ABSTRACT
The term regulon has been coined in the genetic model plant Arabidopsis thaliana, denoting a structural and physiological defense apparatus defined genetically through the identification of the penetration (pen) mutants. The regulon is composed partially by the soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) syntaxin PEN1. PEN1 has homology to a Saccharomyces cerevisae gene that regulates a Secretion (Sec) protein, Suppressor of Sec 1 (Sso1p). The regulon is also composed of the β-glucosidase (PEN2) and an ATP binding cassette (ABC) transporter (PEN3). While important in inhibiting pathogen infection, limited observations have been made regarding the transcriptional regulation of regulon genes until now. Experiments made using the model agricultural Glycine max (soybean) have identified co-regulated gene expression of regulon components. The results explain the observation of hundreds of genes expressed specifically in the root cells undergoing the natural process of defense. Data regarding additional G. max genes functioning within the context of the regulon are presented here, including Sec 14, Sec 4 and Sec 23. Other examined G. max homologs of membrane fusion genes include an endosomal bromo domain-containing protein1 (Bro1), syntaxin6 (SYP6), SYP131, SYP71, SYP8, Bet1, coatomer epsilon (ε-COP), a coatomer zeta (ζ-COP) paralog and an ER to Golgi component (ERGIC) protein. Furthermore, the effectiveness of biochemical pathways that would function within the context of the regulon ave been examined, including xyloglucan xylosyltransferase (XXT), reticuline oxidase (RO) and galactinol synthase (GS). The experiments have unveiled the importance of the regulon during defense in the root and show how the deposition of callose relates to the process.
KEYWORDS: α-SNAP, β-glucosidase, ABC transporter, callose, pathogen, regulon, SNARE
Abbreviations
- SNARE
soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor
- ERGIC
ER to Golgi component
Introduction
The ability of eukaryotic membranes to fuse is important to many biological processes, including secretion.1 Owing to its importance, the genetically regulated process is ancient with gene homologs found in all eukaryotes.2 Subsequent biochemical experiments have determined the affinity that these proteins have for each other under various conditions.3,4 These experiments have provided foundational knowledge while also revealing new roles for some of the proteins. In A. thaliana, the secretory apparatus functions in defense responses.5 Furthermore, this secretion system has been expanded to include structural, biochemical and physiological components and is referred to as a regulon, a binary system composed of two parallel pathways converging on defense.6,7,8 Subsequent experiments have shown that the regulon components have the ability to coordinately regulate their expression (co-regulate) other regulon genes.7,9 In the review presented here, details are provided that describe the components of the regulon. The review examines how observations first made over a century ago regarding ecological plant variants capable of warding off pathogens have provided clues as to how the plant defense process functions.10 The review subsequently intercalates more recent genetic evidence that puts those original ecological observations into the context of the regulon.5,6,8-12 The review describes co-regulation of the regulon genes. The review then describes the identification of other genes expressed in the cells undergoing the process of defense that would be expected to function within the regulon platform.
Early observations of a genetic basis for the regulon
Observations made over a century ago identified the capability of certain ecological variants of the legume Trifolium repens to engage a successful defense response in their shoot that acts against various herbivores.10 This genetic system is defined by two loci.10-12 One diploid parent provides the dominant allele (Ac) driving α-hydroxynitrile glycoside production and one parent provides a dominant allele (Li) encoding for its hydrolyzing α-hydroxynitrile glycosidase.10-12 These observations implicate two different modes of secretion would converge during the process of defense because α-hydroxynitrile glycosidases have a signal peptide allowing it to enter the secretion system while glycosides can be mobilized by ABC transporters.13,14 Subsequent experiments performed in the plant genetic model A. thaliana have led to the identification of this genetic framework, referred to as a regulon.6 The framework of the regulon is defined by three genes including the syntaxin PENETRATION1 (PEN1), β-glucosidase (PEN2) and an ATP binding cassette (ABC) transporter (PEN3) (Fig. 1).5,6,15,16
Figure 1.
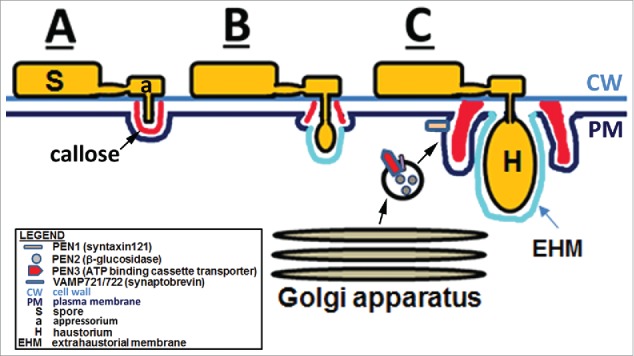
The regulon. In experiments involving Blumeria graminis f. sp. hordei, Erysiphe cichoracearum, Golovinomyces orontii, a structural, biochemical and physiological apparatus referred to as the regulon has been designated to refer to a defense apparatus involving the PENETRATION1-3 proteins. (A) The spore has germinated producing a neck that is attached to the appressorium which begins to push through the cell wall resulting in a plant cell response that leads to callose deposition. (B) The haustorium develops and pushes through the callose during a susceptible response. (C) Golgi-drived vesicles deliver the PEN1-3 proteins to the infection site, leading to a successful defense response that includes increased deposition of callose.5,6,8,15,16,254-266
Knowledge of how regulon components function clearly implicated other genes would have roles within the context of the regulon. Using the Glycine max-Heterodera glycines pathosystem as an experimental model, Klink et al.17 performed gene expression studies examining RNA isolated from G. max root cells parasitized by H. glycines. The experiments led to the determination of gene expression patterns in cells undergoing the process of defense. In those and related studies, Klink et al.17-21 identified that the defense apparatus may be quite extensive, possibly composed of hundreds to thousands of genes. Experiments then examined gene expression patterns occurring at the major H. glycines resistance locus, resistance to heterodera glycines 1 (rhg1).22,23 Transcriptional mapping studies of the rhg1 locus have identified α-SNAP/Sec 17 as a highly expressed component.23,24 α-SNAP is homologous to the Saccharomyces cerevisae Secretion (Sec) gene, Sec 17 (α-SNAP/Sec 17) that functions in secretion.1 In S. cerevisiae, mutants of Sec 17 (sec 17), accumulate 50 nm vesicles that cannot fuse with a target membrane leading to the failure of the cells to transport the cargo protein carboxypeptidase Y.1,25 This result demonstrates that α-SNAP/Sec 17p functions in membrane fusion during anterograde transport. Invaluable to the study of the rhg1 locus in G. max has been the availability of the genetic mapping data and its sequenced genome.22,26-29 Subsequent work has shown the rhg1 locus contains multiple copies of α-SNAP/Sec 17, but no clear understanding of a defense role had been obtained.30,31 However, Matsye et al.23,24 and Sharma et al.9 demonstrated a clear role for Gm-α-SNAP in defense of G. max to H. glycines parasitism. The identification of α-SNAP/Sec 17 as a resistance gene in G. max strengthens the observation that identified SNARE functioning in defense in the plant genetic model A. thaliana.5,9,24
Genetic identification of the importance of secretion
Secretion is an orderly stepwise process that has been demonstrated through the genetic identification of the Sec and related genes in S. cerevisiae (Fig. 2).1,32-35 Functional equivalents (homologs) subsequently have been identified in all eukaryotes.2,36-43 A core set of proteins, known as Soluble NSF Attachment Protein REceptor (SNARE) is one macromolecular part of the membrane fusion apparatus (Fig. 2). The components of SNARE include syntaxin (SYP)/Suppressor of sec 1 (SSO1), a gene homologous to A. thaliana PEN1. 5 Other SNARE components include synaptobrevin (SYB)/YKT6/SEC22 and SNAP-25/SEC9.44-49 The SNARE proteins tether the vesicle to the target membrane. Mammalian uncoordinated-18 (MUNC18/SEC1), also known as SM, may inhibit or facilitate fusion.50,51 Synaptotagmin (SYT)/Tricalbin-3 (TCB3) is believed to serve as a calcium sensor. SNARE metabolism, including its disassembly is mediated by two additional proteins including α-SNAP/Sec 17p and the ATPase N-ethylmaleimide-sensitive factor (NSF)/Sec 18p.1,3 The entire SNARE complex can be biochemically isolated as part a larger 20 S particle, including α-SNAP/Sec 17p and NSF/Sec 18p, that mediates secretion.52,53 Complimentary studies in animal systems investigating pathogenesis have identified botulinum and tetanus microbial neurotoxin effectors that target SNARE components and thus inhibit secretion.54-61 The effect of the neurotoxins is paralysis. Similar types of effectors are also being identified in plants leading to impaired functionality of 20 S components during defense, confirming its importance in the process of defense.62,63
Figure 2.
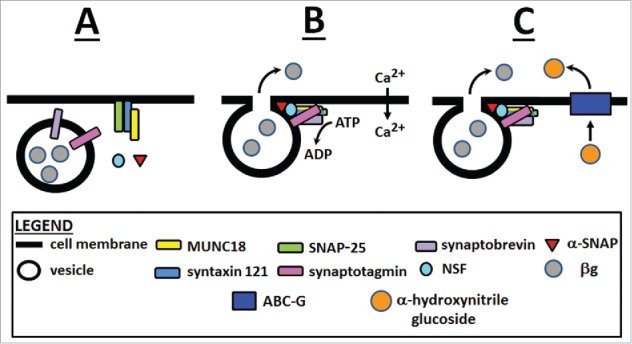
The process of membrane fusion involving the regulon. The involved proteins include SYP121 (PEN1/Sso1p), MUNC18/Sec 1p, SNAP-25/Sec 9p, SYB/VAMP/Ykt6p/Sec 22p), SYT/Tcb3p, NSF/Sec 18p and α-SNAP/Sec 17p, composing the 20 S particle. (A) the secretory vesicle, containing membrane fusion proteins comes into close association with target membrane proteins. (B) membrane fusion and release of cargo. (C) conjugated glucosides are transported through an ABC transporter in the vicinity of its secreted glucosidase resulting in activation of the glucosidase.3
α-SNAP/Sec 17 relates to plant defense by its association with SNARE during membrane fusion
The observation that Gm-α-SNAP functions in G. max defense to H. glycines has introduced questions regarding its activity. The observations that have been made regarding α-SNAP/Sec 17p is that it possesses diverse roles based on the cellular milieu. Through biochemical experiments Söllner et al.52 have identified a number of SNARE-related proteins that are specific for a certain vesicle or target membrane, but requires α-SNAP/Sec 17p for fusion. This work has indicated α-SNAP/Sec 17p is a universal component of constitutive and regulated membrane fusion with variations in SNARE composition dictating specificity.52 In examining neuronal exocytosis, Barszczewski et al.62 have identified clusters of α-SNAP/Sec 17p at the plasma membrane revealing the addition of NSF/Sec 18p facilitates exocytosis. Schwartz and Merz64 have shown α-SNAP/Sec 17p can rescue a SNARE complex that is stalled in its ability to complete fusion, identifying the central role of α-SNAP/Sec 17p in membrane fusion. Lobinger et al.65 have examined α-SNAP/Sec 17p further. The experiments have demonstrated α-SNAP/Sec 17p, along with Sec 1p (Munc18) (SM), actually accelerates fusion that is engaged by SNARE while also protecting it from disassembly by NSF/Sec 18p. Yu et al.66 have determined that the amount of SNARE-SM components in the cell are important for faithful fusion to occur. Yu et al.66 have also determined that for secretion to occur efficiently, the components require macromolecular crowding which is a condition known to influence the thermodynamic and kinetic behaviors of macromolecules.66 Experiments presented by Zick et al.67-69 have revealed the high concentration of purified components can overcome the requirement of lipids for progression to fusion under physiological conditions. This point is important because even in other systems including Drosophila, the balance of α-SNAP/Sec 17p and NSF/Sec 18p concentrations are important for secretion even when overexpressed.42 In other experimental systems, α-SNAP/Sec 17p works with NSF/Sec 18p to facilitate and disassemble all types of SNARE complexes after membrane fusion.70-73 Therefore, the experimental evidence demonstrates α-SNAP/Sec 17p has multiple functions prior to, during and after the process of membrane fusion and that it works at all sites of fusion of membrane-bound structures that utilize SNARE.
Regulon components homologous to the SNARE component PEN1 exhibit co-regulation
A number membrane-bound structures exist within the vesicle transport system.4 These structures include the endoplasmic reticulum (ER), conserved oligomeric Golgi (COG) complex, trans-Golgi network/early endosome (TGN/EE), homotypic fusion and protein sorting (HOPS) complex, the class C core vacuole/endosome tethering (CORVET), exocyst, trafficking protein particle (TRAPP) I–III complexes, Golgi-associated retrograde protein (GARP) complex, endosome-associated retrograde protein (EARP) complex, depends on SLY1-20 (Dsl1) complex and plasma membrane (PM). For details, please refer to Vukašinovi´ and Žárský.4 These structures utilize different macromolecular protein complexes to facilitate their interactions and fusion events.
SNARE, benefitting from having the longest experimental history, has had its core components identified decades ago (Fig. 2).3 While the proteins function effectively to mediate fusion, comparatively little is understood regarding whether the genes influence each other's expression (co-regulation). An early attempt made in S. cerevisiae attempted to understand whether the SNARE genes exhibited co-regulation.74 In this study, it has been shown that sec mutants did not exhibit coordinated suppressed transcriptional activity of the other Sec genes.74 However, technical limits of the time may have complicated such observation. In contrast, engineering SNARE components for constitutive induced expression has been shown to lead to induced expression of other SNARE components.75 Furthermore, experiments in other biological systems have shown ε-COP overexpression can overcome a temperature-dependent suppression of α-COP expression.76 Similar results have been shown for the ER to Golgi component (ERGIC).77 The observation of co-regulated gene expression of vesicle fusion components has held up to genomics-level scrutiny.78 These observations are consistent with those made in the G. max-H. glycines pathosystem.7,9 The question in relation to SNARE became how extensive is co-regulated gene expression during plant defense?
Regulon components homologous to β-glucoside PEN2 exhibit co-regulation
Experiments have been presented showing that the co-regulation of defense-related genes is not limited to the vesicle transport system components. Work done in Lotus japonicus has shown that LjBGD7, a root expressed PEN2 β-glycosidase homolog, is related to α-hydroxynitrile glucosidase.79 LjBGD7 acts to produce hydrogen cyanide (HCN), functioning effectively in defense.79 Furthermore, LjBGD7 is co-regulated with the cytochrome P450 protein LjCYP79D4. 79. In L. japonicus, LjCYP79D4 has increased relative levels of expression exclusively in the roots where LjBGD7 occurs.79 In related studies, Morant et al.80 demonstrated the co-expression of α-hydroxynitrile glucoside and their cognate hydrolyzing α-hydroxynitrile glucosidase. Furthermore, the heterologous expression of a Manihot esculenta (cassava) CYP79D2 in L. japonicus, results in cyanogenic α-hydroxynitrile glucoside accumulation.79 The experiments have clearly identified that genes functioning in α-hydroxynitrile glucoside production and metabolism are co-regulated.
Regulon components homologous to the ABC transporter PEN3 exhibit co-regulation
The ATP binding cassette (ABC) transporter regulon component PEN3 delivers metabolites through various membranes. While comparatively little is known about ABC transporters in plants, work in animal systems have demonstrated they are capable of co-regulating the expression of other genes relating to their own function.81,82 However, while evidence exists for the ability of the regulon components to effect co-regulation the scope and breadth had not been examined in detail. The co-regulation of pathway components leading to the production of secondary metabolites is not limited to PEN2. For example, The Nicotiana benthamiana terpene synthases TPS10 and TPS14 are co-regulated with the two cytochrome P450s (CYP71B31 and CYP76C3) in flowers at anthesis.83 Furthermore, the protein products are found in the endomembrane system resulting in monoterpene alcohol linalool production.83 Therefore, it is likely that other secondary metabolite pathways are important to defense in the G. max-H. glycines pathosystem (Fig. 3).
Figure 3.
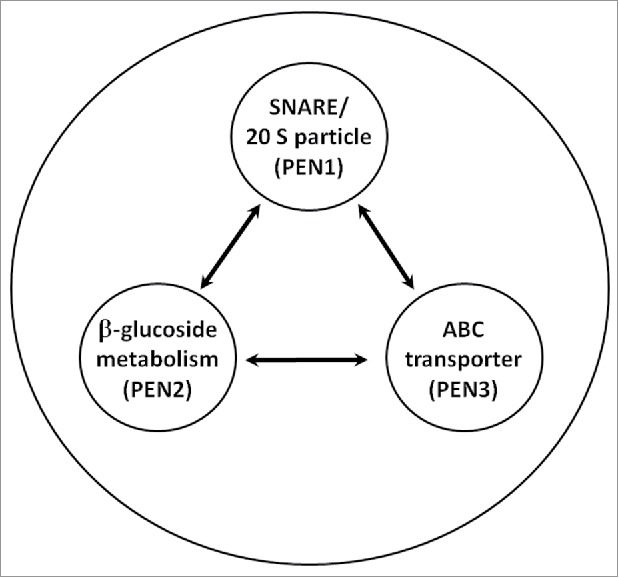
Components of the regulon, composed of PEN1, PEN2 and PEN3, are under co-regulation.9 Other components functioning within this apparatus are represented by the outer ring.
Callose is a structural defense element whose genetic components exhibit co-regulation
The recent identification of callose existing at sites of resistance in G. max roots parasitized by incompatible H. glycines is consistent with numerous observations of its presence at defense sites in many plant pathosystems.9,84-110 These studies have provided extensive support that callose performs an active role in defense. Consistent with these observations, other functional studies have demonstrated the active role callose plays as it participates in defense.111-117 However, in contrast, very limited studies have shown that callose may not accumulate to appreciable levels at defense sites or participate in defense at all.118,119
Callose is a glucose-derived polysaccharide polymerized mainly by callose synthase (CaLS) or GLUCAN SYNTHASE–LIKE (GSL) enzymes, forming β-1,3- and lesser amounts of β-1,6-branches. Conversely, β-1,3-glucanases depolymerize callose. During defense, plants deposit a plate-like structure referred to as a cell wall apposition that is also known as a papillae. It is these papillae that contain callose, among other materials, that are thought to provide a physical barrier to the establishment of an invasion site for the pathogen.120 The development of papillae at the molecular level requires the vesicle transport system.115 For example, in A. thaliana, papillae formation as a consequence of infection by Blumeria graminis f.sp. hordei is mediated by PEN1.115 The transport of callose has been suggested to occur through multivesicular bodies. However, conclusive evidence has not presented.114,115,120 These observations directly link vesicle transport to the delivery of callose at infection sites, consistent with the observations of Sharma et al. 9 in the G. max-H. glycines pathosystem.
Co-regulated gene expression occurring during defense is extensive
The regulon is likely to be an apparatus having a broad structural, biochemical and physiological basis. This concept is supported through gene expression experiments. For example, gene expression experiments examining nematode-parasitized cells undergoing a defense response identified many expressed genes that would be defense candidates in G. max that would be predicted to associate with the regulon (Fig. 4; Table 1; Table S1).23 To examine this concept further, experiments have been done examining candidate defense genes functioning in different defense pathways. These genes include G. max homologs of Sec 14-1, Sec 4-6, Sec 23-5, Bro1-1, SYP6-1, SYP131-1, SYP71-6, SYP8-2, Bet1-1, ε-COP-1, ζ-COP-3 and ERGIC-3-3. Genetic pathways that associate with the regulon have also been examined, including those incorporating reticulon oxidase-40 (Fig. 5; Table S2), xyloglucan xylosyltransferase 2-5 (Fig. 6; Table S2) and galactinol synthase-3 (Fig. 7; Table S2). As a control to show the expression of any gene will not result in engineered resistance, we have selected a G. max homolog of the A. thaliana APETALA2 (AP2) Gm-Apetala2-1 (Glyma01g44130).121 In A. thaliana, APETALA2 (AP2) is the founding member of a family of transcription factors originally identified to regulate flower development.121,122 Further analysis has shown Gm-Apetala2-1 exhibits homology to the A. thaliana AP2 homolog TINY.123 Ectopic expression of A. thaliana TINY, caused by a semidominant dissociation insertion mutation, affects fertility, plant height and the elongation of hypocotyls.123,124 However, Gm-Apetala2-1 is not expressed within the control pericycle cells or syncytia undergoing the process of defense and therefore it would not be expected to contribute to the process (Table S1). A qPCR analysis shows the genetically engineered plants exhibit induced candidate gene expression (Fig. S1). An analysis of genetically engineered roots infected with H. glycines reveals suppressed parasitism as compared to control plants (Fig. 8; Table S3).
Figure 4.
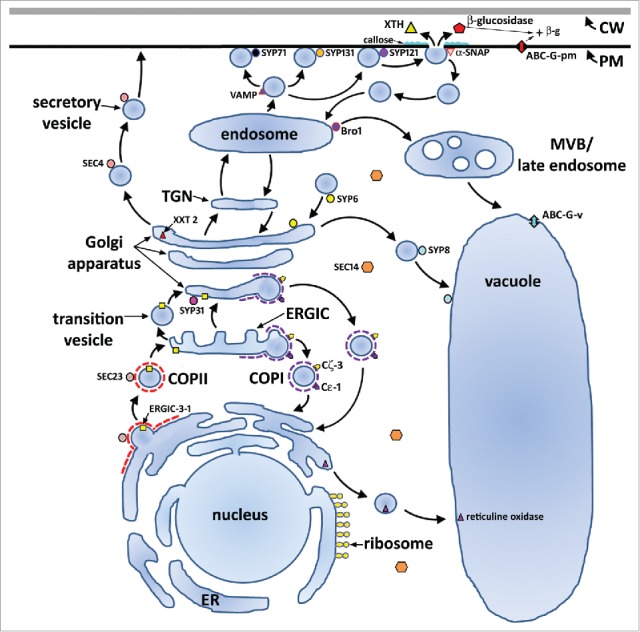
Proteins shown to function during the defense process G. max has to H. glycines parasitism. The previously identified regulon components including SYP31/Sed5p, PEN1 (SYP121/Sso1p), MUNC18/Sec 1p, SNAP-25/Sec 9p, SYB/VAMP/Ykt6p/Sec 22p, SYT/Tcb3p, NSF/Sec 18p and α-SNAP/Sec 17p, β-glucosidase, ABC-G. Presented here, Sec14, Sec 4, Sec 23, Bro1, SYP6, SYP131, SYP71, SYP8, Bet1, Cε, Cζ, ERGIC3.7,9,24
Table 1.
A summary of G. max candidate gene expression.
| Time point (dpi) |
||||
|---|---|---|---|---|
| Gene | Accession | 0* | 3 | 6 |
| Sec14-1 | Glyma14g08180 | N/M | N/M | M |
| Sec4-6 | Glyma20g23210 | N/M | N/M | M |
| Sec23-5 | Glyma18g00670 | N/M | M | M |
| Bro1-1 | Glyma02g10910 | N/M | M | M |
| SYP6-1 | Glyma17g07100 | N/M | M | M |
| SYP131-1 | Glyma12g32100 | N/M | M | M |
| SYP71-6 | Glyma19g36410 | N/M | N/M | M |
| SYP8-2 | Glyma14g02120 | M | M | M |
| Bet1-1 | Glyma06g10370 | M | M | M |
| Cε−1 | Glyma09g03500 | M | M | M |
| Cζ−3 | Glyma15g01150 | M | M | M |
| ERGIC-3-3 | Glyma13g01920 | M | M | M |
| RO-40 | Glyma15g14040 | N/M | N/M | M |
| XXT 2-5 | Glyma19g39760 | N/M | N/M | M |
| GS-3 | Glyma19g41550 | N/M | N/M | M |
| AP2-1 | Glyma01g44130 | N/M | N/M | N/M |
Footnote. Laser microdissection has been used to collect control cells (pericycle) at 0 days post infection (dpi) and Heterodera glycines-induced syncytia undergoing the process of resistance at 3 and 6 dpi. * Uninoculated roots have been used to obtain pericycle cells which have served as the source of the control mRNA samples. M (red) probe has been measured. N/M (blue), probe has not been measured. For details, please see Table S1.
Figure 5.
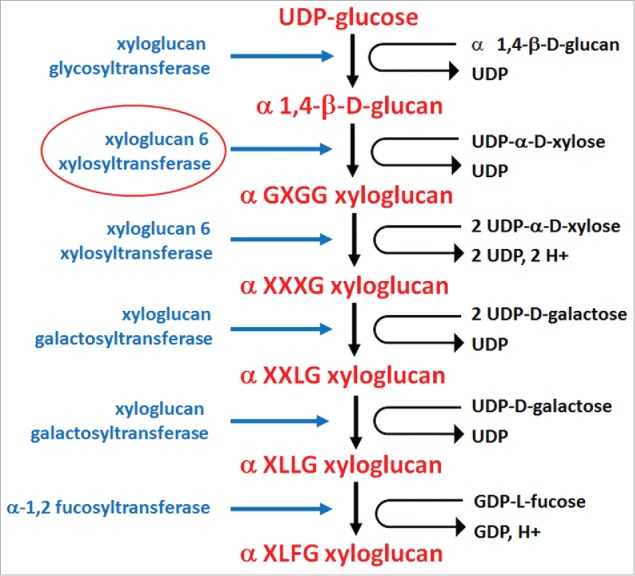
Enzymes functioning in xyloglucan biosynthesis are xyloglucan glycosyltransferase (CSLCS4) (E.C. 2.4.1.168), xyloglucan 6 xylosyltransferase (XXT1, XXT2) (E.C. 2.4.2.39), xyloglucan 6 xylosyltransferase (XXT5) (E.C. 2.4.2.39), xyloglucan galactosyltransferase (KATMARI1) (no E.C. #), xyloglucan galactosyltransferase (no E.C. #), α-1,2 fucosyltransferase (E.C. 2.4.1.69). Xyloglucan terminology: X, glucose residues substituted by α-D-xylose; G, non-substituted glucosyl units within a xylosylglycan; L, glucose residues substituted by α-D-xylose and β-D-galactose; F, glucose residues substituted by α-D-xylose, β-D-galactose and α-L-fucose. Supplemental data is provided (Table S2).267-275
Figure 6.
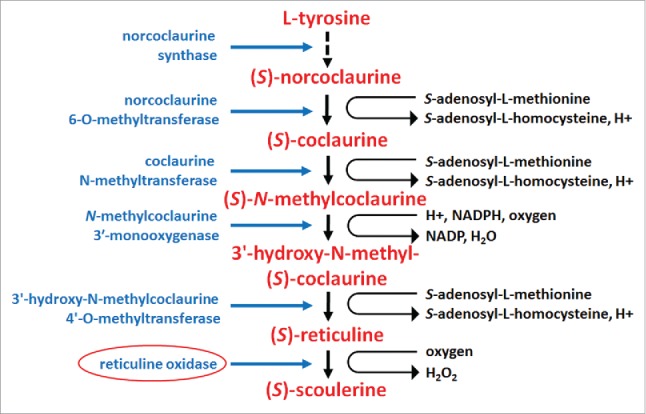
Enzymes functioning in reticuline metabolism. The biogenesis of (S)-reticuline begins with (L)-tyrosine. While a number of subsequent intermediates are made through different pathways, they converge on the production of (S)-noroclaurine via the enzymatic activity of noroclaurine synthase (E.C. 4.2.1.78). Subsequent enzymatic steps include norcoclaurine 6-O-methyltransferase (E.C. 2.1.1.128), coclaurine N-methyltransferase (E.C. 2.1.1.140), the CYP80B1 N-methylcoclaurine 3′-monooxygenase (E.C. 1.14.13.71), 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase (E.C. 2.1.1.116), berberine bridge enzyme tetrahydroprotoberberine synthase (reticuline oxidase) (EC 1.5.3.9). (S)-reticuline is an intermediate whose subsequent metabolism results in the generation of a wide number of isoquinoline alkaloids that are an important response to pathogen attack. Supplemental data is provided (Table S2).229-237
Figure 7.
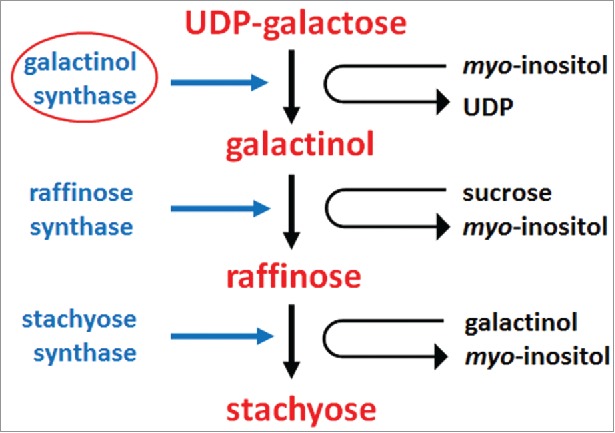
Enzymes functioning in stachyose metabolism. The enzymes involved include galactinol synthase (E.C. 2.4.1.123), raffinose synthase (E.C. 2.4.1.82) and stachyose synthase (E.C. 2.4.1.67). Supplemental data is provided (Table S2).248-253
Figure 8.
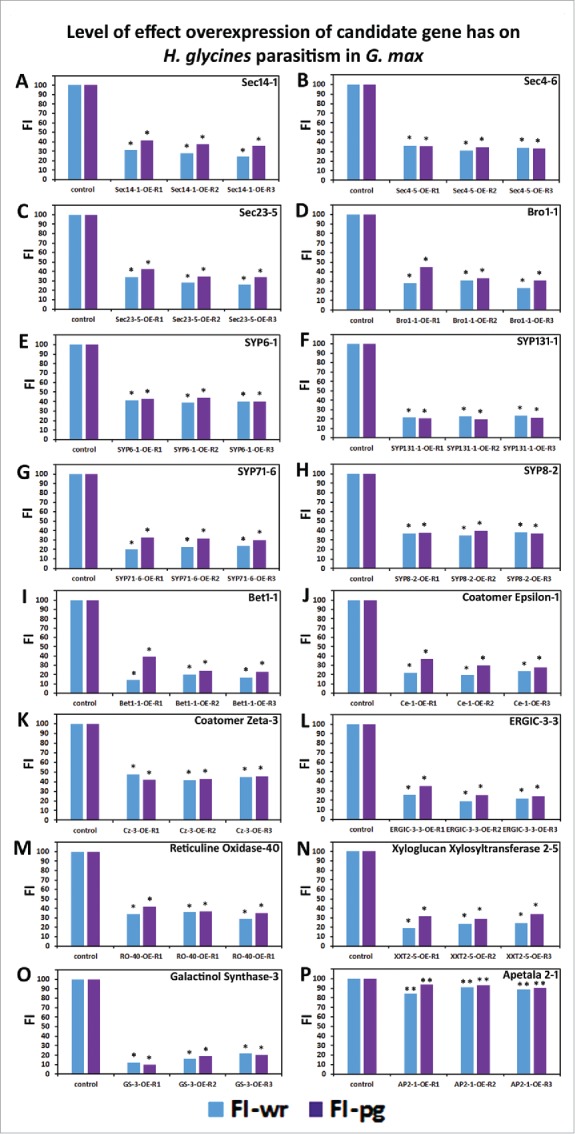
The overexpression of candidate defense genes in G. max leads to suppressed parasitism by H. glycines. (A) Sec 14-1; (B) Sec 4-6; (C) Sec 23-5; (D) Endosomal Targeting BRO1-Domain-1; (E) SYP6; (F) SYP131-1; (G) SYP71-6; (H) SYP8-2; (I) Bet1-1; (J) Coatomer ε-1; (K) Coatomer ζ-3; (L) ERGIC-3-3; (M) RO-40; (N) XXT 2-5; (O) GS-3; (P) AP2-1. The overexpression experiments present the outcome as the female index (FI) which is a percent of infection in relation to the control plants. wr, analysis examining cysts per whole root sample. pg, analysis examining cysts per gram of root tissue. * Statistically significant, p < 0.05; ** Not statistically significant, p > 0.05. Analyzed by Mann–Whitney–Wilcoxon [MWW] Rank-Sum Test. 276 Supplemental data is provided (Table S3).
The regulon apparatus functions broadly in defense
It is not surprising that the presented genes, identified to be expressed within the syncytium undergoing the process of defense, have roles in defense. The S. cerevisae Sec 14p is a cytoplasmic protein required for secretory vesicle formation from the Golgi apparatus and is encoded by a phosphatidylinositol (PtdIns) transfer protein (PITP).25,33,125,126 Sec 14p also functions as a sensor of the PtdIns/phosphatidylcholine (PtdCho) ratio in Golgi by directly regulating PtdCho biosynthesis.127 The S. cerevisae Sec 4p is a Rab GTPase that functions to regulate the assembly of the exocyst through its interaction with Sec 15p.128,129 The exocyst is a protein complex functioning in vesicle transport and membrane fusion between post-Golgi secretory vesicles to the plasma membrane. This interaction provides a regulatory function, occurring upstream of SNARE-mediated membrane fusion.130
The S. cerevisae Sec 23p is a cytosolic protein that functions in concert with Sar1p, Sec 24p, Sec 13p and Sec 14p as the minimal unit for COPII carrier formation.131-133 An important part of this process is phosphorylation of COPII coats.131-133 During vesicle formation, Sec 12p-mediated activation of Sar1p results in its recruitment to the ER membrane which results in recruitment of Sec 23p-Sec 24p. This action results in the formation of the inner COPII prebudding complex which allows export protein capture.131-133
The S. cerevisae Bro1p is endosome-associated, functioning in the multivesicular body134,135 Bro1p is a cytoplasmic protein named for its ∼160 aa domain that interacts with the endosomal sorting complex required for transport complexes (ESCRT).136-138 Mutation of residues within this Bro1p domain interferes with its ability to localize to endosomes.139 An analysis of Bro1p has been performed, revealing it contains a tetratricopeptide repeat (TPR)-like structure that is known to function as a putative ESCRT-III binding site.139 Mutant analysis of Bro1p binds sucrose non-fermenting 7 protein (Snf7p) (ESCRT-III). 135 Bro1p-like proteins also function in plants.140-143
A number of syntaxins that would function in related ways during membrane fusion function in defense. SYP6 has been shown to localize to the trans-Golgi network (TGN) and binds to α-SNAP.144-152 SYP131 is related to t-SNARE that functions in plant-arbuscular mycorrhizal interactions.153 SYP131 is closely related to SYP132, shown to function in symbiotic interactions.154 SYP71 localizes mainly to the plasma membrane and also the cell plate, endosome and ER.155,156 SYP71 can also be subverted during virus infection to facilitate its pathogenicity.157 SYP8 (At-SYP51) has been shown to accumulate on tonoplast and small prevacuolar compartments and co-localize with TGN markers and only partially with endocytic compartments with roles in vacuolar sorting, exocytosis and endocytosis.158
The S. cerevisae Bet1p functions in anterograde transport from the ER to Golgi, found on Golgi membranes and is recruited to COPII vesicles by Sec 24p. Bet1p activates Bos1p with this interaction regulated by the small GTP-binding protein Ypt1p. Bet1p overexpression overcomes mutants of sec 35.159-169
Sec 35p is novel, implicated in the tethering ER-derived vesicles.170 This tethering occurs at the Golgi apparatus. 170 Sec 35p binds to the hydrophilic Sec 34p to form a 480 kD complex that facilitates vesicle traffic.170
The ε-COP and ζ-COP proteins are part of the COPI coat, a 700 kD structure composed of α, β, β’ γ, δ, ε and ζ subunits that function in retrograde transport between the Golgi and ER. Furthermore, COPI functions in the maturation of endosomes and autophagy.76,171-178 Among these intermolecular interactions, ε-COP (Sec 28p) has been shown to stabilize α-COP (Ret1p).76 This function was observed out of work done in a number of studies that showed that the proteins composing COPI (Ret1p [α-COP], Sec 26p [β-COP], Sec 27p [β’-COP], Sec 21p [γ-COP], Ret2p [δ-COP], Ret3p [ζ-COP]) are essential for viability at all temperatures with ε-COP being temperature sensitive.76,172,179,180
The tubulovesicular ERGIC structure performs an important role in the sorting of proteins destined to be delivered to various cellular compartments. The ERGIC appears to be a stable compartment for COPII-dependent delivery of materials that are subsequently transported to the Golgi. ERGIC3 is a protein involved in this process.77,181-191
Secretion functions to drive a defense response
The demonstrated function of the secreted XTH and α-hydroxynitrile glucosidase proteins in defense indicated that the physiological needs of G. max resistance to H. glycines parasitism ran deep into the central metabolic processes of the parasitized cell. From genomics analyses performed in A. thaliana, a fundamental shift in metabolism during plant defense to pathogen attack has been long known and is consistent with observations made in the G. max-H. glycines pathosystem.17,23,192 In agreement with these observations is the demonstration of the involvement of the membrane fusion apparatus that would deliver XTH and α-hydroxynitrile glucosidase to the cell periphery and an ABC transporter that would likely deliver conjugated metabolites to the site of parasitism.7,9 Therefore, it appears that the defense of G. max to H. glycines parasitism involves altering the structure of the cell wall through the enzymatic activities of XTH, impeding the ability of H. glycines to make a functional syncytium. The defense process also involves the delivery of secondary metabolites to combat pathogen effectiveness. Furthermore, other metabolites are probably important to the defense process.
Hemicellulose: xyloglucan metabolism functions in defense in the root
Cytological observations of G. max undergoing the process of resistance to H. glycines have shown the cells engaged in the resistant reaction are limited in their ability to expand.193-196 These observations have indicated cell wall composition and modification are important to the defense process. A large proportion of the cell wall is composed of hemicellulose, a network that functions as a structural stabilizer. The majority of hemicellulose is composed of xyloglucan. Xyloglucan biosynthesis occurs by the stepwise activities of Golgi-localized enzymes and the delivery of these complex polysaccharides are generally believed to occur through bulk flow mediated by secretory vesicles (Fig. 5).197-199 The location of these materials in the Golgi, along with the secretion of these materials into the apoplast strenghtens the importance of the vesicle transport system in defense. Upon polymerization, the hemicellulose strand can be modified by xyloglucan endotransglycosylase/hydrolase (XTH) (E.C. 2.4.1.207), an ancient gene family found in all plants that may predate land colonization.200-204 A G. max XTH has been shown to be highly expressed in H. glycines parasitized root cells undergoing defense and functions in the defense process.7,23 XTHs function to restructure cell walls through cell wall loosening or intercalation of new xyloglucan.205-209 These two different functions of XTH are possible because it serves as a xyloglucan endo-transglucosylase (XET) where xyloglucan polymers are cleaved and joined to different xyloglucan chains.210 In contrast, the XEH activity of XTH hydrolyzes xyloglucan polymers.209 Regarding rapid cell wall expansion, auxin is known to regulate this process.211-213 XTH has a signal peptide and the protein can be N-glycosylated, indicating processing through the secretory pathway.214-220 The overexpression of XTH in Populus sp. results in the initial shortening of xyloglucan chain length, providing mechanistic insight into how plants can use XTH to limit cellular expansion during a defense reaction.7,221
The demonstration that XTH is highly expressed during the G. max defense process toward H. glycines parasitism has indicated that the metabolic processes that lead to the generation of the xyloglucan would also be important.23 In examining this result further, we have identified that mRNA of 4 genes encoding the enzymes that function in xyloglucan biogenesis and metabolism are present in syncytia undergoing he process of defense (Table S2). The enzymes that function in xyloglucan biogenesis and metabolism have been shown to localize to the Golgi apparatus, clearly implicating the importance of a functional secretion system to defense. To test this hypothesis, we have cloned a G. max homolog of XXT and overexpressed it in a susceptible genotype, resulting in impaired parasitism (Fig. 7; Table S3). The results emphasize the importance of hemicellulose metabolism to defense. Furthermore, the importance of a functional secretion system is demonstrated since the proteins are processed through the secretory pathway with some functioning within the Golgi apparatus.
Reticuline metabolism functions in defense in the root
Plants produce an astonishing number of secondary metabolites, numbering over a hundred thousand in total.222 One of these important secondary metabolites, whose subsequent metabolism leads to the production of defense molecules, is (S)-reticuline. (S)-reticuline is a benzylisoquinoline alkaloid belonging to a family of secondary metabolites consisting of more than 2,500 different known small molecules.223,224 Some of the early biochemistry regarding reticuline has been worked out by understanding its basic structure.225-228 The biogenesis of (S)-reticuline begins with (L)-tyrosine. While a number of subsequent intermediates are made through different pathways, they converge on the production of (S)-noroclaurine.229-237 (S)-reticuline is an intermediate whose subsequent metabolism results in the generation of a wide number of isoquinoline alkaloids that are an important response to pathogen attack.230 A second pathway that leads to (S)-reticuline biosynthesis is also known to have its origins with (L)-tyrosine and leading to (S)-norlaudanosoline production, but the pathway has been described in mammalian cells.238,239
Reticuline oxidase (RO) has also been described as berberine bridge enzyme (BBE).227,229,240 RO originally had been shown to be associated with a particle within the cell.228 RO has a signal peptide, indicating it is targeted to the ER.230 In Berberis wilsoniae var. subcaulialata, RO has been shown to localize within vesicles with other enzymes involved in the biosynthesis and metabolism of (S)-reticuline.241 The RO-containing vesicles have been shown to compose a low number of detectable polypeptides (∼20), indicating that vesicles with different types of cargo function in defense. 241 In opium poppy (Papaver somniferum cv Marianne), RO has been shown to localize in the companion cells of phloem.242 Furthermore, the subcellular localization of RO is the ER.223,241,243,244 This observation is consistent with the presence of a signal peptide for the G. max RO-40 (Fig. S2). After elicitor treatment, the ER undergoes a major ultrastructural arrangement becoming dilated and producing vesicles that later fuse with the vacuole.241,243,244 Notably, a vacuolar sorting determinant is present in the N-terminus of the RO protein.243 The production of the metabolic intermediates leading to the production of to (S)-reticuline and the structural requirements appear to be shared between different plant groups.230,243-246 We have identified G. max homologs of genes functioning in reticuline biogenesis and metabolism (Table S2). Further work using Coptis japonica as a model revealed that an ABC transporter was responsible for delivering berberine to its site of function.247 This observation provided mechanistic insight into the delivery of secondary metabolites synthesized by RO to the vacuole. As observed by Sharma et al.,9 it is likely that a number of different ABC transporters function during the defense of G. max to parasitic nematodes.
Stachyose metabolism functions in defense in the root
Stachyose is produced in an enzymatic process beginning with galactinol synthase (E.C. 2.4.1.123) action involving myo-inositol and UDP-D-galactose.248 The process leads to the production of galactinol. 248 The only known role for galactinol is for the production of larger soluble oligosaccharides such as raffinose, stachyose, and verbascose.249 However, galactinol has been shown to be induced in its production during a defense response in roots and functions in signaling during systemic acquired resistance.250 Subsequent enzymatic activities of raffinose synthase (E.C. 2.4.1.82) and stachyose synthase (E.C. 2.4.1.67) result in the production of stachyose (Fig. 7).251-253 The enzymes for the biogenesis of stachyose have been identified in G. max (Table S2).
The framework of defense involving the co-regulation of regulon in roots
The framework of defense as G. max protects itself from its major pathogen, H. glycines is based off of cytological observations and genetic experiments relating its rhg1 locus.7,9,22,24,28,30,31,193 These results put into context the identification of an H. glycines effector that binds G. max α-SNAP/Sec 17 that would have the function of disarming its defense response.63 Evidence has been presented showing that the transcriptional activity of these genes influences each other, indicative of co-regulation.7,9,24 The co-regulation of components of a structural and biochemical element helps in explaining why so many genes are transcriptionally active in the cells destined to undergo the process of resistance. Additional experimental evidence is presented here to support those observations. The experiments show how the expression of these genes recapitulates the natural defense process that leads to suppressed parasitism by H. glycines in G. max. Furthermore, the identification of callose at the site of defense expands the biochemical and structural environment pertaining to defense, unifying work done in other plant pathosystems.5,6,8,15,16,115,254,255 The work likely has identified a genetic program that is functional in other plants against other pathogens or could be altered to facilitate symbiotic interactions. As a polymer, callose has the capacity of rapid building block mobilization to infection sites with their subsequent polymerization into structures that can impede pathogenesis.9,255-257
Methods
The data that has been presented in this review has been obtained through the published methods in Sharma et al. 9 Laser microdissection (LM) has been used to collect control cells (pericycle) at 0 days post infection (dpi) and Heterodera glycines-induced syncytia undergoing the process of resistance at 3 and 6 dpi. Uninoculated roots have been used to obtain pericycle cells which serve as the source of the control mRNA samples. Microarray hybridizations have been run in triplicate (arrays 1-3). The hybridizations have used probe derived from RNA isolated from LM-collected syncytia obtained from 3 independent replicate experiments. For robustness, the experiments have been run independently in two different H. glycines-resistant genotypes. For the gene to be considered expressed at a given time point (3 or 6 days post infection [dpi]), probe signal must have been measurable above threshold on all three arrays for both G. max[Peking/PI 548402] and G. max[PI 88788] (6 total arrays), p < 0.05. The Bioconductor implementation of the standard Affymetrix® detection call methodology (DCM) analysis consists of four steps, including (1) removal of saturated probes, (2) calculation of discrimination scores, (3) p-value calculation using the Wilcoxon's rank test, and (4) making the detection call (present [p < 0.05]/marginal [p = 0.05]/absent [p > 0.05]). In the analysis presented here, the probe set is accepted as detecting probe if p < 0.05. The PCR and qPCR primers used in the studies are provided (Table S4). The qPCR data has been obtained using 2−ΔΔCT to calculate fold change (Fig. S2).9,258 The female index (FI), is the community-accepted method to determine the effect a condition has on H. glycines parasitism.259 The results have been presented as female cysts per whole root mass and female cysts per gram (Table S3).
Supplementary Material
Diclosure of Potential Conflicts of Interest
The authors claim no potential conflicts of interest.
Acknowledgements
The authors thank Dr. Nancy Reichert for providing culture room space that is essential for the transgenic experiments. The Department of Biological Sciences is acknowledged for providing start-up support and teaching assistantships. Support from the Mississippi Soybean Promotion Board, Cotton Incorporated and Mississippi Agricultural and Forestry Experimental Station (MAFES) is acknowledged. Graduate students from the Department of Biological Sciences and Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology contributing to this project and the genomics analyses but are not authors include Prachi Matsye, Prakash Niruala, Tineka Burkhead, Bisho Lawaju and Weasam Aljaafri. Undergraduate students greatly complimenting the research include Christina Jones, Suchit Salian, Nishi Sunthwal, Priyanka Gadre, Dollie Welch, Kim Anderson, Brittany Ginn, John Clune, Hannah Burson, Chase Robinson, Meghan Calhoun, Katherine McCracken, Kayla Moore, Madison Milhoan, Erin Curran, Neil Shannon, Austin Martindale, Keigero Fergusen, Ashley Dowdy, Taylor Henry, Hannah Stimson, Hannah Whitlock, Erin Ball, Henry Pittman, Shelby Janeski, Ashlee Vargason, Annedrea McMillan, Courtney Gagliano, Leslie Canale, Alison Antee, Hannah Miller, Adam Crittenden, Lauren Langston, Jamelle Vance, Carolyn Chacon, Emily Carter, Natalie Rentrop, Eileen Modzeleski, Erica Sowell, Chrissy Miller, Anna Bailey Britt, Chelsea Tittle, Robyn Beattie, Dejanie Dilworth, Aishwarya Dikshit, Harshini Sampathkumar, Samantha Rushing, Meagan Young, Kathryn Stiglet, Makenzie Miller, Morgan Urich, Chase Nash, Kyle Winston, Olivia Long, Jody Clark, Ana Simal, Katherine Thrash, Adrienne McMorris, Rebecca Waters, Haleigh Smith, Kelvin Blade, Jesse Austin, Alex Hammett and Maggie Kuhn. Amanda Lawrence, Imaging and Analytical Technologies Center, Mississippi State University is acknowledged. Dr. Wes Burger (MAFES) and Dr. Giselle Thibaudeau (College of Arts and Sciences) are acknowledged for their interest.
References
- 1.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 1980; 21:205-15; PMID:6996832; http://dx.doi.org/ 10.1016/0092-8674(80)90128-2 [DOI] [PubMed] [Google Scholar]
- 2.Clary DO, Griff IC, Rothman JE. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell 1990; 61:709-21; PMID:2111733; http://dx.doi.org/ 10.1016/0092-8674(90)90482-T [DOI] [PubMed] [Google Scholar]
- 3.Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature 2012; 490:201-7; PMID:23060190; http://dx.doi.org/ 10.1038/nature11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vukasinovi N, Zarsky V. Tethering complexes in the Arabidopsis endomembrane system. Front Cell Dev Biol 2016; 4:46; PMID:27243010; http://doi.org/ 10.3389/fcell.2016.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al.. SNARE-protein mediated disease resistance at the plant cell wall. Nature 2003; 425:973-7; PMID:14586469; http://dx.doi.org/ 10.1038/nature02076 [DOI] [PubMed] [Google Scholar]
- 6.Humphry M, Bednarek P, Kemmerling B, Koh S, Stein M, Gobel U, Stuber K, Pislewska-Bednarek M, Loraine A, Schulze-Lefert P, et al.. A regulon conserved in monocot and dicot plants defines a functional module in antifungal plant immunity. Proc Natl Acad Sci U S A 2010; 107:21896-901; PMID:21098265; http://dx.doi.org/ 10.1073/pnas.1003619107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pant SR, Matsye PD, McNeece BT, Sharma K, Krishnavajhala A, Lawrence GW, Klink VP. Syntaxin 31 functions in Glycine max resistance to the plant parasitic nematode Heterodera glycines. Plant Mol Bio 2014; 85:107-21; http://dx.doi.org/ 10.1007/s11103-014-0172-2 [DOI] [PubMed] [Google Scholar]
- 8.Johansson ON, Fantozzi E, Fahlberg P, Nilsson AK, Buhot N, Tor M, Andersson MX. Role of the penetration-resistance genes PEN1, PEN2 and PEN3 in the hypersensitive response and race specific resistance in Arabidopsis thaliana. Plant J 2014; 79:466-76; PMID:24889055; http://dx.doi.org/ 10.1111/tpj.12571 [DOI] [PubMed] [Google Scholar]
- 9.Sharma K, Pant SR, McNeece BT, Lawrence GW, Klink VP. Co-regulation of the Glycine max soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE)-containing regulon occurs during defense to a root pathogen. J Plant Interactions 2016; 11:74-93; http://dx.doi.org/ 10.1080/17429145.2016.1195891 [DOI] [Google Scholar]
- 10.Armstrong HE, Armstrong EF, Horton E. Herbage studies. II. Variation in Lotus corniculatus and Trifolium repens: (cyanophoric plants). Proc R Soc Lond Ser B 1913; 86:262-9. [Google Scholar]
- 11.Ware WM. Experiments and observations on forms and strains of Trifolium repens. J Agric Sci 1925; 15:47-67. [Google Scholar]
- 12.Hughes MA. The cyanogenic polymorphism in Trifolium repens L. (white clover). Heredity 1991; 66:105-15; http://dx.doi.org/ 10.1038/hdy.1991.13 [DOI] [Google Scholar]
- 13.Kakes P. Linamarase and other beta-glucosidases are present in the cell walls of Trifolium repens L. leaves. Planta 1985; 166:156-60; PMID:24241426; http://dx.doi.org/ 10.1007/BF00397342 [DOI] [PubMed] [Google Scholar]
- 14.Francisco RM, Regalado A, Ageorges A, Burla BJ, Bassin B, Eisenach C, Zarrouk O, Vialet S, Marlin T, Chaves MM, et al.. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-Glucosides.Plant Cell 2013; 25:1840-54; PMID:23723325; http://dx.doi.org/ 10.1105/tpc.112.102152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipka L, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al.. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 2005; 310:1180-3; PMID:16293760; http://dx.doi.org/ 10.1126/science.1119409 [DOI] [PubMed] [Google Scholar]
- 16.Stein M, Dittgen J, Sanchez-Rodriguez C, Hou B-H, Molina A, Schulze-Lefert P, Lipka V, Somerville S. Arabidopsis PEN3/PDR8, an ATP Binding Cassette Transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 2006; 18:731-46; PMID:16473969; http://dx.doi.org/ 10.1105/tpc.105.038372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klink VP, Overall CC, Alkharouf N, MacDonald MH, Matthews BF. Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean roots infected by soybean cyst nematode (Heterodera glycines). Planta 2007; 226:1389-409; PMID:17668236; http://dx.doi.org/ 10.1007/s00425-007-0578-z [DOI] [PubMed] [Google Scholar]
- 18.Klink VP, Hosseini P, Matsye PD, Alkharouf NW, Matthews BF. A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode). Plant Mol Biol 2009; 71:525-67; PMID:19787434; http://dx.doi.org/ 10.1007/s11103-009-9539-1 [DOI] [PubMed] [Google Scholar]
- 19.Klink VP, Hosseini P, Matsye PD, Alkharouf NW, Matthews BF. Syncytium gene expression in Glycine max[PI 88788] roots undergoing a resistant reaction to the parasitic nematode Heterodera glycines. Plant Physiol Biochem 2010a; 48:176-93; PMID:20138530; http://dx.doi.org/ 10.1016/j.plaphy.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 20.Klink VP, Overall CC, Alkharouf NW, MacDonald MH, Matthews BF. Microarray detection calls as a means to compare transcripts expressed within syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines). J Biomed Biotechnol 2010b; 1-30; http://dx.doi.org/ 10.1155/2010/491217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klink VP, Hosseini P, Matsye PD, Alkharouf NW, Matthews BF. Differences in gene expression amplitude overlie a conserved transcriptomic program occurring between the rapid and potent localized resistant reaction at the syncytium of the Glycine max genotype Peking (PI 548402) as compared to the prolonged and potent resistant reaction of PI 88788. Plant Mol Biol 2011; 75:141-65; PMID:21153862; http://dx.doi.org/ 10.1007/s11103-010-9715-3 [DOI] [PubMed] [Google Scholar]
- 22.Caldwell BE, Brim CA, Ross JP. Inheritance of resistance of soybeans to the soybean cyst nematode, Heterodera glycines. Agron J 1960; 52:635-6. [Google Scholar]
- 23.Matsye PD, Kumar R, Hosseini P, Jones CM, Tremblay A, Alkharouf NW, Matthews BF, Klink VP. Mapping cell fate decisions that occur during soybean defense responses. Plant Mol Bio 2011; 77:513-28; http://dx.doi.org/ 10.1007/s11103-011-9828-3 [DOI] [PubMed] [Google Scholar]
- 24.Matsye PD, Lawrence GW, Youssef RM, Kim KH, Matthews BF, Lawrence KS, Klink VP. The expression of a naturally occurring, truncated allele of an α-SNAP gene suppresses plant parasitic nematode infection. Plant Mol Bio 2012; 80:131-5; http://dx.doi.org/ 10.1007/s11103-012-9932-z [DOI] [PubMed] [Google Scholar]
- 25.Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell 1982; 30:439-48; PMID:6754086; http://dx.doi.org/ 10.1016/0092-8674(82)90241-0 [DOI] [PubMed] [Google Scholar]
- 26.Concibido VC, Diers BW, Arelli PR. A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci 2004; 44:1121-31; http://dx.doi.org/ 10.2135/cropsci2004.1121 [DOI] [Google Scholar]
- 27.Grant D, Nelson RT, Cannon SB, Shoemaker RC. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucl Acids Res 2010; 38:D843-6; PMID:20008513; http://dx.doi.org/ 10.1093/nar/gkp798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim M, Hyten DL, Bent AF, Diers BW. Fine mapping of the SCN resistance locus rhg1-b from PI 88788. Plant Genome 2010; 3:81-9; http://dx.doi.org/ 10.3835/plantgenome2010.02.0001 [DOI] [Google Scholar]
- 29.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al.. Genome sequence of the palaeopolyploid soybean. Nature 2010; 463:178-83; PMID:20075913; http://dx.doi.org/ 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- 30.Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless A, Wang J, Hughes TJ, Willis DK, Clemente T, et al.. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 2012; 338:1206-9; PMID:23065905; http://dx.doi.org/ 10.1126/science.1228746 [DOI] [PubMed] [Google Scholar]
- 31.Cook DE, Bayless AM, Wang K, Guo X, Song Q, Jiang J, Bent AF. Distinct copy number, coding sequence and locus methylation patterns underlie Rhg1-mediated soybean resistance to soybean cyst nematode. Plant Physiol 2014; 165:630-47; PMID:24733883; http://dx.doi.org/ 10.1104/pp.114.235952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 1979; 76:1858-62; PMID:377286; http://dx.doi.org/ 10.1073/pnas.76.4.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell 1981; 25:461-69; PMID:7026045; http://dx.doi.org/ 10.1016/0092-8674(81)90064-7 [DOI] [PubMed] [Google Scholar]
- 34.Esmon B, Novick P, Schekman R. Compartmentalized assembly of oligosaccharides on exported glycoproteins in yeast. Cell 1981; 25:451-60; PMID:7026044; http://dx.doi.org/ 10.1016/0092-8674(81)90063-5 [DOI] [PubMed] [Google Scholar]
- 35.Kaiser CA, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 1990; 61:723-33; PMID:2188733; http://dx.doi.org/ 10.1016/0092-8674(90)90483-U [DOI] [PubMed] [Google Scholar]
- 36.Griff IC, Schekman R, Rothman JE, Kaiser CA. The yeast SEC17 gene product is functionally equivalent to mammalian alpha-SNAP protein. J Biol Chem 1992; 267:12106-15; PMID:1601878 [PubMed] [Google Scholar]
- 37.Gerst JE. Conserved alpha-helical segments on yeast homologs of the synaptobrevin/VAMP family of v-SNAREs mediate exocytic function. J Biol Chem 1997; 272:16591-8; PMID:9195971; http://dx.doi.org/ 10.1074/jbc.272.26.16591 [DOI] [PubMed] [Google Scholar]
- 38.Sanderfoot AA, Farhah F, Assaad FF, Raikhel NV. The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol 2001a; 124:1558-69; http://dx.doi.org/ 10.1104/pp.124.4.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. Interactions between syntaxins identify at least 5 SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell 2001b; 12:3733-43; PMID:11739776; http://dx.doi.org/ 10.1091/mbc.12.12.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Payne WE, Kaiser CA, Bevis BJ, Soderholm J, Fu D, Sears IB, Glick BS. Isolation of Pichia pastoris genes involved in ER-to-Golgi transport. Yeast 2000; 16:979-93; PMID:10923020; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 41.Hong K–K, Chakravarti A, Takahashi JS. The gene for soluble N-ethylmaleimide sensitive factor attachment protein alpha is mutated in hydrocephaly with hop gait (hyh) mice. Proc Natl Acad Sci U S A 2004; 101:1748-53; PMID:14755058; http://dx.doi.org/ 10.1073/pnas.0308268100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babcock M, Macleod GT, Leither J, Pallanck L. Genetic analysis of soluble N-ethylmaleimide-sensitive factor attachment protein function in Drosophila reveals positive and negative secretory roles. J Neurosci 2004; 24:3964-73; PMID:15102912; http://dx.doi.org/ 10.1523/JNEUROSCI.5259-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodríguez F, Bustos MA, Zanetti MN, Ruete MC, Mayorga LS, Tomes CN. α-SNAP prevents docking of the acrosome during sperm exocytosis because it sequesters monomeric syntaxin. PLoS One 2011; 6:e21925; http://dx.doi.org/ 10.1371/journal.pone.0021925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oyler GA, Higgins GA, Hart RA, Battenberg E, Billingsley M, Bloom FE, Wilson MC. The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J Cell Biol 1989; 109:3039-52; PMID:2592413; http://dx.doi.org/ 10.1083/jcb.109.6.3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baumert M, Maycox PR, Navone F, De Camilli P, Jahn R. Synaptobrevin: an integral membrane protein of 18,000 daltons present in small synaptic vesicles of rat brain. EMBO J 1989; 8:379-84; PMID:2498078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennett MK, Calakos N, Scheller RH. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 1992; 257:255-9; PMID:1321498; http://dx.doi.org/ 10.1126/science.1321498 [DOI] [PubMed] [Google Scholar]
- 47.Aalto MK, Ronne H, Keränen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J 1993; 12:4095-104; PMID:8223426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Søgaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Söllner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell 1994; 78:937-48; PMID:7923363; http://dx.doi.org/ 10.1016/0092-8674(94)90270-4 [DOI] [PubMed] [Google Scholar]
- 49.McNew JA, Søgaard M, Lampen NM, Machida S, Ye RR, Lacomis L, Tempst P, Rothman JE, Sollner TH. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J Biol Chem 1997; 272:17776-83; PMID:9211930; http://dx.doi.org/ 10.1074/jbc.272.28.17776 [DOI] [PubMed] [Google Scholar]
- 50.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008; 27:923-33; PMID:18337752; http://dx.doi.org/ 10.1038/emboj.2008.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science 2009; 323:474-7; PMID:19164740; http://dx.doi.org/ 10.1126/science.1161748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature 1993a; 362:318-24; PMID:8455717; http://dx.doi.org/ 10.1038/362318a0 [DOI] [PubMed] [Google Scholar]
- 53.Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 1993b; 75:409-18; PMID:8221884; http://dx.doi.org/ 10.1016/0092-8674(93)90376-2 [DOI] [PubMed] [Google Scholar]
- 54.Schiavo G, Poulain B, Rossetto O, Benfenati F, Tauc L, Montecucco C. Tetanus toxin is a zinc protein and its inhibition of neurotransmitter release and protease activity depend on zinc. EMBO J 1992a; 11:3577-83; PMID:1396558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta BR, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature 1992b; 359:832-5; PMID:1331807; http://dx.doi.org/ 10.1038/359832a0 [DOI] [PubMed] [Google Scholar]
- 56.Schiavo G, Rossetto O, Montecucco C. Clostridial neurotoxins as tools to investigate the molecular events of neurotransmitter release. Semin Cell Biol 1994; 5:221-9; PMID:7994006 [DOI] [PubMed] [Google Scholar]
- 57.Pellegrini LL, O'Connor V, Lottspeich F, Betz H. Clostridial neurotoxins compromise the stability of a low energy SNARE complex mediating NSF activation of synaptic vesicle fusion. EMBO J 1995; 14:4705-13; PMID:7588600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature 2006; 444:1096-100; PMID:17167418; http://dx.doi.org/ 10.1038/nature05411 [DOI] [PubMed] [Google Scholar]
- 59.Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature 2006; 444:1092-5; PMID:17167421; http://dx.doi.org/ 10.1038/nature05387 [DOI] [PubMed] [Google Scholar]
- 60.Strotmeier J, Willjes G, Binz T, Rummel A. Human synaptotagmin-II is not a high affinity receptor for botulinum neurotoxin B and G: increased therapeutic dosage and immunogenicity. FEBS Lett 2012; 586:310-3; PMID:22265973; http://dx.doi.org/ 10.1016/j.febslet.2011.12.037 [DOI] [PubMed] [Google Scholar]
- 61.Bennett TL, Kraft SM, Reaves BJ, Mima J, O'Brien KM, Starai VJ. LegC3, an effector protein from Legionella pneumophila, inhibits homotypic yeast vacuole fusion in vivo and in vitro. PLoS One 2013; 82:e56798; http://dx.doi.org/ 10.1371/journal.pone.0056798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barszczewski M, Chua JJ, Stein A, Winter U, Heintzmann R, Zilly FE, Fasshauer D, Lang T, Jahn R. A novel site of action for alpha-SNAP in the SNARE conformational cycle controlling membrane fusion. Mol Biol Cell 2008; 19:776-84; PMID:18094056; http://dx.doi.org/ 10.1091/mbc.E07-05-0498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bekal S, Domier LL, Gonfa B, Lakhssassi N, Meksem K, Lambert KN. A SNARE-like protein and biotin are implicated in soybean cyst nematode virulence. PLoS One 2015; 10:e0145601; PMID:26714307; http://dx.doi.org/ 10.1371/journal.pone.0145601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartz ML, Merz AJ. Capture and release of partially zipped trans-SNARE complexes on intact organelles. J Cell Biol 2009; 185:535-49; PMID:19414611; http://dx.doi.org/ 10.1083/jcb.200811082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lobingier BT, Nickerson DP, Lo SY, Merz AJ. SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec 18. Elife 2014; 3:e02272; PMID:24837546; http://dx.doi.org/ 10.7554/eLife.02272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu H, Rathore SS, Shen C, Liu Y, Ouyang Y, Stowell MH, Shen J. Reconstituting intracellular vesicle fusion reactions: the essential role of macromolecular crowding. J Am Chem Soc 2015; 137:12873-83; PMID:26431309; http://dx.doi.org/ 10.1021/jacs.5b08306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zick M, Stroupe C, Orr A, Douville D, Wickner WT. Membranes linked by trans-SNARE complexes require lipids prone to non-bilayer structure for progression to fusion. Elife 2014; 3:e01879; PMID:24596153; http://dx.doi.org/ 10.7554/eLife.01879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zick M, Stroupe C, Orr A, Douville D, Wickner WT. Correction: Membranes linked by trans-SNARE complexes require lipids prone to non-bilayer structure for progression to fusion. Elife 2015b; 4:e08843; http://dx.doi.org/ 10.7554/eLife.08843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zick M, Orra A, Schwartz ML, Merz AJ, Wickner WT. Sec17 can trigger fusion of trans-SNARE paired membranes without Sec 18. Proc Natl Acad Sci USA 2015a; 112:E2290-E2297; PMID:25902545; http://dx.doi.org/ 10.1073/pnas.1506409112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Block MR, Glick BS, Wilcox CA, Wieland FT, Rothman JE. Purification of an N-ethylmaleimide-sensitiveprotein catalyzing vesicular transport. Proc Natl Acad Sci USA 1988; 85:7852-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malhotra V, Orci L, Glick BS, Block MR, Rothman JE. Role of an N-ethylmaleimide-sensitive transport component in promoting fusion of transport vesicles with cisternae of the Golgi stack. Cell 1988; 54:221-7; http://dx.doi.org/ 10.1016/0092-8674(88)90554-5 [DOI] [PubMed] [Google Scholar]
- 72.Vivona S, Cipriano DJ, O'Leary S, Li YH, Fenn TD, Brunger AT. Disassembly of all SNARE complexes by N-ethylmaleimide-sensitive factor (NSF) is initiated by a conserved 1:1 interaction between α-soluble NSF attachment protein (SNAP) and SNARE complex. J Biol Chem 2013; 288:24984-91; PMID:23836889; http://dx.doi.org/ 10.1074/jbc.M113.489807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, Brunger AT. Mechanistic insights into the recycling machine of the SNARE complex. Nature 2015; 518:61-7; PMID:25581794; http://dx.doi.org/ 10.1038/nature14148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vahlensieck Y, Riezman H, Meyhack B. Transcriptional studies on yeast SEC genes provide no evidence for regulation at the transcriptional level. Yeast 1995; 11:901-11; PMID:8533466; http://dx.doi.org/ 10.1002/yea.320111002 [DOI] [PubMed] [Google Scholar]
- 75.Shanks SG, Carpp LN, Struthers MS, McCann RK, Bryant NJ. The Sec1/Munc18 protein Vps45 regulates cellular levels of its SNARE binding partners Tlg2 and Snc2 in Saccharomyces cerevisiae. PLoS One 2012; 7: e49628; PMID:23166732; http://dx.doi.org/ 10.1371/journal.pone.0049628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duden R, Kajikawa L, Wuestehube L, Schekman R. ε-COP is a structural component of coatomer that functions to stabilize α-COP. EMBO J 1998; 17:985-95; PMID:9463377; http://dx.doi.org/ 10.1093/emboj/17.4.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Otte S, Barlowe C. The Erv41p-Erv46p complex: multiple export signals are required in trans for COPII-dependent transport from the ER. EMBO J 2002; 21:6095-104; PMID:12426381; http://dx.doi.org/ 10.1093/emboj/cdf598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liberali P, Snijder B, Pelkmans L. A hierarchical map of regulatory genetic interactions in membrane trafficking. Cell 2014; 157:1473-87; PMID:24906158; http://dx.doi.org/ 10.1016/j.cell.2014.04.029 [DOI] [PubMed] [Google Scholar]
- 79.Forslund K, Morant M, Jørgensen B, Olsen CE, Asamizu E, Sato S, Tabata S, Bak S. Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiol 2004; 135:71-84; PMID:15122013; http://dx.doi.org/ 10.1104/pp.103.038059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morant AV, Bjarnholt N, Kragh ME, Kjærgaard CH, Jørgensen K, Paquette SM, Piotrowski M, Imberty A, Olsen CE, Møller BL, et al.. The β-glucosidases responsible for bioactivation of hydroxynitrile glucosides in Lotus japonicus. Plant Physiol 2008; 147:1072-91; PMID:18467457; http://dx.doi.org/ 10.1104/pp.107.109512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmitz G, Langmann T. Transcriptional regulatory networks in lipid metabolism control ABCA1 expression. Biochim Biophys Acta 2005; 1735:1-19; PMID:15922656; http://dx.doi.org/ 10.1016/j.bbalip.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 82.Tarr PT, Edwards PA. ABCG1 and ABCG4 are coexpressed in neurons and astrocytes of the CNS and regulate cholesterol homeostasis through SREBP-2. J Lipid Res 2008; 49:169-82; PMID:17916878; http://dx.doi.org/ 10.1194/jlr.M700364-JLR200 [DOI] [PubMed] [Google Scholar]
- 83.Ginglinger JF, Boachon B, Höfer R, Paetz C, Köllner TG, Miesch L, Lugan R, Baltenweck R, Mutterer J, Ullmann P, et al.. Gene coexpression analysis reveals complex metabolism of the monoterpene alcohol linalool in Arabidopsis flowers. Plant Cell 2013; 25:4640-57; PMID:24285789; http://dx.doi.org/ 10.1105/tpc.113.117382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu JH, Dimitman JE. Leaf structure and callose formation as determinants of TMV movement in bean leaves as revealed by UV irradiation studies. Virology 1970; 40:820-7; PMID:5432280; http://dx.doi.org/ 10.1016/0042-6822(70)90127-3 [DOI] [PubMed] [Google Scholar]
- 85.Aist JR. Papillae and related wound plugs of plant cells. Annu Rev Phytopathol 1976; 14:145-63; http://dx.doi.org/ 10.1146/annurev.py.14.090176.001045 [DOI] [Google Scholar]
- 86.Bonhoff A, Rieth B, Golecki J, Grisebach H. Race cultivar-specific differences in callose deposition in soybean roots following infection with Phytophthora megasperma f.sp. glycinea. Planta 1987; 172:101-5; PMID:24225793; http://dx.doi.org/ 10.1007/BF00403034 [DOI] [PubMed] [Google Scholar]
- 87.Jahnen W, Hahlbrock K. Cellular localization of nonhost resistance reactions of parsley (Petroselinum crispum) to fungal infection. Planta 1988; 173:197-204; PMID:24226400; http://dx.doi.org/ 10.1007/BF00403011 [DOI] [PubMed] [Google Scholar]
- 88.Kováts K, Binder A, Hohl HR. Cytology of induced systemic resistance of cucumber to Colletotrichum lagenarium. Planta 1991; 183:484-90; http://dx.doi.org/ 10.1007/BF00194268 [DOI] [PubMed] [Google Scholar]
- 89.Storti E, Latil C, Salti S, Bettini P, Bogani P, Pellegrini MG, Simeti C, Molnar A, Buiatti M. The in vitro physiological phenotype of tomato resistance to Fusarium oxysporum f. sp. lycopersici. Theor Appl Genet 1992; 84:123-8; PMID:24203038; http://dx.doi.org/ 10.1007/BF00223991 [DOI] [PubMed] [Google Scholar]
- 90.Beffa RS, Neuhaus JM, Meins F Jr. Physiological compensation in antisense transformants: specific induction of an “ersatz” glucan endo-1,3-beta-glucosidase in plants infected with necrotizing viruses. Proc Natl Acad Sci U S A 1993; 90:8792-6; PMID:8415609; http://dx.doi.org/ 10.1073/pnas.90.19.8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beffa RS, Hofer RM, Thomas M, Meins F Jr. Decreased susceptibility to viral disease of [beta]-1,3 glucanase-deficient plants generated by antisense transformation. Plant Cell 1996; 8:1001-11; PMID:12239410; http://dx.doi.org/ 10.1105/tpc.8.6.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rodríguez-Gálvez E, Mendgen K. Cell wall synthesis in cotton roots after infection with Fusarium oxysporum. The deposition of callose, arabinogalactans, xyloglucans, and pectic components into walls, wall appositions, cell plates and plasmodesmata. Planta 1995; 197:535-45; http://dx.doi.org/ 10.1007/BF00196676 [DOI] [PubMed] [Google Scholar]
- 93.Bestwick CS, Bennett MH, Mansfield JW. Hrp mutant of Pseudomonas syringae pv phaseolicola Induces cell wall alterations but not membrane damage leading to the hypersensitive reaction in lettuce. Plant Physiol 1995; 108:503-16; PMID:12228488; http://dx.doi.org/ 10.1104/pp.108.2.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benhamou N, Rey P, Chérif M, Hockenhull J, Tirilly Y. Treatment with the mycoparasite Pythium oligandrum triggers induction of defense-related reactions in tomato roots when challenged with Fusarium oxysporum f. sp. radicis-lycopersici. Phytopathology 1997; 87:108-22; PMID:18945162; http://dx.doi.org/ 10.1094/PHYTO.1997.87.1.108 [DOI] [PubMed] [Google Scholar]
- 95.Benhamou N, Kloepper JW, Quadt-Hallman A, Tuzun S. Induction of defense-related ultrastructural modifications in pea root tissues inoculated with endophytic bacteria. Plant Physiol 1996; 112:919-29; PMID:12226427; http://dx.doi.org/ 10.1104/pp.112.3.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Herbers K, Meuwly P, Frommer WB, Metraux JP, Sonnewald U. Systemic acquired resistance mediated by the ectopic expression of invertase: possible hexose sensing in the secretory pathway. Plant Cell 1996; 8:793-803; PMID:12239401; http://dx.doi.org/ 10.1105/tpc.8.5.793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang X, Xie M, Kim YJ, Zhou J, Klessig DF, Martin GB. Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 1999; 11:15-29; PMID:9878629; http://dx.doi.org/ 10.1105/tpc.11.2.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McCormack BA, Gregory AC, Kerry ME, Smith C, Bolwell GP. Purification of an elicitor-induced glucan synthase (callose synthase) from suspension cultures of French bean (Phaseolus vulgaris L.): purification and immunolocation of a probable M(r)-65,000 subunit of the enzyme. Planta 1997; 203:196-203; PMID:9362565; http://dx.doi.org/ 10.1007/s004250050182 [DOI] [PubMed] [Google Scholar]
- 99.Takahashi A, Kawasaki T, Henmi K, ShiI K, Kodama O, Satoh H, Shimamoto K. Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J 1999; 17:535-45; PMID:10205906; http://dx.doi.org/ 10.1046/j.1365-313X.1999.00405.x [DOI] [PubMed] [Google Scholar]
- 100.Vleeshouwers VG, van Dooijeweert W, Govers F, Kamoun S, Colon LT. The hypersensitive response is associated with host and nonhost resistance to Phytophthora infestans. Planta 2000; 210:853-64; PMID:10872215; http://dx.doi.org/ 10.1007/s004250050690 [DOI] [PubMed] [Google Scholar]
- 101.Zimmerli L, Jakab G, Metraux JP, Mauch-Mani B. Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta -aminobutyric acid. Proc Natl Acad Sci U S A 2000; 97:12920-5; PMID:11058166; http://dx.doi.org/ 10.1073/pnas.230416897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stone JM, Heard JE, Asai T, Ausubel FM. Simulation of fungal-mediated cell death by fumonisin B1 and selection of fumonisin B1-resistant (fbr) Arabidopsis mutants. Plant Cell 2000; 12:1811-22; PMID:11041878; http://dx.doi.org/ 10.1105/tpc.12.10.1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Donofrio NM, Delaney TP. Abnormal callose response phenotype and hypersusceptibility to Peronospoara parasitica in defence-compromised arabidopsis nim1-1 and salicylate hydroxylase-expressing plants. Mol Plant Microbe Interact 2001; 14:439-50; PMID:11310731; http://dx.doi.org/ 10.1094/MPMI.2001.14.4.439 [DOI] [PubMed] [Google Scholar]
- 104.Mellersh DG, Heath MC. Plasma membrane-cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell 2001; 13:413-24; PMID:11226194; http://dx.doi.org/ 10.1105/tpc.13.2.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rate DN, Greenberg JT. The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J 2001; 27:203-11; PMID:11532166; http://dx.doi.org/ 10.1046/j.0960-7412.2001.1075umedoc.x [DOI] [PubMed] [Google Scholar]
- 106.Iwano M, Che FS, Goto K, Tanaka N, Takayama S, Isogai A. Electron microscopic analysis of the H(2)O(2) accumulation preceding hypersensitive cell death induced by an incompatible strain of Pseudomonas avenae in cultured rice cells. Mol Plant Pathol 2002; 3:1-8; PMID:20569303; http://dx.doi.org/ 10.1046/j.1464-6722.2001.00087.x [DOI] [PubMed] [Google Scholar]
- 107.Veit S, Wörle JM, Nürnberger T, Koch W, Seitz HU. A novel protein elicitor (PaNie) from Pythium aphanidermatum induces multiple defense responses in carrot, Arabidopsis, and tobacco. Plant Physiol 2001; 127:832-41; PMID:11706166; http://dx.doi.org/ 10.1104/pp.010350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mithöfer A, Müller B, Wanner G, Eichacker LA. Identification of defence-related cell wall proteins in Phytophthora sojae-infected soybean roots by ESI-MS/MS. Mol Plant Pathol 2002; 3:163-166; PMID:20569322; http://dx.doi.org/ 10.1046/j.1364-3703.2002.00109.x [DOI] [PubMed] [Google Scholar]
- 109.Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci U S A 2003; 100:8577-82; PMID:12817082; http://dx.doi.org/ 10.1073/pnas.1431173100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fridborg I, Grainger J, Page A, Coleman M, Findlay K, Angell S. TIP, a novel host factor linking callose degradation with the cell-to-cell movement of Potato virus X. Mol Plant Microbe Interact 2003; 16:132-40; PMID:12575747; http://dx.doi.org/ 10.1094/MPMI.2003.16.2.132 [DOI] [PubMed] [Google Scholar]
- 111.Iglesias VA, Meins F Jr. Movement of plant viruses is delayed in a beta-1,3 glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J 2000; 21:157-66; PMID:10743656; http://dx.doi.org/ 10.1046/j.1365-313x.2000.00658.x [DOI] [PubMed] [Google Scholar]
- 112.Bucher GL, Tarina C, Heinlein M, Di Serio F, Meins F Jr, Iglesias VA. Local expression of enzymatically active class I beta-1, 3 glucanase enhances symptoms of TMV infection in tobacco. Plant J 2001; 28:361-69; PMID:11722778; http://dx.doi.org/ 10.1046/j.1365-313X.2001.01181.x [DOI] [PubMed] [Google Scholar]
- 113.Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB. An Arabidopsis Callose Synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 2003; 15:2503-13; PMID:14555698; http://dx.doi.org/ 10.1105/tpc.016097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Böhlenius H, Mørch SM, Godfrey D, Nielsen ME, Thordal-Christensen H. The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell 2010; 22:3831-44; PMID:21057060; http://dx.doi.org/ 10.1105/tpc.110.078063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nielsen ME, Feechan A, Böhlenius H, Ueda T, Thordal-Christensen H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc Natl Acad Sci U S A 2012; 109:11443-8; PMID:22733775; http://dx.doi.org/ 10.1073/pnas.1117596109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang L, Qin L, Liu G, Peremyslov VV, Dolja VV, Wei Y. Myosins XI modulate host cellular responses and penetration resistance to fungal pathogens. Proc Natl Acad Sci U S A 2014; 111: 13996-4001; PMID:25201952; http://dx.doi.org/ 10.1073/pnas.1405292111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Caillaud MC, Wirthmueller L, Sklenar J, Findlay K, Piquerez SJ, Jones AM, Robatzek S, Jones JD, Faulkner C. The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog 2014; 10:e1004496; PMID:25393742; http://dx.doi.org/ 10.1371/journal.ppat.1004496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM. Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 2000; 24:205-18; PMID:11069695; http://dx.doi.org/ 10.1046/j.1365-313x.2000.00870.x [DOI] [PubMed] [Google Scholar]
- 119.Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC. Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 2003; 301:969-72; PMID:12920300; http://dx.doi.org/ 10.1126/science.1086716 [DOI] [PubMed] [Google Scholar]
- 120.An Q, Hückelhoven R, Kogel KH, van Bel AJ. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol 2006; 8:1009-19; PMID:16681841; http://dx.doi.org/ 10.1111/j.1462-5822.2006.00683.x [DOI] [PubMed] [Google Scholar]
- 121.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell 1989; 1:37-52; PMID:2535466; http://dx.doi.org/ 10.1105/tpc.1.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994; 6:1211-25; PMID:7919989; http://dx.doi.org/ 10.1105/tpc.6.9.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wilson K, Long D, Swinburne J, Coupland G. A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 1996; 8:659-71; PMID:8624440; http://dx.doi.org/ 10.1105/tpc.8.4.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci U S A 1997; 94:1035-40; PMID:9023378; http://dx.doi.org/ 10.1073/pnas.94.3.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bankaitis VA, Malehorn DE, Emr SD, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol 1989; 108:1271-81; PMID:2466847; http://dx.doi.org/ 10.1083/jcb.108.4.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bankaitis VA, Aitken FR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 1990; 347:561-2; PMID:2215682; http://dx.doi.org/ 10.1038/347561a0 [DOI] [PubMed] [Google Scholar]
- 127.Skinner HB, McGee TP, McMaster CR, Fry MR, Bell RM, Bankaitis VA. The Saccharomyces cerevisiae phosphatidylinositol-transfer protein effects a ligand-dependent inhibition of choline-phosphate cytidylyltransferase activity. Proc Natl Acad Sci U S A 1995; 92:112-6; PMID:7816798; http://dx.doi.org/ 10.1073/pnas.92.1.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec 4p, targeting secretory vesicles to sites of exocytosis. EMBO J 1999; 18:1071-80; PMID:10022848; http://dx.doi.org/ 10.1093/emboj/18.4.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem 2012; 81:637-59; PMID:22463690; http://dx.doi.org/ 10.1146/annurev-biochem-052810-093700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jahn R, Scheller RH. SNAREs-engines for membrane fusion. Nat Rev Mol Cell Biol 2006; 7:631-43; PMID:16912714; http://dx.doi.org/ 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- 131.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 1994; 77:895-907; PMID:8004676; http://dx.doi.org/ 10.1016/0092-8674(94)90138-4 [DOI] [PubMed] [Google Scholar]
- 132.Lord C, Ferro-Novick S, Miller EA. The highly conserved COPII coat complex sorts cargo from the endoplasmic reticulum and targets it to the golgi. Cold Spring Harb Perspect Biol 2013; 5:a013367; PMID:23378591; http://dx.doi.org/ 10.1101/cshperspect.a013367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chung KP, Zeng Y, Jiang L. COPII paralogs in plants: functional redundancy or diversity? Trends Plant Sci 2016; 21:758-69; PMID:27317568; http://dx.doi.org/ 10.1016/j.tplants.2016.05.010 [DOI] [PubMed] [Google Scholar]
- 134.Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci 2003; 116: 1893-903; PMID:12668726; http://dx.doi.org/ 10.1242/jcs.00395 [DOI] [PubMed] [Google Scholar]
- 135.Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell 2005; 8:937-47; PMID:15935782; http://dx.doi.org/ 10.1016/j.devcel.2005.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001; 106:145-55; PMID:11511343; http://dx.doi.org/ 10.1016/S0092-8674(01)00434-2 [DOI] [PubMed] [Google Scholar]
- 137.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell 2002a; 3:271-82; PMID:12194857; http://dx.doi.org/ 10.1016/S1534-5807(02)00220-4 [DOI] [PubMed] [Google Scholar]
- 138.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 2002b; 3:283-9; PMID:12194858; http://dx.doi.org/ 10.1016/S1534-5807(02)00219-8 [DOI] [PubMed] [Google Scholar]
- 139.Xu WJ, Smith FJ, Subaran R, Mitchell AP. Multivesicular body-ESCRT components function in pH response regula tion in Saccharomyces cerevisiae and Candida albicans. Mol Biol Cell 2004; 15:5528-37; PMID:15371534; http://dx.doi.org/ 10.1091/mbc.E04-08-0666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gao CJ, Luo M, Zhao Q, Yang RZ, Cui Y, Zeng YL, Xia J, Jiang LW. A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol 2014; 24:2556-63; PMID:25438943; http://dx.doi.org/ 10.1016/j.cub.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 141.Gao CJ, Zhuang XH, Cui Y, Fu X, He YL, Zhao Q, Zeng YL, Shen JB, Luo M, Jiang LW. Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc Natl Acad Sci U S A 2015; 112:1886-91; PMID:25624505; http://dx.doi.org/ 10.1073/pnas.1421271112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cardona-Lopez X, Cuyas L, Marı´n E, Rajulu C, Irigoyen ML, Gil E, Puga MI, Bligny R, Nussaume L, Geldner N, et al.. ESCRT-III-associated protein ALIX mediates high affinity phosphate transporter trafficking to maintain phosphate homeostasis in Arabidopsis. Plant Cell 2015; 27:2560-81; PMID:26342016; http://dx.doi.org/ 10.1105/tpc.15.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shen J, Gao C, Zhao Q, Lin Y, Wang X, Zhuang X Jiang L. AtBRO1 functions in ESCRT-I complex to regulate multivesicular body protein sorting molecular plant 2016; 9:760-3; http://dx.doi.org/ 10.1016/j.molp.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 144.Bock JB, Lin RC, Scheller RH. A new syntaxin family member implicated in targeting of intracellular transport vesicles. J Biol Chem 1996; 271:17961-5; PMID:8663448; http://dx.doi.org/ 10.1074/jbc.271.30.17961 [DOI] [PubMed] [Google Scholar]
- 145.Bock JB, Klumperman J, Davanger S, Scheller RH. Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol Biol Cell 1997; 8:1261-71; PMID:9243506; http://dx.doi.org/ 10.1091/mbc.8.7.1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Holthuis JC, Nichols BJ, Dhruvakumar S, Pelham HR. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J 1998a; 17:113-26; PMID:9427746; http://dx.doi.org/ 10.1093/emboj/17.1.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Holthuis JCM, Nichols BJ, Pelham HRB. The syntaxin Tlg1p mediates trafficking of chitin synthase iii to polarized growth sites in yeast. Mol Biol Cell 1998b; 9:3383-97; PMID:9843576; http://dx.doi.org/ 10.1091/mbc.9.12.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Coe JGS, Lim ACB, Xu J, Hong W. A Role for Tlg1p in the transport of proteins within the Golgi apparatus of Saccharomyces cerevisiae. Mol Biol Cell 1999; 10:2407-23; PMID:10397773; http://dx.doi.org/ 10.1091/mbc.10.7.2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Watson RT, Pessin JE. Functional cooperation of 2 independent targeting domains in syntaxin 6 is required for its efficient localization in the trans-Golgi network of 3T3L1 adipocytes. J Biol Chem 2000; 275:1261-8; PMID:10625671; http://dx.doi.org/ 10.1074/jbc.275.2.1261 [DOI] [PubMed] [Google Scholar]
- 150.Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, Sudhof TC, Rizo J. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J 2002; 21:3620-31; PMID:12110575; http://dx.doi.org/ 10.1093/emboj/cdf381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Choudhury A, Marks DL, Proctor KM, Gould GW, Pagano RE. Regulation of caveolar endocytosis by syntaxin 6-dependent delivery of membrane components to the cell surface. Nat Cell Biol 2006; 8:317-28; PMID:16565709; http://dx.doi.org/ 10.1038/ncb1380 [DOI] [PubMed] [Google Scholar]
- 152.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J 2008; 27:923-33; PMID:18337752; http://dx.doi.org/ 10.1038/emboj.2008.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Huisman R, Hontelez J, Mysore KS, Wen J, Bisseling T, Limpens E. A symbiosis-dedicated SYNTAXIN OF PLANTS 13II isoform controls the formation of a stable host-microbe interface in symbiosis. New Phytol 2016; 211:1338-51; PMID:27110912; http://dx.doi.org/ 10.1111/nph.13973 [DOI] [PubMed] [Google Scholar]
- 154.Pan H, Oztas O, Zhang X, Wu X, Stonoha C, Wang E, Wang B, Wang D. A symbiotic SNARE protein generated by alternative termination of transcription. Nat Plants 2016; 2:15197; PMID:27249189; http://dx.doi.org/ 10.1038/nplants.2015.197 [DOI] [PubMed] [Google Scholar]
- 155.Suwastika IN, Uemura T, Shiina T, Sato MH, Takeyasu K. SYP71, a plant-specific Qc-SNARE protein, reveals dual localization to the plasma membrane and the endoplasmic reticulum in Arabidopsis. Cell Struct Funct 2008; 33:185-92; PMID:18827404; http://dx.doi.org/ 10.1247/csf.08024 [DOI] [PubMed] [Google Scholar]
- 156.El Kasmi F, Krause C, Hiller U, Stierhof YD, Mayer U, Conner L, Kong L, Reichardt I, Sanderfoot AA, Jürgens G. SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol Biol Cell 2013; 24:1593-601; PMID:23515225; http://dx.doi.org/ 10.1091/mbc.E13-02-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wei T, Zhang C, Hou H, Sanfacon H, Wang A. The SNARE protein Syp71 is essential for turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. Plos Pathogens 2013; 9:e1003378; PMID:23696741; http://dx.doi.org/ 10.1371/journal.ppat.1003378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.De Benedictis M, Bleve G, Faraco M, Stigliano E, Grieco F, Piro G, Dalessandro G, Di Sansebastiano GP. AtSYP51/52 functions diverge in the post-Golgi traffic and differently affect vacuolar sorting. Mol Plant 2013; 6:916-30; PMID:23087325; http://dx.doi.org/ 10.1093/mp/sss117 [DOI] [PubMed] [Google Scholar]
- 159.Wuestehube LJ, Duden R, Eun A, Hamamoto S, Korn P, Ram R, Schekman R. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics 1996; 142:393-406; PMID:8852839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hay JC, Hirling H, Scheller RH. Mammalian vesicle trafficking proteins of the endoplasmic reticulum and Golgi apparatus. J Biol Chem 1996; 271:5671-9; PMID:8621431; http://dx.doi.org/ 10.1074/jbc.271.10.5671 [DOI] [PubMed] [Google Scholar]
- 161.Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell 1997; 89:149-58; PMID:9094723; http://dx.doi.org/ 10.1016/S0092-8674(00)80191-9 [DOI] [PubMed] [Google Scholar]
- 162.Zhang T, Wong SH, Tang BL, Xu Y, Peter F, Subramaniam VN, Hong W. The mammalian protein (rbet1) homologous to yeast Bet1p is primarily associated with the pre-Golgi intermediate compartment and is involved in vesicular transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol 1997; 139:1157-68; PMID:9382863; http://dx.doi.org/ 10.1083/jcb.139.5.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stone S, Sacher M, Mao Y, Carr C, Lyons P, Quinn AM, Ferro-Novick S. Bet1p activates the v-SNARE Bos1p. Mol Biol Cell 1997; 8:1175-81; PMID:9243499; http://dx.doi.org/ 10.1091/mbc.8.7.1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Springer S, Schekman R. Nucleation of COPII vesicular coat complex by endoplasmic reticulum to Golgi vesicle SNAREs. Science 1998; 281:698-700; PMID:9685263; http://dx.doi.org/ 10.1126/science.281.5377.698 [DOI] [PubMed] [Google Scholar]
- 165.Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JR 3rd, Abeliovich H, Ferro-Novick S. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J 1998; 17:2494-503; PMID:9564032; http://dx.doi.org/ 10.1093/emboj/17.9.2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rowe T, Dascher C, Bannykh S, Plutner H, Balch WE. Role of vesicle-associated syntaxin 5 in the assembly of pre-Golgi intermediates. Science 1998; 279:696-700; PMID:9445473; http://dx.doi.org/ 10.1126/science.279.5351.696 [DOI] [PubMed] [Google Scholar]
- 167.Kuehn MJ, Herrmann JM, Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature 1998; 391:187-90; PMID:9428766; http://dx.doi.org/ 10.1038/34438 [DOI] [PubMed] [Google Scholar]
- 168.VanRheenen SM, Cao X, Lupashin VV, Barlowe C, Waters MG. Sec 35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J Cell Biol 1998; 141:1107-19; PMID:9606204; http://dx.doi.org/ 10.1083/jcb.141.5.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Adolf F, Rhiel M, Reckmann I, Wieland FT. Sec 24C/D-isoform-specific sorting of the preassembled ER-Golgi Q-SNARE complex. Mol Biol Cell 2016; 27:2697-707; PMID:27413010; http://dx.doi.org/ 10.1091/mbc.E16-04-0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kim DW, Sacher M, Scarpa A, Quinn AM, Ferro-Novick S. High-copy suppressor analysis reveals a physical interaction between Sec 34p and Sec 35p, a protein implicated in vesicle docking. Mol Biol Cell 1999; 10:3317-29; PMID:10512869; http://dx.doi.org/ 10.1091/mbc.10.10.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kuge O, Hara-Kuge S, Orci L, Ravazzola M, Amherdt M, Tanigawa G, Wieland FT, Rothman JE. zeta-COP, a subunit of coatomer, is required for COP-coated vesicle assembly. J Cell Biol 1993; 123:1727-34; PMID:8276893; http://dx.doi.org/ 10.1083/jcb.123.6.1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cosson P, Demolliere C, Hennecke S, Duden R, Letourneur F. γ- and ζ-COP, 2 coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. EMBO J 1996; 15:1792-8; PMID:8617224 [PMC free article] [PubMed] [Google Scholar]
- 173.Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol 2009; 185:305-21; PMID:19364919; http://dx.doi.org/ 10.1083/jcb.200810098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Beck R, Rawet M, Wieland FT, Cassel D. The COPI system: Molecular mechanisms and function. FEBS Lett 2009; 583:2701-9; PMID:19631211; http://dx.doi.org/ 10.1016/j.febslet.2009.07.032 [DOI] [PubMed] [Google Scholar]
- 175.Hsia KC, Hoelz A. Crystal structure of alpha-COP in complex with epsilon-COP provides insight into the architecture of the COPI vesicular coat. Proc Natl Acad Sci U S A 2010; 107:11271-6; PMID:20534429; http://dx.doi.org/ 10.1073/pnas.1006297107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Presley JF, Ward TH, Pfeifer AC, Siggia ED, Phair RD, Lippincott-Schwartz J. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature 2002; 417:187-93; PMID:12000962; http://dx.doi.org/ 10.1038/417187a [DOI] [PubMed] [Google Scholar]
- 177.Yu X, Breitman M, Goldberg J. A structure-based mechanism for arf1-dependent recruitment of coatomer to membranes. Cell 2012; 148:530-42; PMID:22304919; http://dx.doi.org/ 10.1016/j.cell.2012.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ahn HK, Kang YW, Lim HM, Hwang I, Pai HS. Physiological functions of the COPI complex in higher plants. Mol Cells 2015; 38:866-75; PMID:26434491; http://dx.doi.org/ 10.14348/molcells.2015.0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hosobuchi M, Kreis TE, Schekman R. SEC21 is a gene required for ER to Golgi protein transport that encodes a subunit of a yeast coatomer. Nature 1992; 360:603-5; PMID:1461285; http://dx.doi.org/ 10.1038/360603a0 [DOI] [PubMed] [Google Scholar]
- 180.Letourneur F, Gaynor EC, Hennecke S, Démollière C, Duden R, Emr SD, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 1994; 79:1199-207; PMID:8001155; http://dx.doi.org/ 10.1016/0092-8674(94)90011-6 [DOI] [PubMed] [Google Scholar]
- 181.Schweizer A, Ericsson M, Bächi T, Griffiths G, Hauri HP. Characterization of a novel 63 kDa membrane protein. Implications for the organization of the ER-to-Golgi pathway. J Cell Sci 1993; 104:671-83. [DOI] [PubMed] [Google Scholar]
- 182.Appenzeller C, Andersson H, Kappeler F, Hauri HP. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat Cell Biol 1999; 1:330-4; PMID:10559958; http://dx.doi.org/ 10.1038/14020 [DOI] [PubMed] [Google Scholar]
- 183.Appenzeller-Herzog C, Hauri HP. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J Cell Sci 2006; 119:2173-83; PMID:16723730; http://dx.doi.org/ 10.1242/jcs.03019 [DOI] [PubMed] [Google Scholar]
- 184.Breuza L, Halbeisen R, Jenö P, Otte S, Barlowe C, Hong W, Hauri HP. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J Biol Chem 2004; 279:47242-53; PMID:15308636; http://dx.doi.org/ 10.1074/jbc.M406644200 [DOI] [PubMed] [Google Scholar]
- 185.Ben-Tekaya H, Miura K, Pepperkok R, Hauri HP. Live imaging of bidirectional traffic from the ERGIC. J Cell Sci 2005; 118:357-67; PMID:15632110; http://dx.doi.org/ 10.1242/jcs.01615 [DOI] [PubMed] [Google Scholar]
- 186.Welsh LM, Tong AH, Boone C, Jensen ON, Otte S. Genetic and molecular interactions of the Erv41p-Erv46p complex involved in transport between the endoplasmic reticulum and Golgi complex. J Cell Sci 2006; 119:4730-40; PMID:17077122; http://dx.doi.org/ 10.1242/jcs.03250 [DOI] [PubMed] [Google Scholar]
- 187.Lorente-Rodrıguez A, Barlowe C. Entry and exit mechanisms at the cis-face of the Golgi complex. Cold Spring Harb Perspect Biol 2011; 3:a005207; PMID:21482742; http://dx.doi.org/ 10.1101/cshperspect.a005207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Donohoe BS, Kang BH, Gerl MJ, Gergely ZR, McMichael CM, Bednarek SY, Staehelin LA. Cis-Golgi cisternal assembly and biosynthetic activation occur sequentially in plants and algae. Traffic 2013; 14:551-67; PMID:23369235; http://dx.doi.org/ 10.1111/tra.12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wu M, Tu T, Huang Y, Cao Y. Suppression subtractive hybridization identified differentially expressed genes in lung adenocarcinoma: ERGIC3 as a novel lung cancer-related gene. BMC Cancer 2013; 13:44; PMID:23374247; http://dx.doi.org/ 10.1186/1471-2407-13-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shibuya A, Margulis N, Christiano R, Walther TC, Barlowe C. The Erv41-Erv46 complex serves as a retrograde receptor to retrieve escaped ER proteins. J Cell Biol 2015; 208:197-209; PMID:25583996; http://dx.doi.org/ 10.1083/jcb.201408024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hong SH, Chang SH, Cho KC, Kim S, Park S, Lee AY, Jiang HL, Kim HJ, Lee S, Yu KN, et al.. Endoplasmic reticulum-Golgi intermediate compartment protein 3 knockdown suppresses lung cancer through endoplasmic reticulum stress-induced autophagy. Oncotarget 2016; 7(40):65335-47; http://dx.doi.org/ 10.18632/oncotarget.11678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, Hoheisel JD. Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J Biol Chem 2001; 277:10555-61; PMID:11748215; http://dx.doi.org/ 10.1074/jbc.M104863200 [DOI] [PubMed] [Google Scholar]
- 193.Ross JP. Host-Parasite relationship of the soybean cyst nematode in resistant soybean roots. Phytopathology 1958; 48:578-9. [Google Scholar]
- 194.Endo BY. Histological responses of resistant and susceptible soybean varieties, and backcross progeny to entry development of Heterodera glycines. Phytopathology 1965; 55:375-81. [Google Scholar]
- 195.Endo BY. Ultrastructure of initial responses of susceptible and resistant soybean roots to infection by Heterodera glycines. Revue Nématology 1991; 14:73-84. [Google Scholar]
- 196.Endo BY, Veech JA. Morphology and histochemistry of soybean roots infected with Heterodera glycines. Phytopathology 1970; 60:1493-8; http://dx.doi.org/ 10.1094/Phyto-60-1493 [DOI] [Google Scholar]
- 197.White AR, Xin Y, Pezeshk V. Xyloglucan glucosyltransferase in Golgi membranes from Pisum sativum (pea). Biochem J 1993; 294:231-8; PMID:8363577; http://dx.doi.org/ 10.1042/bj2940231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sherrier DJ, VandenBosch KA. Secretion of cell wall poly- saccharides in Vicia root hairs. Plant J 1994; 5:185-95; http://dx.doi.org/ 10.1046/j.1365-313X.1994.05020185.x [DOI] [Google Scholar]
- 199.Driouich A, Follet-Gueye ML, Bernard S, Kousar S, Chevalier L, Vicré-Gibouin M, Lerouxel O. Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Front Plant Sci 2012; 3:79; PMID:22639665; http://dx.doi.org/ 10.3389/fpls.2012.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 1992; 282:821-8; PMID:1554366; http://dx.doi.org/ 10.1042/bj2820821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fry SC, Mohler KE, Nesselrode BH, Frankova L. Mixed-linkage beta-glucan: xyloglucan endotransglucosylase, a novel wall-remodelling enzyme from Equisetum (horsetails) and charophytic algae. Plant J 2008; 55:240-52; PMID:18397375; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03504.x [DOI] [PubMed] [Google Scholar]
- 202.Van Sandt VS, Guisez Y, Verbelen JP, Vissenberg K. Analysis of a xyloglucan endotransglycosylase/hydrolase (XTH) from the lycopodiophyte Selaginella kraussiana suggests that XTH sequence characteristics and function are highly conserved during the evolution of vascular plants. J Exp Bot 2006; 57:2909-22; PMID:16873447; http://dx.doi.org/ 10.1093/jxb/erl064 [DOI] [PubMed] [Google Scholar]
- 203.Van Sandt VS, Guisez Y, Verbelen JP, Vissenberg K. Xyloglucan endotransglycosylase/hydrolase (XTH) is encoded by a multi-gene family in the primitive vascular land plant Selaginella kraussiana. Plant Biol (Stuttg) 2007; 9:142-6; PMID:17099842; http://dx.doi.org/ 10.1055/s-2006-924661 [DOI] [PubMed] [Google Scholar]
- 204.Eklof JM, Brumer H. The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol 2010; 153:456-66; PMID:20421457; http://dx.doi.org/ 10.1104/pp.110.156844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.de Vries H. Plasmalytische studien die wand der vakuolen. Jb f wies Bot 1885; 16:465-598 [Google Scholar]
- 206.Whaley WG, Mollenhauer HH. The Golgi apparatus and cell plate formation-a postulate. J Cell Biol 1963; 17:216-21; PMID:14000199; http://dx.doi.org/ 10.1083/jcb.17.1.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mollenhauer HH, Whaley WG. An observation on the functioning of the Golgi apparatus. J Cell Biol 1963; 17:222-5; PMID:13935874; http://dx.doi.org/ 10.1083/jcb.17.1.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Matile P, Moor H. Vacuolation: Origin and development of the lysosomal apparatus in root-tip cells. Planta 1968; 80:159-75; http://dx.doi.org/ 10.1007/BF00385592 [DOI] [Google Scholar]
- 209.Rose JK, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 2002; 43:1421-35; PMID:12514239; http://dx.doi.org/ 10.1093/pcp/pcf171 [DOI] [PubMed] [Google Scholar]
- 210.Nishitani K, Tominaga R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 1992; 267:21058-64; PMID:1400418 [PubMed] [Google Scholar]
- 211.Potter I, Fry SC. Xyloglucan endotransglycosylase activity in pea internodes. Effects of applied gibberellic acid. Plant Physiol 1993; 103:235-41; PMID:8208849; http://dx.doi.org/ 10.1104/pp.103.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pritchard J, Hetherington PR, Fry SC, Tomos AD. Xyloglucan endotransglycosylase activity, microfibril orientation and the profiles of cell wall properties along growing regions of maize roots. J Exp Bot 1993; 265:1281-9; http://dx.doi.org/ 10.1093/jxb/44.8.1281 [DOI] [Google Scholar]
- 213.Nishitani K. The role of endoxyloglucan transferase in the organization of plant cell walls. Int Rev Cytol 1997; 173:157-206; PMID:9127953; http://dx.doi.org/ 10.1016/S0074-7696(08)62477-8 [DOI] [PubMed] [Google Scholar]
- 214.Campbell P, Braam J. Co- and/or post-translational modifications are critical for TCH4 XET activity. Plant J 1998; 15:553-61; PMID:9753780; http://dx.doi.org/ 10.1046/j.1365-313X.1998.00239.x [DOI] [PubMed] [Google Scholar]
- 215.Yokoyama R, Nishitani K. Endoxyloglucan transferase is localized both in the cell plate and in the secretory pathway destined for the apoplast in tobacco cells. Plant Cell Physiol 2001; 42:292-300; PMID:11266580; http://dx.doi.org/ 10.1093/pcp/pce034 [DOI] [PubMed] [Google Scholar]
- 216.Henriksson H, Denman SE, Campuzano ID, Ademark P, Master ER, Teeri TT, Brumer H 3rd. N-linked glycosylation of native and recombinant cauliflower xyloglucan endotransglycosylase 16A. Biochem J 2003; 375:61-73; PMID:12826015; http://dx.doi.org/ 10.1042/bj20030485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Albert M, Werner M, Proksch P, Fry SC, Kaldenhoff R. The cell wall-modifying xyloglucan endotransglycosylase/hydrolase LeXTH1 is expressed during the defence reaction of tomato against the plant parasite Cuscuta reflexa. Plant Biol (Stuttg) 2004; 6:402-7; PMID:15248122; http://dx.doi.org/ 10.1055/s-2004-817959 [DOI] [PubMed] [Google Scholar]
- 218.Kallas AM, Piens K, Denman SE, Henriksson H, Fäldt J, Johansson P, Brumer H, Teeri TT. Enzymatic properties of native and deglycosylated hybrid aspen (Populus tremulaxtremuloides) xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochem J 2005; 390:105-13; PMID:15804235; http://dx.doi.org/ 10.1042/BJ20041749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Genovesi V, Fornalé S, Fry SC, Ruel K, Ferrer P, Encina A, Sonbol FM, Bosch J, Puigdomènech P, Rigau J, et al.. ZmXTH1, a new xyloglucan endotransglucosylase/hydrolase in maize, affects cell wall structure and composition in Arabidopsis thaliana. J Exp Bot 2008; 59:875-89; PMID:18316315; http://dx.doi.org/ 10.1093/jxb/ern013 [DOI] [PubMed] [Google Scholar]
- 220.Maris A, Suslov D, Fry SC, Verbelen JP, Vissenberg K. Enzymic characterization of 2 recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J Exp Bot 2009; 60:3959-72; PMID:19635745; http://dx.doi.org/ 10.1093/jxb/erp229 [DOI] [PubMed] [Google Scholar]
- 221.Nishikubo N, Takahashi J, Roos AA, Derba-Maceluch M, Piens K, Brumer H, Teeri TT, Stålbrand H, Mellerowicz EJ. Xyloglucan endo-transglycosylase-mediated xyloglucan rearrangements in developing wood of hybrid aspen. Plant Physiol 2011; 155:399-413; PMID:21057113; http://dx.doi.org/ 10.1104/pp.110.166934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wink M. Annual Plant Reviews, Volume 40, 2nd Edition, Biochemistry of Plant Secondary Metabolism. Wiley; 2011:464; http://dx.doi.org/ 10.1002/9781444320503.ch1 [DOI] [Google Scholar]
- 223.Facchini PJ. Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu Rev Plant Physiol Plant Mol Biol 2001; 52:29-66; PMID:11337391; http://dx.doi.org/ 10.1146/annurev.arplant.52.1.29 [DOI] [PubMed] [Google Scholar]
- 224.Beaudoin GA, Facchini PJ. Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta 2014; 240:19-32; PMID:24671624; http://dx.doi.org/ 10.1007/s00425-014-2056-8 [DOI] [PubMed] [Google Scholar]
- 225.Barton DHR, Hesse RH, Kirby GW. The origin of the berberine carbon. Proc Chem Soc (London) 1963; 189:267-68. [Google Scholar]
- 226.Battersby AR, Francis RJ, Hirst M, Staunton J. Biosynthesis of the berberine bridge. Proc Chem Soc (London) 1963; 189:268-8. [Google Scholar]
- 227.Rink E, Bohm H. Conversion of reticuline into scoulerine by a cell free preparation from Macleaya microcarpa cell suspension cultures. FEBS Lett 1973; 49:396-9; http://dx.doi.org/ 10.1016/0014-5793(75)80794-0 [DOI] [PubMed] [Google Scholar]
- 228.Steffens P, Nagakura N, Meinhart H. Purification and characterization of the berberine bridge enzyme from Berberis beaniana cell cultures. Phytochemistry 1985; 24:2577-83; http://dx.doi.org/ 10.1016/S0031-9422(00)80672-X [DOI] [Google Scholar]
- 229.Loeffler S, Zenk MH. The hydroxylation step in the biosynthetic pathway leading from norcoclaurine to reticuline. Phytochemistry 1990; 29:3499-503; http://dx.doi.org/ 10.1016/0031-9422(90)85264-G [DOI] [Google Scholar]
- 230.Dittrich H, Kutchan TM. Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc Natl Acad Sci U S A 1991; 88:9969-73; PMID:1946465; http://dx.doi.org/ 10.1073/pnas.88.22.9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kutchan TM, Dittrich H. Characterization and mechanism of the berberine bridge enzyme, a covalently flavinylated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J Biol Chem 1995; 270:24475-81; PMID:7592663; http://dx.doi.org/ 10.1074/jbc.270.41.24475 [DOI] [PubMed] [Google Scholar]
- 232.Hauschild K, Pauli HH, Kutchan TM. Isolation and analysis of a gene bbe1 encoding the berberine bridge enzyme from the California poppy Eschscholzia californica. Plant Mol Biol 1998; 36:473-8; PMID:9484487; http://dx.doi.org/ 10.1023/A:1005917808232 [DOI] [PubMed] [Google Scholar]
- 233.Pauli HH, Kutchan TM. Molecular cloning and functional heterologous expression of 2 alleles encoding (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P-450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J 1998; 13:793-801; PMID:9681018; http://dx.doi.org/ 10.1046/j.1365-313X.1998.00085.x [DOI] [PubMed] [Google Scholar]
- 234.Morishige T, Tsujita T, Yamada Y, Sato F. Molecular characterization of the S-adenosyl-L-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J Biol Chem 2000; 275:23398-405; PMID:10811648; http://dx.doi.org/ 10.1074/jbc.M002439200 [DOI] [PubMed] [Google Scholar]
- 235.Samanani N, Facchini PJ. Purification and characterization of norcoclaurine synthase. The first committed enzyme in benzylisoquinoline alkaloid biosynthesis in plants. J Biol Chem 2002; 277:33878-83; PMID:12107162; http://dx.doi.org/ 10.1074/jbc.M203051200 [DOI] [PubMed] [Google Scholar]
- 236.Choi KB, Morishige T, Shitan N, Yazaki K, Sato F. Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J Biol Chem 2002; 277:830-5; PMID:11682473; http://dx.doi.org/ 10.1074/jbc.M106405200 [DOI] [PubMed] [Google Scholar]
- 237.Winkler A, Puhl M, Weber H, Kutchan TM, Gruber K, Macheroux P. Berberine bridge enzyme catalyzes the 6 electron oxidation of (S)-reticuline to dehydroscoulerine. Phytochemistry 2009; 70:1092-7; PMID:19570558; http://dx.doi.org/ 10.1016/j.phytochem.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 238.Poeaknapo C, Schmidt J, Brandsch M, Drager B, Zenk MH. Endogenous formation of morphine in human cells. Proc Natl Acad Sci U S A 2004; 101:14091-6; PMID:15383669; http://dx.doi.org/ 10.1073/pnas.0405430101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Boettcher C, Fellermeier M, Boettcher C, Drager B, Zenk MH. How human neuroblastoma cells make morphine. Proc Natl Acad Sci U S A 2005; 102:8495-500; PMID:15937106; http://dx.doi.org/ 10.1073/pnas.0503244102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Frenzel T, Beale JM, Kobayashi M, Zenk MH, Floss HG. Stereochemistry of enzymatic formation of the berberine bridge in protoberberine alkaloids. J Am Chem Soc 1988; 110:7878-80; http://dx.doi.org/ 10.1021/ja00231a050 [DOI] [Google Scholar]
- 241.Amann M, Wanner G, Zenk MH. Intracellular compartmentation of 2 enzymes of berberine biosynthesis in plant cell cultures. Planta 1986; 167:310-20; PMID:24240298; http://dx.doi.org/ 10.1007/BF00391333 [DOI] [PubMed] [Google Scholar]
- 242.Bird DA, Franceschi VR, Facchini PJ. A tale of 3 cell types: Alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 2003; 15:2626-35; PMID:14508000; http://dx.doi.org/ 10.1105/tpc.015396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Bird DA, Facchini PJ. Berberine bridge enzyme, a key branch-point enzyme in benzylisoquinoline alkaloid biosynthesis, contains a vacuolar sorting determinant. Planta 2001; 213:888-97; PMID:11722125; http://dx.doi.org/ 10.1007/s004250100582 [DOI] [PubMed] [Google Scholar]
- 244.Alcantara J, Bird DA, Franceschi VR, Facchini PJ. Sanguinarine biosynthesis is associated with the endoplasmic reticulum in cultured opium poppy cells after elicitor treatment. Plant Physiol 2005; 138:173-83; PMID:15849302; http://dx.doi.org/ 10.1104/pp.105.059287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Galneder E, Rueffer M, Wanner G, Tabata M, Zenk MH. Alternative final steps in berberine biosynthesis in Coptis japonica cell cultures. Plant Cell Rep 1988; 7:1-4; PMID:24241402; http://dx.doi.org/ 10.1007/BF00272964 [DOI] [PubMed] [Google Scholar]
- 246.Bock A, Wanner G, Zenk MH. Immunocytological localization of 2 enzymes involved in berberine biosynthesis. Planta 2002; 216:57-63; PMID:12430014; http://dx.doi.org/ 10.1007/s00425-002-0867-5 [DOI] [PubMed] [Google Scholar]
- 247.Shitan N, Bazin I, Dan K, Obata K, Kigawa K, Ueda K, Sato F, Forestier C, Yazaki K. Involvement of CjMDR1, a plant multidrug-resistance-type ATP-binding cassette protein, in alkaloid transport in Coptis japonica. Proc Natl Acad Sci U S A 2003; 100:751-6; PMID:12524452; http://dx.doi.org/ 10.1073/pnas.0134257100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Liu JJ, Odegard W, de Lumen BO. Galactinol synthase from kidney bean cotyledon and zucchini leaf. Purification and N-terminal sequences. Plant Physiol 1995; 109:505-11; PMID:7480343; http://dx.doi.org/ 10.1104/pp.109.2.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Saravitz DM, Pharr DM, Carter TE. Galactinol synthase activity and soluble sugars in developing seeds of 4 soybean genotypes. Plant Physiol 1987; 83:185-9; PMID:16665199; http://dx.doi.org/ 10.1104/pp.83.1.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kim MS, Cho SM, Kang EY, Im YJ, Hwangbo H, Kim YC, Ryu C-M, Yang KY, Chung GC, Cho BH. Galactinol is a signaling component of the induced systemic resistance caused by Pseudomonas chlororaphis O6 root colonization. Mol Plant Microbe Interact 2008; 21:1643-53; PMID:18986260; http://dx.doi.org/ 10.1094/MPMI-21-12-1643 [DOI] [PubMed] [Google Scholar]
- 251.Bachmann M, Matile P, Keller F. Metabolism of the raffinose family oligosaccharides in leaves of Ajuga reptans L. Cold acclimation, translocation, and sink to source transition: discovery of a chain elongation enzyme. Plant Physiol 1994; 105:1335-45; PMID:12232288; http://dx.doi.org/ 10.1104/pp.105.4.1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hoch G, Peterbauer T, Richter A. Purification and characterization of stachyose synthase from lentil (Lens culinaris) seeds: galactopinitol and stachyose synthesis. Arch Biochem Biophys 1999; 366:75-81; PMID:10334866; http://dx.doi.org/ 10.1006/abbi.1999.1212 [DOI] [PubMed] [Google Scholar]
- 253.Peterbauer T, Mach L, Mucha J, Richter A. Functional expression of a cDNA encoding pea (Pisum sativum L.) raffinose synthase, partial purification of the enzyme from maturing seeds, and steady-state kinetic analysis of raffinose synthesis. Planta 2002; 215:839-46; PMID:12244450; http://dx.doi.org/ 10.1007/s00425-002-0804-7 [DOI] [PubMed] [Google Scholar]
- 254.Meyer D, Pajonk S, Micali C, O'Connell R, Schulze-Lefert P. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J 2009; 57:986-99; PMID:19000165; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03743.x [DOI] [PubMed] [Google Scholar]
- 255.Ellinger D, Naumann M, Falter C, Zwikowics C, Jamrow T, Manisseri C, Somerville SC, Voigt CA. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol 2013; 161:1433-44; PMID:23335625; http://dx.doi.org/ 10.1104/pp.112.211011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al.. Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 2011; 21:1144-55; http://dx.doi.org/ 10.1016/j.devcel.2011.10.006 [DOI] [PubMed] [Google Scholar]
- 257.Eggert D, Naumann M, Reimer R, Voigt CA. Nanoscale glucan polymer network causes pathogen resistance. Scientific Reports 2014; 4:4159; PMID:24561766; http://dx.doi.org/ 10.1038/srep04159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/ 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 259.Golden AM, Epps JM, Riggs RD, Duclos LA, Fox JA, Bernard RL. Terminology and identity of infraspecific forms of the soybean cyst nematode (Heterodera glycines). Plant Dis Rep 1970; 54:544-46 [Google Scholar]
- 260.Assaad FF, Qiu JL, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, Wanner G, Peck SC, Edwards H, Ramonell K, et al.. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell 2004; 15:5118-29; PMID:15342780; http://dx.doi.org/ 10.1091/mbc.E04-02-0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lipka V, Kwon C, Panstruga R. SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu Rev Cell Dev Biol 2007; 23:147-74; PMID:17506694; http://dx.doi.org/ 10.1146/annurev.cellbio.23.090506.123529 [DOI] [PubMed] [Google Scholar]
- 262.Kalde M, Nühse TS, Findlay K, Peck SC. The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc Natl Acad Sci USA 2007; 104:11850-5; PMID:17592123; http://dx.doi.org/ 10.1073/pnas.0701083104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wang W, Devoto A, Turner JG, Xiao S. Expression of the membrane-associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Mol Plant Microbe Interact 2007; 20: 966-76; PMID:17722700; http://dx.doi.org/ 10.1094/MPMI-20-8-0966 [DOI] [PubMed] [Google Scholar]
- 264.Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al.. Co-option of a default secretory pathway for plant immune responses. Nature 2008; 451:835-40; PMID:18273019; http://dx.doi.org/ 10.1038/nature06545 [DOI] [PubMed] [Google Scholar]
- 265.Pajonk S, Kwon C, Clemens N, Panstruga R, Schulze-Lefert P. Activity determinants and functional specialization of Arabidopsis PEN1 syntaxin in innate immunity. J Biol Chem 2008; 283:26974-84; PMID:18678865; http://dx.doi.org/ 10.1074/jbc.M805236200 [DOI] [PubMed] [Google Scholar]
- 266.Kim H, O'Connell R, Maekawa-Yoshikawa M, Uemura T, Neumann U, Schulze-Lefert P. The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes. Plant J 2014; 79:835-47; PMID:24941879; http://dx.doi.org/ 10.1111/tpj.12591 [DOI] [PubMed] [Google Scholar]
- 267.Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 1999; 284:1976-9; PMID:10373113; http://dx.doi.org/ 10.1126/science.284.5422.1976 [DOI] [PubMed] [Google Scholar]
- 268.Faik A, Price NJ, Raikhel NV, Keegstra K. An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci U S A 2002; 99:7797-802; PMID:12032363; http://dx.doi.org/ 10.1073/pnas.102644799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci U S A 2002; 99:3340-5; PMID:11854459; http://dx.doi.org/ 10.1073/pnas.052450699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Cavalier DM, Keegstra K. Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J Biol Chem 2006; 281:34197-207; PMID:16982611; http://dx.doi.org/ 10.1074/jbc.M606379200 [DOI] [PubMed] [Google Scholar]
- 271.Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc Natl Acad Sci U S A 2007; 104:8550-5; PMID:17488821; http://dx.doi.org/ 10.1073/pnas.0703133104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Tamura K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I. KATAMARI1/MURUS3 is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 2005; 17:1764-76; PMID:15863516; http://dx.doi.org/ 10.1105/tpc.105.031930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou YH, Eberhard S, Danhof L, Keegstra K, Hahn MG. Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol 2012; 159:1367-84; PMID:22696020; http://dx.doi.org/ 10.1104/pp.112.198119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Chou YH, Pogorelko G, Zabotina OA. Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol 2012; 159:1355-6; PMID:22665445; http://dx.doi.org/ 10.1104/pp.112.199356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Chou YH, Pogorelko G, Young ZT, Zabotina OA. Protein-protein interactions among xyloglucan-synthesizing enzymes and formation of Golgi-localized multiprotein complexes. Plant Cell Physiol 2015; 56:255-67; PMID:25392066; http://dx.doi.org/ 10.1093/pcp/pcu161 [DOI] [PubMed] [Google Scholar]
- 276.Mann HB, Whitney DR. On a test of whether one of 2 random variables is stochastically larger than the other. Annals Mathematical Statistics 1947; 18:50-60; http://dx.doi.org/ 10.1214/aoms/1177730491 [DOI] [Google Scholar]
- 277.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 2011; 8:785-6; PMID:21959131; http://dx.doi.org/ 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


