Abstract
Mutations in the gene encoding transcription factor TFAP2A result in pigmentation anomalies in model organisms and premature hair graying in humans. However, the pleiotropic functions of TFAP2A and its redundantly-acting paralogs have made the precise contribution of TFAP2-type activity to melanocyte differentiation unclear. Defining this contribution may help to explain why TFAP2A expression is reduced in advanced-stage melanoma compared to benign nevi. To identify genes with TFAP2A-dependent expression in melanocytes, we profile zebrafish tissue and mouse melanocytes deficient in Tfap2a, and find that expression of a small subset of genes underlying pigmentation phenotypes is TFAP2A-dependent, including Dct, Mc1r, Mlph, and Pmel. We then conduct TFAP2A ChIP-seq in mouse and human melanocytes and find that a much larger subset of pigmentation genes is associated with active regulatory elements bound by TFAP2A. These elements are also frequently bound by MITF, which is considered the “master regulator” of melanocyte development. For example, the promoter of TRPM1 is bound by both TFAP2A and MITF, and we show that the activity of a minimal TRPM1 promoter is lost upon deletion of the TFAP2A binding sites. However, the expression of Trpm1 is not TFAP2A-dependent, implying that additional TFAP2 paralogs function redundantly to drive melanocyte differentiation, which is consistent with previous results from zebrafish. Paralogs Tfap2a and Tfap2b are both expressed in mouse melanocytes, and we show that mouse embryos with Wnt1-Cre-mediated deletion of Tfap2a and Tfap2b in the neural crest almost completely lack melanocytes but retain neural crest-derived sensory ganglia. These results suggest that TFAP2 paralogs, like MITF, are also necessary for induction of the melanocyte lineage. Finally, we observe a genetic interaction between tfap2a and mitfa in zebrafish, but find that artificially elevating expression of tfap2a does not increase levels of melanin in mitfa hypomorphic or loss-of-function mutants. Collectively, these results show that TFAP2 paralogs, operating alongside lineage-specific transcription factors such as MITF, directly regulate effectors of terminal differentiation in melanocytes. In addition, they suggest that TFAP2A activity, like MITF activity, has the potential to modulate the phenotype of melanoma cells.
Author summary
Identifying the elements and structure of the gene regulatory network governing melanocyte differentiation may yield insight into the mechanisms of pigmentation diseases and melanoma progression. Pigmentation is abnormal in Tfap2a mutants, but deciphering the exact role of TFAP2A in the network has been complicated by pleiotropic requirements for TFAP2A during development and the redundant function of TFAP2 paralogs in melanocytes. In this study, we find that TFAP2A directly regulates genes involved in melanocyte differentiation and melanin synthesis by binding at both promoters and enhancers associated with these genes. Furthermore, we report evidence that TFAP2A shares many targets with the melanocyte “master regulator” MITF. These findings indicate that TFAP2A drives melanocyte differentiation in parallel with MITF and affects the net pro-differentiation activity that is lost in melanoma.
Introduction
Melanocytes are responsible for pigment deposition in skin and hair follicles, and the dysregulation of melanocyte differentiation underlies both pigmentation disorders and melanoma. Because melanocytes are dispensable for life, the melanocyte lineage can also serve as a model for investigation of developmental processes important in all cell types. Many transcription factors and other regulatory molecules that drive melanocyte development have been identified through genetic analyses of patients with congenital pigmentation disorders, including piebaldism (SNAI2) [1], Waardenburg syndrome types I and III (PAX3) [2], Waardenburg syndrome type II (SNAI2, MITF, SOX10) [3–5], and Waardenburg-Shah syndrome (EDN3/EDNRB, SOX10) [6,7]. Epistasis experiments in model organisms have begun to assemble these genes into functional hierarchies, also called gene regulatory networks (GRNs), that govern specific processes in the development of melanocytes from the neural crest. For example, work in mouse and chick indicates that during melanocyte lineage specification, PAX3 and SOX10 activate expression of MITF [8] and FOXD3 represses it [9–11], while SOX2 and MITF appear to cross-regulate expression of each other [12,13]. A recent integrated analysis of ChIP-seq and expression profile data in mouse found that SOX10 directly activates expression of many genes implicated in melanocyte differentiation, and suppresses those that promote pluripotency [14]. Similarly, MITF ChIP-seq and enhancer deletion studies in human cell lines have shown that in addition to its role in melanocyte fate specification, MITF directly stimulates the expression of many genes encoding effectors of melanin synthesis, including Dopachrome Tautomerase (DCT) [15, reviewed in 16,17,18]. Because mice with loss-of-function mutations in Mitf lack melanocytes, and ectopic expression of MITF activates expression of melanin synthesis genes in heterologous cell types, MITF is considered a “master regulator” of melanocyte development [19–21]. MITF activity has also been described as a rheostat that regulates melanoma phenotype by driving senescence at low levels, an invasive phenotype at mid-levels, and melanocyte proliferation and differentiation at higher levels [22]. Continued exploration of the GRNs controlling melanocyte differentiation will add to the value of the melanocyte as a model cell type, and may also guide the design of differentiation-promoting therapies in melanoma.
Mutations in Transcription Factor Activating Enhancer-Binding Protein 2 Alpha (TFAP2A) result in pigmentation phenotypes similar to those caused by mutations in established members of the melanocyte differentiation GRN. In humans, a variety of missense mutations in TFAP2A cause branchio-oculo-facial syndrome, which frequently includes premature hair graying due to dysregulation of melanocyte stem cells [23]. Mice with Wnt1-Cre-mediated deletion of Tfap2a in neural crest usually die from exencephaly, but rare surviving animals exhibit a white belly spot analogous to the phenotype of mutants heterozygous for a null allele of Kit [24], which is thought to be a direct target of TFAP2A [25,26]. We also observe a greater-than-additive belly-spot phenotype in Tfap2a and Kit double heterozygous mice, signifying a genetic interaction between these genes (TW, unpublished observations). In zebrafish embryos homozygous for strong loss-of-function alleles of tfap2a, melanocytes are fewer in number and exhibit reduced migration relative to melanocytes in wildtype embryos [27–29]. This phenotype resembles zebrafish kita mutants, and there is also evidence of genetic interaction between tfap2a and kita in zebrafish [29]. However, zebrafish tfap2a mutants also have a phenotype of delayed melanization that is not present in zebrafish kita mutants [27–29], and we previously showed that tfap2a and its paralog tfap2e are cell-autonomously required for melanocyte differentiation in zebrafish [30]. These phenotypes imply that TFAP2A contributes to the GRN governing melanocyte migration, possibly upstream of KIT, as well as to a GRN governing melanocyte differentiation by mediating expression of unknown targets.
The precise contribution of TFAP2A to the melanocyte differentiation GRN has been obscured by pleiotropic functions of TFAP2A and its redundantly-acting paralogs during earlier steps in neural crest development. TFAP2A belongs to a family of five paralogs, TFAP2A-E, of which all but TFAP2D have an identical sequence binding preference [reviewed in 31]. In all species thus far analyzed, TFAP2A and one or more additional TFAP2 paralogs with potential for redundant activity are expressed in the neural plate border, premigratory neural crest, and melanocytes, but the identity of the additional paralogs varies among species. Zebrafish melanocytes express tfap2a, tfap2c, and tfap2e [32], and embryos depleted of both tfap2a and tfap2e display a greater-than-additive reduction in both melanocyte number and pigmentation compared to embryos depleted of either gene alone [30]. However, it has not yet been possible to examine the consequence of removing all three Tfap2 paralogs in melanocytes due to another example of redundancy, the lack of neural crest in zebrafish depleted of both Tfap2a and Tfap2c [33,34]. This is also true in mouse, where Tfap2a and Tfap2b are expressed in early neural crest [35] as well as the melanocyte lineage, resulting in almost complete loss of migrating trunk neural crest prior to specification of the melanocyte lineage in Tfap2a/Tfap2b double mutants [36]. Thus, the specific contributions of these factors to the GRN governing melanocyte differentiation have not been thoroughly evaluated.
In this study, we investigate the role of TFAP2A in melanocyte differentiation, utilizing the different advantages of zebrafish, mouse, and cell line models. While pigmentation is clearly reduced in zebrafish tfap2a mutants, mitfa expression levels appear to be normal in the remaining melanocytes [30]. Likewise, it was reported that in 501mel melanoma cells depleted of TFAP2A, the expression levels of MITF were unchanged compared to control cells, while expression of TYR, encoding the rate limiting enzyme of melanin synthesis, was decreased [37]. Pigmentation phenotypes in zebrafish tfap2a mutants are therefore unlikely to be an effect of altered Mitf expression levels. However, MITF activity is regulated by post-translational modifications [38–40], and the expression of enzymes mediating these modifications may depend on TFAP2A. Alternatively, TFAP2A and MITF could directly co-regulate expression of melanocyte differentiation effectors. In support of this model, there is evidence that both proteins regulate expression of CDKN1A/p21 [41,42] and IRF4 [37]. Furthermore, a recent integrative analysis of chromatin mark data in 111 cell types indicated that enhancers active in melanocytes are enriched in the TFAP2A binding site [43], although other, non-TFAP2 family transcription factors may bind similar sites. Co-occupancy of enhancers by MITF and TFAP2A was also reported in human melanoma cell lines [44]. However, melanomas often have reduced TFAP2A expression, accompanied by methylation of the TFAP2A promoter, and so are not ideal systems to study the role of TFAP2A in normal melanocyte development and function [26,45,46]. Here we examine the relationship between TFAP2A and MITF in the context of melanocytes using a combination of molecular, genetic, and bioinformatic analyses in human, mouse, and zebrafish systems. The results confirm that TFAP2A frequently co-occupies regulatory elements with MITF, identify genes underlying pigmentation phenotypes in model organisms and patients with TFAP2A mutations, and reveal TFAP2A as a candidate locus to modify diseases associated with MITF, including melanoma.
Results
TFAP2A is required for normal expression of melanocyte differentiation genes
Zebrafish homozygous for a strong loss-of-function mutation in tfap2a (i.e., lockjaw allele, hereafter tfap2a-/- mutants) lack detectable anti-TFAP2A immunoreactivity [28,47] and exhibit approximately one-third reduction of embryonic melanocytes, impaired melanocyte migration, and delayed melanization relative to wildtype and tfap2a+/- siblings (Fig 1A and 1B) [27,30,48]. In order to better characterize the melanization phenotype, we compared tfap2a-/- mutants to tfap2a+/- siblings over a ten-hour period, starting with the first emergence of melanocytes around 28 hours post fertilization (hpf) (S1 Fig). While melanocytes are initially pale in both genotypes, individual melanocytes in the tfap2a+/- siblings pigmented more quickly than individual melanocytes in tfap2a-/- mutants. This supports earlier evidence that, in addition to reduced numbers and migration, melanocytes in tfap2a-/- mutants have defects in differentiation [30].
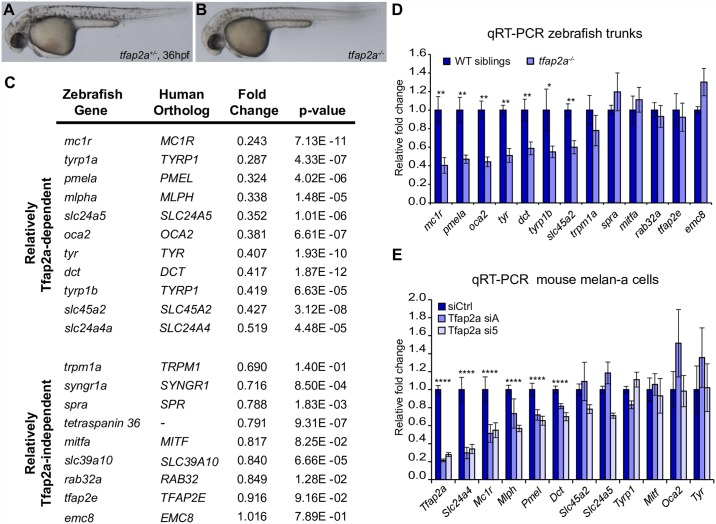
Fig 1. Melanocyte differentiation genes are sensitive to the loss of TFAP2A.
(A, B) Lateral images of tfap2a+/- and tfap2a-/- mutant zebrafish at 36 hpf. tfap2a-/- mutants have fewer melanocytes and delayed pigmentation relative to their wildtype or tfap2a+/- siblings. (C) Fold changes calculated from microarray expression levels for 20 zebrafish melanocyte genes. Top: 11 genes with expression levels between 0.2–0.55-fold in tfap2a-/- mutants compared to WT siblings. Bottom: 9 genes with expression levels reduced no further than the number of melanocytes in tfap2a-/- mutants. (D) qRT-PCR validation of zebrafish microarray results for 13 melanocyte genes (Mann-Whitney U, *p<0.05, **p<0.01). (E) qRT-PCR validation of microarray results for 12 melanocyte genes in mouse melan-a cells transfected with control siRNA or each of two siRNAs targeting Tfap2a (siA, si5). Significance evaluated only when both siA and si5 altered gene expression in the same direction relative to siCtrl (one-way ANOVA with Bonferroni correction, ****p<0.0001).
To extend previous analyses of gene expression in the melanocytes of tfap2a-/- mutants [30,34,48,49], we generated expression profiles of tfap2a-/- mutant zebrafish and their wildtype siblings at 36 hpf. Prior to harvesting RNA, we decapitated animals to eliminate the retinal pigmented epithelium, which appears to be normally pigmented in tfap2a-/- mutants (Fig 1A and 1B). Generating cDNA and probing microarrays revealed that 2,337 unique Ensembl transcripts (corresponding to 2,324 genes) are differentially expressed in the trunks of tfap2a-/- mutants versus siblings (FDR p<0.05). Of these, the expression levels of 124 transcripts in tfap2a-/- mutants are decreased to ≤0.7-fold of wildtype levels and 358 transcripts are increased to ≥1.25-fold (expression profile in S1 Table). We referred to zebrafish gene expression patterns at an online database (ZFIN) to identify 19 genes annotated as “melanoblast,” “melanocyte,” or “pigment cell” [50]. An additional gene, slc24a4a, is annotated as “neural crest,” but is expressed in a pattern resembling that of dct [51]. Most of these 20 genes encode proteins that have known roles in melanocyte differentiation (Fig 1C, S2 Table). In tfap2a-/- mutants, 11 of these genes were expressed between 0.2- to 0.55-fold of wildtype levels, a much greater fraction of melanocyte genes than expected by chance (hypergeometric test, p<0.0001), and were therefore considered Tfap2a-dependent (Fig 1C). The levels of several others, including trpm1a, were not significantly changed or were reduced by no more than expected from the one-third decrease in melanocyte cell number, and we considered these to be Tfap2a-independent. qRT-PCR validation of microarray results confirmed that expression of multiple pigmentation genes depends on Tfap2a in zebrafish (Fig 1D).
Because TFAP2A is expressed in multiple tissues, including the skin where melanocytes reside, the Tfap2a-dependent gene expression in zebrafish melanocytes could reflect an indirect requirement for Tfap2a rather than a cell-autonomous one. To clarify this issue, we conducted a microarray expression profile of immortalized mouse melanocytes (melan-a cells) [52] depleted of Tfap2a. We identified two independent siRNAs that reduced expression of Tfap2a to below 0.25-fold of the level in cells transfected with control siRNAs. Comparison of microarray profiles showed that the expression levels of 30 genes were consistently decreased (≤0.7-fold) and the expression levels of 38 genes were consistently increased (≥1.4-fold) in cells transfected with either Tfap2a-targeted siRNA relative to control siRNAs (S3 Table). Among the 30 genes with decreased expression, Mc1r and Slc24a4 are implicated in pigmentation. qRT-PCR verified significant reduction of these two genes, as well as Dct, Mlph, and Pmel, which narrowly missed the threshold cut-off in the array analysis (Fig 1E). Of note, several of the other genes with decreased expression upon knockdown of Tfap2a have been reported in the literature as activated (Aldh1a3, Igf2bp1, Ephb2, Pbk) or downregulated (Qpct, Wfdc1) in melanoma [53–59]. In summary, expression of a subset of melanocyte differentiation genes, including Slc24a4, Mc1r, Mlph, Pmel, and Dct, was TFAP2A-dependent in both mouse melanocytes and zebrafish trunks, while expression of Mitf orthologs was TFAP2A-independent. Certain pigmentation genes, including oca2 and slc45a2, appeared Tfap2a-dependent in zebrafish but not in mouse melanocytes, perhaps reflecting a more thorough depletion of Tfap2a in the former, or potentially distinct homeostatic mechanisms induced by siRNA versus gene mutation. Nonetheless, these results indicate that TFAP2A, whether directly or indirectly, regulates the expression of genes involved in melanocyte differentiation.
TFAP2A occupancy in mouse and human melanocytes
To determine the direct transcriptional targets of TFAP2A in melanocytes, we conducted chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) in mouse melan-a cells (two replicates) and in human primary melanocytes (one replicate). Anti-TFAP2A immunoreactivity is strong and concentrated in the nucleus of human primary melanocytes (S2A Fig) but appears weaker and more diffuse in several melanoma cell lines (S2B–S2E Fig), consistent with a reduction of TFAP2A RNA levels in melanoma [45]. ChIP-seq detected 16,305 TFAP2A-bound loci in mouse melanocytes, and 13,690 TFAP2A-bound loci in human melanocytes (hereafter, TFAP2A peaks). De novo motif analysis [60] of sequences precipitated by the anti-TFAP2A antibody from mouse or human melanocytes revealed that a known TFAP2A binding site (MA0003.2, JASPAR) is strongly enriched and tends to be centrally located within peaks (S3A and S3C Fig). Anti-TFAP2A ChIP followed by quantitative PCR (ChIP-qPCR) confirmed TFAP2A binding enrichment for selected peaks at genes of interest in mouse or human melanocytes, as well as in the M21 melanoma cell line, which has detectable TFAP2A expression (S3B, S3D and S3E Fig).
Published comparisons of ChIP-seq results for a given transcription factor in mouse and human melanocytes have suggested rapid divergence of binding events during evolution [61–63]. Consistent with these studies, we found that only about 11% of TFAP2A peaks in human primary melanocytes coincide with the orthologs of TFAP2A peaks lifted over from mouse (S4 Table), and conversely, about 9% of the TFAP2A peaks identified in mouse melanocytes coincide with peaks lifted over from human (S5 Table). TFAP2A peaks shared between species are enriched near promoters (S6 Table). In contrast to the modest concordance of peaks and orthologous sequences, the concordance of genes associated with TFAP2A peaks in the two species is very high, as shown below.
Relationship between TFAP2A occupancy and defined regulatory elements
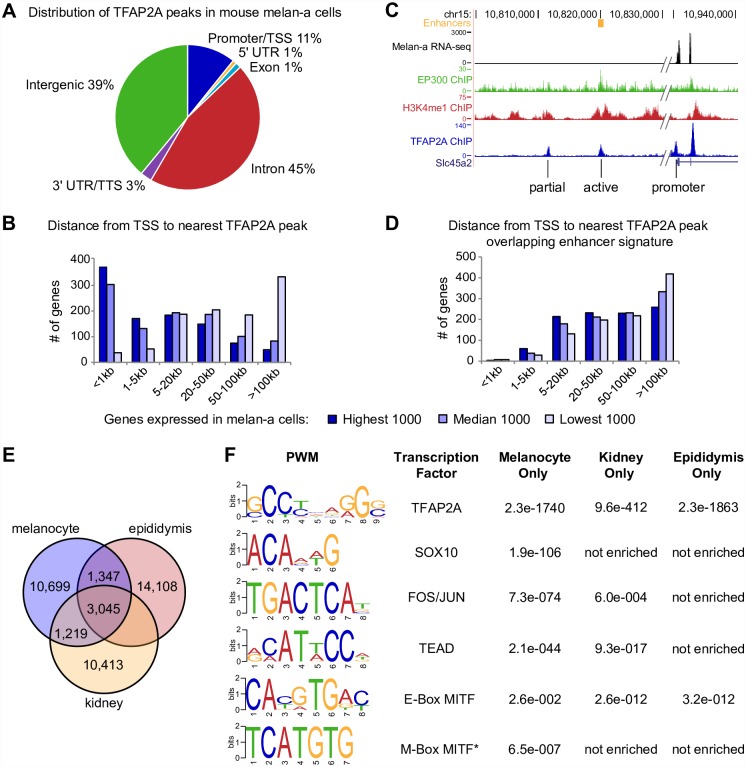
In both mouse and human melanocytes, TFAP2A peaks are more likely to be found within genes, including introns, than in intergenic regions (Fig 2A, S4A Fig). Overall, genes with higher expression in melanocytes are enriched for promoter-proximal peaks of TFAP2A (mouse GSE87051, human GSM958174 [64]) (Fig 2B, S4B Fig). To assess patterns of TFAP2A binding at enhancers, we compared the TFAP2A ChIP-seq data from mouse melan-a cells to a published profile of candidate enhancers also in these cells, which were defined by H3K4me1 peaks flanking a p300 peak [65]. Remarkably, 70% (1,752 of 2,489) of enhancer elements marked in this way overlap with a TFAP2A peak (hypergeometric test, p<0.0001). Conversely, about 10% of TFAP2A peaks are fully marked as enhancers, while another 35% are partially marked (i.e., either overlapping p300 or flanked by at least one H3K4me1 peak). Fig 2C illustrates this pattern upstream of the melanocyte differentiation gene Slc45a2, as well as a TFAP2A peak at the promoter (additional examples in S5A–S5F Fig). In agreement with estimates that the median distance between promoters and cis-acting enhancers is 15kb, genes with high expression are enriched for a TFAP2A peak overlapping the active enhancer signature at a distance of 5–50kb from the TSS (Fig 2D) [66]. However, the promoter-proximal TFAP2A peaks are just as likely to possess a TFAP2A binding site as the promoter-distal peaks (about 50% in both cases). This is in contrast to observations from mouse chondrocytes, where promoter-distal SOX9 peaks are more likely to contain a SOX9 binding site than promoter-proximal ones [67]. Altogether, the binding profile of TFAP2A indicates that it acts at both enhancers and promoters of melanocyte genes.
Fig 2. TFAP2A binds active enhancers and promoters in mouse melanocytes.
(A) Pie chart showing distribution of mouse TFAP2A peaks with respect to genomic features. TSS, transcription start site; TTS, transcription termination site. (B) Distance from TSS to the nearest TFAP2A peak for genes in three expression categories: highest 1000, median 1000, or lowest 1000. Promoter-proximal TFAP2A peaks are enriched at highly expressed genes (RNA-seq on mouse melan-a cells at GSE87051). (C) Examples of active enhancer signatures defined by H3K4me1 peaks flanking a p300 peak [65], and partial enhancer signatures, overlapping TFAP2A peaks upstream of the melanocyte differentiation gene Slc45a2. (D) Distance from TSS to the nearest TFAP2A peaks that overlap the active enhancer signature for genes in three expression categories: highest 1000, median 1000, or lowest 1000. (E) Overlap of TFAP2A ChIP-seq peaks in mouse melanocytes with published TFAP2A ChIP-seq peaks in mouse kidney and epididymis cells [68]. 34% of melanocyte peaks were shared with one or both of the other cell types. (F) MEME-ChIP analysis of unique peaks from each cell type. Melanocyte-unique peaks are significantly enriched for SOX10 and M-Box MITF binding motifs, while kidney-unique and epididymis-unique peaks are not. *All motifs shown are a result of de novo MEME-ChIP enrichment analysis except the M-Box, which we specifically searched using the Analysis of Motif Enrichment (AME) tool [109].
TFAP2A-bound chromatin elements are enriched for distinct binding motifs in different cell types
Because TFAP2A is widely expressed, the extent to which TFAP2A peaks would be melanocyte-specific was unclear. We found that about 15% of TFAP2A peaks in mouse melanocytes overlap those reported in either primary mouse kidney or primary mouse epididymis cells (but not both), and 19% are shared by all three cell types [68] (Fig 2E). MEME-ChIP analysis [69] showed that melanocyte-unique TFAP2A peaks are enriched for the binding motifs of transcription factors active in melanocytes, including SOX10, FOS/JUN, TEAD, and the M-box binding site for MITF [70] (Fig 2F). These transcription factors are also highly enriched in candidate melanocyte enhancers [65]. Kidney-unique and epididymis-unique peaks showed less significant or no enrichment for these binding motifs, although the E-box, recognized by MITF and many other bHLHZip transcription factors, was enriched in all three cell types. The enrichment of binding sites for melanocyte transcription factors like SOX10 and MITF in melanocyte-unique peaks suggests that TFAP2A binds cell-type specific loci in addition to generic ones; binding to any particular locus is presumably a function of cofactor availability and chromatin accessibility [reviewed in 71].
Many TFAP2A-dependent melanocyte genes are direct targets of TFAP2A
We used Genomic Regions Enrichment of Annotations Tool (GREAT) [72] to identify genes associated with TFAP2A peaks in mouse and human melanocytes, with an assignment rule of basal promoter plus 100kb distal, and found that these genes are enriched for ontology terms relevant to melanocyte differentiation, including “pigmentation”, “melanosome”, and “melanoma” (S6A and S6B Fig). However, TFAP2A can both activate and repress gene expression [73]. To find genes likely to be activated by TFAP2A, we integrated our TFAP2A ChIP-seq with H3K27ac ChIP-seq data from human melanocytes (GSM1127072 [64]) and mouse melanocytes [14], as H3K27ac marks active regulatory elements [reviewed in 74,75]. We found that 55% of TFAP2A peaks in human melanocytes and 58% of TFAP2A peaks in mouse melanocytes either overlap H3K27ac peaks or are flanked by H3K27ac peaks, and we refer to them hereafter as active TFAP2A peaks (human Fig 3A, mouse S6C Fig, S7 Table). Analysis with GREAT revealed that genes associated with active TFAP2A peaks include 15 of the 30 genes with significantly decreased expression in Tfap2a-depleted mouse melanocytes, among them Mc1r and Slc24a4 (S8 Table). Similarly, orthologs of eight out of ten Tfap2a-dependent melanocyte genes in zebrafish tfap2a-/- mutants (excepting slc24a5 and tyr) are associated with active TFAP2A peaks in mouse or human melanocytes. These genes are candidates to be direct transcriptional targets of TFAP2A, suggesting that the phenotype of delayed melanization in zebrafish tfap2a-/- mutants can be explained in part by a direct effect on certain melanocyte differentiation effector genes (e.g., dct, mlpha, mc1r, and pmela), and indirect regulation of others (e.g., slc24a5 and tyr).
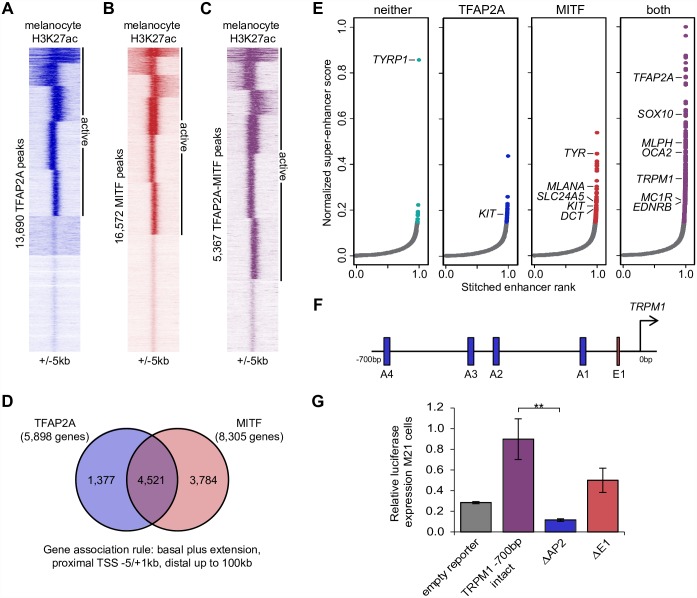
Fig 3. TFAP2A peaks are associated with genes involved in pigmentation.
(A-C) Density-based clustering of H3K27ac signal at (A) TFAP2A peaks, (B) MITF peaks, and (C) TFAP2A peaks that overlap MITF peaks in human melanocytes (H3K27ac data from GSM1127072 [64]), MITF peaks from [18]). (D) Overlap between genes associated with active TFAP2A peaks and genes associated with active MITF peaks in human melanocytes. (E) Typical enhancers (gray) and super-enhancers (colored) in human melanocytes that overlap neither TFAP2A nor MITF peaks, TFAP2A peaks only, MITF peaks only, or both TFAP2A and MITF peaks. Labels identify melanocyte genes of interest. (F) Diagram of the TRPM1 -700 bp promoter element depicting the positions of four TFAP2A binding sites (A1–A4) and the previously reported E-box 1 MITF binding site (E1). (G) Luciferase assays in M21 melanoma cells. Deletion of all four TFAP2A binding sites (ΔAP2) significantly reduced reporter activity compared to the intact TRPM1 -700bp element (Student’s t-test, **p = 0.01).
TFAP2A and MITF co-occupy regulatory elements associated with pigmentation genes
The presence of TFAP2A peaks at a majority of active melanocyte enhancers, as well as the enrichment of the MITF binding motif in TFAP2A peaks, implies that TFAP2A binds many of the same regulatory elements as MITF in melanocytes. To evaluate overlap between TFAP2A and MITF across the genome, we compared our human TFAP2A ChIP-seq results to a set of 16,572 MITF ChIP-seq peaks also from human primary melanocytes [18]. 5,367 (39%) of TFAP2A peaks are shared with MITF, overlapping by at least one base pair. Integrating MITF and H3K27ac ChIP-seq data yielded 61% of MITF peaks (Fig 3B) and 76% of TFAP2A/MITF shared peaks (Fig 3C) that overlap, or lie between, H3K27ac peaks and are thus considered to be active. Using GREAT, we found that about 77% of the genes associated with active TFAP2A peaks are also associated with active MITF peaks, a highly significant overlap (hypergeometric test, p<0.0001) (Fig 3D). Furthermore, 79% of these genes are associated with active TFAP2A/MITF shared peaks, suggesting that TFAP2A and MITF are co-bound at many, but not all, shared targets. GO term analysis [76,77] revealed that the subset of genes associated with both TFAP2A and MITF peaks are enriched for the terms “melanosome” and “pigment granule” (p = 9.08E-07), as well as “DNA repair” (p = 5.98E-08), “mitotic cell cycle process” (p = 5.59E-09), “regulation of cell proliferation” (p = 2.40E-03), and “regulation of cell differentiation” (p = 2.60E-03) (all p-values Bonferroni corrected, S9 Table). This supports a regulatory role for TFAP2A not only in differentiation, but across other categories of genes proposed to be regulated by MITF in melanocytes and melanoma, as with the MITF rheostat [22].
To assess the overlap between targets of TFAP2A and MITF with respect to pigmentation, we focused on a list of 170 genes that cause coat color phenotypes in mice [78], adding TRPM1 based on its role in the coat color phenotype of appaloosa horses [79–82]. Orthologs of 97 genes on this list are associated with active TFAP2A peaks in human melanocytes and/or active TFAP2A peaks in mouse melanocytes (Table 1, asterisks). Of these, 72 genes are also associated with active MITF peaks (Table 1), 46 being active shared TFAP2A/MITF peaks (Table 1, bold). We then examined overlap of MITF and TFAP2A binding at clusters of closely spaced enhancers, sometimes called stretch or super-enhancers (SEs) [83], that are linked to cell type-specific gene expression [84,85]. Following published methods, we used H3K27ac data to identify 652 SEs in human primary melanocytes [85] (S7 Fig). Of these, 530 (81%) are bound by both MITF and TFAP2A (Fig 3E, S10 Table). Interestingly, genes involved in melanocyte differentiation, including those encoding proteins expressed in the melanosome, are associated with SEs bound by MITF only (e.g. TYR, MLANA, SLC24A5, DCT) and SEs bound by both MITF and TFAP2A (MLPH, OCA2, TRPM1, MC1R). Exceptions to this pattern include KIT, which is associated with one SE bound solely by TFAP2A and one SE bound solely by MITF, and TYRP1, which is associated with an SE bound by neither (Fig 3E). Taken together, these results show that TFAP2A and MITF bind regulatory elements associated with melanocyte differentiation effectors. Notably, 409 (63%) of all SEs are bound by TFAP2A peaks that overlap with MITF peaks. It remains to be determined whether TFAP2A and MITF exhibit cooperative binding at these loci.
Table 1. Mouse coat color genes associated with peaks of TFAP2A and MITF that overlap active enhancer marks (H3K27ac).
| TFAP2A | Both | MITF | ||||
|---|---|---|---|---|---|---|
| Melanosome | MYO7A* | SHROOM2* | AP3B1 | HPS6 | PMEL | AP3D1 |
| DCT* | LYST | RAB38 | BLOC1S3 | |||
| DTNBP1* | MLPH | RABGGTA* | HPS1 | |||
| GPNMB | MREG | SLC45A2* | RAB27A | |||
| HPS3* | MYO5A* | TYRP1* | SLC24A5 | |||
| HPS4 | OCA2 | VPS33A | TRAPPC6A | |||
| HPS5* | TYR | |||||
| Eu-/pheo- melanin | EDARADD | POMC* | ATRN | MGRN1 | SMARCA5 | GGT1 |
| MC1R | OSTM1 | SMCHD1 | ||||
| MCHR1 | ||||||
| Development | ADAMTS20* | KITLG* | ADAM17 | GPR161 | PHACTR4 | EGFR |
| ARCN1 | MITF* | APC | ITGB1 | PYGO1* | FOXD3 | |
| DOCK7 | NF1 | BMPR1A | JMJD6 | RB1* | FZD4 | |
| EED | NOTCH2* | BRCA1* | KIT | SEMA3C | ||
| FOXN1* | RECQL4 | CITED1 | LEF1* | SEMA4A | ||
| GNA11 | S1PR2* | DPH1 | MAP2K1 | SNAI2 | ||
| GRLF1* | SUFU | ECE1 | MCOLN3* | SOX10 | ||
| HELLS* | UNC119* | EDNRB | MYC | TFAP2A | ||
| GAS1 | NOTCH1 | TIMP3 | ||||
| GLI3 | PAX3 | TRAF6 | ||||
| GNAQ | PDGFB | ZBTB17* | ||||
| GNPAT | ||||||
| Systemic effects | ATOX1 | FAS | BCL2* | PDPK1 | RXRA | ATP7A |
| ATP7B | RBP1* | CASP3 | POLG | TRPM1 | RPS20 | |
| DST* | HS2ST1 | POLH* | TRPM7 | SLC31A1 | ||
| OAT | RPL24 | VLDLR | ||||
* = associated with a TFAP2A peak in mouse but not human melanocytes
bold = TFAP2A and MITF peaks associated with these genes overlap
While several of the pigmentation genes associated with active TFAP2A and MITF peaks showed TFAP2A-dependent expression in both zebrafish and mouse, we also noted many apparently TFAP2A-independent genes on this list. One possible explanation is that the presence of a TFAP2A peak does not signify contribution of TFAP2A to the activity of a given regulatory element. Alternatively, activity of a redundantly-expressed TFAP2 paralog may compensate for the loss of TFAP2A. To rule out the first possibility, we focused on the gene TRPM1, which has a promoter-proximal TFAP2A peak in both mouse and human melanocytes but is expressed at high levels in melanocytes in a TFAP2A-independent manner. In addition, expression of TRPM1 is a sensitive readout of MITF activity levels [79], and a minimal TRPM1 promoter, which has an MITF peak in melanoma cells [17] and primary melanocytes [18], has activity in melanoma cells that is lost upon deletion of the MITF binding sites [86]. We engineered the TRPM1 promoter into a vector suitable for quantitative luciferase reporter assays and created variants with mutations in either the E-Box MITF binding site shown previously to be most important for promoter activity (ΔE1) [86], or four predicted TFAP2 binding sites (ΔAP2) (Fig 3F). These constructs, together with a control vector, were transfected into M21 melanoma cells, which express TFAP2A (S2C Fig). The intact TRPM1 promoter drove much higher luciferase expression than the empty reporter vector, while the ΔE1 promoter variant drove about 50% of intact, and the ΔAP2 variant drove about 10% of intact (p = 0.01) (Fig 3G). These results show that TFAP2A directly activates the TRPM1 promoter, supporting the hypothesis that other TFAP2 paralogs are able to compensate for the absence of TFAP2A at certain melanocyte genes.
Mouse Tfap2a / Tfap2b double conditional mutants are depleted of melanocytes
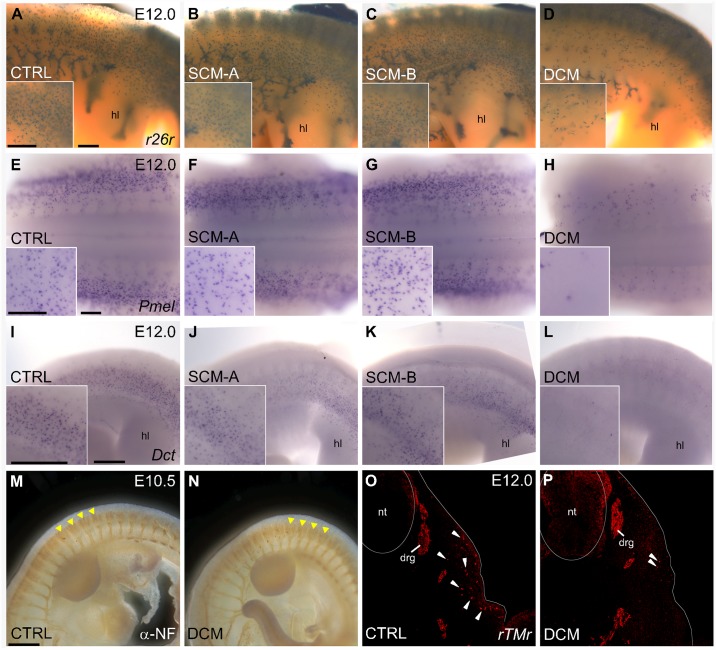
Testing whether TFAP2 paralogs function redundantly in melanocyte development requires simultaneous depletion of all such paralogs expressed in melanocytes. In mouse melanocytes, Tfap2a and Tfap2b have the highest and second highest expression, respectively, while Tfap2c and Tfap2e are undetectable [87,88]. To determine whether Tfap2 paralogs function redundantly in murine melanocyte development, we generated double conditional mutants (DCM) using a previously published Wnt1-Cre transgenic line [89] and conditional alleles of Tfap2a [24] and Tfap2b (EVO and TW, in preparation). We then utilized two approaches to assess melanocyte development in DCM embryos, corresponding single conditional mutant (SCM) embryos, and control embryos. First, embryos in which the Rosa26-reporter (r26r)-allele [90] was also incorporated were dissected at embryonic day 12.0 (E12.0) and subsequently stained for β-galactosidase (β-gal) activity. The r26r-allele used in combination with the Wnt1-Cre transgene results in β-gal positive staining of premigratory neural crest cells and subsequent derivatives (Fig 4A–4D, S8A–S8D Fig). β-gal-positive melanoblasts and corresponding melanocytes migrate ventrolaterally from the dorsal neural tube and can be identified by their position just below the developing surface ectoderm, most easily observed dorsal to the hindlimb. Examination of control (Fig 4A), Tfap2a SCM (Fig 4B), and Tfap2b SCM (Fig 4C) embryos revealed roughly equivalent numbers of stained cells with a similar distribution. In contrast, Tfap2a/Tfap2b DCM embryos have many fewer β-gal-positive cells in this location (Fig 4D). Second, embryos were processed for in situ hybridization with Pmel [91] and Dct [92,93] riboprobes, detecting melanoblasts and differentiated melanocytes (Fig 4E–4L). As with the r26r experiments, this staining labeled similar numbers of Pmel-positive and Dct-positive cells in control (Fig 4E and 4I), Tfap2a SCM (Fig 4F and 4J), and Tfap2b SCM (Fig 4G and 4K) embryos, but far fewer cells in Tfap2a/Tfap2b DCM embryos (Fig 4H and 4L). The absence of Pmel-positive and Dct-positive melanoblasts in DCMs was evident from the time these cells emerged in control embryos at E10.5 and E11.5, suggesting that the reduced melanoblast number in DCMs is not the result of impaired melanoblast migration (S8E–S8L Fig). Because TFAP2 paralogs have been shown to function during the early stages of neural crest induction [33,34], we next tested whether the observed reduction in melanocytes could be explained by a disruption in this step. Both lineage tracing with the r26r-reporter line (S9A–S9F Fig) and in situ hybridization with a Sox10 riboprobe at E9.5 (S9G and S9H Fig) revealed relatively normal neural crest induction in DCMs, as in controls. Consistent with this observation, α-neurofilament immunostaining (Fig 4M and 4N) and lineage tracing (Fig 4O and 4P, S10A–S10D Fig) identified the initial formation of an alternate trunk neural crest derivative, dorsal root ganglia (DRG), in both DCMs and controls. However, similar to the melanocyte lineage, the neural crest-derived enteric nervous system (ENS) was disrupted in Tfap2a SCM embryos and completely failed to populate the gastrointestinal tract of DCM embryos (S10E–S10L Fig). Broadly, these analyses suggest that in mouse embryos, paralogous proteins TFAP2A and TFAP2B act redundantly subsequent to neural crest induction within distinct neural crest lineages. The virtual absence of melanoblasts in Tfap2a/Tfap2b DCM embryos implies that they contribute to specification and differentiation of the melanocyte lineage, similar to the functions of MITF.
Fig 4. TFAP2A and TFAP2B redundantly regulate murine melanocyte development.
(A-D) Lateral views of E12.0 control (A), Tfap2a SCM (B), Tfap2b SCM (C), or Tfap2a/Tfap2b DCM (D) mouse embryos processed for β-galactosidase (β-gal) staining. β-gal+ cells are a result of recombination of the r26r-allele by the Wnt1-Cre transgene, labeling neural crest cells and derivatives. (E-L) In situ hybridization expression patterns for Pmel (E-H, dorsal views) or Dct (I-L, lateral views) in control (E, I), Tfap2a SCM (F, J), Tfap2b SCM (G, K), or Tfap2a/Tfap2b DCM (H, L) E12.0 mouse embryos. Insets in (A-L) show higher magnification images just above the hindlimb in a similar viewing plane as the low magnification image. (M, N) Lateral views of the trunk processed for ɑ-neurofilament immunoreactivity in an E10.5 control (M) or Tfap2a/Tfap2b DCM (N), revealing developing dorsal root ganglia (a subset labeled with arrowheads). (O, P) Immunofluorescence of cryosections (cross-sectional plane at the level of the hindlimb) of an E12.0 control (O) or Tfap2a/Tfap2b DCM (P), containing a Tomato-reporter (rTMr) allele labeling neural crest cells and derivatives (arrowheads highlight a subset of melanocytes in the ventrolateral pathway). Abbreviations: DCM, double conditional mutant; drg, dorsal root ganglia; hl, hindlimb; nt, neural tube; SCM, single conditional mutant (A = Tfap2a or B = Tfap2b). Scale bars = 250μM, except M, N = 500μM.
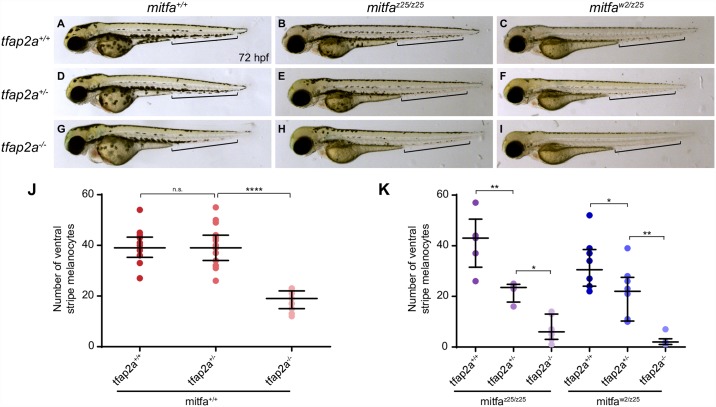
Zebrafish mitfa;tfap2a double mutants display a synergistic genetic interaction
Given the widespread co-occupancy of regulatory elements by TFAP2A and MITF, we predicted that mitfa;tfap2a double mutant zebrafish would have a greater-than-additive melanocyte phenotype consistent with synergistic interaction between these genes. To test this, we used the hypomorphic allele mitfaz25 to reduce Mitfa levels without eliminating the melanocyte lineage, as occurs with total loss-of-function alleles such as mitfaw2 [94,95]. Compared to wildtype embryos at 72 hpf (Fig 5A), mitfaz25/z25 homozygotes have fewer and less dendritic melanocytes (Fig 5B). Melanocytes are further reduced, punctate, and noticeably paler in mitfaw2/z25 trans-heterozygous embryos (Fig 5C). Incrosses of mitfaw2/z25;tfap2a+/- double heterozygous adults and mitfa+/+;tfap2a+/- single heterozygous adults yielded various combinations of mitfa;tfap2a genotypes. Embryos were sorted into phenotypic bins at 72 hpf, photographed individually, and genotyped for both mitfaz25 (when appropriate) and tfap2a. Relative to wildtype embryos (Fig 5A), mitfa+/+;tfap2a-/- homozygous mutants exhibited a clear reduction of melanocytes in the ventral stripe caudal to the tail, reflecting decreased melanocyte migration and/or proliferation (Fig 5G). However, mitfa+/+;tfap2a+/- heterozygous mutants could not be distinguished from wildtypes (compare Fig 5A and 5D). Quantification of ventral tail melanocytes (count area indicated by brackets in Fig 5A–5I) revealed that there is some variation within each genotype, but we detected no genotype/phenotype correlation between tfap2a+/+ and tfap2a+/- embryos (Fig 5J, one-way ANOVA). In contrast, both the mitfaz25/z25 (Fig 5B, 5E and 5H) and mitfaw2/z25 (Fig 5C, 5F and 5I) backgrounds had three distinguishable phenotypic groups corresponding to the tfap2a genotype, with tfap2a+/- embryos showing significant differences in the number of ventral tail melanocytes compared to both tfap2a+/+ and tfap2a-/- embryos (Fig 5K, one-way ANOVA). Thus, the phenotype of double mutants appears to be more severe than the combination of the phenotypes in single mutants. The biochemical underpinning of this genetic interaction is unknown; Mitfa and Tfap2a may interact synergistically at one or more regulatory elements, or Mitfa may regulate tfap2a expression in melanocytes.
Fig 5. tfap2a and mitfa display a genetic interaction in zebrafish.
(A-I) Compared to wildtype embryos at 72 hpf (A), melanocytes in mitfaz25/z25 mutant embryos (B) are dark but punctate, and melanocytes in mitfaw2/z25 trans-heterozygous embryos (C) are pale, punctate, and reduced in number. While heterozygous mutation of tfap2a in the wildtype background (tfap2a+/-) (D) does not result in a melanocyte phenotype, mitfaz25/z25;tfap2a+/- mutants (E) and mitfaw2/z25;tfap2a+/- mutants (F) display a loss of melanocytes in the ventral stripe (brackets). Similarly, the well-characterized phenotype of fewer, paler melanocytes in tfap2a-/- null mutants (G) is more pronounced in both the mitfaz25/z25 mutant background (H) and the mitfaw2/z25 mutant background (I). (J, K) Counts of ventral stripe melanocytes (see brackets in A-I) showed no difference between wildtype and tfap2a+/- embryos (J, n = 10+), while tfap2a+/- embryos in both mitfa mutant backgrounds had significantly fewer melanocytes in the ventral stripe (K, n = 4–8). One-way ANOVA: *p<0.05, **p<0.01, ****p<0.0001.
Forced expression of tfap2a does not rescue melanocytes in mitfa mutant zebrafish
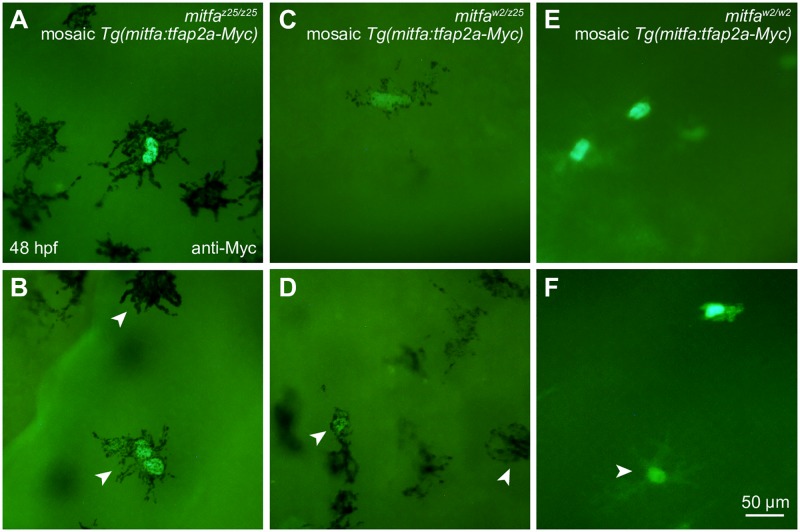
Genetic interaction between tfap2a and mitfa supports the idea that the factors encoded by these genes regulate shared targets in melanocytes, possibly within single or converging pathways. Previously, we found that elevating expression of mitfa (under the sox10 promoter) partially compensates for the reduction of Tfap2 in tfap2a/tfap2e-depleted zebrafish embryos [30]. Thus, we next investigated whether the reciprocal experiment, artificially elevating expression of tfap2a, could rescue melanocyte development in the absence or reduction of Mitfa. To test this, we engineered the tfap2a cDNA to encode 6 Myc epitopes at the carboxy terminus. To confirm that the epitope-tagged Tfap2a was still functional, we injected tfap2a-Myc mRNA into embryos depleted of tfap2a and tfap2c and observed differentiated melanocytes, which are otherwise completely absent from such animals [33]. We then fused tfap2a-Myc cDNA downstream of the mitfa promoter, which is active in melanocytes (mitfa promoter described in [96]), and injected the mitfa:tfap2a-Myc plasmid into embryos derived from an incross of mitfaw2/z25 adults. At 48 hpf, embryos were sorted into three groups based on the mitfa mutant phenotype, fixed, and subsequently processed for anti-Myc immunoreactivity. Ten plasmid-injected embryos and ten uninjected control embryos were documented in each group. We detected brightly-labeled cells, including several melanocytes, in both mitfaz25/z25 (Fig 6A and 6B) and mitfaw2/z25 (Fig 6C and 6D) plasmid-injected embryos, whereas uninjected controls had no labeled cells. However, the anti-Myc immunoreactive melanocytes were phenotypically indistinguishable from neighboring unlabeled melanocytes by morphology, dendricity, or pigmentation (Fig 6B and 6D, arrows). The mitfaw2/w2 plasmid-injected embryos had similar numbers of anti-Myc immunoreactive cells to the other genotypes, but none showed any hint of pigmentation (Fig 6E and 6F). We did observe a highly dendritic but wholly unmelanized cell, which is likely to be a xanthophore (Fig 6F, arrow). In summary, elevating tfap2a expression under the mitfa promoter was not sufficient to rescue melanocytes in mitfaw2/w2 null mutants, nor did it improve the quality or pigmentation of melanocytes in mitfaz25/z25 and mitfaw2/z25 mutants. We conclude that, despite regulating many of the same targets, Tfap2a is unable to replace Mitfa in the melanocyte lineage, at least at the dose of over-expression tested here.
Fig 6. Forced expression of tfap2a does not rescue melanocytes in mitfa mutant zebrafish.
(A-D) Nuclei of melanocytes expressing mosaic Tg(mitfa:tfap2a-Myc) are brightly labeled after anti-Myc immunostaining. At 48 hpf, labeled melanocytes in mitfaz25/z25 mutant embryos (A, B) and mitfaw2/z25 mutant embryos (C, D) display no apparent improvement in pigmentation or dendricity compared to adjacent unlabeled cells (B and D, white arrowheads). (E, F) In mitfaw2/w2 mutants, all labeled cells are unpigmented and resemble other cell types including xanthophores (F, white arrowhead).
Discussion
The current work presents an explanation for the basis of pigmentation phenotypes in zebrafish and mouse Tfap2a mutants, suggests that redundant activity of TFAP2 paralogs is responsible for the relative mildness of these phenotypes, and reveals a previously unappreciated role for TFAP2 alongside MITF in melanocyte differentiation. Microarray analysis on the trunks of zebrafish tfap2a-/- mutants showed that several genes with Tfap2a-dependent expression in vivo are known to be important for melanization of embryonic melanophores (oca2 [97], tyr [98], and slc45a2/albino [99]). The phenotype of delayed melanization in these mutants is plausibly explained by reduction of these genes, as well as others that may have escaped detection due to Tfap2a-dependent expression in melanocytes but not in other cell types (one example of this is kita [27,29]). Analysis of mouse melanocytes depleted of Tfap2a revealed an overlapping but shorter list of TFAP2A-dependent genes, potentially due to incomplete Tfap2a knockdown in these cells. Species dependent differences are also possible. For example, expression of Dct, but not of Irf4 or Tyr, was TFAP2A-dependent in mouse melanocytes, while the opposite trend was seen in the human 501mel cell line [37]. TFAP2A ChIP-seq results from mouse and human melanocytes integrated with H3K27ac ChIP-seq, marking active regulatory elements, revealed that the majority of TFAP2A-dependent pigmentation genes were direct transcriptional targets of TFAP2A. Indeed, far more genes, including most of those mutated in mice with coat color phenotypes, were associated with TFAP2A-bound active regulatory elements than were found to have TFAP2A-dependent expression (at the level of detection of our assays). Three results indicate that compensation by other TFAP2 paralogs is the most likely explanation for why more genes do not appear to be TFAP2A-dependent. First, deletion of TFAP2 binding sites reduced the promoter activity of a gene that was relatively unaffected by loss of TFAP2A in the expression analyses. Second, depletion of tfap2e in zebrafish delays melanocyte differentiation, but only in the context of tfap2a-/- mutants [30], suggesting that these paralogs are at least partially redundant in function. Third, we find here that in the mouse embryo, neural crest-specific deletion of either Tfap2a or Tfap2b alone does not greatly impact embryonic development of the melanocyte lineages, but the combined knockout of both genes causes a significant loss of melanocytes. Thus, TFAP2 paralogs promote induction of the neural crest lineage and subsequently promote differentiation of one of its derivatives; they have a similar feed-forward quality in epidermis [100]. It is important to note the possibility that TFAP2 paralogs act throughout the specification, proliferation, and differentiation of both neural crest and melanocyte lineages such that disruption at any of these steps ultimately results in a melanocyte phenotype. For example, almost complete loss of melanoblasts in Tfap2a/Tfap2b DCM mice could reflect a requirement for TFAP2 in neural crest survival or lineage specification of certain derivatives. Similarly, the genetic interaction between mitfa and tfap2a in zebrafish appears to be primarily due to reduced cell number, as well as defects in melanoblast migration. While further study will be required to uncouple these various functions of TFAP2 paralogs at each step of melanocyte development, the results described here strongly support a role for TFAP2A in the terminal differentiation of melanocytes. Overall, our findings suggest that TFAP2A, acting in partial redundancy with other TFAP2 paralogs, joins MITF, SOX10, YY1, LEF1, and IRF4 in directly regulating the expression of melanocyte differentiation effector genes [20,37,101].
We observed widespread co-occupancy of TFAP2A and MITF at active regulatory elements, but it is unknown whether the two transcription factors bind such elements cooperatively. In support of this possibility, we observed a genetic interaction between mitfa and tfap2a affecting melanocyte development in zebrafish. However, an interaction would also be expected if Tfap2a and Mitfa act in a single pathway. Although mitfa expression in melanocytes is not strongly Tfap2a-dependent, it is possible that expression of tfap2a in melanocytes is Mitfa-dependent, as levels of TFAP2A protein were reduced in 501mel cells depleted of MITF with an shRNA [37]. In a published mass-spectrometric analysis of proteins that immunoprecipitate with MITF, TFAP2A peptides were not identified [44]. However, another similar experiment did identify low levels of TFAP2A peptides, although TFAP2A did not detectably co-immunoprecipitate with an epitope-tagged MITF (J. P. Lambert, A. C. Gingras, personal communication). The strength and importance of any physical interaction among TFAP2A, MITF, SOX10, and other transcription factors bound at regulatory elements active in melanocytes requires further investigation. There is evidence that TFAP2A serves as a pioneer transcription factor for androgen receptor in epididymis cells [68], and as TFAP2A expression precedes MITF expression in the melanocyte lineage, it is conceivable that TFAP2A plays a similar role for MITF. Conversely, the inability of Tfap2a to substitute for loss of Mitfa in zebrafish, at least at the doses tested here, is consistent with MITF serving as a pioneer factor for TFAP2A. It is also possible that TFAP2A and MITF bind independently, but nonetheless have a cooperative effect on gene expression, as indicated by in vitro tests of an intronic enhancer of the IRF4 gene [37]. Analysis of TFAP2A chromatin binding in cells depleted of MITF, and of MITF chromatin binding in cells depleted of relevant TFAP2 paralogs, might address whether either protein is required to make the chromatin accessible for the other.
In metastatic melanoma, the levels of MITF activity have been proposed to control the cellular phenotype: high levels promote melanocyte proliferation and differentiation, while lower levels confer an invasive state [22,102]. It is notable that TFAP2A levels are decreased in advanced-stage melanoma tumors versus earlier stage melanoma and nevi, whereas MITF expression levels are relatively constant (data from The Cancer Genome Atlas [46]). Furthermore, elevating levels of TFAP2A in A375SM cells was reported to inhibit tumorigenicity and metastatic potential in nude mice [103]. Based on this evidence, it is possible that the level of MITF activity is adjusted through loss or gain of an essential collaborator, TFAP2A (and perhaps its paralogs). Here, we find that TFAP2A peaks are associated with many genes encoding regulators not only of melanocyte differentiation, but also of other cellular phenotypes purported to be governed by MITF, such as growth and senescence. The melanoma subtype that is most difficult to target therapeutically, and which is thought to depend on a relatively low level of MITF activity, has stem cell qualities, an invasive phenotype, and an expression profile resembling that of neural crest [104]. Thus, early requirements for Tfap2a and its paralog Tfap2c observed in zebrafish neural crest induction [33,34] may suggest a role for TFAP2 in this invasive subtype of melanoma as well. Further investigation will be necessary to determine the potential tumor-promoting or tumor-inhibiting consequences of TFAP2A expression (or activity) levels in melanoma.
Materials and methods
Cell culture
Mouse melan-a cells
Mouse immortalized melanocytes, Ink4a-ARF double null melan-a cells, were maintained as described previously [105].
Human primary melanocytes
Primary melanocytes were regularly maintained in culture as described previously [106], using complete growth medium consisting of Minimum Essential Medium Eagle (MCDB) 153 supplemented with 4% fetal bovine serum, insulin (5 μg/ml), α-tocopherol (1 μg/ml), 1% penicillin/streptomycin/amphotericin, human basic fibroblast growth factor (bFGF, 0.6 ng/ml), phorbol 12-myristate 13-acetate (PMA, 8 nM), and bovine pituitary extract (BPE, 13 μg/ml). Primary melanocytes were grown in T-75 cm2 flasks. To split flasks, cells were washed for 2 min in EDTA solution and trypsinized for 2 min in 500 μl of a 0.25% trypsin solution.
Human melanoma cells
The M21 melanoma cell line was obtained from the American Type Culture Collection (ATCC, Manassas, VA), and maintained in RPMI containing 10% FBS and 1% PenStrep.
Microarrays and qRT-PCR
Zebrafish
tfap2a-/- mutant [lockjaw allele, 28] and non-mutant siblings (i.e. wildtype and tfap2a+/- heterozygous) zebrafish larvae were sorted at 36 hpf based on pigmentation phenotype, and heads were removed to eliminate the RPE. Pools of 25–30 embryos were collected in TRIzol (Invitrogen), and RNA was extracted according to the manufacturer protocol. RNA was further purified using the RNeasy Mini Kit (Qiagen) and tested for quality using an Experion bioanalyzer (Bio-Rad). The SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) was used to generate ds-cDNA, which was also tested for quality on the Experion bioanalyzer. ds-cDNA was labeled using Cy3-coupled random nanomers (Nimblegen). Biological replicates were hybridized to sub-arrays of Roche Nimblegen gene expression microarrays (12 x 135K, design number 090505_Zv7_EXPR). The arrays were scanned on an Axon GenePix 4200A microarray scanner (Molecular Devices). Raw data (Pair files) were normalized in ArrayStar software, version 12.0.0 (DNASTAR, Inc). Background correction, quantile normalization, and median polish summarization were performed by applying the robust multichip analysis (RMA) algorithm. A Student’s t-test, corrected for multiple testing by the Benjamini and Hochberg false discovery rate (FDR) method, was performed for each pair-wise comparison. Transcripts were considered differentially expressed if the FDR-adjusted P-value was less than 0.05. Primers for qRT-PCR were designed to amplify fragments of 150–200 bp spanning exon junctions (listed in S11 Table). qRT-PCR was carried out using iQ SYBR Green Supermix (Bio-Rad) on a CFX96 Real-Time PCR Detection system (Bio-Rad) according to the default settings. Three technical replicates were averaged for each sample, and the 2-ΔΔCt method was used to calculate the difference in gene expression between samples, after normalization to the reference gene b2m. Statistical significance was evaluated via a Mann-Whitney U test for each gene.
Mouse
Melan-a cells were transfected in triplicate with either of two independent siTfap2a double strand duplex RNAs, siA = sure select siRNA #s74846 (Ambion), si5 = MMS.RNAIN011547.12.5 (Integrated DNA Technologies), or a non-silencing control siRNA NC-1 (Integrated DNA Technologies) using Lipofectamine RNAimax protocol (Invitrogen) with 25pmol of oligo per 6 well dish. Total RNA isolated from each bioreplicate was used to synthesize cDNA for hybridization to Mouse Gene 2.0 ST Arrays (Affymetrix). cDNA from each bioreplicate was then assayed by qRT-PCR with 3X technical replicates using TAQMAN qPCR probes (ABI, listed in S11 Table), on a StepOne machine (Thermo Fisher) with standard fast protocol conditions. Relative sample quantifications were performed using a standard curve method, where gene expression for each bioreplicate was normalized to Actb, then plotted relative to non-silencing control samples. Statistical significance was evaluated via one-way ANOVA with Bonferroni multiple test correction for each gene when response to both siA and si5 agreed.
ChIP-seq and ChIP-qPCR
Mouse
Melan-a cell ChIP-seq experiments (2 replicates) were performed as described by Gorkin and colleagues with the following alterations [65]: sonication was performed using a Q500 horn sonicator (Qsonica), 70% amplitude, 25 minute sonication cycle occurring with a 10 second pulse, 20 second off setting. Peaks that overlap by at least 1 bp between replicates were used for subsequent analyses. Anti-TFAP2A and IgG ChIP samples from mouse melan-a cells were validated by ChIP-qPCR with primer sets listed in S11 Table. Quantification of PCR products was performed as per manufacturer’s instructions using FAST SYBR Green master mix (ABI) and the Step-One Plus Real-time PCR machine (ABI) under FAST standard cycling conditions.
Human
ChIP was performed on human primary melanocytes that had not exceeded 10 passages using 10 μg anti-TFAP2A (3B5, Santa Cruz Biotechnology, Inc.) or control Ab-108C polyclonal Goat IgG control (R&D Systems). Over 45 million mapped reads were obtained for one biological replicate and one input control sample. Approximately 40 million human primary melanocytes were fixed at room temperature for 10 minutes with 1% formaldehyde diluted in cell culture media (v/v). Following fixation, formaldehyde was quenched with 1.25M glycine for 5 minutes at room temperature. Fixed/quenched cells were subsequently spun at 1,200 rpm for 5 minutes, washed with 1x PBS, and respun. Pelleted cells were then resuspended in 1mL cell lysis buffer (150mM NaCl, 10mM Hepes pH 7.4, 1.5mM MgCl2, 10 mM KCl, 0.5% NP-40, 0.5mM DTT, 1mM EDTA, plus protease inhibitors) and incubated on ice 10 minutes. Following cell lysis, nuclei were pelleted by spinning at 5,000 rpm, for 5 minutes at 4°C. Nuclei were then resuspended in 1 part nuclear lysis buffer (50mM Tris pH 8.0, 10mM EDTA, 1% SDS plus protease inhibitors) and 2 parts ChIP-dilution buffer (150mM NaCl, 16.7mM Tris pH 7.5, 3.3mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% Na-Doc, plus protease inhibitors), and sonicated using either a Covaris instrument (Model S220) or with a standard probe-tip sonicator (VirTis Virsonic 600). Sonication settings were determined empirically to generate fragments ranging from 200–500 bp. Following sonication samples were spun at 14,000 rpm for 15 minutes, and supernatant was saved for immunoprecipitation. For immunoprecipitation, Dynabeads (Life Technologies) were first washed 3x with ChIP-dilution buffer and subsequently incubated with equal amounts (5–10ug) of either anti-TFAP2A (3B5, Santa Cruz Biotechnology, Inc.) or Mouse IgG (Millipore) for 2 hours to overnight at 4°C. Following incubation, antibody-bound beads were washed 3x with ChIP-dilution buffer. Sonicated chromatin was equally split between two tubes, brought to approximately 1 mL with ChIP-dilution buffer, and used to resuspend antibody bound beads. Samples were incubated at 4°C overnight. Antibody/bead/chromatin complexes were then washed with a series of solutions 3x and eluted with 100 μL elution buffer. Crosslinks were reversed overnight at 65°C with 0.2 M NaCl. DNA was further cleaned using a Qiagen PCR purification kit as per manufacturer’s instructions (Qiagen). Total enriched DNA was quantified using a PicoGreen dsDNA assay kit (Life Technologies) and shearing efficiency was confirmed using a High Sensitivity DNA Analysis Kit and a 2100 Bioanalyzer (Agilent Technologies) on the input sample. Libraries were then constructed using ABI's SOLiD ChIP-seq kit with barcoding (rev. 08/06/2010), as per manufacturer’s instructions (Life Technologies). Following library construction, samples were sequenced using the ABI SOLiD 3.0 platform, using a 50bp sequencing run. LifeScope Genomic Analysis Software was used for basecalling and alignment to hg19 genome with default specifications for each program. Quality assessment was performed using Phantompeak tools as per the guidelines of the Encode project. Peaks were called using the SPP R program with an FDR setting of 0.05.
ChIP-qPCR was performed to validate chromatin enrichment at known TFAP2A targets using primer sets listed in S11 Table. Quantification of PCR products was performed as per manufacturer’s instructions using iQ SYBR Green Supermix (Bio-Rad) on a CFX96 Real-Time PCR Detection system (Bio-Rad) according to the default settings. Three technical replicates were averaged for each sample, and the 2-ΔΔCt method was used to calculate the differences in gene expression.
ChIP-seq data analysis
The ChIP-seq Tool Set in Galaxy was used for all peak overlap analyses [107]. For comparison of human ChIP-seq peaks to gene expression, we used a published RNA-seq expression profile of human penis foreskin melanocytes from the Roadmap Epigenomics Project (GEO accession number, GSM958174) [64]. Motif enrichment analysis was carried out using the MEME-ChIP suite [108], including CentriMo [60], MEME-ChIP [69], and AME [109] tools. For gene set enrichment analysis, we used the Genomic Regions Enrichment of Annotations Tool (GREAT), with the association rule basal plus extension, proximal: 5 kb upstream, 1 kb downstream, plus distal: up to 100 kb [72]. ChIP-seq read density clustering analysis and quantitative comparisons were performed using k-means linear enrichment cluster function in seqMINER with the following parameters: window size = -5K to +5K, read extension = 200bp, seed = 12 [110] (http://bips.u-strasbg.fr/). The Panther Classification System was used for GO term enrichment analysis on gene lists [76,77].
Super-enhancer analysis
Human foreskin melanocyte H3K27ac ChIP-seq alignment results were obtained from the Roadmap Epigenomics Project (GEO accession number, GSM1127072) [64]. Typical enhancers and super-enhancers were called using HOMER [111]. Briefly, all the stitched enhancer peaks were generated and sorted based on normalized tag count in descending order. Super-enhancers were defined by slope>1 (slope = Δ(normalized tag count) / Δrank) [84]. All stitched enhancers were compared with human melanocyte TFAP2A ChIP-seq peaks and MITF ChIP-seq peaks [18] using BEDTools (v. 2.24.0) [112]. We then normalized the rank by the total number of stitched enhancers and plotted this against normalized super-enhancer score (obtained by calculating normalized tag counts / highest normalized tag count).
RNA-seq in mouse melan-a cells
RNA sequencing (RNA-seq) libraries were prepared with the TrueSeq stranded mRNA kit (Illumina) and sequenced on the Illumina HiSeq 2000 platform. The ten 5’-most bases were trimmed from all of the raw RNA-seq reads. Reads were aligned to the mouse reference genome sequence (mm9) using the STAR alignment software (v. 2.3.0e). RNA-seq reads derived from rRNAs were removed using the split_bam.py script available in RSeQC (v. 2.3.7), using genomic locations of known rDNAs that were downloaded from UCSC. Counts for RNA-seq reads mapping to Ensembl-annotated transcripts (release 67) were calculated using the htseq-count software (v. 0.5.3p3). These raw RNA-seq counts were used for differential gene expression analysis that was performed using DESeq2 (v. 1.10.1).
Promoter deletion analysis
The following versions of the TRPM1 promoter sequence (748 bp) were obtained as double stranded gBlocks gene fragments from IDT: intact sequence, ΔAP2A with mutations in four TFAP2A binding sites, and ΔE1 with a mutation in the main MITF binding site. The consensus TFAP2A binding site GCCNNNGG was disrupted by changing the two underlined bases to T, whereas the E1 MITF site was changed as previously published by [86]. Fragments were cloned into a Tol2-cfos-FFluc vector via Gibson assembly and confirmed with Sanger sequencing.
M21 melanoma cells were grown to 70–90% confluency in a 24-well culture plate. In each well, the reporter plasmid (1μg) and a β-galactosidase plasmid (100ng) were transfected using Lipofectamine 3000 (Life Technologies). Approximately 48 hours after transfection, luciferase assays were conducted using the Dual-Light System from Applied Biosystems, and 20/20n Luminometer (Turner Biosystems, Sunnyvale, CA) according to the manufacturer protocols. Briefly, cells were washed with cold PBS and incubated in 40 μL lysis solution on a shaker for 15 minutes. Cell lysates were transferred to tubes and centrifuged at 12000 rpm for 2 minutes at 4°C, and supernatant was transferred to a clean tube. 10 μL of cell lysate was added to 25 μL of Buffer A and placed in the luminometer, where the injector adds 100 μL Buffer B/Galacton-Plus substrate and reads the signal after 1 second. Samples were incubated in the dark for 30 minutes before injection of 100 μL Accelerator-II and measurement of the β-gal signal after 1 second. For each version of the TRPM1 promoter element, transfection was carried out in triplicate. Firefly luciferase reads from each sample were normalized to the respective β-gal reads, and an average signal was calculated within groups.
Mouse genetics and immunostaining
Experiments utilizing mice in this study were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Colorado Denver. Noon on the day a copulatory plug was present was denoted as embryonic day 0.5. Mice used in this study included males that were heterozygous for a Tfap2a-null allele [113], heterozygous for a newly generated Tfap2b-null allele (EVO/TW, in preparation), and hemizygous for the Wnt1-Cre transgene [89] (Tfap2anull/wt;Tfap2bnull/wt;Wnt1-Cre). These males were crossed with females that were homozygous for both a Tfap2a-conditional [24] and Tfap2b-conditional (EVO/TW, in preparation) allele, resulting in a 1:8 frequency of generating single or double conditional mutants, as well as various other genotype combinations. Mice were maintained on an outbred Black Swiss background. Of note, by virtue of this breeding scheme, the single conditional mutants would always be conditionally heterozygous for the alternate paralog. For r26r [90] and ‘tomato’ [114] experiments, the female was also homozygous for the reporter allele. Yolk sacs or tail clips were used for genotyping. DNA for PCR was extracted using DirectPCR Lysis Reagent (Viagen Biotech. Inc) plus 10 ug/ml Proteinase K (Roche) followed by heat inactivation at 85°C for 45 min. Samples were then used directly for PCR-based genotyping using allele specific primers (available upon request) at a concentration of 200 nM using the Qiagen DNA polymerase kit, including the optional Q Buffer solution (Qiagen).
With the exception of whole-mount β-galactosidase (β-gal) staining, procedures used for mouse embryo analysis (in situ hybridization, ɑ-neurofilament immunostaining, and immunofluorescence) have all been previously described [115]. Following euthanasia, embryos were collected at the indicated time-points in DEPC-PBS and subsequently processed. Briefly, for in situ hybridization, after embryo collection, trunks were removed, fixed overnight in 4% paraformaldehyde (PFA), and stained using the indicated riboprobe (Pmel, Dct, Sox10). For immunostaining, embryos were processed using an ɑ-neurofilament primary antibody [116] (IgG clone 2H3, obtained from the Developmental Studies Hybridoma Bank–University of Iowa), followed by colorimetric staining using a standard secondary antibody and DAB-detection. For β-gal immunofluorescence, collected embryos underwent a short fixation in 0.25% glutaraldehyde, were taken through a series of sucrose/O.C.T. solutions (Tissue-Tek O.C.T. Compound, Electron Microscopy Sciences), until being embedded in 100% O.C.T. and frozen on dry-ice. Cryosections were then cut at ~12μM on a Leica CM 1900 cryostat (Leica Biosystems Inc.) using the hindlimb as a delimiting rostral-caudal boundary between samples. Following sectioning, slides were washed in PBS, 2 x 10 min, blocked in 3% BSA (in PBS) 1 hr at room temperature, incubated over-night with a rabbit polyclonal anti-β-gal antibody (Product 55976, MP Biomedicals, LLC) diluted 1:200 in block solution. Subsequently, sections were washed 2 x 10 min in PBS, followed by a 1 hr incubation in goat-anti-rabbit Alexa Fluor 488 (Thermo Fisher) and counterstained with DRAQ5 (Abcam), washed again 2 x 10 min in PBS, and then cover-slipped. Processed samples were imaged on a Leica TCS SP5 II confocal microscope and representative images taken. Finally, for β-gal staining, embryos were collected at appropriate time points and fixed ~1hr at room temperature with 0.25% glutaraldehyde in PBS. Subsequently, embryos were washed 3 x 30 minutes in a ‘lacZ rinse buffer’ (0.2M sodium phosphate, 2mM magnesium chloride, 0.02% NP40, and 0.01% sodium deoxycholate), and then incubated overnight in a ‘lacZ staining solution’ (lacZ rinse buffer plus 5mM potassium ferricyanide, 5mM potassium ferrocyanide, and 1 mg/ml X-gal) at 37°C. Following adequate staining, embryos were post-fixed in 4% PFA overnight, moved to PBS, and subsequently imaged.
Zebrafish genetics and immunostaining
Zebrafish alleles used in this study include tfap2alow [lockjaw allele, 28], mitfaw2 [94], and mitfaz25 [95]. Lines were maintained as mitfa trans-heterozygous and tfap2a heterozygous (mitfaw2/z25;tfap2a+/-) or mitfa wildtype and tfap2a heterozygous (tfap2a+/-). For genetic interaction experiments, mitfaw2/z25;tfap2a+/- animals were incrossed to generate tfap2a+/+, tfap2a+/-, and tfap2a-/- genotypes in each of mitfaz25/z25 and mitfaw2/z25 backgrounds. To obtain control animals in the mitfa+/+ background, mitfa+/+;tfap2a+/- animals were also incrossed. At 72 hpf, embryos were binned according to melanocyte phenotype, photographed, and genotyped via High Resolution Melt Analysis (HRMA) using Precision Melt Supermix (Bio-Rad) on a CFX96 Real-Time PCR Detection system (Bio-Rad) according to the default settings. Precision Melt Analysis Software (Bio-Rad) was used to analyze melt curves. HRMA primer sequences are as follows: tfap2alow (forward: GTA GCT ATG TTT CGT GGT TA; reverse: ACA ATA AGC AGC TGC TTT AC), mitfaz25 (forward: GCA GAA GTC AGA GCC CTG GC; reverse: ACG GAT CAT TTG ACT TGG GAA TTA AAG). Ventral stripe melanocytes were counted from lateral-view images taken at 72 hpf and statistical significance between groups was tested using a one-way ANOVA with multiple test correction.
For zebrafish rescue experiments, mitfaw2/z25 trans-heterozygotes were incrossed to obtain mitfaz25/z25, mitfaw2/z25, and mitfaw2/w2 genotypes, which were clearly distinguishable based on melanocyte phenotype at 48 hpf. 3-way Gateway cloning technology was used to generate a pDestTol2CG2 plasmid [117] containing 5’ entry mitfa promoter [96], middle entry tfap2a cDNA, and 3’ entry 6x Myc epitope tag followed by polyA, resulting in the presence of Myc epitope tags on the carboxy terminus of Tfap2a. This construct was then injected into the above cross along with Tol2 mRNA, both at a concentration of 30 ng/uL. Due to the presence of a cmlc2:GFP reporter in the vector, injected embryos were screened for successful plasmid integration based on expression of GFP in the heart at 28–30 hpf. GFP positive embryos and uninjected control embryos were fixed overnight in 4% paraformaldehyde at 48 hpf. After fixation, embryos were rinsed 3x in PBS, blocked in PBDT+2.5% goat serum for 1 hr at room temperature, and incubated in anti-Myc primary overnight at 4°C (9E10, obtained from the Developmental Studies Hybridoma Bank—University of Iowa, 1:100 diluted in block solution). The following day, embryos were rinsed 4x in PBS + 0.1% Triton X-100 (PBS-Tx) and incubated overnight at 4°C in Alexa Fluor goat-anti-mouse 488 secondary (Thermo Fisher) diluted in block solution. Embryos were then rinsed 4x15 minutes in PBS-Tx and mounted on slides. Ten individual embryos of each genotype with plasmid injection or uninjected control were viewed at 40x and photographed.
Immunostaining in cell lines
Cells were grown to approximately 75% confluency in 24-well plate on poly-lysine coated disks and fixed with 4% paraformaldehyde for 1 hr at 4°C. Following fixation, cells were rinsed 3x20 minutes with 1x PBS, permeabilized in 1x PBS + 0.3% Triton X-100 for 30 min at 37°C, rinsed 3x10 minutes in PBS-Tx, and blocked (1% BSA in PBS-Tx) overnight at 4°C. Cells were then incubated in anti-TFAP2A primary overnight at 4°C (3B5, Santa Cruz Biotechnology, Inc., 1:100, 250ug/mL stock diluted in block solution). The following day, cells were rinsed 3x20 minutes in PBS-Tx and incubated for 2 hr at room temperature in Alexa Fluor goat-anti-mouse 488 secondary (Thermo Fisher) diluted in block solution. Cells were then rinsed 4x15 minutes in PBS-Tx and disks mounted in Prolong Gold antifade reagent and DAPI SlowFade solution (Life Technologies). Slides were imaged on a Zeiss LSM 700 Flexible Confocal Microscope (Carl Zeiss Microscopy). Briefly, for quantification of immunofluorescent intensity approximately three 20x images were taken of each slide, and three slides of each cell line processed (including a no primary control). Each image included both the antibody of interest as well as DAPI staining to identify cell nuclei.
Ethics statement
All experiments were approved by the University of Iowa or University of Colorado Institutional Animal Care and Use Committee (IACUC). We abide by PHS Policy, USDA-Animal Welfare regulations, the Guide for the Care and Use of Laboratory Animals, University of Iowa policies and regulations and any state and local laws and regulations.
Supporting information
(A-L) Lateral views of live tfap2a+/- (left column) and tfap2a-/- (right column) zebrafish embryos at the indicated age from 28-38 hpf. (C, I) Melanocytes in a tfap2a+/- zebrafish (C) appear darkly pigmented by 32 hpf, whereas melanocytes at the same location in a tfap2a-/- mutant (I) at this stage remain pale and punctate. (F, L) At 38 hpf, there is still a detectable difference in the level of pigmentation in tfap2a+/- (F) and tfap2a-/- (L) mutant animals.
(PDF)
(A-E) Nuclear anti-TFAP2A immunoreactivity (green fluorescence) in human primary melanocytes (A) and four human melanoma cell lines (B-E). Inset shows DAPI counter-stain for nuclei. TFAP2A expression is low but detectable in M21s (C).
(PDF)
(A, B) Validation of TFAP2A ChIP-seq in mouse melan-a cells. (A) CentriMo analysis shows that mouse TFAP2A ChIP-seq peaks are centered on the TFAP2A/TFAP2C binding motif (TFAP2A p = 1.8e-2332, TFAP2C p = 2.7e-2186) (B) Four of five sites tested by ChIP-qPCR confirmed enrichment of TFAP2A binding over the IgG control, whereas an off-target site (Nos2 -50kb) showed no enrichment. (C, D) Validation of TFAP2A ChIP-seq in human primary melanocytes. (C) CentriMo analysis shows that human TFAP2A ChIP-seq peaks are centered on the TFAP2A/TFAP2C binding motif (TFAP2A p = 8.1e-820, TFAP2C p = 8.2e-785). (D) Five of six sites tested by ChIP-qPCR confirmed enrichment of TFAP2A binding over the IgG control, whereas an off-target site (chrm17) showed no enrichment. (E) Validation of TFAP2A ChIP-seq in M21 melanoma cells. Four of five sites tested by ChIP-qPCR confirmed enrichment of TFAP2A binding over the IgG control, whereas an off-target site (chrm17) showed no enrichment. Notably, DCT showed enrichment for TFAP2A binding in the mouse melan-a cells, but not in human primary melanocytes or M21s, which is consistent with the ChIP-seq results for mouse and human, respectively.
(PDF)
(A) Pie chart showing distribution of human TFAP2A peaks with respect to genomic features. TSS, transcription start site; TTS, transcription termination site. (B) Distance from TSS to the nearest TFAP2A peak for genes in three expression categories: highest 1000, median 1000, or lowest 1000. Highly expressed genes are more likely to have promoter-proximal TFAP2A peaks. Human expression data from the Roadmap Epigenomics Project GSM958174 [64].
(PDF)
(A-F) UCSC genome browser tracks showing mouse TFAP2A ChIP-seq peaks that overlap an active enhancer signature (p300 flanked by H3K4me1) or a partial enhancer signature near the promoters of (A) Pmel, (B) Irf4, (C) Mc1r, (D) Mitf, (E) Oca2, and (F) Tyr.
(PDF)
(A, B) GO term enrichment analysis of genes associated with (A) 13,690 TFAP2A peaks in human melanocytes and (B) 16,305 TFAP2A peaks in mouse melanocytes. Gene association was identified using the GREAT algorithm with an assignment rule of basal plus extension, proximal TSS -5/+1kb, distal up to 100kb. For each category, the top five most significant terms are listed, followed by select terms of interest. Significance is charted as -log10(binomial p-value). (C) Density-based clustering of H3K27ac signal at TFAP2A peaks in mouse melanocytes (H3K27ac data from [14]).
(PDF)
Chart depicting typical enhancers (gray) and super-enhancers (black) in human melanocytes.
(PDF)
(A-D) Dorsal views of E12.5 control (A), Tfap2a SCM (B), Tfap2b SCM (C), or Tfap2a/Tfap2b DCM (D) mouse embryos processed for β-galactosidase (β-gal) staining, labeling neural crest cells and their derivatives, including melanocytes migrating on the ventrolateral pathway (as in Fig 4A–4D). (E-L) Lateral views of E10.5 control (E, I) or Tfap2a/Tfap2b DCM (F, J) and E11.5 control (G, K) or Tfap2a/Tfap2b DCM (H, L) mouse embryos processed for Pmel (E-H) or Dct (I-L) expression by in situ hybridization. Yellow lines (in E, I, G, K) indicate extent of rostral-caudal regions of in situ signal, rostral left. Abbreviations: DCM, double conditional mutant; fl, forelimb; hl, hindlimb; SCM, single conditional mutant (A = Tfap2a or B = Tfap2b). Scale bars = 500μM.
(PDF)
(A-D) Dorsal (A, B) or lateral (C, D) views of the trunk of an E9.5 control (A, C) or Tfap2a/Tfap2b DCM (B, D) mouse embryo processed for β-gal staining (as in Fig 4A–4D). (E, F)Transverse immunofluorescent cryosections through the trunk of an E9.5 control (E) or Tfap2a/Tfap2b DCM (F) embryo immunostained with an anti:β-gal antibody (green), revealing migrating neural crest cells (tissue counter stained with Draq5, blue). (G, H) Lateral trunk views of E9.5 control (G) and Tfap2a/Tfap2b DCM (H) embryos processed by in situ hybridization with a Sox10 riboprobe. Abbreviations: DCM, double conditional mutant; nt, neural tube. Scale bars = 500μM.
(PDF)
(A-H) Transverse (A-D) or ventral (E-H) trunk views of E12.5 control (A, E), Tfap2a SCM (B, F), Tfap2b SCM (C, G), or Tfap2a/Tfap2b DCM (D, H) mouse embryos processed for β-galactosidase (β-gal) staining, labeling neural crest cells and their derivatives (as in Fig 4A–4D). (A-D) highlights the dorsal root ganglia while (E-H) highlights a portion of the enteric nervous system (ENS) populating the gastrointestinal (GI) tract. (I, J) Transverse cryosections through the GI-tract of an E12.5 control (I) or Tfap2a/Tfap2b DCM (J) embryo in which a fluorescent Tomato-reporter (rTMr) has been incorporated, labeling neural crest cells contributing to the ENS. (K, L) Late embryonic stage (E17.5) control (K) or Tfap2a/Tfap2b DCM (L) intestines processed for β-gal staining, revealing neural crest-derived ENS components (note, the ENS includes ‘striated’ surface staining in (K) whereas internal β-gal staining in (L) is background). Abbreviations: DCM, double conditional mutant; drg, dorsal root ganglia; SCM, single conditional mutant (A = Tfap2a, B = Tfap2b). Scale bars = 500μM.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank the NIH Intramural Sequencing Center for ChIP-seq library preparation and high-throughput sequencing. We acknowledge access to expression profile and chromatin mark data catalogued at: http://nihroadmap.nih.gov/epigenomics/.
Data Availability
TFAP2A ChIP-seq data in human primary melanocytes are available at GEO, accession number GSE67555. TFAP2A ChIP-seq data in mouse immortalized melanocytes is available at GEO, accession number GSE72953.
Funding Statement
This work was supported by: National Institutes of Health (AR062547, RAC) (DE12728, TW), the National Science Foundation (IOS-1147221, RAC), the University of Iowa Melanoma Research Fund (RAC), and fellowship F32DE023709 from the National Institute of Dental and Craniofacial Research (EVO). This work was supported in part by the National Human Genome Research Institute Intramural Research Program at the National Institutes of Health (DEG, SKL, and WJP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sanchez-Martin M, Perez-Losada J, Rodriguez-Garcia A, Gonzalez-Sanchez B, Korf BR, et al. (2003) Deletion of the SLUG (SNAI2) gene results in human piebaldism. Am J Med Genet A 122A: 125–132. 10.1002/ajmg.a.20345 [DOI] [PubMed] [Google Scholar]
- 2.Hoth CF, Milunsky A, Lipsky N, Sheffer R, Clarren SK, et al. (1993) Mutations in the paired domain of the human PAX3 gene cause Klein-Waardenburg syndrome (WS-III) as well as Waardenburg syndrome type I (WS-I). Am J Hum Genet 52: 455–462. [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez-Martin M, Rodriguez-Garcia A, Perez-Losada J, Sagrera A, Read AP, et al. (2002) SLUG (SNAI2) deletions in patients with Waardenburg disease. Hum Mol Genet 11: 3231–3236. [DOI] [PubMed] [Google Scholar]
- 4.Tassabehji M, Newton VE, Read AP (1994) Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet 8: 251–255. 10.1038/ng1194-251 [DOI] [PubMed] [Google Scholar]
- 5.Bondurand N, Dastot-Le Moal F, Stanchina L, Collot N, Baral V, et al. (2007) Deletions at the SOX10 gene locus cause Waardenburg syndrome types 2 and 4. Am J Hum Genet 81: 1169–1185. 10.1086/522090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofstra RM, Osinga J, Tan-Sindhunata G, Wu Y, Kamsteeg EJ, et al. (1996) A homozygous mutation in the endothelin-3 gene associated with a combined Waardenburg type 2 and Hirschsprung phenotype (Shah-Waardenburg syndrome). Nat Genet 12: 445–447. 10.1038/ng0496-445 [DOI] [PubMed] [Google Scholar]
- 7.Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Prehu MO, et al. (1998) SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet 18: 171–173. 10.1038/ng0298-171 [DOI] [PubMed] [Google Scholar]
- 8.Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ (2000) Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet 107: 1–6. [DOI] [PubMed] [Google Scholar]
- 9.Curran K, Lister JA, Kunkel GR, Prendergast A, Parichy DM, et al. (2010) Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev Biol 344: 107–118. 10.1016/j.ydbio.2010.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curran K, Raible DW, Lister JA (2009) Foxd3 controls melanophore specification in the zebrafish neural crest by regulation of Mitf. Dev Biol 332: 408–417. 10.1016/j.ydbio.2009.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas AJ, Erickson CA (2009) FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development 136: 1849–1858. 10.1242/dev.031989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adameyko I, Lallemend F, Furlan A, Zinin N, Aranda S, et al. (2012) Sox2 and Mitf cross-regulatory interactions consolidate progenitor and melanocyte lineages in the cranial neural crest. Development 139: 397–410. 10.1242/dev.065581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimadamore F, Shah M, Amador-Arjona A, Navarro-Peran E, Chen C, et al. (2012) SOX2 modulates levels of MITF in normal human melanocytes, and melanoma lines in vitro. Pigment Cell Melanoma Res 25: 533–536. 10.1111/j.1755-148X.2012.01012.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fufa TD, Harris ML, Watkins-Chow DE, Levy D, Gorkin DU, et al. (2015) Genomic analysis reveals distinct mechanisms and functional classes of SOX10-regulated genes in melanocytes. Hum Mol Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang D, Lu MM, Huang L, Engleka KA, Zhang M, et al. (2005) Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 433: 884–887. 10.1038/nature03292 [DOI] [PubMed] [Google Scholar]
- 16.Vachtenheim J, Borovansky J (2010) "Transcription physiology" of pigment formation in melanocytes: central role of MITF. Exp Dermatol 19: 617–627. 10.1111/j.1600-0625.2009.01053.x [DOI] [PubMed] [Google Scholar]
- 17.Strub T, Giuliano S, Ye T, Bonet C, Keime C, et al. (2011) Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene 30: 2319–2332. 10.1038/onc.2010.612 [DOI] [PubMed] [Google Scholar]
- 18.Webster DE, Barajas B, Bussat RT, Yan KJ, Neela PH, et al. (2014) Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition. Genome Res 24: 751–760. 10.1101/gr.166231.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koludrovic D, Davidson I (2013) MITF, the Janus transcription factor of melanoma. Future Oncol 9: 235–244. 10.2217/fon.12.177 [DOI] [PubMed] [Google Scholar]
- 20.Wan P, Hu Y, He L (2011) Regulation of melanocyte pivotal transcription factor MITF by some other transcription factors. Mol Cell Biochem. [DOI] [PubMed] [Google Scholar]
- 21.Levy C, Khaled M, Fisher DE (2006) MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 12: 406–414. 10.1016/j.molmed.2006.07.008 [DOI] [PubMed] [Google Scholar]
- 22.Goding CR (2011) Commentary. A picture of Mitf in melanoma immortality. Oncogene 30: 2304–2306. 10.1038/onc.2010.641 [DOI] [PubMed] [Google Scholar]
- 23.Milunsky JM, Maher TM, Zhao G, Wang Z, Mulliken JB, et al. (2010) Genotype-phenotype analysis of the branchio-oculo-facial syndrome. Am J Med Genet A 155A: 22–32. [DOI] [PubMed] [Google Scholar]
- 24.Brewer S, Feng W, Huang J, Sullivan S, Williams T (2004) Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Dev Biol 267: 135–152. 10.1016/j.ydbio.2003.10.039 [DOI] [PubMed] [Google Scholar]
- 25.Baldi A, Santini D, Battista T, Dragonetti E, Ferranti G, et al. (2001) Expression of AP-2 transcription factor and of its downstream target genes c-kit, E-cadherin and p21 in human cutaneous melanoma. J Cell Biochem 83: 364–372. [DOI] [PubMed] [Google Scholar]
- 26.Huang S, Jean D, Luca M, Tainsky MA, Bar-Eli M (1998) Loss of AP-2 results in downregulation of c-KIT and enhancement of melanoma tumorigenicity and metastasis. Embo J 17: 4358–4369. 10.1093/emboj/17.15.4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knight RD, Javidan Y, Nelson S, Zhang T, Schilling T (2004) Skeletal and pigment cell defects in the lockjaw mutant reveal multiple roles for zebrafish tfap2a in neural crest development. Dev Dyn 229: 87–98. 10.1002/dvdy.10494 [DOI] [PubMed] [Google Scholar]
- 28.Knight RD, Nair S, Nelson SS, Afshar A, Javidan Y, et al. (2003) lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development 130: 5755–5768. 10.1242/dev.00575 [DOI] [PubMed] [Google Scholar]
- 29.O'Brien EK, d'Alencon C, Bonde G, Li W, Schoenebeck J, et al. (2004) Transcription factor Ap-2alpha is necessary for development of embryonic melanophores, autonomic neurons and pharyngeal skeleton in zebrafish. Dev Biol 265: 246–261. [DOI] [PubMed] [Google Scholar]
- 30.Van Otterloo E, Li W, Bonde G, Day KM, Hsu MY, et al. (2010) Differentiation of zebrafish melanophores depends on transcription factors AP2 alpha and AP2 epsilon. PLoS Genet 6: e1001122 10.1371/journal.pgen.1001122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckert D, Buhl S, Weber S, Jager R, Schorle H (2005) The AP-2 family of transcription factors. Genome Biol 6: 246 10.1186/gb-2005-6-13-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higdon CW, Mitra RD, Johnson SL (2013) Gene expression analysis of zebrafish melanocytes, iridophores, and retinal pigmented epithelium reveals indicators of biological function and developmental origin. PLoS One 8: e67801 10.1371/journal.pone.0067801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Cornell RA (2007) Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev Biol 304: 338–354. 10.1016/j.ydbio.2006.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF (2007) Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zool B Mol Dev Evol 308: 679–691. 10.1002/jez.b.21189 [DOI] [PubMed] [Google Scholar]
- 35.Moser M, Ruschoff J, Buettner R (1997) Comparative analysis of AP-2 alpha and AP-2 beta gene expression during murine embryogenesis. Developmental Dynamics 208: 115–124. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt M, Huber L, Majdazari A, Schutz G, Williams T, et al. (2011) The transcription factors AP-2beta and AP-2alpha are required for survival of sympathetic progenitors and differentiated sympathetic neurons. Dev Biol 355: 89–100. 10.1016/j.ydbio.2011.04.011 [DOI] [PubMed] [Google Scholar]
- 37.Praetorius C, Grill C, Stacey SN, Metcalf AM, Gorkin DU, et al. (2013) A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell 155: 1022–1033. 10.1016/j.cell.2013.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartman ML, Czyz M (2015) MITF in melanoma: mechanisms behind its expression and activity. Cell Mol Life Sci 72: 1249–1260. 10.1007/s00018-014-1791-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE (1998) MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature 391: 298–301. 10.1038/34681 [DOI] [PubMed] [Google Scholar]
- 40.Price ER, Ding HF, Badalian T, Bhattacharya S, Takemoto C, et al. (1998) Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. J Biol Chem 273: 17983–17986. [DOI] [PubMed] [Google Scholar]
- 41.Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, et al. (2005) Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature 433: 764–769. 10.1038/nature03269 [DOI] [PubMed] [Google Scholar]
- 42.Zeng YX, Somasundaram K, el-Deiry WS (1997) AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat Genet 15: 78–82. 10.1038/ng0197-78 [DOI] [PubMed] [Google Scholar]
- 43.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laurette P, Strub T, Koludrovic D, Keime C, Le Gras S, et al. (2015) Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berger AJ, Davis DW, Tellez C, Prieto VG, Gershenwald JE, et al. (2005) Automated quantitative analysis of activator protein-2alpha subcellular expression in melanoma tissue microarrays correlates with survival prediction. Cancer Res 65: 11185–11192. 10.1158/0008-5472.CAN-05-2300 [DOI] [PubMed] [Google Scholar]
- 46.Hallberg AR, Vorrink SU, Hudachek DR, Cramer K, Milhem MM, et al. (2015) Aberrant CpG methylation of the TFAP2A gene constitutes a mechanism for loss of TFAP2A expression in human metastatic melanoma. Epigenetics (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Otterloo E, Li W, Garnett A, Cattell M, Medeiros DM, et al. (2012) Novel Tfap2-mediated control of soxE expression facilitated the evolutionary emergence of the neural crest. Development 139: 720–730. 10.1242/dev.071308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barrallo-Gimeno A, Holzschuh J, Driever W, Knapik EW (2004) Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development 131: 1463–1477. 10.1242/dev.01033 [DOI] [PubMed] [Google Scholar]
- 49.Knapik EW (2000) ENU mutagenesis in zebrafish—from genes to complex diseases. Mamm Genome 11: 511–519. [DOI] [PubMed] [Google Scholar]
- 50.Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, et al. (2013) ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res 41: D854–860. 10.1093/nar/gks938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thisse B, Thisse C. (2004) Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. [Google Scholar]
- 52.Bennett DC, Cooper PJ, Hart IR (1987) A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int J Cancer 39: 414–418. [DOI] [PubMed] [Google Scholar]
- 53.Luo Y, Dallaglio K, Chen Y, Robinson WA, Robinson SE, et al. (2012) ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells 30: 2100–2113. 10.1002/stem.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Craig EA, Weber JD, Spiegelman VS (2012) Involvement of the mRNA binding protein CRD-BP in the regulation of metastatic melanoma cell proliferation and invasion by hypoxia. J Cell Sci 125: 5950–5954. 10.1242/jcs.115204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elcheva I, Tarapore RS, Bhatia N, Spiegelman VS (2008) Overexpression of mRNA-binding protein CRD-BP in malignant melanomas. Oncogene 27: 5069–5074. 10.1038/onc.2008.141 [DOI] [PubMed] [Google Scholar]
- 56.Tworkoski K, Singhal G, Szpakowski S, Zito CI, Bacchiocchi A, et al. (2011) Phosphoproteomic screen identifies potential therapeutic targets in melanoma. Mol Cancer Res 9: 801–812. 10.1158/1541-7786.MCR-10-0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryu B, Kim DS, Deluca AM, Alani RM (2007) Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS One 2: e594 10.1371/journal.pone.0000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muthusamy V, Duraisamy S, Bradbury CM, Hobbs C, Curley DP, et al. (2006) Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res 66: 11187–11193. 10.1158/0008-5472.CAN-06-1274 [DOI] [PubMed] [Google Scholar]
- 59.Liu S, Howell P, Ren S, Fodstad O, Zhang G, et al. (2009) Expression and functional analysis of the WAP four disulfide core domain 1 gene in human melanoma. Clin Exp Metastasis 26: 739–749. 10.1007/s10585-009-9273-8 [DOI] [PubMed] [Google Scholar]
- 60.Bailey TL, Machanick P (2012) Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40: e128 10.1093/nar/gks433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, et al. (2010) Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040. 10.1126/science.1186176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng Y, Ma Z, Kim BH, Wu W, Cayting P, et al. (2014) Principles of regulatory information conservation between mouse and human. Nature 515: 371–375. 10.1038/nature13985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, et al. (2007) Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39: 730–732. 10.1038/ng2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, et al. (2010) The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048. 10.1038/nbt1010-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorkin DU, Lee D, Reed X, Fletez-Brant C, Bessling SL, et al. (2012) Integration of ChIP-seq and machine learning reveals enhancers and a predictive regulatory sequence vocabulary in melanocytes. Genome Res 22: 2290–2301. 10.1101/gr.139360.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He B, Chen C, Teng L, Tan K (2014) Global view of enhancer-promoter interactome in human cells. Proc Natl Acad Sci U S A 111: E2191–2199. 10.1073/pnas.1320308111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohba S, He X, Hojo H, McMahon AP (2015) Distinct Transcriptional Programs Underlie Sox9 Regulation of the Mammalian Chondrocyte. Cell Rep 12: 229–243. 10.1016/j.celrep.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pihlajamaa P, Sahu B, Lyly L, Aittomaki V, Hautaniemi S, et al. (2014) Tissue-specific pioneer factors associate with androgen receptor cistromes and transcription programs. EMBO J 33: 312–326. 10.1002/embj.201385895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Machanick P, Bailey TL (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pogenberg V, Ogmundsdottir MH, Bergsteinsdottir K, Schepsky A, Phung B, et al. (2012) Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev 26: 2647–2658. 10.1101/gad.198192.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Glahs A, Zinzen RP (2016) Putting chromatin in its place: the pioneer factor NeuroD1 modulates chromatin state to drive cell fate decisions. EMBO J 35: 1–3. 10.15252/embj.201593324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, et al. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ren Y, Liao WS (2001) Transcription factor AP-2 functions as a repressor that contributes to the liver-specific expression of serum amyloid A1 gene. J Biol Chem 276: 17770–17778. 10.1074/jbc.M010307200 [DOI] [PubMed] [Google Scholar]
- 74.Kimura H (2013) Histone modifications for human epigenome analysis. J Hum Genet 58: 439–445. 10.1038/jhg.2013.66 [DOI] [PubMed] [Google Scholar]
- 75.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD (2016) PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res 44: D336–342. 10.1093/nar/gkv1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mi H, Muruganujan A, Casagrande JT, Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8: 1551–1566. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Montoliu L, Oetting WS, Bennett DC Color Genes. In: Web ESfPCRWW, editor.
- 79.Devi S, Kedlaya R, Maddodi N, Bhat KM, Weber CS, et al. (2009) Calcium homeostasis in human melanocytes: role of transient receptor potential melastatin 1 (TRPM1) and its regulation by ultraviolet light. Am J Physiol Cell Physiol 297: C679–687. 10.1152/ajpcell.00092.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oancea E, Vriens J, Brauchi S, Jun J, Splawski I, et al. (2009) TRPM1 forms ion channels associated with melanin content in melanocytes. Sci Signal 2: ra21 10.1126/scisignal.2000146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bellone RR, Forsyth G, Leeb T, Archer S, Sigurdsson S, et al. (2010) Fine-mapping and mutation analysis of TRPM1: a candidate gene for leopard complex (LP) spotting and congenital stationary night blindness in horses. Brief Funct Genomics 9: 193–207. 10.1093/bfgp/elq002 [DOI] [PubMed] [Google Scholar]
- 82.Bellone RR, Holl H, Setaluri V, Devi S, Maddodi N, et al. (2013) Evidence for a retroviral insertion in TRPM1 as the cause of congenital stationary night blindness and leopard complex spotting in the horse. PLoS One 8: e78280 10.1371/journal.pone.0078280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pott S, Lieb JD (2015) What are super-enhancers? Nat Genet 47: 8–12. 10.1038/ng.3167 [DOI] [PubMed] [Google Scholar]
- 84.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, et al. (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, et al. (2013) Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, et al. (2004) Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res 64: 509–516. [DOI] [PubMed] [Google Scholar]
- 87.Sennett R, Wang Z, Rezza A, Grisanti L, Roitershtein N, et al. (2015) An Integrated Transcriptome Atlas of Embryonic Hair Follicle Progenitors, Their Niche, and the Developing Skin. Dev Cell 34: 577–591. 10.1016/j.devcel.2015.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rezza A, Wang Z, Sennett R, Qiao W, Wang D, et al. (2016) Signaling Networks among Stem Cell Precursors, Transit-Amplifying Progenitors, and their Niche in Developing Hair Follicles. Cell Rep 14: 3001–3018. 10.1016/j.celrep.2016.02.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP (1998) Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol 8: 1323–1326. [DOI] [PubMed] [Google Scholar]
- 90.Soriano P (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71. 10.1038/5007 [DOI] [PubMed] [Google Scholar]
- 91.Baxter LL, Pavan WJ (2003) Pmel17 expression is Mitf-dependent and reveals cranial melanoblast migration during murine development. Gene Expr Patterns 3: 703–707. [DOI] [PubMed] [Google Scholar]
- 92.Loftus SK, Baxter LL, Buac K, Watkins-Chow DE, Larson DM, et al. (2009) Comparison of melanoblast expression patterns identifies distinct classes of genes. Pigment Cell Melanoma Res 22: 611–622. 10.1111/j.1755-148X.2009.00584.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Steel KP, Davidson DR, Jackson IJ (1992) TRP-2/DT, a new early melanoblast marker, shows that steel growth factor (c-kit ligand) is a survival factor. Development 115: 1111–1119. [DOI] [PubMed] [Google Scholar]
- 94.Johnson SL, Nguyen AN, Lister JA (2011) mitfa is required at multiple stages of melanocyte differentiation but not to establish the melanocyte stem cell. Dev Biol 350: 405–413. 10.1016/j.ydbio.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zeng Z, Johnson SL, Lister JA, Patton EE (2015) Temperature-sensitive splicing of mitfa by an intron mutation in zebrafish. Pigment Cell Melanoma Res 28: 229–232. 10.1111/pcmr.12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lister JA, Close J, Raible DW (2001) Duplicate mitf Genes in Zebrafish: Complementary Expression and Conservation of Melanogenic Potential. Dev Biol 237: 333–344. 10.1006/dbio.2001.0379 [DOI] [PubMed] [Google Scholar]
- 97.Beirl AJ, Linbo TH, Cobb MJ, Cooper CD (2014) oca2 Regulation of chromatophore differentiation and number is cell type specific in zebrafish. Pigment Cell Melanoma Res 27: 178–189. 10.1111/pcmr.12205 [DOI] [PubMed] [Google Scholar]
- 98.Ota S, Hisano Y, Ikawa Y, Kawahara A (2014) Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes Cells 19: 555–564. 10.1111/gtc.12154 [DOI] [PubMed] [Google Scholar]
- 99.Dooley CM, Schwarz H, Mueller KP, Mongera A, Konantz M, et al. (2013) Slc45a2 and V-ATPase are regulators of melanosomal pH homeostasis in zebrafish, providing a mechanism for human pigment evolution and disease. Pigment Cell Melanoma Res 26: 205–217. 10.1111/pcmr.12053 [DOI] [PubMed] [Google Scholar]
- 100.Wang X, Pasolli HA, Williams T, Fuchs E (2008) AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J Cell Biol 183: 37–48. 10.1083/jcb.200804030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pavan WJ, Raible DW (2012) Specification of neural crest into sensory neuron and melanocyte lineages. Dev Biol 366: 55–63. 10.1016/j.ydbio.2012.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, et al. (2006) Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev 20: 3426–3439. 10.1101/gad.406406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jean D, Gershenwald JE, Huang S, Luca M, Hudson MJ, et al. (1998) Loss of AP-2 results in up-regulation of MCAM/MUC18 and an increase in tumor growth and metastasis of human melanoma cells. J Biol Chem 273: 16501–16508. [DOI] [PubMed] [Google Scholar]
- 104.Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, et al. (2016) A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351: aad2197 10.1126/science.aad2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sviderskaya EV, Hill SP, Evans-Whipp TJ, Chin L, Orlow SJ, et al. (2002) p16(Ink4a) in melanocyte senescence and differentiation. J Natl Cancer Inst 94: 446–454. [DOI] [PubMed] [Google Scholar]
- 106.Abdel-Malek Z, Swope VB, Suzuki I, Akcali C, Harriger MD, et al. (1995) Mitogenic and melanogenic stimulation of normal human melanocytes by melanotropic peptides. Proc Natl Acad Sci U S A 92: 1789–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, et al. (2016) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44: W3–W10. 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McLeay RC, Bailey TL (2010) Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11: 165 10.1186/1471-2105-11-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, et al. (2011) seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res 39: e35 10.1093/nar/gkq1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, et al. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, et al. (1996) Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature 381: 238–241. 10.1038/381238a0 [DOI] [PubMed] [Google Scholar]
- 114.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, et al. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Van Otterloo E, Feng W, Jones KL, Hynes NE, Clouthier DE, et al. (2016) MEMO1 drives cranial endochondral ossification and palatogenesis. Dev Biol 415: 278–295. 10.1016/j.ydbio.2015.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dodd J, Morton SB, Karagogeos D, Yamamoto M, Jessell TM (1988) Spatial regulation of axonal glycoprotein expression on subsets of embryonic spinal neurons. Neuron 1: 105–116. [DOI] [PubMed] [Google Scholar]
- 117.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, et al. (2007) The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 236: 3088–3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A-L) Lateral views of live tfap2a+/- (left column) and tfap2a-/- (right column) zebrafish embryos at the indicated age from 28-38 hpf. (C, I) Melanocytes in a tfap2a+/- zebrafish (C) appear darkly pigmented by 32 hpf, whereas melanocytes at the same location in a tfap2a-/- mutant (I) at this stage remain pale and punctate. (F, L) At 38 hpf, there is still a detectable difference in the level of pigmentation in tfap2a+/- (F) and tfap2a-/- (L) mutant animals.
(PDF)
(A-E) Nuclear anti-TFAP2A immunoreactivity (green fluorescence) in human primary melanocytes (A) and four human melanoma cell lines (B-E). Inset shows DAPI counter-stain for nuclei. TFAP2A expression is low but detectable in M21s (C).
(PDF)
(A, B) Validation of TFAP2A ChIP-seq in mouse melan-a cells. (A) CentriMo analysis shows that mouse TFAP2A ChIP-seq peaks are centered on the TFAP2A/TFAP2C binding motif (TFAP2A p = 1.8e-2332, TFAP2C p = 2.7e-2186) (B) Four of five sites tested by ChIP-qPCR confirmed enrichment of TFAP2A binding over the IgG control, whereas an off-target site (Nos2 -50kb) showed no enrichment. (C, D) Validation of TFAP2A ChIP-seq in human primary melanocytes. (C) CentriMo analysis shows that human TFAP2A ChIP-seq peaks are centered on the TFAP2A/TFAP2C binding motif (TFAP2A p = 8.1e-820, TFAP2C p = 8.2e-785). (D) Five of six sites tested by ChIP-qPCR confirmed enrichment of TFAP2A binding over the IgG control, whereas an off-target site (chrm17) showed no enrichment. (E) Validation of TFAP2A ChIP-seq in M21 melanoma cells. Four of five sites tested by ChIP-qPCR confirmed enrichment of TFAP2A binding over the IgG control, whereas an off-target site (chrm17) showed no enrichment. Notably, DCT showed enrichment for TFAP2A binding in the mouse melan-a cells, but not in human primary melanocytes or M21s, which is consistent with the ChIP-seq results for mouse and human, respectively.
(PDF)
(A) Pie chart showing distribution of human TFAP2A peaks with respect to genomic features. TSS, transcription start site; TTS, transcription termination site. (B) Distance from TSS to the nearest TFAP2A peak for genes in three expression categories: highest 1000, median 1000, or lowest 1000. Highly expressed genes are more likely to have promoter-proximal TFAP2A peaks. Human expression data from the Roadmap Epigenomics Project GSM958174 [64].
(PDF)
(A-F) UCSC genome browser tracks showing mouse TFAP2A ChIP-seq peaks that overlap an active enhancer signature (p300 flanked by H3K4me1) or a partial enhancer signature near the promoters of (A) Pmel, (B) Irf4, (C) Mc1r, (D) Mitf, (E) Oca2, and (F) Tyr.
(PDF)
(A, B) GO term enrichment analysis of genes associated with (A) 13,690 TFAP2A peaks in human melanocytes and (B) 16,305 TFAP2A peaks in mouse melanocytes. Gene association was identified using the GREAT algorithm with an assignment rule of basal plus extension, proximal TSS -5/+1kb, distal up to 100kb. For each category, the top five most significant terms are listed, followed by select terms of interest. Significance is charted as -log10(binomial p-value). (C) Density-based clustering of H3K27ac signal at TFAP2A peaks in mouse melanocytes (H3K27ac data from [14]).
(PDF)
Chart depicting typical enhancers (gray) and super-enhancers (black) in human melanocytes.
(PDF)
(A-D) Dorsal views of E12.5 control (A), Tfap2a SCM (B), Tfap2b SCM (C), or Tfap2a/Tfap2b DCM (D) mouse embryos processed for β-galactosidase (β-gal) staining, labeling neural crest cells and their derivatives, including melanocytes migrating on the ventrolateral pathway (as in Fig 4A–4D). (E-L) Lateral views of E10.5 control (E, I) or Tfap2a/Tfap2b DCM (F, J) and E11.5 control (G, K) or Tfap2a/Tfap2b DCM (H, L) mouse embryos processed for Pmel (E-H) or Dct (I-L) expression by in situ hybridization. Yellow lines (in E, I, G, K) indicate extent of rostral-caudal regions of in situ signal, rostral left. Abbreviations: DCM, double conditional mutant; fl, forelimb; hl, hindlimb; SCM, single conditional mutant (A = Tfap2a or B = Tfap2b). Scale bars = 500μM.
(PDF)
(A-D) Dorsal (A, B) or lateral (C, D) views of the trunk of an E9.5 control (A, C) or Tfap2a/Tfap2b DCM (B, D) mouse embryo processed for β-gal staining (as in Fig 4A–4D). (E, F)Transverse immunofluorescent cryosections through the trunk of an E9.5 control (E) or Tfap2a/Tfap2b DCM (F) embryo immunostained with an anti:β-gal antibody (green), revealing migrating neural crest cells (tissue counter stained with Draq5, blue). (G, H) Lateral trunk views of E9.5 control (G) and Tfap2a/Tfap2b DCM (H) embryos processed by in situ hybridization with a Sox10 riboprobe. Abbreviations: DCM, double conditional mutant; nt, neural tube. Scale bars = 500μM.
(PDF)
(A-H) Transverse (A-D) or ventral (E-H) trunk views of E12.5 control (A, E), Tfap2a SCM (B, F), Tfap2b SCM (C, G), or Tfap2a/Tfap2b DCM (D, H) mouse embryos processed for β-galactosidase (β-gal) staining, labeling neural crest cells and their derivatives (as in Fig 4A–4D). (A-D) highlights the dorsal root ganglia while (E-H) highlights a portion of the enteric nervous system (ENS) populating the gastrointestinal (GI) tract. (I, J) Transverse cryosections through the GI-tract of an E12.5 control (I) or Tfap2a/Tfap2b DCM (J) embryo in which a fluorescent Tomato-reporter (rTMr) has been incorporated, labeling neural crest cells contributing to the ENS. (K, L) Late embryonic stage (E17.5) control (K) or Tfap2a/Tfap2b DCM (L) intestines processed for β-gal staining, revealing neural crest-derived ENS components (note, the ENS includes ‘striated’ surface staining in (K) whereas internal β-gal staining in (L) is background). Abbreviations: DCM, double conditional mutant; drg, dorsal root ganglia; SCM, single conditional mutant (A = Tfap2a, B = Tfap2b). Scale bars = 500μM.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
TFAP2A ChIP-seq data in human primary melanocytes are available at GEO, accession number GSE67555. TFAP2A ChIP-seq data in mouse immortalized melanocytes is available at GEO, accession number GSE72953.