Abstract
Osteopontin (Opn), a multifunctional extracellular matrix protein, is implicated in the pathogenesis of various inflammatory disorders. Under physiologic conditions, its expression is restricted to certain tissues including bone and kidney tubule. However, cellular activation during disease development induces Opn expression in various immune cells. In this study, using Opn-EGFP knock-in (KI) mice we found that CD8α+ T cells in the intestinal tissues, including Peyer’s patch, lamina propria and epithelium, express Opn under steady state conditions. Therefore, we examined the role of Opn-expressing CD8α+ T cells in intestinal homeostasis. Interestingly, Opn knockout (KO) mice had altered fecal microflora concordant with a reduction of TCRγδ+ intraepithelial lymphocytes (IELs). Consistent with this result, both treatment with anti-Opn blocking antibody and deficiency of Opn resulted in decreased survival of TCRγδ+ and TCRαβ+ IELs. This data suggests that a possibility that Opn may function as a survival factor for IELs in the intestinal tissue. Collectively, these data suggest the possibility that Opn might regulate the homeostasis of intestinal microflora through maintenance of TCRγδ+ IELs, possibly by support of IEL survival.
Introduction
Osteopontin (Opn), a multifunctional extracellular matrix protein, contains at least two distinct cell-binding domains; Arg159-Gly-Asp161 (RGD), which binds to the RGD-recognizing integrin αvβ3, and Ser162-Val-Val-Tyr-Gly-Leu-Arg168 (SVVYGLR), which binds to α4 and α9 integrins. Under physiologic conditions, expression of Opn is known to be restricted to tissues such as bone and kidney. In these organs, Opn has been shown to be involved in various physiological functions including biomineralization of bone [1,2] and the regulation of renal crystal formation [3]. In contrast, Opn is upregulated in inflamed and injured tissues, and is implicated in the pathogenesis of various inflammatory disorders [4], tissue remodeling [5], wound healing [6], tumor invasion [7], and metastasis [8].
Of note, Opn was shown by immunohistochemistry to be distributed on epithelial cells and plasma cells in normal human colon tissue [9]. Several groups reported that Opn is involved in inflammatory bowel diseases (IBDs) including Crohn’s disease (CD) and ulcerative colitis (UC), which are caused by excessive responses to commensal microbiota and other intestinal antigens [10]. Further, colon tissues from CD and UC showed upregulation of Opn [9] and Opn-deficient mice were resistant to 2, 4, 6-trinitro benzene sulfonic acid (TNBS) [11] and dextran sulfate sodium (DSS)-induced colitis [12], which are the models for CD and UC, respectively. During disease development, Opn was markedly upregulated in various immune cells, such as lymphocytes and DCs. Among these, CD103- dendritic cells highly express Opn and transfer of these cells induces severe acute colitis concordantly with increases of IL-17 and IFN-γ-producing CD4+ T cells [13]. As described above, the role of Opn in inflamed conditions has been demonstrated, whereas the significance of Opn in the normal gut remains unclear. Intestinal tissue is continuously exposed to antigens from microbiota, and Opn expression is increased in response to some infections, such as pneumococcal pneumonia and dengue virus [14,15]. Therefore, it has been speculated that Opn might be induced in immune cells following exposure to antigens from these microbes.
Commensal microbiota are critical for maintenance of the intestinal immune system [16] and the intestinal environment is controlled by 1) immunoglobulin A (IgA) from B cells present in the gut associated lymphoid tissue (GALT), particularly in Peyer’s patch (PP) and lamina propria (LP) [16], and 2) antimicrobial peptides produced by intraepithelial lymphocytes (IEL) and epithelial cells [16,17]. Microbes bind to toll-like receptors (TLRs) on intestinal epithelial cells leading to the production of B cell activating factor (BAFF) and a proliferation-inducing ligand (APRIL), which promote IgA-producing B cell generation and subsequent plasma cell differentiation. Consequently, IgA is secreted into the lumen and regulates microbiota composition [18]. On the other hand, recognition of microbes by epithelial cells through MyD88-pathways is required for production of antimicrobial peptides from IELs [17]. These antimicrobial peptides kill bacteria by various mechanisms, such as inhibition of bacteria growth [19,20], interference with metabolism [21–23], and generation of reactive nitrogen species and reactive oxygen species [24]. Deficiency and/or dysfunction of IgA or IELs result in alteration of the bacterial community and increased entry of bacteria into the intestinal epithelial tissue [17,25].
To clarify the role of Opn in the normal intestinal tissues, we first used reporter mice in which the Opn gene was replaced by EGFP (EGFP-Opn knock-in mice) to monitor Opn expression [26]. We found that in normal mice, CD8α+ T cells in the PP, LP, and intestinal epithelium expressed Opn. Notably, in Opn-deficient mice, there was a reduction of frequency and number in TCRγδ+ IELs and the microbiota of fecal samples was drastically altered as compared with WT mice. Thus, our data suggest the possibility that Opn might contribute to maintenance of homeostasis of intestinal commensal bacteria, possibly by TCRγδ+ IEL survival.
Materials and methods
Ethics statement
Mice were maintained under specific pathogen-free conditions at the Center for Experimental Animals of Kyoto University, and all procedures were performed according to the protocols approved by the Institutional Committee for Use and Care of Laboratory Animals of Kyoto University, which was granted by Kyoto University Ethics Review Board (MedKyo14064), and the Guide for Care and Use of Laboratory Animals published by the U.S. National Institutes of Health (NIH publication 85–23, revised 1996). For collection of tissue samples, mice were sacrificed by cervical dislocation. All efforts were made to minimize suffering.
Mice
C57BL/6 mice and Opn Knock out (Opn KO) mice were purchased from Japan SLC and Jackson Laboratory, respectively. EGFP-Opn knock-in (KI) mice were generated as described previously [26]. These mice were bred and maintained at least three generations at the Center for Experimental Animals of Kyoto University.
Antibodies and reagents
PE anti-CD3, Pacific Blue anti-CD8α, biotin-conjugated anti-CD8β, PE Cy7 anti-CD4, APC Cy7 anti-CD62L, APC anti-CD103, PerCP Cy5.5 anti-TCRγδ, PE Cy7 anti-TCRβ, APC Cy7-B220, PerCP Cy5.5 anti-B220, FITC-EpCAM, PE Cy7 anti-CD11b, APC anti-F4/80, biotin-conjugated anti-IgA antibodies were purchased from BioLegend. Streptavidin-PE was purchased from BD Biosciences.
Preparation of cells from intestinal epithelium and lamina propria
Small intestines were collected and Peyer’s Patches and fat tissue were carefully excised. After pushing out the intestinal contents, the intestine was opened longitudinally. Then, pieces of intestine were stirred with 5% FCS, 5 mM EDTA, and 1 mM DTT-containing PBS for 30 min at 37°C. Intestinal tissues were placed on a cell strainer, and the cells that passed through the strainer were collected. Remnant tissues were collected, further cut into small pieces, and then stirred with 0.15% collagenase D (Roche) / RPMI for 40 min at 37°C. After stirring, lamina propria cells were collected in the supernatant. This collagenase digestion step was repeated twice. For isolation of lymphocytes and epithelial cells, cell suspensions were separated with a 40/70% Percoll gradient and the intermediate layer and top layer were collected for analysis of lymphocytes and epithelial cells, respectively.
Flow cytometric analysis
Lymphocytes from lamina propria (LP) and intraepithelium were isolated as described above. Peyer’s patches (PP), inguinal lymph nodes (iLN), mesenteric lymph nodes (mLN), and spleen (Spl) were isolated, and erythrocytes were lysed with ACK lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 1 mM EDTA). Single cell suspensions were stained with primary antibodies for CD3, CD8α, CD8β, CD62L, CD103, TCRγδ, TCRβ, B220, CD11b, and F4/80. To evaluate IgA-producing cells, cells were stained with biotin-conjugated anti IgA antibody, followed by Streptavidin-PE. In some experiments, IELs (CD3+CD8α+TCRβ+ or TCRγδ+ cells) and intestinal epithelial cells (CD103-EpCAM+ cells) were sorted by a FACS Aria II or III (BD Biosciences) from isolated intestinal epithelium. All analyses were performed on a FACS Canto II (BD Biosciences) with FlowJo Software (Tree Star).
Evaluation of IgA-coated fecal bacteria
Flow cytometric analysis of bacteria was performed as described previously [27] with some modification. In brief, feces were collected and mashed with syringe plunger in PBS. After filtration with 70 μm nylon mesh, bacteria were separated by 20% / 80% Percoll gradient. After washing with PBS, bacteria were stained with biotin-conjugated anti-IgA antibody as described above. For detection of bacteria, feces were stained with Propidium Iodide (PI).
Histology
Intestinal tissues of KI mice were obtained and fixed with 2% paraformaldehyde for 2 hours at room temperature, and then tissues were placed in 30% sucrose overnight at 4°C. Tissues were then embedded with OCT compound and frozen by liquid nitrogen. After sectioning (6 μm) and drying, sections were blocked with 1% BSA/PBS, and then tissues were incubated with a primary antibody against CD8α for 1 hour at room temperature, followed by Cy3-conjugated anti-rat IgG antibody. Finally, tissues were stained with APC-anti B220 antibody for 1 hour and sections were analyzed by laser confocal microscopy (LSM710, Zeiss).
Fecal DNA preparation and analysis of fecal microbiota (Illumina MiSeq)
Feces were collected and genome DNA was extracted using QIAamp DNA stool mini kit (QIAGEN). Microbiome analysis was conducted by Illumina MiSeq. In brief, the V3 and V4 region of 16S rRNA was amplified by PCR and the amplicon was cleaned using the QIAquick Gel Extraction kit (QIAGEN). Cleaned amplicon was then used to prepare a library by limited cycle PCR, which was then d on an Illumina MiSeq (Illumina).
RNA extraction and quantitative real-time PCR analysis
TCRγδ+ IELs, defined as CD3+CD8α+ TCRγδ+, and Epithelial cells, defined as EpCAM+CD103- from Opn KO and WT mice were sorted as described above and were lysed with TRIzol reagent (Invitrogen) to extract total RNA. For quantitative PCR analysis, extracted RNAs were utilized to synthesize cDNA using a first strand cDNA synthesis kit (Roche). Real-time PCR was performed using LightCycler-FastStart DNA Master SYBR Green I Systems (Roche). All specific primers used in this study are shown in S1 Table. Expression levels were determined using the -⊿⊿Ct method of real-time PCR. Data were normalized against the expression of cyclophillin.
Survival assay
Each IEL subset (TCRαβ and TCRγδ) was sorted from 6–10 wk old female Opn KO and WT mice as described above. Twenty-thousand cells of each type were seeded into 96-well round-bottom plate (FALCON) in 10% FCS/RPMI. Cells were cultured for 12 hours in the presence of anti-murine Opn antibody (35B6) [3] or control antibody (10 μg/ml). Surviving cells were counted using a conventional counting method with Trypan Blue and Cell Titer Glo (Promega) according to the manufactures’ instructions.
Statistical analyses
All data are presented as the mean ±S.E.M or ±S.D as described in Figure legend. Significance of the difference between two groups was determined using Student’s t-test. * and ** denote p<0.05 and p<0.01, respectively, against control. N.S., not significant.
Results
Intestinal CD8+ T cells express Opn
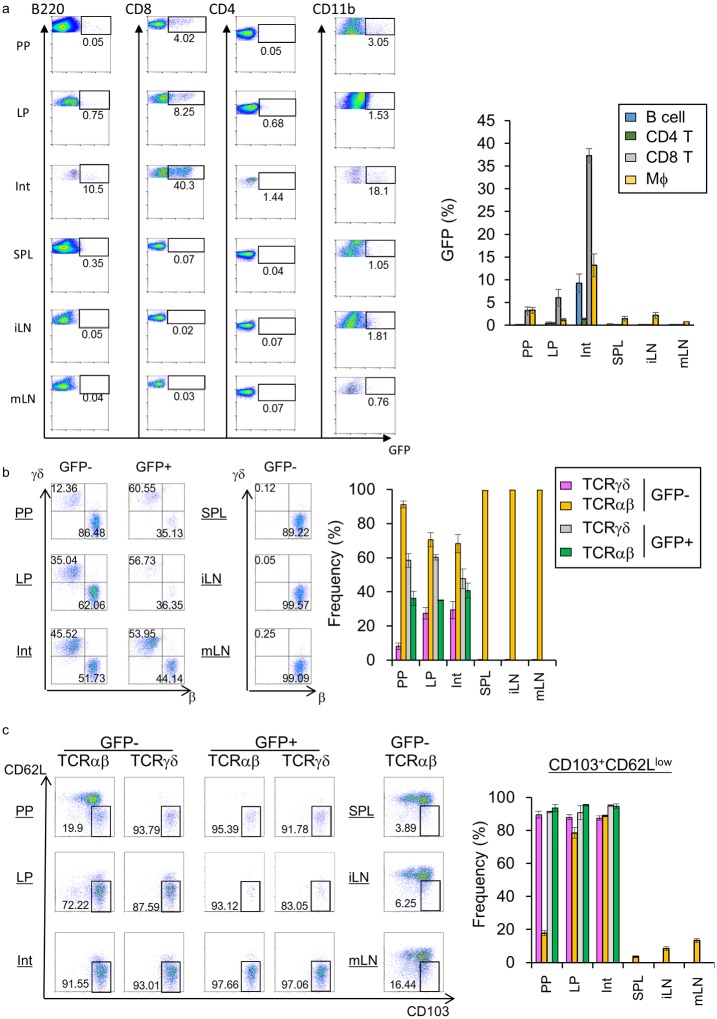
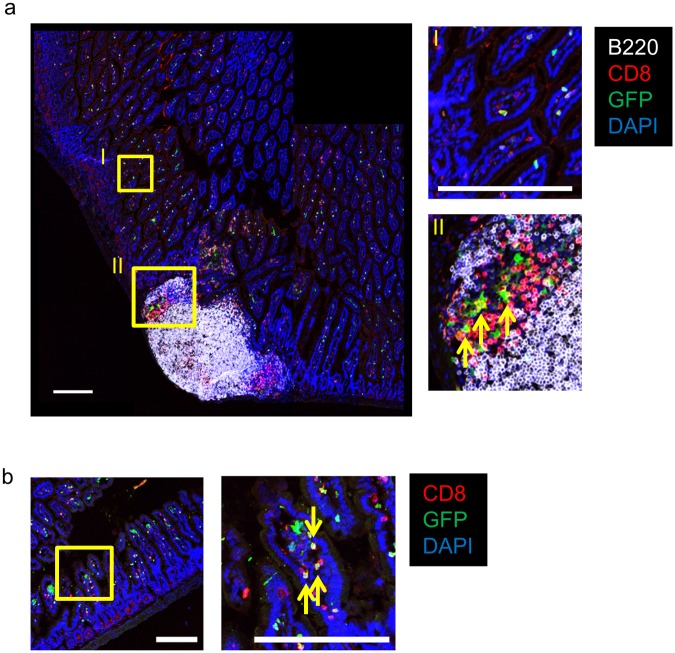
In a previous report Opn was detected in epithelial cells and plasma cells in the normal colon [9]. Therefore, we first identified the source of Opn in the intestinal tissue using Opn-reporter (EGFP-Opn knock-in (KI)) mice, which harbor the EGFP construct in the place of Opn [26]. Using hemizygous KI mice, we performed flow cytometric analysis to examine the source of Opn. Of note, CD8α+ T cells in the intestine, including those in Peyer’s Patches (PPs), lamina propria (LP), and intestinal epithelial lymphocytes (IELs), expressed GFP (Fig 1a). We also examined the expression of Opn on plasma cells and epithelial cells in the intestinal tissue, as reported previously; however, we did not observe Opn expression on epithelial cells (S1 Fig). As Opn has been reported to be expressed on activated T cells [26,28], we further characterized these cells. Interestingly, in GFP+ CD8α+ T cells of the PP and LP, TCRγδ+ cells were the majority (Fig 1b). In the intestinal epithelium, GFP+TCRγδ+ cells were equally exist as compared with TCRαβ+ cells (Fig 1b). In contrast, in the GFP- CD8α+ T cell population, TCRαβ+ cells were dominant in the PP, LP, and IEL (Fig 1b). Almost none of the GFP+ TCRγδ+ cells in the epithelium expressed CD8β, whereas CD8α+ T cells in the spleen expressed CD8β (S2 Fig), indicating that TCRγδ+ IELs are CD8αα IEL and not CD8αβ+ T cells immigrated from the peripheral lymphoid organs. In addition, these GFP+ CD8α+ T cells were highly enriched in the CD103+CD62Llow fraction regardless of TCR expression (Fig 1c), indicating that these GFP+ CD8α+ T cells were activated. In a previous report, CD103+CD62LlowCD69+ CD8α+ T cells were shown to be generated at the site of viral or bacterial infection and retained until secondary infection [29,30]. These CD103+CD62LlowCD69+CD8α+ T cells are referred to as resident memory T cells (Trm cells), and are able to intercept and eradicate pathogens immediately at sites of infections (skin or intestinal mucosa) [29,31]. As the Trm were reported to be localized to the non-lymphoid tissue [29], we also assessed localization of GFP-expressing CD8α T cells. Intestinal tissues of KI mice were immunostained for CD8α and B220 and analyzed by laser confocal microscopy. We found that GFP-expressing CD8α T cells were broadly distributed in the intestine (Fig 2a and 2b), while in the PP, GFP+ CD8α+ T cells were localized to the T cell zone.
Fig 1. Expression of Opn in specific CD8+ T cell subsets in the intestine, but not in other secondary lymphoid organs.
Six-week-old female EGFP-Opn mice were analyzed by flow cytometry. Cells obtained from Peyer’s patches (PP), lamina propria (LP), intestinal epithelium (Int), spleen (SPL), inguinal (iLN), and mesenteric lymph nodes (mLN) were stained for detection of (a) CD4 (CD3+CD4+), CD8 T cells (CD3+CD8α+), B cells (B220+), and macrophages (CD11b+) (b) TCRαβ or TCRγδ+ cells in CD3+CD8α+ cells (c) CD103+CD62Llow resident memory-type CD8 T cells. Graphs show (a) GFP+ cells, (b) TCRαβ+ and TCRγδ+, and (c) CD103+CD62Llow TCRαβ+ or TCRγδ+ cells in GFP+ or GFP- populations. Bars indicate ±S.E.M (n = 3~5). Data are representative of two independent experiments.
Fig 2. Localization of Opn-expressing CD8 T cells in the intestine.
Representative pictures of longitudinal section of intestine (a) including PP and (b) distal to PP obtained from EGFP-Opn mice and stained with B220 (white), CD8α (red), DAPI (blue). Inset shows magnified images. Arrows indicate GFP-expressing CD8α+ cells. Scale bars indicate 200 μm. The photograph is representative of 5 mice.
Opn deficiency affects intestinal bacterial communities
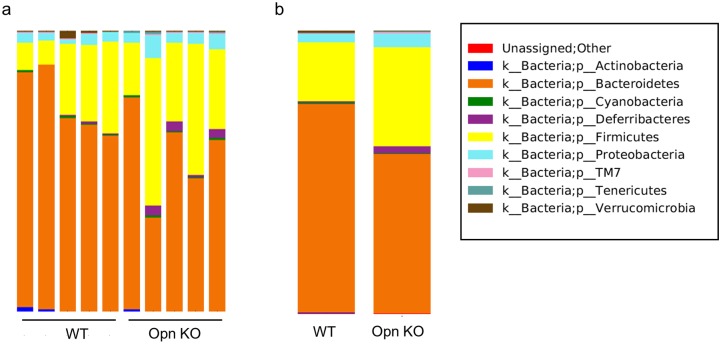
Intestinal tissues such as Peyer’s patches and epithelia are critical sites for the maintenance of intestinal microbiota through production of IgA and antimicrobial peptides, respectively [16]. To examine whether Opn-producing cells play a role in the homeostasis of commensal microflora, we examined the composition of intestinal bacteria using Illumina MiSeq analysis of DNA extracted from fecal samples collected from co-housed WT and Opn KO mice. In order to insure uniform exposure to same condition, these mice were maintained for at least three generations in the same facility. We found that Bacteroidetes (WT 49500.2 ± 3092.8 vs KO 38587.2 ± 4731.1) were reduced whereas Firmicutes (WT 13969.8 ± 3357.2 vs KO 24107.2 ± 4536.7), Deferribacteres (WT 291.4 ± 90.8 vs KO 1600.4 ± 524.2), Proteobacteria (WT 1964.6 ± 238.2 vs KO 3337.4 ± 669.6), and Tenericutes (WT 68.6 ± 12.7 vs 298.6 ± 111.0) were increased in fecal samples of Opn KO mice (Fig 3 and S2 Table), suggesting that deficient of Opn affected intestinal bacterial communities. In spite of co-housing, Opn KO mice showed alterations in fecal microflora, suggesting that Opn exerts a regulatory mechanism for controlling microflora.
Fig 3. Alteration of fecal bacterial communities in Opn KO mice.
Eight-week-old female Opn KO and WT mice were co-housed for 3 weeks prior to collection of feces and extraction of DNA. These mice were bred and maintained in the same facility for at least three generations. Illumina MiSeq analysis of 16S rRNA was then performed. Data are from (a) each sample and (b) the average of the groups. Data was normalized against common bacterial 16S rRNA expression and is presented as a ratio to the value obtained in WT mice (n = 5, per group).
Opn is not essential for generation of IgA-producing B cells
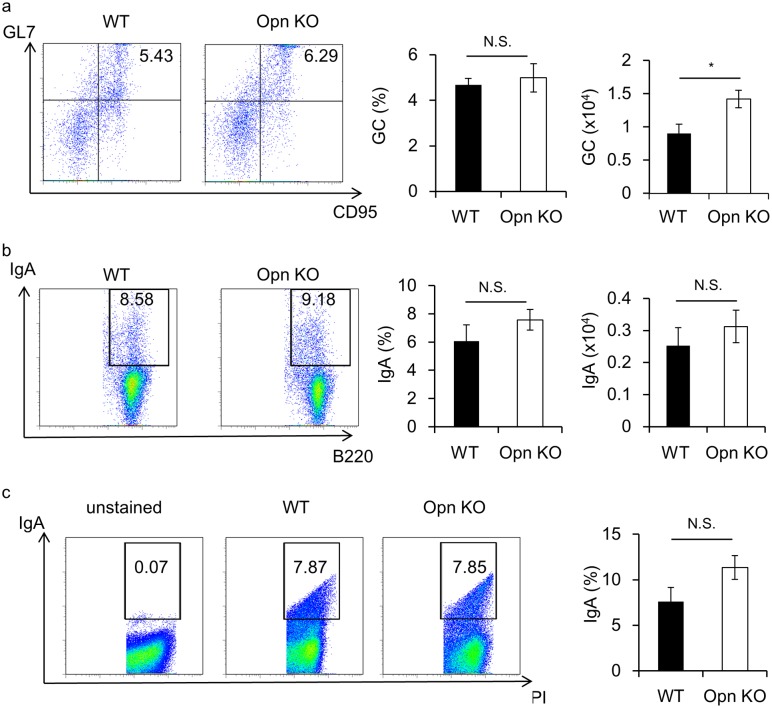
Intestinal microflora is regulated by 1) functional IgA production in the PP and LP and 2) antimicrobial peptide production by IELs and epithelial cells [16,17]. It has been reported that binding of Opn to B cells is crucial for their survival and that Opn-producing CD4+ T cells play a role in the development of the spontaneous germinal center (GC), and subsequent production of autoantibodies [26]. To evaluate the molecular mechanisms underlying the role of Opn in the regulation of microflora, we first examined whether Opn plays any role in generation of IgA-producing cells in the PP, which is the inductive site of IgA-producing B cells (Fig 4a and 4b). We observed that in Opn KO mice the total GC number was increased as compared with WT mice (Fig 4a). Of note, IgA-producing B cells were not affected in the PP of Opn KO mice as compared with WT mice (Fig 4b), suggesting that Opn-producing cells in the PP are dispensable for the differentiation of IgA-producing B cells. To further examine the function of Opn in IgA production, we investigated the binding affinity of IgA to bacteria in Opn KO mice (Fig 4c). Consistent with Fig 4a and 4b, the frequency of IgA-coated bacteria in feces of Opn KO mice was comparable to that of WT mice (Fig 4c). These results confirm that Opn is not essential for production of functional IgA.
Fig 4. Opn is dispensable for germinal center generation and IgA-producing B cells in PPs.
Eight-week-old female Opn KO and WT mice were cohoused for 3 weeks as described for Fig 3. Cells obtained from PPs were analyzed for detection of (a) GC B cell (GL7+CD95+ B220+ cells) and (b) IgA-producing B cells. Graphs show frequency and numbers of GC and IgA+ cells gated on B220+ cells. (c) Analysis of IgA-coating bacteria in the feces. Bacteria isolated from feces were stained with IgA and PI. PI+ events were considered to be bacteria. Graph shows the frequency of IgA+ cells with gating on PI+ cells. Bars indicate mean ±S.E.M. (n = 5; per group). Data are representative of four independent experiments.
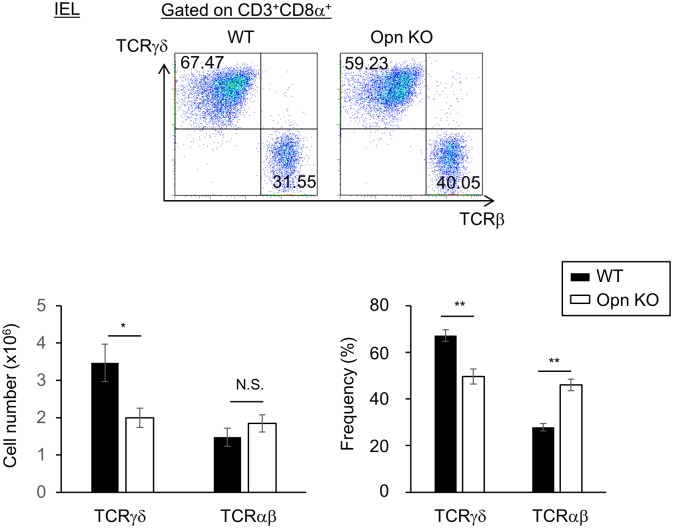
Reduction of TCRγδ+ T cells in the IEL of Opn KO mice
Antimicrobial peptides are produced by TCRγδ+ IELs and intestinal epithelial cells, resulting in the maintenance of commensal microbiota. Thus, we next attempted to determine the significance of Opn expression in TCRγδ+ IELs. To this end, we first examined whether deficiency of Opn affects the frequency of these cells. Eight-week-old, female Opn KO and WT mice were co-housed for three weeks prior to the analysis. Interestingly, Opn KO mice showed decreases in the frequency and number of CD8α+TCRγδ+ IELs compared to WT mice (Fig 5), suggesting the possibility that maintenance of CD8α+TCRγδ+ IELs in the intestine is disrupted by the loss of Opn.
Fig 5. Maintenance of TCRγδ+ IELs in the intestinal epithelium by Opn.
Analysis of TCRγδ+ and TCRαβ+ cells in Opn KO and WT mice. Opn KO and WT mice were cohoused as described for Fig 3 and the number and frequency of CD3+ CD8α+TCRγδ+ T cells and CD3+ CD8α+TCRαβ+ T cells in the IEL were analyzed by flow cytometric analysis. (top) The representative dot plots of FACS analysis gated on CD3+ CD8α+. Graph shows the mean of (lower left) absolute numbers and (lower right) frequency of TCRγδ+ and TCRαβ+ cells. Bars indicate ±S.E.M. (n = 4~5; per group). Data are representative of four independent experiments.
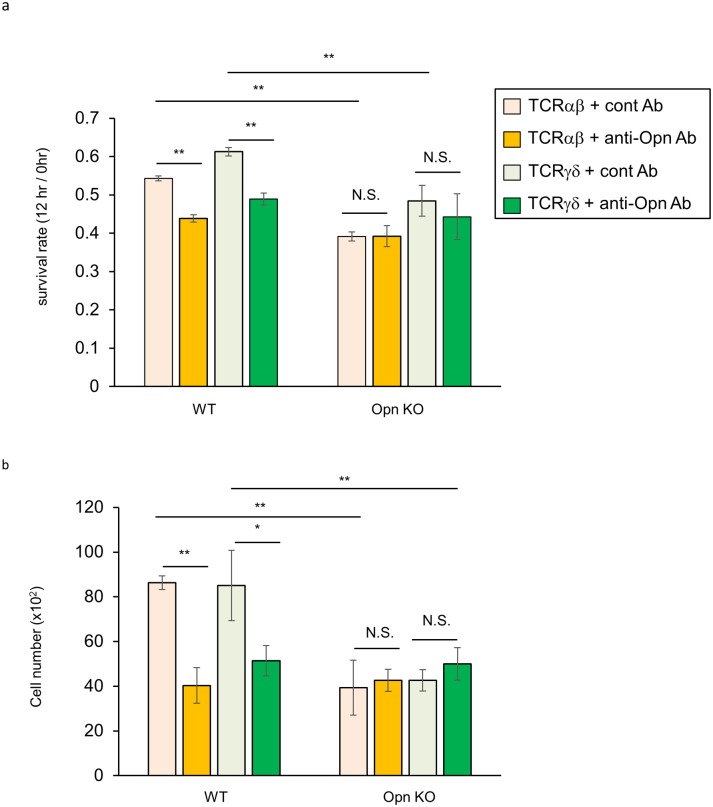
The binding of Opn to activated T cells induced prolonged survival and exacerbation of EAE [32]. Therefore, to examine whether Opn contributes to IEL survival, we sorted TCRαβ+ and TCRγδ+ IELs from WT and Opn KO mice and cells were cultured in the presence of anti-Opn blocking antibody (35B6) [3] for 12 hours. As shown in Fig 6, treatment with anti-Opn antibody inhibited the survival of TCRαβ+ and TCRγδ+ IELs. In addition, the inhibitory effect of anti-Opn antibody was comparable with that of Opn KO mice, suggesting that the survival effect of Opn is evident both in TCRαβ+ and TCRγδ+ IELs.
Fig 6. Secreted Opn functions as a survival factor for IELs.
In vitro survival assay. TCRαβ+ or TCRγδ+ cells were sorted from WT and Opn KO mice and each of 2 x104 cells was cultured for 12 hours in the presence of 10 μg/ml of anti-Opn blocking antibody (35B6) or control antibody. After cultivation, live cells were counted by (a) Cell Titer Glo and (b) conventional cell counting methods (see Materials and methods). Results were normalized to the values at 0 hours. The graphs show the means from triplicate wells. Bars indicate ±S.D. Data is representative of four independent experiments.
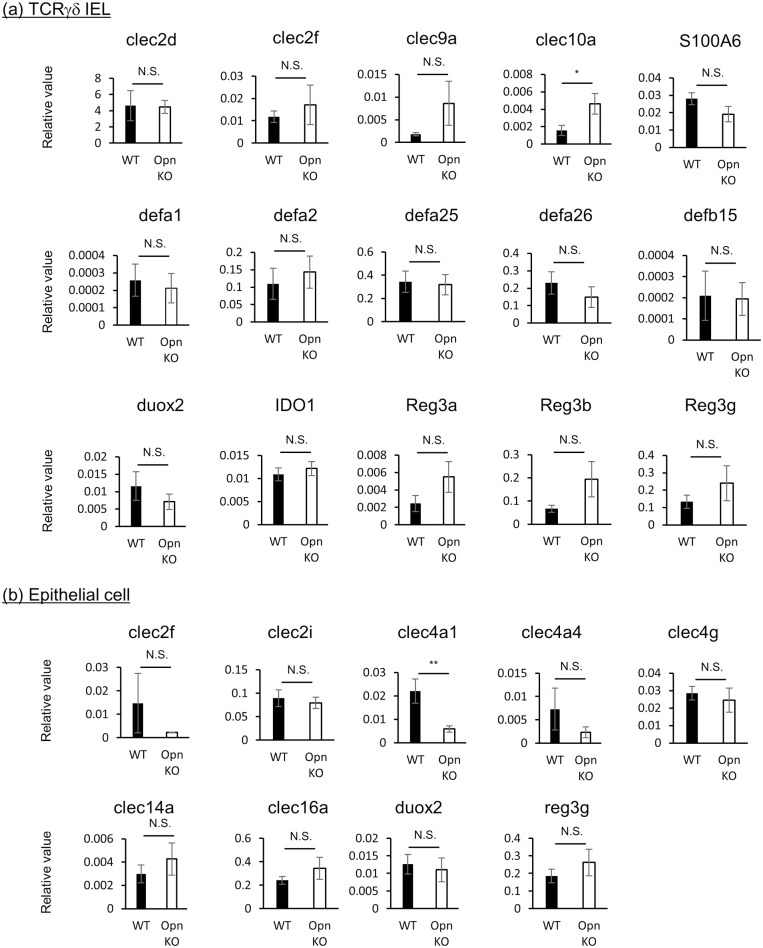
Expression profiles of antimicrobial proteins on TCRγδ+ IELs and epithelial cells between Opn KO mice and WT mice
Finally, we examined the expression profile of antimicrobial proteins in Opn KO mice. To this end, we sorted CD8α+TCRγδ+IELs and epithelial cells (CD103-EpCAM+) from Opn KO or WT mice (sorting schema and purity achieved are shown in S3 Fig), and performed quantitative real-time PCR analysis (Fig 7). In IELs from Opn KO mice, the expression of Clec10a was upregulated (Fig 7a), while in epithelial cells from Opn KO mice, the expression of Clec4a1 was downregulated (Fig 7b) compared to WT mice. However, the expression of most genes was found to be comparable between WT and Opn KO mice.
Fig 7. Comparison of antimicrobial factor gene expression in TCRγδ+ IELs and epithelial cells from Opn KO and WT mice.
Quantitative PCR analysis of antimicrobial factors in (a) TCRγδ IELs and (b) epithelial cells from Opn KO and WT mice. Bars indicate means ±S.E.M. (n = 4). Data is representative of two independent experiments.
Discussion
In this study, we examined the role of Opn in the normal intestine. To explore the source of Opn, we analyzed EGFP-Opn knock-in (KI) mice [26]. In a previous study, it has been suggested that Opn is distributed on epithelial cells and plasma cells [9]. However, we found that epithelial cells and plasma cells were not major sources of Opn (S1 Fig). Moreover, using KI mice, we found that CD8α T cells residing in intestinal tissue, including PP, LP, and IEL, express Opn (Fig 1a). Further, of the cells in the IEL, CD8α T cells showed the highest expression of Opn (Fig 1a). Previously, it was reported that Opn expression on T cells is found in ex vivo TCR-stimulated cells and aged mice, involving IFN-γ expression and spontaneous GC formation, respectively [26,28]. IFN-γ protects from colitis via upregulation of MHC class II on intestinal epithelial cells [33] and GC formation in PPs is critical for IgA production and subsequent maintenance of commensal bacteria [34]. Although TCRγδ+ IELs are known to be critical for the maintenance of microflora and prevent Salmonella infection via production of antimicrobial peptides [17], the contribution of Opn in TCRγδ+ IELs remains unclear. These issues collectively lead us to examine the role of Opn-expressing T cells in normal intestinal tissue. Interestingly, Opn KO mice had altered bacterial communities in their feces (Fig 3). It has been demonstrated that PD-1-/- mice, which exhibit an increased number of follicular helper T cells with a dysfunctional phenotype, have altered microflora as a con of production of dysfunctional IgA [25]. PPs are the main inductive site of IgA [34] and GC formation occurs spontaneously in response to gut bacteria. Specifically, follicular helper T cells are recruited into the PP B cell follicle and interact with B cells, resulting in class switch recombination to IgA production due to cytokine conditions. In a recent report, Tahir et al., demonstrated that Opn-producing CD153+PD-1+CD44hi CD4 T cells promote spontaneous GC formation in the spleen of lupus-prone New Zealand Black x White F1 mice [26]. In addition, a small fraction of CD8 T cells contributes to the generation of IgA-producing B cells [35]. Nevertheless, we observed that deficiency in Opn did not reduce GC formation or IgA production in the PP (Fig 4a and 4b) or binding of IgA to fecal bacteria (Fig 4c). These data demonstrate that Opn production is dispensable for GC formation as well as IgA production and function. In contrast to what has been reported for Opn-expressing CD4 T cells [26], intestinal Opn-expressing CD8 T cells were found to reside at a location distal to the B cell area (Fig 2a). Thus, it is likely that the anatomical distribution of Opn-expressing CD8 T cells is the causative factor for their failure to affect GC formation and IgA production.
In this study, we found that the frequency and number of CD8α+TCRγδ+ IELs was reduced in Opn KO mice (Fig 5). Opn is known to be a survival factor for activated T cells in the central nervous systems of mice with EAE [32]. Consistent with this report, when we examined the survival of IELs using anti-Opn blocking antibody and Opn KO mice, loss of Opn function resulted in the inhibition of IEL survival (Fig 6), suggesting that Opn is involved in IEL survival. In addition, there is a reduction in the frequency and number of TCRγδ+ cells, but not in TCRαβ+ cells (Fig 5). These data collectively suggest that the contribution of Opn to cell survival is higher in TCRγδ+ IELs in vivo. However, in our study, the inhibitory effect on IEL survival was only partial when we used anti-Opn antibody or in Opn KO mice. IL-7 and IL-15 are critical factors for survival of TCRγδ+ IEL cells [36–38]. Of note, Cantor et al., recently reported that intracellular Opn (iOpn) promotes the ability of IL-15Rα to bind its ligand, resulting in the prolonged survival of NK cells [39]. This report suggests the possibility that iOpn also contributes to the survival of TCRγδ+ IELs, and thus, we will need further investigation of the mechanisms underlying Opn-mediated survival of TCRγδ+ IELs.
Antimicrobial peptides are produced from TCRγδ+ IELs and intestinal epithelium, resulting in the regulation of microflora. Therefore, we finally examined whether expression of antimicrobial factors by CD8α+ TCRγδ+ IELs and epithelial cells is affected by the expression of Opn. In IELs and epithelial cells, almost all of the antimicrobial factors that we examined were found to be comparable between WT and Opn KO mice, with the exception of CLEC10a and CLEC4a1 (Fig 7). Thus, Opn has little effect on the expression of antimicrobial factors in the intestinal tissue.
Here we propose a unique function of Opn in the regulation of intestinal homeostasis. As illustrated in S4 Fig, TCRγδ+ IELs express Opn. Moreover, Opn expression, possibly both of the secreted form and the intracellular form, increases survival of these cells. In Opn KO mice, TCRγδ+ IELs are reduced due to a failure to survive compared to their WT counterparts. As a con, the amounts of antimicrobial factors produced are reduced, resulting in the subsequent alteration of the microflora. However, in this study, we compared fecal microbiota from Opn KO mice and WT mice purchased from different suppliers and maintained for at least three generations in same facility. As microbiota are largely dependent on the nursing mother [40], cohousing of adult mice as performed in this study may not be sufficient to insure uniformity of microbiota. Thus, although our data suggest that Opn is involved in the regulation of microbiota, further investigation using littermates will be required in the future to confirm this.
It has been demonstrated that changes in the microbiota composition are associated with allergy and inflammatory/autoimmune diseases [41]. For instance, it has been reported that obese patients show higher ratios of Firmicutes / Bacteroidetes than healthy individuals [42]. In the current study, in Opn-deficient mice Firumicutes were increased while Bacteroidetes were decreased. Interestingly, a recent paper shows an increased ratio of fat to body weight in Opn deficient mice [43]. Collectively, our data suggest Opn might be involved in the regulation of gut microflora via maintenance of TCRγδ+ IELs, and therefore, may be implicated in obesity and associated inflammatory diseases and further study of Opn in this context is warranted.
Supporting information
GFP expression in plasma cells and epithelial cells (B220-CD138+CD38+ and EpCAM+CD103-, respectively) from intestinal epithelial tissues (Int) and spleens (SPL). Histograms are a representative of three mice. The graph shows the mean of frequency of GFP+ plasma cells, epithelial cells, or CD8α cells. Bars indicates ±S.E.M (n = 3, per group). Data are representative of two independent experiments.
(PDF)
The expression of CD8β on CD8α+ IEL or splenocytes was analyzed using eight-week-old female KI mice. IELs and cells obtained from spleens were stained with CD3, CD8α, TCRγδ, TCRβ, and CD8β. (Upper panels) Histograms showing CD8β expression on GFP- TCRαβ, GFP- TCRγδ, GFP+ TCRαβ, and GFP+ TCRγδ or splenic CD8α+ cells. (Lower panel) The graph shows the mean of frequency of CD8β+ cells. Bars indicate ±S.E.M (n = 3, per group). Data are representative of three independent experiments.
(PDF)
TCRγδ or TCRαβ IELs were sorted by gating on CD3+CD8α+TCRγδ or CD3+CD8α+TCRαβ cells. Intestinal epithelial cells were sorted by gating on EpCAM+CD103- cells. Data are representative of 8 mice from two independent experiments.
(PDF)
(Left) In the normal intestine of WT mice, TCRγδ+CD8 T cells express Opn. Intracellular Opn contributes to the survival of these cells. TCRγδ+CD8 T cells also express various types of antimicrobial factors resulting in the regulation of intestinal microflora. (Right) In Opn KO mice, IEL TCRγδ+CD8 T cells were decreased due to a lack of Opn-mediated survival signals. As a consequence, the total amounts of antimicrobial factors were reduced, resulting in the alteration of the microflora.
(PDF)
(PDF)
The means (±S.E.M) of the data obtained in Fig 3 using Illumina MiSeq.
(PDF)
Acknowledgments
We thank M. Kawahara and M. Chihara (Kyoto University) for technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported from grants from Astellas Pharma Inc. in the Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program and from Japan Societies for the Promotion of Sciences, Grant-in-Aid for Young Scientists (B) (Grant Number: 15K19075 to K. I. https://kaken.nii.ac.jp/grant/KAKENHI-PROJECT-15K19075/). Publication of this article was approved by an intellectual property committee composed of representatives from Kyoto University and Astellas Pharma Inc. The funders had no role in study design, data collection and analysis, decision to publish.
References
- 1.Holm E, Gleberzon JS, Liao Y, Sorensen ES, Beier F, Hunter GK, et al. (2014) Osteopontin mediates mineralization and not osteogenic cell development in vitro. Biochem J 464: 355–364. 10.1042/BJ20140702 [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Singh K, Mukherjee BB, Sodek J (1993) Developmental expression of osteopontin (OPN) mRNA in rat tissues: evidence for a role for OPN in bone formation and resorption. Matrix 13: 113–123. [DOI] [PubMed] [Google Scholar]
- 3.Hamamoto S, Yasui T, Okada A, Hirose M, Matsui Y, Kon S, et al. (2011) Crucial role of the cryptic epitope SLAYGLR within osteopontin in renal crystal formation of mice. J Bone Miner Res 26: 2967–2977. 10.1002/jbmr.495 [DOI] [PubMed] [Google Scholar]
- 4.Diao H, Kon S, Iwabuchi K, Kimura C, Morimoto J, Ito D, et al. (2004) Osteopontin as a mediator of NKT cell function in T cell-mediated liver diseases. Immunity 21: 539–550. 10.1016/j.immuni.2004.08.012 [DOI] [PubMed] [Google Scholar]
- 5.Liaw L, Lombardi DM, Almeida MM, Schwartz SM, deBlois D, Giachelli CM (1997) Neutralizing antibodies directed against osteopontin inhibit rat carotid neointimal thickening after endothelial denudation. Arterioscler Thromb Vasc Biol 17: 188–193. [DOI] [PubMed] [Google Scholar]
- 6.Liaw L, Birk DE, Ballas CB, Whitsitt JS, Davidson JM, Hogan BL (1998) Altered wound healing in mice lacking a functional osteopontin gene (spp1). J Clin Invest 101: 1468–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotoh M, Sakamoto M, Kanetaka K, Chuuma M, Hirohashi S (2002) Overexpression of osteopontin in hepatocellular carcinoma. Pathol Int 52: 19–24. [DOI] [PubMed] [Google Scholar]
- 8.Rangaswami H, Bulbule A, Kundu GC (2006) Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol 16: 79–87. 10.1016/j.tcb.2005.12.005 [DOI] [PubMed] [Google Scholar]
- 9.Sato T, Nakai T, Tamura N, Okamoto S, Matsuoka K, Sakuraba A, et al. (2005) Osteopontin/Eta-1 upregulated in Crohn's disease regulates the Th1 immune response. Gut 54: 1254–1262. 10.1136/gut.2004.048298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heilmann K, Hoffmann U, Witte E, Loddenkemper C, Sina C, Schreiber S, et al. (2009) Osteopontin as two-sided mediator of intestinal inflammation. J Cell Mol Med 13: 1162–1174. 10.1111/j.1582-4934.2008.00428.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oz HS, Zhong J, de Villiers WJ (2012) Osteopontin ablation attenuates progression of colitis in TNBS model. Dig Dis Sci 57: 1554–1561. 10.1007/s10620-011-2009-z [DOI] [PubMed] [Google Scholar]
- 12.Zhong J, Eckhardt ER, Oz HS, Bruemmer D, de Villiers WJ (2006) Osteopontin deficiency protects mice from Dextran sodium sulfate-induced colitis. Inflamm Bowel Dis 12: 790–796. [DOI] [PubMed] [Google Scholar]
- 13.Kourepini E, Aggelakopoulou M, Alissafi T, Paschalidis N, Simoes DC, Panoutsakopoulou V (2014) Osteopontin expression by CD103- dendritic cells drives intestinal inflammation. Proc Natl Acad Sci U S A 111: E856–865. 10.1073/pnas.1316447111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Windt GJ, Hoogendijk AJ, Schouten M, Hommes TJ, de Vos AF, Florquin S, et al. (2011) Osteopontin impairs host defense during pneumococcal pneumonia. J Infect Dis 203: 1850–1858. 10.1093/infdis/jir185 [DOI] [PubMed] [Google Scholar]
- 15.Chagan-Yasutan H, Lacuesta TL, Ndhlovu LC, Oguma S, Leano PS, Telan EF, et al. (2014) Elevated levels of full-length and thrombin-cleaved osteopontin during acute dengue virus infection are associated with coagulation abnormalities. Thromb Res 134: 449–454. 10.1016/j.thromres.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki K, Nakajima A (2014) New aspects of IgA synthesis in the gut. Int Immunol 26: 489–494. 10.1093/intimm/dxu059 [DOI] [PubMed] [Google Scholar]
- 17.Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL, et al. (2011) Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci U S A 108: 8743–8748. 10.1073/pnas.1019574108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fagarasan S, Honjo T (2003) Intestinal IgA synthesis: regulation of front-line body defences. Nat Rev Immunol 3: 63–72. 10.1038/nri982 [DOI] [PubMed] [Google Scholar]
- 19.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, et al. (2004) Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432: 917–921. 10.1038/nature03104 [DOI] [PubMed] [Google Scholar]
- 20.Cash HL, Whitham CV, Behrendt CL, Hooper LV (2006) Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313: 1126–1130. 10.1126/science.1127119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, et al. (2008) Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319: 962–965. 10.1126/science.1152449 [DOI] [PubMed] [Google Scholar]
- 22.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, et al. (2013) Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A 110: 3841–3846. 10.1073/pnas.1220341110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, et al. (2012) Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11: 227–239. 10.1016/j.chom.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muhl H, Bachmann M, Pfeilschifter J (2011) Inducible NO synthase and antibacterial host defence in times of Th17/Th22/T22 immunity. Cell Microbiol 13: 340–348. 10.1111/j.1462-5822.2010.01559.x [DOI] [PubMed] [Google Scholar]
- 25.Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, et al. (2012) The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science 336: 485–489. 10.1126/science.1217718 [DOI] [PubMed] [Google Scholar]
- 26.Tahir S, Fukushima Y, Sakamoto K, Sato K, Fujita H, Inoue J, et al. (2015) A CD153+CD4+ T Follicular Cell Population with Cell-Senescence Features Plays a Crucial Role in Lupus Pathogenesis via Osteopontin Production. J Immunol 194: 5725–5735. 10.4049/jimmunol.1500319 [DOI] [PubMed] [Google Scholar]
- 27.Chu VT, Beller A, Rausch S, Strandmark J, Zanker M, Arbach O, et al. (2014) Eosinophils promote generation and maintenance of immunoglobulin-A-expressing plasma cells and contribute to gut immune homeostasis. Immunity 40: 582–593. 10.1016/j.immuni.2014.02.014 [DOI] [PubMed] [Google Scholar]
- 28.Shinohara ML, Jansson M, Hwang ES, Werneck MB, Glimcher LH, Cantor H (2005) T-bet-dependent expression of osteopontin contributes to T cell polarization. Proc Natl Acad Sci U S A 102: 17101–17106. 10.1073/pnas.0508666102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. (2013) The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol 14: 1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- 30.Beura LK, Masopust D (2014) SnapShot: resident memory T cells. Cell 157: 1488–1488 e1481. 10.1016/j.cell.2014.05.026 [DOI] [PubMed] [Google Scholar]
- 31.Sheridan BS, Pham QM, Lee YT, Cauley LS, Puddington L, Lefrancois L (2014) Oral infection drives a distinct population of intestinal resident memory CD8(+) T cells with enhanced protective function. Immunity 40: 747–757. 10.1016/j.immuni.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hur EM, Youssef S, Haws ME, Zhang SY, Sobel RA, Steinman L (2007) Osteopontin-induced relapse and progression of autoimmune brain disease through enhanced survival of activated T cells. Nat Immunol 8: 74–83. 10.1038/ni1415 [DOI] [PubMed] [Google Scholar]
- 33.Thelemann C, Eren RO, Coutaz M, Brasseit J, Bouzourene H, Rosa M, et al. (2014) Interferon-gamma induces expression of MHC class II on intestinal epithelial cells and protects mice from colitis. PLoS One 9: e86844 10.1371/journal.pone.0086844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lycke NY, Bemark M (2012) The role of Peyer's patches in synchronizing gut IgA responses. Front Immunol 3: 329 10.3389/fimmu.2012.00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagoo AS, Eldridge JH, Lagoo-Deenadaylan S, Black CA, Ridwan BU, Hardy KJ, et al. (1994) Peyer's patch CD8+ memory T cells secrete T helper type 1 and type 2 cytokines and provide help for immunoglobulin secretion. Eur J Immunol 24: 3087–3092. 10.1002/eji.1830241226 [DOI] [PubMed] [Google Scholar]
- 36.Lai YG, Hou MS, Lo A, Huang ST, Huang YW, Yang-Yen HF, et al. (2013) IL-15 modulates the balance between Bcl-2 and Bim via a Jak3/1-PI3K-Akt-ERK pathway to promote CD8alphaalpha+ intestinal intraepithelial lymphocyte survival. Eur J Immunol 43: 2305–2316. 10.1002/eji.201243026 [DOI] [PubMed] [Google Scholar]
- 37.Shitara S, Hara T, Liang B, Wagatsuma K, Zuklys S, Hollander GA, et al. (2013) IL-7 produced by thymic epithelial cells plays a major role in the development of thymocytes and TCRgammadelta+ intraepithelial lymphocytes. J Immunol 190: 6173–6179. 10.4049/jimmunol.1202573 [DOI] [PubMed] [Google Scholar]
- 38.Laky K, Lefrancois L, Lingenheld EG, Ishikawa H, Lewis JM, Olson S, et al. (2000) Enterocyte expression of interleukin 7 induces development of gammadelta T cells and Peyer's patches. J Exp Med 191: 1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leavenworth JW, Verbinnen B, Wang Q, Shen E, Cantor H (2015) Intracellular osteopontin regulates homeostasis and function of natural killer cells. Proc Natl Acad Sci U S A 112: 494–499. 10.1073/pnas.1423011112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daft JG, Ptacek T, Kumar R, Morrow C, Lorenz RG (2015) Cross-fostering immediately after birth induces a permanent microbiota shift that is shaped by the nursing mother. Microbiome 3: 17 10.1186/s40168-015-0080-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamada N, Seo SU, Chen GY, Nunez G (2013) Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 13: 321–335. 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- 42.Gerard P (2016) Gut microbiota and obesity. Cell Mol Life Sci 73: 147–162. 10.1007/s00018-015-2061-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Q, Shou P, Zhang L, Xu C, Zheng C, Han Y, et al. (2014) An osteopontin-integrin interaction plays a critical role in directing adipogenesis and osteogenesis by mesenchymal stem cells. Stem Cells 32: 327–337. 10.1002/stem.1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GFP expression in plasma cells and epithelial cells (B220-CD138+CD38+ and EpCAM+CD103-, respectively) from intestinal epithelial tissues (Int) and spleens (SPL). Histograms are a representative of three mice. The graph shows the mean of frequency of GFP+ plasma cells, epithelial cells, or CD8α cells. Bars indicates ±S.E.M (n = 3, per group). Data are representative of two independent experiments.
(PDF)
The expression of CD8β on CD8α+ IEL or splenocytes was analyzed using eight-week-old female KI mice. IELs and cells obtained from spleens were stained with CD3, CD8α, TCRγδ, TCRβ, and CD8β. (Upper panels) Histograms showing CD8β expression on GFP- TCRαβ, GFP- TCRγδ, GFP+ TCRαβ, and GFP+ TCRγδ or splenic CD8α+ cells. (Lower panel) The graph shows the mean of frequency of CD8β+ cells. Bars indicate ±S.E.M (n = 3, per group). Data are representative of three independent experiments.
(PDF)
TCRγδ or TCRαβ IELs were sorted by gating on CD3+CD8α+TCRγδ or CD3+CD8α+TCRαβ cells. Intestinal epithelial cells were sorted by gating on EpCAM+CD103- cells. Data are representative of 8 mice from two independent experiments.
(PDF)
(Left) In the normal intestine of WT mice, TCRγδ+CD8 T cells express Opn. Intracellular Opn contributes to the survival of these cells. TCRγδ+CD8 T cells also express various types of antimicrobial factors resulting in the regulation of intestinal microflora. (Right) In Opn KO mice, IEL TCRγδ+CD8 T cells were decreased due to a lack of Opn-mediated survival signals. As a consequence, the total amounts of antimicrobial factors were reduced, resulting in the alteration of the microflora.
(PDF)
(PDF)
The means (±S.E.M) of the data obtained in Fig 3 using Illumina MiSeq.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.