Abstract
Our 13C- and 1H-chemical exchange saturation transfer (CEST) experiments previously revealed a dynamic exchange between partially closed and open conformations of the SAM-II riboswitch in the absence of ligand. Here, all-atom structure-based molecular simulations, with the electrostatic effects of Manning counter-ion condensation and explicit magnesium ions are employed to calculate the folding free energy landscape of the SAM-II riboswitch. We use this analysis to predict that magnesium ions remodel the landscape, shifting the equilibrium away from the extended, partially unfolded state towards a compact, pre-organized conformation that resembles the ligand-bound state. Our CEST and SAXS experiments, at different magnesium ion concentrations, quantitatively confirm our simulation results, demonstrating that magnesium ions induce collapse and pre-organization. Agreement between theory and experiment bolsters microscopic interpretation of our simulations, which shows that triplex formation between helix P2b and loop L1 is highly sensitive to magnesium and plays a key role in pre-organization. Pre-organization of the SAM-II riboswitch allows rapid detection of ligand with high selectivity, which is important for biological function.
Author summary
The presence of positively charged metal ions is essential to maintain the structural fold and function of RNA. Among different metal ions, magnesium is particularly important for the stability of RNA because it can efficiently support a close assembly of negatively charged phosphate groups in an RNA fold. The SAM-II riboswitch is an example of a classical pseudoknot fold, which binds S-adenosyl methionine, stabilizing an alternate folded form to inhibit gene expression. In our early 13C- and 1H-chemical exchange saturation transfer (CEST) experiments, we found a conformational transition between a minor, partially closed and a major, open state conformation in the absence of ligand. Our CEST experiments at different magnesium concentrations now suggest that magnesium ions can induce a conformational pre-organization in the apo SAM-II riboswitch, which is expected to facilitate ligand binding. To understand the microscopic details of this magnesium-induced transition, we perform all-atom structure-based molecular simulations including electrostatics and explicit magnesium ions. Our free energy calculations reveal that the partially closed pre-organized state is further stabilized with increasing magnesium concentration. This is in excellent agreement with our 13C-CEST profile, SAXS, and size-exclusion chromatographic data, and with recent single molecule FRET experiments. Our results suggest that a sufficiently high concentration of magnesium is essential to pre-organize the apo SAM-II riboswitch.
Introduction
Non-coding RNAs are currently thought to account for over 75% of the human genome [1]. In bacteria, non-coding RNAs play important roles in gene regulation. One such class of RNAs, riboswitches, regulates metabolite production. Here, a single RNA sequence folds into one of two or more mutually exclusive folds depending on the metabolite concentration [2,3]. In some cases, such as the S-adenosylmethionine-I (SAM-I) riboswitch, the RNA contains a transcriptional terminator that forms when ligand is present, in effect silencing genes important for ligand production [4–6]. When the ligand is not present, the terminator does not form, allowing gene expression, and therefore ligand production, to continue efficiently. In other cases, such as the SAM-II riboswitch, ligand binding may lead to sequestration of the Shine/Dalgarno sequence, likely blocking ribosome binding and, as a consequence, protein synthesis [7]. While these examples of ligand-dependent secondary structure switches have been known for some time, a detailed thermodynamic understanding at the atomistic level, including the indispensable effect of the RNA’s ion-atmosphere, has not been achieved. In recent years, riboswitches have become canonical systems for studies of diverse RNA behaviors, as they possess quintessential characteristics of many RNA systems: ligand binding, Magnesium ion (Mg2+) sensitivity, conformational changes, secondary structure remodeling, and regulatory functions. Chemical footprinting, NMR, small-angle X-ray scattering (SAXS) and single molecule FRET techniques are being exploited to elucidate the folding kinetics, thermodynamics and the magnesium ion sensitivity in RNA systems such as the TPP riboswitch [8], glycine-dependent riboswitches [9], different variants of P4-P6 RNA [10–12] and P5abc subdomain of the Tetrahymena group I intron ribozyme [13]. Other work focuses on more complex functions, such as splicing and ligand recognition and their associations with proteins or different metabolites [14,15]. Success in understanding the structural, dynamical and functional aspects of riboswitch systems requires an integrated experimental and theoretical approach. Traditional crystallographic techniques produce static snapshots of the riboswitch. SmFRET, NMR, and SAXS methods obtain kinetic information and overall distributions of conformations. Molecular simulation allows one to integrate disparate experimental data into a single coherent picture, characterizing transitions in atomistic detail and the free energy landscape with fine resolution. A large number of riboswitches have been crystallized and have also been investigated via fluorescence and single molecule techniques [16–28].
Molecular simulations have also been used to study a number of riboswitches, including but not limited to the SAM-I, SAM-II, pre-Q, and adenine riboswitches [29–32]. Some of these studies replaced the essential atmosphere of divalent Mg2+ ions with other, monovalent ions. Others used a repulsive Debye-Hückel interaction between the phosphate groups of the RNA. Such implicit treatments of the ions, however, neglect important near-field effects that occur inside the core of the riboswitch, where the ions are strongly coupled to the RNA.
Although the role of Mg2+ in stabilizing the RNA tertiary structure has long been realized [33,34], the molecular basis of ion-RNA interactions, in terms of structure and function, is not well understood. In a pioneering study, Draper and co-workers distinguished three classes of ion environments: (i) the diffuse ions, which are not restrained to any particular region, (ii) water-surrounded ions separated from the RNA by a single hydration layer, which we call outer-sphere ions, and (iii) chelated ions in the inner sphere, which form direct contacts with at least two different phosphate groups of the RNA [34,35]. While the potential role of chelated ions has been emphasized in many studies, recent work using explicit solvent molecular dynamics instead highlights a dense layer of outer-sphere Mg2+ ions, which are primarily responsible for anchoring the RNA structure [36]. These outer-sphere Mg2+ ions are only transiently bound but nonetheless strongly coupled to the RNA dynamics. Such highly correlated Mg2+ ions may even reside in the core region of riboswitch RNAs [36,37]. This dynamic cloud of Mg2+ has also been simulated in all-atom reduced models that combine Manning theory with a background of monovalent ions, represented by Debye-Hückel interactions [37,38]. In addition to these native basin simulations, studies of metabolite recognition and specificity have also been initiated using conformational ensemble sampling, again in the absence of Mg2+ ions [39].
While most riboswitch studies have focused on riboswitches in the 5’-UTR of mRNA that control transcription, less attention has been paid to translational control by riboswitches through ligand-dependent sequestration of the ribosome binding site (i.e., the Shine/Dalgarno sequence). The SAM-II riboswitch is one such relatively small RNA element which regulates methionine and SAM biosynthesis. A single hairpin, classic H-type pseudoknot and triplex interaction near the ligand binding site make this RNA an interesting system to study RNA control over the translation initiation process [21,40–44].
A previous single molecule fluorescence resonance energy transfer (smFRET) [21] imaging study sheds light on the dynamic nature of ligand-free SAM-II riboswitch, which becomes conformationally restrained upon ligand binding. The flexibility of such highly transient conformations is tuned to ensure a viable time scale for conformational transitions in the absence of ligand. More rapid sensing could, however, be achieved if the riboswitch adopted a binding competent conformation in the ligand-free state. Mg2+ ions can act as effective anchors, aiding in the preservation of the structural integrity of the RNA. The emergence of two distinct FRET configurations in the presence of 2 mM Mg2+ in the ligand free system suggests that Mg2+ has the ability to compress the RNA structure, in such a way that it might pre-organize the RNA to form a binding competent conformation. In a series of small angle X-ray scattering (SAXS) experiments, we observed an analogous signature of Mg2+ induced structural collapse that can facilitate subsequent ligand binding [40]. Our studies provided direct insight into the global rearrangement induced by both Mg2+ and ligand. The compaction of RNA by Mg2+ was also studied by size-exclusion chromatography (SEC): changes in the measured elution volume suggested a decrease in the particles’ hydrodynamic radius [40].
Pre-organization by Mg2+ has also been observed in other riboswitches [21,28,45]. In the SAM-I system, our biochemical studies have shown that addition of Mg2+ yields the pre-organized partially folded state. In addition, we have shown that, in the absence of Mg2+, the fully folded state cannot be achieved, even at high ligand concentrations [23,28,46]. The presence of both, Mg2+ and ligand are required for the stabilization of the fully folded ligand-bound configuration.
While previous smFRET and SAXS data revealed that ligand free RNA undergoes substantial structural changes upon variation of Mg2+ concentration, these structural changes often remained undetected by traditional NMR and X-ray crystallography techniques because of the transient nature and low population levels of such intermediates [47]. The newly developed Chemical Exchange Saturation Transfer (CEST) measurements are now capable of probing these sparsely and transiently populated RNA conformations. Earlier we studied NMR dynamics of the SAM-II system with this new method [47]. The data indeed confirmed that SAM-II riboswitch can access a sparsely populated but bound-like pre-organized state even in the absence of ligand [40,47].
In the present study, we performed molecular simulations to predict the effect of Mg2+ on the conformational landscape of the SAM-II riboswitch. We then tested these predictions with 13C-CEST data. Analysis of our simulations yields the free energy landscape of the SAM-II riboswitch, the effect of Mg2+ on this landscape and insight into the microscopic origins of these effects. More specifically, reappraisal of the 13C-CEST data for the ligand-free SAM-II riboswitch at different Mg2+ concentrations enabled us to probe the influence of Mg2+ on sparsely populated bound-like pre-organized states. We then revisited earlier smFRET, SAXS and SEC elution profiles and compared with our present equilibrium simulation results to integrate these data into a unified scenario of Mg2+-induced collapse. We calculated the free energy landscape of the SAM-II riboswitch using our recently developed all-atom structure-based model (SBM) that includes explicit Mg2+ ions and the effects of Manning condensation and Debye-Hückel Potassium and Chloride interactions. We specifically predict that, as Mg2+ concentration is increased from 0.25 mM to 2 mM, the SAM-II riboswitch collapses from an extended, partially unfolded state to a highly compact, pre-organized state, in agreement with the 13C-CEST studies, where we observe a shift in population towards a bound-like conformation. In addition, our simulations characterize this collapse transition in terms of the radius of gyration as a function of Mg2+ concentration, which is qualitatively similar to previous SAXS measurements. This agreement gives us confidence in the microscopic details of our simulations, showing that the triplex formation between helix P2b and loop L1 plays an important role in the collapse process.
Results
Simulations of the SAM-II riboswitch were performed with and without SAM at different Mg2+ concentrations. The simulation started from the crystal structure of SAM-II riboswitch (pdb accession code: 2QWY) [20]. A global view of this structure and details of the secondary structure are presented in Fig 1. As ligand-free conformations of SAM-II are still under investigation in experiments, we explore the full folding free energy landscape both in the presence of ligand (SAM) and in the absence of ligand to characterize the entire accessible conformational space. To cover the folding landscape, we employed umbrella sampling over the reaction coordinate, Q, which is the fraction of intra-molecular native contacts, present in both the free and bound states, formed by the riboswitch.
Fig 1. Secondary and tertiary structure of SAM-II riboswitch in ligand-bound state (pdb:2QWY).
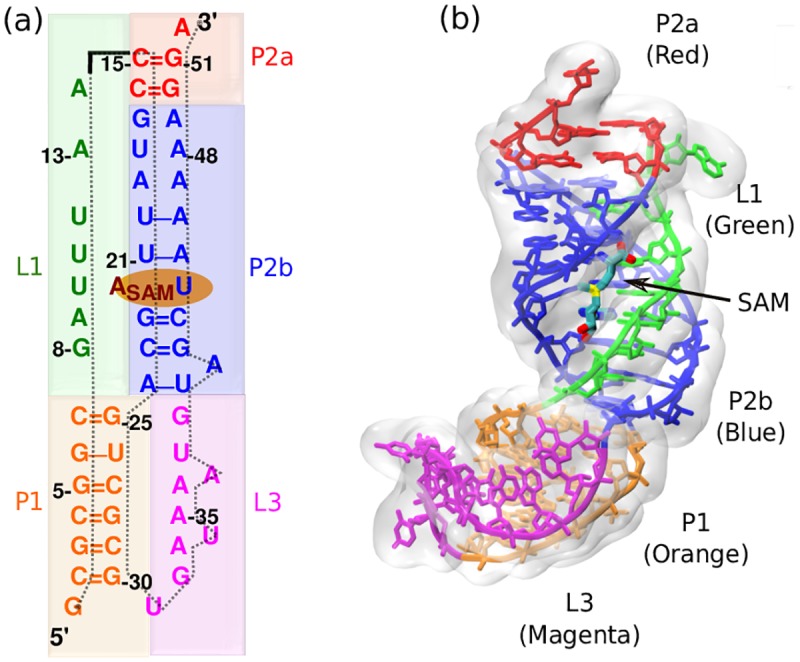
(a) Sequence-aligned secondary structure of the SAM-II riboswitch where base pair and stacking interactions are indicated. There are three helices and two strands highlighted with different colors. (P1: Orange, P2a: Red, P2b: Blue, L1: Green, L3: Magenta). (b) Tertiary structure displays triple helix between helix P2b and loop L1.
Detailed agreement between 13C-CEST, smFRET, SAXS, SEC elution profile and generalized Manning model corroborates pre-organization by Mg2+
As mentioned earlier, CEST experiments are able to capture transiently populated dynamic conformations [13]. This strategy was applied to the ligand-free SAM-II riboswitch in the presence of 0.25 mM and 2 mM Mg2+. The 13C-CEST profiles of the labeled ribose C1’ and base C6 carbons of C43 were recorded at three different B1-fields (17.5, 27.9, 37.8 Hz) with a mixing time of 0.3 s at 298 K [47]. We compare the data for 0.25 mM and 2 mM Mg2+ concentrations at B1-field of 17.5 Hz (Fig 2a). The data were fit with a two-state model where the low population of the partially closed state (peaks around 300 Hz spin-lock offset) appears to increase with addition of 2 mM Mg2+. Consistent results were obtained from CEST profiles for other B1-fields of 27.9 and 37.8 Hz (Fig S1 in the S1 Text). Trajectory plots of the fraction of native contacts extracted from the generalized Manning equilibrium simulations of ligand-free SAM-II at these two concentrations clearly show the hopping between different conformations (Fig 2b). Furthermore, the dynamic transitions between the two major states (bound-like: Q≈0.9 and open: Q≈0.7) visit native-like conformations (Fig 2c) more frequently at 2 mM than at 0.25 mM Mg2+, as summarized in the corresponding contact histograms, P(Q) (Fig 2d). Both CEST experiments and simulation data indicate that the equilibrium shifts from the open conformations toward the native bound-like state as we increase Mg2+ concentration.
Fig 2. Detailed agreement between 13C CEST, smFRET, SAXS, and SEC elution profiles with the generalized Manning model corroborates pre-organization by Mg2+.
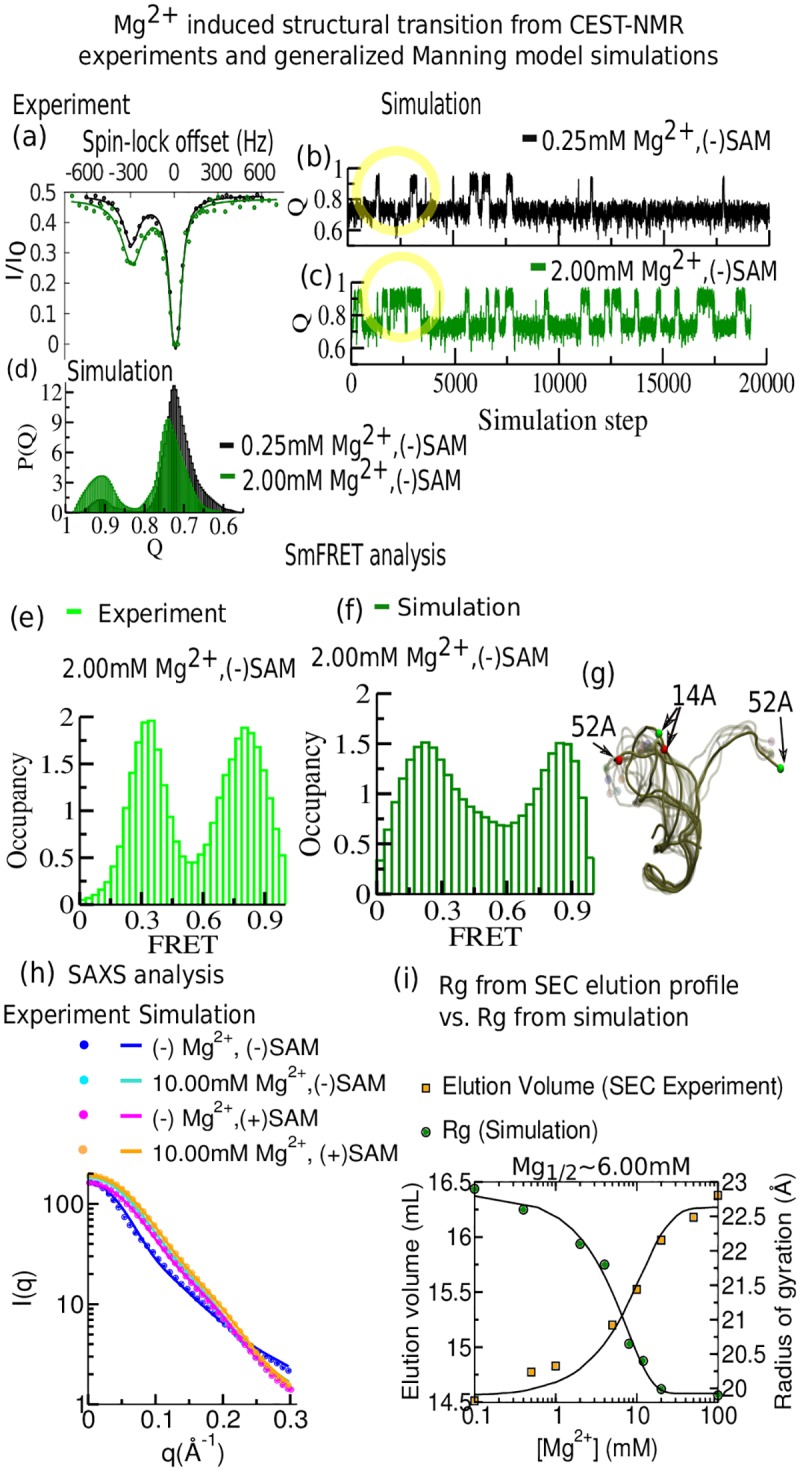
(a)-(d) Comparison between CEST-NMR experiments and Manning model simulations. (a) 13C CEST profiles for C43-C6 of SAM-II riboswitch in the ligand-free situation in the presence of two different concentrations of Mg2+ (black dots: 0.25 mM, green dots: 2.0 mM) at B1 field of 17.5 Hz. The population distribution involving two major states was obtained by fitting each 13C CEST profile (solid lines) into a two-state model. (b) The fraction of native contacts (Q) dynamics extracted from simulations indicate the transition between two major states at 0.25 mM. (c) Same as (b) for 2.0 mM Mg2+. (d) The fraction of native contact population histograms at these two concentrations. (e)-(f) Steady state smFRET analysis from fluorescently labeled experiments and theoretical prediction from simulations. (e) Experimental smFRET occupancy histogram in the presence of 2.0 mM Mg2+ (adapted from ref. 22) [22]. (f) Theoretical prediction of FRET occupancy in the presence of 2.0 mM Mg2+. (g) Superposition of a set of different conformations at 2.0 mM Mg2+ depicts the prevalence of two distinct sets of ensembles. (h) Comparison of experimental scattering profiles of SAM-II collected from four different buffer conditions. Theoretical SAXS predictions are depicted by solid lines and the experimental data by dots (experimental data adapted from ref. 40). (i) Compaction of SAM-II as a function of [Mg2+] obtained from SEC elution profiles is consistent with a decrease in hydrodynamic radius (experimental data adapted from ref. 40), [40] in qualitative agreement with the observed folding transition in the average radius of gyration (Rg) obtained from equilibrium simulations at Mg1/2≈6.0 mM.
The signature of the existence of such Mg2+ induced bound-like states has also been reported in previous smFRET experiments (Fig 2e) [22]. To support our observations we have revisited some of these smFRET efficiency assessments [22] and compared them with theoretical FRET predictions obtained from our generalized Manning model simulations under similar buffer conditions. The equation used for theoretical FRET prediction is described in section S1 in the S1 Text. We tracked the dynamics of positions 14 and 52, where acceptor (cy5) and donor (cy3) fluorophore labels were placed in the smFRET experiments (Fig 2f). Both experimental and simulation FRET confirm the coexistence of two states at 2 mM Mg2+ (Fig 2g) [22].
Previous SAXS data corroborates well the existing smFRET observations [22,40]. The SAXS data also indicated both ligand and Mg2+ ions are required to effectively fold this riboswitch. To microscopically understand their mutual and stand-alone effects from the present simulations, we studied the conformational differences of this riboswitch in four extreme buffer conditions and compared our computational results with experimental SAXS data. For this comparison, we extracted multiple snapshots from several long trajectories and computed ensemble averaged SAXS profiles using the Debye formula for spherical scatterers parameterized in the FoXS web server [48,49] as described in section S2 in the S1 Text. The predicted SAXS curves here show qualitative agreement with experiments (Fig 2h) [40]. The Kratky representation of SAXS data presented in Fig s2 in the S1 Text shows a pronounced peak, indicating the emergence of more extended conformations with decreasing Mg2+ concentrations. We note that capturing the entire conformational heterogeneity of an extended state is computationally challenging. This mostly applies for the extreme case where neither ligand nor Mg2+ is present. In this case, the correlation between theoretical and experimental SAXS profiles leaves room for improvement. Values for chi-square reflect that and are shown in Table S1 in the S1 Text. These analyses indeed suggest the potential impact of both, ligand and Mg2+, stabilizing the closed conformations, which we characterize further below with contact data to describe the pre-organization and the ligand-organized closing.
A significant Mg2+ induced collapse transition, as indicated by the SAXS data, has been followed over a wide concentration range (up to 100 mM) of Mg2+ in SEC elution volume profile (Fig 2i). Here RNA elutes after longer retention times with increasing Mg2+ concentration ([Mg2+]) in the mobile phase [40]. Bigger elution volume signifies decreasing hydrodynamic radius of a monomeric RNA molecule. The folding transitions, both from experimental elution volume data and from average Rg measured from the equilibrium simulation analysis as functions of [Mg2+], follow sigmoid curves with transition midpoint, Mg1/2 at 6 mM (Fig 2i).
Mg2+ remodels the free energy landscape, favoring pre-organized states
At this point, a range of experimental techniques and simulation data support the existence of pre-organized states. Here we aim to obtain a thermodynamic description of how Mg2+ governs the energy landscape of RNA from our model simulation study. In Fig 3a, we show the free energy landscape for the folding transition of SAM-II riboswitch in SAM-bound (in the presence of explicit ligand) and SAM-free (in the absence of ligand) conditions near the physiological concentration of Mg2+ ([Mg2+] = 2.0 mM). During this folding transition, each secondary structural segment folds sequentially illuminating the pathway of folding (Fig 3b). The free energy profile, in the presence of explicit SAM has a distinct bound-state-well, reflecting the ligand-induced stabilization of the closed conformations (designated as (i) in Fig 3c). In the apo-form of the riboswitch, the fully closed bound state does not correspond to a minimum in the landscape. At lower Q than this ligand-bound state, the free energy profile for apo-SAM-II riboswitch reveals three distinct minima. They involve: a ligand-free partially closed state (state (ii) in Fig 3c), which has a substantial overlap with the ligand-bound closed conformation. In this state, the nonlocal contacts (involving base-pairing contacts) including base-stacking contacts in P1, the P1-L3 pseudo-knot interaction, and major segments of P2b and the L1-P2b triplex interactions remain secured, while the contacts involved in Shine/Dalgarno sequence (AAAG50G51A523´), and in the part of L1-P2b are disrupted (state (ii) in Fig 3c).
Fig 3. Magnesium dependence of the SAM-II riboswitch free energy landscape demonstrates Mg2+ pre-organization of bound-like state.
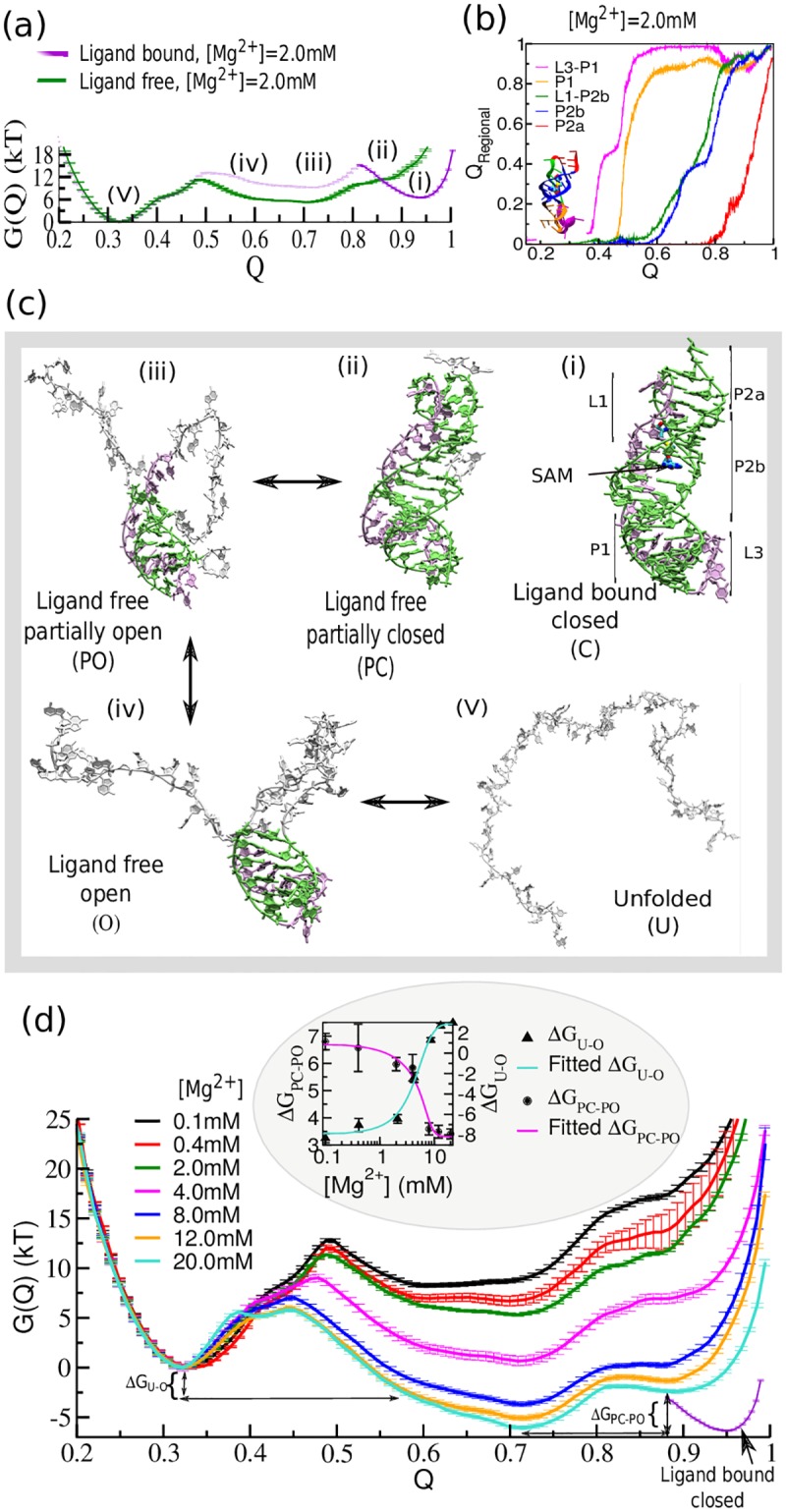
(a) The free energy landscape as a function of the fraction of native contacts (Q) of SAM-II near physiological Mg2+ concentration ([Mg2+] = 2.0 mM). The system explores three distinct barrier-separated minima on the free energy landscape. (b) The order of secondary structure formation as a function of the fraction of all native contacts (Q) as measured by the fraction of non-local regional contacts (QRegional). The transition displays high cooperativity. (c) Representative structures. The representative structure corresponding to each region of the energy landscape is designated as follows: (i) ligand-bound closed (C), (ii) ligand-free partially closed (PC) including triplex interactions, (iii) ligand-free partially open (PO), (iv) ligand-free open (O), and (v) the unfolded (U) state. Green/mauve, bases in native conformations. The ligand-bound closed minimum has been characterized from the free energy profile of the folding transition of SAM-bound RNA which indicates the ligand-free partially closed conformation has a substantial resemblance with the ligand-bound closed state. (d) Mg2+ concentration dependence of the folding transition of apo-SAM-II riboswitch. Average free energy profiles of folding transition at different [Mg2+] show significant stability difference between the ligand-free partially closed (PC) and the partially open (PO) minima (ΔGPC-PO); and the unfolded (U) and the extended open (O) state minima (ΔGU-O). In the inset ΔGPC-PO and ΔGU-O are plotted together as a function of [Mg2+]. Both sigmoid curves follow the same transition midpoint, Mg1/2 ≈6.0 mM, as found in the SEC profile.
Recent fluorescence and NMR spectroscopic data also indicated that C16 in P2a helix remains mostly unpaired in the absence of SAM [22]. The data also suggested that formation of the pseudoknot in the absence of SAM is highly transient in nature. Intermediate states, (iii) and (iv) in Fig 3c, although marginally separated by a small barrier, effectively belong to a broad, flat basin which involves an ensemble of partially folded open configurations. A representative unfolded structure ((v) in Fig 3c) is shown to describe the unfolded minimum. As we increase the concentration of Mg2+ we find enhanced stabilization of the pre-organized partially closed conformations (state (ii) in Fig 3d) relative to the open conformations. Our latest 13C-CEST chemical exchange data anticipated that the emergence of Mg-induced pre-organization can have immense consequences for rapid ligand recognition [47]. In the context of the simulation, Mg2+ induced thermodynamic stabilization is reflected by the difference in stability, ΔGPC-PO between the bound-like partially closed (PC) conformation and the partially open (PO) conformation and by ΔGU-O between the unfolded (U) and the open conformation (O). These two stability differences, ΔGPC-PO and ΔGU-O vary upon increasing [Mg2+] until they reach their saturation limits. Both ΔGPC-PO and ΔGU-O plotted as functions of [Mg2+], are fitted well to sigmoid curves with a Mg2+1/2 value around 6 mM (inset of Fig 3d) which again correlates well with the SEC elution volume data (Fig 2i) [40].
Mg2+-induced phosphate interactions facilitate pre-organization
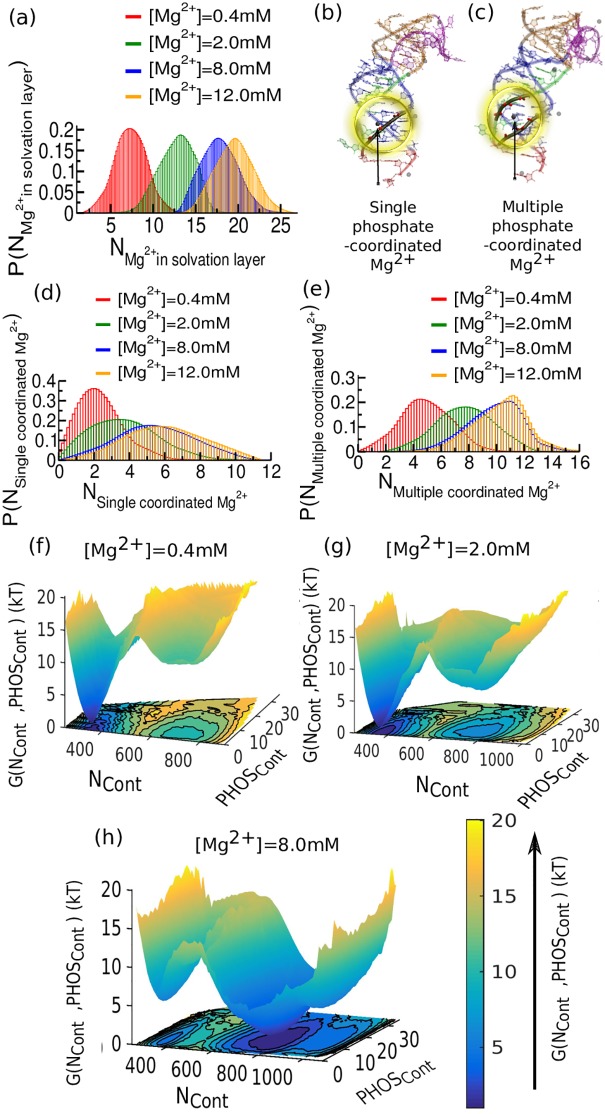
To address the open question of how Mg2+ ions regulate structural collapse, we have determined the Mg2+ distribution in the ion-solvation layer of SAM-II, which accommodates increasing numbers of Mg2+ up to 8 mM Mg2+ content (Fig 4a). Subsequent additions of Mg2+ beyond 8 mM do not effectively add to the 1st layer of Mg2+ solvation. How we characterize the ion-solvation layer from our simulated trajectories is described in section S3 in the S1 Text (Fig s3 in S1 Text). We have further classified the outer sphere Mg2+ present in the ion-solvation layer into two categories based on their number of associated phosphate groups: (i) Single phosphate coordinating Mg2+ (Fig 4b), which efficiently neutralize the negative charge of the adjacent phosphate (Fig 4d), and (ii) multiple phosphate coordinating Mg2+ (Fig 4c). The key role in stabilizing the structure is played by such Mg2+ bridging multiple phosphates, which can act as glue in compact structures by holding a number of negatively charged phosphates together in close proximity.
Fig 4. Probability distribution and free energy landscape of the SAM-II riboswitch as a function of the number of Mg2+ions in the ion solvation layer and Mg2+ mediated phosphate contacts (PHOSCont).
(a) The distribution of Mg2+ in the ion solvation layer shows gradual shift accommodating more number of Mg2+ with increasing Mg2+ concentration. Beyond 8.0 mM the first ion-solvation layer appears saturated. Subsequent additions of Mg2+ beyond 8.0 mM do not effectively participate in the 1st layer of solvation by ionic interaction. (b) Snapshot extracted from the simulation at buffer condition [Mg2+] = 0.4 mM has single phosphate coordinated Mg2+. (c) Snapshot extracted from the simulation at buffer condition [Mg2+] = 8.0 mM has multiple phosphate coordinated Mg2+. (d) Single phosphate coordinated Mg2+ as a function of Mg concentration. (e) Multiple phosphate coordinated Mg2+ as a function of [Mg2+]. The population shift of such multiple coordinated Mg2+ serves to connect negatively charged phosphate groups. (f) Free energy landscape as a function of Ncont and PHOSCont for [Mg2+] = 0.4 mM. (g) Same as (f) for [Mg2+] = 2.0 mM. (h) Same as (f) for [Mg2+] = 8.0 mM. Increasing [Mg2+] reshapes the landscape, shifting the equilibrium from a basin at lower Ncont to a basin at higher Ncont.
The population shift coincident with multiple coordinated Mg2+ ions with increasing Mg2+ concentration directly supports their role in stabilizing the structure (Fig 4e). We have also investigated the thermodynamic impact of Mg2+-mediated phosphate contacts (PHOSCont: total number of pair-wise phosphate-phosphate contacts) on the energy landscape as a function of overall folding progress, expressed by the number of native contacts (NCont), as shown in Fig 4f–4h. As we increase the Mg2+ concentration the broad minimum that appeared around NCont~800, involving partially folded open conformations, gradually becomes more stabilized. Concurrent enrichment of phosphate-phosphate contacts extends the contour of the minimum asymmetrically toward higher PHOSCont. Additionally, by 8 mM [Mg2+], the bound-like pre-organized state grows with substantial population, stabilized again by phosphate connections (Fig 4h).
Mg2+-induced triplex interactions between helix P2b and L1 loop help pre-organize the SAM-II riboswitch
We analyzed long equilibrium trajectories of the apo- and bound-forms of SAM-II slightly below the folding temperature in order to capture the essential characteristics of the pre-organized state, and also to compare this state with the fully folded ligand bound state. We have evaluated the distribution of native contact formation in each segment of secondary structure as a function of the total Q at different Mg2+ concentrations. Plots show contact formation in P2b (Fig 5a–5d) and the triplex interaction between helix P2b and loop L1 (Fig 5e–5h), which are most affected by Mg2+ concentration. Data for the nonlocal contacts of P1, L3-P1, P2a, which appear only marginally affected by Mg2+ concentration, are shown in Fig s4 in S1 Text. The two distinct basins visible at low [Mg2+], for the P2b helix and L1-P2b triplex contacts correspond to the pre-organized (at higher Q) and open states (at lower Q). At increasing [Mg2+], the populations gradually shift towards the pre-organized state.
Fig 5. Thermodynamics of triplex formation.
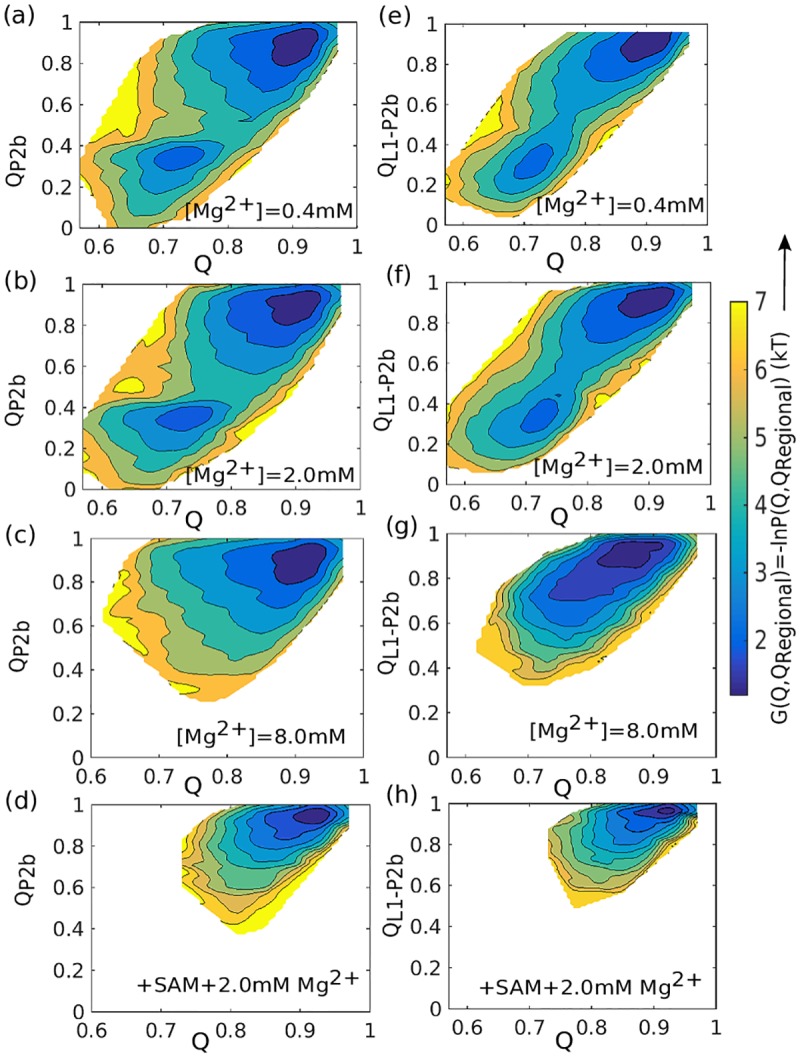
Free energy landscape of the SAM-II riboswitch as a function of total native contacts (x-axis) and native contacts for an individual structural element (y-axis). (a)-(c) Free energy landscapes for helix P2b formation for increasing values of [Mg2+], without SAM. (d) Free energy landscapes for helix P2b formation for the case of [Mg2+] = 2.0 mM, with SAM. (e)-(g) Free energy landscapes for triplex formation (L1-P2b contacts) for increasing values of [Mg2+], without SAM. (h) Free energy landscapes for triplex formation for the case of [Mg2+] = 2.0 mM, with SAM. (a)-(c) and (e)-(g) Collective integrity of triplex interaction (involving both P2b and P2b-L1) appears most sensitive RNA element to [Mg2+].
Around 8 mM Mg2+, the dominant contribution arising from this pre-organized triplex to the conformational space is evident from Fig 5c and 5g. Ligand binding also strongly favors structure formation, even at moderate [Mg2+], as the ligand bridges the gap between L1 strand and P2b helix, producing the fully formed triplex.
Discussion
Motivated by our 13C-CEST profiles for SAM-II and their [Mg2+] dependence we have explored the free energy landscape of the SAM-II riboswitch using a recently developed all-atom SBM that includes explicit Mg2+ ions, Debye-Hückel treatment of implicit KCl interactions, and the effects of Manning condensation to accurately account for the ion atmosphere around the RNA. Our results support a mechanism involving Mg2+ induced pre-organization followed by conformational selection by the ligand, SAM, as we speculated in an early study [47]. The free energy analysis validates the observations of that pre-organization, providing an atomistic and thermodynamic basis for the enhanced population of a partially collapsed, pre-organized ensemble at sufficiently high Mg2+ concentration in the absence of ligand. We observe three distinct sets of conformations in the folding free energy landscape of ligand-free SAM-II riboswitch: (i) an ensemble of unfolded conformations, (ii) a broad ensemble of partially folded open conformations, and (iii) an ensemble of pre-organized bound-like conformations. As we increase magnesium concentration beyond 2–4 mM, the bound-like ensemble is further stabilized, shifting the equilibrium toward the pre-organized states. All the experimental results from our 13C-CEST profile, recent SAXS, single molecule FRET, and size-exclusion chromatographic studies are assembled and found to be in good agreement with the present simulation results (Fig 2). At higher concentrations, Mg2+ stabilizes compact structures by coordinating multiple charged phosphate groups of RNA in close proximity. The experimental results, together with free energy landscapes confirm that sufficient Mg2+ can indeed promote stable ligand binding in the SAM-II riboswitch, and is likely the structural basis for the switching control of protein translation.
While this structural pre-organization of SAM-II can assist in rapid ligand recognition, our study suggests that a sufficiently high concentration of Mg2+ is necessary to capture those pre-organized states. Only when the system achieves a well-organized ion solvation layer at high [Mg2+], the effect of additional Mg2+ seems limited. This layer involves a number of Mg2+ ions, each coordinating with multiple phosphate groups. Mg2+ ions thus serve as glue to the negatively charged phosphates and facilitate the structural compaction. We note that while chelated Mg2+ may play an important role in other riboswitch RNAs, no specific chelated ions have been reported so far in the SAM-II system.
Our molecular simulation trajectories also allow us to pinpoint the structural basis of the effect, revealing that triplex interaction between the helix P2b and its association with the L1 strand dominate the process of pre-organization as summarized in Fig 5a–5c and 5e–5g, showing the gain of structure with increasing [Mg2+]. In the final step, ligand binding firmly bridges the extended gap between L1 and P2b, which seems otherwise not achievable through the addition of small, dynamic Mg2+ alone. But although P2b and its connection with L1 can be secured by the ligand, its presence again alone cannot fully stabilize the overall structure without addition of significant amount of Mg2+ (Fig 2h). These findings suggest that a sufficiently high concentration of Mg2+ is necessary to stabilize the pre-organized triplex and then the presence ligand promotes the native triplex formation, as summarized in Fig 5d and 5h. We note that triplexes have recently emerged as important players in gene regulation by non-coding RNAs [50–53]. Base triples also play a role in RNase P and the Diels-Alder ribozyme [54]. Heroic calculations, as such recent microsecond explicit solvent simulations of riboswitches, will also shed light on these effects, especially regarding the role of solvation [55].
Nucleic acid-ion interactions make a substantial energetic contribution in the stabilization of the native state of RNAs, including complex formation with proteins and other macromolecules [56]. The dynamics of nucleic acids are also found to be strongly influenced by the motion of their ion atmospheres. Relative to other ionic species, Mg2+ can efficiently support a close assembly of negatively charged phosphates by mediating favorable interactions among them. Other earth alkali metals/divalent ions (e.g. Ca2+) and even monovalent ions are also able to induce similar transitions, albeit at higher concentration. Our early SEC elution profiles for SAM-II show that the transition midpoint in presence of Potassium (K+) alone occurs only at [K]1/2 ≈ 25 mM. The midpoint for Calcium (Ca2+) is [Ca]1/2 ≈ 8 mM, compared to 6 mM for the Mg2+ ion [40]. This is a direct result of the larger charge/radius ratio of magnesium [40,57].
Thus, having these special characteristics, Mg2+ efficiently helps pre-organize the system and enables access to the partially collapsed states that are further stabilized by ligand binding. The general importance of Mg2+ for the stability of compact RNA structures supports a possibly universal role of conformational selection in ligand-binding RNAs, such as riboswitches, aptamers, and possibly protein-binding RNAs. A detailed thermodynamic understanding of the underlying landscape will indeed enable greater control of riboswitch regulation, highly sought after by researchers in synthetic biology who are currently employing riboswitches as ligand-dependent ‘knobs’ to control desired gene expression [58].
Methods
RNA electrostatic model
Our all-atom structure-based model (SBM) has proven successful in describing the dynamics of numerous proteins and macromolecular complexes [59–63]. To elucidate RNA free energy landscapes under the influence of Mg2+, models capable of quantitatively describing the ion atmosphere are needed, including ionic condensation around the negatively charged phosphate groups of RNA. Early studies have simply included electrostatic effects in SBM of RNA via repulsive Debye-Hückel interactions, thus treating all ions implicitly [29,30]. Recently, our group developed a more detailed model of RNA electrostatics and applied it within all-atom structure-based molecular dynamics simulations. Our model treats Mg2+ ions explicitly to account for ion-ion correlations neglected by mean-field theories [38]. The KCl buffer, which completes the experimental setup, is treated implicitly by a generalized Manning counter ion condensation model [38,64], since mean-field theories correctly assess the charge densities of monovalent K+ and Cl- ions. Classical Manning counter-ion condensation theory was originally developed for understanding the low concentration limiting behavior of polyelectrolyte chains modeled with an infinite line of charge. Folded RNA, however, is not a line of charge. To account for the compact and irregular structures of RNAs and the effects of varying ion concentrations, we improve the Manning counter ion condensation model to handle electrostatic heterogeneity, making the condensed charge density a dynamical function of each phosphate coordinate. KCl screening is characterized by a Debye-Hückel potential. Removal of the continuum screening ions from the inaccessible volume of RNA is a substantial extension to Manning counter-ion condensation. The model has been tested against experimental measurements of excess Mg2+ associated with RNA, characterizing the Mg2+-RNA interaction free energy. This hybrid SBM has opened up new possibilities to study various structural and functional processes of RNA that are essentially controlled by ions [38]. In the present study we used this recently developed all-atom hybrid SBM to understand the conformational transition of SAM-II and the corresponding Mg2+ sensitivity. The energy function used in this model is given below,
| (1) |
where, ΦSBM is the all-atom SBM potential ensuring a global minimum in the landscape for the native state of RNA. The SBM potential is composed of two general types of interactions:
| (2) |
where, Φlocal characterizes the local interactions that encode covalent bonds and torsional angles, maintaining the correct local geometry and chirality. Φnon-local comprises two non-local contributions: (i) an attractive term that is applied specifically to all tertiary interactions determined from the native structure, (ii) the general repulsive interactions, that describe the excluded volume by symmetric hard potentials (to avoid any unwanted chain crossing). ΦMg-Size adds the excluded volume interactions involving the explicit Mg2+ ions, regulating RNA-Mg2+ and Mg2+-Mg2+ interactions. Φion-effect accounts for all interactions between charges in the system which consist of the fixed charge distribution of the RNA and the dynamic contribution from the ions. Mg2+ and phosphate charges interact via a Debye-Hückel potential with a screening term that depends, in turn, on the distribution of the monovalent ions. The monovalent ions, K+ and Cl- from the added salt, fall into two categories: screening ions and Manning condensed ions. The screening ion density is obtained using Debye-Hückel electrostatics. The density of the Manning-condensed ions is modeled as the sum of two normalized Gaussian distributions where the center of each Gaussian is located on the position of the negatively charged phosphate group. All the condensation variables along with the explicit Mg2+ and RNA coordinates are evolved with Langevin dynamics [38]. The mathematical formulations of all the terms and the related parameterizations are discussed in depth in section S4 in the S1 Text.
Free energy simulations
The umbrella sampling method [65] was used to sample the conformational space of SAM-II riboswitch along the reaction coordinate, Q, which is the fraction of intra-molecular native contacts in the riboswitch. The Weighted Histogram Analysis Method [66] was then used to calculate the thermodynamic quantity, G(Q). The detail is described in section S5 in the S1 Text.
13C-CEST experiments
CEST data were collected using a pseudo-3D HSQC experiment with the B1 field offsets (-600 to 600 Hz) incremented in an interleaved manner with 3 references (no CEST period) [47]. A total of 1024x16 complex points were recorded [40] with 32 transients with a recovery delay of 1.5 s for a total experimental time of approximately 12 hr for each spin-lock field. A CEST saturation period of 100 ms was used for the base and 200 ms for ribose. The pulse program used was an adaptation of a previously published one without the need for selective pulses [47].
Analysis of relaxation parameters
We used a two-state model to fit each of the three profiles of the selectively labeled carbon (ribose C1’ and base C6) and quantitatively extracted the carbon chemical shift (Δω), the exchange rate, and the population of the minor state based on the Bloch−McConnell 7x7 matrix [47]. The CEST data was plotted as I(t)/I(0) versus spin-lock offset (Hz) and was fit by numerically solving the matrix exponential for the CEST spin-lock period based on this 7x7 two-state Bloch-McConnell equation as described earlier [47,67].
FRET
In experiments, Fluorescence Resonance Energy Transfer (FRET) efficiency is the quantum yield of the energy transfer where a donor chromophore from its excited electronic state may transfer its energy to an acceptor chromophore through a non-radiative dipole-dipole coupling. The FRET efficiency varies with the separation between donor and acceptor fluorophores following the Fӧrster relation. For theoretical FRET predictions we use the Fӧrster relation where the value of Fӧrster radius is taken as 53Å [68]. We described it in detail in section S1 in the S1 Text.
SAXS analysis
In SAXS experiments, the scattering intensity is measured from the electron density difference between the purified sample and that of the solvent/buffer. FoXs is a method that uses the Debye formula by which a theoretical scattering profile of a structure can be computed [48,49]. The detail is discussed in section S2 in the S1 Text.
Supporting information
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Institute of Health Grant (R01-GM110310 to JNO, R01- GM110310 to KYS, P50 GM103297 to TKD) and National Science Foundation (NSF) (DBI1040158 and NSF CHE1213668 to TKD). We acknowledge technical support and resources from the Center for Theoretical Biological Physics. Work at the Center for Theoretical Biological Physics was also supported by the NSF (Grant PHY-1427654 and NSF-CHE-1614101) and by the Cancer Prevention and Research Institute of Texas (CPRIT - grant R1110). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Huarte M (2015) The emerging role of lncRNAs in cancer. Nat Med 21: 1253–1261. 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 2.Montange RK, Batey RT (2008) Riboswitches: Emerging Themes in RNA Structure and Function. Annu Rev Biophys 37: 117–133. 10.1146/annurev.biophys.37.032807.130000 [DOI] [PubMed] [Google Scholar]
- 3.Roth A, Breaker RR (2009) The Structural and Functional Diversity of Metabolite-Binding Riboswitches. Annu Rev Biochem 78: 305–334. 10.1146/annurev.biochem.78.070507.135656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin JC, Yoon J, Hyeon C, Thirumalai D (2015) Using simulations and kinetic network models to reveal the dynamics and functions of riboswitches. Methods Enzymol 553: 235–258. 10.1016/bs.mie.2014.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serganov A, Patel DJ (2007) Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nature Reviews Genetics 8: 776–790. 10.1038/nrg2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR (2003) An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Structural Biology 10: 701–707. 10.1038/nsb967 [DOI] [PubMed] [Google Scholar]
- 7.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, et al. (2005) Genome Biology 6: R70 10.1186/gb-2005-6-8-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamauchi T, Miyoshi D, Kubodera T, Nishimura A, Nakai S, et al. (2005) Roles of Mg2+ in TPP-dependent riboswitch. FEBS Lett 579: 2583–2588. 10.1016/j.febslet.2005.03.074 [DOI] [PubMed] [Google Scholar]
- 9.Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, et al. (2007) Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol 365: 1393–1406. 10.1016/j.jmb.2006.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisaria N, Greenfeld M, Limouse C, Pavlichin DS, Mabuchi H, et al. (2016) Kinetic and thermodynamic framework for P4-P6 RNA reveals tertiary motif modularity and modulation of the folding preferred pathway. Proc Natl Acad Sci U S A 113: E4956–4965. 10.1073/pnas.1525082113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bisaria N, Herschlag D (2015) Probing the kinetic and thermodynamic consequences of the tetraloop/tetraloop receptor monovalent ion-binding site in P4-P6 RNA by smFRET. Biochem Soc Trans 43: 172–178. 10.1042/BST20140268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takamoto K, Das R, He Q, Doniach S, Brenowitz M, et al. (2004) Principles of RNA compaction: insights from the equilibrium folding pathway of the P4-P6 RNA domain in monovalent cations. J Mol Biol 343: 1195–1206. 10.1016/j.jmb.2004.08.080 [DOI] [PubMed] [Google Scholar]
- 13.Xue Y, Gracia B, Herschlag D, Russell R, Al-Hashimi HM (2016) Visualizing the formation of an RNA folding intermediate through a fast highly modular secondary structure switch. Nat Commun 7: ncomms11768 10.1038/ncomms11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bocobza SE, Aharoni A (2014) Small molecules that interact with RNA: riboswitch-based gene control and its involvement in metabolic regulation in plants and algae. Plant J 79: 693–703. 10.1111/tpj.12540 [DOI] [PubMed] [Google Scholar]
- 15.Russell R (2015) Unwinding the mechanisms of a DEAD-box RNA helicase in cancer. J Mol Biol 427: 1797–1800. 10.1016/j.jmb.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batey RT, Gilbert SD, Montange RK (2004) Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415. 10.1038/nature03037 [DOI] [PubMed] [Google Scholar]
- 17.Edwards AL, Reyes FE, Heroux A, Batey RT (2010) Structural basis for recognition of S-adenosylhomocysteine by riboswitches. RNA 16: 2144–2155. 10.1261/rna.2341610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garst AD, Heroux A, Rambo RP, Batey RT (2008) Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem 283: 22347–22351. 10.1074/jbc.C800120200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert SD, Montange RK, Stoddard CD, Batey RT (2006) Structural Studies of the Purine and SAM Binding Riboswitches. Cold Spring Harbor Symposia on Quantitative Biology 71: 259–268. 10.1101/sqb.2006.71.015 [DOI] [PubMed] [Google Scholar]
- 20.Gilbert SD, Rambo RP, Van Tyne D, Batey RT (2008) Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol 15: 177–182. 10.1038/nsmb.1371 [DOI] [PubMed] [Google Scholar]
- 21.Haller A, Altman RB, Souliere MF, Blanchard SC, Micura R (2013) Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proceedings of the National Academy of Sciences 110: 4188–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haller A, Rieder U, Aigner M, Blanchard SC, Micura R (2011) Conformational capture of the SAM-II riboswitch. Nature Chemical Biology 7: 393–400. 10.1038/nchembio.562 [DOI] [PubMed] [Google Scholar]
- 23.Hennelly SP, Sanbonmatsu KY (2010) Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Research 39: 2416–2431. 10.1093/nar/gkq1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heppell B, Mulhbacher J, Penedo JC, Lafontaine DA (2009) Application of Fluorescent Measurements for Characterization of Riboswitch–Ligand Interactions Methods in Molecular Biology: Springer Science + Business Media; pp. 25–37. [DOI] [PubMed] [Google Scholar]
- 25.Lemay J-F, Penedo JC, Tremblay R, Lilley DMJ, Lafontaine Daniel A (2006) Folding of the Adenine Riboswitch. Chemistry & Biology 13: 857–868. [DOI] [PubMed] [Google Scholar]
- 26.Montange RK, Batey RT (2006) Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441: 1172–1175. 10.1038/nature04819 [DOI] [PubMed] [Google Scholar]
- 27.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, et al. (2010) Free State Conformational Sampling of the SAM-I Riboswitch Aptamer Domain. Structure 18: 787–797. 10.1016/j.str.2010.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wacker A, Buck J, Mathieu D, Richter C, Wohnert J, et al. (2011) Structure and dynamics of the deoxyguanosine-sensing riboswitch studied by NMR-spectroscopy. Nucleic Acids Research 39: 6802–6812. 10.1093/nar/gkr238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho SS, Pincus DL, Thirumalai D (2009) Assembly mechanisms of RNA pseudoknots are determined by the stabilities of constituent secondary structures. Proceedings of the National Academy of Sciences 106: 17349–17354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyeon C, Thirumalai D (2005) Mechanical unfolding of RNA hairpins. Proceedings of the National Academy of Sciences 102: 6789–6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, et al. (2009) Nonlocal Helix Formation Is Key to Understanding S-Adenosylmethionine-1 Riboswitch Function. Biophysical Journal 96: L7–L9. 10.1016/j.bpj.2008.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue X, Yongjun W, Zhihong L (2015) Folding of SAM-II riboswitch explored by replica-exchange molecular dynamics simulation. Journal of Theoretical Biology 365: 265–269. 10.1016/j.jtbi.2014.10.022 [DOI] [PubMed] [Google Scholar]
- 33.Di Palma F, Colizzi F, Bussi G (2014) Role of Magnesium Ions and Ligand Stacking in the Adenine Riboswitch Folding. Biophysical Journal 106: 284a–284a. [Google Scholar]
- 34.Draper DE (2004) A guide to ions and RNA structure. RNA 10: 335–343. 10.1261/rna.5205404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misra VK, Draper DE (2001) A thermodynamic framework for Mg2+ binding to RNA. Proceedings of the National Academy of Sciences 98: 12456–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayes RL, Noel JK, Mohanty U, Whitford PC, Hennelly SP, et al. (2012) Magnesium Fluctuations Modulate RNA Dynamics in the SAM-I Riboswitch. J Am Chem Soc 134: 12043–12053. 10.1021/ja301454u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayes Ryan L, Noel Jeffrey K, Whitford Paul C, Mohanty U, Sanbonmatsu Karissa Y, et al. (2014) Reduced Model Captures Mg2+-RNA Interaction Free Energy of Riboswitches. Biophysical Journal 106: 1508–1519. 10.1016/j.bpj.2014.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayes RL, Noel JK, Mandic A, Whitford PC, Sanbonmatsu KY, et al. (2015) Generalized Manning Condensation Model Captures the RNA Ion Atmosphere. Phys Rev Lett 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quarta G, Kim N, Izzo JA, Schlick T (2009) Analysis of Riboswitch Structure and Function by an Energy Landscape Framework. Journal of Molecular Biology 393: 993–1003. 10.1016/j.jmb.2009.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen B, Zuo X, Wang YX, Dayie TK (2011) Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Research 40: 3117–3130. 10.1093/nar/gkr1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doshi U, Kelley JM, Hamelberg D (2011) Atomic-level insights into metabolite recognition and specificity of the SAM-II riboswitch. RNA 18: 300–307. 10.1261/rna.028779.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuchs RT, Grundy FJ, Henkin TM (2007) S-adenosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA. Proceedings of the National Academy of Sciences 104: 4876–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu C, Smith AM, Ding F, Chowdhury A, Henkin TM, et al. (2011) Variable Sequences outside the Sam-Binding Core Critically Influence the Conformational Dynamics of the SAM-III/SMK Box Riboswitch. Journal of Molecular Biology 409: 786–799. 10.1016/j.jmb.2011.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith AM, Fuchs RT, Grundy FJ, Henkin TM (2010) The SAM-responsive SMK box is a reversible riboswitch. Molecular Microbiology 78: 1393–1402. 10.1111/j.1365-2958.2010.07410.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suddala KC, Wang J, Hou Q, Walter NG (2015) Mg 2+ Shifts Ligand-Mediated Folding of a Riboswitch from Induced-Fit to Conformational Selection. J Am Chem Soc 137: 14075–14083. 10.1021/jacs.5b09740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hennelly SP, Novikova IV, Sanbonmatsu KY (2012) The expression platform and the aptamer: cooperativity between Mg2+ and ligand in the SAM-I riboswitch. Nucleic Acids Research 41: 1922–1935. 10.1093/nar/gks978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen B, LeBlanc R, Dayie TK (2016) SAM-II Riboswitch Samples at least Two Conformations in Solution in the Absence of Ligand: Implications for Recognition. Angewandte Chemie International Edition 55: 2724–2727. 10.1002/anie.201509997 [DOI] [PubMed] [Google Scholar]
- 48.Pelikan M, Hura GL, Hammel M (2009) Structure and flexibility within proteins as identified through small angle X-ray scattering. General Physiology and Biophysics 28: 174–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneidman-Duhovny D, Hammel M, Sali A (2010) FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Research 38: W540–W544. 10.1093/nar/gkq461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergmann JH, Spector DL (2014) Long non-coding RNAs: modulators of nuclear structure and function. Current Opinion in Cell Biology 26: 10–18. 10.1016/j.ceb.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA (2012) Formation of triple-helical structures by the 3'-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A 109: 19202–19207. 10.1073/pnas.1217338109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conrad NK (2014) The emerging role of triple helices in RNA biology. Wiley Interdiscip Rev RNA 5: 15–29. 10.1002/wrna.1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, et al. (2012) A triple helix stabilizes the 3 ' ends of long noncoding RNAs that lack poly(A) tails. Genes & Development 26: 2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lescoute A, Westhof E (2006) The interaction networks of structured RNAs. Nucleic Acids Research 34: 6587–6604. 10.1093/nar/gkl963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villa A, Wohnert J, Stock G (2009) Molecular dynamics simulation study of the binding of purine bases to the aptamer domain of the guanine sensing riboswitch. Nucleic Acids Research 37: 4774–4786. 10.1093/nar/gkp486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lipfert J, Doniach S, Das R, Herschlag D (2014) Understanding Nucleic Acid-Ion Interactions. Annual Review of Biochemistry, Vol 83 83: 813–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahmadi MS, Fattahi A (2011) On the binding of Mg2+, Ca2+, Zn2+ and Cu+ metal cations to 2 '-deoxyguanosine: Changes on sugar puckering and strength of the N-glycosidic bond. Scientia Iranica 18: 1343–1352. [Google Scholar]
- 58.Espah Borujeni A, Mishler DM, Wang J, Huso W, Salis HM (2016) Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Research 44: 1–13. 10.1093/nar/gkv1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jana B, Morcos F, Onuchic JN (2014) From structure to function: the convergence of structure based models and co-evolutionary information. Physical Chemistry Chemical Physics 16: 6496–6507. 10.1039/c3cp55275f [DOI] [PubMed] [Google Scholar]
- 60.Noel JK, Whitford PC, Onuchic JN (2012) The shadow map: a general contact definition for capturing the dynamics of biomolecular folding and function. Journal of Physical Chemistry B 116: 8692–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noel JK, Whitford PC, Sanbonmatsu KY, Onuchic JN (2010) SMOG@ctbp: simplified deployment of structure-based models in GROMACS. Nucleic Acids Research 38: W657–661. 10.1093/nar/gkq498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Onuchic JN, Luthey-Schulten Z, Wolynes PG (1997) Theory of protein folding: the energy landscape perspective. Annu Rev Phys Chem 48: 545–600. 10.1146/annurev.physchem.48.1.545 [DOI] [PubMed] [Google Scholar]
- 63.Wolynes PG (2015) Evolution, energy landscapes and the paradoxes of protein folding. Biochimie 119: 218–230. 10.1016/j.biochi.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manning GS (1978) The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys 11: 179–246. [DOI] [PubMed] [Google Scholar]
- 65.Torrie GM, Valleau JP (1974) Monte-Carlo Free-Energy Estimates Using Non-Boltzmann Sampling—Application to Subcritical Lennard-Jones Fluid. Chemical Physics Letters 28: 578–581. [Google Scholar]
- 66.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM (1992) The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules .1. The Method. Journal of Computational Chemistry 13: 1011–1021. [Google Scholar]
- 67.Garcia-Mayoral MF, Trevino MA, Perez-Pinar T, Caballero ML, Knaute T, et al. (2014) Relationships between IgE/IgG4 epitopes, structure and function in Anisakis simplex Ani s 5, a member of the SXP/RAL-2 protein family. PLoS Negl Trop Dis 8: e2735 10.1371/journal.pntd.0002735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobitski AY, Nierth A, Helm M, Jaschke A, Nienhaus GU (2007) Mg2+-dependent folding of a Diels-Alderase ribozyme probed by single-molecule FRET analysis. Nucleic Acids Research 35: 2047–2059. 10.1093/nar/gkm072 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.