Abstract
By use of the nearly perfectly colinear genomes of Rickettsia conorii and Rickettsia prowazekii, we compared the usefulness of three types of sequences for typing of R. conorii isolates: (i) 5 variable coding genes comprising the 16S ribosomal DNA, gltA, ompB, and sca4 (gene D) genes, which are present in both genomes, and the ompA gene, which is degraded in R. prowazekii; (ii) 28 genes degraded in R. conorii but intact in R. prowazekii, including 23 split and 5 remnant genes; and (iii) 27 conserved and 25 variable intergenic spacers. The 4 conserved and 23 split genes as well as the 27 conserved intergenic spacers each had identical sequences in 34 human and 5 tick isolates of R. conorii. Analysis of the ompA sequences identified three genotypes of R. conorii. The variable intergenic spacers were significantly more variable than conserved genes, split genes, remnant genes, and conserved spacers (P < 10−2 in all cases). Four of the variable intergenic spacers (dksA-xerC, mppA-purC, rpmE-tRNAfMet, and tRNAGly-tRNATyr) had highly variable sequences; when they were combined for typing, multispacer typing (MST) identified 27 different genotypes in the 39 R. conorii isolates. Two batches from the same R. conorii strain, Malish (Seven), with different culture passage histories were found to exhibit the same MST type. MST was more discriminatory for strain genotyping than multiple gene sequencing (P < 10−2). Phylogenetic analysis based on MST sequences was concordant with the geographic origins of R. conorii isolates. Our study supports the usefulness of MST for strain genotyping. This tool may be useful for tracing a strain and identifying its source during outbreaks, including those resulting from bioterrorism.
The growing challenge presented by strains of bacterial pathogens with increased virulence and/or transmissibility, strains with antibiotic resistance, and strains used in bioterrorist attacks has highlighted the requirement for effective methods to identify such strains and track their spread (26). Key factors in the control of these strains are rapid detection, appropriate therapy, and contact tracing to arrest further transmission. To date, molecular characterization of bacterial strains has been pursued for two different objectives (57, 60). The first is global or long-term epidemiology studies that monitor how microbial populations change over time. Much of the current knowledge of bacterial population genetics (38, 52, 54) has been obtained by methods such as multilocus enzyme electrophoresis and multilocus sequence typing (15) of housekeeping genes. The second objective is the tracing of strains causing outbreaks of disease in hospitals or a local community. Multilocus enzyme electrophoresis and multilocus sequence typing are not suitable for such investigations (23), so methods with increased discriminatory power are used. These include whole-genome analysis by pulsed-field gel electrophoresis (20), arbitrarily primed PCR, randomly amplified polymorphic DNA, amplified fragment length polymorphism analysis (34), enterobacterial repetitive intergenic consensus PCR, repetitive element sequence-based PCR (20), insertion sequence typing (37, 50), spacer oligonucleotide typing (spoligotyping) (14, 21), ribotyping (9), and study of microsatellites (35). These methods are highly discriminatory and rely on uncharacterized genomic differences between isolates of bacterial species. However, the procedures are laborious, may require large amounts of purified DNA (for pulsed-field gel electrophoresis), are somewhat subjective (arbitrarily primed PCR, ribotyping), and produce results that may not always be reproducible and are difficult to share between laboratories.
Another drawback of the methods listed above is the empirical choice of target sequences. Over recent years, full-genome sequencing has been performed on an increasing number of bacteria. This has provided useful information for taxonomic, evolutionary, and phylogenic purposes, but although it is the most detailed form of genotyping, full-genome sequencing is clearly not adapted to strain typing. The availability of full-genome sequences, however, enables the rational selection of target sequences in molecular studies. We speculated that the areas of the genome containing the most-variable sequences in closely related bacterial species would also be the most variable among strains of the species and hence the most useful for differentiation of strains. In plants, it has been demonstrated that noncoding sequences were superior to genes for the phylogenetic and genotypic classification of species (7, 10, 17, 18, 56, 61). We speculated that noncoding sequences, which should not be subject to selection pressure, would be appropriate for typing bacterial strains.
To test our hypotheses, we compared the genome sequences of two closely related bacteria and attempted to develop a rational method for selecting sequences suitable for strain typing. We used Rickettsia conorii in our study for several reasons, including (i) the availability and colinearity of the R. conorii and Rickettsia prowazekii genome sequences, which facilitate comparisons of various types of sequences, (ii) the absence of effective phenotypic (43) or genotypic (47-49, 51) typing methods at the strain level, and (iii) the availability in our laboratory of a large collection of R. conorii isolates. Because our purpose was not a taxonomic study of rickettsiae closely related to R. conorii but rather the development of a genotyping tool at the strain level for R. conorii sensu stricto (type strain, Malish), we did not include in our study the Astrakhan fever rickettsia, Israeli spotted fever rickettsia, and Indian tick typhus rickettsia, whose taxonomic status is uncertain and which can easily be differentiated from each other and from R. conorii sensu stricto on the basis of specific nucleotide substitutions within the ompA, ompB, and sca4 nucleotide sequences (45, 48, 51).
MATERIALS AND METHODS
Selection of target sequences.
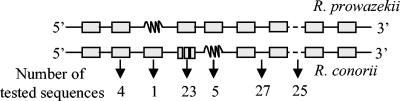
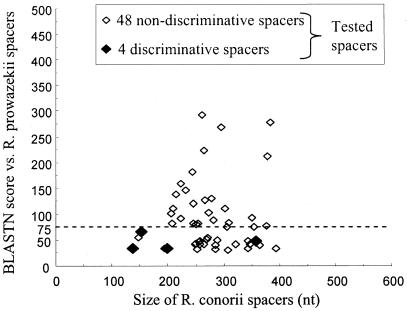
We aligned the genome sequences of R. conorii (GenBank accession number NC_003103) and R. prowazekii (NC_000963) by using BLAST (1) and identified conserved or degraded fragments within both coding and noncoding sequences. To test whether noncoding sequences would be better candidates for R. conorii strain genotyping than coding sequences, we studied four conserved genes present in the genomes of both R. conorii and R. prowazekii (the 16S ribosomal DNA [rDNA], gltA, ompB, and sca4 [also referred to as gene D] genes [Table 1; Fig. 1] [45, 47-49, 51]); ompA, a coding gene present in R. conorii and used for species identification of tick-borne rickettsiae, which is present as a remnant in R. prowazekii (Table 1; Fig. 1) (45); 23 split genes in R. conorii which had statistical coding potential but were fragmented compared to their orthologs in R. prowazekii (Table 1; Fig. 1); 5 remnant genes in R. conorii which were identified as genomic regions exhibiting significant sequence similarity with bona fide genes in R. prowazekii but were too degraded to be identified as bona fide genes by gene-finding programs; 25 variable intergenic spacers, selected from 150- to 500-bp sequences separating 2 genes consecutive in both genomes, which had BLASTN scores of <75 between the two genomes; and 27 conserved intergenic spacers with BLASTN scores of >75 (Fig. 2).
TABLE 1.
Sequences of primers, amplicon sizes, annealing temperatures, and accession numbers used in this study
| DNA target namea | Forward primer (5′-3′) | Reverse primer (5′-3′) | Amplicon size (bp) | Annealing temp (°C) | GenBank accession no. |
|---|---|---|---|---|---|
| Genes conserved in both genomes | |||||
| 16S rDNA | fD1, SFG3, SFG4b | Rp2, SFG2, SFG5, SFG6b | 1,400 | See reference 47 | AF541999 |
| gltA | CS1d, CS535d, Rp877pc | CS428r, CS890r, Rp1258nc | 1,047 | See reference 49 | U59730 |
| ompB | 120M59, 120-607, 120-1378, 120AA2235, 120-2788, 120-3462, 120-4232d | 120-807, 120-1497, 120-2399, 120-2988, 120-3599, 120-4346, 120-4879d | 4,682 | See reference 48 | AF123721 |
| sca4 (gene D) | D1f, D767f, D1219f, D1738f, D2338fe | D928r, D1390r, D1876r, D2482r, D3069re | 2,725 | See reference 51 | AF163008 |
| Genes degraded in one of the two genomes | |||||
| ompA | 190-70f | 190-701f | 632 | 52 | U43806 (Malish), U45244 (Moroccan), U43798 (M1) |
| Remnant of Rp352 | GCAAAACGGTTGACATTTGA | CTCAAGACAAAATGGGGAAAA | 221 | 50 | AY515514 |
| Remnant of Rp543 | ATTTTGTTAAAACATATGTAGCGGTAT | TTCAATAGACCTCTTCCCAAGC | 250 | 50 | AY515515 |
| Remnant of Rp550 | CGGCATTAGATTTTCTGCTTG | CCATGTCATTCTTGCTTTCG | 230 | 50 | AY515516 |
| Remnant of Rp723 | CTCTACTAAAGCAGATGTCGAAGG | AACGAGCAATACACCTGCAC | 250 | 50 | AY515517 |
| Remnant of Rp820 | AACCGATGCCTTTTGATTTG | CAACTGCTACATGCCCTGAA | 288 | 50 | AY515518 |
| Split Rc148 | TCTTTTTGAAGATTGGTGGAAAA | AAGGTTGATGCAAGGGACTTT | 166 | 52 | AY518488 |
| Split Rc149 | TTTGCAGGTACTGGCTTTATTTC | CCCTGAAAACCGTAACGAAT | 159 | 52 | AY518489 |
| Split Rc215 | CAAACCGGCTTGAATGATTT | AAGTATCGGCACTTCACATGC | 174 | 52 | AY518490 |
| Split Rc217 | ATGGAAGCGAGGAGAACCTA | TGCAGAAATATTATCGGTAAAAGC | 105 | 52 | AY518491 |
| Split Rc269 | CACAGGGGGCTTAAGTAGGTC | ACGGCAAACCAAATACTGTAAA | 150 | 52 | AY518492 |
| Split Rc630 | GGCATCATTATGCGGGAACAT | CCTCCTACAAGACCGACACC | 106 | 52 | AY518493 |
| Split Rc653 | AAGCATGCCGATGATTTACC | TGCAATTTCTTGAAGGCTTTT | 156 | 52 | AY518494 |
| Split Rc654 | TGCAAATCAAGGAGTAATGGTG | TTGCGTTATGCTTTGTATATTCG | 157 | 52 | AY518495 |
| Split Rc655 | GTCAAAACCGGAAATTGCAC | AAATGAAGGTTCGTTAAAAGCA | 103 | 52 | AY518496 |
| Split Rc704 | GAGACGATGGAATAGGAGTACTGAA | TTTTCTGCCCCAAATTTTTCC | 157 | 52 | AY518497 |
| Split Rc721 | TGAGACTCAAAGCCCTTATTTTTC | GGTGTTATTTCTTTATATCGCCAGT | 150 | 52 | AY518498 |
| Split Rc776 | GGAGGAGCTAAGGGAGCAAT | AAATTTCTTCAAATCGCTCAGG | 160 | 52 | AY518499 |
| Split Rc777 | TGGCTGATATAAAGGTAAGAAAACA | TCCTGCAATGTTTTTGTTTGA | 156 | 52 | AY518500 |
| Split Rc837 | GCTTGTGGTCTTGGTGTGG | AGCAAACATTTCGGTAACACC | 150 | 52 | AY518501 |
| Split Rc838 | GCCGAAAATGACGGTAAAAA | AGTGGAGCATGGTTTGCTGT | 156 | 52 | AY518502 |
| Split Rc839 | TTGGTTACAGCTGGAAATATGG | CACAAAGATATTTTACAAGAAAGTCA | 187 | 52 | AY518503 |
| Split Rc840 | GAGAAGCAGTTAATTGGACGTAAA | CAAGAAGCGCATCAAGATCA | 150 | 52 | AY518504 |
| Split Rc925 | ATTCCGGCATGGATGCTT | CCAAAGCACAAACCATAGAAAA | 155 | 52 | AY518505 |
| Split Rc1042 | GGTAGGGGCAAACAAAAGCTG | TGCCTTAAGTTTGAGTTGCTTGAA | 196 | 52 | AY518506 |
| Split Rc1100 | TGCTGTTTCTAGCTTAATGTG | CCTGTTTCTTTTTCCAACTTTTG | 159 | 52 | AY518507 |
| Split Rc1101 | TGCAGCTCAGGTTATTCATCA | TTAAAAGGTATGGTGCAAATAGAA | 160 | 52 | AY518508 |
| Split Rc1102 | AGGATTTAATTGGCGTCTTGC | TTTTTGTCGATATTTGCTTTTTCA | 150 | 52 | AY518509 |
| Split Rc1137 | CGCTTTCGTGAGAATGACAA | TCACCGTAGCTTCTCAAAGTTTC | 161 | 52 | AY518510 |
| Intergenic spacers | |||||
| nusG-rplK | CAGTTGCAATATTGGTAAAGCA | CAGCAGCTGGAATTATCAAGTT | 270 | 54 | AY345058 |
| rpoB-rpoC | CAGGCATTCCTGAATCATTT | TCCGTAAAAATTTACTACGCTCA | 257 | 54 | AY345057 |
| yqiX-gatB | CTGCGGCAGTACCGACTATT | ATCCGACGCTTGTGAATCAG | 258 | 54 | AY345059 |
| rrf-pyrH | GAGCTTTCTCCATCTTTTCTTG | AAAGGGGAATATACGACAATTGAG | 238 | 54 | AY345060 |
| RC0241-RC0242 | AGCTCAAATTGTGGTGTTTCC | GGGATCCCTATTACAGCAAAA | 343 | 54 | AY345061 |
| rne-coxW | CGGAAAAGAATGCAGAGTCTTG | CCATTTTGTAATTAAACTTTTCTGC | 244 | 54 | AY345062 |
| RC0432-hsIV | TGTGTGGAGTTAATGTATATTGCGAT | CGAGACTTGTCCATCTGCTG | 283 | 54 | AY345063 |
| asmA-rimM | TTAAAGGAATAAAGAAAGGAATAACAA | TTTTCAAATCAAACAACCGAAT | 281 | 54 | AY345064 |
| murG-RC0563 | GAAGAAAAGAAGGGCATAAGCTA | CAAGCTGAAAGTAAAAACATTCC | 293 | 54 | AY345065 |
| dnaN-RC0584 | TCGTCATGCCTGTTAAGGTG | TTGGATAATCACCCGCTAAGA | 354 | 54 | AY518297 |
| lig-tgt | TTTTTGTGCTTCCTCTTCAGAT | CCAAAATCTCATGAGCCGTA | 285 | 54 | AY345066 |
| rpsA-cmk | GCTGCAAGTTGTGGAACAAA | TTACCGGCTTCAGAGATGCTTT | 254 | 54 | AY345067 |
| Rho-RC0760 | CGGTQTTGTTAAGTTCTGCTGTG | TGCATGCCATTACTTATTACAAATG | 385 | 54 | AY345068 |
| folC-bioY | AGGTCGGCACCGGAAAAT | TACGGCGGCGTATTACCTT | 269 | 54 | AY354069 |
| tRNALeu-mgtE | AGCATTGAGGGTGCTGTTCT | TTCAGCAAATTGATCGTGATG | 267 | 54 | AY345072 |
| pth-rplY | TTCCTGGATTACCAAGACCAA | GAAGCTGAAGGGGAACAACA | 322 | 54 | AY518299 |
| ntrY-rpsU | AGCTGCTGTTGCTAAAGTAAAAA | CAAGAAGCAGCAAGAAGACAGA | 363 | 54 | AY345070 |
| cspA-tRNALeu | CGCCATTGTCCTGTTCAATA | TCCGTTATGTCTACCATTCCA | 375 | 54 | AY345073 |
| tmk-proP4 | TTCCCCTCCCTCAAATGTAA | CGGAGCAAGAAACCCATAAA | 258 | 54 | AY354074 |
| msbA2-RC1074 | TCGAAATATTTGCAGAGAGCAG | TGAGCTCGCGAAAGTTAGAA | 302 | 54 | AY518298 |
| proP5-RC1171 | TGCGTGATTTTGTTTGTTTCA | GCACGTAAAATGGGAAAGTGA | 261 | 54 | AY345075 |
| dksA-xerC | TCCCATAGGTAATTTAGGTGTTTC | TACTACCGCATATCCAATTAAAAA | 416 | 54 | See Fig. 4 |
| serS-virB4 | CGGATGTCTTGATAAATTACATGG | TCAAATTTTCGTAAACCACTAAACA | 344 | 54 | AY345076 |
| tRNAIle-pal | GCGTGCTCTAACCAACTGAG | GAAGAAGCTTTTGCCTATAATCG | 307 | 54 | AY345079 |
| pbpA2-RC0856 | AGGTTTCCATTTTTCCCAAA | CGAGTAGAGTGZZGGZTZCTCGATG | 343 | 54 | AY518300 |
| tRNAPhe-nifR3 | TTGAACCAACGACACAAGGA | CCGTAACACCTGACATTGGA | 251 | 54 | AY345080 |
| spo0J-abcT1 | AAAGATTTGGAAGAATTAGACTTGAT | TTTGCTTAAACCAACCATTTCA | 259 | 54 | AY345081 |
| RC0098-dcd | CCGATGCAAGGCAAATAATA | CGCAAAGGGCCTTATCATAC | 288 | 54 | AY345083 |
| RC0102-RC0103 | GCGATAAGCGATTTATTAGGC | GAAAGCCTAAAGCCTCCACA | 240 | 54 | AY345084 |
| tRNAfMet-RC0138 | GGTCGTTGGTTCAAATCCAG | AAGTCGTCATTGCGAGAAGG | 265 | 54 | AY521231 |
| 23S rRNA-5S rRNA | GTTGATAGGTCGGGTGTGGA | GGGATCGTGTGTTTCACTCA | 200 | 54 | AY345100 |
| mppA-purC | GCAATTATCGGTCCGAATG | TTTCATTTATTTGTCTCAAAATTCA | 160 | 54 | AY345089 (type A), AY345087 (type B), AY345086 (type C), AY345085 (type D), AY465118 (type E) |
| tRNAGly-tRNATyr | AGCTTGGAAGGCTGGAACTC | ATCCTTCTCCCTCCACCACT | 148 | 54 | AY345097 (type A), AY345099 (type B) |
| rpmE-tRNAfMet | TTCCGGAAATGTAGTAAATCAATC | TCAGGTTATGAGCCTGACGA | 144 | 54 | AY345091 (type A), AY345092 (type B) |
| fabZ-lpxD | TGTTAGGATCGATTTTAAGTACTCTATCT | TGGATTGGCATAGACAATCTATTA | 195 | 54 | AY518302 |
| fusA-tRNATrp | TGATCAAGTGCCGAGTCAAG | GCGCTCTACCAATTGAGCTAC | 149 | 54 | AY518303 |
| RC0669-RC0670 | TTTAATACCGTTAAACTTATCCAAGTG | TGTTCAACGCCATCATCTTC | 295 | 54 | AY518481 |
| RC0280-23S RNA | CAAAAAGCCGACAAAGCCTA | CCTTCATCGCCTTCTAGTGC | 258 | 54 | AY518484 |
| acrD-hupA | GGGCGTTTAATACAAATTTTAGACA | CAATTCTCCTTTGATAGGTTAATATGT | 388 | 54 | AY518473 |
| RC1137-tlc-5 | CGGGATAACGCCGAGTAATA | ATGCCGCTCTGAATTTGTTT | 264 | 54 | AY518475 |
| pal-RC1201 | TGCAAGCACACATAATGCAA | TCAAAATCGATTCCTCTTTTCC | 216 | 54 | AY518472 |
| lgt-RC073 | CCTCCGACTATTATGCCTATAACG | ATGACATTTCCTAATATCAATCCAA | 244 | 54 | AY518479 |
| udg-RC1213 | AATCCCACATATCCGCTACC | AGCCAAAGATAATGAAATCAGAA | 248 | 54 | AY518483 |
| secB-czcR | ATGCAGGATTCCAGCCTTTA | GGCTCGCCTTCAATTAACAA | 224 | 54 | AY518486 |
| RC0230-RC0231 | TGCACCCGCCTAAAACTAAC | ATGGTCGGCCGTAGAAAAA | 232 | 54 | AY518480 |
| groES-RC0970 | CTTGCATCGGCTTTTCTTTT | AGCTTTGAGCTGATGGGCTA | 215 | 54 | AY518487 |
| secA-prsA | GCAGGTTCAAGCGAGTTAATTT | AAAAGCAATACCGGAAAGCA | 209 | 54 | AY518485 |
| RC1282-fdxA | CATGCCCTCAGCAAATGATA | GGTTCTGTGAAGATTGCTAATTGA | 289 | 54 | AY518482 |
| RC0604-RC0605 | AAAGGCAATAACGGCAAAAA | AGCTCGCCAGTTCATTCATC | 353 | 54 | AY518474 |
| RC0409-trmU | AACCTTGACGTGCATATTCTAAA | GCCTGACATTGCGACAACTA | 270 | 54 | AY518477 |
| RC0272-gyrA | AACAAGAATAGAGCAGCGTTCA | TTTCATCTCATCTTCGATATTTACC | 368 | 54 | AY518478 |
| RC1027-xthA2 | GGTATGTAAATGAGCCTTATCAATACT | TCAGTAGTATAAGTAGCTCCTGCTGTC | 351 | 54 | AY518476 |
Intergenic spacer designations consist of the name of the 5′ ORF- the name of the 3′ ORF. ORFs encoding putative proteins of unknown function have designations beginning with RC and are numbered with reference to the R. conorii Malish (Seven) genome (GenBank accession no. NC_003103).
See reference 47.
See reference 49.
See reference 48.
See reference 51.
See reference 45.
FIG. 1.
Genome fragments selected for comparison of variability between R. prowazekii and R. conorii. Open boxes, ORFs present in both genomes; wavy lines, remnant in one Rickettsia species from a gene present in the other species; trisected box, gene split in R. conorii but not in R. prowazekii; solid lines between boxes, intergenic spacers exhibiting low interspecies variability; dashed lines between boxes, intergenic spacers with high interspecies variability.
FIG. 2.
Degree of intergenic spacer variability between R. conorii and R. prowazekii genomes as estimated by BLASTN scores. The most variable spacers exhibited low scores, whereas the most conserved spacers had high scores. White diamonds represent the spacers amplified and sequenced in our study that were not discriminatory among R. conorii strains; black diamonds represent the discriminatory spacers. Dashed horizontal line, BLASTN score of 75.
Rickettsial strains.
The 38 R. conorii strains studied were from the collection of our laboratory and are listed in Table 2. These strains had been isolated from ticks or clinical specimens from France, Spain, Portugal, Croatia, Russia, Turkey, Algeria, Tunisia, Morocco, Kenya, and Zimbabwe and had been identified as R. conorii by use of partial ompA sequencing (45). Sequences obtained from these 38 strains were also compared to the genome of R. conorii strain Malish (Seven) (accession number NC_003103). By comparison with R. conorii strain Malish (Seven) used for genome sequencing in 1999 (41), the R. conorii strain Malish (Seven) used in this study was the same strain but had been passaged in cell culture 60 times over the last 5 years. Overall, we compared the sequences from 39 R. conorii strains, including R. conorii strain Malish (Seven) with 2 different passage histories. We focused our study on strains of R. conorii sensu stricto.
TABLE 2.
R. conorii strains included in our study
| Straina | Origin | Geographical origin | Reference | Supplier or source |
|---|---|---|---|---|
| Moroccan, ATCC VR-141T | Unknown | Morocco | 6 | ATCCb |
| Malish (Seven), ATCC VR-613Tc | Human | South Africa | Geard | ATCC |
| M1 | Rhipicephalus sanguineus | Georgia, former Soviet Union | 22 | Gamaleya Institute, Moscow, Russia |
| Zim1 | Haemaphysalis leachi | Zimbabwe | 30 | P. J. Kelly |
| ZimA | Rhipicephalus simus | Zimbabwe | 30 | P. J. Kelly |
| Kenya | Haemaphysalis leachi | Kenya | 30 | G. Dasch |
| Spain96 | Human | Spain | 8 | N. Cardenosa |
| 16-B | Human | Spain | 8 | N. Cardenosa |
| SV9 | Human | Spain | N. Cardenosa | |
| Portugal4S | Human | Portugal | F. Bacellar | |
| Portugal454 | Human | Portugal | F. Bacellar | |
| Portugal821 | Human | Portugal | F. Bacellar | |
| URRCFrance1 | Human | France | ||
| URRCFrance2 | Human | France | ||
| URRCSpain3 | Human | Spain | ||
| URRCFranceFEe4 | Human | France | ||
| URRCFranceFEe5 | Human | France | ||
| URRCFranceFEe6 | Human | France | ||
| URRCFranceE7 | Human | France | ||
| URRCFranceFEe8 | Human | France | ||
| URRCFranceF9 | Human | France | ||
| URRCFrance10 | Human | France | ||
| URRCFranceFE11 | Human | France | ||
| URRCFranceFE17 | Human | France | ||
| URRCAlgeria18 | Human | Algeria | ||
| URRCFranceFEe25 | Human | France | ||
| URRCTunisia28 | Human | Tunisia | ||
| URRCCroatia29 | Human | Croatia | ||
| URRCFranceFEe31 | Human | France | ||
| URRCFranceFEe32 | Human | France | ||
| URRCFranceTick46 | Rhipicephalus sanguineus | France | ||
| URRCFranceFEe48 | Human | France | ||
| URRCFranceFEe49 | Human | France | ||
| URRCFranceFEe53 | Human | France | ||
| URRCFranceFEe57 | Human | France | ||
| URRCTurkey58 | Human | Turkey | ||
| URRCTurkey59 | Human | Turkey | ||
| URRCTurkey61 | Human | Turkey |
Strain designations include the country in which the patient was infected or from which the tick was collected.
ATCC, American Type Culture Collection.
Strain Malish (Seven) used for PCR amplification and sequencing in this study was the same as that used for genome sequencing in 1999 (GenBank accession no. NC_003103) but had undergone 60 cell culture passages since that date.
J. H. Gear, personal communication.
Rickettsiae were propagated at 32°C on Vero cell (ATCC CRL-1587) monolayers in Eagle's minimal essential medium (Seromed, Berlin, Germany) supplemented with 4% fetal bovine serum (Seromed) and 2 mM glutamine. When cells stained with Gimenez stain were heavily infected (3 to 5 days), the cultures were harvested, centrifuged (12,000 × g for 10 min), resuspended in Eagle's minimal essential medium, and stored at −70°C until further processing.
PCR amplification and sequencing.
The primers used to amplify the fragments described were obtained from Eurogentec (Seraing, Belgium) and are listed in Table 1. Their specificity was predicted by using the BLAST software (1). Genomic DNA was extracted from the rickettsial cultures by using the QIAamp tissue kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. PCRs were carried out in a PTC-200 automated thermal cycler (MJ Research, Waltham, Mass.). Two microliters of the DNA preparation was amplified in a 50-μl reaction mixture containing 50 pM each primer, 200 μM (each) dATP, dCTP, dGTP, and dTTP (Invitrogen, Gaithersburg, Md.), 1 U of eLONGase polymerase (Invitrogen), 2 μl of eLONGase buffer A, and 8 μl of eLONGase buffer B. The following conditions were used for amplification: an initial 3 min of denaturation at 94°C was followed by 40 cycles of denaturation for 30 s at 94°C, annealing for 30 s at various temperatures given in Table 1, and extension for 1 min at 68°C. Amplification was completed by holding the reaction mixture for 3 min at 68°C to allow complete extension of the PCR products. PCR products were purified by using a QIAquick Spin PCR purification kit (QIAGEN) as described by the manufacturer. Sequencing reactions were carried out by using the dRhodamine Terminator cycle sequencing ready reaction kit with Amplitaq polymerase FS (Perkin-Elmer, Coignieres, France) as described by the manufacturer. For all PCR products, sequences from both DNA strands were determined twice. Sequencing products were resolved by using an ABI 3100 automated sequencer (Perkin-Elmer). Sequence analysis was performed by using the ABI Prism DNA sequencing analysis software package (version 3.0; Perkin-Elmer). Sterile water was used as a negative control in each assay.
Sequence analysis.
In order to estimate “in silico” (by computer simulation) the relative variability of coding and intergenic sequences between the two genomes, we selected 102 pairs of putative orthologous intergenic sequences and 770 pairs of orthologous coding sequences that were reciprocal best hits with a significant BLASTN E-value of <0.001 and exhibited small size differences (<20%). Then we calculated the mean (± standard deviation [SD]) nucleotide sequence similarity for noncoding and coding sequences.
DNA sequences obtained in our study and those from the genome of R. conorii strain Malish (accession no. NC_003103) were aligned by using CLUSTALW software, version 1.81 (1). For split and remnant genes, for which the lengths of the sequences compared could be quite different, we considered consecutive gaps as a single mismatch. Percentages of similarity among sequences were determined by using the MEGA 2.1 software package (31). Phylogenetic relationships among R. conorii strains were inferred from both the intergenic spacer sequences and the multigene sequences by using the MEGA 2.1 software package (31). Distance matrices were determined under the assumptions of Kimura by using complete deletion analysis and were used to infer dendrograms by the unweighted pair group method with arithmetic means (UPGMA) available in the MEGA 2.1 software package (31).
Statistical analysis.
Student's t test was used to compare the nucleotide sequence similarity means of noncoding and coding sequences and of coding genes, split genes, remnant genes, and intergenic spacers. Multispacer typing (MST) and multigene typing were compared by using the χ2 test. STATA software (version 7.0; Stata Corporation, College Station, Tex.) was used for statistical analysis.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in GenBank (accession no. AY515514 to AY515518, AY518488 to AY518510, AY345057 to AY345070, AY345072 to AY345076, AY345079 to AY345081, AY345083, AY345084, AY518297 to AY518300, AY521231, AY345100, AY345085 to AY345087, AY345089, AY465118, AY345091, AY345092, AY345097, AY345099, AY518302, AY518303, AY518472 to AY518487, AY428738 to AY428750, AY462116, and AY497559).
RESULTS
In comparing the R. conorii and R. prowazekii genome sequences, we observed that the mean nucleotide sequence similarity (± SD) of the 25 variable spacers was 68.3% ± 7.8%. This was significantly lower than those of the 4 conserved coding genes (89.3% ± 7.8%; P < 10−2), the 23 split genes (83.1% ± 4.6%; P < 10−2), the 5 remnant genes (84.6% ± 7.5%; P < 10−2), and the conserved spacers (84.9% ± 2.3%; P < 10−2) but was similar to that of ompA (66.3%).
In the 38 strains of R. conorii for which we determined sequences in this study, the sequences of all 4 conserved coding genes, 23 split genes, and 5 remnant genes were identical to those of the genome of R. conorii strain Malish (Seven) (Table 1). Analysis of ompA sequences enabled us to identify three genotypes: one comprising 36 strains which had 100% similarity with the genome of R. conorii strain Malish (Seven), one containing R. conorii strain Moroccan, and one consisting of R. conorii strain M1 (Table 1). Overall, by using multigene typing with coding genes in R. conorii (16S rDNA, gltA, ompB, sca4, and ompA), we were able to classify the 39 strains of R. conorii we compared into three genotypes.
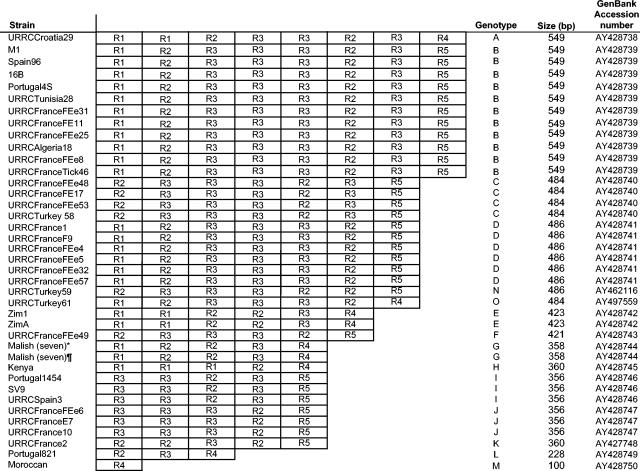
PCR amplification of the 52 intergenic spacers we studied in the 38 test strains of R. conorii yielded product sizes consistent with those of the genome of R. conorii strain Malish (Seven) (Table 1) except for the dksA-xerC spacer, for which the amplicon length ranged from 100 to 549 bp in different strains of R. conorii (Fig. 3). Only 4 spacers, all of which belonged to the 25 variable spacers (Fig. 2), had nucleotide differences in the R. conorii strains we studied: dksA-xerC, mppA-purC, rpmE-tRNAfMet, and tRNAGly-tRNATyr (Table 3). Seven nucleotide differences within the mppA-purC spacer enabled R. conorii strains to be classified into five genotypes (Table 3). By using differences at two positions in the rpmE-tRNAfMet spacer, strains could be classified into two genotypes (Table 3). The tRNAGly-tRNATyr spacer enabled the identification of two genotypes based on three nucleotide mutations. The dksA-xerC spacer was found to contain variable numbers of 63- to 102-bp repeat units designated R1 to R5, depending on the strain (Fig. 3 and 4). The percentage of nucleotide sequence similarity between repeats ranged from 50% between R1 and R4 to 98% between R1 and R2. Their G+C contents ranged from 11.8% for R5 to 17.5% for R3 (Fig. 3). Differences observed in the dksA-xerC spacers enabled us to classify the R. conorii strains we studied into 15 genotypes (Table 3). The number, type, and arrangement of repeat units for all strains tested are detailed in Fig. 4. The two batches of R. conorii strain Malish (Seven) exhibited identical spacer sequences. By combining the results obtained from analysis of the four variable spacers, we were able to identify 27 genotypes of R. conorii (Table 3). However, identification of these 27 genotypes was obtained by combining the results from the dksA-xerC, mppA-purC, and rpmE-tRNAfMet spacers only. The tRNAGly-tRNATyr-based typing did not provide any additional genotype. Therefore, this spacer was removed from the MST analysis. Twenty-three of the 27 genotypes contained only a single strain, while the remainder contained four strains (2 genotypes), three strains (2 genotypes), or two strains (1 genotype). MST identified a significantly greater number of genotypes than multigene typing (27 of 39 versus 3 of 39; P < 10−2).
FIG. 3.
Repeated sequences making up the dksA-xerC intergenic spacers of R. conorii strains.
TABLE 3.
Results of MST for R. conorii strains
| Straina | Genotype derived from:
|
|||
|---|---|---|---|---|
| dksA-xerC | mppA-purC | rpmE-tRNAfMet | MSTb | |
| URRCCroatia29 | A | B | B | 1 |
| 16B | B | B | A | 2 |
| M1 | B | B | B | 3 |
| Spain96 | B | B | B | 3 |
| URRCTunisia28 | B | B | B | 3 |
| URRCFranceFEe31 | B | B | B | 3 |
| Portugal4S | B | D | A | 4 |
| URRCFranceFEe8 | B | D | B | 5 |
| URRCFranceTick46 | B | D | B | 5 |
| URRCFranceFE11 | B | A | B | 6 |
| URRCAlgeria18 | B | A | B | 6 |
| URRCFranceFEe25 | B | A | B | 6 |
| URRCFranceFE17 | C | A | B | 7 |
| URRCFranceFEe48 | C | E | B | 8 |
| URRCFranceFEe53 | C | A | B | 9 |
| URRCTurkey58 | C | B | A | 10 |
| URRCFrance1 | D | A | B | 11 |
| URRCFranceFEe5 | D | A | B | 11 |
| URRCFranceF9 | D | A | B | 11 |
| URRCFranceFEe32 | D | A | B | 11 |
| URRCFranceFEe4 | D | B | B | 12 |
| URRCFranceFEe57 | D | A | A | 13 |
| ZimA | E | B | B | 14 |
| Zim1 | E | C | B | 15 |
| URRCFranceFEe49 | F | A | B | 16 |
| Malish (Seven)* | G | B | B | 17 |
| Malish (Seven)¶ | G | B | B | 17 |
| Kenya | H | B | B | 18 |
| URRCSpain3 | I | A | B | 19 |
| Portugal1454 | I | B | A | 20 |
| SV9 | I | B | B | 21 |
| URRCFranceFEe6 | J | A | B | 22 |
| URRCFranceFE7 | J | A | B | 22 |
| URRCFrance10 | J | A | B | 22 |
| URRCFrance2 | K | A | B | 23 |
| Portugal82I | L | B | A | 24 |
| Moroccan | M | B | B | 25 |
| URRCTurkey59 | N | B | A | 26 |
| URRCTurkey61 | O | B | A | 27 |
*, strain Malish (Seven) used for genome sequencing; ¶, strain Malish (Seven) with 60 more culture passages.
MST uses combined results from the three spacers.
FIG. 4.
Number and repartition of VNTRs in the dksA-xerC intergenic spacers of 39 R. conorii strains. *, strain Malish (Seven) used for genome sequencing; ¶, strain Malish (Seven) with 60 more culture passages.
By comparing coding and intergenic sequences between the two genomes in silico, we found that the 102 intergenic sequences studied had a mean nucleotide sequence similarity (± SD) of 79.4% ± 13.5%, significantly lower than that of the 770 open reading frames (ORFs) (89.1% ± 3.9%; P < 10−2).
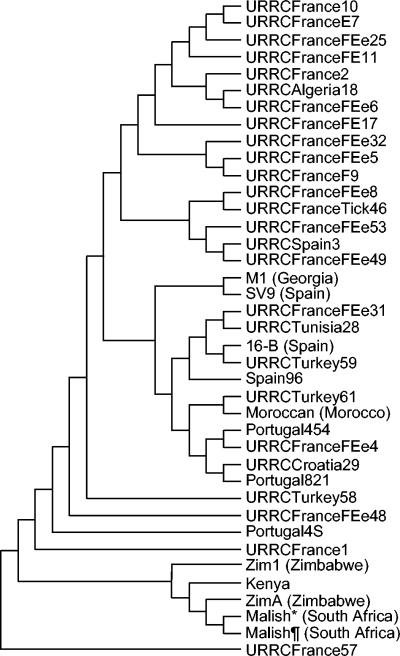
When sequences from the dksA-xerC, mppA-purC, and rpmE-tRNAfMet variable spacers were concatenated, three clusters could be differentiated. One contained all 5 sub-Saharan African strains; another was composed of 16 strains, mostly from France; and the third cluster included 13 strains from various geographical locations, including strains Moroccan and M1 (Fig. 5). Five strains of R. conorii did not group within any of the clusters.
FIG. 5.
Unrooted tree showing the phylogenetic relationships among the 39 R. conorii strains studied, inferred from sequence analysis of the combination of the dksA-xerC, mppA-purC, and rpmE-tRNAfMet intergenic spacers using the UPGMA method. The geographic origins of strains are given either in their names or in parentheses. *, strain Malish (Seven) used for genome sequencing; ¶, strain Malish (Seven) with 60 more culture passages.
DISCUSSION
We tested our hypothesis that the most suitable sequences for genotyping of bacterial strains are those that are found to be most variable when the genomes of two closely related bacteria are aligned. Using R. conorii and R. prowazekii, we found that the most variable sequences at the species level, variable intergenic spacers in this case, were also the most variable at the strain level and that suitable typing sequences were found in noncoding zones. Using a combination of sequences from three variable spacers in a multispacer tool, we identified 27 genotypes among the 39 strains of R. conorii we studied, including R. conorii strain Malish (Seven), for which we tested two batches with different passage histories.
Prior to our work, there was no genotyping method described for rickettsiae at the strain level. The development of a typing method for rickettsial strains has become crucial with the classification of R. prowazekii as a potential agent of bioterrorism. We used a rational technique rather than an empirical strategy to search for the most suitable genome fragments for this purpose. Comparison of the R. conorii and R. prowazekii genomes, which exhibit a high degree of colinearity, enabled us to compare the interspecies variability of coding genes, degraded genes, conserved intergenic spacers, and variable spacers. By in silico analysis, we found that variable intergenic sequences were more variable than coding genes, degraded genes, and conserved spacers (P < 10−2 in all cases). It has been suggested that intergenic spacer sequences are an important source of genome plasticity because they do not undergo selection pressure (13). For the rickettsiae, it has been suggested that most of the intergenic sequences of R. prowazekii and R. conorii consist of decayed genes that are no longer active but have not yet been totally eliminated from the genome (42). To date, the intergenic spacer used most widely in work with bacteria has been the 16S-23S rDNA spacer. In many bacterial species (24, 25, 32, 36, 46, 55) this spacer has been shown to have great variability, not only in its sequence and length but also in the number of alleles per genome (33, 53). However, studying the 16S-23S rDNA spacer of rickettsiae is not possible, because the 16S rDNA gene is separated from the 23S and 5S rDNA genes, which are tightly linked together (5).
When we compared the overall variability of intergenic spacers and coding sequences between the genomes of R. prowazekii and R. conorii, we found that spacers were significantly more variable than genes (P < 10−2). We were surprised, however, to observe that there were both highly variable spacers and spacers that were as conserved as coding sequences. R. conorii and R. prowazekii are estimated to have diverged from a common ancestor 80 million years ago (42), and it would seem probable that sequences which were not under selection pressure during this period would have been more variable than coding sequences. For prokaryotes, variations in the conservation of spacers has been reported, but the role of conserved spacers is incompletely understood (39, 44). For eukaryotes, recent studies on comparative genomics have shown that in yeasts (29) and mammals (11) some intergenic spacers are highly conserved at the species level. Some of these spacers include regulatory motifs, but the function of many remains unidentified. The factors responsible for the heterogeneity of intergenic spacers in rickettsial genomes have yet to be determined. Nevertheless, our results emphasize the importance of comparing genomes in order to select variable sequences instead of targeting sequences presumed to be variable.
At the intraspecies level, we confirmed that the 25 variable spacers we studied were the best targets, with 4 being highly variable and a combination of 3 enabling us to identify 27 genotypes among the 39 strains of R. conorii we studied. As predicted by in silico analysis, conserved genes were highly conserved among the R. conorii strains in our experiments. Only one of the coding genes we studied exhibited interstrain variability. This was ompA, which encodes a high-molecular-weight, surface-exposed protein and is one of the most variable coding genes in the spotted-fever-group rickettsiae (16). The variability we found in the gene was limited, however, and enabled us to differentiate only three genotypes. ompA-based genotyping was not congruent with MST, which classified strain M1 in a genotype with other strains. However, because the phylogenetic analysis using MST sequences was congruent with the geographic origins of strains, MST may be more relevant than ompA for R. conorii strain typing. Even when all five coding genes were used in multigene typing, there was less variability than that found with MST (P < 10−2). One could expect that genes which have undergone degradation since the divergence of R. conorii and R. prowazekii would exhibit higher levels of sequence divergence than conserved genes (3, 4) and thus would be better candidates for strain typing. We found, however, that split genes and remnant genes in R. conorii were also highly conserved and were not suitable for genotyping at the strain level. These findings support our strategy of selecting intergenic sequences with the greatest interspecies variability as targets for strain typing.
One of the 34 human isolates studied, URRCFranceFEe48, was obtained from a patient with malignant boutonneuse fever from Marseilles. Although it is likely that host factors play a key role in the development of severe forms of Mediterranean spotted fever (12), the specific role of strain variation in R. conorii has not been evaluated. The demonstration that this strain was a specific genotype by MST highlights the usefulness of our genotyping method for isolates associated with particular clinical presentations. In addition, the phylogenetic classification inferred from all four spacers was consistent with the geographic distribution of strains (Fig. 5).
In order to estimate the effect of culture passage on MST variation, we compared the intergenic sequences of two batches of R. conorii strain Malish (Seven) with different passage histories. No difference in spacer sequence was found between the two batches. Therefore, MST may be valuable for tracing rickettsial isolates from a single source with a difference in culture history of at least 60 passages.
Among the four variable spacers, the dksA-xerC spacer was composed of 63- to 102-bp repeat units (Fig. 3). One salient feature of the R. conorii genome is the high density of repeated sequences (40). Six hundred fifty-six interspersed repeated sequences, named Rickettsia palindromic elements (RPE), have been identified in R. conorii and represent 3.2% of the genome (2, 41, 42). Such interspersed repeats are usually confined to the intergenic regions of bacterial genomes (59), but in R. conorii the repeated sequences are also present within protein-coding regions (41). The repeats we found in the dksA-xerC intergenic spacer were highly conserved and were present only in this locus. Repeats representing a single locus and showing interindividual length variability are designated VNTRs (variable number of tandem repeats) (59). Changes in the number of repeats in a given genetic locus are an important source of DNA variability in eukaryotes (58). Such markers are well-established molecular targets for pedigree analysis in humans (27) and have also been used for bacteria (28). The VNTRs we found within the dksA-xerC intergenic spacer of R. conorii had two peculiar features. First, they occurred within an intergenic spacer, whereas VNTRs usually appear to be mainly involved in implementing size variation in cell wall- or membrane-associated proteins. This may cause enhanced or diminished exposure of active protein domains on bacterial surfaces (59). In Rickettsia species, VNTRs are known to occur within the ompA gene (59), and their number and arrangement vary in Rickettsia species (19). Second, the G+C content of the dksA-xerC VNTRs was low in contrast with that of the GC-rich RPEs common in intergenic regions of spotted-fever-group rickettsiae (3). In view of the fact that the overall G+C content of Rickettsia species is low, the dksA-xerC VNTRs may be remnants of a decaying rickettsial gene and not imported elements.
Our study demonstrated that in silico identification of the most variable sequences between two closely related bacterial genomes enables the selection of target sequences for strain genotyping. For rickettsiae, we demonstrated that intergenic spacers, in particular those that showed the greatest variability between the genomes of two closely related species, are more suitable for strain genotyping than coding sequences and degraded genes. The combined use of variable spacer sequences, which we named multispacer typing, is significantly more discriminatory than multigene sequencing. The advantages of MST include high discrimination, reproducibility, simplicity of interpretation (because one technique is used rather than a combination of techniques), and ease of incorporation of the data generated into databases that are directly comparable and readily shared by laboratories via the Internet. This technique may be applied for tracking isolates obtained from a wide variety of sources, including isolates from a single strain with different passage histories, and may even be applied directly to clinical specimens. Moreover, this technique may be applicable to other bacteria, in particular those considered potential agents of bioterrorism.
Acknowledgments
We thank Patrick J. Kelly for assistance with the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amiri, H., C. M. Alsmark, and S. G. Andersson. 2002. Proliferation and deterioration of Rickettsia palindromic elements. Mol. Biol. Evol. 19:1234-1243. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, J. O., and S. G. E. Andersson. 1999. Genome degradation is an ongoing process in Rickettsia. Mol. Biol. Evol. 16:1178-1191. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, J. O., and S. G. E. Andersson. 2001. Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol. Biol. Evol. 18:829-839. [DOI] [PubMed] [Google Scholar]
- 5.Andersson, S. G., A. Zomorodipour, H. H. Winkler, and C. G. Kurland. 1995. Unusual organization of the rRNA genes in Rickettsia prowazekii. J. Bacteriol. 177:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, E. J., and H. G. Stoenner. 1960. Immunologic relationships among the spotted fever group of rickettsias determined by toxin neutralization tests in mice with convalescent animal serums. J. Immunol. 84:171-182. [PubMed] [Google Scholar]
- 7.Bremer, B., K. Bremer, N. Heidari, P. Erixon, R. G. Olmstead, A. A. Anderberg, M. Kallersjo, and E. Barkhordarian. 2002. Phylogenetics of asterids based on 3 coding and 3 non-coding chloroplast DNA markers and the utility of non-coding DNA at higher taxonomic levels. Mol. Phylogenet. Evol. 24:274-301. [DOI] [PubMed] [Google Scholar]
- 8.Cardenosa, N., V. Roux, B. Font, I. Sanfeliu, D. Raoult, and F. Segura. 2000. Isolation and identification of two spotted fever group rickettsial strains from patients in Catalonia, Spain. Am. J. Trop. Med. Hyg. 62:142-144. [DOI] [PubMed] [Google Scholar]
- 9.Clark, C. G., T. M. Kruk, L. Bryden, Y. Hirvi, R. Ahmed, and F. G. Rodgers. 2003. Subtyping of Salmonella enterica serotype Enteritidis strains by manual and automated PstI-SphI ribotyping. J. Clin. Microbiol. 41:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demesure, B., N. Sodzi, and R. J. Petit. 1995. A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol. Ecol. 4:129-131. [DOI] [PubMed] [Google Scholar]
- 11.Dermitzakis, E. T., A. Reymond, N. Scamuffa, C. Ucla, E. Kirkness, C. Rossier, and S. E. Antonarakis. 2003. Evolutionary discrimination of mammalian conserved non-genic sequences (CNGs). Science 302:1033-1035. [DOI] [PubMed] [Google Scholar]
- 12.Dignat-George, F., H. Tissot-Dupont, G. Grau, L. Camoin-Jau, D. Raoult, and J. Sampol. 1999. Differences in levels of soluble E-selectin and VCAM-1 in malignant versus non-malignant Mediterranean spotted fever. Thromb. Haemost. 82:1610-1613. [PubMed] [Google Scholar]
- 13.Dobrindt, U., and J. Hacker. 2001. Whole genome plasticity in pathogenic bacteria. Curr. Opin. Microbiol. 4:550-557. [DOI] [PubMed] [Google Scholar]
- 14.Driscoll, J. R., P. J. Bifani, B. Mathema, M. A. McGarry, G. M. Zickas, B. N. Kreiswirth, and H. W. Taber. 2002. Spoligologos: a bioinformatic approach to displaying and analyzing Mycobacterium tuberculosis data. Emerg. Infect. Dis. 8:1306-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 16.Fournier, P. E., V. Roux, and D. Raoult. 1998. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int. J. Syst. Bacteriol. 48:839-849. [DOI] [PubMed] [Google Scholar]
- 17.Gielly, L., and P. Taberlet. 1994. The use of chloroplast DNA to resolve plant phylogenies: noncoding versus rbcL sequences. Mol. Biol. Evol. 11:769-777. [DOI] [PubMed] [Google Scholar]
- 18.Gielly, L., Y. M. Yuan, P. Kupfer, and P. Taberlet. 1996. Phylogenetic use of noncoding regions in the genus Gentiana L.: chloroplast trnL (UAA) intron versus nuclear ribosomal internal transcribed spacer sequences. Mol. Phylogenet. Evol. 5:460-466. [DOI] [PubMed] [Google Scholar]
- 19.Gilmore, R. D. 1993. Comparison of the rompA gene repeat regions of Rickettsiae reveals species-specific arrangements of individual repeating units. Gene 125:97-102. [DOI] [PubMed] [Google Scholar]
- 20.Goering, R. V. 2004. Pulsed-field gel electrophoresis, p. 185-196. In D. H. Persing, F. C. Tenover, J. Versalovic, Y. W. Tang, E. R. Unger, D. A. Relman, and T. J. White (ed.), Molecular microbiology: diagnostic principles and practice. ASM Press, Washington, D.C.
- 21.Goguet de la Salmoniere, Y. O., H. M. Li, G. Torrea, A. Bunschoten, J. van Embden, and B. Gicquel. 1997. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J. Clin. Microbiol. 35:2210-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golinevitch, H. 1960. A propos de la différenciation de quelques rickettsies du groupe de la fièvre pourprée à tiques. Arch. Inst. Pasteur Tunis 37:13-22. [Google Scholar]
- 23.Hanage, W. P., E. J. Feil, A. B. Brueggemann, and B. G. Spratt. 2004. Multilocus sequence typing: strain characterization, population biology, and patterns of evolutionary descent, p. 235-243. In D. H. Persing, F. C. Tenover, J. Versalovic, Y. W. Tang, E. R. Unger, D. A. Relman, and T. J. White (ed.), Molecular microbiology: diagnostic principles and practice. ASM Press, Washington, D.C.
- 24.Hassan, A. A., I. U. Khan, A. Abdulmawjood, and C. Lammler. 2003. Inter- and intraspecies variations of the 16S-23S rDNA intergenic spacer region of various streptococcal species. Syst. Appl. Microbiol. 26:97-103. [DOI] [PubMed] [Google Scholar]
- 25.Hill, K. E., C. E. Davies, M. J. Wilson, P. Stephens, M. A. Lewis, V. Hall, J. Brazier, and D. W. Thomas. 2002. Heterogeneity within the gram-positive anaerobic cocci demonstrated by analysis of 16S-23S intergenic ribosomal RNA polymorphisms. J. Med. Microbiol. 51:949-957. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmaster, A. R., C. C. Fitzgerald, E. Ribot, L. W. Mayer, and T. Popovic. 2002. Molecular subtyping of Bacillus anthracis and the 2001 bioterrorism-associated anthrax outbreak, United States. Emerg. Infect. Dis. 8:1111-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffreys, A. J., V. Wilson, S. L. Thein, D. J. Weatherall, and B. A. Ponder. 1986. DNA “fingerprints” and segregation analysis of multiple markers in human pedigrees. Am. J. Hum. Genet. 39:11-24. [PMC free article] [PubMed] [Google Scholar]
- 28.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 30.Kelly, P. J., and P. R. Mason. 1990. Serological typing of spotted fever group rickettsia isolates from Zimbabwe. J. Clin. Microbiol. 28:2302-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Department of Biology, Arizona State University, Tempe, Ariz. [DOI] [PubMed]
- 32.Maiwald, M., P. W. Lepp, and D. A. Relman. 2003. Analysis of conserved non-rRNA genes of Tropheryma whipplei. Syst. Appl. Microbiol. 26:3-12. [DOI] [PubMed] [Google Scholar]
- 33.McClelland, M., C. Petersen, and J. Welsh. 1992. Length polymorphisms in tRNA intergenic spacers detected by using the polymerase chain reaction can distinguish streptococcal strains and species. J. Clin. Microbiol. 30:1499-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meijer, A., S. A. Morre, A. J. van den Brule, P. H. Savelkoul, and J. M. Ossewaarde. 1999. Genomic relatedness of Chlamydia isolates determined by amplified fragment length polymorphism analysis. J. Bacteriol. 181:4469-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metzgar, D., E. Thomas, C. Davis, D. Field, and C. Wills. 2001. The microsatellites of Escherichia coli: rapidly evolving repetitive DNAs in a non-pathogenic prokaryote. Mol. Microbiol. 39:183-190. [DOI] [PubMed] [Google Scholar]
- 36.Mijs, W., P. de Haas, R. Rossau, T. Van der Laan, L. Rigouts, F. Portaels, and D. van Soolingen. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp. hominissuis ’ for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52:1505-1518. [DOI] [PubMed] [Google Scholar]
- 37.Motin, V. L., A. M. Georgescu, J. M. Elliott, P. Hu, P. L. Worsham, L. L. Ott, T. R. Slezak, B. A. Sokhansanj, W. M. Regala, R. R. Brubaker, and E. Garcia. 2002. Genetic variability of Yersinia pestis isolates as predicted by PCR-based IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD). J. Bacteriol. 184:1019-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musser, J. M. 1996. Molecular population genetic analysis of emerged bacterial pathogens: selected insights. Emerg. Infect. Dis. 2:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolaou, C., and Y. Almirantis. 2002. A study of the middle-scale nucleotide clustering in DNA sequences of various origin and functionality, by means of a method based on a modified standard deviation. J. Theor. Biol. 217:479-492. [DOI] [PubMed] [Google Scholar]
- 40.Ogata, H., S. Audic, C. Abergel, P. E. Fournier, and J. M. Claverie. 2002. Protein coding palindromes are a unique but recurrent feature in Rickettsia. Genome Res. 12:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogata, H., S. Audic, V. Barbe, F. Artiguenave, P. E. Fournier, D. Raoult, and J. M. Claverie. 2000. Selfish DNA in protein-coding genes of Rickettsia. Science 290:347-350. [DOI] [PubMed] [Google Scholar]
- 42.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 43.Raoult, D., and V. Roux. 1997. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10:694-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogozin, I. B., K. S. Makarova, D. A. Natale, A. N. Spiridonov, R. L. Tatusov, Y. I. Wolf, J. Yin, and E. V. Koonin. 2002. Congruent evolution of different classes of non-coding DNA in prokaryotic genomes. Nucleic Acids Res. 30:4264-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roux, V., P. E. Fournier, and D. Raoult. 1996. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J. Clin. Microbiol. 34:2058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roux, V., and D. Raoult. 1995. Inter- and intraspecies identification of Bartonella (Rochalimaea) species. J. Clin. Microbiol. 33:1573-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roux, V., and D. Raoult. 1995. Phylogenetic analysis of the genus Rickettsia by 16S rDNA sequencing. Res. Microbiol. 146:385-396. [DOI] [PubMed] [Google Scholar]
- 48.Roux, V., and D. Raoult. 2000. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int. J. Syst. Evol. Microbiol. 50:1449-1455. [DOI] [PubMed] [Google Scholar]
- 49.Roux, V., E. Rydkina, M. Eremeeva, and D. Raoult. 1997. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int. J. Syst. Bacteriol. 47:252-261. [DOI] [PubMed] [Google Scholar]
- 50.Sampson, S. L., R. M. Warren, M. Richardson, T. C. Victor, A. M. Jordaan, G. D. van der Spuy, and P. D. van Helden. 2003. IS6110-mediated deletion polymorphism in the direct repeat region of clinical isolates of Mycobacterium tuberculosis. J. Bacteriol. 185:2856-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekeyova, Z., V. Roux, and D. Raoult. 2001. Phylogeny of Rickettsia spp. inferred by comparing sequences of ‘gene D,’ which encodes an intracytoplasmic protein. Int. J. Syst. Evol. Microbiol. 51:1353-1360. [DOI] [PubMed] [Google Scholar]
- 52.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaver, Y. J., M. L. Nagpal, K. F. Fox, R. Rudner, and A. Fox. 2001. Variations in the 16S-23S rRNA intergenic spacer regions among Bacillus subtilis 168 isolates. Mol. Microbiol. 42:101-109. [DOI] [PubMed] [Google Scholar]
- 54.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stamm, L. V., H. L. Bergen, and R. L. Walker. 2002. Molecular typing of papillomatous digital dermatitis-associated Treponema isolates based on analysis of 16S-23S ribosomal DNA intergenic spacer regions. J. Clin. Microbiol. 40:3463-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taberlet, P., L. Gielly, G. Pautou, and J. Bouvet. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17:1105-1109. [DOI] [PubMed] [Google Scholar]
- 57.Tenover, F. C., R. D. Arbeit, and R. V. Goering. 1997. How to select and interpret molecular strain typing methods for epidemiologic studies of bacterial infections: a review for healthcare epidemiologists. Infect. Control Hosp. Epidemiol. 18:493. [DOI] [PubMed] [Google Scholar]
- 58.Turner, B. J., J. F. Elder, Jr., T. F. Laughlin, and W. P. Davis. 1990. Genetic variation in clonal vertebrates detected by simple-sequence DNA fingerprinting. Proc. Natl. Acad. Sci. USA 87:5653-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Belkum, A., M. Struelens, A. de Visser, H. Verbrugh, and M. Tibayrenc. 2001. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin. Microbiol. Rev. 14:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, Y. W., P. Y. Tai, Y. Chen, and W. H. Li. 2002. A study of the phylogeny of Brassica rapa, B. nigra, Raphanus sativus, and their related genera using noncoding regions of chloroplast DNA. Mol. Phylogenet. Evol. 23:268-275. [DOI] [PubMed] [Google Scholar]