ABSTRACT
A previous study showed that Nitrogen-Fixing-subunit-U-type protein NFU3 may act an iron-sulfur scaffold protein in the assembly and transfer of 4Fe-4S and 3Fe-4S clusters in the chloroplast. Examples of 4Fe-4S and 3Fe-4S-requiring proteins and complexes include Photosystem I (PSI), NAD(P)H dehydrogenase, and ferredoxin-dependent glutamine oxoglutarate aminotransferases. In this paper, the authors provided additional evidence for the role of NFU3 in 4Fe-4S and 3Fe-4S cluster assembly and transfer, as well as its role in overall plant fitness. Confocal microscopic analysis of the fluorescently-tagged NFU3 protein confirmed the chloroplast localization of the NFU3 protein. Detailed analysis of chlorophyll fluorescence data revealed that a substantial increase in minimal fluorescence is the primary contributor to the decrease in PSII maximum photochemical efficiency observed in the nfu3 mutants. The substantial increase in minimal fluorescence in the nfu3 mutants is probably the result of an impaired PSI function, blockage of electron flow from PSII to PSI, and over-accumulation of reduced plastoquinone at the acceptor side of PSII. Analyses of seed morphology and germination showed that NFU3 is essential to seed development and germination, in addition to plant growth, development, and flowering. In summary, NFU3 has wide-ranging effects on many biologic processes and is therefore important to overall plant fitness. NFU3 may exert these effects by modulating the availability of 4Fe-4S and 3Fe-4S clusters to 4Fe-4S and 3Fe-4S-requiring proteins and complexes involved in various biologic processes.
KEYWORDS: Arabidopsis thaliana, assembly and transfer of iron-sulfur clusters, iron-sulfur scaffold protein, overall plant fitness, nitrogen fixation subunit, seed development, seed germination
Introduction
Iron-sulfur clusters are involved in various vital biologic processes including; photosynthesis, respiration, sulfur and nitrogen assimilation, amino acid and purine metabolism, chlorophyll and cofactor metabolism, hormone synthesis, DNA replication and repair, ribosome biogenesis, tRNA maturation, and translation.1-3 Iron-sulfur clusters may contain different numbers of iron and sulfur ions. The 4 most common iron-sulfur clusters in plants are: classic 2Fe-2S, Rieske-type 2Fe-2S, 3Fe-4S, and 4Fe-4S clusters.1-3 Three iron-sulfur assembly and transfer pathways have been identified in plants: the SUF (sulfur mobilization) pathway in plastids, the ISC (iron-sulfur cluster) pathway in mitochondria, and the CIA (cytosolic iron-sulfur) assembly pathway.1-3 The SUF pathway is responsible for the assembly and transfer of classic 2Fe-2S, Rieske-type 2Fe-2S, 3Fe-4S, and 4Fe-4S clusters in plastids; the ISC pathway is in charge of assembly and transfer of classic 2Fe-2S, 3Fe-4S, and 4Fe-4S clusters in mitochondria; and the CIA pathway assembles and transfers classic 2Fe-2S and 4Fe-4S clusters in the cytosol.1 In all 3 pathways, the process can be divided into 2 steps: (1) assembly of iron-sulfur clusters on an iron-sulfur scaffold protein by cysteine desulfurase, and (2) subsequent transfer of iron-sulfur clusters from the iron-sulfur scaffold protein to recipient apoproteins.1-3
Recently, it was found that Nitrogen-Fixing-subunit-U-type protein NFU3 acts as a scaffold protein in the assembly and transfer of 4Fe-4S and 3Fe-4S clusters in the chloroplast.4 The recombinant NFU3 protein showed characteristics of 4Fe-4S and 3Fe-4S clusters and an in vitro activity of iron-sulfur cluster reconstitution. The loss-of-function nfu3 mutants of Arabidopsis (Arabidopsis thaliana) had reduced amounts of 4Fe-4S-containing PSI core subunits PsaA, PsaB, and PsaC (where Psa stands for PSI) and 3Fe-4S-containing ferredoxin-dependent glutamine oxoglutarate aminotransferases (Fd-GOGATs). The ∼90% reduction in the levels of 4Fe-4S-containing PSI core subunits resulted in a near-complete loss of PSI activity and a substantial decrease in Fv/Fm, the maximum quantum efficiency of PSII photochemistry.
In this paper, the authors confirmed the chloroplast localization of the NFU3 protein, identified the causal parameters for reduced Fv/Fm in the nfu3 mutants, and analyzed the morphology and germination rate of the nfu3 seeds. These results demonstrated that NFU3 is essential to plant growth and development, flowering, seed development, and germination.
Analysis of the NFU3-Cerulean Fluorescent Protein (CFP) fusion protein confirmed the chloroplast localization of NFU3
According to the Arabidopsis eFP browser, NFU3 is highly expressed in leaves, flowers, and early stage embryos (Fig. S1).5 Based on TargetP prediction,6 NFU3 is located in the plastid. To test this prediction, the full-length NFU3 coding region without the stop codon (NFU31–708 bp, corresponding to NFU31–236 AA) was inserted between the cauliflower mosaic virus 35S RNA promoter and the CFP coding sequence of a binary vector (pPH5ADEST-CFP) suitable for transient expression. The Agrobacterium tumefaciens culture containing the fusion protein construct (pPH5ADEST-NFU3-CFP) was used to inoculate Nicotiana benthamiana leaves, as described previously.7 Transformed leaf tissues were observed using a Nikon C2+ confocal laser scanning microscope and a CFI Apochromat 60x oil immersion objective. Chlorophyll red autofluorescence and NFU3-CFP fusion protein analysis confirmed that NFU3 is indeed located in the chloroplast (Fig. 1). Consistent with our results, confocal microscopic analysis of a fusion protein containing the N-terminal region of NFU3 and the full-length Green Fluorescent Protein (GFP) expressed in Arabidopsis protoplasts also demonstrated the chloroplast localization of NFU3.8
Figure 1.
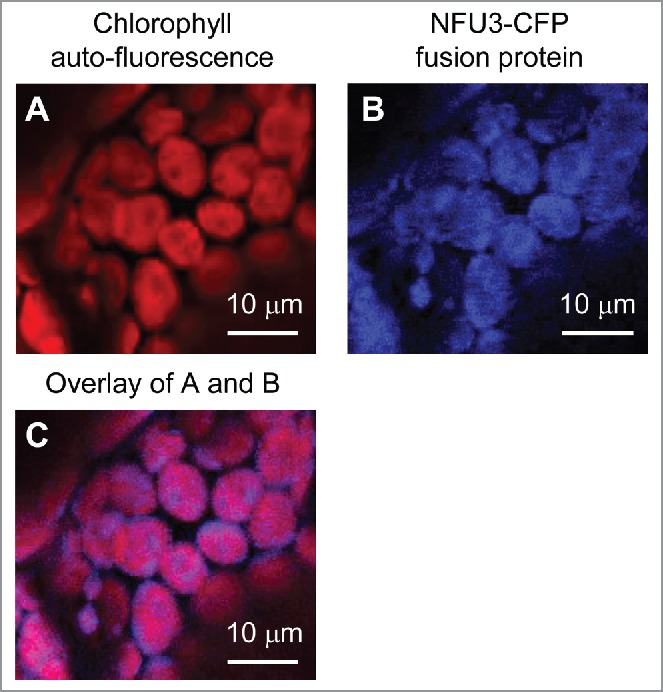
Localization of an NFU3–CFP fusion protein during transient expression in Nicotiana benthamiana leaf cells. (A) Chlorophyll auto-fluorescence of N. benthamiana mesophyll cells transiently expressing the NFU3-CFP fusion protein. Bars = 10 μm. (B) CFP fluorescence of N. benthamiana mesophyll cells transiently expressing the NFU3-CFP fusion protein. (C) Overlay of chlorophyll auto-fluorescence and CFP fluorescence of N. benthamiana mesophyll cells transiently expressing the NFU3-CFP fusion protein.
The nfu3 mutants had increased minimal, maximal and variable fluorescence
Previous studies have found that the calculated Fv/Fm ratio in the nfu3 mutants was 13% lower than that observed in the wild type.4 A low Fv/Fm (Fv/Fm = [Fm – Fo]/Fm = 1 – Fo/Fm) value could be the result of a low Fv (variable fluorescence) or a high Fo (minimal fluorescence) value. A low Fv value is indicative of an inability of PSII to perform primary photochemistry.9 A high Fo value could be the result of an over-accumulation of reduced QA (bound primary plastoquinone electron acceptor of PSII) in the dark.10 To identify the causal parameter(s) for reduced Fv/Fm in the nfu3 mutants, we determined the minimal, maximal, and variable fluorescence of dark-adapted wild-type and nfu3 plants (Fig. 2). We found that Fo in the nfu3 mutants (0.21 on average) was 190% higher than that in the wild type (0.07), Fm in the nfu3 mutants (0.75 on average) was 80% higher than that in the wild type (0.42), and Fv in the nfu3 mutants (0.54 on average) was 57% higher than that in the wild type (0.35). These data indicate that PSII in the nfu3 mutants is not defective and that the substantial increase in Fo is a major contributor of the observed decrease in Fv/Fm in the nfu3 mutants. The significant increase of the Fo value observed in the nfu3 mutants suggests that reduced QA might be over-accumulated.10 Consistent with this hypothesis, the redox state of PSII acceptor side (1-qP) in the nfu3 mutants was much higher than that in the wild type.4 Over-accumulation of reduced QA and increases in the redox state of the PSII acceptor side could be caused by a defect in intersystem electron transport or PSI.11-13 The near-complete loss of PSI activity in the nfu3 mutants strongly suggests that these observations are initially caused by the defects in PSI. In summary, these data provide additional support that, in the nfu3 mutants, PSI suffers greatly from the lack of the 4Fe-4S cluster and the reduction in PSII activity is likely a secondary effect of the near-complete loss of PSI activity.4
Figure 2.
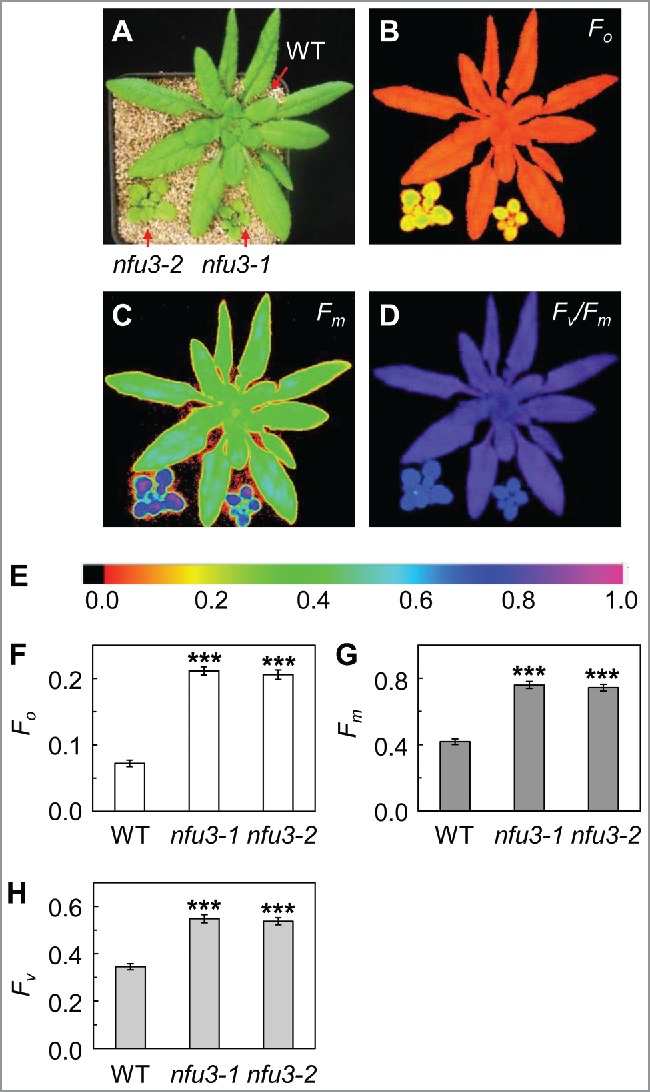
Minimal and maximal fluorescence were increased in the nfu3 mutants. (A) Images of 4-week-old wild-type, nfu3–1, and nfu3–2 plants. (B-D) False-color Fo, Fm, and Fv/Fm images of 4-week-old plants, respectively. (E) Color scale for chlorophyll fluorescence parameters F0, Fm, and Fv/Fm in B-D. F-H, Fo, Fm, and Fv values of 4-week-old plants. Data are presented as mean ± SE (n = 4). The asterisk indicates a significant difference between the mutant and the wild type (WT) (Student's t test; *, p < 0.05; **, p < 0.01; ***, p < 0.001). Plants used for chlorophyll fluorescence analysis were grown on a 12-h light/12-h dark photoperiod with an irradiance of 150 μmol photons m−2 s−1 during the light period.
The nfu3 mutants displayed retarded growth and delayed flowering and seeding
The nfu3 mutants were much smaller than wild-type plants at the same age (Fig. 2A), indicative of retarded growth. After 6 weeks under a 12-h-light /12-h-dark photoperiod, the Columbia wild-type plants flowered, while the nfu3 mutants continued to stay in a vegetative growth stage (see Fig. 1E in ref. 4). Consistent with retarded growth and delayed flowering, the amount of seeds produced by the nfu3 mutants was approximately 15% of those produced by the wild type (see Fig. 1G in ref. 4). These observations strongly suggest that NFU3 is essential to the normal growth, flowering, and seeding of Arabidopsis plants. Similar defects in growth, flowering, and seeding were also observed in the loss-of-function Arabidopsis mutants of NFU2, which have previously been shown to be required for the assembly and transfer of 4Fe-4S and classic 2Fe-2S clusters in the chloroplast.14,15
The nfu3 seeds had reduced weight and reduced germination
Loss-of-function mutants of several other genes involved in the assembly and transfer of iron-sulfur clusters showed abnormal seed morphology and germination;2,3,16-18 therefore we analyzed the morphology and germination of the nfu3 seeds (Fig. 3). The tip of the nfu3 siliques tended to be thin and filled with small, dark brown, and wrinkled seeds (Fig. 3A–B). Consequently, the average nfu3 seed weight was approximately 48% of wild-type seed weight (Fig. 3C). The small, dark brown, and wrinkled seed morphology has also been observed in other Arabidopsis mutants with defects in iron-sulfur assembly and transfer. For example, loss-of-function mutants of plastid-targeted iron-sulfur scaffold protein SufD/NAP6 and dual (mitochondrion and cytosol)-targeted DnaJ-like co-chaperone protein HscB displayed the same seed morphology.16,17 SufD/NAP6 forms a SufBC2D complex with SufB and SufC and fulfills the scaffold function in the assembly and transfer of 4Fe-4S, 3Fe-4S, and Rieske-type 2Fe-2S clusters in the chloroplast.1-3,16 HscB interacts with DnaK-like chaperone protein HscA and iron-sulfur scaffold protein IscU1; therefore, HscB was proposed to participate in the assembly and transfer of iron-sulfur clusters in mitochondria and the cytosol.1-3,17
Figure 3.
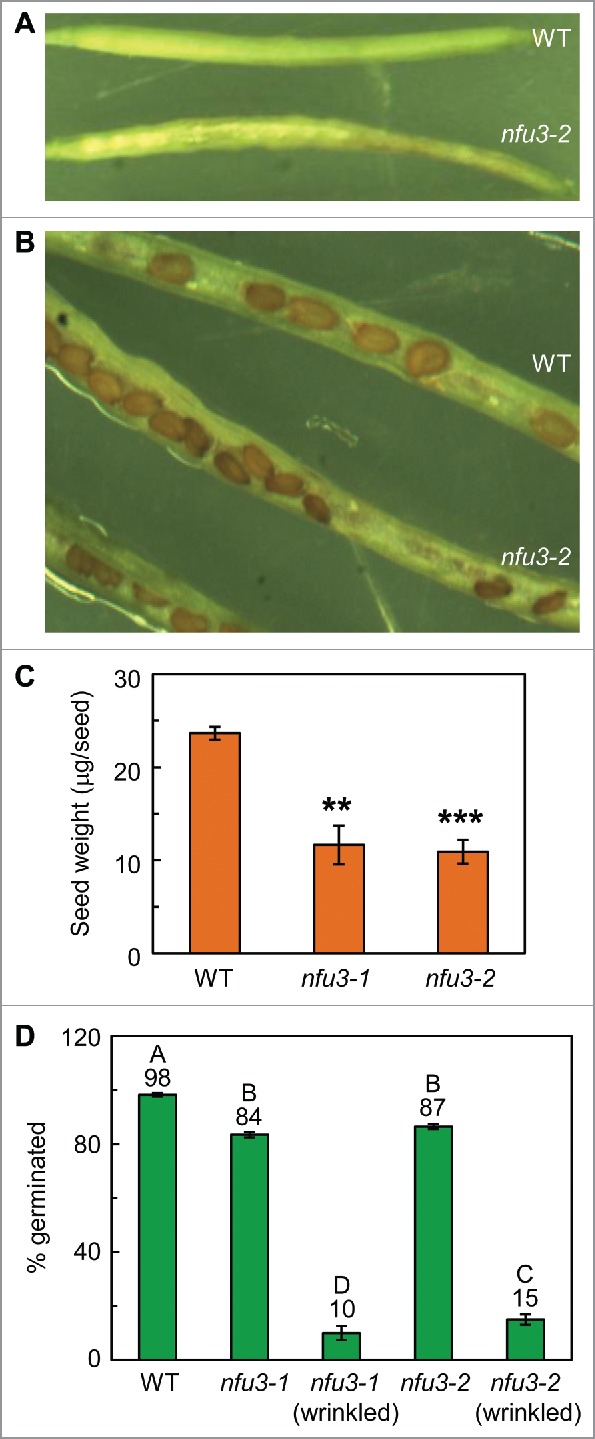
Analyses of seeds from wild-type and nfu3 plants.(A) Intact desiccating siliques from wild-type and nfu3–2 plants. (B) Opened desiccating siliques from wild-type and nfu3–2 plants. (C) Average weight of a seed from wild-type and nfu3 plants. Data are presented as mean ± SE (n = 5–7). The asterisk indicates a significant difference between the mutant and the wild type (WT) (Student's t test; *, p < 0.05; **, p < 0.01; ***, p < 0.001). (D) Germination of seeds from wild-type and nfu3 plants. Data are presented as mean ± SE (n = 4). Values not connected by the same letter are significantly different (Student's t test, p < 0.05). Plants used for seed analyses were grown on a 12-h light/12-h dark photoperiod with an irradiance of 150 μmol photons m−2 s−1 during the light period.
We separated the wrinkled nfu3 seeds from the non-wrinkled nfu3 seeds, determined their germination rates, and compared the germination rates between the 2 groups as well as with the wild type. The germination rate of the wrinkled nfu3 seeds (10–15%) was substantially lower than those of the wild-type seeds (98%) and the non-wrinkled nfu3 seeds (84–87%) (Fig. 3D). It is not surprising that the small, dark brown and wrinkled nfu3 seeds germinated poorly, as the wrinkled sufd/nap6 seeds also had a poor germination rate.16 It is interesting that the germination rate of the non-wrinkled nfu3 seeds was 11–14% lower than that of the wild-type seeds.
NFU3 is essential to overall plant fitness
The nfu3 mutants suffer from a series of growth and developmental defects, likely explained by the role of NFU3 in modulating the availability of 4Fe-4S and 3Fe-4S clusters to 4Fe-4S and 3Fe-4S-requiring proteins and complexes. 4Fe-4S and 3Fe-4S clusters are required in various vital biologic processes, many of which directly affect plant growth and development.
For example, PSI core subunits PsaA, PsaB, and PsaC require 4Fe-4S clusters to be functional.1-4 Loss-of-function mutations in NFU3 resulted in substantial reductions in the abundances of PsaA, PsaB, and PsaC, a near-complete loss of PSI activity, and a secondary reduction in PSII activity.4 PSI and PSII are essential to photosynthesis and the products of photosynthesis are required for plant growth and development. Therefore, the growth and developmental defects in the nfu3 mutants could be secondary effects of the near-complete loss of PSI activity due to limited availability of the 4Fe-4S cluster to PSI. Indeed, other mutants with reduced photosynthetic efficiency also displayed reduced growth and development.14,19-21 NFU2 is involved in assembly and transfer of 4Fe-4S and classic 2Fe-2S clusters and the nfu2 mutants displayed similar trends as the nfu3 mutants.14 The nfu2 mutants had a ∼30% reduction in PSI activity (calculated from Fig. 3 in reference14) and this is accompanied by delayed growth, flowering, and seeding.
The NAD(P)H dehydrogenase (NDH) complex in the chloroplast is another example of 4Fe-4S-containing protein complexes.1,3,22 The NdhI and NdhK subunits of this complex bind one and 2 4Fe-4S clusters, respectively.1,3,22 Although the abundances of NdhI and NdhK proteins were not determined, the levels of 2 other chloroplastic NDH subunits (NdhB and NdhH) were substantially reduced in the nfu3 mutants.4 This suggests that the activity of chloroplastic NDH might be compromised in the nfu3 mutants. In addition to NAD(P)H dehydrogenation, the chloroplastic NDH complex also acts as ferrodoxin-dependent plastoquinone reductase, mediating a minor pathway of PSI cyclic electron transport.22,23 PSI cyclic electron transport is essential for balancing ATP/NADPH production, which is required in various growth and developmental processes.24-26 The NDH-dependent PSI cyclic electron transport is specifically important for stress tolerance and photoprotection.24
A third example is 3Fe-4S-containing Fd-GOGAT1 and Fd-GOGAT21-4, which are expressed in leaf chloroplasts and root plastids, respectively.27 Fd-GOGATs have 2 functions: nitrogen assimilation and re-assimilation of photorespiratory ammonia.27 Under ambient CO2 conditions, loss-of-function mutants of Fd-GOGAT1 had a smaller plant size with small and pale green leaves,27 similar to the nfu3 mutants (Fig. 2A). Interestingly, the abundance of Fd-GOGAT1 was 45% lower in the nfu3 leaves than in wild-type leaves.4
In addition to PSI, NDH, and Fd-GOGATs, other 4Fe-4S- (e.g., nitrite reductase and sulfite reductase) and 3Fe-4S- (e.g., NADH-GOGAT) containing proteins and complexes exist in the chloroplast.1-3 Although not determined, the abundances of these proteins and complexes might also be reduced in the nfu3 mutants, due to limited availability of 4Fe-4S and 3Fe-4S clusters. Taken together, NFU3 is required for the assembly and transfer of 4Fe-4S and 3Fe-4S clusters in the chloroplast. NFU3 influences plant growth and development by modulating the availability of 4Fe-4S and 3Fe-4S clusters to 4Fe-4S- and 3Fe-4S-requiring proteins and complexes involved in various growth and developmental processes. Therefore, NFU3 is essential to overall plant fitness.
Materials and methods
Plant materials and growth conditions
The 2 Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines (nfu3–1 [GABI_381H10] and nfu3–2 [GABI_791C01], both in Columbia ecotype) used in this study were obtained from the Arabidopsis Biological Resource Center. Plants were grown in a growth chamber on a 12-h-light/12-h-dark photoperiod, as described previously.28 The light intensity was 150 μmol photons m−2 s−1, the temperature was 20°C, and the relative humidity was 50%.
Transient Expression of NFU3-CFP in Nicotiana benthamiana
Transient Expression of the NFU3-CFP fusion protein in N. benthamiana was performed as described previously.7 The full-length NFU3 coding region without the stop codon (NFU31–708 bp, corresponding to NFU31–236 AA) was amplified using the mRNA:cDNA hybrid, Phusion High-Fidelity DNA Polymerase (New England Biolabs), a forward primer NFU3_F (5′-CACCATGGGTTCTG TTTCGGGTCAA-3′), and a reverse primer NFU3_R (5′-CTCTAGAAGCTGGACTGCAC-3′). The resulting PCR products were gateway-cloned into the pENTR™/D-TOPO® vector (Thermo Fisher) and sequenced to confirm the absence of errors. The confirmed NFU31–711 bp fragment was sub-cloned into the pPH5ADEST-CFP vector (unpublished; provided by Jian Yao, Department of Biological Sciences, Western Michigan University, Kalamazoo, MI), using the Gateway® LR Clonase® II enzyme mix (Thermo Fisher). The resulting pPH5ADEST-NFU3-CFP construct, which is under the control of cauliflower mosaic virus 35S RNA promoter, was introduced into Agrobacterium tumefaciens strain C58C1. Transformed A. tumefaciens was cultured over-night at 30°C in Luria-Bertani medium containing appropriate antibiotics, harvested by centrifugation at 3,500 rpm at room temperature for 10 min, washed once in infiltration buffer (10 mM MES pH 5.8; 10 mM MgCl2; 0.2% sucrose), and resuspended to OD600 = 0.2 in infiltration buffer containing 300 μM acetosyringone. The A. tumefaciens cultures were inoculated into mature leaves of N. benthamiana and transient expression of the NFU3-CFP fusion protein was analyzed by confocal microscopy at 36–48 hours after inoculation.
Chlorophyll fluorescence analysis
Minimal fluorescence (Fo) and maximal fluorescence (Fm) were measured on dark-adapted plants at room temperature with the MAXI version of the IMAGING-PAM M-Series chlorophyll fluorescence system (Heinz Walz GmbH), as described previously.29,30 Maximum photochemical efficiency of PSII (Fv/Fm) is calculated using the following equation: Fv/Fm = (Fm – Fo)/Fm, where Fv is variable fluorescence of dark-adapted leaves.4
Seed morphology
Desiccating siliques were harvested and imaged under a Nikon SMZ-U dissecting scope and a QIMAGING RETIGA EXi Fast 1394 color camera. After the intact silique was imaged, one valve was removed to expose and image seeds in the silique. The average seed weight was determined by measuring the precise weight of ∼2.0 mg seeds and subsequent counting of the seed number under a dissecting microscope.
Seed germination
Harvested seeds were mixed well and aliquots of ∼100 seeds were imbibed in 0.01% Triton for 30 min, surface-sterilized with 95% ethanol for 5 min and 10% bleach for 5 min, and rinsed with sterile water 5 times. Sterilized seeds were sown in 25×150 mm Petri dishes containing 25-mL of solid germination media (1X Murashige and Skoog Basal Salt Mixture, 1X Gamborg's Vitamin Solution, 10 g/L sucrose, 0.25 g/L MES, 7 g/L agar, pH 5.65–5.8). Sown seeds were stratified at 4°C for 2–3 d and placed in a growth chamber on a 12-h-light/12-h-dark photoperiod for 7 d. The germination rate was determined by counting the numbers of germinated seedlings and un-germinated seeds.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Ryan L. Wessendorf for plant care, Jian Yao for sharing Gateway expression vector pPH5ADEST-CFP, and Christopher D. Jackson for growth chamber management.
Funding
This work was supported by the US. National Science Foundation under Grant Number MCB-1244008.
References
- 1.Balk J, Pilon M. Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci 2011; 16(4):218-26; PMID:21257336; http://dx.doi.org/ 10.1016/j.tplants.2010.12.006 [DOI] [PubMed] [Google Scholar]
- 2.Couturier J, Touraine B, Briat JF, Gaymard F, Rouhier N. The iron-sulfur cluster assembly machineries in plants: current knowledge and open questions. Front Plant Sci 2013; 4:259; PMID:23898337; http://dx.doi.org/ 10.3389/fpls.2013.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balk J, Schaedler TA. Iron cofactor assembly in plants. Annu Rev Plant Biol 2014; 65:125-53; PMID:24498975; http://dx.doi.org/ 10.1146/annurev-arplant-050213-035759 [DOI] [PubMed] [Google Scholar]
- 4.Nath K, Wessendorf RL, Lu Y. A nitrogen-fixing subunit essential for accumulating 4Fe-4S-containing Photosystem I core proteins. Plant Physiol 2016; 172(4):2459-70; PMID:27784767; http://dx.doi.org/ 10.1104/pp.16.01564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2007; 2(8):e718; PMID:17684564; http://dx.doi.org/ 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 2000; 300(4):1005-16; PMID:10891285; http://dx.doi.org/ 10.1006/jmbi.2000.3903 [DOI] [PubMed] [Google Scholar]
- 7.Withers J, Yao J, Mecey C, Howe GA, Melotto M, He SY. Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc Nat Acad Sci USA 2012; 109(49):20148-53; PMID:23169619; http://dx.doi.org/12553879 10.1073/pnas.1210054109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Léon S, Touraine B, Ribot C, Briat JF, Lobréaux S. Iron-sulphur cluster assembly in plants: distinct NFU proteins in mitochondria and plastids from Arabidopsis thaliana. Biochem J 2003; 371:823-30; PMID:12553879; http://dx.doi.org/ 10.1042/bj20021946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker NR, Harbinson J, Kramer DM. Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant Cell Environ 2007; 30(9):1107-25; PMID:17661750; http://dx.doi.org/ 10.1111/j.1365-3040.2007.01680.x [DOI] [PubMed] [Google Scholar]
- 10.Briantais JM, Dacosta J, Goulas Y, Ducruet JM, Moya I. Heat stress induces in leaves an increase of the minimum level of chlorophyll fluorescence, Fo: A time-resolved analysis. Photosynth Res 1996; 48(1-2):189-96; PMID:24271298; http://dx.doi.org/ 10.1007/BF00041008 [DOI] [PubMed] [Google Scholar]
- 11.Lennartz K, Plucken H, Seidler A, Westhoff P, Bechtold N, Meierhoff K. HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b(6)f complex in Arabidopsis. Plant Cell 2001; 13(11):2539-51; PMID:11701887; http://dx.doi.org/ 10.1105/tpc.010245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amann K, Lezhneva L, Wanner G, Herrmann RG, Meurer J. ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 2004; 16(11):3084-97;PMID:15494558; http://dx.doi.org/15173568 10.1105/tpc.104.024935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walters RG, Ibrahim DG, Horton P, Kruger NJ. A mutant of Arabidopsis lacking the triose-phosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiol 2004; 135(2):891-906; PMID:15173568; http://dx.doi.org/ 10.1104/pp.104.040469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Touraine B, Boutin JP, Marion-Poll A, Briat JF, Peltier G, Lobreaux S. Nfu2: a scaffold protein required for [4Fe-4S] and ferredoxin iron-sulphur cluster assembly in Arabidopsis chloroplasts. Plant J 2004; 40(1):101-11; PMID:15361144; http://dx.doi.org/15031412 10.1111/j.1365-313X.2004.02189.x [DOI] [PubMed] [Google Scholar]
- 15.Yabe T, Morimoto K, Kikuchi S, Nishio K, Terashima I, Nakai M. The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and Photosystem I. Plant Cell 2004; 16(4):993-1007; PMID:15031412; http://dx.doi.org/ 10.1105/tpc.020511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hjorth E, Hadfi K, Zauner S, Maier UG. Unique genetic compartmentalization of the SUF system in cryptophytes and characterization of a SufD mutant in Arabidopsis thaliana. FEBS Lett 2005; 579(5):1129-35; PMID:15710401; http://dx.doi.org/ 10.1016/j.febslet.2004.12.084 [DOI] [PubMed] [Google Scholar]
- 17.Xu XM, Lin H, Latijnhouwers M, Moller SG. Dual localized AtHscB involved in iron sulfur protein biogenesis in Arabidopsis. PLoS One 2009; 4(10):e7662; PMID:19865480; http://dx.doi.org/ 10.1371/journal.pone.0007662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wydro MM, Sharma P, Foster JM, Bych K, Meyer EH, Balk J. The evolutionarily conserved iron-sulfur protein INDH is required for complex I assembly and mitochondrial translation in Arabidopsis. Plant Cell 2013; 25(10):4014-27; PMID:24179128; http://dx.doi.org/ 10.1105/tpc.113.117283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Yang H, Lu Q, Wen X, Chen F, Peng L, Zhang L, Lu C. PSBP-DOMAIN PROTEIN1, a nuclear-encoded thylakoid lumenal protein, is essential for Photosystem I assembly in Arabidopsis. Plant Cell 2012; 24(12):4992-5006; PMID:23221595; http://dx.doi.org/ 10.1105/tpc.112.106542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng LW, Ma JF, Chi W, Guo JK, Zhu SY, Lu QT, Lu CM, Zhang LX. LOW PSII ACCUMULATION1 is involved in efficient assembly of Photosystem II in Arabidopsis thaliana. Plant Cell 2006; 18(4):955-69; PMID:16531500; http://dx.doi.org/ 10.1105/tpc.105.037689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y. Identification and roles of Photosystem II assembly, stability, and repair factors in Arabidopsis. Front Plant Sci 2016; 7:168; PMID:26909098; http://dx.doi.org/ 10.3389/fpls.2016.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng L, Yamamoto H, Shikanai T. Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim Biophys Acta 2011; 1807(8):945-53; PMID:21029720; http://dx.doi.org/ 10.1016/j.bbabio.2010.10.015; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 23.Shikanai T. Chloroplast NDH: A different enzyme with a structure similar to that of respiratory NADH dehydrogenase. Biochim Biophys Acta 2016; 1857(7):1015-22; PMID:26519774; http://dx.doi.org/ 10.1016/j.bbabio.2015.10.013 [DOI] [PubMed] [Google Scholar]
- 24.Munekage Y, Hashimoto M, Miyake C, Tomizawa K-I, Endo T, Tasaka M, Shikanai T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 2004; 429(6991):579-82; PMID:15175756; http://dx.doi.org/ 10.1038/nature02598 [DOI] [PubMed] [Google Scholar]
- 25.Wang C, Yamamoto H, Shikanai T. Role of cyclic electron transport around photosystem I in regulating proton motive force. Biochim Biophys Acta 2015; 1847(9):931-8; PMID:25481109; http://dx.doi.org/ 10.1016/j.bbabio.2014.11.013 [DOI] [PubMed] [Google Scholar]
- 26.Yamori W, Shikanai T. Physiological functions of cyclic electron transport around Photosystem I in sustaining photosynthesis and plant growth. Annu Rev Plant Biol 2016; 67:81-106; PMID:26927905; http://dx.doi.org/ 10.1146/annurev-arplant-043015-112002 [DOI] [PubMed] [Google Scholar]
- 27.Coschigano KT, Melo-Oliveira R, Lim J, Coruzzi GM. Arabidopsis gls mutants and distinct Fd-GOGAT genes. Implications for photorespiration and primary nitrogen assimilation. Plant Cell 1998; 10(5):741-52; PMID:9596633; http://dx.doi.org/ 10.1105/tpc.10.5.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark TJ, Lu Y. Analysis of loss-of-function mutants in aspartate kinase and homoserine dehydrogenase genes points to complexity in the regulation of aspartate-derived amino acid contents. Plant Physiol 2015; 168(4):1512-26; PMID:26063505; http://dx.doi.org/ 10.1104/pp.15.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, Hall DA, Last RL. A small zinc finger thylakoid protein plays a role in maintenance of Photosystem II in Arabidopsis thaliana. Plant Cell 2011; 23(5):1861-75; PMID:21586683; http://dx.doi.org/ 10.1105/tpc.111.085456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y. The occurrence of a thylakoid-localized small zinc finger protein in land plants. Plant Signal Behav 2011; 6(12):1181-5; PMID:22080791; http://dx.doi.org/ 10.4161/psb.6.12.18022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


