Wang et al. discuss the intricate processes required during embryogenesis for the formation of the branched architecture of organs such as the lung, kidney, and blood vessels.
Abstract
Many embryonic organs undergo branching morphogenesis to maximize their functional epithelial surface area. Branching morphogenesis requires the coordinated interplay of multiple types of cells with the extracellular matrix (ECM). During branching morphogenesis, new branches form by “budding” or “clefting.” Cell migration, proliferation, rearrangement, deformation, and ECM dynamics have varied roles in driving budding versus clefting in different organs. Elongation of the newly formed branch and final maturation of the tip involve cellular mechanisms that include cell elongation, intercalation, convergent extension, proliferation, and differentiation. New methodologies such as high-resolution live imaging, tension sensors, and force-mapping techniques are providing exciting new opportunities for future research into branching morphogenesis.
Introduction: Overview of branching morphogenesis
During embryonic development, simple bud-like organ precursors undergo dramatic branching to generate the complex branched architectures of many organs. This process expands an organ’s epithelial surface area by orders of magnitude to maximize secretion or absorption by epithelia located in thousands of acini or alveoli and their ducts (Davies, 2005).
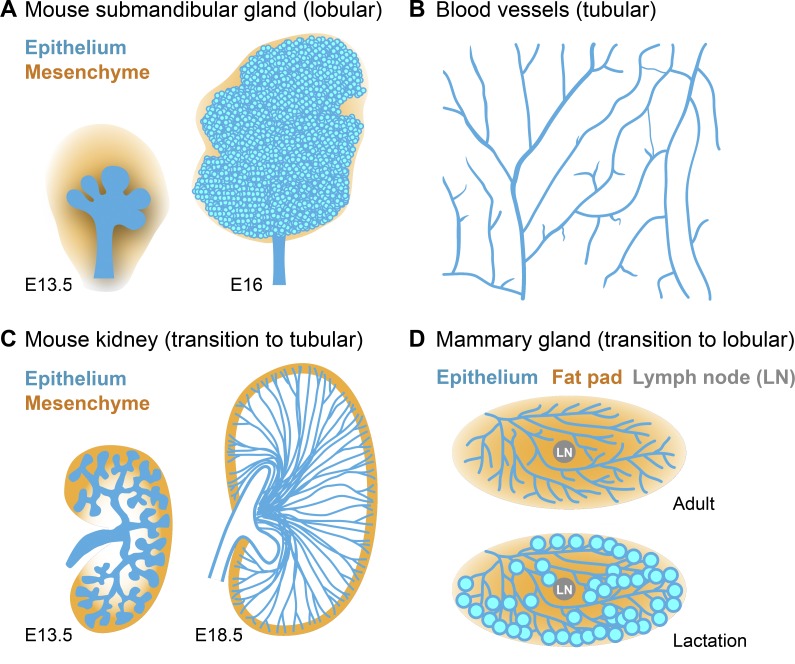
Branched structures are common in both plants and animals. For example, plants have tree branches and leaf veins. In animals, branching morphogenesis has been studied extensively in Drosophila melanogaster trachea (Ghabrial et al., 2003; Affolter and Caussinus, 2008), mammalian lung (Morrisey and Hogan, 2010; Warburton et al., 2010; Varner and Nelson, 2017), blood vessels (Meadows and Cleaver, 2015), kidney (Costantini and Kopan, 2010), pancreas (Shih et al., 2013), mammary gland (Gray et al., 2010; Inman et al., 2015), and salivary gland (Patel et al., 2006; Larsen et al., 2010). The morphology of these organs can differ dramatically, ranging from predominantly “lobular” in salivary glands to completely “tubular” in blood vessels (Fig. 1, A and B). Their appearance largely depends on the aspect ratio (length-to-diameter) of branches between branch points, as well as the relative size of branch tips compared with stalks.
Figure 1.
Varying morphology of branched organs. (A) Schematics of mouse submandibular glands from embryonic day (E) 13.5 and E16 stage embryos. Submandibular gland morphology is predominantly lobular. (B) Schematic of blood vessels from a mouse retina. The branched vascular network is completely tubular. (C) Schematics of the ureteric bud of mouse kidney from E13.5 and E18.5 stage embryos. At E13.5, mouse kidney is relatively lobular. At E18.5, the elongated collecting ducts convert kidney morphology to predominantly tubular. (D) Schematics of mouse mammary gland at young adult stage and lactation stage. At the young adult stage, the mammary gland is a tubular network. During pregnancy and lactation, dramatic remodeling occurs in the mammary gland so that lactating alveoli form at branch tips, which transforms the structure to primarily lobular.
During development, the lobular versus tubular appearance of some organs can change. For example, the ureteric bud of the mouse kidney is relatively lobular at early stages of branching morphogenesis, with bulging tips connected by relatively short branches (Fig. 1 C, left). However, later in development, the branches (future collecting ducts) elongate dramatically to become very tubular (Costantini and Kopan, 2010), resembling a collection of lines radiating from the ureter (Fig. 1 C, right). In contrast, the mouse mammary gland begins as a tubular network (Fig. 1 D, top) that is later remodeled to become lobular during pregnancy and lactation (Hennighausen and Robinson, 2005; Fig. 1 D, bottom).
Branching morphogenesis can be stereotypic or stochastic. The branching patterns of both mammalian lung and Drosophila tracheal system are highly stereotyped (Ghabrial et al., 2003; Metzger et al., 2008), suggesting their branching programs are genetically hardwired. The topology of kidney branches appear stereotyped, though individual kidneys differ in branching patterns (Short et al., 2014; Sampogna et al., 2015). In contrast, branching of blood vessels and mammary gland appears stochastic without defined patterns, presumably because of the absence of space constraints (Andrew and Ewald, 2010). Other organs, such as the salivary gland, also exhibit stochastic branching patterns, at least in organ culture.
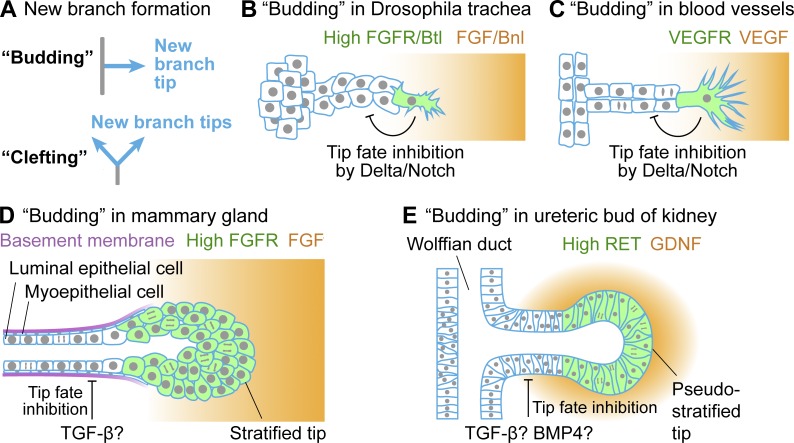
Morphogenesis of a branched organ requires both the formation of new branches and remodeling of existing branches. Forming new branches in mammalian organs occurs through two geometrically distinct processes, “budding” or “clefting” (Fig. 2 A and Table 1). In this paper, we define budding as de novo branching from the surface of a primordial epithelium or from the lateral side of a preexisting branch, whereas clefting splits a preexisting branch tip into several tips (usually two or three). Topologically, each budding event generates a new branch tip, whereas each clefting event simultaneously eliminates an old branch tip and generates at least two new tips (Fig. 2 A).
Figure 2.
New branch formation by budding. (A) New branches can form through two geometrically distinct processes, budding or clefting. Schematics of budding in Drosophila trachea (B) and mouse retina blood vessels (C), where budding occurs by an invasive form of collective migration. The identity of the protrusive tip cell is specified by high RTK signaling. A Delta/Notch-mediated lateral inhibition mechanism prevents follower cells from becoming leader cells. Schematics of budding in mouse mammary gland (D) and the ureteric bud of mouse kidney (E), where budding occurs by a noninvasive form of collective migration and regionalized cell proliferation. The stratified tip of the mammary gland, or TEB, has high FGF receptor (FGFR) activity and higher proliferation rate than the stalk, which contains two cell layers. The tip of the ureteric bud at this early stage is pseudostratified, and its identity is specified by high RET signaling activity. EGFR, EGF receptor; VEGFR, VEGF receptor.
Table 1. New branch formation by budding versus clefting.
| Cellular mechanisms | Budding | Clefting |
|---|---|---|
| Initiation of new branches | Protrusion of a group of cells outward from an epithelium to generate a new branch tip (in blood vessels, Drosophila trachea, mammary gland, lung, kidney,a and pancreasb) | Separation of cells in a preexisting branch tip to generate two or more new branch tips (in salivary gland,c lung, kidney, and pancreasb) |
| Collective cell migration | ||
| Invasive | Drosophila trachea | — |
| Blood vessels | ||
| Noninvasive | Mammary gland | — |
| Lung (?) | ||
| Kidney (?) | ||
| Patterned cell proliferation | ||
| Regionalized proliferation | Lung | — |
| Kidney | ||
| Oriented cell division | — | Lung |
| Kidney (?) | ||
| Actomyosin contractility | Not required in blood vessels and lung | Salivary gland, kidney, |
| Required in mammary gland (?) and pancreasb | lung, pancreasb | |
| Differential cell motilities | Mammary gland | Salivary gland |
| Pancreasb | Pancreasb | |
| Physical constraint | ||
| By basement membrane | — | Salivary gland |
| By smooth muscle cells | — | Lung |
—, not yet described or established.
Ureteric bud in kidney.
Mechanisms of pancreas branching may be complex, including a unique process involving the formation of microlumens.
Submandibular salivary gland.
At the cellular level, new branch formation can be driven by collective cell migration, patterned cell proliferation, coordinated cell deformation, and/or cell rearrangement. Plant tissues can lack cell migration and cell shape plasticity, so they rely on patterned cell proliferation and growth for branching. In contrast, animal organs often use multiple cell and tissue remodeling strategies.
In this review, we examine the complex cell biology underlying budding and clefting in various branched organs. Budding in blood vessels and Drosophila trachea occurs by an invasive form of collective cell migration, by which a leader cell forms extensive protrusions and migrates outward, followed by other cells (Fig. 2, B and C; Scarpa and Mayor, 2016). Budding in mammalian epithelial organs, however, appears to be powered by a noninvasive form of collective cell migration along with cell proliferation (Fig. 2, D and E; Ewald et al., 2008).
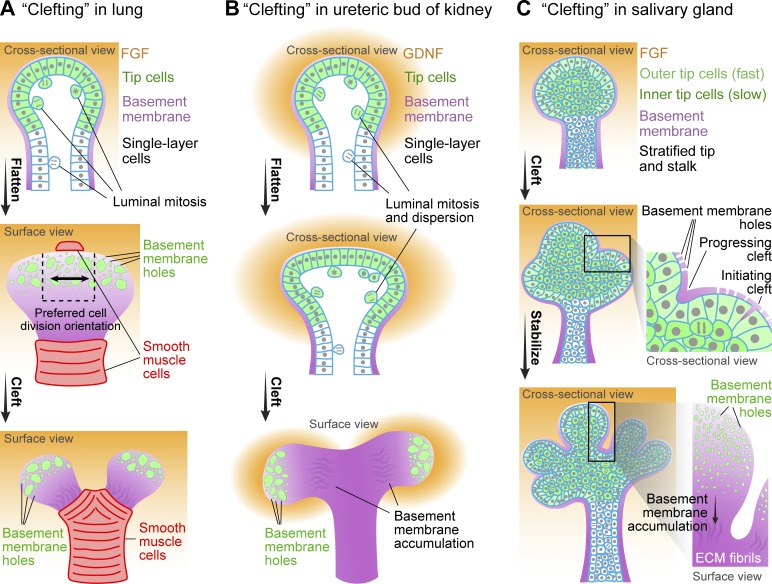
In contrast, clefting at the branch tip in lung and kidney requires proliferation to enlarge the tip, which deforms and splits (Fig. 3, A and B; Watanabe and Costantini, 2004; Schnatwinkel and Niswander, 2013). In salivary gland, clefting can occur without cell proliferation, and cytoskeletal and ECM remodeling mediate cell rearrangement and epithelial bud deformation (Fig. 3 C; Daley and Yamada, 2013; Harunaga et al., 2014).
Figure 3.
New branch formation by clefting. (A) Schematics of clefting (or terminal bifurcation) in mouse lung. The clefting tip of developing lung contains a single layer of cells that flattens before clefting. Within the tip, cells toward the center (dashed box in the center panel) divide preferentially parallel to the axis of flattening (black arrow in the center panel). Before clefting, smooth muscle cells differentiate at the future clefting site, which helps to deform the flattened tip to complete clefting. (B) Schematics of clefting in the ureteric bud of mouse kidney. The clefting tip of kidney contains a single layer of cells that also flattens before clefting. For cell proliferation, premitotic cells delaminate from the single-layered epithelium, complete cell division in the lumen, and then reinsert into the epithelium. (C) Schematics of clefting in mouse salivary gland. In these images, the clefting tip (bud) is stratified. The outer tip cells are more columnar and more regularly arranged than the inner tip cells. The outer tip cells also move much faster than the inner tip cells. Shallow clefts form stochastically with ECM invasion into the outer layer of epithelium, and they widen and stabilize to complete clefting. Clefting in all three systems is accompanied by microscopic perforations in the basement membrane toward the tip and accumulation of basement membrane components away from the tip. For each organ, a critically essential growth factor regulator is listed, although others contribute. GDNF, glial cell–derived neurotrophic factor.
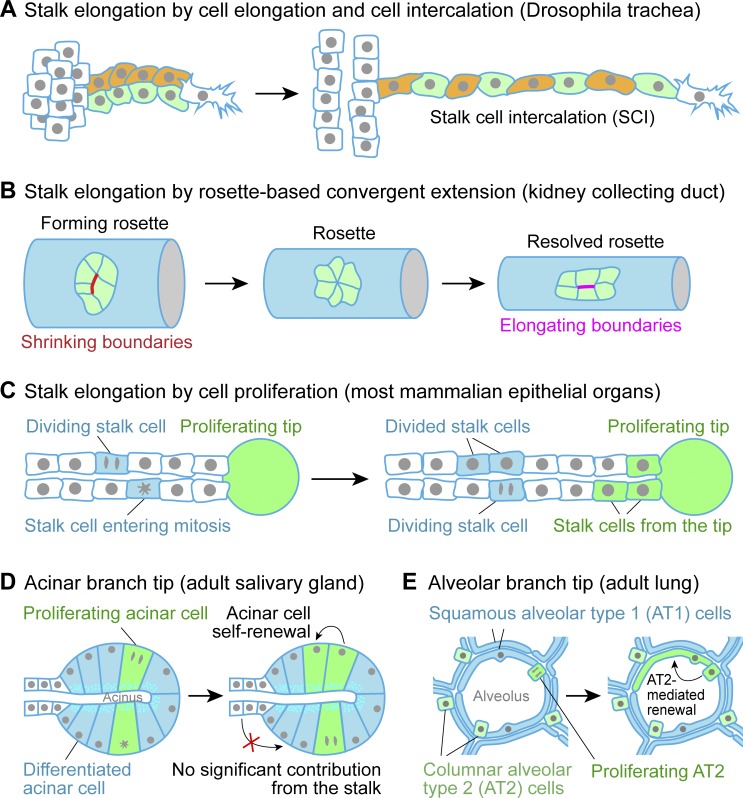
Other processes, such as branch elongation, lumen formation, and tip maturation, help to shape branched organs by adjusting the aspect ratio of branch stalks and tips. At the cellular level, branch elongation can occur through cell rearrangement by intercalation or convergent extension, cell elongation, and/or cell proliferation (Fig. 4, A–C; Andrew and Ewald, 2010). Lumen formation occurs concomitantly with branching in many organs, though is substantially delayed in salivary glands (Andrew and Ewald, 2010). Tip maturation in lung and glandular organs is driven by cell proliferation and deformation to form alveoli or acini (Fig. 4, D and E), whereas the kidney represents a special case in which tips of ureteric bud branches or collecting ducts need to fuse with nephrons developed from the surrounding metanephric mesenchyme (Costantini and Kopan, 2010).
Figure 4.
Refining organ architecture by branch elongation and maturation. (A–C) Schematics of stalk elongation by cell elongation and intercalation in Drosophila trachea. (A) In response to the pulling force of the migrating tip cell, stalk cells elongate and intercalate, resulting in >2-fold increased stalk length. (B) Schematics of stalk elongation by rosette-based convergent extension in vertebrate kidney collecting duct. (C) Schematics of branch stalk elongation by cell proliferation in most mammalian epithelial organs. Some cells divide within the stalk, whereas some cells are deposited by the proliferating tip to elongate the stalk. (D) Schematics of the acinar branch tip in adult mouse salivary gland. During maturation, cells at the branch tip differentiate to become columnar cells that surround a relatively small cavity to form an acinus for saliva secretion. Homeostasis of the acinar cells is maintained by self-renewal with little contribution from the stalk. (E) Schematics of the alveolar branch tip in adult mouse lung. During maturation, lung cells at the branch tip differentiate into flat, squamous AT1 cells and columnar AT2 cells. AT2 cell–mediated self-renewal can be triggered by AT1 injury.
Patterning of cell behavior in branching morphogenesis
Branched organs contain multiple cell types plus their ECM. The core structure of all branched organs consists of tightly associated epithelial cells. The epithelium is surrounded by the basement membrane, a dense network of ECM glycoproteins and proteoglycans. Beyond the basement membrane is usually a loosely organized mixture of mesenchymal cells and their ECM. In addition to these common structures, the presence and morphogenetic roles of other cell types including smooth muscle cells, neurons, and blood vessels have been described in multiple epithelial organs. During branching morphogenesis, the epithelial cells actively interact with the ECM and these other cell types, both biochemically and biophysically.
Patterning of cell behavior is thought to involve patterned signaling. To date, numerous growth factors and regulatory signaling pathways have been implicated in branching morphogenesis. We have cataloged the loss-of-function phenotypes of major signaling pathway genes in Table S1 and transcription factors in Table S2. Despite this extensive knowledge, how these different regulators are integrated into patterns governing the various branching mechanisms remains poorly understood. A fundamental requirement for one or more receptor tyrosine kinase (RTK) signaling pathways has been established for most branched organs (Schuchardt et al., 1994; Luetteke et al., 1999; De Moerlooze et al., 2000; Olsson et al., 2006). The ligands of these core RTK signaling modules are usually produced by the mesenchyme, whereas the corresponding RTK receptors are expressed in the epithelium. Modulators of RTK signaling have been identified in the mesenchyme, ECM, and epithelium itself. The regulation and cross talk of these pathways has been reviewed extensively (Hennighausen and Robinson, 2005; Costantini and Shakya, 2006; Olsson et al., 2006; Patel et al., 2006; Jørgensen et al., 2007; Lu and Werb, 2008; Affolter et al., 2009; Costantini and Kopan, 2010; Blake and Rosenblum, 2014). In this review, we will limit our discussion of signaling pathways to their roles in pattern formation.
New branch formation by invasive collective cell migration
An invasive form of collective cell migration can drive budding of new branches in Drosophila trachea (Samakovlis et al., 1996) and mouse retina blood vessels (Gerhardt et al., 2003). In both cases, a protrusive leader cell migrates outward at the vanguard of a group of follower cells that rearrange to form a new branch (Fig. 2, B and C; Scarpa and Mayor, 2016). The identity of the leader cell is specified by RTK signaling in both systems: FGF (Branchless/Bnl) in Drosophila trachea and VEGF in blood vessels (Gerhardt et al., 2003; Ghabrial and Krasnow, 2006). In addition, a Delta-Notch–mediated lateral inhibition mechanism prevents the follower cells from becoming leader cells in both systems (Ghabrial and Krasnow, 2006; Hellström et al., 2007; Suchting et al., 2007; Lebreton and Casanova, 2014).
Differences exist in the responses of stalk cells to RTK signaling. In Drosophila trachea, FGF/Bnl activation is only important in the leader cell at the tip, whereas the follower stalk cells respond passively to pulling forces generated by leader cells (Caussinus et al., 2008; Lebreton and Casanova, 2014). During blood vessel branching, both tip and stalk cells respond to VEGF, which regulates tip cell migration and stalk cell proliferation (Gerhardt et al., 2003).
New branch formation by noninvasive collective cell migration
A noninvasive form of collective migration has been described in budding of the mammary gland, where multilayered terminal end buds (TEBs) grow out from an epithelial cyst ex vivo (Ewald et al., 2008). However, unlike blood vessels and Drosophila trachea, cells at the leading edge of the TEB lack invasive protrusions penetrating the basement membrane (Fig. 2 D; Ewald et al., 2008; Huebner et al., 2016). Moreover, cells within the TEB can dynamically switch positions at the migration front. Whether outward migration of the TEB is driven by pushing forces from the stalk (Friedl and Gilmour, 2009) could be tested by analyzing TEB behavior after laser ablation of the stalk versus the tip.
Cell behavior similar to mammary gland TEB budding occurs in budding of mouse lung (Schnatwinkel and Niswander, 2013) and ureteric bud of kidney (Fig. 3 B; Chi et al., 2009; Riccio et al., 2016), although the term “collective migration” was not used. Live imaging of sparsely labeled lung and ureteric bud cells reveal a group of cells that remain at the tip and are apparently pushed outward by cell division and rearrangement of the forming stalk. Unlike mammary gland TEB, branch tips in both lung and ureteric bud (kidney) are both single-layered, although the newly formed ureteric bud tip is pseudostratified (Chi et al., 2009; Schnatwinkel and Niswander, 2013).
Tip identity establishment and maintenance by cell competition
A recurring theme in branching morphogenesis by budding is that RTK signaling-dependent cell competition appears to maintain the identity of the tip cells (Fig. 2, B–E). As mentioned in the New branch formation by invasive collective cell migration section, leader cell identity for invasive branching in Drosophila trachea and blood vessels is mediated by FGF and VEGF signaling, respectively, whereas the leader cell inhibits the followers through Notch signaling in both systems. Strikingly, chimeric analysis in Drosophila trachea reveals that mutant cells defective for FGF signaling never take the leader cell position (Ghabrial and Krasnow, 2006). Similarly, genetic mosaic analysis in mammary glands shows that Fgfr2-null cells are progressively outcompeted by Fgfr2-positive cells in the TEBs (Lu et al., 2008), whereas Ret-null cells (Ret encodes the RTK receptor important for kidney branching; Schuchardt et al., 1994) are excluded from the tips of chimeric ureteric buds (Chi et al., 2009). Furthermore, this RTK-dependent cell competition is dose dependent, because ureteric bud cells with up-regulated RTK signaling (Sprouty1 null cells) outcompete wild-type cells to occupy the branch tips (Chi et al., 2009). How tip cells inhibit the stalk cells in mammalian epithelial organs is not entirely clear, though TGF-β is a candidate for mammary gland (Nelson et al., 2006; Lu and Werb, 2008) and kidney (Rivitos et al., 1995). Taken together, a conserved role for RTK signaling appears to establish, as well as to maintain, the identity of cells at the branch tip.
New branch formation by patterned cell proliferation
Patterned cell proliferation operates at two levels: regionalized proliferation and oriented cell division. Regionalized proliferation at the branch tip appears important for formation of new branches by budding in most cases, except for Drosophila trachea and blood vessels (Fig. 2, B–E). Studies using BrdU or radioactive thymidine labeling reveal a consistent pattern of higher proliferation in epithelial buds than in the stalks of many vertebrate organs (Bernfield et al., 1972; Goldin and Wessells, 1979; Goldin and Opperman, 1980; Michael and Davies, 2004; Lu et al., 2008; Schnatwinkel and Niswander, 2013). This pattern of high proliferative activity often mirrors the RTK activity (Costantini and Shakya, 2006; Patel et al., 2006) and could drive budding; in fact, elevated proliferation often precedes or coincides with budding (Nogawa et al., 1998; Michael and Davies, 2004; Schnatwinkel and Niswander, 2013). In contrast, localized proliferation activity has never been observed to precede clefting at branch tips. Consistent with this difference between budding and clefting, blocking cell proliferation abolishes budding in cultured mouse lung (Goldin et al., 1984) and mammary gland (Ewald et al., 2008), whereas clefting in salivary gland still proceeds (Nakanishi et al., 1987; Spooner et al., 1989).
Another type of patterned cell proliferation—oriented cell division—appears to contribute to clefting of lung branch tips. In mouse embryonic lung, live imaging of GFP–Histone 2B revealed no regional differences in rates of cell division inside the tip before clefting. However, cells toward the center divide preferentially along the axis of flattening (Fig. 3 A, black arrow in the center panel), which helps expand the bud in the same direction in preparation for clefting (Schnatwinkel and Niswander, 2013). It is not clear how the orientation of cell divisions in different regions of the branching lung tip is regulated; however, planar cell polarity (PCP) regulators and external compression by smooth muscle cells are possible candidates (see Physical constraint by basement membrane or smooth muscle cells in clefting; Yates and Dean, 2011; Kim et al., 2015).
A similar homogeneous distribution of mitotic cells within branch tips before clefting has also been described in mouse kidney (Michael and Davies, 2004), which undergoes clefting by a similar sequence of enlarging, flattening, and clefting (Fig. 3, A and B; Watanabe and Costantini, 2004; Schnatwinkel and Niswander, 2013). Interestingly, cell division in kidney during ureteric bud branching occurs in a manner similar to lung, where premitotic cells first delaminate from the single-layered epithelium, complete cell division in the lumen, and reinsert back to the epithelium (Fig. 3, A and B; Packard et al., 2013; Schnatwinkel and Niswander, 2013). However, whether preferential orientation of cell division occurs in the clefting tips of kidney requires further examination. In sum, both regionalized proliferation and orientation of cell divisions can contribute to new branch formation, depending on the organ.
New branch formation by cell rearrangement and coordinated deformation
Besides proliferation, recent studies have highlighted the importance in branch tip clefting of actomyosin contractility, differential cell motility, ECM remodeling, and external physical restraints. These studies reveal unifying themes and organ-specific differences.
Contributions of actomyosin contractility to budding and clefting
One emerging theme is the contrasting roles of actomyosin contractility in the budding versus clefting mechanisms of new branch formation: budding usually does not rely on actomyosin contractility, whereas clefting always does. A caveat for this process is that complex, sometimes contradictory, results exist in the earlier literature, which can often be explained by specific choices of inhibitors or organ culture conditions. We have omitted reviewing results using problematic inhibitors (e.g., the myosin inhibitor 2,3-butanedione monoxime; Ostap, 2002) and will discuss results that appear contradictory superficially, but not fundamentally.
Inhibition of actomyosin contractility in clefting-dominant branching organs (Fig. 3, B and C) consistently inhibits branch formation. In mouse salivary gland, inhibiting myosin II activity by blebbistatin, or Rho kinase (ROCK) activity by Y-27632 or ROCK I siRNA (but not ROCK II siRNA; Daley et al., 2009; Hsu et al., 2013), all inhibit branching, even though shallow initial clefts still form (Daley et al., 2009). Similarly, mouse ureteric bud branching is strongly suppressed by inhibiting ROCK by Y-27632 or actin polymerization by cytochalasin D (Michael et al., 2005). A conflicting study found Y-27632 stimulates rat kidney branching, but in mesenchyme-free Matrigel culture (Meyer et al., 2006).
Importantly, studies inhibiting actomyosin activity in the mouse lung, an organ using both budding (domain branching) and clefting (Fig. 3 A; terminal bifurcation), demonstrate differential requirements for actomyosin activity in budding versus clefting (Moore et al., 2005; Schnatwinkel and Niswander, 2013). Although Moore et al. (2005) initially reported overall inhibition of lung branching by inhibiting ROCK or actin polymerization, deeper analyses by Schnatwinkel and Niswander (2013) revealed that either blebbistatin inhibition of myosin activity or ML7 inhibition of myosin light chain kinase (MLCK) disrupt bifurcation (clefting) without blocking domain branch formation (budding). Inhibition of myosin, but not MLCK, further altered domain branch position and the shape of bifurcating buds.
Inhibiting actomyosin activity in budding-dominant branching systems often fails to prevent new branch formation. In a 3D culture model of blood vessels, where budding is driven by the invasive form of collective migration (Fig. 2 C), Fischer et al. (2009) elegantly demonstrated that a local loss of cortical myosin preceded filopodia protrusion and tip cell outgrowth. In fact, global myosin inhibition by blebbistatin greatly enhanced blood vessel branching, and local application of blebbistatin triggered filopodial extension of tip cells.
However, the mechanisms appear more complicated in mouse mammary gland organoid culture (Fig. 2 D). Although inhibiting ROCK by Y-27632 did not prevent branching, inhibiting either Rac (NSC23766) or MLCK (ML7) did block branching of mammary gland organoids (Ewald et al., 2008). Because these ex vivo organoids undergo a characteristic luminal clearance and filling process to become a “branching-competent” complex cyst (Ewald et al., 2008), treating the organoids at the complex cyst stage might help distinguish roles of Rac and MLCK in branching versus organoid remodeling. A study of budding in embryonic chick lung described prominent enrichment of actin filaments at the apical surface of forming buds, hinting at an apical constriction–mediated contractile budding mechanism (Kim et al., 2013). However, inhibition of new bud formation by the myosin II inhibitor blebbistatin was marginal and did not rule out a role for regionalized proliferation (Figs. 2 I and 4 D with different y axes in Kim et al., 2013). We suggest that live imaging could distinguish the different possibilities.
To summarize, the published literature appears to support the concept that new branch formation by clefting requires actomyosin contractility, whereas budding may not. As discussed in New branch formation by patterned cell proliferation, regionalized proliferation instead seems to be more crucial for budding (Fig. 2, D and E). Interestingly, recent work in mouse pancreas and salivary gland suggests that patterned actomyosin contractility, rather than global contractility, is more important for pancreas branching. Either globally activating or inactivating contractility blocked pancreatic branching (Shih et al., 2016), suggesting that either a certain level of contractility is critical or a pattern of contractility is important for branching. In both pancreas and salivary gland, outer (peripheral) cells at the branch tip have a characteristic columnar shape (Fig. 3 C); importantly, this columnar shape becomes either cuboidal or misshapen upon actomyosin perturbation to resemble inner cell shape (Hsu et al., 2013; Shih et al., 2016), suggesting possible differential contractility of outer versus inner cells. Whether these specific differences between outer and inner cells and locally tuned differences in contractility are crucial for branching remains to be determined.
Differential cell motilities in branching morphogenesis
In addition to differences in shape, differential motilities of outer and inner cells at branch tips (or epithelial “buds”) that could contribute to branching have been quantified in both mouse salivary gland and pancreas, but there are several differences. Outer cells touching the basement membrane move more than twice as fast as inner cells in tips (buds) of the salivary gland and show greater displacement (Fig. 3 C; Larsen et al., 2006; Hsu et al., 2013). Cells in these two regions move at similar speeds in the pancreas, and the major regional difference in pancreas involves directionality, with outer cells undergoing twofold greater displacement than inner cells (Shih et al., 2016). Although both systems require integrins to branch, integrin inhibition has different effects. Although inhibiting β1 integrin to disrupt cell–ECM interactions slows down outer cells in the salivary gland to rates comparable to inner cells (Hsu et al., 2013), it reduces the directionality of pancreatic outer cells without altering their velocity (Shih et al., 2016).
The most dramatic difference between salivary gland and pancreas involves opposite effects of perturbing the cell–cell adhesion molecule E-cadherin. In salivary gland, blocking E-cadherin slightly slows the outer cells and significantly accelerates the inner cells, which eliminates the differential velocity and inhibits branching (Walker et al., 2008; Hsu et al., 2013). In contrast, pancreatic deletion of E-cadherin almost doubles the branching index and partially rescues the nonbranching phenotype of pancreatic β1-integrin deletion (Shih et al., 2016). Lastly, although both pancreas and salivary gland seem to down-regulate E-cadherin in the outer cells, they seem to do so through distinct pathways. Although a fibronectin–Btbd7 axis is important for this regulation in the salivary gland (Sakai et al., 2003; Onodera et al., 2010), an integrin–Src axis appears to operate in the pancreas (Martinez-Rico et al., 2010; Shih et al., 2016). Taken together, these studies highlight ECM-associated differential cell behavior involved in branching, but the specific mechanisms can differ significantly in different organs.
Physical constraint by basement membrane or smooth muscle cells in clefting
In addition to its functions in cell motility, the basement membrane plays other biochemical and biophysical roles in branching morphogenesis. For example, modulation of growth factor signaling by heparan sulfate proteoglycans and cleaved collagen IV fragments has been described in many cases (Patel et al., 2006, 2014; Rebustini et al., 2009; Crawford et al., 2010; Shah et al., 2011). A fundamental requirement for the ECM protein fibronectin in branching morphogenesis has been demonstrated in several branched organs (Sakai et al., 2003). Whether their mechanisms of action differ in clefting versus budding deserves further examination.
Classical studies established the importance of basement membrane in organ branching, with a general pattern of thinner basement membrane at branch tips during remodeling (Grobstein and Cohen, 1965; Bernfield and Banerjee, 1982; Moore et al., 2005). Recently, a deeper examination by live imaging reveals that the basement membrane at branch tips is remarkably dynamic (Harunaga et al., 2014). Surprisingly, the basement membrane ECM at branch tips of salivary gland, lung, and kidney becomes perforated by numerous well-defined microscopic holes that facilitate pliability (Fig. 3). Additionally, the basement membrane as a whole constantly moves rearward away from the tip. Both the microscopic holes and the rearward movement require protease activity and actomyosin contractility (Harunaga et al., 2014). The rearward movement of basement membrane hints at a potentially important mechanism for cleft formation: a stochastically formed nascent cleft (Sakai et al., 2003) could initiate, progress deeper, and become stabilized by basement membrane components (Kadoya and Yamashina, 2010). This combination of pliability at the tip/bud with basement membrane translocation and thickening at the cleft bottom to stabilize morphology may permit both rapid tip outgrowth and stabilization of clefts (Fig. 3 C). Laser ablation studies of the basement membrane should test these predictions.
Besides the ECM-based basement membrane, physical constraint can also be provided by cells. Kim et al. (2015) reported recently that tip clefting in lung is always preceded by the local appearance of mesenchyme-derived smooth muscle cells at the future cleft. The differentiated smooth muscle cells locally deform and cleft the tip (Fig. 3 A). Globally inhibiting or promoting smooth muscle differentiation disrupted lung bifurcation or clefting (Kim et al., 2015). Collectively, local physical constraints seem to be particularly important for clefting-mediated branch formation, whether by ECM or cells.
What remains mysterious in salivary gland and lung is the origin of pattern formation. A key characteristic of a pattern generator is that either global activation or inhibition should disrupt the pattern. The inhibitor studies by Kim et al. (2015) point to a key role for sonic hedgehog (SHH), because both activating SHH signaling by smoothened agonist (SAG) or inhibiting SHH by cyclopamine disrupt the stereotyped differentiation of smooth muscle and bifurcation. Interestingly, reaction-diffusion modeling using only three components—FGF10, SHH, and the SHH receptor—is sufficient to generate bifurcation patterns (Menshykau et al., 2012). Further efforts are clearly required to generate quantitative models with sufficient predictive power to explain cleft patterning.
Beyond epithelium: Contributions of other cell types
As we have seen, the epithelium of branching organs is crucial for branching morphogenesis. However, establishing the architecture of branched organs requires contributions from many other cell types, including the surrounding mesenchyme, as well as blood vessels and neurons.
The encapsulating mesenchyme adjacent to the basement membrane contributes to epithelial branching both biochemically and biophysically. A crucial function of the mesenchyme is to produce the ligands for key RTK receptors, including VEGF, FGFs, and glial cell–derived neurotrophic factor (Fig. 2, B–E; and Fig. 3; Lu and Werb, 2008). If certain growth factors are provided, mesenchyme-free cultures of many embryonic organs can generate branched structures (Shamir and Ewald, 2014), demonstrating that the epithelia themselves are able to branch. However, these branched structures often appear morphologically different from in vivo organs or whole-organ explant cultures—for the most part, they appear to recapitulate budding and stalk elongation much more effectively than clefting (Morita and Nogawa, 1999; Qiao et al., 1999; Meyer et al., 2004; Steinberg et al., 2005; Ewald et al., 2008; Ghosh et al., 2011; Greggio et al., 2013; Huch et al., 2013). Thus, the epithelium may require interactions with the mesenchyme to prepattern local clefting signals. Importantly, studies using heterotypic recombinations of epithelium and mesenchyme from different organs have found the pattern of branching is often dictated by the tissue origin of the mesenchyme rather than the epithelium (Lubkin, 2008). For example, recombination of mammary gland epithelium with salivary gland mesenchyme yields an epithelial structure morphologically similar to salivary gland even though it produces milk (Kratochwil, 1969; Sakakura et al., 1976). Lubkin (2008) suggests that viscosity of the mesenchyme largely determines epithelial morphology, which will be interesting to test experimentally. To summarize, the epithelium appears competent to generate branches autonomously by budding, likely through RTK signaling–based cell competition, but it requires epithelial–mesenchymal cross talk to generate local cleft patterning.
Branched epithelial organs are also often “copatterned” with blood vessels and neurons, consistent with requirements for nutrition and neuronal regulation (Nelson and Larsen, 2015). Interestingly, diametrically opposed perfusion-independent roles for blood vessels in branching morphogenesis of lung and pancreas have been described. Although expression of a decoy VEGF receptor to reduce blood vessels inhibits branching and alters branching stereotypy of the lung (Lazarus et al., 2011), blood vessels in the pancreas instead seem to restrict its growth and branching (Magenheim et al., 2011). It is not clear, though, whether vascularization itself requires signals from the epithelial cells. Consistently positive roles for parasympathetic innervation have been described in branching of both salivary gland and lung, though through different pathways. In the salivary gland, parasympathetic innervation maintains epithelial progenitor cells through acetylcholine/muscarinic M1 and EGF receptor signaling (Knox et al., 2010). Removing the parasympathetic ganglion reduced but did not abolish branching (Knox et al., 2010). In contrast, laser ablation of parasympathetic neurons on one side of the mouse lung completely halted branching on that side, and acetylcholine signaling does not appear to be required (Bower et al., 2014).
Interestingly, parasympathetic innervation in both salivary gland and lung initiates after branching has started. In fact, salivary gland epithelial cells first secrete Wnt ligands to guide parasympathetic innervation, which in turn promotes epithelial growth and branching at later stages (Knosp et al., 2015). For other branching organs, the temporal sequence and interdependence of epithelial branching compared with recruitment of neurons and blood vessels need clarification.
Refining organ architecture by branch elongation and maturation
The overall morphology of a branched organ depends not only on the pattern of new branch formation, but also on the aspect ratio of branch stalks and tips. Diverse cellular mechanisms control branch stalk elongation and tip maturation, including cell rearrangement, elongation, proliferation, and differentiation.
Branch stalk elongation can occur by several mechanisms: cell rearrangement by intercalation or convergent extension, cell elongation, and cell proliferation (Fig. 4, A–C; Andrew and Ewald, 2010). Elongation of Drosophila tracheal branches occurs without cell division; instead, the stalk cells elongate along the outgrowth axis and intercalate to elongate the branch (Fig. 4 A; Caussinus et al., 2008). Another type of cell rearrangement, rosette-based convergent extension, plays an important role in elongation of both mouse and Xenopus laevis kidney tubules (Fig. 4 B; Lienkamp et al., 2012). This convergent extension depends on both Wnt and PCP signaling (Karner et al., 2009; Lienkamp et al., 2012). Additionally, PCP signaling directs the orientation of cell divisions to further promote kidney tubule elongation (Fischer et al., 2006; Saburi et al., 2008). Oriented cell division is also important for regulating branch shape in mouse lung, where cell division is thought to orient longitudinally by default or to be modulated by the activity of extracellular signal–regulated kinase (Tang et al., 2011). In mammary gland and branching ureteric bud, mitosis at branch tips increases stalk length (Ewald et al., 2008; Chi et al., 2009), though it is not yet known how proliferation preferentially increases stalk length rather than diameter. Overall, different organs clearly employ different mechanisms to elongate branch stalks.
Maturation of branched organs often involves cell differentiation at branch tips. At this stage, epithelial cells at the branch tips of salivary gland are slightly wedge-shaped cuboidal cells surrounding a small lumen to form an acinus (Fig. 4 D; Aure et al., 2015). However, tip epithelial cells in lung have differentiated into two types of cells: flat, squamous alveolar type (AT) 1 cells and cuboidal, surfactant-secreting AT2 cells, which together surround a comparatively large lumen to form an alveolus (Fig. 4 E; Morrisey and Hogan, 2010). Branch tips of the mammary gland also form alveoli for lactation, which can regress and reform between pregnancies (Hennighausen and Robinson, 2005; Akhtar et al., 2009, 2016; Akhtar and Streuli, 2013). For maturation of other branched organs, there are several excellent recent reviews for lung, kidney, and pancreas (Costantini and Kopan, 2010; Morrisey and Hogan, 2010; Shih et al., 2013).
Future directions
Branching morphogenesis has been studied intensively, not only because it is an intrinsically beautiful, dynamic process, but also because it holds promise for regenerative medicine. Two important and related future goals for studying branching morphogenesis include achieving a quantitative understanding of this process and bringing regenerative medicine to fruition by replacing or regenerating impaired branched organs.
To achieve a full understanding of branching morphogenesis, we predict that recursive applications of quantitative observation, mathematical modeling, and precisely controlled experimental perturbations will be crucial. For quantitative observation, live organ imaging at high spatiotemporal resolution as well as local force measurements will be valuable. Recently, several high-resolution 3D imaging methods with low phototoxicity have been developed, including dual-view inverted selective plane illumination microscope (Wu et al., 2013; Kumar et al., 2014) and lattice light-sheet microscopy (Chen et al., 2014).
For tracking the behavior of individual cells in the crowded cellular environment of developing organs, valuable tools will include chimeric labeling strategies such as Brainbow (Livet et al., 2007), photo-convertible dyes such as KikGR (Tsutsui et al., 2005; Hsu et al., 2013), and analysis software such as RACE (real-time accurate cell-shape extractor; Stegmaier et al., 2016). Application of advanced microscopy methods to chimerically labeled organs should help characterize cell dynamics and shape changes in more detail to catalyze new hypotheses. For quantitative modeling of branching morphogenesis, recently developed intracellular tension sensors will be useful (Kumar et al., 2016). For measuring tension and compression forces in live tissues, approaches such as oil microdroplet force-mapping will also be valuable (Campàs et al., 2014). For example, mapping actomyosin contractile forces during branch formation will help elucidate the different roles of actomyosin contractility in clefting versus budding. Complementary descriptive gene expression data can be obtained using single-cell or laser microdissection transcriptomic sequencing (RNA sequencing).
Once a speculative model is derived from descriptive observations, mathematical modeling can simulate the process, followed by tests of its experimental predictions to guide further model refinement. Modeling signaling pattern formation by reaction-diffusion (Turing, 1952; Kondo and Miura, 2010) and mechanical interactions between different cells (Lubkin, 2008; Varner and Nelson, 2017) may be particularly illuminating. For example, a simplified model focusing on FGF and SHH has provided initial limited success in explaining lung branching (Menshykau et al., 2012; Iber and Menshykau, 2013), and a cell-based model simulates cleft progression in salivary gland (Ray et al., 2013).
Besides further application of gene ablation and RNAi approaches, precise perturbations of specific proteins and local forces will be valuable. Recent advances in the temporal control of protein depletion using an auxin-inducible protein degradation method (Nishimura et al., 2009) could be added to the Cre-LoxP gene ablation toolkit. For spatial control, laser-based inactivation and activation will be useful. For example, laser ablation can be used to measure tensile forces at specific locations or to ablate specific cells or ECM structures. Laser micropatterned activation or inactivation based on optogenetic switches (Zhang and Cui, 2015) and chromophore-assisted laser inactivation (Sano et al., 2014) will help test hypotheses and models. Photo-caged inhibitors and activators may also be useful (Lee et al., 2009). The rapid evolution of CRISPR-Cas9–mediated genome-editing methods (Doudna and Charpentier, 2014; Wright et al., 2016) will accelerate development of novel perturbation methods.
Regenerative therapies could be achieved by transplantation of tissue-specific stem cells/progenitors or gene therapy to restore impaired organs or by implanting a functional bioengineered organ to replace damaged or lost organs. Both approaches are active areas of research. For stem cell therapy to be developed as a regenerative therapy, the locations of potentially useful progenitor cells need to be identified. Although progenitor cells can reside in duct regions, recent studies reveal that such cells involved in homeostasis of mature lung and salivary gland are located in terminal alveoli or acini (Fig. 4, D and E; Desai et al., 2014; Aure et al., 2015; Hauser and Hoffman, 2015). Alternatively, successful restoration of both salivary gland and lacrimal gland function by transplantation of bioengineered embryonic organ germs has been described in mice (Hirayama et al., 2013; Ogawa et al., 2013). Potential future embryonic stem cell or induced pluripotent stem cell redifferentiation to organ-specific progenitors, organoids, or even functional organs can be envisioned (Shamir and Ewald, 2014). All of these approaches will benefit from applying new knowledge about the mechanisms of normal branching morphogenesis to guide successful regenerative therapy.
Supplementary Material
Acknowledgments
Work in the authors’ laboratory is supported by the Intramural Research Program of the National Institutes of Health, National Institute of Dental and Craniofacial Research (ZIA DE000524 and ZIA DE000525).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AT1
- alveolar type1
- MLCK
- myosin light chain kinase
- PCP
- planar cell polarity
- ROCK
- Rho kinase
- RTK
- receptor tyrosine kinase
- SHH
- sonic hedgehog
- TEB
- terminal end bud
References
- Affolter M., and Caussinus E.. 2008. Tracheal branching morphogenesis in Drosophila: New insights into cell behaviour and organ architecture. Development. 135:2055–2064. 10.1242/dev.014498 [DOI] [PubMed] [Google Scholar]
- Affolter M., Zeller R., and Caussinus E.. 2009. Tissue remodelling through branching morphogenesis. Nat. Rev. Mol. Cell Biol. 10:831–842. 10.1038/nrm2797 [DOI] [PubMed] [Google Scholar]
- Akhtar N., and Streuli C.H.. 2013. An integrin-ILK-microtubule network orients cell polarity and lumen formation in glandular epithelium. Nat. Cell Biol. 15:17–27. 10.1038/ncb2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar N., Marlow R., Lambert E., Schatzmann F., Lowe E.T., Cheung J., Katz E., Li W., Wu C., Dedhar S., et al. 2009. Molecular dissection of integrin signalling proteins in the control of mammary epithelial development and differentiation. Development. 136:1019–1027. 10.1242/dev.028423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar N., Li W., Mironov A., and Streuli C.H.. 2016. Rac1 controls both the secretory function of the mammary gland and its remodeling for successive gestations. Dev. Cell. 38:522–535. 10.1016/j.devcel.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew D.J., and Ewald A.J.. 2010. Morphogenesis of epithelial tubes: Insights into tube formation, elongation, and elaboration. Dev. Biol. 341:34–55. 10.1016/j.ydbio.2009.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aure M.H., Konieczny S.F., and Ovitt C.E.. 2015. Salivary gland homeostasis is maintained through acinar cell self-duplication. Dev. Cell. 33:231–237. 10.1016/j.devcel.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernfield M., and Banerjee S.D.. 1982. The turnover of basal lamina glycosaminoglycan correlates with epithelial morphogenesis. Dev. Biol. 90:291–305. 10.1016/0012-1606(82)90378-5 [DOI] [PubMed] [Google Scholar]
- Bernfield M.R., Banerjee S.D., and Cohn R.H.. 1972. Dependence of salivary epithelial morphology and branching morphogenesis upon acid mucopolysaccharide-protein (proteoglycan) at the epithelial surface. J. Cell Biol. 52:674–689. 10.1083/jcb.52.3.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake J., and Rosenblum N.D.. 2014. Renal branching morphogenesis: Morphogenetic and signaling mechanisms. Semin. Cell Dev. Biol. 36:2–12. 10.1016/j.semcdb.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Bower D.V., Lee H.-K., Lansford R., Zinn K., Warburton D., Fraser S.E., and Jesudason E.C.. 2014. Airway branching has conserved needs for local parasympathetic innervation but not neurotransmission. BMC Biol. 12:92 10.1186/s12915-014-0092-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campàs O., Mammoto T., Hasso S., Sperling R.A., O’Connell D., Bischof A.G., Maas R., Weitz D.A., Mahadevan L., and Ingber D.E.. 2014. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods. 11:183–189. 10.1038/nmeth.2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E., Colombelli J., and Affolter M.. 2008. Tip-cell migration controls stalk-cell intercalation during Drosophila tracheal tube elongation. Curr. Biol. 18:1727–1734. 10.1016/j.cub.2008.10.062 [DOI] [PubMed] [Google Scholar]
- Chen B.-C., Legant W.R., Wang K., Shao L., Milkie D.E., Davidson M.W., Janetopoulos C., Wu X.S., Hammer J.A. III, Liu Z., et al. 2014. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science. 346:1257998 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Michos O., Shakya R., Riccio P., Enomoto H., Licht J.D., Asai N., Takahashi M., Ohgami N., Kato M., et al. 2009. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev. Cell. 17:199–209. 10.1016/j.devcel.2009.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F., and Kopan R.. 2010. Patterning a complex organ: Branching morphogenesis and nephron segmentation in kidney development. Dev. Cell. 18:698–712. 10.1016/j.devcel.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F., and Shakya R.. 2006. GDNF/Ret signaling and the development of the kidney. BioEssays. 28:117–127. 10.1002/bies.20357 [DOI] [PubMed] [Google Scholar]
- Crawford B.E., Garner O.B., Bishop J.R., Zhang D.Y., Bush K.T., Nigam S.K., and Esko J.D.. 2010. Loss of the heparan sulfate sulfotransferase, Ndst1, in mammary epithelial cells selectively blocks lobuloalveolar development in mice. PLoS One. 5:e10691 10.1371/journal.pone.0010691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley W.P., and Yamada K.M.. 2013. ECM-modulated cellular dynamics as a driving force for tissue morphogenesis. Curr. Opin. Genet. Dev. 23:408–414. 10.1016/j.gde.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley W.P., Gulfo K.M., Sequeira S.J., and Larsen M.. 2009. Identification of a mechanochemical checkpoint and negative feedback loop regulating branching morphogenesis. Dev. Biol. 336:169–182. 10.1016/j.ydbio.2009.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.A. 2005. Branching Morphogenesis. Springer US, Boston, MA. 246 pp. [Google Scholar]
- De Moerlooze L., Spencer-Dene B., Revest J.M., Hajihosseini M., Rosewell I., and Dickson C.. 2000. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 127:483–492. [DOI] [PubMed] [Google Scholar]
- Desai T.J., Brownfield D.G., and Krasnow M.A.. 2014. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 507:190–194. 10.1038/nature12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A., and Charpentier E.. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 346:1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Ewald A.J., Brenot A., Duong M., Chan B.S., and Werb Z.. 2008. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell. 14:570–581. 10.1016/j.devcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E., Legue E., Doyen A., Nato F., Nicolas J.-F., Torres V., Yaniv M., and Pontoglio M.. 2006. Defective planar cell polarity in polycystic kidney disease. Nat. Genet. 38:21–23. 10.1038/ng1701 [DOI] [PubMed] [Google Scholar]
- Fischer R.S., Gardel M., Ma X., Adelstein R.S., and Waterman C.M.. 2009. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr. Biol. 19:260–265. 10.1016/j.cub.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., and Gilmour D.. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10:445–457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Gerhardt H., Golding M., Fruttiger M., Ruhrberg C., Lundkvist A., Abramsson A., Jeltsch M., Mitchell C., Alitalo K., Shima D., and Betsholtz C.. 2003. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell Biol. 161:1163–1177. 10.1083/jcb.200302047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A.S., and Krasnow M.A.. 2006. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 441:746–749. 10.1038/nature04829 [DOI] [PubMed] [Google Scholar]
- Ghabrial A., Luschnig S., Metzstein M.M., and Krasnow M.A.. 2003. Branching morphogenesis of the Drosophila tracheal system. Annu. Rev. Cell Dev. Biol. 19:623–647. 10.1146/annurev.cellbio.19.031403.160043 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Lau H., Simons B.W., Powell J.D., Meyers D.J., De Marzo A.M., Berman D.M., and Lotan T.L.. 2011. PI3K/mTOR signaling regulates prostatic branching morphogenesis. Dev. Biol. 360:329–342. 10.1016/j.ydbio.2011.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin G.V., and Opperman L.A.. 1980. Induction of supernumerary tracheal buds and the stimulation of DNA synthesis in the embryonic chick lung and trachea by epidermal growth factor. J. Embryol. Exp. Morphol. 60:235–243. [PubMed] [Google Scholar]
- Goldin G.V., and Wessells N.K.. 1979. Mammalian lung development: The possible role of cell proliferation in the formation of supernumerary tracheal buds and in branching morphogenesis. J. Exp. Zool. 208:337–346. 10.1002/jez.1402080310 [DOI] [PubMed] [Google Scholar]
- Goldin G.V., Hindman H.M., and Wessells N.K.. 1984. The role of cell proliferation and cellular shape change in branching morphogenesis of the embryonic mouse lung: Analysis using aphidicolin and cytochalasins. J. Exp. Zool. 232:287–296. 10.1002/jez.1402320216 [DOI] [PubMed] [Google Scholar]
- Gray R.S., Cheung K.J., and Ewald A.J.. 2010. Cellular mechanisms regulating epithelial morphogenesis and cancer invasion. Curr. Opin. Cell Biol. 22:640–650. 10.1016/j.ceb.2010.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio C., De Franceschi F., Figueiredo-Larsen M., Gobaa S., Ranga A., Semb H., Lutolf M., and Grapin-Botton A.. 2013. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 140:4452–4462. 10.1242/dev.096628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobstein C., and Cohen J.. 1965. Collagenase: Effect on the morphogenesis of embryonic salivary epithelium in vitro. Science. 150:626–628. 10.1126/science.150.3696.626 [DOI] [PubMed] [Google Scholar]
- Harunaga J.S., Doyle A.D., and Yamada K.M.. 2014. Local and global dynamics of the basement membrane during branching morphogenesis require protease activity and actomyosin contractility. Dev. Biol. 394:197–205. 10.1016/j.ydbio.2014.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser B.R., and Hoffman M.P.. 2015. Regulatory mechanisms driving salivary gland organogenesis. Curr. Top. Dev. Biol. 115:111–130. 10.1016/bs.ctdb.2015.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellström M., Phng L.-K., Hofmann J.J., Wallgard E., Coultas L., Lindblom P., Alva J., Nilsson A.-K., Karlsson L., Gaiano N., et al. 2007. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 445:776–780. 10.1038/nature05571 [DOI] [PubMed] [Google Scholar]
- Hennighausen L., and Robinson G.W.. 2005. Information networks in the mammary gland. Nat. Rev. Mol. Cell Biol. 6:715–725. 10.1038/nrm1714 [DOI] [PubMed] [Google Scholar]
- Hirayama M., Ogawa M., Oshima M., Sekine Y., Ishida K., Yamashita K., Ikeda K., Shimmura S., Kawakita T., Tsubota K., and Tsuji T.. 2013. Functional lacrimal gland regeneration by transplantation of a bioengineered organ germ. Nat. Commun. 4:2497 10.1038/ncomms3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.C., Koo H., Harunaga J.S., Matsumoto K., Doyle A.D., and Yamada K.M.. 2013. Region-specific epithelial cell dynamics during branching morphogenesis. Dev. Dyn. 242:1066–1077. 10.1002/dvdy.24000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Bonfanti P., Boj S.F., Sato T., Loomans C.J.M., van de Wetering M., Sojoodi M., Li V.S.W., Schuijers J., Gracanin A., et al. 2013. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32:2708–2721. 10.1038/emboj.2013.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner R.J., Neumann N.M., and Ewald A.J.. 2016. Mammary epithelial tubes elongate through MAPK-dependent coordination of cell migration. Development. 143:983–993. 10.1242/dev.127944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iber D., and Menshykau D.. 2013. The control of branching morphogenesis. Open Biol. 3:130088 10.1098/rsob.130088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman J.L., Robertson C., Mott J.D., and Bissell M.J.. 2015. Mammary gland development: Cell fate specification, stem cells and the microenvironment. Development. 142:1028–1042. 10.1242/dev.087643 [DOI] [PubMed] [Google Scholar]
- Jørgensen M.C., Ahnfelt-Rønne J., Hald J., Madsen O.D., Serup P., and Hecksher-Sørensen J.. 2007. An illustrated review of early pancreas development in the mouse. Endocr. Rev. 28:685–705. 10.1210/er.2007-0016 [DOI] [PubMed] [Google Scholar]
- Kadoya Y., and Yamashina S.. 2010. Cellular dynamics of epithelial clefting during branching morphogenesis of the mouse submandibular gland. Dev. Dyn. 239:1739–1747. 10.1002/dvdy.22312 [DOI] [PubMed] [Google Scholar]
- Karner C.M., Chirumamilla R., Aoki S., Igarashi P., Wallingford J.B., and Carroll T.J.. 2009. Wnt9b signaling regulates planar cell polarity and kidney tubule morphogenesis. Nat. Genet. 41:793–799. 10.1038/ng.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.Y., Varner V.D., and Nelson C.M.. 2013. Apical constriction initiates new bud formation during monopodial branching of the embryonic chicken lung. Development. 140:3146–3155. 10.1242/dev.093682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.Y., Pang M.-F., Varner V.D., Kojima L., Miller E., Radisky D.C., and Nelson C.M.. 2015. Localized smooth muscle differentiation is essential for epithelial bifurcation during branching morphogenesis of the mammalian lung. Dev. Cell. 34:719–726. 10.1016/j.devcel.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knosp W.M., Knox S.M., Lombaert I.M.A., Haddox C.L., Patel V.N., and Hoffman M.P.. 2015. Submandibular parasympathetic gangliogenesis requires sprouty-dependent Wnt signals from epithelial progenitors. Dev. Cell. 32:667–677. 10.1016/j.devcel.2015.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox S.M., Lombaert I.M.A., Reed X., Vitale-Cross L., Gutkind J.S., and Hoffman M.P.. 2010. Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science. 329:1645–1647. 10.1126/science.1192046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., and Miura T.. 2010. Reaction-diffusion model as a framework for understanding biological pattern formation. Science. 329:1616–1620. 10.1126/science.1179047 [DOI] [PubMed] [Google Scholar]
- Kratochwil K. 1969. Organ specificity in mesenchymal induction demonstrated in the embryonic development of the mammary gland of the mouse. Dev. Biol. 20:46–71. 10.1016/0012-1606(69)90004-9 [DOI] [PubMed] [Google Scholar]
- Kumar A., Wu Y., Christensen R., Chandris P., Gandler W., McCreedy E., Bokinsky A., Colón-Ramos D.A., Bao Z., McAuliffe M., et al. 2014. Dual-view plane illumination microscopy for rapid and spatially isotropic imaging. Nat. Protoc. 9:2555–2573. 10.1038/nprot.2014.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Ouyang M., Van den Dries K., McGhee E.J., Tanaka K., Anderson M.D., Groisman A., Goult B.T., Anderson K.I., and Schwartz M.A.. 2016. Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity. J. Cell Biol. 213:371–383. (published erratum appears in J. Cell Biol. 2016. 214:231) 10.1083/jcb.201510012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M., Wei C., and Yamada K.M.. 2006. Cell and fibronectin dynamics during branching morphogenesis. J. Cell Sci. 119:3376–3384. 10.1242/jcs.03079 [DOI] [PubMed] [Google Scholar]
- Larsen M., Yamada K.M., and Musselmann K.. 2010. Systems analysis of salivary gland development and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2:670–682. 10.1002/wsbm.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus A., Del-Moral P.M., Ilovich O., Mishani E., Warburton D., and Keshet E.. 2011. A perfusion-independent role of blood vessels in determining branching stereotypy of lung airways. Development. 138:2359–2368. 10.1242/dev.060723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton G., and Casanova J.. 2014. Specification of leading and trailing cell features during collective migration in the Drosophila trachea. J. Cell Sci. 127:465–474. 10.1242/jcs.142737 [DOI] [PubMed] [Google Scholar]
- Lee H.-M., Larson D.R., and Lawrence D.S.. 2009. Illuminating the chemistry of life: design, synthesis, and applications of “caged” and related photoresponsive compounds. ACS Chem. Biol. 4:409–427. 10.1021/cb900036s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienkamp S.S., Liu K., Karner C.M., Carroll T.J., Ronneberger O., Wallingford J.B., and Walz G.. 2012. Vertebrate kidney tubules elongate using a planar cell polarity-dependent, rosette-based mechanism of convergent extension. Nat. Genet. 44:1382–1387. 10.1038/ng.2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J., Weissman T.A., Kang H., Draft R.W., Lu J., Bennis R.A., Sanes J.R., and Lichtman J.W.. 2007. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 450:56–62. 10.1038/nature06293 [DOI] [PubMed] [Google Scholar]
- Lu P., and Werb Z.. 2008. Patterning mechanisms of branched organs. Science. 322:1506–1509. 10.1126/science.1162783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Ewald A.J., Martin G.R., and Werb Z.. 2008. Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev. Biol. 321:77–87. 10.1016/j.ydbio.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubkin S.R. 2008. Branched organs: Mechanics of morphogenesis by multiple mechanisms. Curr. Top. Dev. Biol. 81:249–268. 10.1016/S0070-2153(07)81008-8 [DOI] [PubMed] [Google Scholar]
- Luetteke N.C., Qiu T.H., Fenton S.E., Troyer K.L., Riedel R.F., Chang A., and Lee D.C.. 1999. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 126:2739–2750. [DOI] [PubMed] [Google Scholar]
- Magenheim J., Ilovich O., Lazarus A., Klochendler A., Ziv O., Werman R., Hija A., Cleaver O., Mishani E., Keshet E., and Dor Y.. 2011. Blood vessels restrain pancreas branching, differentiation and growth. Development. 138:4743–4752. 10.1242/dev.066548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rico C., Pincet F., Thiery J.-P., and Dufour S.. 2010. Integrins stimulate E-cadherin-mediated intercellular adhesion by regulating Src-kinase activation and actomyosin contractility. J. Cell Sci. 123:712–722. 10.1242/jcs.047878 [DOI] [PubMed] [Google Scholar]
- Meadows S.M., and Cleaver O.. 2015. Vascular patterning: Coordinated signals keep blood vessels on track. Curr. Opin. Genet. Dev. 32:86–91. 10.1016/j.gde.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Menshykau D., Kraemer C., and Iber D.. 2012. Branch mode selection during early lung development. PLOS Comput. Biol. 8:e1002377 10.1371/journal.pcbi.1002377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger R.J., Klein O.D., Martin G.R., and Krasnow M.A.. 2008. The branching programme of mouse lung development. Nature. 453:745–750. 10.1038/nature07005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T.N., Schwesinger C., Bush K.T., Stuart R.O., Rose D.W., Shah M.M., Vaughn D.A., Steer D.L., and Nigam S.K.. 2004. Spatiotemporal regulation of morphogenetic molecules during in vitro branching of the isolated ureteric bud: Toward a model of branching through budding in the developing kidney. Dev. Biol. 275:44–67. 10.1016/j.ydbio.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Meyer T.N., Schwesinger C., Sampogna R.V., Vaughn D.A., Stuart R.O., Steer D.L., Bush K.T., and Nigam S.K.. 2006. Rho kinase acts at separate steps in ureteric bud and metanephric mesenchyme morphogenesis during kidney development. Differentiation. 74:638–647. 10.1111/j.1432-0436.2006.00102.x [DOI] [PubMed] [Google Scholar]
- Michael L., and Davies J.A.. 2004. Pattern and regulation of cell proliferation during murine ureteric bud development. J. Anat. 204:241–255. 10.1111/j.0021-8782.2004.00285.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael L., Sweeney D.E., and Davies J.A.. 2005. A role for microfilament-based contraction in branching morphogenesis of the ureteric bud. Kidney Int. 68:2010–2018. 10.1111/j.1523-1755.2005.00655.x [DOI] [PubMed] [Google Scholar]
- Moore K.A., Polte T., Huang S., Shi B., Alsberg E., Sunday M.E., and Ingber D.E.. 2005. Control of basement membrane remodeling and epithelial branching morphogenesis in embryonic lung by Rho and cytoskeletal tension. Dev. Dyn. 232:268–281. 10.1002/dvdy.20237 [DOI] [PubMed] [Google Scholar]
- Morita K., and Nogawa H.. 1999. EGF-dependent lobule formation and FGF7-dependent stalk elongation in branching morphogenesis of mouse salivary epithelium in vitro. Dev. Dyn. 215:148–154. [DOI] [PubMed] [Google Scholar]
- Morrisey E.E., and Hogan B.L.M.. 2010. Preparing for the first breath: Genetic and cellular mechanisms in lung development. Dev. Cell. 18:8–23. 10.1016/j.devcel.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi Y., Morita T., and Nogawa H.. 1987. Cell proliferation is not required for the initiation of early cleft formation in mouse embryonic submandibular epithelium in vitro. Development. 99:429–437. [DOI] [PubMed] [Google Scholar]
- Nelson D.A., and Larsen M.. 2015. Heterotypic control of basement membrane dynamics during branching morphogenesis. Dev. Biol. 401:103–109. 10.1016/j.ydbio.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.M., Vanduijn M.M., Inman J.L., Fletcher D.A., and Bissell M.J.. 2006. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 314:298–300. 10.1126/science.1131000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Nogawa H., Morita K., and Cardoso W.V.. 1998. Bud formation precedes the appearance of differential cell proliferation during branching morphogenesis of mouse lung epithelium in vitro. Dev. Dyn. 213:228–235. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Oshima M., Imamura A., Sekine Y., Ishida K., Yamashita K., Nakajima K., Hirayama M., Tachikawa T., and Tsuji T.. 2013. Functional salivary gland regeneration by transplantation of a bioengineered organ germ. Nat. Commun. 4:2498 10.1038/ncomms3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A.-K., Dimberg A., Kreuger J., and Claesson-Welsh L.. 2006. VEGF receptor signalling - in control of vascular function. Nat. Rev. Mol. Cell Biol. 7:359–371. 10.1038/nrm1911 [DOI] [PubMed] [Google Scholar]
- Onodera T., Sakai T., Hsu J.C., Matsumoto K., Chiorini J.A., and Yamada K.M.. 2010. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science. 329:562–565. 10.1126/science.1191880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostap E.M. 2002. 2,3-Butanedione monoxime (BDM) as a myosin inhibitor. J. Muscle Res. Cell Motil. 23:305–308. 10.1023/A:1022047102064 [DOI] [PubMed] [Google Scholar]
- Packard A., Georgas K., Michos O., Riccio P., Cebrian C., Combes A.N., Ju A., Ferrer-Vaquer A., Hadjantonakis A.K., Zong H., et al. 2013. Luminal mitosis drives epithelial cell dispersal within the branching ureteric bud. Dev. Cell. 27:319–330. 10.1016/j.devcel.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V.N., Rebustini I.T., and Hoffman M.P.. 2006. Salivary gland branching morphogenesis. Differentiation. 74:349–364. 10.1111/j.1432-0436.2006.00088.x [DOI] [PubMed] [Google Scholar]
- Patel V.N., Lombaert I.M.A., Cowherd S.N., Shworak N.W., Xu Y., Liu J., and Hoffman M.P.. 2014. Hs3st3-modified heparan sulfate controls KIT+ progenitor expansion by regulating 3-O-sulfotransferases. Dev. Cell. 29:662–673. 10.1016/j.devcel.2014.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Sakurai H., and Nigam S.K.. 1999. Branching morphogenesis independent of mesenchymal-epithelial contact in the developing kidney. Proc. Natl. Acad. Sci. USA. 96:7330–7335. 10.1073/pnas.96.13.7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S., Yuan D., Dhulekar N., Oztan B., Yener B., and Larsen M.. 2013. Cell-based multi-parametric model of cleft progression during submandibular salivary gland branching morphogenesis. PLOS Comput. Biol. 9:e1003319 10.1371/journal.pcbi.1003319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebustini I.T., Myers C., Lassiter K.S., Surmak A., Szabova L., Holmbeck K., Pedchenko V., Hudson B.G., and Hoffman M.P.. 2009. MT2-MMP-dependent release of collagen IV NC1 domains regulates submandibular gland branching morphogenesis. Dev. Cell. 17:482–493. 10.1016/j.devcel.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio P., Cebrian C., Zong H., Hippenmeyer S., and Costantini F.. 2016. Ret and Etv4 promote directed movements of progenitor cells during renal branching morphogenesis. PLoS Biol. 14:e1002382 (published erratum appears in PLoS Biol. 2016. 14:e1002488) 10.1371/journal.pbio.1002382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivitos O., Tuuri T., Eramaa M., Sainio K., Hilden K., Saxen L., and Gilbert S.F.. 1995. Activin disrupts epithelial branching morphogenesis in developing glandular organs of the mouse. Mech. Dev. 50:229–245. 10.1016/0925-4773(94)00342-K [DOI] [PubMed] [Google Scholar]
- Saburi S., Hester I., Fischer E., Pontoglio M., Eremina V., Gessler M., Quaggin S.E., Harrison R., Mount R., and McNeill H.. 2008. Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat. Genet. 40:1010–1015. 10.1038/ng.179 [DOI] [PubMed] [Google Scholar]
- Sakai T., Larsen M., and Yamada K.M.. 2003. Fibronectin requirement in branching morphogenesis. Nature. 423:876–881. 10.1038/nature01712 [DOI] [PubMed] [Google Scholar]
- Sakakura T., Nishizuka Y., and Dawe C.J.. 1976. Mesenchyme-dependent morphogenesis and epithelium-specific cytodifferentiation in mouse mammary gland. Science. 194:1439–1441. 10.1126/science.827022 [DOI] [PubMed] [Google Scholar]
- Samakovlis C., Hacohen N., Manning G., Sutherland D.C., Guillemin K., and Krasnow M.A.. 1996. Development of the Drosophila tracheal system occurs by a series of morphologically distinct but genetically coupled branching events. Development. 122:1395–1407. [DOI] [PubMed] [Google Scholar]
- Sampogna R.V., Schneider L., and Al-Awqati Q.. 2015. Developmental programming of branching morphogenesis in the kidney. J. Am. Soc. Nephrol. 26:2414–2422. 10.1681/ASN.2014090886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y., Watanabe W., and Matsunaga S.. 2014. Chromophore-assisted laser inactivation--towards a spatiotemporal-functional analysis of proteins, and the ablation of chromatin, organelle and cell function. J. Cell Sci. 127:1621–1629. 10.1242/jcs.144527 [DOI] [PubMed] [Google Scholar]
- Scarpa E., and Mayor R.. 2016. Collective cell migration in development. J. Cell Biol. 212:143–155. 10.1083/jcb.201508047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnatwinkel C., and Niswander L.. 2013. Multiparametric image analysis of lung-branching morphogenesis. Dev. Dyn. 242:622–637. 10.1002/dvdy.23961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchardt A., D’Agati V., Larsson-Blomberg L., Costantini F., and Pachnis V.. 1994. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature. 367:380–383. 10.1038/367380a0 [DOI] [PubMed] [Google Scholar]
- Shah M.M., Sakurai H., Gallegos T.F., Sweeney D.E., Bush K.T., Esko J.D., and Nigam S.K.. 2011. Growth factor-dependent branching of the ureteric bud is modulated by selective 6-O sulfation of heparan sulfate. Dev. Biol. 356:19–27. 10.1016/j.ydbio.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir E.R., and Ewald A.J.. 2014. Three-dimensional organotypic culture: Experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 15:647–664. 10.1038/nrm3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H.P., Wang A., and Sander M.. 2013. Pancreas organogenesis: From lineage determination to morphogenesis. Annu. Rev. Cell Dev. Biol. 29:81–105. 10.1146/annurev-cellbio-101512-122405 [DOI] [PubMed] [Google Scholar]
- Shih H.P., Panlasigui D., Cirulli V., and Sander M.. 2016. ECM signaling regulates collective cellular dynamics to control pancreas branching morphogenesis. Cell Reports. 14:169–179. 10.1016/j.celrep.2015.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short K.M., Combes A.N., Lefevre J., Ju A.L., Georgas K.M., Lamberton T., Cairncross O., Rumballe B.A., McMahon A.P., Hamilton N.A., et al. 2014. Global quantification of tissue dynamics in the developing mouse kidney. Dev. Cell. 29:188–202. 10.1016/j.devcel.2014.02.017 [DOI] [PubMed] [Google Scholar]
- Spooner B.S., Bassett K.E., and Spooner B.S. Jr. 1989. Embryonic salivary gland epithelial branching activity is experimentally independent of epithelial expansion activity. Dev. Biol. 133:569–575. 10.1016/0012-1606(89)90059-6 [DOI] [PubMed] [Google Scholar]
- Stegmaier J., Amat F., Lemon W.C., McDole K., Wan Y., Teodoro G., Mikut R., and Keller P.J.. 2016. Real-time three-dimensional cell segmentation in large-scale microscopy data of developing embryos. Dev. Cell. 36:225–240. 10.1016/j.devcel.2015.12.028 [DOI] [PubMed] [Google Scholar]
- Steinberg Z., Myers C., Heim V.M., Lathrop C.A., Rebustini I.T., Stewart J.S., Larsen M., and Hoffman M.P.. 2005. FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development. 132:1223–1234. 10.1242/dev.01690 [DOI] [PubMed] [Google Scholar]
- Suchting S., Freitas C., le Noble F., Benedito R., Bréant C., Duarte A., and Eichmann A.. 2007. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc. Natl. Acad. Sci. USA. 104:3225–3230. 10.1073/pnas.0611177104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., Marshall W.F., McMahon M., Metzger R.J., and Martin G.R.. 2011. Control of mitotic spindle angle by the RAS-regulated ERK1/2 pathway determines lung tube shape. Science. 333:342–345. 10.1126/science.1204831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H., Karasawa S., Shimizu H., Nukina N., and Miyawaki A.. 2005. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 6:233–238. 10.1038/sj.embor.7400361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turing A.M. 1952. The chemical basis of morphogenesis. Philos. Trans. R. Soc. B Biol. Sci. 237:37–72. 10.1098/rstb.1952.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varner V.D., and Nelson C.M.. 2017. Computational models of airway branching morphogenesis. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.L., Menko A.S., Khalil S., Rebustini I., Hoffman M.P., Kreidberg J.A., and Kukuruzinska M.A.. 2008. Diverse roles of E-cadherin in the morphogenesis of the submandibular gland: Insights into the formation of acinar and ductal structures. Dev. Dyn. 237:3128–3141. 10.1002/dvdy.21717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D., El-Hashash A., Carraro G., Tiozzo C., Sala F., Rogers O., De Langhe S., Kemp P.J., Riccardi D., Torday J., et al. 2010. Lung organogenesis. Curr. Top. Dev. Biol. 90:73–158. 10.1016/S0070-2153(10)90003-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., and Costantini F.. 2004. Real-time analysis of ureteric bud branching morphogenesis in vitro. Dev. Biol. 271:98–108. 10.1016/j.ydbio.2004.03.025 [DOI] [PubMed] [Google Scholar]
- Wright A.V., Nuñez J.K., and Doudna J.A.. 2016. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell. 164:29–44. 10.1016/j.cell.2015.12.035 [DOI] [PubMed] [Google Scholar]
- Wu Y., Wawrzusin P., Senseney J., Fischer R.S., Christensen R., Santella A., York A.G., Winter P.W., Waterman C.M., Bao Z., et al. 2013. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat. Biotechnol. 31:1032–1038. 10.1038/nbt.2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates L.L., and Dean C.H.. 2011. Planar polarity: A new player in both lung development and disease. Organogenesis. 7:209–216. 10.4161/org.7.3.18462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., and Cui B.. 2015. Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 33:92–100. 10.1016/j.tibtech.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.