, 2004), the canonical histone H3 is replaced with a histone variant initially identified in humans and named CENP-A (Earnshaw and Rothfield, 1985; Palmer et al., 1987). Paradoxically, except in budding yeast, this basic unit of chromosome inheritance is not defined by the DNA sequence, as α-satellite DNA sequences are neither sufficient nor essential for centromere identity (Karpen and Allshire, 1997; Cleveland et al., 2003; Stimpson and Sullivan, 2010). Rather, centromeres are defined and maintained epigenetically by CENP-A–containing chromatin that has been shown to identify, maintain, and propagate centromere function indefinitely in human cells and fission yeast (Fachinetti et al., 2013).
The chromatin composition of human centromeres has proven challenging to elucidate given their large (1–4 Mb) size and highly repetitive nature (Sullivan et al., 2001). Although it is known that centromeric chromatin is composed of both CENP-A– and H3-containing chromatin (Blower et al., 2002; Sullivan and Karpen, 2004), the precise nature and composition of CENP-A–containing chromatin in vivo has remained highly controversial. There is agreement that CENP-A readily assembles in vitro into octameric nucleosomes containing two molecules each of CENP-A and histones H2A, H2B, and H4 (Black et al., 2007; Falk et al., 2016) and forms a structure similar to that assembled with histone H3 (Tachiwana et al., 2011). However, the precise nature and composition of CENP-A–containing chromatin in vivo at multiple cell cycle points has not been settled. One prominent proposal is that human CENP-A chromatin oscillates in a cell cycle–dependent manner between two forms: a hemisome (containing only one molecule of histones H2A, H2B, CENP-A, and H4) that is maintained throughout most of the cell cycle but transitions at the end of G1 to octameric nucleosomes and then transitions back to hemisomes at the end of S phase (Bui et al., 2012). Contrasting this, sequential photobleaching combined with total internal reflection microscopy provided some evidence that at all cell cycle points CENP-A complexes contain CENP-A dimers (Padeganeh et al., 2013), albeit it is not established if these are assembled at centromeres, onto chromosome arms, or unassembled complexes with the CENP-A chaperone HJURP. Evidence in flies has also supported that some centromeric chromatin contains dimers of CENP-A (CID in flies; Zhang et al., 2012). There is agreement that in fly and human cells, CENP-A–containing chromatin assembly occurs only after exit from mitosis (Jansen et al., 2007; Schuh et al., 2007; Mellone et al., 2011; Nechemia-Arbely et al., 2012), whereas the CENP-A initially centromere bound is quantitatively redistributed to each daughter centromere during DNA replication (Jansen et al., 2007). However, none of the in vivo evidence to date has used genome-wide analyses to comprehensively test potential cell cycle–dependent changes in CENP-A–containing chromatin assembled at each human centromere.
Here, we use newly constructed reference models of the centromeres of all 22 human autosomes (unpublished data) and a recently published model for the X chromosome (Miga et al., 2014) to identify CENP-A and histone H3.1 binding within these megabase-sized domains. The overwhelming majority (97%) of α-satellite DNA is found at each human centromere to be assembled with histone H3.1–containing nucleosomes with wrapped DNA termini that protect 147 bp of DNA. The 2–4% of α-satellite DNA bound by CENP-A is shown to protect α-satellite DNA lengths centered on 133 bp in both G1 and G2 phases of the cell cycle, which is consistent with the idea that CENP-A chromatin at endogenous human centromeres is composed of octameric nucleosomes with DNA unwrapping at entry and exit. These findings provide quantitative support for a model in which α-satellite DNA sequences at all cell cycle phases are composed of rare, homotypic nucleosomes assembled with CENP-A that are interspersed among predominant H3.1 octameric nucleosomes.
Results
Centromeric CENP-A chromatin is a homotypic particle with two molecules of CENP-A and no H3 across the cell cycle
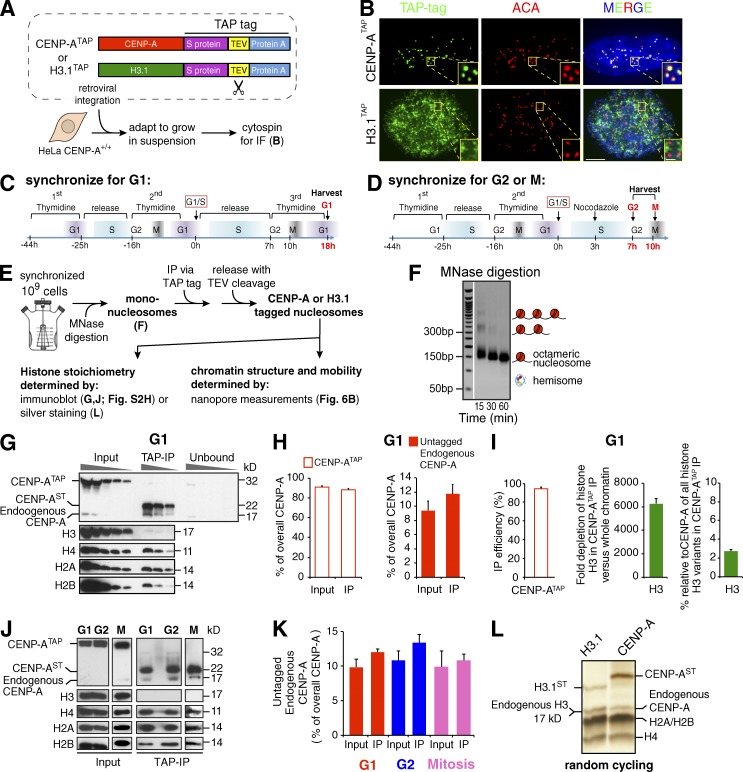
We previously established CENP-ATAP and H3.1TAP HeLa cell lines that stably express carboxy-terminally tandem affinity purification (TAP)–tagged (the TAP tag consists of S protein and protein A domains separated by a tobacco etch virus [TEV] protease cleavage site) variants of CENP-A or H3.1 (Foltz et al., 2006). CENP-ATAP was first determined to be fully functional in supporting long-term centromere identity and high-fidelity chromosome segregation in the absence of endogenous CENP-A. Starting from an RPE-1 cell line (CENP-A−/Flox; Fachinetti et al., 2013) with one endogenous CENP-A allele disrupted and the other one “floxed,” a CENP-ATAP (or EYFPCENP-A) encoding gene was stably integrated and expressed, and then the remaining endogenous CENP-A allele was converted to a null allele by the action of Cre recombinase (Fig. S1). Complete loss of endogenous CENP-A was confirmed in surviving colonies (Fig. S1, A–C), and accumulation of CENP-ATAP (or EYFPCENP-A) was determined to be comparable to CENP-A in the parental cells (Fig. S1 C). Importantly, both CENP-ATAP and EYFPCENP-A rescued long-term cell viability (Fig. S1 D). CENP-ATAP also maintained high-fidelity chromosome segregation to a level comparable with that found in the parental cells, as determined by a low rate (<6%) of micronuclei formation assayed after >100 generations (Fig. S1 E).
Next, HeLa cells expressing CENP-ATAP or H3.1TAP (as well as the endogenous untagged CENP-A and histone H3) were adapted to growth in suspension (Fig. 1 A). Total CENP-A (tagged and endogenous) accumulated to a level higher than in the original parental cells (Fig. S2 A). To measure total CENP-A (tagged and endogenous) accumulation in CENP-ATAP cells, we performed immunoblotting using a stack of three membranes, as a fraction of untagged CENP-A has been reported to pass through an initial immunoblot membrane (Bodor et al., 2014). Approximately 30% of untagged endogenous CENP-A passed through the first membrane onto the second and third, consistent with a previous study (Bodor et al., 2014), whereas CENP-ATAP was almost completely retained on the first membrane (Fig. S2, B and C). After correcting for the underscoring of untagged endogenous CENP-A, total CENP-A (tagged and endogenous) accumulated to 4.5-fold compared with the original endogenous CENP-A (Fig. S2 D). In parallel with CENP-ATAP accumulation, endogenous CENP-A was reduced to ∼60% of its normal level, presumably reflecting competition of CENP-ATAP and CENP-A for recognition and loading by its chaperone HJURP (Dunleavy et al., 2009; Foltz et al., 2009; Fig. S2, A and D). The final ratio of endogenous CENP-A:CENP-ATAP was 1:11 (Fig. S2, A–D). Importantly, and as expected for its ability to complement absence of CENP-A, CENP-ATAP was localized to centromeres (Fig. 1 B).
Figure 1.
Purified CENP-A chromatin is a homotypic octamer at G1 and G2 and in mitosis and does not contain H3. (A) HeLa cells stably expressing CENP-ATAP or H3.1TAP (Foltz et al., 2006) were adapted to growth in suspension. The TAP tag includes S protein, TEV protease cleavage site, and protein A. (B) Immunofluorescence (IF) for the TAP-tagged proteins using FITC-IgG. Bar: 5 µm; (inset) 1 µm. (C and D) Double thymidine block synchronization protocol for enriching cells at G1 (C) or at G2 and M phase (D). (E) Experimental design for obtaining mononucleosome pools and TAP-tagged monochromatin particles. (F) Titration of MNase digestion time identifies conditions to generate a pool of bulk soluble mononucleosomes. (G) SDS-PAGE of serial 1:2 dilutions of input, immunoprecipitation, and unbound fractions of affinity-purified CENP-ATAP at G1. (H) Quantification of CENP-ATAP and untagged endogenous CENP-A levels in the input and immunoprecipitation of the blot shown in G, after correcting for the CENP-A fraction that passes through the immunoblot membrane as shown in Fig. S2 (B and C). n = 3 from three independent loadings in the gel shown in G. Error bars represent SEM. (I, left) Efficiency of CENP-ATAP recovery in the immunoprecipitation shown in G. (I, middle) Fold depletion of histone H3 after CENP-ATAP immunoprecipitation. (I, right) Percentage of histone H3 relative to CENP-A in the CENP-ATAP immunoprecipitate. For each panel, n = 3 from three independent loadings in the gel shown in G. Error bars represent SEM. (J) Purified CENP-ATAP monochromatin particles from G1, G2, and M synchronized cells run on SDS-PAGE and blotted for the different histones. (K) Quantification of untagged endogenous CENP-A levels in the input and immunoprecipitation of the blot shown in J. n = 2 from two independent replicates. Error bars represent SEM. (L) Silver-stained SDS-PAGE of purified H3.1TAP and CENP-ATAP monochromatin particles from random cycling cells reveals all four histones at comparable levels.
To determine the state of CENP-A chromatin across the cell cycle, CENP-ATAP cells were synchronized at G1, G2, or mitosis using a double thymidine block and release protocol (Fig. 1, C and D). Synchronization achieved enriched populations of 80%, 85%, and 98% for G1, G2, and mitosis, respectively, as measured by analysis of DNA content with FACS. Similar measures for synchronization were obtained by assessing levels of cyclin B1 and phosphorylated histone H3 (Fig. S2, F and G).
Recognizing that all of the evidence for hemisomes (Dalal et al., 2007; Wang et al., 2008; Dimitriadis et al., 2010; Bui et al., 2012; Krassovsky et al., 2012; Shivaraju et al., 2012; Henikoff et al., 2014) has used chromatin isolation after DNA digestion with micrococcal nuclease under conditions that also produce octameric nucleosomes, in vivo CENP-A chromatin composition at each cell cycle phase was determined at the single-chromatin particle level after digestion of chromatin with micrococcal nuclease. In the bulk DNA, this produced the 147–150 bp of protected DNA length expected for mononucleosomes assembled with histone H3 (Fig. 1, E and F; and Fig. 2 B, top). Chromatin complexes were then affinity purified using a rabbit-IgG antibody coupled to magnetic beads. TEV protease cleavage produced CENP-AST or H3.1ST chromatin from their bead-bound CENP-ATAP and H3.1TAP progenitors (Fig. 1 E) and eluted chromatin from the beads in its native state.
Figure 2.
High-throughput sequencing and mapping to α-satellite DNA reveals that centromeric CENP-A chromatin is an octameric nucleosome with transient DNA unwrapping of the DNA entry and exit sites at G1, G2, and mitosis. (A) Experimental design for obtaining CENP-ATAP– and H3.1TAP-bound DNA sequences. (B) Microcapillary electrophoresis of MNase-digested bulk input mononucleosomes (top) and CENP-ATAP or H3.1TAP or NH2H3CATD native ChIP (middle and bottom, immunoprecipitation). Purified CENP-A chromatin is nucleosome-like but protects a shorter DNA length than does H3.1-containing nucleosomes. (C) Quantitative real-time PCR for α-satellite DNA extracted from CENP-ATAP chromatin in random cycling (RC; magenta), G1 (red), and G2 (blue). n = 2 from two independent replicates. Error bars represent SEM. (D) CENP-ATAP– and H3.1TAP-bound DNA was sequenced using paired-end 100-bp ChIP sequencing and then mapped to the human genome 38 assembly (hg38), which contains α-satellite sequence models for each centromere. Approximately 50% of CENP-ATAP–bound DNA is centromeric at G1 and G2. (E) CENP-ATAP–bound DNA sequences that mapped to α-satellite DNA were analyzed for their nucleosomal DNA length and overlaid on the microcapillary electrophoresis data (black line) shown in B. (F) CENP-ATAP–bound DNA sequences that mapped to α-satellite DNA on chromosome X were analyzed for their nucleosomal DNA length and overlaid on the microcapillary electrophoresis data (black line) shown in B. (G) Distribution of the chromatin DNA lengths into two length bins: subnucleosomal (60–85 bp, as predicted for tetrasomes and hemisomes; Hasson et al., 2013) and nucleosomal (100–170 bp) for bulk input chromatin and affinity-purified CENP-ATAP and H3.1TAP chromatin mapped to α-satellite DNA. n = 2 from two independent replicates. Error bars represent SEM.
The purified chromatin was analyzed for composition by immunoblotting (Fig. 1, G–K; and Fig. S2 H) and silver staining (Fig. 1 L). At G1, G2, and M, untagged endogenous CENP-A comprised between 9% and 13% of the total CENP-A in the affinity-purified complexes (Fig. 1, G, H, J, and K), consistent with its level relative to CENP-ATAP in the initial randomly cycling cell lysates (Fig. S2, A and D). Combining our determination that 97% of CENP-ATAP was recovered by the affinity purification (Fig. 1, G and I) and the retention at each cell cycle point of the same proportion of CENP-A:CENP-ATAP in the affinity purification that was present in the initial lysates (Fig. 1, J and K; and Fig. S2, A and D), almost all of CENP-A must be assembled into complexes containing at least two CENP-A molecules (Fig. S2 E). This outcome is inconsistent with hemisomes or heterotypic CENP-A/histone H3 nucleosomes comprising more than a small proportion of CENP-A chromatin. In addition, immunoblotting (Fig. 1, G and J) and silver staining (Fig. 1 L) of the affinity-purified complexes from all cell cycles stages revealed that H2A, H2B, and H4 were copurified at levels relative to CENP-AST that matched their levels relative to histone H3 in H3.1ST chromatin (Fig. S2 H), findings that rule out a significance presence of CENP-A tetrasomes (which lack H2A/H2B) at any cell cycle stage.
Immunoblotting for histone H3 revealed that its level had been depleted ∼6,250-fold in the purified CENP-AST complexes compared with its level in the input (Fig. 1, G and I, middle). Although the CENP-ATAP comprises at most ∼0.5% of the total level of histone H3 variants (CENP-A assembled into chromatin when expressed at normal levels has been measured to be 0.1% of total mammalian cell chromatin; Bodor et al., 2014), the remaining histone H3 after affinity purification was only ∼2.5% of the amount of CENP-AST in the affinity purified material (Fig. 1, G and I, right). Consequently, even in cells in which CENP-A accumulated to levels up to 4.5 times the normal level, heterotypic complexes of CENP-A with histone H3 can comprise at most 2.5% of CENP-A chromatin.
Collectively, our results demonstrate that before (G1) and after (G2 and M) DNA replication, including the reduction by half in the amount of CENP-A assembled at an individual centromere (Jansen et al., 2007), almost all CENP-A is assembled into complexes without histone H3 but with at least two molecules of CENP-A.
Genome-wide mapping of CENP-A and histone H3.1 chromatin onto 23 human centromere reference models
Affinity purified CENP-A– or H3.1TAP-containing chromatin was isolated from random cycling and/or synchronized H3.1TAP or CENP-ATAP cells, respectively. Assay with quantitative PCR of affinity-purified CENP-A containing chromatin from random cycling or G1 CENP-ATAP cells determined that α-satellite DNA sequences from chromosome 1, 3, 5, and 10 were enriched by 30-fold (Fig. 2, A and C) relative to their level in the initial total chromatin. As expected from the known redistribution of previously bound centromeric CENP-A onto the two DNA daughter strands after DNA replication without loading of new CENP-A (Jansen et al., 2007), α-satellite DNA enrichment decreased by half in CENP-ATAP chromatin from G2 cells (Fig. 2 C).
Libraries of DNA were prepared from micrococcal nuclease–resistant CENP-ATAP or H3.1TAP-bound chromatin and sequenced using paired-end 100-bp deep sequencing. Paired reads were merged and then mapped (Fig. 2, A and D–F; Fig. 3; and Tables S1 and S2) onto reference models for the α-satellite DNA sequences on the X chromosome (Miga et al., 2014) or newly assembled models that represent the observed variation in α-satellite sequences and repeat order found in a database of α-satellite DNA reads for the centromeres of chromosomes 1 through 22 (unpublished data; Tables S3 and S4). These reference models contain all α-satellite sequences (centromeric and pericentromeric) in the human genome assembly 38, including the second and third α-satellite arrays that exist in some of the chromosomes. Specifically, these sequences contain all higher-order repeat arrays as well as the highly divergent monomeric α-satellite sequences. We also mapped these sequence reads onto a previously published whole-genome sequence database of α-satellite sequences (Levy et al., 2007; Hayden et al., 2013). With either mapping approach, approximately half (44% and 56%, respectively, in G1 and G2) of the CENP-ATAP–bound DNA mapped to α-satellite DNA (Fig. 2 D).
Figure 3.
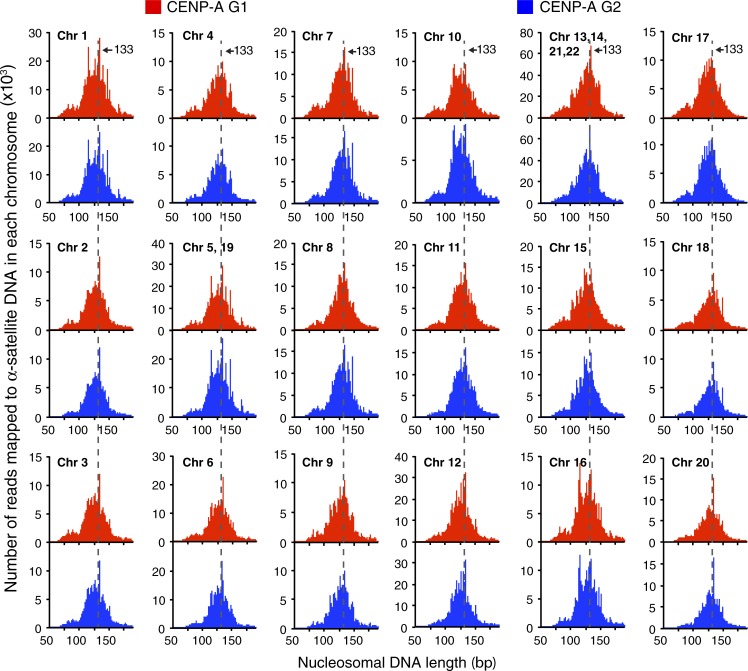
CENP-A chromatin is nucleosome-like but with transient DNA unwrapping of the DNA entry and exit sites before (G1) and after (G2) DNA replication at all human centromeres. CENP-ATAP–bound DNA sequences that mapped, using human centromere reference models for the centromeres for the 22 human autosomes (unpublished data), to α-satellite DNA throughout each centromere were analyzed for DNA lengths protected from micrococcal nuclease digestion.
Purified chromatin from histone H3.1TAP cells or the bulk chromatin from the CENP-ATAP cells revealed that the lengths of histone H3.1–associated or bulk micrococcal nuclease–resistant DNAs centered on 147 bp (Fig. 2 B, top and middle), as expected for DNA protected by conventional histone H3–containing nucleosomes. In contrast, the length distributions of DNAs protected by CENP-ATAP chromatin and mapped to total α-satellite sequences (Fig. 2 E) or to α-satellite sequences at each of 23 human centromeres (Figs. 2 F and 3) were centered on 133 bp, irrespective of cell cycle position (Fig. 2, E and F; and Fig. 3; compare red and blue) and mimicked what was observed with microcapillary electrophoresis (Fig. 2, B, E, and F, black line). The distribution of sizes was consistent with the large majority of CENP-A assembled into nucleosomes with unwinding of the DNA at entry and exit, as previously established for octameric CENP-A nucleosomes assembled in vitro (Conde e Silva et al., 2007).
Mapping of the sequences bound by CENP-A onto individual α-satellite arrays in each centromere reference model provided an unbiased test of the nature of CENP-A containing chromatin. Nucleosome-like DNA lengths were protected by CENP-A chromatin at each human centromere both before (G1) and after (G2) DNA replication (Fig. 2, E–G; and Fig. 3). Indeed, 86% and 89% of α-satellite bound CENP-A protected DNA larger than 100 bp in G1 and G2, respectively (Fig. 2 G). A minority (7.0% or 5.5%) of CENP-ATAP purified DNA in G1 or G2, respectively, migrated at DNA lengths of 60–85 bp (Fig. 2, E–G). Although these sizes are consistent with wrapping either of tetrasomes or hemisomes (Hasson et al., 2013), this minority may also represent products of microccocal nuclease digestion of DNAs initially assembled into octameric nucleosomes, a possibility strongly supported by a comparable percentage (5.5% or 2.5%, respectively) of similarly sized products in bulk input or affinity purified H3.1TAP chromatin. Interestingly, the micro-capillary electrophoresis fluorescence peak representing the total CENP-A chromatin at these smaller DNA sizes was more pronounced than the CENP-A reads mapped to α-satellite reads (Fig. 2 E), suggesting that the smaller particles are noncentromeric.
Almost all (>97%) of α-satellite chromatin is assembled with nucleosomes containing histone H3.1
Mapping of histone H3.1 bound to α-satellite DNAs at each human centromere also identified that in random cycling cells 2.15% of total H3.1TAP-bound DNAs aligned to α-satellite sequences (Fig. 2 D and Tables S1 and S2) and yielded micrococcal nuclease resistant DNA sizes centered on 147 bp (Fig. 2 E), as expected for a conventional octameric nucleosome assembled with histone H3 (Shaw et al., 1976; Luger et al., 1997) and with wrapped DNAs at entry/exit (Hasson et al., 2013). Because there are only ∼400 molecules of CENP-A loaded at each centromere (Bodor et al., 2014), these 400 hemisomes or 200 nucleosomes can cover 40kb (assuming each chromatin particle maximally to span 100 or 200 bp of DNA for hemisomes or nucleosomes, respectively). Since the mean centromere size is of ∼2.5 Mb (Lee et al., 1997), CENP-A is maximally assembled onto only ∼2% of centromeric chromatin. Indeed, our genome wide analysis in conjunction with use of the centromere reference models revealed that histone H3.1 is assembled into nucleosomes onto almost all (97.7%; that is, 2.15% of total H3.1 sequence reads mapped to α-satellite DNA, which comprises 2.2% of the genome) pericentromeric and centromeric α-satellite DNA–containing chromatin.
The CENP-A CATD and amino-terminal tail mediate assembly of nucleosomes with DNA unwinding
We next tested if assembly into nucleosomes with uncrossed DNAs at entry/exit was conferred to CENP-A chromatin by the CENP-A centromere-targeting domain (CATD), a region spanning loop 1 and α2 helix of CENP-A and (a) that confers compaction and unique structural rigidity to (CENP-A/H4)2 tetramers in solution (Black et al., 2004) and (b) is required for its centromere targeting (Black et al., 2004; Fachinetti et al., 2013). For this, we used the RPE-1 cell line in which both endogenous CENP-A alleles were disrupted and long-term viability and high-fidelity centromere function were rescued by expression of a chimeric histone H3 (NH2H3CATD) in which both the CATD and the CENP-A amino-terminal tail have been substituted for the corresponding regions of histone H3 (Fachinetti et al., 2013). Immunoprecipitation and microcapillary electrophoresis of DNAs from NH2H3CATD chromatin yielded a distribution of protected DNAs centered on 133 bp that was indistinguishable from the corresponding distribution for CENP-A chromatin (Fig. 2 B, bottom). Thus, the CATD and the amino-terminal tail of CENP-A are sufficient to mediate assembly of histone H3 into nucleosomes with the DNA unwrapping found for CENP-A chromatin.
Endogenous CENP-A is assembled into homotypic particles with two molecules of CENP-A and no histone H3
To measure CENP-A chromatin composition without overexpression, we exploited a cell line (Mata et al., 2012) in which homologous recombination was used to carboxy-terminally tag an endogenous allele of CENP-A with a localization (EYFP) and affinity (His) purification (LAP) affinity tag (Fig. 4 A). Carboxy-terminal tagging of CENP-A with YFP has been shown to produce a completely functional CENP-A variant that functions in the absence of wild-type CENP-A (Bodor et al., 2014). The CENP-A+/LAP cell line was adapted to growth in suspension, and centromeric localization of CENP-ALAP was confirmed (Fig. 4, A and B).
Figure 4.
Affinity purified CENP-ALAP expressed from an endogenous CENP-A locus is assembled into chromatin as a homotypic octamer and does not contain H3. (A) Schematic for construction and use of a HeLa cell line in which CENP-A was tagged at the endogenous allele with a LAP-tag consisting of Hisx6, a Precission protease cleavage site and EYFP (Mata et al., 2012). The resulting CENP-A+/LAP cell line was adapted to growth in suspension. (B) Localization of endogenously tagged CENP-ALAP determined with indirect immunofluorescence using anti-GFP antibody. Bars: 5 µm; (inset) 1 µm. (C) Experimental design for affinity purification of endogenously tagged CENP-ALAP monochromatin particles. (D) SDS-PAGE of serial 1:2 dilutions of input, immunoprecipitation, and unbound fractions of CENP-ALAP affinity purification from cells synchronized in G2. (E, left) Quantification of CENP-ALAP and nontagged CENP-A levels in the blot shown in D before correcting for the fraction of CENP-A that passes through the membrane. (right) Quantification of CENP-ALAP and nontagged CENP-A levels in the input and immunoprecipitation (IP) in the blot shown in D after correcting for the underscoring of CENP-A as shown in Fig. S3 (A and B). For each panel, n = 3 from three independent loadings in the gel shown in D. Error bars represent SEM. (F) CENP-ALAP immunoprecipitation efficiency (left) and levels of unbound nontagged endogenous CENP-A (right) were quantified in the blot shown in D. For each panel, n = 3 from three independent loadings in the gel shown in D. Error bars represent SEM.
CENP-ALAP chromatin was affinity purified from cells synchronized to be in G2 and eluted by protease cleavage of the tag (Fig. 4 C). Immunoblot signals for CENP-ALAP:CENP-A were quantified to be 1.5:1 (Fig. 4 E, left). After correcting for the retention of only two thirds of untagged CENP-A on the immunoblot filter (Fig. S3, A and B; Bodor et al., 2014), CENP-ALAP and CENP-A were calculated to accumulate to equivalent levels, as expected for equal synthetic rates from the unmodified and LAP-tagged endogenous alleles (Fig. 4 E, right, input). After affinity elution with Precission cleavage to produce CENP-AHis from CENP-ALAP, CENP-AHis:CENP-A in the final immunopurified fraction was 2:1 (Fig. 4, D and E, right, IP), as expected for two molecules of CENP-A in each complex and inconsistent with a significant proportion of complexes with a single CENP-A molecule (Fig. S3 C). Despite >95% recovery of the input CENP-ALAP (Fig. 4 F), histone H3 remaining in the affinity-purified material was <1% of CENP-ALAP, indicating the near absence of heterotypic nucleosomes (Fig. 4 D) and in agreement with what we observed with CENP-ATAP (Fig. 1). In addition, the level of untagged CENP-A in the unbound fraction was 22.5% of the total CENP-A levels in the input, nearly an exact match to the 25% value predicted to remain after removal of a population of CENP-A chromatin particles with two molecules of CENP-A in each complex (Fig. 4, D and F; and Fig. S3 C).
CENP-A chromatin has the physical characteristics of nucleosomes
Next, recognizing that evidence from atomic force microscopy has been used to support a model in which CENP-A chromatin transitions from hemisomes in G1 to octasomes before DNA replication and then reverts back to hemisomes after replication (Bui et al., 2012), we extended our test for the nature of in vivo derived CENP-A chromatin by examining its physical properties after isolation from cell populations enriched at G1, G2, or mitotic cell cycle positions. We first determined the sedimentation velocity of in vitro–reconstituted CENP-A and H3 octameric nucleosomes wrapped with 145 bp of template DNA (using the “601” nucleosome positioning sequence; Lowary and Widom, 1998) as well as (H3/H4)2 and (CENP-A/H4)2 tetrasomes wrapped with 101 bp of template DNA (Fig. 5, A and B; and Fig. S4 A). As expected, in vitro reconstituted histone H3 or CENP-A octameric mononucleosomes sedimented more rapidly (peaking in fraction 17) than did either (H3/H4)2 or (CENP-A/H4)2 tetrasomes (peaking in fraction 9; Fig. 5 B).
Figure 5.
CENP-A chromatin has the physical characteristics of nucleosomes. (A) Experimental design for the separation of in vitro–reconstituted octameric nucleosomes or tetrasomes over a 5–25% sucrose gradient. (B) After sedimentation of in vitro–reconstituted H3 or CENP-A octameric nucleosomes or (CENP-A/H4)2 or (H3/H4)2 tetrasomes, fractions were immunoblotted for CENP-A or H3. (C) Experimental design for the sedimentation of in vivo bulk nucleosomes and short polynucleosomes at different points of the cell cycle. (D) Moderate MNase digestion profile of bulk chromatin from random cycling (RC) cells expressing CENP-ATAP or H3.1TAP. (E) Sucrose gradient sedimentation and fractionation of bulk nucleosomes from CENP-ATAP–expressing cells synchronized at G1, G2, and mitosis. (top) Ethidium bromide stained DNA agarose gel revealing the DNA length extracted from the different fractions. (bottom) Immunoblot for CENP-ATAP. (F) Real-time quantitative PCR for α-satellite DNA in the different fractions (colored). Second axis shows quantification of CENP-A immunoblot shown in E. No CENP-A or α-satellite DNA was detected in fractions 7 and 9.
Next, chromatin obtained from synchronized cells was digested with micrococcal nuclease to produce primarily mononucleosomes (85%) with the remaining 15% composed of short stretches of up to three nucleosomes (Fig. 5, C and D). After sedimentation, fractions were analyzed for their α-satellite DNA content and presence of CENP-A. Sedimentation of CENP-ATAP–containing chromatin closely tracked both the bulk nucleosomal DNA and in vitro–reconstituted, CENP-A– or H3-containing octameric nucleosomes (Fig. 4 E, bottom panels). CENP-A fractionation was indistinguishable in chromatin isolated from G1, G2, and mitotic-synchronized cells (Fig. 4 E), as well as from H3.1TAP randomly cycling cells (Fig. S4 B). Importantly, in vivo–purified CENP-A chromatin did not appear in the slower-migrating fractions that represent sedimentation positions of in vitro–assembled subnucleosomal structures (Fig. 5, B and E; and Fig. S4 C). Similarly, alphoid DNA content peaked in mononucleosome fractions in random cycling or G2 synchronized cells and was undetectable in fractions corresponding to positions of tetrasomes (Figs. 5 F and S4 D).
Solid-state nanopore measurements of single CENP-A chromatin particles
Finally, we used solid-state nanopores to measure in vivo–derived CENP-A chromatin. Solid-state nanopores have been shown to discriminate between nucleosome and subnucleosomal structures at the single-complex level (Soni and Dekker, 2012). In this approach, a flow cell is assembled with a nanometer-size hole as the only opening that connects fluids on either side of a thin solid-state membrane. Under an applied potential, ions that flow from one side of the membrane to the other through the pore are measured as open pore current. Translocation of biological molecules through the pore under the applied voltage is detected as transient blockade events in pore conductance (ΔG), which in turn is a measure of the molecular volume of the translocating biomolecule. Statistical analysis of many hundreds of such single-molecule translocation events can be used to distinguish and characterize biomolecular populations of different molecular sizes (Fig. 6 A).
Figure 6.
Solid-state nanopore analysis of CENP-A chromatin reveals it to have the physical characteristics of nucleosomes. (A) Experimental design of chromatin translocation through a solid-state 20-nm-wide nanopore. (B) Conductance drop (ΔG) measurements during the translocation of (top, red) in vivo affinity-purified CENP-ATAP–containing chromatin particles (total events = 3,096), in vivo affinity-purified H3.1TAP-containing chromatin particles (middle, green; total events = 4,141), and (bottom, purple) in vitro–reconstituted (CENP-A/H4)2 tetrasomes (total events = 383). n = 2 from two independent datasets. Error bars represent SD. A fitted Gaussian distribution is overlaid in black. Fits were performed by the Levenberg–Marquardt algorithm to reduce the χ2 value. The reduced R2 values for the three fits were 0.94, 0.954, and 0.961. ΔG values represent the mean of the fitted Gaussian distribution, and the error bars are the width of the Gaussian fits. Vertical dashed line is the peak ΔG value for the tetrasome, as measured by the Gaussian fits. (C) Summary of the chromatin types measured in this study and previously (Soni and Dekker, 2012) using solid-state nanopore, along with their predicted molecular masses (with 4.5 kD added to each tagged histone to account for the S-peptide tag remaining after TEV cleavage). In vitro H3.1 nucleosomes were reconstituted on a 344-bp 601 nucleosome positioning sequence.
In vitro–reconstituted (CENP-A/H4)2 tetrasomes driven through a nanopore had a mean conductance drop of 5.4 ± 0.4 nS, in agreement with 4.8 nS that was measured previously (Soni and Dekker, 2012) for in vitro tetramers without any wrapping DNA (Fig. 6 C). Wrapping with 101 bp DNA (MW of 121.5 kD) adds ∼2 nm of DNA around the tetramer and is expected to add ∼1 nS of conductance drop (Kowalczyk et al., 2011). In vivo–purified CENP-ATAP chromatin from two independent experiments produced a normal distribution of conductance drops (ΔG) with a mean of 7.0 ± 1.0 nS (Fig. 6, B and C), a value similar to that measured for purified H3.1TAP nucleosomes (mean ΔG of 6.7 ± 1.5 nS). Notably these values are consistently larger than the 5.4 nS value for the smaller tetrasomes and comparable to the 8.0 nS previously measured (Soni and Dekker, 2012) on histone H3–containing nucleosomes assembled in vitro on a 344-bp DNA template, as the longer DNA template used previously is estimated to result in an additional ∼1 nS (Kowalczyk et al., 2011; Fig. 6 C). These measurements clearly indicate a comparable volume for the CENP-A and histone H3.1–containing single chromatin particles, an outcome consistent with the idea that a large majority of in vivo CENP-A chromatin consists of octameric nucleosomes.
Discussion
By using high-throughput sequencing of DNAs bound by CENP-A and reference models for 23 human centromeres, combined with biochemical, hydrodynamic, and solid-state nanopore analyses, we have determined that at least 86% of human CENP-A chromatin bound to α-satellite DNA at each human centromere is composed of homotypic octameric nucleosomes at all cell cycles phases with (a) two molecules of CENP-A; (b) comparable stoichiometries of CENP-A and histones H2A, H2B, and H4; and (c) an unchanging DNA wrapping length centered on 133 bp, consistent with assembly into an octameric nucleosome with unwound DNA termini.
A parallel DNA sequencing effort also revealed that almost all (>97%) of the α-satellite DNA is bound with histone H3.1–containing nucleosomes that protect 147 bp of α-satellite DNA, consistent with nucleosomes that have DNA termini completely wrapped around the histone core (Hasson et al., 2013). The structural difference between centromeric and pericentromeric CENP-A nucleosomes (with their shorter DNA length protected) and those assembled with histone H3 was further shown to be mediated by the CENP-A amino-terminal tail and CATD, the latter of which (a) enables centromere targeting (Black et al., 2004), (b) creates a rigid interface between CENP-A and H4 that results in a more compact nucleosome (Black et al., 2004, 2007; Sekulic et al., 2010), (c) enables templating of the centromeric chromatin through its recognition by HJURP at the exit of mitosis (Foltz et al., 2009; Fachinetti et al., 2013), and (d) is also the domain responsible at all cell cycle phases for unwinding of DNA in CENP-A nucleosomes (Fig. 2 B), as we show in this study. Thus, the CATD differentiates CENP-A chromatin from the rest of the chromosome, including centromeric/pericentromeric chromatin containing histone H3.
Although evidence from successive immunoprecipitations was previously interpreted to support that a large majority of CENP-A chromatin assembled in vivo at noncentromeric sites in asynchronously cycling cells is composed of heterotypic CENP-A/histone H3-containing nucleosomes (Lacoste et al., 2014), our experiments have revealed that histone H3 is virtually absent from CENP-A affinity-purified chromatin and at all cell cycle stages nontagged CENP-A is coaffinity purified with tagged CENP-A and in the ratio expected for a homotypic particle with two molecules of CENP-A. We show this to be true for CENP-A when expressed at normal or elevated levels. Thus, the overwhelming majority of CENP-A at all cell cycle points is assembled at both centromeric and noncentromeric sites into homotypic chromatin particles with two molecules of CENP-A, as predicted by a nucleosomal model.
Although we found no evidence at any cell cycle point for CENP-A–containing hemisomes assembled onto α-satellite DNA, a small percentage of CENP-A particles display properties consistent with subnucleosomal complexes. Although they could indicate the presence of a minor tetrasome or hemisome component, it should be noted that they could arise as an artifact of the isolation procedure or simply represent octameric nucleosomes that are more sensitive to nuclease digestion. It is important to point out, however, that all prior studies supporting the hemisome model have relied on comparable nuclease digestion paradigms but claimed a much higher percentage of hemisomes in cells randomly cycling (Dalal et al., 2007; Wang et al., 2008; Dimitriadis et al., 2010; Krassovsky et al., 2012; Henikoff et al., 2014) or before or after DNA replication (Bui et al., 2012; Shivaraju et al., 2012), compared with the very rare “subnucleosomal particles” reported here.
Our evidence establishes that one feature that differentiates centromeric/pericentromeric CENP-A and histone H3 chromatin at all cell cycle points is a shorter wrapping DNA length of 133 bp (versus 147 bp for histone H3) after nuclease digestion, a feature likely the result of DNA unwrapping at nucleosome entry and exit. Although one group previously interpreted DNA lengths of 100–150 bp as hemisomal and 150–200 bp as octameric (Bui et al., 2012), molecular modeling has produced much shorter (60 to 80 bp) estimated lengths of DNA wrapping by either a tetrasome or hemisome (Hasson et al., 2013). CENP-A nucleosomes have been shown previously to have 7 bp unwinding of the DNA termini, resulting in a structure that is more nuclease accessible and protecting a shorter DNA length both in vitro and in random cycling cells (Conde e Silva et al., 2007; Tachiwana et al., 2011; Hasson et al., 2013; Lacoste et al., 2014). Although a normal human centromere has only ∼400 molecules of CENP-A (only ∼200 molecules after DNA replication; Bodor et al., 2014), only enough for ∼2–4% of all nucleosomes on α-satellite DNA, our evidence has demonstrated that this minority has distinctive chromatin wrapping (protecting 133 bp of DNA), but this does not spread to histone H3–containing nucleosomes assembled onto α-satellite DNA, which retain the typical 147 bp of protected DNA. Finally, although micrococcal nuclease digestion results in DNA peak lengths of 133 bp and 147 bp for CENP-A and histone H3 chromatin assembled onto α-satellite DNA, respectively, there is also a small “shoulder” at ∼80 to 110 bp for all chromatin types in our study, as well as in many prior studies (Hasson et al., 2013; Lacoste et al., 2014). Although these may reflect the presence of a small minority of hemisomes, they seem more likely simply to be a byproduct of nuclease digestion.
More broadly, construction and use of the centromere reference models represent a first step in defining the relationship between the complex repetitive sequence structure and the chromatin composition of endogenous human centromeres and pericentromeres. Recognizing that evidence using SNAP tagging has supported deposition of histone H3.3 at centromeres during S phase as a placeholder for the CENP-A to be added in G1 (Dunleavy et al., 2011), a key future direction will be to use the genome-wide approach and reference centromeres to test this model on a genome-wide basis, albeit this will be technically very challenging given the small proportion (0.04%) of total cell histone H3.3 that is expected to be centromere bound after DNA replication. Nevertheless, our evidence for CENP-A and histone H3.1 presented here provides quantitative support for a model in which α-satellite DNAs at all cell cycle phases are composed of rare, homotypic nucleosomes assembled with CENP-A interspersed among what are predominantly histone H3.1–containing octameric nucleosomes.
Materials and methods
Constructs
The full length of the human CENP-A open reading frame tagged with EYFP at the amino terminus or with TAP at the carboxy terminus was cloned into a pBabe-based vector for retrovirus generation.
Cell culture
Adherent HeLa cells stably expressing CENP-ATAP or H3.1TAP by retrovirus infection (Foltz et al., 2006) or endogenously tagged CENP-ALAP by infection of a rAAV harboring an LAP targeting construct containing homology arms for CENP-A (Mata et al., 2012; gifts from D.R. Foltz [Northwestern University, Chicago, IL] and L.E.T. Jansen [Gulbenkian Institute, Oeiras, Portugal], respectively) were adapted to grow in suspension by selecting surviving cells and were maintained in DMEM containing 10% FBS (Omega), 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM l-glutamine at 37°C in a 5% CO2 atmosphere with 21% oxygen. hTERT RPE-1 CENP-A−/− stably expressing CENP-ATAP, EYFPCENP-A, or NH2H3CATD cells were maintained in DMEM/F12 medium containing 10% FBS (Omega Scientific), 0.348% sodium bicarbonate, 100 U/ml hygromicin, 100 U/ml streptomycin, and 2 mM l-glutamine.
Generation of stable cell lines
The CENP-A transgenes (CENP-ATAP and EYFPCENP-A) used in this study were introduced by retroviral delivery. Retroviral plasmids (pBABEblast-EYFP or TAP fusions) were cotransfected into 293-GP cells along with the vesicular stomatitis virus G pseudotyping plasmid for the production of amphotropic retrovirus. The resulting retroviral supernatant was mixed with 8 µg/ml hexadimethrine bromide (Polybrene; Sigma-Aldrich) and incubated with RPE-1 cells for 12 h. Media were replaced, and the cells were split for selection in 5 µg/ml puromycin or 10 µg/ml blasticidin S after 48 h. After 2 weeks of selection, cells were subjected to flow cytometry (FACSVantage; BD), and single clones were isolated.
Clonogenic colony assay and adeno-Cre treatment
Cells were plated in a 12-well plate at 4 × 104. The next day, cells were washed three times in DMEM/F12 medium containing 2% FBS. Adeno-Cre virus was added at MOI 250 in 400 µl DMEM/F12 medium containing 2% FBS. After 3.5 h, cells were washed three times with DMEM/F12 medium containing 10% FBS. After 2 d, 500 cells were plated in triplicate on a 10-cm2 dish. After an additional 14 d, colonies were fixed 10 min in methanol and stained for 10 min using a crystal violet staining solution (1% crystal violet and 20% EtOH).
Cell synchronization
Suspension HeLa cells were treated with 2 mM thymidine in complete medium for 19 h, pelleted and washed twice in PBS, and released in complete medium containing 24 µM deoxycytidine for 9 h followed by addition of thymidine to a final concentration of 2 mM for 16 h, after which cells were released again into complete medium containing 24 µM deoxycytidine and assayed. Nocodazole was used at100 ng/ml.
Cell cycle analysis
106 cells were harvested, washed in PBS, and fixed in 70% ethanol. Cells were stained for phosphorylated histone H3 by blocking cells for 10 min in PBS with 0.5% BSA and then incubating cells with rabbit anti–pH3 antibody conjugated with Alexa Fluor 488 (9708S, 1:200; Cell Signaling Technology). Cells were then washed and DNA was stained by incubating cells for 30 min with 1% FBS, 10 µg/ml propidium iodide, and 0.25 mg/ml RNase A in PBS followed by FACS analysis for DNA content using a BD LSR II Flow Cytometer (BD).
Chromatin extraction
Nuclei from 109 HeLa cells were prepared by pelleting and resuspending cells in buffer containing 3.75 mM Tris, pH 7.5, 20 mM KCl, 0.5 mM EDTA, 0.5 mM DTT, 0.05 mM spermidine, 0.125 mM spermine, 1 mM PMSF, and 0.1% digitotin. Cells were homogenized with 10 strokes, and nuclei were pelleted at 300 g. Nuclei were then washed once in wash buffer (20 mM Hepes, pH 7.7, 20 mM KCl, 0.5 mM EDTA, 0.5 mM DTT, and 0.5 mM PMSF), followed by wash buffer containing 150 mM NaCl. Nuclei were resuspended in wash buffer supplemented with 150 mM NaCl and 3 mM CaCl. Chromatin was digested at room temperature using 140 U/ml micrococcal nuclease (10107921001; Roche) for 20 min to produce mononucleosomes and short oligonucleosomes of up to three nucleosomes or for 35 min to produce a pool of mononucleosomes. After micrococcal nuclease treatment, extracts were supplemented with 5 mM EGTA and 0.05% NP-40 and centrifuged at 10,000 g for 15 min at 4°C. The supernatant was then used as the starting material for all sedimentation velocity experiments and immunopurifications.
Affinity purification
TAP- or LAP-tagged chromatin was purified in two steps. In the first step, native TAP-tagged chromatin was immunoprecipitated by incubating the bulk soluble mononucleosome pool with rabbit IgG (Sigma-Aldrich) coupled to Dynabeads M-270 Epoxy (14301; Thermo Fisher Scientific). Alternatively, CENP-ALAP chromatin was immunoprecipitated using mouse anti–GFP antibody (clones 19C8 and 19F7; Monoclonal Antibody Core Facility, Memorial Sloan-Kettering Cancer Center; Heiman et al., 2008) coupled to Dynabeads M-270 Epoxy. Chromatin extracts were incubated with antibody-bound beads for 16 h at 4°C. Bound complexes were washed once in buffer A (20 mM Hepes, pH 7.7, 20 mM KCl, 0.4 mM EDTA, and 0.4 mM DTT), once in buffer A with 300 mM KCl, and finally twice in buffer A with 300 mM KCl, 1 mM DTT, and 0.1% Tween 20. In the second step, TAP–chromatin complexes were incubated 16 h in final wash buffer with 50 µl recombinant TEV protease, resulting in cleavage of the TAP tag and elution of the chromatin complexes from the beads. Alternatively, CENP-ALAP chromatin was eluted from the beads by cleaving the LAP tag using PreScission protease (4 h, 4°C).
DNA extraction
After elution of the chromatin from the beads, 100 µg/ml proteinase K was added and samples were incubated for 2 h at 55°C. DNA was purified from proteinase K–treated samples using a DNA purification kit following the manufacturer instructions (A9282; Promega) and was subsequently analyzed either by running a 2% low melting agarose (APEX) gel or by an Agilent Technologies 2100 Bioanalyzer by using the DNA 1000 kit. The Bioanalyzer determines the quantity of DNA on the basis of fluorescence intensity.
Quantitative real-time PCR (qPCR)
Quantitative real-time PCR (qPCR) was performed using SYBR Green mix (Bio-Rad Laboratories) with CFX384 Bio Rad Laboratories Real Time System. The following primer sequences were used to amplify α-satellite DNA from chromosomes 1, 3, 5, and 10: 5′-CTAGACAGAAGAATTCTCAG-3′ (forward) and 5′-CTGAAATCTCCACTTGC-3′ (reverse; Alonso et al., 2007). Melting curve analysis was used to confirm primer specificity. To ensure linearity of the standard curve, reaction efficiencies over the appropriate dynamic range were calculated. Using the dCt method, we calculated fold-enrichment of α-satellite DNA after immunopurification of CENP-ATAP chromatin, compared with its level in the bulk input chromatin. Reported values are the means of two independent biological replicates with technical duplicates that were averaged for each experiment. Error bars represent SEM.
Immunoblotting
For immunoblot analysis, protein samples were separated by SDS–PAGE, transferred onto PVDF membranes (EMD Millipore) and then probed with the following antibodies: rabbit anti–CENP-A (2186s, 1:1,000; Cell Signaling Technology), mouse anti–α-tubulin (DM1A, 1:5,000; Abcam), rabbit anti-H3 (1:5,000, H0164; Sigma-Aldrich), rabbit anti-H2A (Ab18255, 1:500; Abcam), rabbit anti-H2B (IMG-359, 1:250; Imgenex), rabbit anti-H4 (Ab10158, 1:250; Abcam), or GAPDH (2118, 1:5,000; Cell Signaling Technology). After incubation with HRP-labeled antibody (NA931V or NA934V; GE Healthcare), HRP was detected using enhanced chemiluminescence substrate (34080 or 34096; Thermo Fisher Scientific). Band intensity was quantified using ImageJ. Immunoprecipitation efficiency was determined by measuring the levels of CENP-ATAP or CENP-ALAP remaining in the unbound fractions after immunoprecipitation and subtracting the result from 1.
Immunofluorescence and micronuclei quantification
106 suspension cells were centrifuged and resuspended with PBS. 105 cells were immobilized on glass slide by cytospin centrifugation for 3 min at 800 rpm. Cells were then fixed using ice-cold methanol at −20°C for 10 min, followed by washing with cold PBS and then incubated in Triton Block (0.2 M glycine, 2.5% FBS, 0.1% Triton X-100, and PBS) for 1 h. The following primary antibodies were used in Triton Block for 1 h and washed with 0.1% Triton X-100 in PBS: human anti–centromere antibodies (1:500; 15-234-0001; Antibodies Inc.), mouse anti–cyclin B1 (1:50; sc-245; Santa Cruz Biotechnology, Inc.), mouse anti-GFP (11814460001, 1:500; Roche), and rabbit anti–CENP-B (1:1,000; 25734; Abcam). The following secondary antibodies (from Jackson ImmunoResearch Laboratories, Inc.) were used for 45 min: donkey anti–human Texas red (1:300), anti–mouse FITC (1:250), and anti–rabbit Cy5 (1:250). TAP fusion proteins were visualized by incubation with FITC–rabbit IgG (1:200; Jackson ImmunoResearch Laboratories, Inc.). Cells were then washed with 0.1% Triton X-100 in PBS, counterstained with DAPI, and mounted with mounting medium (P36934; Molecular Probes). Immunofluorescent images were acquired on a Deltavision Core system at 60–100×. 0.2-µm Z-stack deconvolved projections were generated using the softWoRx program. To quantify the number of cells stained for cyclin B1, positive cells were counted in 20 images of 60×, in each time point in five replicates synchronization experiments. To estimate the percentage of cells entering mitosis, cells were scored also for their DNA condensation state by counting cells with condensed DNA (DAPI) in 20 images per time point in each experiment. To quantify the percentage of cells with micronuclei, cells were stained with DAPI. Cells and micronuclei were counted in 20 images of 60×.
Chromatin immunoprecipitation (ChIP) sequencing library generation and sequencing
ChIP libraries were prepared following Illumina protocols with minor modifications (Illumina). To reduce biases induced by PCR amplification of a repetitive region, libraries were prepared from 80–100 ng input or ChIP DNA. The DNA was end-repaired and A-tailed, and Illumina Truseq adaptors were ligated. Libraries were run on a 2% agarose gel. Because the chromatin was digested to mononucleosomes, after adaptor ligation, the libraries size was 250 to 280 bp. The libraries were size selected for 200 to 375 bp. The libraries were then PCR-amplified using only five or six PCR cycles, because the starting DNA amount was high. Resulting libraries were sequenced using 100-bp, paired-end sequencing on a HiSeq 2000 instrument per the manufacturer’s instructions with some modifications (Illumina). Sequence reads are summarized in Tables S1 and S2.
DNA sequences mapping to an α-satellite database
Illumina paired-end reads were merged to determine CENP-A or H3 containing target fragments of varying length using PEAR software (Zhang et al., 2014), with standard parameters (P = 0.01; minimum overlap, 10 bases; minimum assembly length, 50 bp). Merged reads were determined to contain α-satellite by two methods. First, reads were mapped (Bwa-Mem, standard parameters; Li and Durbin, 2010; Li, 2013) to the human genome 38 (hg38) assembly (including alternative assemblies), which contains α-satellite sequence models in each centromeric region (Miga et al., 2014; BioProject: PRJNA193213; Table S3). Reads were identified as containing an α-satellite if they overlapped sites (BEDTools: intersect; Quinlan and Hall, 2010) in the genome previously annotated as α-satellite (UCSC table browser was used to obtain a bed file of all sites annotated as ALR/α-satellite; Karolchik et al., 2004). Total α-satellite DNA content in the human genome 38 assembly was estimated using UCSC RepeatMasker Annotation (Karolchik et al., 2004; Rosenbloom et al., 2015). In addition to our mapping strategy, we determined merged sequences containing α-satellite using a previously published whole-genome sequence read database of α-satellite, representing 2.6% of sequences from the HuRef genome (Levy et al., 2007; Hayden et al., 2013). To do so, we identified a listing of ∼8 million α-satellite–specific 18-mers (i.e., 18-mers that did not contain an exact match with any sequence in the HuRef genome, or hg38 reference assembly outside of sequences of known α-satellites). Merged sequences were defined as containing an α-satellite if they contained an exact match to at least two 18-mers specific to α-satellites. Comparisons between the mapping and k-mer–based strategies were highly concordant. Mapped sequence reads are summarized in Tables S1, S2, and S4.
In vitro reconstitution of chromatin
Human histones H3, H4, H2A, and H2B were purified as monomers and mixed to form (H3–H4)2 tetramer and (H2A–H2B) dimer complexes, and human (CENP-A–H4)2 was expressed from a bicistronic plasmid (Black et al., 2004) and then purified as a soluble tetramer. The DNA template for reconstitutions was a 145-bp 601 nucleosome positioning sequence (5′-ATCAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCGAT-3′; Lowary and Widom, 1998). The DNA template for (CENP-A–H4)2 or (H3–H4)2 was a 101-bp sequence (underlined above). The indicated histone complexes were combined with DNA and assembled in nucleosomes by gradual dialysis from high to low salt. After reconstitution, nucleosomes and tetrasomes were incubated for 2 h at 55°C to achieve uniform positioning. Nucleosome assembly and positioning on the DNA was assessed by 5% native PAGE.
Sucrose gradient fractionation
In vitro–reconstituted chromatin or in vivo bulk chromatin containing short oligonucleosomes was loaded onto a prechilled 5–25% sucrose gradient containing 10 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 2 mM EDTA and centrifuged for 22 h at 26,000 rpm (4°C). 1-ml fractions were collected and analyzed for DNA and protein content. The following molecular mass markers were used: thyroglobulin, 670 kD; β amylase, 200 kD; and anhydrase, 29 kD. Peak fractions of the molecular mass markers are indicated in Fig. 5 (B and E) and Fig. S4 (B and C).
Nanopore assay
Affinity-purified TAP-tagged chromatin eluted from beads was concentrated using amicon ultra 0.5 ml (UFC503024; EMD Millipore) and dialyzed against TE buffer. Chromatin was then driven through solid-state nanopores (Soni and Dekker, 2012). To fabricate nanopores, silicon wafers with triple coating of 20 nm silicon nitride, 100 nm silicon oxide, and 500 nm silicon nitride were processed with standard photolithography and e-beam lithography to fabricate freestanding silicon nitride membranes (with 20 µm square shape and 20 nm thickness; Storm et al., 2003; Wu et al., 2009; van den Hout et al., 2010). A focused transmission EM beam was used to drill 20-nm-wide pores in these membranes. We mounted these chips onto a fluid cell with fluid chambers with separate access to the cis and trans sides of the nanopore membrane. The chambers were filled with buffer (1 M KCl and 10 mM Tris-EDTA, pH 8), and a 100-mV potential was applied across the nanopore membrane using Ag/AgCl electrodes. Nanopore current was measured using Axopatch 200B (Molecular Devices) set at full bandwidth (100 KHz). To detect the sample at single-molecule resolution, ∼1 pM of the nucleosome sample was introduced in the flow chamber with negative bias. Voltage-dependent translocation of individual nucleosomes through the nanopores was recorded as temporary drops in the nanopore conductance. This drop in conductance is directly proportional to the molecular volume of the nucleosome complex (Soni and Dekker, 2012). Data were recorded using a DAQ card (NI-PCI 6251; National Instruments) at acquisition rate of 200 kHz. For analysis, data were filtered at 35 kHz. All data acquisition and event detection in current traces was done by custom-written software in LabVIEW (National Instruments) and MATLAB (Mathworks Inc.), respectively (Storm et al., 2005; Soni and Dekker, 2012).
Statistical analysis
For all experiments shown, n is indicated in the figure legends. Percentages of tagged and untagged CENP-A chromatin, as well as H3 depletion and levels of unbound CENP-A, were calculated in immunoblots by densitometric analysis using ImageJ software. Values represent the mean ± SEM (as indicated in the figure legends). For nanopore experiments, a fitted Gaussian distribution was used. ΔG data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows that CENP-ATAP maintains long-term centromere function and viability in the absence of endogenous CENP-A. Fig. S2 is a characterization of CENP-ATAP cells and synchronization efficiency for producing CENP-A chromatin at multiple cell cycle points. Fig. S3 shows the transfer efficiency of untagged endogenous CENP-A and endogenously tagged CENP-ALAP. Fig. S4 shows that the sedimentation of CENP-A is inconsistent with a hemisome/tetrasome structure as the major form of CENP-A chromatin. Tables S1 and S2 show the read statistics for ChIP-sequencing experiments. Table S3 shows the array sizes of the centromeric reference models. Table S4 shows a summary of the mapping data of CENP-ATAP and H3.1TAP sequence reads onto α-satellite DNAs in human centromere reference models for each autosome and the X chromosome.
Supplementary Material
Acknowledgments
The authors would like to thank A. Desai for critical discussion and helpful suggestions; B. Cetin (Sloan-Kettering, New York, NY) for technical help; L.E.T. Jansen and D.R. Foltz for providing reagents; and A. Gurwitz, J.S. Han, M. McMahon, and S. Sun (Ludwig Institute for Cancer Research) for helpful suggestions.
C. Dekker was supported by H2020 European Research Council Advanced Grant SynDiv (669598). This work was supported by a National Institutes of Health grant (R01 GM-074150) to D.W. Cleveland, who receives salary support from the Ludwig Institute for Cancer Research, and by a National Institutes of Health grant (R01 GM-082989) to B.E. Black.
The authors declare no competing financial interests.
Author contributions: Y. Nechemia-Arbely and D.W. Cleveland conceived and designed experiments and wrote the manuscript. Y. Nechemia-Arbely performed experiments. D. Fachinetti performed fusion protein functionality experiments and provided key experimental input. K.H. Miga analyzed the sequencing data. N. Sekulic and B.E. Black provided critical reagents and expertise. G.V. Soni and C. Dekker performed nanopore experiments and provided data interpretation. D. Hyun Kim performed experiments. A.Y. Lee and B. Ren prepared sequencing libraries and provided resources. A.K. Wong and K. Nguyen provided experimental technical help.
Footnotes
Abbreviations used:
- CATD
- CENP-A centromere-targeting domain
- CENP-A
- centromere protein A
- ChIP
- chromatin immunoprecipitation
- hg38
- human genome 38
- LAP
- localization and affinity purification
- TAP
- tandem affinity purification
- TEV
- tobacco etch virus
References
- Alonso A., Fritz B., Hasson D., Abrusan G., Cheung F., Yoda K., Radlwimmer B., Ladurner A.G., and Warburton P.E.. 2007. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 8:R148 10.1186/gb-2007-8-7-r148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L. Jr., and Cleveland D.W.. 2004. Structural determinants for generating centromeric chromatin. Nature. 430:578–582. 10.1038/nature02766 [DOI] [PubMed] [Google Scholar]
- Black B.E., Brock M.A., Bédard S., Woods V.L. Jr., and Cleveland D.W.. 2007. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. 104:5008–5013. 10.1073/pnas.0700390104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D., Sullivan B.A., and Karpen G.H.. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2:319–330. 10.1016/S1534-5807(02)00135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D.L., Mata J.F., Sergeev M., David A.F., Salimian K.J., Panchenko T., Cleveland D.W., Black B.E., Shah J.V., and Jansen L.E.. 2014. The quantitative architecture of centromeric chromatin. eLife. 3:e02137 10.7554/eLife.02137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M., Dimitriadis E.K., Hoischen C., An E., Quénet D., Giebe S., Nita-Lazar A., Diekmann S., and Dalal Y.. 2012. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 150:317–326. 10.1016/j.cell.2012.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Mao Y., and Sullivan K.F.. 2003. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell. 112:407–421. 10.1016/S0092-8674(03)00115-6 [DOI] [PubMed] [Google Scholar]
- Conde e Silva N., Black B.E., Sivolob A., Filipski J., Cleveland D.W., and Prunell A.. 2007. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 370:555–573. 10.1016/j.jmb.2007.04.064 [DOI] [PubMed] [Google Scholar]
- Dalal Y., Wang H., Lindsay S., and Henikoff S.. 2007. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 5:e218 10.1371/journal.pbio.0050218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis E.K., Weber C., Gill R.K., Diekmann S., and Dalal Y.. 2010. Tetrameric organization of vertebrate centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. 107:20317–20322. 10.1073/pnas.1009563107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., and Almouzni-Pettinotti G.. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 137:485–497. 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Dunleavy E.M., Almouzni G., and Karpen G.H.. 2011. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus. 2:146–157. 10.4161/nucl.2.2.15211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., and Rothfield N.. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 91:313–321. 10.1007/BF00328227 [DOI] [PubMed] [Google Scholar]
- Fachinetti D., Folco H.D., Nechemia-Arbely Y., Valente L.P., Nguyen K., Wong A.J., Zhu Q., Holland A.J., Desai A., Jansen L.E., and Cleveland D.W.. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15:1056–1066. 10.1038/ncb2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S.J., Lee J., Sekulic N., Sennett M.A., Lee T.H., and Black B.E.. 2016. CENP-C directs a structural transition of CENP-A nucleosomes mainly through sliding of DNA gyres. Nat. Struct. Mol. Biol. 23:204–208. 10.1038/nsmb.3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R. III, and Cleveland D.W.. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R. III, Bassett E.A., Wood S., Black B.E., and Cleveland D.W.. 2009. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 137:472–484. 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson D., Panchenko T., Salimian K.J., Salman M.U., Sekulic N., Alonso A., Warburton P.E., and Black B.E.. 2013. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 20:687–695. 10.1038/nsmb.2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden K.E., Strome E.D., Merrett S.L., Lee H.R., Rudd M.K., and Willard H.F.. 2013. Sequences associated with centromere competency in the human genome. Mol. Cell. Biol. 33:763–772. 10.1128/MCB.01198-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M., Schaefer A., Gong S., Peterson J.D., Day M., Ramsey K.E., Suárez-Fariñas M., Schwarz C., Stephan D.A., Surmeier D.J., et al. . 2008. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 135:738–748. 10.1016/j.cell.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ramachandran S., Krassovsky K., Bryson T.D., Codomo C.A., Brogaard K., Widom J., Wang J.P., and Henikoff J.G.. 2014. The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo. eLife. 3:e01861 10.7554/eLife.01861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L.E., Black B.E., Foltz D.R., and Cleveland D.W.. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805. 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., and Kent W.J.. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32:D493–D496. 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G.H., and Allshire R.C.. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13:489–496. 10.1016/S0168-9525(97)01298-5 [DOI] [PubMed] [Google Scholar]
- Kowalczyk S.W., Grosberg A.Y., Rabin Y., and Dekker C.. 2011. Modeling the conductance and DNA blockade of solid-state nanopores. Nanotechnology. 22:315101 10.1088/0957-4484/22/31/315101 [DOI] [PubMed] [Google Scholar]
- Krassovsky K., Henikoff J.G., and Henikoff S.. 2012. Tripartite organization of centromeric chromatin in budding yeast. Proc. Natl. Acad. Sci. USA. 109:243–248. 10.1073/pnas.1118898109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N., Woolfe A., Tachiwana H., Garea A.V., Barth T., Cantaloube S., Kurumizaka H., Imhof A., and Almouzni G.. 2014. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 53:631–644. 10.1016/j.molcel.2014.01.018 [DOI] [PubMed] [Google Scholar]
- Lee C., Wevrick R., Fisher R.B., Ferguson-Smith M.A., and Lin C.C.. 1997. Human centromeric DNAs. Hum. Genet. 100:291–304. 10.1007/s004390050508 [DOI] [PubMed] [Google Scholar]
- Levy S., Sutton G., Ng P.C., Feuk L., Halpern A.L., Walenz B.P., Axelrod N., Huang J., Kirkness E.F., Denisov G., et al. . 2007. The diploid genome sequence of an individual human. PLoS Biol. 5:e254 10.1371/journal.pbio.0050254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. eprint arXiv Available at: https://arxiv.org/abs/1303.3997 (accessed January 30, 2017).
- Li H., and Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26:589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary P.T., and Widom J.. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19–42. 10.1006/jmbi.1997.1494 [DOI] [PubMed] [Google Scholar]
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., and Richmond T.J.. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 389:251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Mata J.F., Lopes T., Gardner R., and Jansen L.E.. 2012. A rapid FACS-based strategy to isolate human gene knockin and knockout clones. PLoS One. 7:e32646 10.1371/journal.pone.0032646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone B.G., Grive K.J., Shteyn V., Bowers S.R., Oderberg I., and Karpen G.H.. 2011. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 7:e1002068 10.1371/journal.pgen.1002068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga K.H., Newton Y., Jain M., Altemose N., Willard H.F., and Kent W.J.. 2014. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 24:697–707. 10.1101/gr.159624.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechemia-Arbely Y., Fachinetti D., and Cleveland D.W.. 2012. Replicating centromeric chromatin: Spatial and temporal control of CENP-A assembly. Exp. Cell Res. 318:1353–1360. 10.1016/j.yexcr.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeganeh A., Ryan J., Boisvert J., Ladouceur A.M., Dorn J.F., and Maddox P.S.. 2013. Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle. Curr. Biol. 23:764–769. 10.1016/j.cub.2013.03.037 [DOI] [PubMed] [Google Scholar]
- Palmer D.K., O’Day K., Wener M.H., Andrews B.S., and Margolis R.L.. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805–815. 10.1083/jcb.104.4.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., and Hall I.M.. 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. . 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43(D1):D670–D681. 10.1093/nar/gku1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., Lehner C.F., and Heidmann S.. 2007. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 17:237–243. 10.1016/j.cub.2006.11.051 [DOI] [PubMed] [Google Scholar]
- Sekulic N., Bassett E.A., Rogers D.J., and Black B.E.. 2010. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 467:347–351. 10.1038/nature09323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw B.R., Herman T.M., Kovacic R.T., Beaudreau G.S., and Van Holde K.E.. 1976. Analysis of subunit organization in chicken erythrocyte chromatin. Proc. Natl. Acad. Sci. USA. 73:505–509. 10.1073/pnas.73.2.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M., Unruh J.R., Slaughter B.D., Mattingly M., Berman J., and Gerton J.L.. 2012. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell. 150:304–316. 10.1016/j.cell.2012.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni G.V., and Dekker C.. 2012. Detection of nucleosomal substructures using solid-state nanopores. Nano Lett. 12:3180–3186. 10.1021/nl301163m [DOI] [PubMed] [Google Scholar]
- Stimpson K.M., and Sullivan B.A.. 2010. Epigenomics of centromere assembly and function. Curr. Opin. Cell Biol. 22:772–780. 10.1016/j.ceb.2010.07.002 [DOI] [PubMed] [Google Scholar]
- Storm A.J., Chen J.H., Ling X.S., Zandbergen H.W., and Dekker C.. 2003. Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mater. 2:537–540. 10.1038/nmat941 [DOI] [PubMed] [Google Scholar]
- Storm A.J., Storm C., Chen J., Zandbergen H., Joanny J.F., and Dekker C.. 2005. Fast DNA translocation through a solid-state nanopore. Nano Lett. 5:1193–1197. 10.1021/nl048030d [DOI] [PubMed] [Google Scholar]
- Sullivan B.A., and Karpen G.H.. 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11:1076–1083. 10.1038/nsmb845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A., Blower M.D., and Karpen G.H.. 2001. Determining centromere identity: Cyclical stories and forking paths. Nat. Rev. Genet. 2:584–596. 10.1038/35084512 [DOI] [PubMed] [Google Scholar]
- Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., Hayashi-Takanaka Y., Oda T., Sato M., Park S.Y., et al. . 2011. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 476:232–235. 10.1038/nature10258 [DOI] [PubMed] [Google Scholar]
- van den Hout M., Hall A.R., Wu M.Y., Zandbergen H.W., Dekker C., and Dekker N.H.. 2010. Controlling nanopore size, shape and stability. Nanotechnology. 21:115304 10.1088/0957-4484/21/11/115304 [DOI] [PubMed] [Google Scholar]
- Wang H., Dalal Y., Henikoff S., and Lindsay S.. 2008. Single-epitope recognition imaging of native chromatin. Epigenetics Chromatin. 1:10 10.1186/1756-8935-1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard H.F. 1985. Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet. 37:524–532. [PMC free article] [PubMed] [Google Scholar]
- Wu M.Y., Smeets R.M., Zandbergen M., Ziese U., Krapf D., Batson P.E., Dekker N.H., Dekker C., and Zandbergen H.W.. 2009. Control of shape and material composition of solid-state nanopores. Nano Lett. 9:479–484. 10.1021/nl803613s [DOI] [PubMed] [Google Scholar]
- Zhang J., Kobert K., Flouri T., and Stamatakis A.. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 30:614–620. 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Colmenares S.U., and Karpen G.H.. 2012. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol. Cell. 45:263–269. 10.1016/j.molcel.2011.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.