Abstract
MYH9 expression has previously been demonstrated as an independent predictor of clinical outcome in solid tumors. However, the prognostic relevance of MYH9 expression in acute myeloid leukemia is still unclear. Here, we found high MYH9 expressers were seen more frequently in females and more frequently in M4 morphology. We also found high MYH9 expressers had lower percentage of bone marrow blasts. In addition, overexpression of MYH9 was associated with an inferior overall survival. Notably, distinct microRNA signatures were seen in high MYH9 expressers. These results were also validated in an independent cohort of AML patients using the published data. In conclusion, gene of MYH9 expression might serve as a reliable predictor for overall survival in AML patients.
Keywords: MYH9, acute myeloid leukemia, prognosis
INTRODUCTION
Acute myeloid leukemia (AML) is a heterogeneous group of hematologic malignancies characterized by a wide spectrum of prognostically relevant cytogenetic aberrations, oncogenes of mutations and/or abnormal expressions. To date, although chromosomal abnormalities together with NPM1, FLT3-ITD, and CEBPA mutations have been recommended as an effective tool for the risk stratification [1], the reliable prognostic biomarkers for personalized therapy are still required.
In our phase 3 clinical trial, we found favorable cytogenetics can serve as a survival predictor for patients with AML when treated with homoharringtonine (HHT)-based chemotherapy [2]. However, the reason why patients would benefit from HHT-based chemotherapy remains unclear [3]. We and other groups have tried to further investigate the underlying mechanisms. Recent studies suggested HHT could regulate several oncogenes expression and induce blast cells death or apoptosis [3, 4]. As far as we known, HHT has multiple drug targets. Ying Gu et al. reported that p-eIF4E was a drug target of HHT [4]. Instead, we found myosin-9 was one of the critical targets in AML cell lines. Our study further suggested HHT could up-regulate the myosin-9 expression in AML cell lines, and up-regulated myosin-9 expression was associated with the percentage of apoptotic cells treated by HHT. Thus, up-regulated myosin-9 may increase the sensitivity of the leukemia cells to the cytotoxicity of HHT in vitro. Taken together, these results indicated over-expressed MYH9 might associate with favorable prognosis in leukemia. However, these results are from experiments in vitro, and may not always be in line with the results in vivo. Thus, it is urgent for us to explore the prognostic value of MYH9 expression in AML.
The MYH9 gene encodes the non-muscle myosin heavy chain IIA (NMMHC-IIA), a cytoskeletal contractile protein. Several mutations in the MYH9 gene lead to premature release of platelets from the bone marrow, macrothrombocytopenia, and cytoplasmic inclusion bodies within leukocytes. It is worthy to note that myosin-9 plays a key role in cancer cell migration, invasion, and metastasis in solid tumors [5]. With respect to expression value of MYH9, it was reported that reduced expression of myosin-9 was exhibited in CLL samples from high-risk patients. In contrast, high MYH9 expression was associated with poor clinical outcome in patients with several tumors such as lung cancer [6], esophageal [7], bladder [8], gastric cancer [9] and malignant pleural mesothelioma [5]. However, the clinical significance of MYH9 expression in AML is still unclear. Here, we found AML patients with high MYH9 expression had a distinct microRNA signature and poor overall survival. The prognostic value of MYH9 expression was also validated in an independent cohort of AML patients.
RESULTS
Characteristics of patients with MYH9 overexpression
The median expression value of MYH9 in our cohort was 0.64 with the range from 0.02 to 4.87. Of 324 patients, 131 (40%) were classified as high MYH9 expressers using Cutoff Finder software analysis. Clinical characteristics of patients with high MYH9 expression are described in Table 1. High MYH9 expressers were seen more frequently in females (55% vs. 40%, P = 0.008), and more frequently in M4 morphology (P = 0.005). We also found high MYH9 expressers showed lower percentage of bone marrow blasts (median: 58% vs. 72%, P < 0.001). There was no statistically significant correlation between MYH9 expression and other variables including age, white blood cell counts (WBC), hemoglobin levels, platelet counts, karyotype risk groups, genes mutations in FLT3-ITD, NPM1 and CEBPA, and different treatment protocols (Table 1).
Table 1. Characteristics of AML patients by high and low MYH9 expression.
| Variables | Low expression | High expression | P value |
|---|---|---|---|
| Number, (%) | 193 (60) | 131 (40) | |
| Age, median (range), years | 44 (14,84) | 46 (16,79) | 0.262 |
| Female, n (%) | 77 (40) | 72 (55) | 0.008 |
| WBC, median (range), × 10^9/L1 | 26 (0.3,358) | 23 (0.2,230) | 0.442 |
| HB, median (range), g/L2 | 82 (38,137) | 79 (42,141) | 0.11 |
| PLT, median(range), × 10^9/L3 | 41 (3,778) | 40 (2,556) | 0.913 |
| BM blast,median (range), %4 | 72 (20,98) | 58 (21,96) | < 0.001 |
| FAB classification,n (%)5 | 0.005 | ||
| M0 | 10 (5) | 9 (7) | |
| M1 | 23 (12) | 10 (8) | |
| M2 | 96 (50) | 50 (38) | |
| M4 | 6 (3) | 18 (14) | |
| M5 | 53 (28) | 41 (31) | |
| M6 | 5 (3) | 3 (2) | |
| Karyotype risk, n (%) | 0.549 | ||
| Favorable | 7 (4) | 3 (2) | |
| Intermediate | 175 (91) | 117 (89) | |
| Unfavorable | 11 (6) | 11 (8) | |
| Genes mutations, n(%) | |||
| FLT3ITD | 31 (17) | 25 (20) | 0.447 |
| NPM1 | 43 (23) | 22 (17) | 0.199 |
| CEBPADM6 | 26 (15) | 12 (10) | 0.201 |
| Treatment protocols7 | 0.526 | ||
| DA | 52 (27) | 29 (22) | |
| IA | 66 (34) | 44 (34) | |
| HAA | 75 (39) | 58 (44) |
Abbreviations:
WBC, white blood cell;
HB, hemoglobin;
PLT, platelet counts;
BM, bone marrow;
FAB, French–American–British classification systems;
DM: Double-allele.
The protocols used for induction therapy in different groups including donorubicin/Ara-C (DA)-based treatment group, idarubicin/Ara-C (IA)-based, and homoharringtonine/Ara-C/aclarubicin (HAA)-based treatment group.
Association of MYH9 expression with overall survival from the ZIH cohort
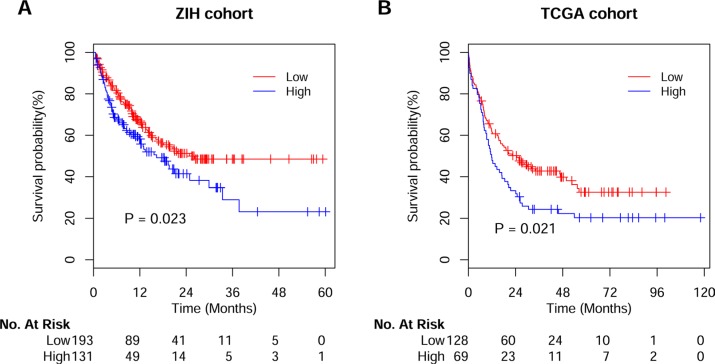
With a median follow-up for living patients of 434 days (95% CI, 479–594 days), high MYH9 expressers (n = 131) had an adverse OS comparing to low expressers (n = 193) (Figure 1A). Importantly, in the subgroup analyses we found high MYH9 expressions were associated with poor OS in patients with the cytogenetic intermediate risk group (P = 0.042) and cytogenetically normal AMLs (P = 0.031; Figure S1A–S1B). In the multivariable analysis for OS, high MYH9 expression was still associated with poor survival after adjusting for age, sex, WBC, percent blasts, karyotype risk groups, genes of FLT3-ITD, NPM1 and CEBPA mutations, and treatment protocols [HR (95% CI), 1.50 (1.03, 2.19); P = 0.034; Table 2].
Figure 1. Kaplan-Meier survival analysis of AML patients.
Overall survival curves of the patients in ZIH cohort (A) and TCGA cohort (B) according to distinct expression value of MYH9 gene.
Table 2. Multivariable analysis for overall survival in AML patients from ZIH cohort.
| Variables | HR (95% CI) | P value |
|---|---|---|
| MYH9 expression (High vs.Low) | 1.50 (1.03,2.19) | 0.034 |
| Age | 1.01 (1.00,1.02) | 0.054 |
| Sex (Female vs.Male) | 0.81 (0.56,1.16) | 0.248 |
| WBC1 | 1.003 (1,1.006) | 0.025 |
| Percent blast | 1.00 (0.99,1.01) | 0.529 |
| Karyotype-risk group | ||
| Intermediate vs. favorable | 4.63 (0.64,33.53) | 0.129 |
| Poor vs. favorable | 13.31 (1.69,105.01) | 0.014 |
| Gene mutations | ||
| FLT3ITD | 1.45 (0.92,2.28) | 0.108 |
| NPM1 | 0.86 (0.54,1.37) | 0.534 |
| CEBPADM2 | 0.31 (0.15,0.64) | 0.002 |
| Treatment protocols3 | ||
| HAA vs.DA | 0.90 (0.56,1.44) | 0.656 |
| IA vs.DA | 0.83 (0.54,1.30) | 0.418 |
Abbreviations:
WBC,white blood cell;
DM: Double-allele.
The protocols used for induction therapy in different groups including homoharringtonine/Ara-C/aclarubicin (HAA)-based treatment group, donorubicin/Ara-C (DA)-based treatment group, and idarubicin/Ara-C (IA)-based; CI, confidence intervals; HR, hazard ratio.
Validation of the impact of MYH9 expression on OS from the TCGA cohort
In order to validate the prognostic value of MYH9 expression derived from our discovery cohort, similarly, we defined high and low MYH9 expressers from TCGA cohort using Cutoff Finder. As a result, out of 197 patients from the TCGA cohort, 69 (35%) were defined as high MYH9 expressers and 128 (65%) as low expressers (Figure 1B). In the univariate analysis, patients with high MYH9 expressers obtained a higher risk of death compared with lower expressers [HR (95% CI), 1.50 (1.06, 2.13); P = 0.02]. Moreover, in the multivariate analysis, high MYH9 expressers were significantly associated with poor OS [HR (95% CI), 1.69 (1.17, 2.43); P = 0.005, Table S1] in the context of age, sex, WBC, percent blasts, karyotypes and genes of FLT3-ITD, NPM1 and CEBPA mutations.
MicroRNA expression profiling
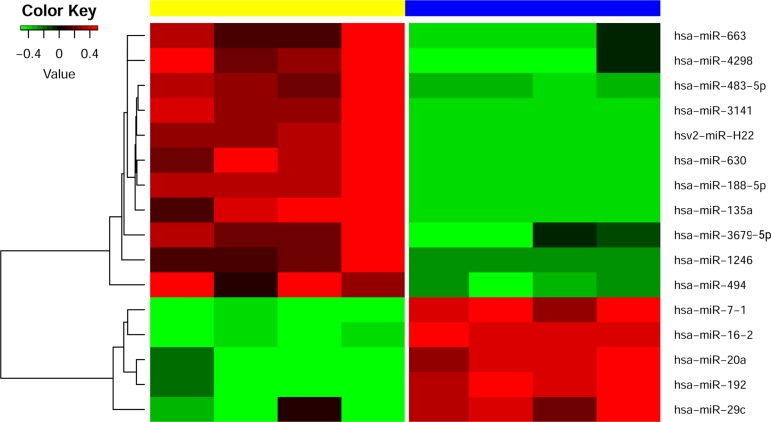
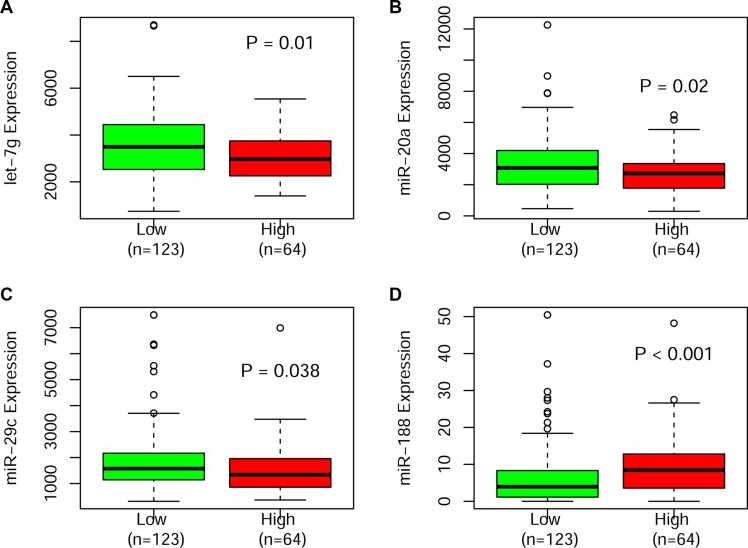
We used four samples with high MYH9 expression and four samples with low expression to assess the differences of microRNA expression. The most significant changes of miRNAs in high expressers included up-regulation of miR-663, miR-4298, miR-483-5p, miR-3141, miR-630, miR-188-5p, miR-135a, miR-3679-5p, miR-1246, miR-494 and hsv2-miR-H22 and down-regulation of let-7-1, miR-16-2, miR-20a,miR-192 and miR-29c (p-value less than 0.005, Figure 2). Notably, we also validated up-regulated miR-188 and down-regulated miRNAs of let-7, miR-20a and miR-29c that were significantly associated with high MYH9 expressers in a large and independent cohort of TCGA patients (Figure 3).
Figure 2. Heatmap visualizing the differentially expressed microRNAs between high and low MYH9 expressers.
MYH9 expressers are color-coded: a yellow bar indicates overexpression and blue bar under-expression in our patients with AML.
Figure 3. Boxplots illustrating the significantly different expression of miRNAs validated in the TCGA cohort.
DISCUSSION
In this study, we reported that high MYH9 expression associated with shorter overall survival in AML. In addition, patients with high MYH9 expression had distinct microRNAs. Our findings were also validated in an independent cohort of AML patients. These results suggest that MYH9 expression will be a reliable biomarker in the clinical practice.
MYH9 gene is located on chromosome 22q13.1 and encodes a conventional non-muscle myosin (myosin-9). Myosin-9 is one part of the myosinIIA protein. There are three forms of myosin II, called myosin IIA, myosin IIB and myosin IIC. The normal function of the myosin II protein is cytokinesis, cell motility and maintenance of cell shape [10]. The clinical significance of MYH9 gene lesions currently are undergoing study. For example, MYH9 gene polymorphisms are associated with cerebrovascular disease or nephropathy in patients with diabetes [11, 12]. The gene mutation in MYH9 is involved in the development of the disorders of inherited macrothrombocytopenias. It was also reported MYH9 expression is associated with inflammation of gastrointestinal diseases [13]. Notably, myosin-9 has been reported to be a novel tumor suppressor and play important roles in cancer progression. In this study, we found high MYH9 expressers were linked to M4 morphology. Blasts with FAB-M4 morphology often present extramedullary disease such as cutaneous involvement. Interestingly, high MYH9 expressers were mostly seen in patients with low percent blasts. These findings imply that high MYH9 expression might enable blasts to migrate away from bone marrow and subsequently reside in other tissues, and escape from chemotherapy. Therefore, AML cases with high MYH9 expression might be more resistant to chemotherapy, and associate with a poorer outcome. Here, we find that high MYH9 expressers harbored poor overall survival in two different cohorts. These results were consistent with several recent reports showing that high expression of MYH9 was correlated with short survival in solid tumors [7, 9, 10].
In order to further identify the biological insights into the aberrant MYH9 expression, we conducted microRNA analysis in AMLs. Among differentially expressed miRNAs, we found 16 miRNAs were dysregulated expression in our patients. Up-regulated expression of miR-188 and down-regulated expression of 3 miRNAs including miR-20a, miR-29c and let-7 were validated in a large cohort of patients. Among these significant microRNAs, most of them have been proven to have an impact on clinical prognosis of AML patients as previous reported. For example, low miR-188-5p expression was associated with longer overall survival and event free survival for CN-AML [14]. miR-16-1 expression was used as a good candidate for prognosis prediction in chronic myeloid leukemia [15]. In addition, miR-29c is of important prognostic value and influences response to azacitidine treatment in older AML patients [16]. These differentially expressed microRNAs may help us to further understand the biological insights of poor survival of patients with high MYH9 expression, and also serve as potential therapy targets in these patients in the future.
In conclusion, we present high MYH9 expression as a reliable and powerful prognostic factor for patients with AML.
MATERIALS AND METHODS
Patients
Clinical data were abstracted from medical records of AML patients in Zhejiang Institute of Hematology (ZIH) in China. Between March 2010 and June 2014, 324 patients with detailed diagnoses and treatment information were enrolled in this study. WHO classification, conventional cytogenetic banding assay, and molecular analyses were performed as previously described in AML diagnosis [17]. Cytogenetic groups of patients were classified as favorable, intermediate, and unfavorable risk according to the NCCN guideline [18]. Favorable subgroups included t(8;21)/AML1-ETO and inv16/CBFβ-MYH11; adverse consisted of t(9;22), inv(3)/t(3;3), -5, -7, del(5q), del(7p), 11q23 and complex translocations; intermediate subtype contained cytogenetically normal and AML with other cytogenetic abnormalities. Patients received HAA (homoharringtonin 2 mg/m2/day for 7 days, cytarabine 100 mg/m2/day for7 days and aclarubicin 20 mg/m2/day for 5 days), DA (daunorubicin 45 mg/m2/day for 3 days and cytarabine 100 mg/m2/day for 7 days) and IA regimen (idarubicin 8–10 mg/m2/day for 3 days and cytarabine 150 mg/m2/day for 7 days) [2, 19]. In the consolidation therapy, younger patients were treated with a high-dose cytarabine-based chemotherapy [19]. The chemotherapy consolidation for elderly patients was decided by the physicians in an individualized manner, as described previously [19]. No patient in our study received allogeneic transplantation. Patients with secondary AML or acute promyelocytic leukemia were excluded. All of the subjects were well informed about the study and provided written informed consent to participate in the study. We used a AML cohort of 197 patients from TCGA (https://tcga-data.nci.nih.gov/tcga/) as a validation cohort, which contains publicly available data of gene microarray expression and clinical information [20]. The study was approved by the Institutional Review boards of the First Affiliated Hospital of Zhejiang University.
Cytogenetic and gene mutation analysis
The bone marrow (BM) samples of de novo AML patients were studied mostly by R-banding analysis. Chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature [21]. DNA and RNA samples of AML patients were obtained from mononuclear cells isolated by Ficoll gradient centrifugation from bone marrow samples at primary diagnosis. Gene mutations of NPM1, FLT3-ITD, and CEBPA were analyzed by whole-gene sequencing as previously described [22]. RNA samples were used to determine PMLRARA, AML1ETO, and CBFβMYH11 fusion genes by reverse transcription polymerase chain reaction (RT-PCR).
Quantitative reverse transcriptase-PCR
RNA was extracted using RNeasy Mini kit (Qiagen, Venlo, Netherlands) and first-strand complementary DNA synthesis was performed using the MMLV systems (Life Technologies). Quantitative PCR was performed in triplicate using SYBR-Green PCR Master Mix kit (Takara, Japan) on an IQ5 real time PCR instrument (Bio-Rad, USA), using standard settings: 95°C (1 min), 40 cycles of 95°C (5s) and 60°C (1 min). mRNA levels were normalized to GAPDH housekeeping gene. The following primers were used for quantitative PCR: MYH9 5′-TTCAGCTCGGCAACATCGTCT-3′ (sense) and 5′-ATTCCTCTGGTGAAATCGGTCA-3′ (antisense); GAPDH (control), 5′-ATGGGGAAGGTGAAGGTCG-3′ (sense) and 5′-GGGTCATTGATGGCAACAATATC-3′ (antisense). PCR reactions were performed in a total volume of 25 μl containing of 1 μl of 100 ng/μl sample cDNA, 12.5 μl of 2 × PCR Mix, 1 μl of 0.5 μM of each primer, and 10.5 μl of ddH2O.
MicroRNA experiments
For miRNA profiling, total RNA was extracted and purified using mirVana™ miRNA Isolation Kit (Ambion, Austin, TX, US) following the manufacturer's instructions. RNA intergrity number (RIN) was assessed by an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, US). miRNA expression was performed using the Agilent Human miRNA Microarray Kit Version 16.0. Total RNA (100 ng) was hybridized per sample and processed according to the manufacturer's instructions. The arrays were scanned by an Agilent Technology G2565BA scanner. The scanned images were gridded and analyzed with Agilent Feature Extraction Software Version 10.7. Raw data were normalized by quantile algorithm, Gene Spring Software 11.0. Each microRNA signature is represented by the average of its expression value of replicate probes. Nonparameter T-test was used to test for the difference of microRNA signatures between high and low MYH9 expressers. P-values < 0.005 demonstrated statistical difference. Hierarchical clustering based on expression levels of these microRNAs was performed and visualized by heatmap.
Definition of clinical end points and statistical analysis
Patient characteristics were summarized using descriptive statistics, which included frequency counts, median, and range. The primary end point of the study was overall survival (OS). OS was measured as time from disease diagnosis to death from any cause, or censoring for patients alive at their last known date of contact. Determination of optimal cutoff value for MYH9 expression in our discovery and the validation TCGA cohort was done with Cutoff Finder using log-rank test (http://molpath.charite.de/cutoff/). The proportional-hazards assumption was checked for each variable before fitting Cox models. Variables with a p-value less than 0.2 were selected as adjustment covariates into the multivariable analyses. All statistical analyses were conducted with R statistic packages, version 2.15.0 (www.r-project.org). The two-sided level of significance was set at p-value less than 0.05.
SUPPLEMENTARY MATERIALS
Acknowledgments
We are very thankful to Ms. Jennifer Strong from University of Cincinnati College of Medicine for carefully reading and polishing the manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors have declared that no competing interests exist.
GRANT SUPPORT
This work was supported by National Natural Science Foundation of China grants (81370643, 81470305, U1404806), National Public Health Grand Research Foundation (201202017) and the Foundation of Innovation Team for Basic and Clinical Research of Zhejiang Province (2011R50015).
REFERENCES
- 1.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin J, Wang JX, Chen FF, Wu DP, Hu J, Zhou JF, Hu JD, Wang JM, Li JY, Huang XJ, Ma J, Ji CY, Xu XP, et al. Homoharringtonine-based induction regimens for patients with de-novo acute myeloid leukaemia: a multicentre, open-label, randomised, controlled phase 3 trial. Lancet Oncol. 2013;14:599–608. doi: 10.1016/S1470-2045(13)70152-9. [DOI] [PubMed] [Google Scholar]
- 3.Zhang T, Shen S, Zhu Z, Lu S, Yin X, Zheng J, Jin J. Homoharringtonine binds to and increases myosin-9 in myeloid leukaemia. Br J Pharmacol. 2016;173:212–221. doi: 10.1111/bph.13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu Y, Zhou H, Gan Y, Zhang J, Chen J, Gan X, Li H, Zheng W, Meng Z, Ma X, Wang X, Xu X, Xu G, et al. Small-molecule induction of phospho-eIF4E sumoylation and degradation via targeting its phosphorylated serine 209 residue. Oncotarget. 2015;6:15111–15121. doi: 10.18632/oncotarget.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Rienzo A, Archer MA, Yeap BY, Dao N, Sciaranghella D, Sideris AC, Zheng Y, Holman AG, Wang YE, Dal Cin PS, Fletcher JA, Rubio R, Croft L, et al. Gender-Specific Molecular and Clinical Features Underlie Malignant Pleural Mesothelioma. Cancer Res. 2016;76:319–328. doi: 10.1158/0008-5472.CAN-15-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katono K, Sato Y, Jiang SX, Kobayashi M, Nagashio R, Ryuge S, Fukuda E, Goshima N, Satoh Y, Saegusa M, Masuda N. Prognostic significance of MYH9 expression in resected non-small cell lung cancer. PLoS One. 2015;10:e0121460. doi: 10.1371/journal.pone.0121460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia ZK, Yuan YC, Yin N, Yin BL, Tan ZP, Hu YR. Nonmuscle myosin IIA is associated with poor prognosis of esophageal squamous cancer. Diseases of the esophagus. 2012;25:427–436. doi: 10.1111/j.1442-2050.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 8.Xiong D, Ye YL, Chen MK, Qin ZK, Li MZ, Zhang H, Xu LH, Xu ZZ, Zeng MS. Non-muscle myosin II is an independent predictor of overall survival for cystectomy candidates with early-stage bladder cancer. Oncol Rep. 2012;28:1625–1632. doi: 10.3892/or.2012.1965. [DOI] [PubMed] [Google Scholar]
- 9.Liu D, Zhang L, Shen Z, Tan F, Hu Y, Yu J, Li G. Clinicopathological significance of NMIIA overexpression in human gastric cancer. Int J Mol Sci. 2012;13:15291–15304. doi: 10.3390/ijms131115291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma X, Adelstein RS. The role of vertebrate nonmuscle Myosin II in development and human disease. Bioarchitecture. 2014;4:88–102. doi: 10.4161/bioa.29766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling C, Cai CY, Chang BC, Shi WT, Wei FJ, Yu P, Chen LM, Li WD. MYH9 gene polymorphisms may be associated with cerebrovascular blood flow in patients with type 2 diabetes. Genet Mol Res. 2015;14:1008–1016. doi: 10.4238/2015.February.6.4. [DOI] [PubMed] [Google Scholar]
- 12.Cooke JN, Bostrom MA, Hicks PJ, Ng MC, Hellwege JN, Comeau ME, Divers J, Langefeld CD, Freedman BI, Bowden DW. Polymorphisms in MYH9 are associated with diabetic nephropathy in European Americans. Nephrol Dial Transplant. 2012;27:1505–1511. doi: 10.1093/ndt/gfr522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao B, Qi Z, Li Y, Wang C, Fu W, Chen YG. The non-muscle-myosin-II heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells. Nat Commun. 2015;6:7166. doi: 10.1038/ncomms8166. [DOI] [PubMed] [Google Scholar]
- 14.Jinlong S, Lin F, Yonghui L, Li Y, Weidong W. Identification of let-7a-2-3p or/and miR-188-5p as prognostic biomarkers in cytogenetically normal acute myeloid leukemia. PLoS One. 2015;10:e0118099. doi: 10.1371/journal.pone.0118099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fallah P, Amirizadeh N, Poopak B, Toogeh G, Arefian E, Kohram F, Hosseini Rad SM, Kohram M, Teimori Naghadeh H, Soleimani M. Expression pattern of key microRNAs in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Int J Lab Hematol. 2015;37:560–568. doi: 10.1111/ijlh.12351. [DOI] [PubMed] [Google Scholar]
- 16.Butrym A, Rybka J, Baczynska D, Poreba R, Kuliczkowski K, Mazur G. Clinical response to azacitidine therapy depends on microRNA-29c (miR-29c) expression in older acute myeloid leukemia (AML) patients. Oncotarget. 2016 doi: 10.18632/oncotarget.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JH, Chen WL, Li JM, Wu SF, Chen TL, Zhu YM, Zhang WN, Li Y, Qiu YP, Zhao AH, Mi JQ, Jin J, Wang YG, et al. Prognostic significance of 2-hydroxyglutarate levels in acute myeloid leukemia in China. Proc Natl Acad Sci U S A. 2013;110:17017–17022. doi: 10.1073/pnas.1315558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Donnell MR, Abboud CN, Altman J, Appelbaum FR, Arber DA, Attar E, Borate U, Coutre SE, Damon LE, Goorha S, Lancet J, Maness LJ, Marcucci G, et al. Acute myeloid leukemia. J Natl Compr Canc Netw. 2012;10:984–1021. doi: 10.6004/jnccn.2012.0103. [DOI] [PubMed] [Google Scholar]
- 19.Ma QL, Wang JH, Wang YG, Hu C, Mu QT, Yu MX, Wang L, Wang DM, Yang M, Yin XF, Chen FF, Lu SS, Chen J, et al. High IDH1 expression is associated with a poor prognosis in cytogenetically normal acute myeloid leukemia. Int J Cancer. 2015;137:1058–65. doi: 10.1002/ijc.29395. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research N. Genomic, epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brothman AR, Persons DL, Shaffer LG. Nomenclature evolution: Changes in the ISCN from the 2005 to the 2009 edition. Cytogenet Genome Res. 2009;127:1–4. doi: 10.1159/000279442. [DOI] [PubMed] [Google Scholar]
- 22.Chen WL, Wang JH, Zhao AH, Xu X, Wang YH, Chen TL, Li JM, Mi JQ, Zhu YM, Liu YF, Wang YY, Jin J, Huang H, et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood. 2014;124:1645–1654. doi: 10.1182/blood-2014-02-554204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.