Abstract
Wild-type p53-induced phosphatase (Wip1) is an established oncogene and is associated with development of multiple forms of human cancer. However, the expression and role of Wip1 in human bladder transitional cell carcinoma (TCC) remains to be elucidated. In the present study, immunohistochemistry demonstrated that Wip1 was overexpressed in bladder TCC tissues compared with corresponding normal bladder tissues in 106 bladder TCC cases (P<0.0001). Furthermore, high expression levels of Wip1 were significantly associated with increasing tumor size (P=0.002), pathological grade (P=0.025), clinical T stage (P=0.001) and lymph nodal metastasis (P=0.003). Kaplan-Meier survival analysis identified that patients with high Wip1 expression levels exhibited a lower overall survival time (P<0.0001), and Cox proportional hazards regression model analysis demonstrated that Wip1 expression was an independent prognostic factor in patients with bladder TCC (P=0.025). In addition, downregulation of Wip1 expression by transfection with small interfering RNA in bladder cancer cells inhibited cell proliferation, invasion and migration (P<0.05), along with the upregulation of p53 protein levels (P<0.05). These findings suggest that Wip1 may function as a potential prognostic marker and therapeutic target in bladder cancer.
Keywords: Wip1, bladder transitional cell carcinoma, prognosis, proliferation, invasion, migration
Introduction
Bladder cancer is the second most common genitourinary malignancy worldwide (1). Over 90% of bladder cancer cases are transitional cell carcinoma (TCC). Approximately 25% of cases are diagnosed as muscle-invasive bladder cancer (MIBC), and ≤15% of patients with non-muscle-invasive bladder cancer will develop an invasive recurrent cancer within 1 year of treatment (2). Radical cystectomy is considered as the gold standard treatment for patients with MIBC. Although the techniques of surgery and chemoradiotherapy have improved, the 5-year survival rate of bladder TCC patients ranges from 48–66% (3). Furthermore, once patients develop distant metastases, the 5-year survival rate drops to only 6% (4). Better understanding of the molecular mechanisms and identifying a novel prognostic marker of bladder cancer may help to treat this disease.
Wip1 was first identified as a phosphatase that is induced by p53 in response to ionizing radiation (5). Wip1 is encoded by protein phosphatase magnesium-dependent 1 Δ, which is located on the 17q22/24 human chromosomal region, and is a member of the protein phosphatase type 2C (PP2C) family. Previous studies have demonstrated that Wip1 is overexpressed in various types of cancer and that Wip1 is associated with cancer development, suggesting that this protein may be a tumor biomarker and therapeutic target (6–9). However, the expression and role of Wip1 in bladder cancer have not yet been investigated.
The purpose of the present study was to determine the levels of Wip1 expression in patients with bladder cancer and the association between Wip1 expression and clinicopathological features. The effects of Wip1 knockdown on proliferation, invasion and migration in T24 (human bladder carcinoma cell line) cells and the underlying signaling pathways were also evaluated.
Materials and methods
Patients and tissue samples
A total of 106 formalin-fixed paraffin-embedded bladder TCC tissues and corresponding normal bladder tissues were collected from patients at the Department of Urology, Beijing Friendship Hospital, Capital Medical University (Beijing, China) who underwent surgical resection between January 2009 and December 2011. All patients were diagnosed with TCC by postoperative pathological examination. No patients were treated with radiotherapy or chemotherapy prior to surgery. Clinical data included age, gender, tumor size, pathological grade, lymph nodal status, tumor-node-metastasis (TNM) classification stage and patient survival time. Tumor staging was based on the seventh edition of the TNM classification system (2009) (10). Follow-up information was obtained using medical records and phone investigations. The date of the last follow-up was October 1, 2015. The study protocol was approved by the Ethics Committee of Beijing Friendship Hospital, Capital Medical University and written informed consent was obtained from all patients.
Immunohistochemistry
Paraffin-embedded tissue sections (4 µm thick) were deparaffinized, rehydrated and subjected to antigen retrieval by immersion in boiling citric acid buffer (pH 6.0; ZSGB-BIO, Beijing, China) for 10 min. Endogenous peroxidase and non-specific conjugation were blocked by 3% hydrogen peroxide and normal goat serum (ZSGB-BIO, Beijing, China). Sections were incubated with anti-Wip1 antibody (#ab31270; dilution 1:1,000; Abcam, Cambridge, UK) at 4°C overnight. Subsequently, sections were incubated with biotinylated anti-rabbit secondary antibody (#ZB-2010; dilution, 1:400; ZSGB-BIO) and streptavidin-horseradish peroxidase complex (ZSGB-BIO) at room temperature for 10 min. Finally, sections were stained with 3,3′-diaminobenzidine and counterstained with hematoxylin, then dehydrated and mounted. An immunoreactivity score was used to evaluate the staining of Wip1 expression in bladder TCC tissues and normal bladder tissues. Four randomly selected fields of each slide were counted using an inverted light microscope (DMi1; Leica Microsystems GmbH, Wetzlar, Germany). Wip1 expression was quantified according to the staining intensity and the percentage of positive cells. The staining intensity was defined as follows: 0 point, no staining; 1 point, weak staining; 2 points, moderate staining; and 3 points, strong staining. The percentage of positive cells was defined as follows: 1 point, ≤10%; 2 points, 11–50%; 3 points, 51–80%; and 4 points, ≥81%. The final points were defined as the points of the staining intensity multiplied by those of the percentage of positive cells. Wip1 expression was divided into the following categories: No expression, 0–2 points; positive expression, ≥3 points (low expression, 3–6 points and high expression, >6 points). Two pathologists (Department of Pathology, Beijing Friendship Hospital, Capital Medical University) assessed these results independently.
Cell culture and small interfering RNA (siRNA) transfection
Human bladder T24 cells were purchased from the Cell Resource Center, Peking Union Medical College (Beijing, China) and were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in a humidified incubator at 37°C with 5% CO2. siRNA for Wip1 and negative control siRNA were designed and synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China), according to the NM_003620 gene sequence in the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov/nuccore/312434022/). The sequences of Wip1-siRNA were as follows: Sense, 5′-AGGUGACACAGGACCAUAAdTdT-3′, antisense, 3′-dTdTUCCACUGUGUCCUGGUAUU-5′ and target sequence, 5′-AGGTGACACAGGACCATAA-3′. T24 cells were transfected using Lipofectamine® 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. When the cells reached 30–50% confluence in 6-well plates, the medium was transferred to Opti-MEM® Reduced-Serum medium (Invitrogen; Thermo Fisher Scientific, Inc.) containing the complexes of Wip1-siRNA and Lipofectamine 2000. Cells were incubated for 6 h, then the medium was switched to DMEM supplemented with 10% FBS.
Reverse transcription quantitative-polymerase chain reaction (RT-qPCR)
Total RNA was extracted with the RNeasy® Mini kit (Qiagen Inc., Valencia, CA, USA) at 24 h post-transfection, according to the manufacturer's protocol. Total RNA (1 µg) was reverse transcribed into cDNA using a PrimeScript™ 1st Strand cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China) on an ABI Veriti™ Thermal Cycler (Applied Biosystems; Thermo Fisher Scientific, Inc.). RT-qPCR was conducted in 20 µl reaction buffer with SYBR® Premix DimerEraser™ (Perfect Real Time) (Takara Biotechnology Co., Ltd., Dalian, China) using the ABI® 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The following primer sequences were used: Wip1 forward, 5′-GGAAGAAACTGGCGGAATG-3′ and reverse, 5′-TGGGAAGTTCTGGCTTATGG-3′; GAPDH, forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse, 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. GAPDH was used as the internal control. RT-qPCR was performed under the following conditions: 95°C for 30 sec, 40 cycles at 95°C for 3 sec, 60°C for 30 sec and 72°C for 30 sec. The melting curve was analyzed for each sample. All samples were performed in triplicate. The 2-ΔΔCq method was used to calculate the relative mRNA levels of Wip1 (11).
Western blot analysis
Western blotting was performed at 48 h post-transfection. The harvested T24 cells were washed with ice-cold PBS and were lysed in radio immunoprecipitation assay lysis buffer containing phenylmethanesulfonyl fluoride (Beyotime Institute of Biotechnology, Haimen, China) at 4°C for 30 min. The cells were subsequently centrifuged at 12,000 × g at 4°C for 10 min, and the supernatant was taken and protein concentration detected using the BCA protein assay kit (Beyotime Institute of Biotechnology). Total protein (40 µg) was separated by 10% SDS-PAGE, then transferred to a nitrocellulose filter membrane. Following blocking with 5% non-fat dry milk for 2 h at room temperature, the membranes were incubated with the following primary antibodies: Anti-Wip1 antibody (#ab31270; dilution, 1:800; Abcam), anti-P53 antibody (#ab179477; dilution, 1:1,000; Abcam) and anti-GAPDH antibody (#ab181602; dilution, 1:1,000; Abcam) at 4°C overnight. Subsequently, the membranes were incubated with peroxidase-conjugated affinipure goat anti-rabbit secondary antibody (#ZB-5301; dilution 1:5,000; Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China) at room temperature for 2 h. Following washing, protein bands were detected using the Bio-Rad ChemiDoc™ XRS+ system (Bio-Rad Laboratories, Co., Ltd, Hercules, CA, USA) using Image Lab™ software (Bio-Rad Laboratories, Co., Ltd.). GAPDH was used as the protein control and all experiments were performed in triplicate.
MTT assay
The T24 cells were plated in 96-well plates at a density of 2,000 cells/well in 100 µl DMEM supplemented with 10% FBS. Following transfection with Wip1-siRNA or negative control siRNA, the cells were cultured for 0, 24, 48 and 72 h. A total of 10 µl 5 mg/ml MTT was added to each well and the cells were incubated at 37°C for 4 h. Next, 100 µl formazan solution was added to each well and the cells were incubated at 37°C for an additional 4 h. The absorption value was measured at 570 nm using a SpectraMax® M3 Multi-Mode Microplate Reader (Molecular Devices, LLC, Sunnyvale, CA, USA). All reactions were performed in triplicate.
Invasion and migration assays
The 24-well (8 µm pore) Transwell® cell culture inserts (Corning Inc., Corning, NY, USA) were coated with 50 µl Matrigel™ Basement Membrane Matrix (BD Biosciences, San Jose, CA, USA). Following transfection with Wip1-siRNA or negative control siRNA, 1×105 cells were plated in the upper chamber, and cultured with serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.). DMEM containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was placed in the lower chamber. Following incubation at 37°C for 24 h, the cells and medium in the upper chamber were removed with a cotton swab. Polyoxymethylene was added to the lower chamber to fix the cells attached to the lower membrane surface, and 0.1% crystal violet was added to the lower chamber to stain the cells. Following this, crystal violet staining was decolorized using 33% acetic acid. The absorption value was measured at 570 nm to indicate cell invasion. Migration assays were performed using the aforementioned method without Matrigel. All experiments were performed in triplicate.
Statistical analysis
All statistical analyses were performed using SPSS version 19.0 (IBM SSPS, Armonk, New York, USA). Values were expressed as the mean ± standard deviation. For the clinical characteristics, P-values were calculated using the χ2 test. Survival outcomes were estimated with the Kaplan-Meier method and then compared using the log-rank test. The Cox proportional hazards regression model was used to define factors predicting prognosis and the Student's t-test was used to evaluate the significance of differences between two groups. P≤0.05 was considered to indicate a statistically significant difference.
Results
Wip1 is overexpressed in bladder TCC tissues
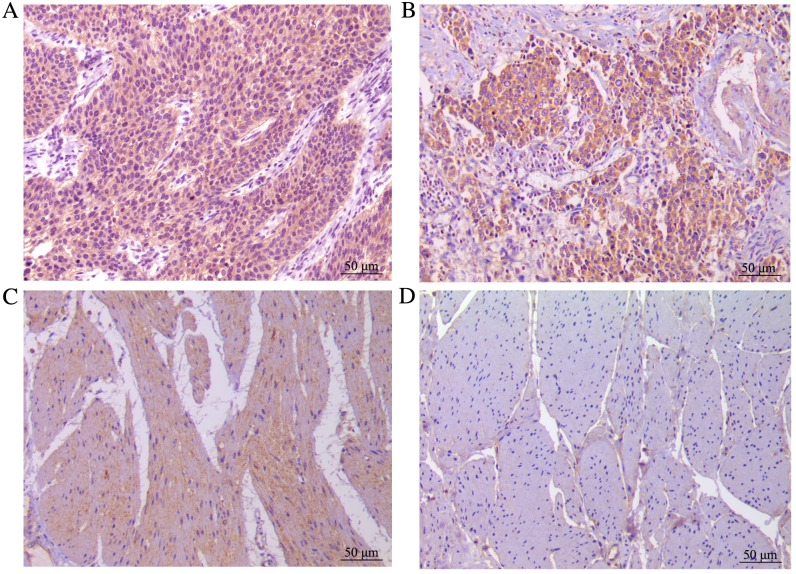
In the present study, 106 bladder TCC tissues and corresponding normal bladder tissues were analyzed using immunohistochemistry. Wip1 expression was significantly higher in the bladder TCC tissues (99/106, 93.4%) than in the normal tissues (13/106, 12.3%; P<0.0001). In addition, the expression rate of Wip1 in the low and high groups was 48.1 and 45.3%, respectively, in the bladder TCC tissues. In normal tissues, Wip1 was expressed at a low level or not expressed at all. Wip1 staining is presented in Fig. 1.
Figure 1.
Immunohistochemical analysis of Wip1 expression in the bladder TCC tissues and corresponding normal bladder tissues. (A) High expression of Wip1 in bladder TCC tissue. Low expression of Wip1 in (B) bladder TCC and (C) normal bladder tissues. (D) No expression of Wip1 in normal bladder tissue. Magnification, ×200; scale bar, 50 µm. TCC, transitional cell carcinoma.
Wip1 expression is associated with clinicopathological features and prognosis
To analyze the correlation between Wip1 expression and clinical data, the patients were divided into low (including no expression) and high expression groups based on immunohistochemical points of bladder TCC tissues. High expression levels of Wip1 were significantly associated with increased tumor size (P=0.002), pathological grade (P=0.025), clinical T stage (P=0.001) and lymph nodal metastasis (P=0.003), but not with gender and age (Table I). The Kaplan-Meier survival curves and the log-rank tests demonstrated that the overall survival time of patients in the high expression group was significantly lower, compared with patients in the low expression group (P<0.0001; Fig. 2). In the Cox proportional hazards regression model analyses for prognosis, clinical T stage (P=0.004), lymph nodal metastasis (P=0.001) and Wip1 expression (P=0.025) were independent prognostic factors in patients with bladder TCC (Table II).
Table I.
Association between wild-type p53-induced phosphatase expression levels and the clinicopathological features of patients with bladder transitional cell carcinoma.
| Clinicopathological features | Total | Low | High | P-value |
|---|---|---|---|---|
| Gender | 0.144 | |||
| Male | 84 | 49 | 35 | |
| Female | 22 | 9 | 13 | |
| Age, years | 0.526 | |||
| ≥60 | 74 | 39 | 35 | |
| <60 | 32 | 19 | 13 | |
| Tumor size, cm | 0.002 | |||
| ≥3 | 51 | 20 | 31 | |
| <3 | 55 | 38 | 17 | |
| Pathological grade | 0.025 | |||
| G1 | 21 | 13 | 8 | |
| G2 | 56 | 33 | 23 | |
| G3 | 29 | 12 | 17 | |
| Clinical T stage | 0.001 | |||
| Ta+Tis+T1 | 43 | 32 | 11 | |
| T2-T4 | 63 | 26 | 37 | |
| Lymph nodal metastasis | 0.003 | |||
| N0 | 73 | 48 | 25 | |
| N1 | 22 | 7 | 15 | |
| N2 | 11 | 3 | 8 |
Figure 2.
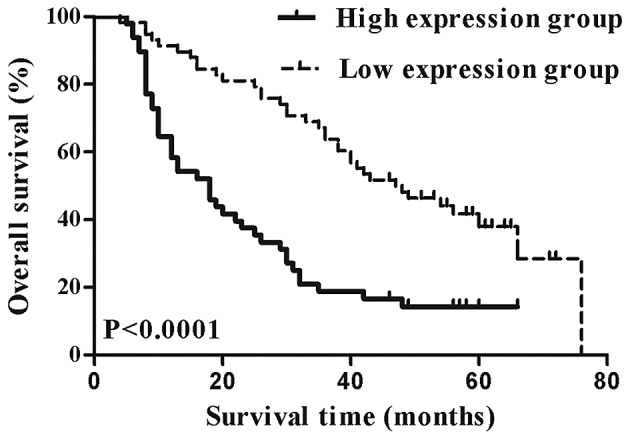
Kaplan-Meier survival analysis of the overall survival time of patients with bladder transitional cell carcinoma. The overall survival time of patients in the high expression group was significantly lower, compared with patients in the low expression group (P<0.0001 vs. low expression group).
Table II.
Cox proportional hazards regression model analyses of Wip1 expression and other clinical prognostic factors in patients with bladder transitional cell carcinoma.
| Cox regression analysis | ||
|---|---|---|
| Variable | HR (95% CI) | P-value |
| Gender (male/female) | 0.74 (0.41–1.32) | 0.302 |
| Age (≥60/<60 years) | 1.06 (0.62–1.79) | 0.842 |
| Tumor size (≥3/<3 cm) | 0.91 (0.55–1.51) | 0.709 |
| Pathological grade (G1+G2/G3) | 1.79 (0.92–3.48) | 0.087 |
| Clinical T stage (Ta+Tis+T1/T2-T4) | 2.49 (1.33–4.64) | 0.004 |
| Lymph nodal metastasis (N0/N+) | 3.34 (1.60–6.98) | 0.001 |
| Wip1 expression (high/low) | 1.86 (1.02–3.18) | 0.025 |
Wip1, wild-type p53-induced phosphatase; HR, hazard ratio; CI, confidence interval.
Downregulation of Wip1 decreases T24 cell proliferation, invasion and migration
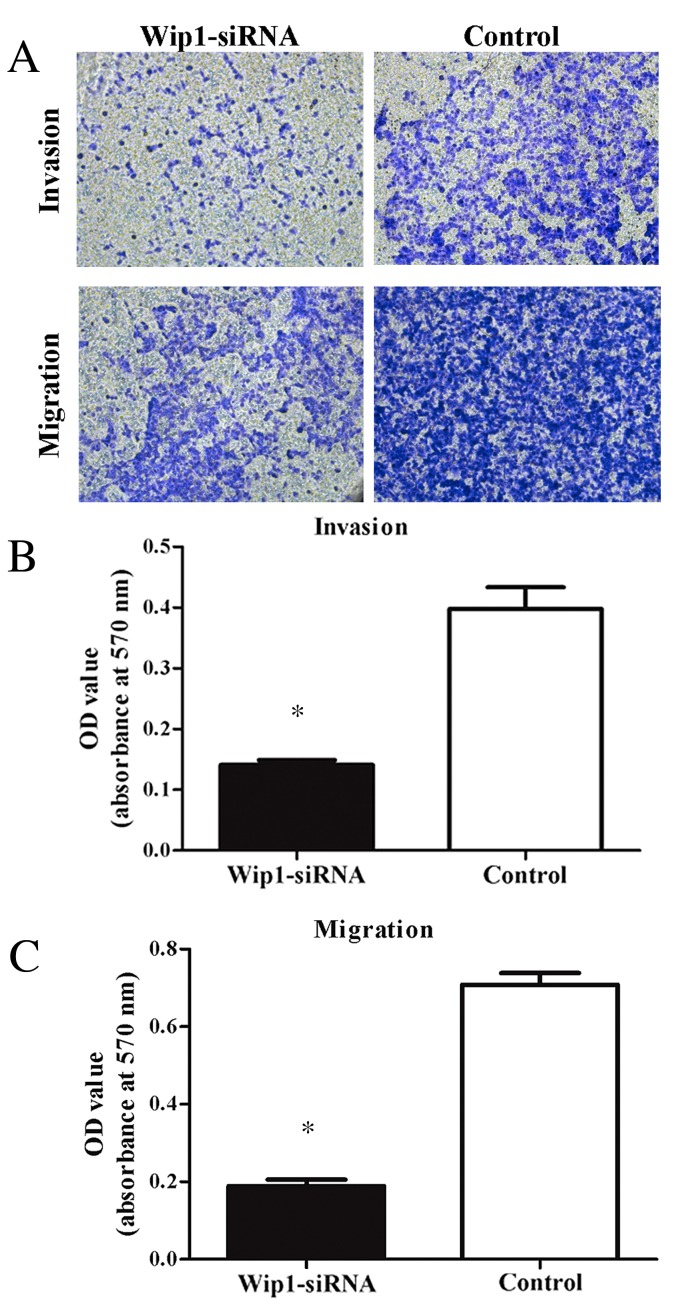
To assess the effects of Wip1 on TCC cell proliferation, invasion and migration, Wip1-siRNA or negative control siRNA was transiently transfected into T24 cells. RT-qPCR and western blot analyses demonstrated that Wip1 mRNA and protein expression levels in the Wip1-siRNA-transfected T24 cells were significantly lower compared with the control group (mRNA, P=0.019; protein, P<0.001; Fig. 3A-C). MTT assays indicated that Wip1-siRNA treatment significantly inhibited T24 cell proliferation at 24 h (P=0.005), 48 h (P=0.002) and 72 h (P<0.001) post-transfection (Fig. 4). Furthermore, invasion and migration abilities were significantly reduced in the Wip1-siRNA-transfected T24 cells when compared with the controls (invasion and migration, P<0.001; Fig. 5). To further examine the potential signaling pathways mediating the aforementioned functions of Wip1, the expression levels of p53 (a tumor suppressor) were analyzed. Western blot analysis demonstrated that p53 protein levels were significantly upregulated in the Wip1-siRNA-transfected T24 cells (P=0.026; Fig. 3B and D).
Figure 3.
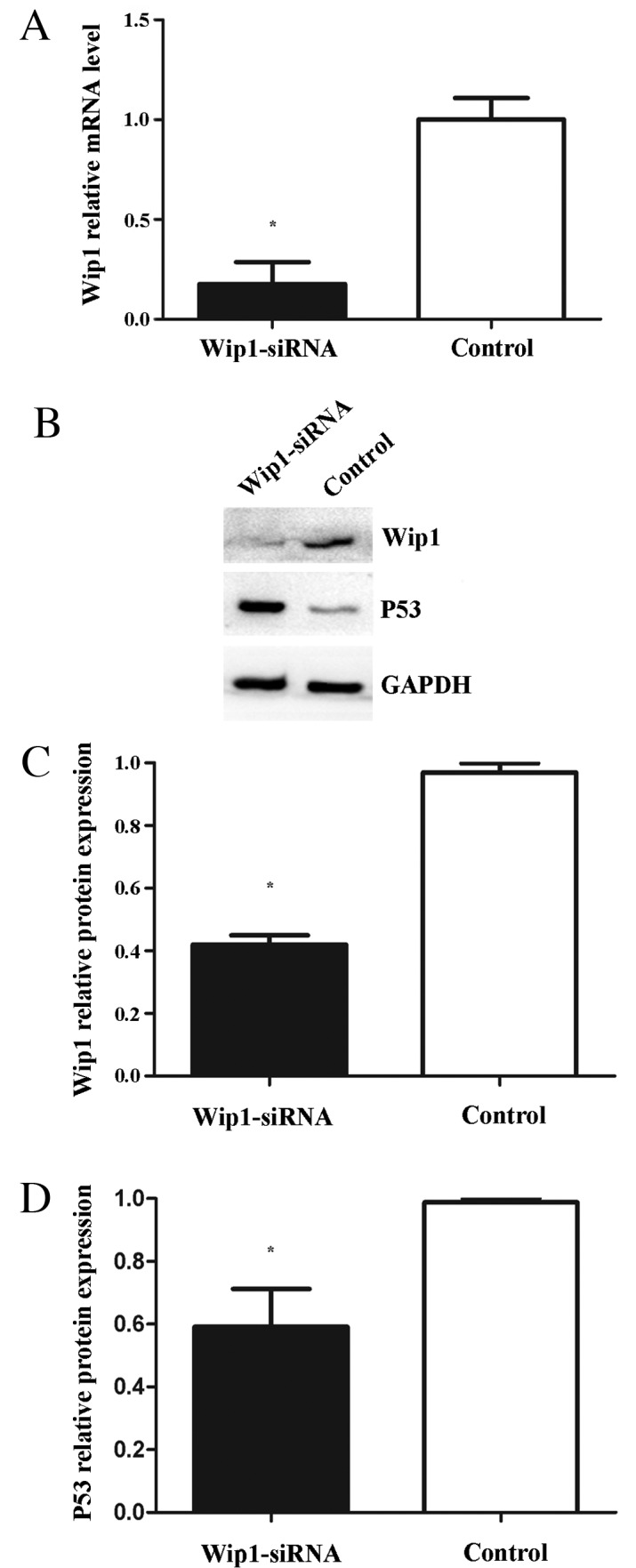
(A) Reverse transcription-quantitative polymerase chain reaction analysis of Wip1 mRNA expression in T24 cells. (B and C) Western blot analysis of Wip1 protein expression levels in T24 cells. (B and D) Western blot analysis of p53 protein expression levels in T24 cells. Wip1 mRNA expression was downregulated in Wip1-siRNA-transfected cells (A, *P<0.05 vs. control). Wip1 protein expression was downregulated in Wip1-siRNA-transfected cells (B and C; *P<0.05 vs. control). P53 protein expression was upregulated in the Wip1-siRNA-transfected cells (B and D; *P<0.05 vs. control). Wip1, wild-type p53-induced phosphatase; siRNA, small interfering RNA.
Figure 4.
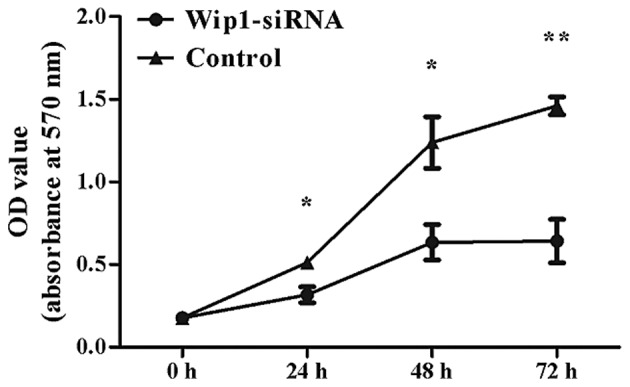
MTT assay analysis of cell proliferation. Downregulation of Wip1 inhibited T24 cells proliferation at 24, 48 and 72 h, compared with the control group (*P<0.05 and **P<0.0001 vs. control). Wip1, wild-type p53-induced phosphatase; siRNA, small interfering RNA; OD, optical density.
Figure 5.
Transwell assay analysis of cell invasion and migration. Downregulation of Wip1 inhibited T24 cell invasion and migration, compared with the control group (*P<0.05 vs. control). Magnification, ×200. Wip1, wild-type p53-induced phosphatase; siRNA, small interfering RNA; OD, optical density.
Discussion
Bladder cancer is a leading global cause of cancer morbidity and mortality (1,12). In 2016, 76,960 new cases and 16,390 mortalities are predicted in the United States (4). However, the molecular mechanisms underlying tumorigenesis and the progression of bladder cancer remain to be elucidated.
Wip1, an established oncogene, has been the subject of a number of studies since it was identified in 1997 (5,13–15). It has been reported that Wip1 negatively regulates DNA damage response pathways by dephosphorylating several key proteins, including p53, p16, ataxia telangiectasia mutated, checkpoint kinase 1 and p38 mitogen-activated protein kinases (MAPKs), resulting in tumorigenesis (16,17). In addition, the Wip1-p53 signaling pathway is well researched in the DNA damage response system. The tumor suppressor p53 serves an essential role in DNA damage response pathways by regulating cell cycle arrest and apoptosis (18). It is known that the dysfunction of p53 has been associated with the progression, prognosis and therapeutic response of tumors (19). Wip1 is not only able to directly dephosphorylate p53 protein at serine 15, but also indirectly inactivates p53 protein via p38, MAPK and Mdm2 (16,20,21). Therefore, restoring p53 function by regulating Wip1 may be a potential therapeutic approach. Furthermore, previous studies on solid tumors demonstrated that Wip1 was associated with poor prognosis in patients with nasopharyngeal carcinoma (8), liver and (9) prostate cancer (22) and lung adenocarcinoma (23). However, little is understood regarding Wip1 expression levels in bladder cancer and its association with clinicopathological features.
To the best of our knowledge, the present study is the first to investigate the expression of Wip1 in patients with bladder TCC. A total of 106 bladder TCC and corresponding normal bladder tissues were analyzed by immunohistochemistry. The data demonstrated that Wip1 was overexpressed in the bladder TCC tissues, compared with the normal tissues. Furthermore, high expression levels of Wip1 were positively associated with more aggressive tumor characteristics, including tumor size, pathological grade, clinical T stage and lymph nodal metastasis. To determine the correlation between Wip1 expression and prognosis, Kaplan-Meier survival curve and the Cox proportional hazards regression model analyses were implemented. The results revealed that high levels of Wip1 expression led to a shorter overall survival time, and that Wip1 expression was an independent prognostic factor. These results suggest that Wip1 expression may be a potential prognostic marker for patients with bladder cancer.
To assess the effects of Wip1 in bladder cancer, further in vitro experiments were performed. Human bladder T24 cells were transfected with Wip1-siRNA to downregulate mRNA and protein expression levels of Wip1. The results demonstrated that cell proliferation, invasion and migration were decreased in the Wip1-siRNA-transfected T24 cells compared with the control group, indicating that Wip1 may be a potential therapeutic target. Furthermore, levels of p53 protein expression were upregulated in the Wip1-siRNA-transfected T24 cells, similar to previous studies (24,25). These results suggest that the downregulation of Wip1 expression may inhibit bladder cancer cell proliferation, invasion and migration via activation of the p53 pathway. However, mutations of p53 were observed in more than half of patients with invasive bladder TCC (26). Whether Wip1 is able to restore functional p53 protein requires elucidation in future studies.
In conclusion, the current study demonstrated that Wip1 is overexpressed in patients with bladder TCC and that high levels of Wip1 expression were positively correlated with more aggressive tumor characteristics and poorer prognosis. Downregulation of Wip1 expression inhibited bladder cancer cell proliferation, invasion and migration by activating the p53 pathway in the T24 cells. Therefore, these findings indicate that Wip1 may serve as a potential prognostic marker and therapeutic target in bladder cancer.
References
- 1.Burger M, Catto J, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La Vecchia C, Shariat S, Lotan Y. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63:234–241. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 2.Boccardo F, Palmeri L. Adjuvant chemotherapy of bladder cancer. Ann Oncol. 2006;17:v129–v132. doi: 10.1093/annonc/mdj967. (Suppl 5) [DOI] [PubMed] [Google Scholar]
- 3.Stein JP, Lieskovsky G, Cote R, Groshen S, Feng AC, Boyd S, Skinner E, Bochner B, Thangathurai D, Mikhail M, et al. Radical cystectomy in the treatment of invasive bladder cancer: Long-term results in 1054 patients. J Clin Oncol. 2001;19:666–675. doi: 10.1200/JCO.2001.19.3.666. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–301. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 5.Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Woude GF Vande, O'Connor PM, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Chen LH, Wan H, Yang R, Wang Z, Feng J, Yang S, Jones S, Wang S, Zhou W, et al. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat Genet. 2014;46:726–730. doi: 10.1038/ng.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter M, Dayaram T, Gilmartin AG, Ganji G, Pemmasani SK, Van Der Key H, Shohet JM, Donehower LA, Kumar R. Wip1 phosphatase as a potential therapeutic target in neuroblastoma. PLoS One. 2015;10:e0115635. doi: 10.1371/journal.pone.0115635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun GG, Zhang J, Ma XB, Wang YD, Chen YJ, Hu WN. Overexpression of wild-type p53-induced phosphatase1 confers poor prognosis of patients with nasopharyngeal carcinoma. Pathol Oncol Res. 2015;21:283–291. doi: 10.1007/s12253-014-9819-1. [DOI] [PubMed] [Google Scholar]
- 9.Li GB, Zhang XL, Yuan L, Jiao QQ, Liu DJ, Liu J. Protein phosphatase magnesium-dependent 1δ (PPM1D) mRNA expression is a prognosis marker for hepatocellular carcinoma. PLoS One. 2013;8:e60775. doi: 10.1371/journal.pone.0060775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobin L, Gospodarowicz M, Wittekind C. UICC International Union Against Cancer. 7th. Wiley-Blackwell; 2009. TNM classification of malignant tumors; pp. 262–265. [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Witjes JA, Compérat E, Cowan NC, De Santis M, Gakis G, Lebret T, Ribal MJ, Van der Heijden AG, Sherif A. European Association of Urology: EAU guidelines on muscle-invasive and metastatic bladder cancer: Summary of the guidelines. Eur Urol. 2014;65:778–792. doi: 10.1016/j.eururo.2013.11.046. [DOI] [PubMed] [Google Scholar]
- 13.Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell. 2006;23:757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto H, Onishi N, Kato N, Takekawa M, Xu XZ, Kosugi A, Kondo T, Imamura M, Oishi I, Yoda A, Minami Y. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ. 2006;13:1170–1180. doi: 10.1038/sj.cdd.4401801. [DOI] [PubMed] [Google Scholar]
- 16.Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: An oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008;27:123–135. doi: 10.1007/s10555-008-9127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe J, Cha H, Lee MO, Mazur SJ, Appella E, Fornace AJ., Jr Regulation of the Wip1 phosphatase and its effects on the stress response. Front Biosci (Landmark Ed) 2012;17:1480–1498. doi: 10.2741/3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, Olivier M, Hall J, Mollereau B, Hainaut P, Bourdon JC. Biological functions of p53 isoforms through evolution: Lessons from animal and cellular models. Cell Death Differ. 2011;18:1815–1824. doi: 10.1038/cdd.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfister C, Flaman JM, Dunet F, Grise P, Frebourg T. p53 mutations in bladder tumors inactivate the transactivation of the p21 and Bax genes, and have a predictive value for the clinical outcome after bacillus Calmette-Guerin therapy. J Urol. 1999;162:69–73. doi: 10.1097/00005392-199907000-00017. [DOI] [PubMed] [Google Scholar]
- 20.Crescenzi E, Raia Z, Pacifico F, Mellone S, Moscato F, Palumbo G, Leonardi A. Down-regulation of wild-type p53-induced phosphatase 1 (Wip1) plays a critical role in regulating several p53-dependent functions in premature senescent tumor cells. J Biol Chem. 2013;288:16212–16224. doi: 10.1074/jbc.M112.435149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–354. doi: 10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 22.Jiao L, Shen D, Liu G, Jia J, Geng J, Wang H, Sun Y. PPM1D as a novel biomarker for prostate cancer after radical prostatectomy. Anticancer Res. 2014;34:2919–2925. [PubMed] [Google Scholar]
- 23.Satoh N, Maniwa Y, Bermudez VP, Nishimura K, Nishio W, Yoshimura M, Okita Y, Ohbayashi C, Hurwitz J, Hayashi Y. Oncogenic phosphatase Wip1 is a novel prognostic marker for lung adenocarcinoma patients survival. Cancer Sci. 2011;102:1101–1106. doi: 10.1111/j.1349-7006.2011.01898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun GG, Wang YD, Liu Q, Hu WN. Expression of Wip1 in kidney carcinoma and its correlation with tumor metastasis and clinical significance. Pathol Oncol Res. 2015;21:219–224. doi: 10.1007/s12253-014-9811-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang HY, Liu ZS, Qiu L, Guo J, Li YF, Zhang J, Wang TJ, Liu XD. Knockdown of Wip1 enhances sensitivity to radiation in HeLa cells through activation of p38 MAPK. Oncol Res. 2014;22:225–233. doi: 10.3727/096504015X14386062091479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner U, Sauter G, Moch H, Novotna H, Epper R, Mihatsch MJ, Waldman FM. Patterns of p53, erbB-2, and EGF-r expression in premalignant lesions of the urinary bladder. Hum Pathol. 1995;26:970–978. doi: 10.1016/0046-8177(95)90086-1. [DOI] [PubMed] [Google Scholar]