Abstract
Recent studies show that the complex genetic architecture of schizophrenia (SZ) is driven in part by polygenic components, or the cumulative effect of variants of small effect in many genes, as well as rare single-locus variants with large effect sizes. Here we discuss genetic aberrations known as copy number variants (CNVs), which fall in the latter category and are associated with a high risk for SZ and other neuropsychiatric disorders. We briefly review recurrent CNVs associated with SZ, and then highlight one CNV in particular, a recurrent 1.6-Mb deletion on chromosome 3q29, which is estimated to confer a 40-fold increased risk for SZ. Additionally, we describe the use of genetic mouse models, behavioral tools, and patient-derived induced pluripotent stem cells as a means to study CNVs in the hope of gaining mechanistic insight into their respective disorders. Taken together, the genomic data connecting CNVs with a multitude of human neuropsychiatric disease, our current technical ability to model such chromosomal anomalies in mouse, and the existence of precise behavioral measures of endophenotypes argue that the time is ripe for systematic dissection of the genetic mechanisms underlying such disease.
Keywords: CNVs, genetics, schizophrenia, CRISPR, behavioral assays
I. Introduction
Overview of Schizophrenia
Approximately seven out of 1000 individuals (0.7%) are afflicted with schizophrenia (SZ) (McGrath et al., 2008), with symptoms being grouped into three categories: positive, negative, and cognitive. Onset is typically between the ages of 18-25 years, although presentation is about five years earlier for males than females (American Psychiatric AssociationAssociation, 2013). Positive symptoms, sometimes collectively referred to as “psychosis,” are characterized by hallucinations, delusions, conceptual disorganization, and thought disorder (Association, 2013). Negative symptoms are characterized by social withdrawal and lack of motivation (Andreasen, 1995, Lewis and Lieberman, 2000). Cognitive deficits are perhaps the most detrimental symptoms and include impaired attention, executive function, declarative memory (ability to recall and use known information), and working memory (ability to process and manipulate information) (Lewis and Lieberman, 2000, Perry et al., 2000, Lett et al., 2014). A vast array of pharmaceuticals is used to treat SZ, but the effectiveness of each drug varies, as shown by a meta-analysis that revealed heterogeneity in both drug response and side effects (Miyamoto et al., 2012, Leucht et al., 2013). While drug treatment can ameliorate the positive symptoms of SZ, the negative and cognitive symptoms remain recalcitrant (Makinen et al., 2008, Fumagalli et al., 2009, Hill et al., 2010, Miyamoto et al., 2012, Tcheremissine et al., 2012, Millan et al., 2016). Furthermore, a recent study suggests that low doses of medication combined with talk therapy and family support are more effective at improving outcome than drug treatment alone (Kane et al., 2015). These studies highlight the complications involved in treating SZ but hold out the hope that a better understanding of the disease's molecular pathology could improve treatment strategies.
Risk factors for SZ include obstetric complications, urban birth, immigrant status, young maternal age, advanced paternal age, and family history (Preti et al., 2000, Jablensky et al., 2005, Sorensen et al., 2014). Family history has a large effect size, with a ~10-fold increase in risk for those with an affected first-degree relative, suggesting a strong role for genetics in the etiology of SZ. Elegant studies of adoptive families in Denmark first confirmed the genetic risk (Kety et al., 1976, Kety et al., 1994). Of 14,000 children who were adopted away from their biological families between the years 1924-1947, 47 were diagnosed with SZ. Forty-seven control adoptees were also identified, and the rate of SZ was examined in both biological and adoptive relatives of SZ cases and control adoptees. The researchers found a 12.5-fold excess of SZ in the biological relatives of those with SZ, supporting genetics as a strong risk factor (Heston, 1966, Kety et al., 1971, Wender et al., 1974, Kety et al., 1976, Kety et al., 1994, Mulle, 2012).
Because genetic factors are a significant contributor to the development of SZ, collaborative efforts have focused on identifying genes disrupted in patients. The history of schizophrenia genetics is long, voluminous, and even infamous (Owen, 1992), with excellent reviews available elsewhere (Sullivan et al., 2012). Here we focus on the most recent findings, including recent genome-wide association studies (GWAS). Results from a 2008 GWAS used 738 cases and 733 controls but could uncover no loci significantly associated with SZ, suggesting this study was underpowered to detect common variants (Sullivan et al., 2008, Mulle, 2012). A larger GWAS conducted in 2009 by the International Schizophrenia Consortium unearthed the first evidence in support of SZ as a polygenic disorder (International Schizophrenia et al., 2009). This discovery prompted a wave of SZ GWAS with larger sample sizes. A 2013 study used a sample size of 21,246 cases and 38,072 controls and identified 13 new loci of interest (Ripke et al., 2013). A follow-up study in 2014 with a sample size of 36,989 cases and 113,075 controls identified 108 schizophrenia-associated single-nucleotide polymorphisms (SNPs) and pointed to multiple biological pathways involved in the onset of SZ (Schizophrenia Working Group of the Psychiatric Genomics, 2014). These data provided further support for polygenic influences in SZ.
In contrast to polygenic influences, copy number variants (CNVs, reviewed below) associated with SZ are rare, often contain multiple genes, and are associated with large effect sizes. While we discuss other CNVs associated with SZ, we focus more in depth on the 3q29 deletion due to the recent discovery of the significant risk it confers for SZ. In addition, we examine the possibility that SZ exists on a continuum with other psychiatric disorders. Finally, we go on to discuss both the molecular and behavioral tools that can be used to study CNVs, allowing us to learn more about the underlying biology of associated conditions.
II. Copy Number Variants
What Are CNVs?
CNVs are defined as a gain or loss of a segment of DNA greater than one kilobase (> 1 kb). Some CNVs may be larger than 100 kb, although CNVs of this size typically occur at low frequency in the population. Multiple genes and regulatory regions can be affected in CNVs of all sizes (Itsara et al., 2009, Dasouki et al., 2011, Torres et al., 2016). In 2008, a study revealed the increased presence of large, rare CNVs in schizophrenic patients versus controls (Walsh et al., 2008). These findings are consistent with the hypothesis that CNVs may be significant genetic risk factors for SZ. Additional studies with larger sample sizes led to the discovery of several recurrent CNVs (Table I) associated with SZ and other psychiatric disorders (International Schizophrenia, 2008, Stefansson et al., 2008, Moreno-De-Luca et al., 2010, Mulle et al., 2010, Mulle, 2012, Rees et al., 2014b). These SZ-associated CNVs share many characteristics. First, these CNVs result in multiple symptoms apart from neuropsychiatric disorders, among them congenital defects and craniofacial abnormalities (Hastings et al., 2009, Torres et al., 2016). Second, they cover large regions of the genome that contain many genes affecting multiple signaling pathways, cell types, and tissues. SZ-associated CNVs are also rare (<0.1% in the general population) and confer high risk for the onset of SZ; the increased risk for SZ associated with CNVs is many times the minimal risk associated with SNPs (Bodmer and Bonilla, 2008, Xu et al., 2008, Mulle, 2012). It is important to emphasize that these associations have been replicated by multiple studies, suggesting they are molecular targets worth dissecting to understand the biology of this disease.
Table I.
Recurrent CNVs Associated with SZ
| Location | Type of CNV | Psychiatric Disorder(s) | Additional Symptoms | Gene(s) of Interest | Refs |
|---|---|---|---|---|---|
| 1q21.1 | Deletion and duplication | ASD, ID, SZ | Microcephaly (deletion) macrocephaly (duplication) | PRKAB2 | (Torres et al., 2016) |
| 3q29 | Deletion | ASD, ID, SZ | Speech delay, facial dysmorphisms | DLG1, PAK2 | (Cox and Butler, 2015b, Glassford et al., 2016) |
| 7q11.2 | Duplication | ASD, ID, SZ | Speech delay | GTF2I | (Somerville et al., 2005, Velleman and Mervis, 2011, Escudero and Johnstone, 2014, Adamo et al., 2015) |
| 15q11.2 | Deletion | ASD, ID, SZ | Motor and speech delay | CYFIP1 | (Cox and Butler, 2015a) |
| 15q11.2-13.1 | Duplication | ASD, ID, SZ | Language impairment, facial dysmorphisms | HBII52 (a small nucleolar RNA) | (Nakatani et al., 2009, Stewart et al., 2011, Coppola et al., 2013) |
| 15q13.3 | Deletion | ASD, ID, SZ | Hypotonia, cardiac defect | CHRNA7 | (Ziats et al., 2016) |
| 16p11.2 | Duplication | ASD, ID, SZ | Decreased body mass index | DOC2A, TAOK2 | (Arbogast et al., 2016) |
| 16p13 | Duplication | ASD, ID, SZ | Heart defects | NDE1 | (Kuang et al., 2011, Ramalingam et al., 2011, Kimura et al., 2015, Torres et al., 2016) |
| 17p12 | Deletion | SZ | Neuropathy | PMP22 | (Kirov et al., 2009, Luigetti et al., 2014) |
| 17q12 | Deletion | ASD, ID, SZ | Kidney abnormalities | LHX1 | (Moreno-De-Luca et al., 2010, Palumbo et al., 2014, Clissold et al., 2016) |
| 22q11 | Deletion | ASD, ID, SZ | Congenital defects, facial dysmorphisms | COMT, PRODH | (Bassett and Chow, 2008, Biswas and Furniss, 2016) |
Multiple CNVs with their respective symptoms, characteristics, and corresponding references.
Recurrent CNVs, including those associated with SZ, arise due to the presence of repetitive sequences within the genome, known as low copy repeats (LCRs) (Sasaki et al., 2010). LCRs are stretches of DNA that are typically 10-500 kb (though their size can vary), with greater than 90% sequence identity to another place in the genome (Bailey et al., 2001). The presence of LCRs puts the genome at risk for chromosomal rearrangements that can cause CNVs, such as deletions and duplications (Stankiewicz and Lupski, 2002). These rearrangements occur because LCRs serve as substrates for non-allelic homologous recombination (NAHR), which is recombination between two highly similar DNA sequences that are not alleles (Shaw and Lupski, 2004). The result of NAHR depends on the orientation of the LCRs: if the repetitive sequences are in the same direction, NAHR causes a deletion or duplication, although not necessarily in similar proportions (Turner et al., 2008). Conversely, if the repetitive sequences are oriented in opposite directions, the result is an inversion (Shaffer and Lupski, 2000). A recent review provides further information regarding the mechanism of NAHR (Sasaki et al., 2010).
What Have CNVs Told Us About SZ?
Given the large size of many CNVs, it is difficult to pinpoint what in the region leads to the phenotype. One way to gain some insight into how CNVs confer high risk for SZ is to perform Gene Set Enrichment Analysis (GSEA). Briefly, GSEA is a statistical tool that allows one to determine the biological pathways associated with a phenotype based on the family of genes grouped with the disorder of interest (Subramanian et al., 2005). Recently, Kirov et al. used GSEA on gene sets assembled from proteomic analyses of the synapse and discovered significant enrichment of genes that code for proteins in the post-synaptic density (PSD) in de novo CNVs associated with SZ (Kirov et al., 2012). Specifically, genes encoding elements of the N-methyl-D-aspartic acid receptors (NMDAR) network (proteins that interact with NMDAR (NMDAR complex)) and proteins that interact with the cytoskeleton-associated protein (ARC) complex were significantly enriched. A replication study was conducted on 7,907 patients with SZ and 10,585 controls, and the results showed significant enrichment of genes of the NMDAR complex, but not the ARC complex, in the CNVs of schizophrenic patients (Kirov et al., 2012). A more recent study analyzed 623 schizophrenia trios (individuals with SZ and their unaffected parents) via exome sequencing and found that small, de novo mutations in NMDAR and ARC complexes were enriched in SZ patients (Fromer et al., 2014). This overlap in proteomic and genomic approaches implicates the disruption of networks associated with NMDARs or ARC as relevant to the disease mechanism behind SZ.
CNVs Associated with SZ
There are multiple recurrent CNVs associated with SZ and other psychiatric disorders, as summarized in Table 1. Here, we discuss three CNVs in particular, the 22q11 deletion, the 16p11.2 duplication, and the 3q29 deletion, due to the high risk they confer for SZ (Rees et al., 2014b). We go on to discuss the 3q29 deletion in more detail because recent data indicate it confers the greatest genetic risk for SZ (Mulle, 2015).
The most common of the recurring CNVs, the 22q11 deletion, occurs in approximately 1 in 4,000 live births. Most of these reported deletions are 3 Mb in size, which consists of ~60 genes; there is also a smaller recurrent deletion, 1.5 Mb consisting of 28 genes, that comprises the remainder of reported deletions. Both deletion sizes are associated with multiple symptoms, such as cardiac defects, learning difficulties, and facial dysmorphisms (Bassett and Chow, 2008, Drew et al., 2011, Torres et al., 2016). Additionally, this deletion is associated with multiple neuropsychiatric disorders, among them autism spectrum disorder (ASD) and intellectual disability (ID). Patients with this deletion are also at risk for SZ (Grozeva et al., 2012, Monks et al., 2014, Rees et al., 2014a, Rees et al., 2014b); notably, this CNV carries the second highest risk for developing the disease (Rees et al., 2014b, Kotlar et al., 2015).
Among the genes located in this region, two candidate genes are proposed to contribute to the SZ phenotype: catechol-o-methyl transferase (COMT) and proline dehydrogenase 1 (PRODH). COMT encodes the postsynaptic enzyme known to regulate the degradation of dopamine (Yavich et al., 2007); the SZ phenotype has been associated previously with dopamine deficits (Yamaguchi et al., 2015). PRODH encodes an enzyme responsible for glutamate production in the mitochondria. Loss of normal activity of PRODH has been linked to psychiatric disorders, such as SZ (Jacquet et al., 2002, Torres et al., 2016).
The 16p11.2 duplication spans a region of 600 kb and has a population prevalence of 1 in 1,000 (Blumenthal et al., 2014). This CNV is associated with a plethora of symptoms, including metabolic dysregulation and microcephaly (Maillard et al., 2015, Arbogast et al., 2016). In addition to SZ, this duplication is associated with neuropsychiatric disorders, such as ASD and ID (Arbogast et al., 2016).
Among the 28 genes in the region, there are multiple candidates that might contribute to the neuropsychiatric disorder (Arbogast et al., 2016). One of these genes is double C2 domain alpha (DOC2A). DOC2A regulates the release of Ca2+-dependent neurotransmitters, and mouse studies indicate that DOC2A functions in memory processes (Mochida et al., 1998, Sakaguchi et al., 1999, Arbogast et al., 2016). A second candidate gene in the 16p11.2 region is TAO kinase 2 (TAOK2). This gene is a kinase that has multiple roles, including the regulation of dendrite formation on cortical neurons (Arbogast et al., 2016); it is also an ASD susceptibility gene (de Anda et al., 2012).
A recently discovered CNV, the 1.6-Mb 3q29 deletion, was first reported in 2005 in six patients with ID and musculoskeletal abnormalities (Willatt et al., 2005). There are now more than 40 reported cases of individuals with this deletion. Most of the reported deletions are de novo, with an incidence rate of one in 30-40,000 births; however, inheritance has been documented (Digilio et al., 2009, Stefansson et al., 2014). While the symptoms of the deletion are heterogeneous, most patients experience varying degrees of ID and speech delay (Cox and Butler, 2015b); the deletion is also associated with anxiety, feeding problems during the first year of life, decreased weight at birth, and dental abnormalities (Glassford et al., 2016). The deletion was first reported to be associated with schizophrenia in 2010, and this association has since been replicated (Mulle et al., 2010, Levinson et al., 2011, Rees et al., 2014b, Szatkiewicz et al., 2014). A recent meta-analysis revealed that the 3q29 deletion increases the risk of developing schizophrenia by 41.1-fold (Mulle, 2015). This deletion is also linked to bipolar disorder (Clayton-Smith et al., 2010, Glassford et al., 2016, Green et al., 2016). Case reports connected the 3q29 deletion to ASD (Cobb et al., 2010, Quintero-Rivera et al., 2010, Sagar et al., 2013), and two recent reports confirm this association (Sanders et al., 2015, Glassford et al., 2016).
Within the 1.6-Mb deletion interval, there are 22 protein-coding genes, but none have been identified definitively as key to the neuropsychiatric sequelae (Mulle, 2015). The three genes in the interval nominated as candidates are: discs large 1 (DLG1), p21 protein (Cdc42/Rac)-activated kinase 2 (PAK2), and F-box protein 45 (FBXO45). Of these, DLG1 has generated the most interest. DLG1 is a scaffold protein (meaning its role is to assist in the localization and organization of multiple proteins) capable of interacting with subunits of NMDARs. Briefly, DLG1 interacts with GluN2 (a subunit of NMDARs) and is thought to assist in the maturation of NMDARs by localizing them to the Golgi complex, suggesting a function in synaptic signaling (Walch, 2013). In addition, there is clinical evidence implicating DLG1 as a candidate gene for SZ. In one study, multiple SNPs within DLG1 were identified in a cohort of Japanese males with SZ. Further statistical analyses indicated that one of those SNPs showed significant genotypic association within this cohort of patients (Uezato et al., 2012). In addition, Purcell et al. found an enrichment of DLG1 mutations in SZ patients using exome sequencing on a cohort of 2,536 cases and 2,543 controls (Purcell et al., 2014). With mounting evidence of its role in NMDAR function and the risk it confers in the onset of SZ, larger cohorts of SZ patients should be assessed for mutations in DLG1.
Until rigorous studies are done, it is strictly speculative to nominate candidate genes within the 22q11, 16p11.2, and 3q29 loci (and those mentioned in Table 1) as associated with SZ. History is littered with failures of the candidate gene approach to unravel complex neuropsychiatric disorders (Sullivan et al., 2012, Farrell et al., 2015). To avoid this, both in vivo and in vitro tools, discussed later in this review, are required to dissect which gene(s) contribute to the phenotype. Once accomplished, this work may lead to an understanding of the pathophysiology of these CNVs, which would give us powerful mechanistic insight into the etiology of SZ.
Neural Disorders on a Spectrum
One observation from CNV studies is that a single CNV may confer risk for multiple neuropsychiatric disorders—ASD, ID, and SZ (Figure 1)—suggesting the possibility of a shared etiology among these conditions (Guilmatre et al., 2009, Crespi and Crofts, 2012, Torres et al., 2016). The idea that these conditions may exist on a continuum is controversial, particularly among clinicians, but genetic evidence to support this hypothesis continues to mount. Here we briefly discuss the evidence that these disorders are not separate entities, but linked.
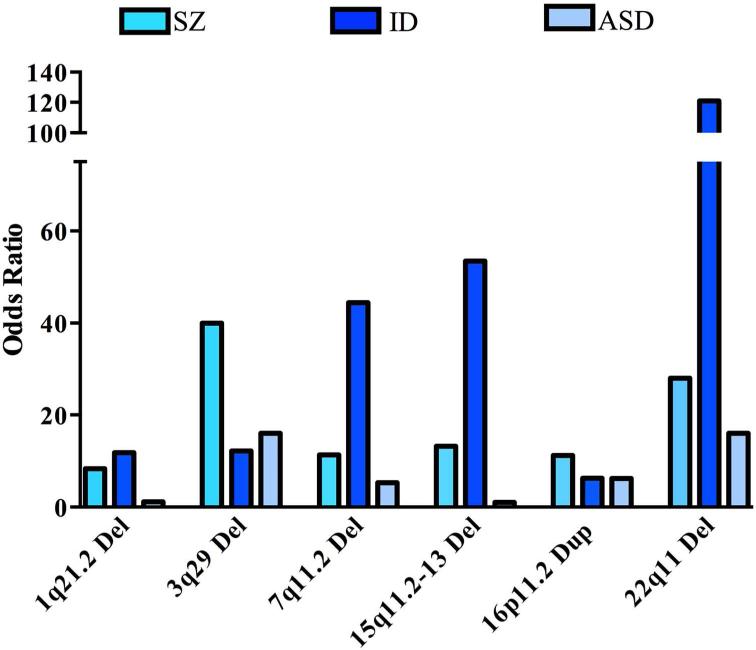
Figure 1. Risk for SZ, ID, and ASD associated with recurrent CNVs.
Odds ratios (ORs) for SZ were obtained from (Rees et al., 2014b, Mulle, 2015). ORs for ID were estimated using ID data from (Kaminsky et al., 2011), when zero CNV were observed in controls the OR was estimated by Haldane's correction (adding 0.5 to each cell in the 2×2 matrix). ORs for ASD were estimated from ASD case data (Sanders et al., 2015), n = 4687 cases) and controls (Cooper et al., 2011), n = 8329 controls) using Fisher's exact test in R; when zero CNV were observed in controls the OR was estimated by Haldane's correction.
Per the DSM-V, ASD is characterized by deficits in social communication and interaction, the presence of repetitive actions, and is often first observed during childhood (Association, 2013). The notion that ASD and SZ are linked is not new, as they were even believed to be one and the same prior to the 1970s (Chisholm et al., 2015). These disorders share several symptoms, including deficits in social reciprocity (reciprocal response in a conversation between two people). One striking difference between these disorders was thought to be psychosis, a specific hallmark of SZ but not ASD (Barneveld et al., 2011, Chisholm et al., 2015); however, multiple studies have shown that individuals diagnosed with ASD during childhood are at a higher risk of also developing psychosis later in life (Joshi et al., 2013, Sullivan et al., 2013). Indeed, in some studies up to 35% of ASD patients experienced psychosis (Konstantareas and Hewitt, 2001, Chisholm et al., 2015). There are also shared molecular pathways implicated separately in the cause of both ASD and SZ (Gao and Penzes, 2015). One such pathway involves the aforementioned NMDARs; genes encoding various NMDAR subunits are disrupted in several CNVs associated with both ASD and SZ. Genetically engineered mice with perturbations of NMDAR subunits show impaired social interaction and phenotypes corresponding to both ASD and SZ, respectively (Belforte et al., 2010, Won et al., 2012, Gao and Penzes, 2015). NMDARs, as well as other pathways, including mammalian target of rapamycin pathway (mTOR) signaling (Gao and Penzes, 2015), hint at molecular similarities between these disorders.
ID is another condition associated with both ASD and SZ. ID is a heterogeneous disorder defined as the inability of an individual to learn new skills, to comprehend new information, and to thrive independently (Ropers, 2008). Diagnosis typically occurs before age 18 years, and the severity of ID depends on a test of one's intelligence quotient (IQ). An IQ between 50-70 is considered mild ID, while an IQ less than 50 indicates severe ID. Studies have shown that roughly 40% of patients with ID also have ASD, and an estimated 50-80% of patients with ASD have ID (Brereton et al., 2006, Matson and Shoemaker, 2009). Furthermore, a separate study found that ID was present in approximately 4% of a cohort of SZ patients (Morgan et al., 2008). The common molecular pathway among the three diagnosed disorders is unclear, but many of the characterized CNVs are known to underlie ASD, ID, and SZ (Torres et al., 2016). Mutations in NRXN1, which maintains proper synaptic activity by modulating calcium, have been found in individual patients who have all three disorders (Vinas-Jornet et al., 2014). Another recent review also highlights the link between calcium signaling and neuropsychiatric disorders, such as ASD and SZ (Kotlar et al., 2015).
If these disorders are on a continuum, suffering from one disorder would suggest a person is more likely to suffer from the others, which seems to be supported by the current data. Confirming the co-occurrence of these three disorders will require large cohorts that can be followed longitudinally. Studying how this occurs in patients is necessary, but will be slow and expensive; however, modeling CNVs in mouse combined with behavioral studies and the use of patient-derived induced pluripotent stem cells (iPSCs) may allow us to dissect the mechanism of this continuum.
III. Tools to Study CNVs
The growing number of CNVs associated with neurodevelopmental and psychiatric disorders demands that we better understand how and why they confer risk, putting our efforts into modeling these CNVs in vivo. The ability to precisely manipulate the mouse genome to recapitulate specific chromosomal lesions enables straightforward generation of useful mouse models. Combined with careful behavioral phenotyping, the consequence of such lesions can be measured. In this section we review the power of using large deletions and duplications to understand disease etiology and focus on how mouse models are generated and behaviorally phenotyped. We also introduce the use of (iPSCs) as another means to understand genomic and molecular changes caused by a CNV, and to improve drug efficacy.
Chromosome Engineering Using Cre-loxP
Manipulation of the mouse genome to generate chromosomal deletions, duplications, inversions, and translocations has a rich history. Starting with George Snell's demonstration that the deleterious effects of X-rays could be inherited (Russell, 1954), classic mouse geneticists realized the power of using irradiation to induce deletions and study the resulting phenotypes. The approach evolved to include additional mutagens and to identify inversions and chromosomal translocations (Russell, 1951, Russell et al., 1989, Davisson et al., 1990, Stubbs et al., 1997, Yu and Bradley, 2001). Indeed, gamma irradiation induced the chromosome 16 duplication (syntenic to part of human chromosome 21) that first modeled Down syndrome (Davisson et al., 1990, Reeves et al., 1995). However, the challenge of using irradiation or chemical mutagens was the inability to control the breakpoints, making it hard to genetically mimic human disease. This challenge was met partly through studies that induced nested deletions, whose phenotypes could be compared to define critical regions (You et al., 1997, Schimenti et al., 2000, Naf et al., 2001). The ultimate solution came with the advent of Cre-loxP technology, which enabled targeting of a loxP site precisely at the desired chromosomal breakpoint (Lewandoski and Martin, 1997).
Cre-lox technology grew popular initially as a way to generate conditional alleles of single genes, enabling spatial or temporal control of deletion (Sauer and Henderson, 1988, Orban et al., 1992). Allan Bradley's group exploited the system for chromosome engineering. By sequentially targeting distinct loxP sites to two points on a chromosome, Cre-mediated recombination results in a deletion or inversion, depending on whether the loxP sites are in the same or opposite orientation. If the two loxP sites are targeted to different chromosomes, chromosomal translocations are possible. In reality, the efficiency of Cre recombining two loxP sites decreases as the distance between the two loxP sites grows. Thus, the Bradley group included a selectable marker whose gene was split between the two loxP sites requiring recombination for its intact expression. They chose to use the hypoxanthine phosphoribosyl transferase (Hprt) minigene split into its5’-Hp and 3’-rt halves. This approach enabled generation of mice with specific chromosomal anomalies using ES cells through six steps: 1) targeting of a 5'Hp-loxP construct, 2) targeting of a loxP-3'rt construct, 3) Cre transfection, 4) HAT selection (for recombined Hprt expression), 5) Southern blot and fluorescence in situ hybridization (FISH) analysis to confirm the desired chromosomal event occurred, and 6) blastocyst injection and chimera production (Yu and Bradley, 2001, Zheng et al., 2001), making for a labor-intensive and expensive, albeit feasible, approach. The Bradley lab greatly improved this approach by generating a resource of premade targeting vectors and mapping them throughout the genome so that researchers could order up the clones necessary for chromosomal engineering (Adams et al., 2004).
Chromosome engineering led to a series of mouse models that precisely mimicked the genetic architecture of human patients. In many cases the approach enabled the etiology of the disease to be linked to individual or small groups of genes. For example, the mouse model of the recurrent 22q11 microdeletion (DiGeorge syndrome) displayed congenital heart disease similar to patients, which was subsequently proven to be due to haploinsufficiency of the Tbx1 gene (Lindsay et al., 1999) (Lindsay et al., 2001). To dissect constitutional chromosome 17 deletions, three distinct mouse lines were made: a 6.9-Mb deletion, the reciprocal duplication, and a smaller 1.8-Mb deletion. Analysis of each line alone and subsequently in combination with each of the other lines made it clear that a small portion of the region led to the heart defects observed in patients, likely due to haploinsufficiency (Yu et al., 2006). Regulatory elements can be uncovered via chromosomal engineering, as well. By generating a large deletion covering most of the HoxD cluster and investigating what aspects of gene expression could be rescued with large human transgenes, Spitz and colleagues identified the location of enhancer sequences (Spitz et al., 2005). Taken together, chromosome engineering revolutionized our ability to molecularly dissect chromosomal regions and anomalies.
Chromosomal engineering has proved useful for generating mouse models of neurodevelopmental and neuropsychiatric disease. The 22q11 deletion mice revealed that Dgcr8, a gene that encodes a component of the microRNA biogenesis pathway, is a significant contributor to the SZ phenotype and not to the cardiac deficits. This highlights the power of deletion analysis to tease apart multiple mechanisms of disease. In addition, disruption of miRNA biogenesis affects genes throughout the genome, emphasizing the complexity underlying CNV-associated psychiatric disorders (Forstner et al., 2013). Patients with recurrent 15q11-13 duplication syndrome display symptoms that include autism, intellectual disability, seizures, and developmental delay. To genetically recapitulate the disease, researchers targeted the loxP sites in trans to generate a mouse carrying a 6.3-Mb duplication of the syntenic region on mouse chromosome 7. These mice exhibit an autistic endophenotype (aspects of a disorder that are quantitative and can be modeled in a rodent; see the SZ-Related Rodent Behaviors section of this review), providing an animal model to study this recurrent CNV (Nakatani et al., 2009). Down syndrome (DS) and monosomy 21, on the other hand, affect chromosomal regions with multiple syntenic regions in the mouse genome. Using Cre-lox, researchers have generated deletions on both mouse chromosome 10 and 17 to model the human CNV, 21q22.3 (monosomy 21). The mouse model displays reduced learning and memory function, consistent with the intellectual deficits in patients (Yu et al., 2010). Similarly, researchers generated duplications on mouse chromosomes 17, 16, and 10 to model the trisomic regions of DS. These mice are allowing distinct DS phenotypes, such as heart abnormalities, to be connected to discrete genomic regions (Liu et al., 2011). In the course of chromosome engineering, both deletions and duplications can be generated concordantly, depending on whether the loxP sites are in cis or trans. Indeed, researchers have created both 16p11.2 deletion and duplication mice and observed aspects of the disorder the same as in patients. The deletion mouse model, but not the reciprocal duplication mouse model, recapitulated the human disorder with an autistic endophenotype. This work underscores the challenge of genetic heterogeneity when using mouse models to study human disorders arising from recurrent CNVs (Horev et al., 2011).
Modulating Double-Strand Breaks to Generate CNVs
While the power of chromosomal engineering is undeniable, so is the technical élan it requires. Recent advances using nucleases appear to be lowering the technical barrier for chromosomal engineering. This transformation began with including transcription activator-like effector nucleases (TALENs) and zinc-finger nucleases (ZFNs), and these in turn are now being superseded by clustered regularly interspaced short palindromic repeats (CRISPRs). All three technologies depend on the same principle George Snell used: induction of double-strand breaks (DSBs) to induce rearrangements (Xiao et al., 2013, Wijshake et al., 2014). But, unlike irradiation and similar to Cre-loxP, all three permit specification of the breakpoint.
TALENs and ZFNs are similar genetic engineering tools in that both consist of a nuclease, which is fused to a protein that binds to the genetic locus of interest. Once bound to DNA, a DSB is generated. Repair of DSBs is often mediated via nonhomologous end joining (NHEJ), which can occur perfectly or imperfectly, resulting in the insertion or deletion of genetic material (indel)(Lee et al., 2012, Sander and Joung, 2014). Depending on the indel, there may be a functional consequence. When TALENs or ZFNs are targeted to two positions on a chromosome, a deletion or (less frequently) an inversion can occur. Indeed, both TALENs and ZFNs are sufficient to generate CNVs. In zebrafish, researchers used TALENs to generate both deletions and inversions up to 1 Mb in size (Xiao et al., 2013). In a human cell line, ZFNs were able to create a deletion and duplication of a 230-kb region, as well as a 15-kb inversion (Lee et al., 2012). Despite such proofs of principle, TALENs and ZFNs never became popular. While the exact reasons are unclear, here at the dawn of the CRISPR age, the issue is now moot.
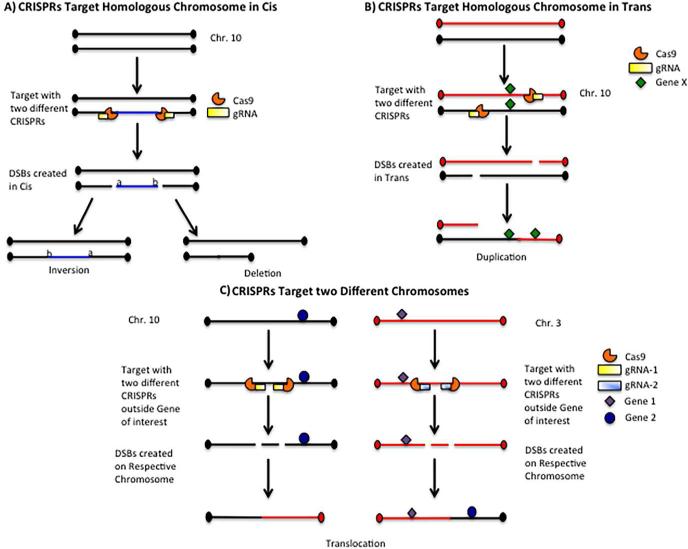
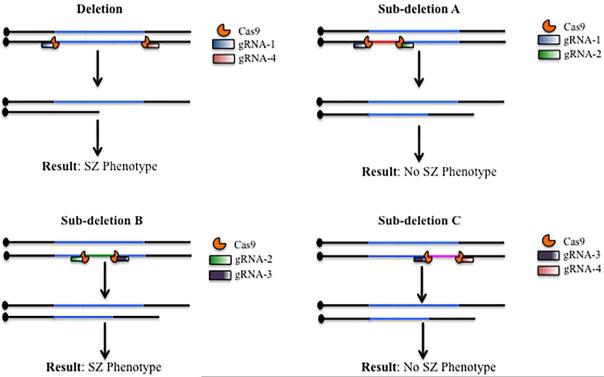
CRISPR is popular because the system is straightforward to design and optimize. CRISPR relies on a short sequence of RNA, known as the guide RNA (gRNA), to target the nuclease (originally Cas9) to the site of DNA cleavage to generate a DSB. The repair mechanisms are identical to those described earlier for TALENs and ZFNs: to create a deletion or inversion using CRISPR, two CRISPRs to the desired breakpoints are needed (Sander and Joung, 2014). If those two CRISPRs target the chromosomes in trans, a duplication is possible. Chromosomal translocations can be made by designing CRISPRs to appropriate breakpoints on distinct chromosomes. The design for creating a variety of CNVs using CRISPR is depicted in Figure 2. The same logic of the original geneticists who exploited deletion analysis can again be used to design nested deletions and define critical regions. Such subdeletions can share a breakpoint with a larger deletion or occur within the larger deletion (Figure 3).
Figure 2. Generating CNVs using CRISPR.
A. To generate an inversion or deletion, CRISPRs are designed at the desired breakpoints to create DSBs. The resulting inversion or deletion is generated only if the DSBs are created on the same homologous chromosome (cis).
B. If the DSBs are generated on different homologous chromosomes (trans), this may result in a duplication.
Generation of DSBs in cis or trans is random and cannot be controlled.
C. As an example of how to generate a translocation, two CRISPRs are needed. The first CRISPR is designed to create a DSB outside the first gene of interest on chromosome 3, and the second CRISPR is designed to create a DSB outside the other gene of interest on chromosome 10.
Figure 3. Generating subdeletions using CRISPR.
Designing subdeletions of a CNV is relatively simple. Briefly, the recurrent deletion is divided into three (or more) segments (A, B, C). Each of these segments is separately deleted using the corresponding CRISPR. Functional studies can then be done on each subdeletion to determine which segment contains the gene(s) that confer risk for a disorder, such as schizophrenia (SZ).
Other rearrangements are possible with this design, as outlined in Figure 2.
In terms of chromosome engineering in mice, CRISPR provides an unparalleled advantage over previous methods: CRISPRs and Cas9 nucleases can be directly injected in the one-cell zygote, eliminating the need for ES cell targeting or blastocyst injection. Within a month, the offspring can be screened directly, and even though some may be mosaic, the fact that editing occurs during the first few cleavages of the developing embryo increases the likelihood of germline transmission. Of critical note, the fact that a DSB is induced increases the risk of unwanted rearrangement, making Southern analysis necessary to ensure that only the desired manipulation occurred (Wang et al., 2013). In addition, FISH is well suited to confirm that the chromosomal rearrangement is correct (Zheng et al., 2001). Such controls will always be needed when manipulating the genome. The advantage of being able to directly edit the genome is that it allows chromosomal engineering to be performed more quickly and cheaply than ever before.
While most CRISPR-generated mouse alleles are for single genes, using two CRISPRs at a time can generate CNVs in vivo (Singh et al., 2015), including the generation of a 30-Mb deletion and the creation of a chromosomal translocation (Essletzbichler et al., 2014, Torres et al., 2014). This also includes the generation of an 11-Mb inversion that modeled a form of lung cancer driven by the EML4-ALK oncogene, recapitulating the human disorder and showing the use of ALK inhibitors blocked tumor formation (Maddalo et al., 2014). Similar manipulations can be created in vitro; one report used CRISPR to model the recurrent CNVs 16p11.2 and 15q13.3 in iPSCs. This study is proof of principle that the breakpoints of a recurrent CNV can be easily controlled using CRISPR and should be possible in mouse (Tai et al., 2016).
One of the initial concerns about CRISPR was off-target cleavage creating second-site mutations (Wang et al., 2013), a problem that has been well addressed with better algorithms for CRISPR target design with the advent of the new generation of nucleases (Kleinstiver et al., 2016, Slaymaker et al., 2016, Tsai and Joung, 2016). While there may still be some concerns in cell culture, when working in vivo, even if second-site mutations are induced, the founder mouse must be bred such that mutations will recombine out of the experimental cohort over time. Thus, researchers can be confident that the phenotype should be attributable to the CNV being interrogated.
There will surely be challenges using CRISPR for chromosomal engineering. In the past two decades, gene targeting demonstrated that some genomic regions are more difficult to manipulate, so it is possible that some CNVs may prove harder to model. In addition to NHEJ, DSBs can also be repaired through homology-directed repair (HDR) from a DNA template donor (Singh et al., 2015). Providing a donor of the desired outcome may make it possible to increase the frequency of a desired manipulation (Chen et al., 2011). Alternatively, one could imagine a hybrid approach using CRISPRs in ES cells and adapting the Hprt minigene selection, or finding an equivalent gene that could be split and then selected for in vivo. Such problems, as well as unforeseen ones, will surely be solved in time; the enormous amount of biology that the approach can reveal will drive solutions.
In terms of studying neuropsychiatric disease, CNVs can now be created in mouse with great precision for less money and in a shorter time than ever before. As explained in the next section, this is especially important when performing behavioral assays, which can be highly strain specific. Due to the relatively low cost and short timeframe of chromosomal engineering using CRISPR, it is reasonable to generate the same deletion (or other manipulation) on multiple strain backgrounds to dissect the mechanism of such complex diseases. CRISPR is now poised to revolutionize our ability to engineer chromosomes, and thus to understand CNV in human variation and in disease states.
SZ-Related Rodent Behaviors
Once a mouse model of SZ is created, it is imperative to choose behavioral tests that reflect this psychiatric disorder. While it is naïve to suppose that a “schizophrenic” mouse could be engineered to encompass all these symptoms, the SZ endophenotype can be modeled and tested individually in rodents. Endophenotypes are not themselves symptoms or signs of the disease, but are rather components of the disease that are quantitative and amenable to assessment in rodents in the laboratory. This reductionist approach has vastly improved our understanding of the mechanisms behind SZ and its potential treatment (McGrath et al., 2004). In the following section, we will address how behavioral tests are chosen that reflect SZ endophenotypes, and how these tests can act as correlates to SZ behavior.
One important aspect of modeling disease in animals is construct validity, which denotes a common underlying neurobiology in both the animal model and the human psychiatric disorder (Nestler and Hyman, 2010). Given the 40-fold increased incidence of SZ in patients with the 3q29 deletion, mice generated to carry the deletion would seem an ideal rodent model capable of providing powerful insight into the underlying genetics and neurobiology of SZ. In addition to construct validity, rodent models should strive to achieve face and predictive validity. Face validity is the extent to which a model “looks like the disease”; if the mouse 3q29 deletion recapitulates the behavioral features of SZ, it will have face validity. If treatments (e.g., antipsychotic drugs) that provide therapeutic benefit in humans with disease also ameliorate the endophenotypes observed in mice carrying the 3q29 deletion, the model then possesses predictive validity and can be used as a preclinical screen for novel compounds.
Mouse behaviors relevant to SZ endophenotypes can be categorized into three main symptom clusters: positive, negative, and cognitive (Powell and Miyakawa, 2006, Nestler and Hyman, 2010). The most common measure thought to reflect positive schizophrenic symptoms in rodents is hyperactivity. Increased locomotor activity at baseline or in a novel environment is seen in multiple proposed SZ rodent models (Lipska and Weinberger, 2000). This parallels the psychomotor agitation observed in some schizophrenic patients. In addition, psychotomimetic drugs, such as ketamine, phencyclidine, and amphetamine, which induce psychosis in human controls and exacerbate symptoms in SZ patients, induce hyperactivity and stereotypic behaviors in rodents, while rodent models of SZ have shown increased sensitivity to the locomotor-activating consequences of psychotomimetic drugs. The finding that SZ patients exhibit enhanced dopamine release following amphetamine injection (Kegeles et al. 2010) validates this assay as a preclinical assessment for rodent models of SZ. These results should be interpreted with caution, however, because hyperactivity is not a defining characteristic of SZ (Kegeles et al., 2010) and is not present in all patients.
Social withdrawal is one of the more debilitating aspects of SZ. This endophenotype is quantified in rats and mice by exposing them to a “stranger” rodent of the same species. There are multiple ways to assess social interaction in rodents, from allowing the stimulus rodent free movement throughout an area to confinement to a specific location. Whatever particular procedure is employed, the amount of time investigating the stimulus rodent(s) serves as the dependent variable. As a note of caution, if experimental subjects are tested in an apparatus other than their home cage, these sociability measures can be confounded by anxiety induced by a novel environment. In situations when this is a concern, subjects should either be tested in their home cage or undergo extensive habituation to the test enclosure. In addition to asociality, a subset of SZ patients suffers from anhedonia, or blunted affect. These negative symptoms can also be measured in rodent behavioral tests. Sucrose preference testing, in which subjects are allowed access to a sweetened solution and the amount consumed is recorded, as well as detailed examination of mouth and tongue movements following sucrose consumption, allow for quantification of motivation and reward. Intracranial self-stimulation, which uses voluntary operant responses to determine the sensitivity of the reward system, can also be employed to assess hedonic state. As is the case with many endophenotypes examined in rodent models of schizophrenia, these symptoms can occur in other psychiatric conditions and thus are not exclusive to SZ.
Cognitive symptoms of schizophrenia have also been assessed in rodent models of the disease. These cognitive endophenotypes can be subcategorized into deficits in working memory, declarative memory, and attention. Working memory can be measured in the eight-arm radial maze, delayed alternation in the T-maze, or spontaneous alternation in the Y-maze. These tasks require subjects to integrate their recent previous behavior into current choice selection. For example, in the eight-arm radial maze, re-entries into a food-baited arm after the reward has been found and consumed are identified as working memory errors. Working memory errors can also result from perseveration and a lack of behavioral flexibility. For this reason, it is difficult to determine whether performance deficits in these tasks are the result of impaired working memory or poor impulse control. This concern is somewhat offset by the knowledge that SZ patients also exhibit reduced behavioral flexibility and perseveration (executive function) (Crider, 1997). In addition to working memory, SZ is also associated with impairments in declarative memory (Saykin et al., 1991, Saykin et al., 1994, Heinrichs and Zakzanis, 1998). The eight-arm radial maze, Morris water maze, and Barnes maze all measure declarative spatial memory in rodents. These tasks require subjects to use the interrelationship between extra-maze cues to locate food reinforcement (radial maze) or escape from an aversive stimulus (Morris water maze and Barnes maze). For example, in the Morris water maze, subjects are trained to locate an invisible escape platform in a pool of water using spatial visual cues in the room. The latency and distance traveled to locate the platform over trials are the primary dependent variables for task acquisition. Recall memory can be assessed in a probe trial, during which the escape platform is removed and the amount of time spent in and around its former location are recorded. Adding an inter-trial interval and concurrent exposure to a previously encountered “stimulus” rodent, paired with a novel conspecific in the social interaction assays mentioned above, allow for assessment of social memory. Mice have an innate preference for novelty, and will normally spend more time interacting with a novel subject compared to a familiar subject, so an equal time spent investigating a novel and familiar animal can reveal social memory impairment. Of particular relevance to the current review, the DF(16)A+/− mouse model of 22q11.2 deletion exhibits strongly impaired social memory (Piskorowski et al., 2016), likely suggesting a similar deficit in mice carrying the 3q29 deletion.
The rodent behavioral task with the best-regarded face validity for assessing SZ endophenotypes is prepulse inhibition (PPI), a measure of sensorimotor gating, which is associated with impaired selective attention in SZ patients (Scholes and Martin-Iverson, 2010). This phenomenon is measured in humans and rodents by exposing subjects to an auditory startle stimulus. This startle stimulus is assessed in the presence or absence of a less intense non-noxious stimulus. In normal humans and control mice, the pre-pulse decreases the response to the startle stimulus. Importantly, SZ patients and some rodent models of SZ, including 22q11.2 microdeletion, demonstrate a deficit in PPI, signifying preattention processing deficits (Prasad et al., 2008). Latent inhibition is a related process in which exposure to a stimulus decreases future associations formed with that stimulus. SZ patients are generally resistant to latent inhibition, particularly when acutely symptomatic (Weiner, 2003, Lubow, 2005), as are some mutant mice models of the disorder (Zeng et al., 2001, Rimer et al., 2005). Additional cognitive deficits commonly seen in SZ patients include attention and executive function deficiencies. These processes can also be modeled by rodent behavioral tasks. The Wisconsin Card Sorting (WCS) task reveals attentional set shifting problems in schizophrenic patients (Owen et al., 1993). Rats and mice can be trained to use odor and texture cues to discriminate for food reinforcement. In a manner similar to the WCS, the rules of the discrimination can be changed during testing, such that subjects must shift their attention amongst the cues to continue receiving reward. The five-choice serial reaction time task (5-CSRTT) requires subjects to attend to five separate locations for presentation of a visual stimulus. They can then report detection of the stimulus by performing an operant response for food reinforcement. The 5-CSRTT measures spatial attention but can also detect impulsivity and perseverative behavior (Robbins, 2002, Chudasama and Robbins, 2004). Very recently, the Novel Methods leading to New Medications in Depression and Schizophrenia (NEWMEDS) initiative developed and validated a battery of tasks intended to examine cognitive SZ endophenotypes (Hvoslef-Eide et al., 2015). The battery includes the 5-CSRTT and an additional test of attention, as well measures of cognitive flexibility, spatial working memory, and long-term memory. The entire test battery can be preformed in the same touchscreen operant chambers, allowing for high-throughput characterization of SZ-associated endophenotypes, and represents the vanguard of behavioral phenotyping. Importantly, the NEWMEDS battery is fully automated, eliminating or reducing confounds produced by experimenter bias or subject anxiety as a result of extensive investigator handling and intervention. Task automation is recommended whenever possible, and fortunately most if not all of the SZ-related tasks described earlier are capable of automation with a video tracking system, and software-controlled and -monitored operant and startle chambers. These tools allow experimenters to analyze multiple different maze and conditioning environments, providing for automated scoring of cognitive tasks, such as the Morris water maze and PPI. Even more advanced tracking systems allow assessment of multiple subjects in the same apparatus, fully automating measurement of social behavior, as well.
When testing mice for behavioral abnormalities, challenges are always likely. One potential challenge is that some phenotypes will be more robust on different strain backgrounds. To overcome this, it is beneficial to study a CNV on more than one background strain. As examples, mice with poor eyesight (e.g., adult FVB mice have retinal degeneration) or coordination have trouble performing in the Morris water maze task, mice with high levels of anxiety typically display low levels of social interaction, and mice with auditory deficiencies can appear deficient in PPI; using mice with overt alterations in brain morphology (e.g., 129 mice have a partial deficit in corpus callosum formation) may introduce more variability in some tests (Wahlsten, 1982, Chang et al., 2002). Thus, it is critical to have proper controls and alternatives for each behavioral test. Many of these controls are built into the test, such as eyesight (visible platform in Morris water maze) and hearing (baseline startle in PPI). It is also important for the studies to be powered adequately to account for intra-strain variability in behavioral response. Thus, testing 8-16 mice per genotype is recommended (depending on whether or not males and females are combined in the analysis), which in the literature is more than sufficient to uncover statistically significant differences between genotypes, even in mixed genetic background populations (Weinshenker et al., 2002, Marino et al., 2005, Grubb et al., 2009). It is also important to note that not all symptoms of SZ are present to the same degree in all patients, indicating that genetic modifiers regulate the expression of these symptoms. Because baseline performance in cognitive and behavioral tasks is highly variable between different strains of mice, the choice of strain background is a critical factor for experimental success. For example, it might be difficult to observe a deficit in PPI caused by the deletion on a strain background that would have difficulty performing PPI due to hearing difficulties. As such, one might choose to investigate the phenotypic consequences of the deletion on four strains of mice initially, and then pare down to one or two strain backgrounds that give the most robust phenotypes for subsequent analysis of subdeletions of the interval and single-gene knockouts. A good resource for understanding which strain to choose for behavioral assays is the Mouse Phenome Database (http://phenome.jax.org), which provides a compilation of mouse behavioral data based on strain background.
As discussed, manipulation of the mouse genome in combination with behavioral assays provides a powerful means to study CNVs associated not only with SZ, but other psychiatric disorders, as well. Combining the ease of using the mouse model in behavioral studies with the availability of tools to investigate the mouse genome will move the field towards a better understanding of causes of the CNV. Coupled with pharmacological studies that can be done on mice carrying these recurrent CNVs, we are more likely to find better ways of alleviating some of the symptoms of SZ.
Using iPSCs to Study CNVs
Another tool to understand how CNVs confer risk for a psychiatric disorder is induced pluripotent stem cells (iPSCs). Briefly, fibroblasts or PBMCs from a patient are isolated and treated with a cocktail of transcription factors (SOX2, OCT4, KLF4, and c-MYC). Treatment with these factors reprograms the cells into iPSCs, which can then be differentiated into the cell type of interest, such as neurons (Takahashi et al., 2007). The neurons are then characterized using a series of analyses, among them cellular morphology, electrophysiology, and molecular characterization (Wen et al., 2014). Thus, the use of iPSCs provides a means to study potential neuronal dysfunction caused from CNVs. For example, iPSCs have been derived from a series of patients with either the deletion or duplication at the 7q11.23 locus. The deletion is the cause of the well-known Williams-Beuren syndrome, and the duplication is also associated with both schizophrenia (Mulle et al., 2014) and autism (Sanders et al., 2015). Study of iPSCs resulted in the identification of additional candidate genes outside the breakpoints that may contribute to the phenotype (Adamo et al., 2015, Urban and Purmann, 2015). In another study using iPSCs from patients with SZ-associated DISC1 mutations, abnormal synaptic signaling was observed, providing molecular insight into the cause of SZ (Wen et al., 2014).
The future is promising for the use of iPSCs in the study of CNVs and psychiatric disorders. As techniques continue to improve, the next steps will include comprehensive, coordinated analysis of iPSCs from multiple patients containing different SZ-associated CNVs for cross-comparison and potential identification of common molecular mechanisms. As mentioned in the CNV section of the review, NMDARs and ARC are two protein families found enriched in CNVs associated with SZ. The use of iPSCs is an ideal system to determine whether the expression of these protein families (or others) is altered in the respective CNV. The discovery of altered biological pathways in the various CNVs could then lead to personalized treatment for the patient groups.
V. Conclusion
Extensive collaboration is necessary for the next phase of SZ research. Collective efforts are what led to the discovery of SZ-associated CNVs, and it will take a comparable effort to uncover the disease mechanism, with the ultimate goal of effective treatments. The first step in this challenging endeavor is to understand how each of these CNVs results in SZ on a molecular level. Modeling the effect of CNVs in animals and iPSCs can be invaluable tools to identify the key genes within CNV intervals that contribute to neurodevelopmental phenotypes and further correlate genotype and phenotype. With the development of CRISPR, not only is generating these CNVs conceivable, but so is generating a series of smaller deletions/duplications for each CNV to narrow the interval and specify which molecular targets contribute to the phenotypes. One resource that may help us find the gene that correlates to the respective phenotype is mousephenotype.org, a database of information on knockout mouse models, created in the hope of one day assigning a biological function to each gene in the mouse genome.
The database also includes behavioral assay results from knockout mice, providing yet another tool for unlocking the molecular complexity of a CNV. Finally, cross-CNV comparisons will be necessary to unravel relevant biological pathways. This is especially true for the 3q29 deletion and 22q11 microdeletion. These two CNVs account for only 0.082% and 0.29%, respectively, of all SZ cases (Rees et al., 2014b); however, of all the CNVs associated with SZ, they confer the highest risk (Grozeva et al., 2012, Mulle, 2015). It is possible that an understanding of how they confer such a high risk for SZ at a molecular level could lead to understanding of the mechanisms of SZ. While the 22q11 deletion has been dissected extensively, there is still more to learn (Forstner et al., 2013); the molecular mechanism of the 3q29 deletion is not known. The better we understand SZ at the molecular level, the more likely we are to find effective treatments.
The discovery of CNVs that cause neuropsychiatric disorders has led to the notion of an etiologic spectrum among ASD, ID, and SZ. Other genetic factors, or modifier loci, for pathogenesis are likely to be important. In the most severe case, some individuals have a “second hit” variant that influences penetrance at the primary CNV (Veltman and Brunner, 2010, Girirajan et al., 2012). With the advent of routine whole-genome sequencing for clinical indications, future studies in humans will investigate genomic background when assessing risk. Mouse models will also help us understand the contribution of background genomic influences associated with a given CNV. This will require the creation of multiple genetic mouse models, generating an identical CNV on different genetic strains. Although this is no small task, the use of CRISPR and standardized behavioral assays put answers within reach. This type of work, albeit involved, might also provide in vivo evidence to support the continuum model. With the tools outlined in this review, the future promises exciting new research that will help us understand the molecular mechanisms behind CNV-associated neuropsychiatric phenotypes, with the ultimate goal of effective treatment for patients’ symptoms.
Significance.
Copy number variants (CNVs) are large genomic aberrations that confer risk for multiple psychiatric disorders, including SZ. Most CNVs affect the function of multiple genes, making it difficult to understand the molecular mechanism of these CNVs. Cre-loxP is one technique that has proved successful to model CNVs and identify individual genes that contribute to a phenotype; however, this genetic tool, while powerful, is both time-consuming and expensive. Fortunately, with recent advances in genomic engineering via the use of CRISPR technology, it is now feasible to dissect CNVs and identify individual genes contributing to the phenotypes of psychiatric disorders.
Acknowledgements
The authors would like to thank Sarah Bay and Cheryl Timms Strauss for editing and the NIGMS for funding (R01 GM097331).
Footnotes
Contributions:
T. R., J.S., and G.G. were the main authors of the review. S.W. advised on projects that contributed to the content of this review. D.W. edited and revised the molecular and behavior content of the review. T.C. edited and revised the genetic content of the review. J.M. edited and revised the clinical and medical content of the review. J.M. generated Figure 1; T.R. generated Figures 2 and 3 with assistance from T.C.
Conflict of Interest:
The authors have nothing to disclose.
References
- Adamo A, Atashpaz S, Germain PL, Zanella M, D'Agostino G, Albertin V, Chenoweth J, Micale L, Fusco C, Unger C, Augello B, Palumbo O, Hamilton B, Carella M, Donti E, Pruneri G, Selicorni A, Biamino E, Prontera P, McKay R, Merla G, Testa G. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat Genet. 2015;47:132–141. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- Adams DJ, Biggs PJ, Cox T, Davies R, van der Weyden L, Jonkers J, Smith J, Plumb B, Taylor R, Nishijima I, Yu Y, Rogers J, Bradley A. Mutagenic insertion and chromosome engineering resource (MICER). Nat Genet. 2004;36:867–871. doi: 10.1038/ng1388. [DOI] [PubMed] [Google Scholar]
- Andreasen NC. Symptoms, signs, and diagnosis of schizophrenia. Lancet. 1995;346:477–481. doi: 10.1016/s0140-6736(95)91325-4. [DOI] [PubMed] [Google Scholar]
- Arbogast T, Ouagazzal AM, Chevalier C, Kopanitsa M, Afinowi N, Migliavacca E, Cowling BS, Birling MC, Champy MF, Reymond A, Herault Y. Reciprocal Effects on Neurocognitive and Metabolic Phenotypes in Mouse Models of 16p11.2 Deletion and Duplication Syndromes. PLoS Genet. 2016;12:e1005709. doi: 10.1371/journal.pgen.1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Association AP. Diagnostic and Statistical Manual of Mental Disorders. American Psychiatric Publishing; Arlington: 2013. [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barneveld PS, Pieterse J, de Sonneville L, van Rijn S, Lahuis B, van Engeland H, Swaab H. Overlap of autistic and schizotypal traits in adolescents with Autism Spectrum Disorders. Schizophr Res. 2011;126:231–236. doi: 10.1016/j.schres.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Bassett AS, Chow EW. Schizophrenia and 22q11.2 deletion syndrome. Curr Psychiatry Rep. 2008;10:148–157. doi: 10.1007/s11920-008-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belforte JE, Zsiros V, Sklar ER, Jiang Z, Yu G, Li Y, Quinlan EM, Nakazawa K. Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nat Neurosci. 2010;13:76–83. doi: 10.1038/nn.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas AB, Furniss F. Cognitive phenotype and psychiatric disorder in 22q11.2 deletion syndrome: A review. Res Dev Disabil. 2016;53-54:242–257. doi: 10.1016/j.ridd.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Blumenthal I, Ragavendran A, Erdin S, Klei L, Sugathan A, Guide JR, Manavalan P, Zhou JQ, Wheeler VC, Levin JZ, Ernst C, Roeder K, Devlin B, Gusella JF, Talkowski ME. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am J Hum Genet. 2014;94:870–883. doi: 10.1016/j.ajhg.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brereton AV, Tonge BJ, Einfeld SL. Psychopathology in children and adolescents with autism compared to young people with intellectual disability. J Autism Dev Disord. 2006;36:863–870. doi: 10.1007/s10803-006-0125-y. [DOI] [PubMed] [Google Scholar]
- Chang B, Hawes NL, Hurd RE, Davisson MT, Nusinowitz S, Heckenlively JR. Retinal degeneration mutants in the mouse. Vision Res. 2002;42:517–525. doi: 10.1016/s0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm K, Lin A, Abu-Akel A, Wood SJ. The association between autism and schizophrenia spectrum disorders: A review of eight alternate models of co occurrence. Neurosci Biobehav Rev. 2015;55:173–183. doi: 10.1016/j.neubiorev.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Chudasama Y, Robbins TW. Psychopharmacological approaches to modulating attention in the five-choice serial reaction time task: implications for schizophrenia. Psychopharmacology (Berl) 2004;174:86–98. doi: 10.1007/s00213-004-1805-y. [DOI] [PubMed] [Google Scholar]
- Clayton-Smith J, Giblin C, Smith RA, Dunn C, Willatt L. Familial 3q29 microdeletion syndrome providing further evidence of involvement of the 3q29 region in bipolar disorder. Clin Dysmorphol. 2010;19:128–132. doi: 10.1097/MCD.0b013e32833a1e3c. [DOI] [PubMed] [Google Scholar]
- Clissold RL, Shaw-Smith C, Turnpenny P, Bunce B, Bockenhauer D, Kerecuk L, Waller S, Bowman P, Ford T, Ellard S, Hattersley AT, Bingham C. Chromosome 17q12 microdeletions but not intragenic HNF1B mutations link developmental kidney disease and psychiatric disorder. Kidney Int. 2016;90:203–211. doi: 10.1016/j.kint.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb W, Anderson A, Turner C, Hoffman RD, Schonberg S, Levin SW. 1.3 Mb de novo deletion in chromosome band 3q29 associated with normal intelligence in a child. Eur J Med Genet. 2010;53:415–418. doi: 10.1016/j.ejmg.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola A, Ruosi P, Santulli L, Striano S, Zara F, Striano P, Sisodiya SM. Neurological features and long-term follow-up in 15q11.2-13.1 duplication. Eur J Med Genet. 2013;56:614–618. doi: 10.1016/j.ejmg.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Cox DM, Butler MG. The 15q11.2 BP1-BP2 microdeletion syndrome: a review. Int J Mol Sci. 2015a;16:4068–4082. doi: 10.3390/ijms16024068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DM, Butler MG. A clinical case report and literature review of the 3q29 microdeletion syndrome. Clin Dysmorphol. 2015b;24:89–94. doi: 10.1097/MCD.0000000000000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi BJ, Crofts HJ. Association testing of copy number variants in schizophrenia and autism spectrum disorders. J Neurodev Disord. 2012;4:15. doi: 10.1186/1866-1955-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crider A. Perseveration in schizophrenia. Schizophr Bull. 1997;23:63–74. doi: 10.1093/schbul/23.1.63. [DOI] [PubMed] [Google Scholar]
- Dasouki MJ, Lushington GH, Hovanes K, Casey J, Gorre M. The 3q29 microdeletion syndrome: report of three new unrelated patients and in silico “RNA binding” analysis of the 3q29 region. Am J Med Genet A. 2011;155A:1654–1660. doi: 10.1002/ajmg.a.34080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson MT, Schmidt C, Akeson EC. Segmental trisomy of murine chromosome 16: a new model system for studying Down syndrome. Prog Clin Biol Res. 1990;360:263–280. [PubMed] [Google Scholar]
- de Anda FC, Rosario AL, Durak O, Tran T, Graff J, Meletis K, Rei D, Soda T, Madabhushi R, Ginty DD, Kolodkin AL, Tsai LH. Autism spectrum disorder susceptibility gene TAOK2 affects basal dendrite formation in the neocortex. Nat Neurosci. 2012;15:1022–1031. doi: 10.1038/nn.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digilio MC, Bernardini L, Mingarelli R, Capolino R, Capalbo A, Giuffrida MG, Versacci P, Novelli A, Dallapiccola B. 3q29 Microdeletion: a mental retardation disorder unassociated with a recognizable phenotype in two mother-daughter pairs. Am J Med Genet A. 2009;149A:1777–1781. doi: 10.1002/ajmg.a.32965. [DOI] [PubMed] [Google Scholar]
- Drew LJ, Crabtree GW, Markx S, Stark KL, Chaverneff F, Xu B, Mukai J, Fenelon K, Hsu PK, Gogos JA, Karayiorgou M. The 22q11.2 microdeletion: fifteen years of insights into the genetic and neural complexity of psychiatric disorders. Int J Dev Neurosci. 2011;29:259–281. doi: 10.1016/j.ijdevneu.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero I, Johnstone M. Genetics of schizophrenia. Curr Psychiatry Rep. 2014;16:502. doi: 10.1007/s11920-014-0502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essletzbichler P, Konopka T, Santoro F, Chen D, Gapp BV, Kralovics R, Brummelkamp TR, Nijman SM, Burckstummer T. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 2014;24:2059–2065. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O'Donovan MC, Corvin A, Cichon S, Sullivan PF. Evaluating historical candidate genes for schizophrenia. Mol Psychiatry. 2015;20:555–562. doi: 10.1038/mp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstner AJ, Degenhardt F, Schratt G, Nothen MM. MicroRNAs as the cause of schizophrenia in 22q11.2 deletion carriers, and possible implications for idiopathic disease: a mini-review. Front Mol Neurosci. 2013;6:47. doi: 10.3389/fnmol.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, Carrera N, Humphreys I, Johnson JS, Roussos P, Barker DD, Banks E, Milanova V, Grant SG, Hannon E, Rose SA, Chambert K, Mahajan M, Scolnick EM, Moran JL, Kirov G, Palotie A, McCarroll SA, Holmans P, Sklar P, Owen MJ, Purcell SM, O'Donovan MC. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F, Frasca A, Racagni G, Riva MA. Cognitive effects of second-generation antipsychotics: current insights into neurochemical mechanisms. CNS Drugs. 2009;23:603–614. doi: 10.2165/00023210-200923070-00005. [DOI] [PubMed] [Google Scholar]
- Gao R, Penzes P. Common mechanisms of excitatory and inhibitory imbalance in schizophrenia and autism spectrum disorders. Curr Mol Med. 2015;15:146–167. doi: 10.2174/1566524015666150303003028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, Nezarati MM, Asamoah A, Jackson KE, Gowans GC, Martin JA, Carmany EP, Stockton DW, Schnur RE, Penney LS, Martin DM, Raskin S, Leppig K, Thiese H, Smith R, Aberg E, Niyazov DM, Escobar LF, El-Khechen D, Johnson KD, Lebel RR, Siefkas K, Ball S, Shur N, McGuire M, Brasington CK, Spence JE, Martin LS, Clericuzio C, Ballif BC, Shaffer LG, Eichler EE. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassford MR, Rosenfeld JA, Freedman AA, Zwick ME, Unique Rare Chromosome Disorder Support G. Mulle JG. Novel features of 3q29 deletion syndrome: Results from the 3q29 registry. Am J Med Genet A. 2016 doi: 10.1002/ajmg.a.37537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EK, Rees E, Walters JT, Smith KG, Forty L, Grozeva D, Moran JL, Sklar P, Ripke S, Chambert KD, Genovese G, McCarroll SA, Jones I, Jones L, Owen MJ, O'Donovan MC, Craddock N, Kirov G. Copy number variation in bipolar disorder. Mol Psychiatry. 2016;21:89–93. doi: 10.1038/mp.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva D, Conrad DF, Barnes CP, Hurles M, Owen MJ, O'Donovan MC, Craddock N, Kirov G, Wtccc Independent estimation of the frequency of rare CNVs in the UK population confirms their role in schizophrenia. Schizophr Res. 2012;135:1–7. doi: 10.1016/j.schres.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb SC, Maddatu TP, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2009;37:D720–730. doi: 10.1093/nar/gkn778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit JM, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frebourg T, Saugier Veber P, Campion D. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs RW, Zakzanis KK. Neurocognitive deficit in schizophrenia: a quantitative review of the evidence. Neuropsychology. 1998;12:426–445. doi: 10.1037//0894-4105.12.3.426. [DOI] [PubMed] [Google Scholar]
- Heston LL. Psychiatric disorders in foster home reared children of schizophrenic mothers. Br J Psychiatry. 1966;112:819–825. doi: 10.1192/bjp.112.489.819. [DOI] [PubMed] [Google Scholar]
- Hill SK, Bishop JR, Palumbo D, Sweeney JA. Effect of second-generation antipsychotics on cognition: current issues and future challenges. Expert Rev Neurother. 2010;10:43–57. doi: 10.1586/ern.09.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horev G, Ellegood J, Lerch JP, Son YE, Muthuswamy L, Vogel H, Krieger AM, Buja A, Henkelman RM, Wigler M, Mills AA. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci U S A. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvoslef-Eide M, Mar AC, Nilsson SR, Alsio J, Heath CJ, Saksida LM, Robbins TW, Bussey TJ. The NEWMEDS rodent touchscreen test battery for cognition relevant to schizophrenia. Psychopharmacology (Berl) 2015;232:3853–3872. doi: 10.1007/s00213-015-4007-x. [DOI] [PubMed] [Google Scholar]
- International Schizophrenia C. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, Mefford H, Ying P, Nickerson DA, Eichler EE. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablensky AV, Morgan V, Zubrick SR, Bower C, Yellachich LA. Pregnancy, delivery, and neonatal complications in a population cohort of women with schizophrenia and major affective disorders. Am J Psychiatry. 2005;162:79–91. doi: 10.1176/appi.ajp.162.1.79. [DOI] [PubMed] [Google Scholar]
- Jacquet H, Raux G, Thibaut F, Hecketsweiler B, Houy E, Demilly C, Haouzir S, Allio G, Fouldrin G, Drouin V, Bou J, Petit M, Campion D, Frebourg T. PRODH mutations and hyperprolinemia in a subset of schizophrenic patients. Hum Mol Genet. 2002;11:2243–2249. doi: 10.1093/hmg/11.19.2243. [DOI] [PubMed] [Google Scholar]
- Joshi G, Wozniak J, Petty C, Martelon MK, Fried R, Bolfek A, Kotte A, Stevens J, Furtak SL, Bourgeois M, Caruso J, Caron A, Biederman J. Psychiatric comorbidity and functioning in a clinically referred population of adults with autism spectrum disorders: a comparative study. J Autism Dev Disord. 2013;43:1314–1325. doi: 10.1007/s10803-012-1679-5. [DOI] [PubMed] [Google Scholar]
- Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST, Richard G, Compton JG, Fuller AE, Gliem TJ, Huang S, Collinson MN, Beal SJ, Ackley T, Pickering DL, Golden DM, Aston E, Whitby H, Shetty S, Rossi MR, Rudd MK, South ST, Brothman AR, Sanger WG, Iyer RK, Crolla JA, Thorland EC, Aradhya S, Ledbetter DH, Martin CL. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane JM, Robinson DG, Schooler NR, Mueser KT, Penn DL, Rosenheck RA, Addington J, Brunette MF, Correll CU, Estroff SE, Marcy P, Robinson J, Meyer-Kalos PS, Gottlieb JD, Glynn SM, Lynde DW, Pipes R, Kurian BT, Miller AL, Azrin ST, Goldstein AB, Severe JB, Lin H, Sint KJ, John M, Heinssen RK. Comprehensive Versus Usual Community Care for First-Episode Psychosis: 2-Year Outcomes From the NIMH RAISE Early Treatment Program. Am J Psychiatry. 2015 doi: 10.1176/appi.ajp.2015.15050632. appiajp201515050632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegeles LS, Abi-Dargham A, Frankle WG, Gil R, Cooper TB, Slifstein M, Hwang DR, Huang Y, Haber SN, Laruelle M. Increased synaptic dopamine function in associative regions of the striatum in schizophrenia. Arch Gen Psychiatry. 2010;67:231–239. doi: 10.1001/archgenpsychiatry.2010.10. [DOI] [PubMed] [Google Scholar]
- Kety SS, Rosenthal D, Wender PH, Schulsinger F. Mental illness in the biological and adoptive families of adpoted schizophrenics. Am J Psychiatry. 1971;128:302–306. doi: 10.1176/ajp.128.3.302. [DOI] [PubMed] [Google Scholar]
- Kety SS, Rosenthal D, Wender PH, Schulsinger F, Jacobsen B. Mental illness in the biological and adoptive families of adopted individuals who have become schizophrenic. Behav Genet. 1976;6:219–225. doi: 10.1007/BF01065721. [DOI] [PubMed] [Google Scholar]
- Kety SS, Wender PH, Jacobsen B, Ingraham LJ, Jansson L, Faber B, Kinney DK. Mental illness in the biological and adoptive relatives of schizophrenic adoptees. Replication of the Copenhagen Study in the rest of Denmark. Arch Gen Psychiatry. 1994;51:442–455. doi: 10.1001/archpsyc.1994.03950060006001. [DOI] [PubMed] [Google Scholar]
- Kimura H, Tsuboi D, Wang C, Kushima I, Koide T, Ikeda M, Iwayama Y, Toyota T, Yamamoto N, Kunimoto S, Nakamura Y, Yoshimi A, Banno M, Xing J, Takasaki Y, Yoshida M, Aleksic B, Uno Y, Okada T, Iidaka T, Inada T, Suzuki M, Ujike H, Kunugi H, Kato T, Yoshikawa T, Iwata N, Kaibuchi K, Ozaki N. Identification of Rare, Single-Nucleotide Mutations in NDE1 and Their Contributions to Schizophrenia Susceptibility. Schizophr Bull. 2015;41:744–753. doi: 10.1093/schbul/sbu147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, International Schizophrenia C, Wellcome Trust Case Control C. Craddock N, Owen MJ, O'Donovan MC. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, van de Lagemaat LN, Bayes A, Fernandez E, Olason PI, Bottcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SG, Purcell S, Sklar P, O'Donovan MC, Owen MJ. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantareas MM, Hewitt T. Autistic disorder and schizophrenia: diagnostic overlaps. J Autism Dev Disord. 2001;31:19–28. doi: 10.1023/a:1005605528309. [DOI] [PubMed] [Google Scholar]
- Kotlar AV, Mercer KB, Zwick ME, Mulle JG. New discoveries in schizophrenia genetics reveal neurobiological pathways: A review of recent findings. Eur J Med Genet. 2015;58:704–714. doi: 10.1016/j.ejmg.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang SQ, Guo DC, Prakash SK, McDonald ML, Johnson RJ, Wang M, Regalado ES, Russell L, Cao JM, Kwartler C, Fraivillig K, Coselli JS, Safi HJ, Estrera AL, Leal SM, LeMaire SA, Belmont JW, Milewicz DM, Gen TACI Recurrent chromosome 16p13.1 duplications are a risk factor for aortic dissections. PLoS Genet. 2011;7:e1002118. doi: 10.1371/journal.pgen.1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kweon J, Kim E, Kim S, Kim JS. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22:539–548. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lett TA, Voineskos AN, Kennedy JL, Levine B, Daskalakis ZJ. Treating working memory deficits in schizophrenia: a review of the neurobiology. Biol Psychiatry. 2014;75:361–370. doi: 10.1016/j.biopsych.2013.07.026. [DOI] [PubMed] [Google Scholar]
- Leucht S, Cipriani A, Spineli L, Mavridis D, Orey D, Richter F, Samara M, Barbui C, Engel RR, Geddes JR, Kissling W, Stapf MP, Lassig B, Salanti G, Davis JM. Comparative efficacy and tolerability of 15 antipsychotic drugs in schizophrenia: a multiple-treatments meta-analysis. Lancet. 2013;382:951–962. doi: 10.1016/S0140-6736(13)60733-3. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, Zhang N, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Kendler KS, Freedman R, Dudbridge F, Pe'er I, Hakonarson H, Bergen SE, Fanous AH, Holmans PA, Gejman PV. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M, Martin GR. Cre-mediated chromosome loss in mice. Nat Genet. 1997;17:223–225. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Lieberman JA. Catching up on schizophrenia: natural history and neurobiology. Neuron. 2000;28:325–334. doi: 10.1016/s0896-6273(00)00111-2. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Botta A, Jurecic V, Carattini-Rivera S, Cheah YC, Rosenblatt HM, Bradley A, Baldini A. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Lipska BK, Weinberger DR. To model a psychiatric disorder in animals: schizophrenia as a reality test. Neuropsychopharmacology. 2000;23:223–239. doi: 10.1016/S0893-133X(00)00137-8. [DOI] [PubMed] [Google Scholar]
- Liu C, Morishima M, Yu T, Matsui S, Zhang L, Fu D, Pao A, Costa AC, Gardiner KJ, Cowell JK, Nowak NJ, Parmacek MS, Liang P, Baldini A, Yu YE. Genetic analysis of Down syndrome-associated heart defects in mice. Hum Genet. 2011;130:623–632. doi: 10.1007/s00439-011-0980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubow RE. Construct validity of the animal latent inhibition model of selective attention deficits in schizophrenia. Schizophr Bull. 2005;31:139–153. doi: 10.1093/schbul/sbi005. [DOI] [PubMed] [Google Scholar]
- Luigetti M, Del Grande A, Conte A, Lo Monaco M, Bisogni G, Romano A, Zollino M, Rossini PM, Sabatelli M. Clinical, neurophysiological and pathological findings of HNPP patients with 17p12 deletion: a single-centre experience. J Neurol Sci. 2014;341:46–50. doi: 10.1016/j.jns.2014.03.046. [DOI] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, Lowe SW, Ventura A. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard AM, Ruef A, Pizzagalli F, Migliavacca E, Hippolyte L, Adaszewski S, Dukart J, Ferrari C, Conus P, Mannik K, Zazhytska M, Siffredi V, Maeder P, Kutalik Z, Kherif F, Hadjikhani N, Beckmann JS, Reymond A, Draganski B, Jacquemont S, p11.2 European C The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol Psychiatry. 2015;20:140–147. doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen J, Miettunen J, Isohanni M, Koponen H. Negative symptoms in schizophrenia: a review. Nord J Psychiatry. 2008;62:334–341. doi: 10.1080/08039480801959307. [DOI] [PubMed] [Google Scholar]
- Marino MD, Bourdelat-Parks BN, Cameron Liles L, Weinshenker D. Genetic reduction of noradrenergic function alters social memory and reduces aggression in mice. Behav Brain Res. 2005;161:197–203. doi: 10.1016/j.bbr.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Matson JL, Shoemaker M. Intellectual disability and its relationship to autism spectrum disorders. Res Dev Disabil. 2009;30:1107–1114. doi: 10.1016/j.ridd.2009.06.003. [DOI] [PubMed] [Google Scholar]
- McGrath J, Saha S, Chant D, Welham J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol Rev. 2008;30:67–76. doi: 10.1093/epirev/mxn001. [DOI] [PubMed] [Google Scholar]
- McGrath J, Saha S, Welham J, El Saadi O, MacCauley C, Chant D. A systematic review of the incidence of schizophrenia: the distribution of rates and the influence of sex, urbanicity, migrant status and methodology. BMC Med. 2004;2:13. doi: 10.1186/1741-7015-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan MJ, Andrieux A, Bartzokis G, Cadenhead K, Dazzan P, Fusar-Poli P, Gallinat J, Giedd J, Grayson DR, Heinrichs M, Kahn R, Krebs MO, Leboyer M, Lewis D, Marin O, Marin P, Meyer-Lindenberg A, McGorry P, McGuire P, Owen MJ, Patterson P, Sawa A, Spedding M, Uhlhaas P, Vaccarino F, Wahlestedt C, Weinberger D. Altering the course of schizophrenia: progress and perspectives. Nat Rev Drug Discov. 2016;15:485–515. doi: 10.1038/nrd.2016.28. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Miyake N, Jarskog LF, Fleischhacker WW, Lieberman JA. Pharmacological treatment of schizophrenia: a critical review of the pharmacology and clinical effects of current and future therapeutic agents. Mol Psychiatry. 2012;17:1206–1227. doi: 10.1038/mp.2012.47. [DOI] [PubMed] [Google Scholar]
- Mochida S, Yokoyama CT, Kim DK, Itoh K, Catterall WA. Evidence for a voltage-dependent enhancement of neurotransmitter release mediated via the synaptic protein interaction site of N-type Ca2+ channels. Proc Natl Acad Sci U S A. 1998;95:14523–14528. doi: 10.1073/pnas.95.24.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks S, Niarchou M, Davies AR, Walters JT, Williams N, Owen MJ, van den Bree MB, Murphy KC. Further evidence for high rates of schizophrenia in 22q11.2 deletion syndrome. Schizophr Res. 2014;153:231–236. doi: 10.1016/j.schres.2014.01.020. [DOI] [PubMed] [Google Scholar]
- Moreno-De-Luca D, Consortium S, Mulle JG, Simons Simplex Collection Genetics C. Kaminsky EB, Sanders SJ, GeneStar. Myers SM, Adam MP, Pakula AT, Eisenhauer NJ, Uhas K, Weik L, Guy L, Care ME, Morel CF, Boni C, Salbert BA, Chandrareddy A, Demmer LA, Chow EW, Surti U, Aradhya S, Pickering DL, Golden DM, Sanger WG, Aston E, Brothman AR, Gliem TJ, Thorland EC, Ackley T, Iyer R, Huang S, Barber JC, Crolla JA, Warren ST, Martin CL, Ledbetter DH. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan VA, Leonard H, Bourke J, Jablensky A. Intellectual disability co-occurring with schizophrenia and other psychiatric illness: population-based study. Br J Psychiatry. 2008;193:364–372. doi: 10.1192/bjp.bp.107.044461. [DOI] [PubMed] [Google Scholar]
- Mulle JG. Schizophrenia genetics: progress, at last. Curr Opin Genet Dev. 2012;22:238–244. doi: 10.1016/j.gde.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Mulle JG. The 3q29 deletion confers >40-fold increase in risk for schizophrenia. Mol Psychiatry. 2015;20:1028–1029. doi: 10.1038/mp.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG, Dodd AF, McGrath JA, Wolyniec PS, Mitchell AA, Shetty AC, Sobreira NL, Valle D, Rudd MK, Satten G, Cutler DJ, Pulver AE, Warren ST. Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet. 2010;87:229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG, Pulver AE, McGrath JA, Wolyniec PS, Dodd AF, Cutler DJ, Sebat J, Malhotra D, Nestadt G, Conrad DF, Hurles M, Barnes CP, Ikeda M, Iwata N, Levinson DF, Gejman PV, Sanders AR, Duan J, Mitchell AA, Peter I, Sklar P, O'Dushlaine CT, Grozeva D, O'Donovan MC, Owen MJ, Hultman CM, Kahler AK, Sullivan PF, Molecular Genetics of Schizophrenia C. Kirov G, Warren ST. Reciprocal duplication of the Williams-Beuren syndrome deletion on chromosome 7q11.23 is associated with schizophrenia. Biol Psychiatry. 2014;75:371–377. doi: 10.1016/j.biopsych.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naf D, Wilson LA, Bergstrom RA, Smith RS, Goodwin NC, Verkerk A, van Ommen GJ, Ackerman SL, Frankel WN, Schimenti JC. Mouse models for the Wolf-Hirschhorn deletion syndrome. Hum Mol Genet. 2001;10:91–98. doi: 10.1093/hmg/10.2.91. [DOI] [PubMed] [Google Scholar]
- Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, Tomonaga S, Watanabe Y, Chung YJ, Banerjee R, Iwamoto K, Kato T, Okazawa M, Yamauchi K, Tanda K, Takao K, Miyakawa T, Bradley A, Takumi T. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci U S A. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen AM, Roberts AC, Hodges JR, Summers BA, Polkey CE, Robbins TW. Contrasting mechanisms of impaired attentional set-shifting in patients with frontal lobe damage or Parkinson's disease. Brain. 1993;116(Pt 5):1159–1175. doi: 10.1093/brain/116.5.1159. [DOI] [PubMed] [Google Scholar]
- Owen MJ. Will schizophrenia become a graveyard for molecular geneticists? Psychol Med. 1992;22:289–293. doi: 10.1017/s0033291700030221. [DOI] [PubMed] [Google Scholar]
- Palumbo P, Antona V, Palumbo O, Piccione M, Nardello R, Fontana A, Carella M, Corsello G. Variable phenotype in 17q12 microdeletions: clinical and molecular characterization of a new case. Gene. 2014;538:373–378. doi: 10.1016/j.gene.2014.01.050. [DOI] [PubMed] [Google Scholar]
- Perry W, Light GA, Davis H, Braff DL. Schizophrenia patients demonstrate a dissociation on declarative and non-declarative memory tests. Schizophr Res. 2000;46:167–174. doi: 10.1016/s0920-9964(99)00229-7. [DOI] [PubMed] [Google Scholar]
- Piskorowski RA, Nasrallah K, Diamantopoulou A, Mukai J, Hassan SI, Siegelbaum SA, Gogos JA, Chevaleyre V. Age-Dependent Specific Changes in Area CA2 of the Hippocampus and Social Memory Deficit in a Mouse Model of the 22q11.2 Deletion Syndrome. Neuron. 2016;89:163–176. doi: 10.1016/j.neuron.2015.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CM, Miyakawa T. Schizophrenia-relevant behavioral testing in rodent models: a uniquely human disorder? Biol Psychiatry. 2006;59:1198–1207. doi: 10.1016/j.biopsych.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad SE, Howley S, Murphy KC. Candidate genes and the behavioral phenotype in 22q11.2 deletion syndrome. Dev Disabil Res Rev. 2008;14:26–34. doi: 10.1002/ddrr.5. [DOI] [PubMed] [Google Scholar]
- Preti A, Cardascia L, Zen T, Marchetti M, Favaretto G, Miotto P. Risk for obstetric complications and schizophrenia. Psychiatry Res. 2000;96:127–139. doi: 10.1016/s0165-1781(00)00185-2. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O'Dushlaine C, Chambert K, Bergen SE, Kahler A, Duncan L, Stahl E, Genovese G, Fernandez E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PK, Banks E, Shakir K, Garimella K, Fennell T, DePristo M, Grant SG, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA, Sklar P. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero-Rivera F, Sharifi-Hannauer P, Martinez-Agosto JA. Autistic and psychiatric findings associated with the 3q29 microdeletion syndrome: case report and review. Am J Med Genet A. 2010;152A:2459–2467. doi: 10.1002/ajmg.a.33573. [DOI] [PubMed] [Google Scholar]
- Ramalingam A, Zhou XG, Fiedler SD, Brawner SJ, Joyce JM, Liu HY, Yu S. 16p13.11 duplication is a risk factor for a wide spectrum of neuropsychiatric disorders. J Hum Genet. 2011;56:541–544. doi: 10.1038/jhg.2011.42. [DOI] [PubMed] [Google Scholar]
- Rees E, Kirov G, Sanders A, Walters JT, Chambert KD, Shi J, Szatkiewicz J, O'Dushlaine C, Richards AL, Green EK, Jones I, Davies G, Legge SE, Moran JL, Pato C, Pato M, Genovese G, Levinson D, Duan J, Moy W, Goring HH, Morris D, Cormican P, Kendler KS, O'Neill FA, Riley B, Gill M, Corvin A, Wellcome Trust Case Control C. Craddock N, Sklar P, Hultman C, Sullivan PF, Gejman PV, McCarroll SA, O'Donovan MC, Owen MJ. Evidence that duplications of 22q11.2 protect against schizophrenia. Mol Psychiatry. 2014a;19:37–40. doi: 10.1038/mp.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL, Mahoney-Davies G, Legge SE, Moran JL, McCarroll SA, O'Donovan MC, Owen MJ, Kirov G. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry. 2014b;204:108–114. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- Rimer M, Barrett DW, Maldonado MA, Vock VM, Gonzalez-Lima F. Neuregulin-1 immunoglobulin-like domain mutant mice: clozapine sensitivity and impaired latent inhibition. Neuroreport. 2005;16:271–275. doi: 10.1097/00001756-200502280-00014. [DOI] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, Kim Y, Lee SH, Magnusson PK, Sanchez N, Stahl EA, Williams S, Wray NR, Xia K, Bettella F, Borglum AD, Bulik-Sullivan BK, Cormican P, Craddock N, de Leeuw C, Durmishi N, Gill M, Golimbet V, Hamshere ML, Holmans P, Hougaard DM, Kendler KS, Lin K, Morris DW, Mors O, Mortensen PB, Neale BM, O'Neill FA, Owen MJ, Milovancevic MP, Posthuma D, Powell J, Richards AL, Riley BP, Ruderfer D, Rujescu D, Sigurdsson E, Silagadze T, Smit AB, Stefansson H, Steinberg S, Suvisaari J, Tosato S, Verhage M, Walters JT, Multicenter Genetic Studies of Schizophrenia C, Levinson DF, Gejman PV, Kendler KS, Laurent C, Mowry BJ, O'Donovan MC, Owen MJ, Pulver AE, Riley BP, Schwab SG, Wildenauer DB, Dudbridge F, Holmans P, Shi J, Albus M, Alexander M, Campion D, Cohen D, Dikeos D, Duan J, Eichhammer P, Godard S, Hansen M, Lerer FB, Liang KY, Maier W, Mallet J, Nertney DA, Nestadt G, Norton N, O'Neill FA, Papadimitriou GN, Ribble R, Sanders AR, Silverman JM, Walsh D, Williams NM, Wormley B, Psychosis Endophenotypes International C, Arranz MJ, Bakker S, Bender S, Bramon E, Collier D, Crespo-Facorro B, Hall J, Iyegbe C, Jablensky A, Kahn RS, Kalaydjieva L, Lawrie S, Lewis CM, Lin K, Linszen DH, Mata I, McIntosh A, Murray RM, Ophoff RA, Powell J, Rujescu D, Van Os J, Walshe M, Weisbrod M, Wiersma D, Wellcome Trust Case Control C. Donnelly P, Barroso I, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin AP, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Spencer CC, Band G, Bellenguez C, Freeman C, Hellenthal G, Giannoulatou E, Pirinen M, Pearson RD, Strange A, Su Z, Vukcevic D, Donnelly P, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Potter SC, Ravindrarajah R, Ricketts M, Tashakkori-Ghanbaria A, Waller MJ, Weston P, Widaa S, Whittaker P, Barroso I, Deloukas P, Mathew CG, Blackwell JM, Brown MA, Corvin AP, McCarthy MI, Spencer CC, Bramon E, Corvin AP, O'Donovan MC, Stefansson K, Scolnick E, Purcell S, McCarroll SA, Sklar P, Hultman CM, Sullivan PF. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins TW. The 5-choice serial reaction time task: behavioural pharmacology and functional neurochemistry. Psychopharmacology (Berl) 2002;163:362–380. doi: 10.1007/s00213-002-1154-7. [DOI] [PubMed] [Google Scholar]
- Ropers HH. Genetics of intellectual disability. Curr Opin Genet Dev. 2008;18:241–250. doi: 10.1016/j.gde.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Russell LB. The effects of radiation on mammalian prenatal development. McGraw-Hill; New York: 1954. [Google Scholar]
- Russell LB, Hunsicker PR, Cacheiro NL, Bangham JW, Russell WL, Shelby MD. Chlorambucil effectively induces deletion mutations in mouse germ cells. Proc Natl Acad Sci U S A. 1989;86:3704–3708. doi: 10.1073/pnas.86.10.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WL. X-ray-induced mutations in mice. Cold Spring Harb Symp Quant Biol. 1951;16:327–336. doi: 10.1101/sqb.1951.016.01.024. [DOI] [PubMed] [Google Scholar]
- Sagar A, Bishop JR, Tessman DC, Guter S, Martin CL, Cook EH. Co-occurrence of autism, childhood psychosis, and intellectual disability associated with a de novo 3q29 microdeletion. Am J Med Genet A. 2013;161A:845–849. doi: 10.1002/ajmg.a.35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi G, Manabe T, Kobayashi K, Orita S, Sasaki T, Naito A, Maeda M, Igarashi H, Katsuura G, Nishioka H, Mizoguchi A, Itohara S, Takahashi T, Takai Y. Doc2alpha is an activity-dependent modulator of excitatory synaptic transmission. Eur J Neurosci. 1999;11:4262–4268. doi: 10.1046/j.1460-9568.1999.00855.x. [DOI] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, Goldberg AP, Jinlu C, Keaney JF, 3rd, Klei L, Mandell JD, Moreno-De-Luca D, Poultney CS, Robinson EB, Smith L, Solli-Nowlan T, Su MY, Teran NA, Walker MF, Werling DM, Beaudet AL, Cantor RM, Fombonne E, Geschwind DH, Grice DE, Lord C, Lowe JK, Mane SM, Martin DM, Morrow EM, Talkowski ME, Sutcliffe JS, Walsh CA, Yu TW, Autism Sequencing C. Ledbetter DH, Martin CL, Cook EH, Buxbaum JD, Daly MJ, Devlin B, Roeder K, State MW. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol. 2010;11:182–195. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci U S A. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Gur RC, Gur RE, Mozley PD, Mozley LH, Resnick SM, Kester DB, Stafiniak P. Neuropsychological function in schizophrenia. Selective impairment in memory and learning. Arch Gen Psychiatry. 1991;48:618–624. doi: 10.1001/archpsyc.1991.01810310036007. [DOI] [PubMed] [Google Scholar]
- Saykin AJ, Shtasel DL, Gur RE, Kester DB, Mozley LH, Stafiniak P, Gur RC. Neuropsychological deficits in neuroleptic naive patients with first-episode schizophrenia. Arch Gen Psychiatry. 1994;51:124–131. doi: 10.1001/archpsyc.1994.03950020048005. [DOI] [PubMed] [Google Scholar]
- Schimenti JC, Libby BJ, Bergstrom RA, Wilson LA, Naf D, Tarantino LM, Alavizadeh A, Lengeling A, Bucan M. Interdigitated deletion complexes on mouse chromosome 5 induced by irradiation of embryonic stem cells. Genome Res. 2000;10:1043–1050. doi: 10.1101/gr.10.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics C Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes KE, Martin-Iverson MT. Disturbed prepulse inhibition in patients with schizophrenia is consequential to dysfunction of selective attention. Psychophysiology. 2010;47:223–235. doi: 10.1111/j.1469-8986.2009.00927.x. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet 13 Spec No. 2004;1:R57–64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-Filas E. A mouse geneticist's practical guide to CRISPR applications. Genetics. 2015;199:1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Perez-Jurado LA, Morris CA, Scherer SW, Osborne LR. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N Engl J Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen HJ, Nielsen PR, Pedersen CB, Benros ME, Nordentoft M, Mortensen PB. Population impact of familial and environmental risk factors for schizophrenia: a nationwide study. Schizophr Res. 2014;153:214–219. doi: 10.1016/j.schres.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Spitz F, Herkenne C, Morris MA, Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat Genet. 2005;37:889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, Bjornsdottir G, Walters GB, Jonsdottir GA, Doyle OM, Tost H, Grimm O, Kristjansdottir S, Snorrason H, Davidsdottir SR, Gudmundsson LJ, Jonsson GF, Stefansdottir B, Helgadottir I, Haraldsson M, Jonsdottir B, Thygesen JH, Schwarz AJ, Didriksen M, Stensbol TB, Brammer M, Kapur S, Halldorsson JG, Hreidarsson S, Saemundsen E, Sigurdsson E, Stefansson K. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Group, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart LR, Hall AL, Kang SH, Shaw CA, Beaudet AL. High frequency of known copy number abnormalities and maternal duplication 15q11-q13 in patients with combined schizophrenia and epilepsy. BMC Med Genet. 2011;12:154. doi: 10.1186/1471-2350-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs L, Carver EA, Cacheiro NL, Shelby M, Generoso W. Generation and characterization of heritable reciprocal translocations in mice. Methods. 1997;13:397–408. doi: 10.1006/meth.1997.0546. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, Liu W, Downing AM, Lieberman J, Close SL. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Rai D, Golding J, Zammit S, Steer C. The association between autism spectrum disorder and psychotic experiences in the Avon longitudinal study of parents and children (ALSPAC) birth cohort. J Am Acad Child Adolesc Psychiatry. 2013;52:806–814. e802. doi: 10.1016/j.jaac.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Szatkiewicz JP, O'Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, Fromer M, Ruderfer D, Akterin S, Bergen SE, Kahler A, Magnusson PK, Kim Y, Crowley JJ, Rees E, Kirov G, O'Donovan MC, Owen MJ, Walters J, Scolnick E, Sklar P, Purcell S, Hultman CM, McCarroll SA, Sullivan PF. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai DJ, Ragavendran A, Manavalan P, Stortchevoi A, Seabra CM, Erdin S, Collins RL, Blumenthal I, Chen X, Shen Y, Sahin M, Zhang C, Lee C, Gusella JF, Talkowski ME. Engineering microdeletions and microduplications by targeting segmental duplications with CRISPR. Nat Neurosci. 2016;19:517–522. doi: 10.1038/nn.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tcheremissine OV, Castro MA, Gardner DR. Targeting cognitive deficits in schizophrenia: a review of the development of a new class of medicines from the perspective of community mental health researchers. Expert Opin Investig Drugs. 2012;21:7–14. doi: 10.1517/13543784.2012.634798. [DOI] [PubMed] [Google Scholar]
- Torres F, Barbosa M, Maciel P. Recurrent copy number variations as risk factors for neurodevelopmental disorders: critical overview and analysis of clinical implications. J Med Genet. 2016;53:73–90. doi: 10.1136/jmedgenet-2015-103366. [DOI] [PubMed] [Google Scholar]
- Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun. 2014;5:3964. doi: 10.1038/ncomms4964. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, Beck S, Hurles ME. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezato A, Kimura-Sato J, Yamamoto N, Iijima Y, Kunugi H, Nishikawa T. Further evidence for a male-selective genetic association of synapse-associated protein 97 (SAP97) gene with schizophrenia. Behav Brain Funct. 2012;8:2. doi: 10.1186/1744-9081-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban AE, Purmann C. Using iPSCs and genomics to catch CNVs in the act. Nat Genet. 2015;47:100–101. doi: 10.1038/ng.3204. [DOI] [PubMed] [Google Scholar]
- Velleman SL, Mervis CB. Children with 7q11.23 Duplication Syndrome: Speech, Language, Cognitive, and Behavioral Characteristics and their Implications for Intervention. Perspect Lang Learn Educ. 2011;18:108–116. doi: 10.1044/lle18.3.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. Understanding variable expressivity in microdeletion syndromes. Nat Genet. 2010;42:192–193. doi: 10.1038/ng0310-192. [DOI] [PubMed] [Google Scholar]
- Vinas-Jornet M, Esteba-Castillo S, Gabau E, Ribas-Vidal N, Baena N, San J, Ruiz A, Coll MD, Novell R, Guitart M. A common cognitive, psychiatric, and dysmorphic phenotype in carriers of NRXN1 deletion. Mol Genet Genomic Med. 2014;2:512–521. doi: 10.1002/mgg3.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlsten D. Deficiency of corpus callosum varies with strain and supplier of the mice. Brain Res. 1982;239:329–347. doi: 10.1016/0006-8993(82)90513-3. [DOI] [PubMed] [Google Scholar]
- Walch L. Emerging role of the scaffolding protein Dlg1 in vesicle trafficking. Traffic. 2013;14:964–973. doi: 10.1111/tra.12089. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner I. The “two-headed” latent inhibition model of schizophrenia: modeling positive and negative symptoms and their treatment. Psychopharmacology (Berl) 2003;169:257–297. doi: 10.1007/s00213-002-1313-x. [DOI] [PubMed] [Google Scholar]
- Weinshenker D, Miller NS, Blizinsky K, Laughlin ML, Palmiter RD. Mice with chronic norepinephrine deficiency resemble amphetamine-sensitized animals. Proc Natl Acad Sci U S A. 2002;99:13873–13877. doi: 10.1073/pnas.212519999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, Makri G, Nauen D, Yu H, Guzman E, Chiang CH, Yoritomo N, Kaibuchi K, Zou J, Christian KM, Cheng L, Ross CA, Margolis RL, Chen G, Kosik KS, Song H, Ming GL. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender PH, Rosenthal D, Kety SS, Schulsinger F, Welner J. Crossfostering. A research strategy for clarifying the role of genetic and experiential factors in the etiology of schizophrenia. Arch Gen Psychiatry. 1974;30:121–128. doi: 10.1001/archpsyc.1974.01760070097016. [DOI] [PubMed] [Google Scholar]
- Wijshake T, Baker DJ, van de Sluis B. Endonucleases: new tools to edit the mouse genome. Biochim Biophys Acta. 2014;1842:1942–1950. doi: 10.1016/j.bbadis.2014.04.020. [DOI] [PubMed] [Google Scholar]
- Willatt L, Cox J, Barber J, Cabanas ED, Collins A, Donnai D, FitzPatrick DR, Maher E, Martin H, Parnau J, Pindar L, Ramsay J, Shaw-Smith C, Sistermans EA, Tettenborn M, Trump D, de Vries BB, Walker K, Raymond FL. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am J Hum Genet. 2005;77:154–160. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, Ha S, Chung C, Jung ES, Cho YS, Park SG, Lee JS, Lee K, Kim D, Bae YC, Kaang BK, Lee MG, Kim E. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, Zu Y, Li W, Huang P, Tong X, Zhu Z, Lin S, Zhang B. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013;41:e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Lee YA, Goto Y. Dopamine in socioecological and evolutionary perspectives: implications for psychiatric disorders. Front Neurosci. 2015;9:219. doi: 10.3389/fnins.2015.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavich L, Forsberg MM, Karayiorgou M, Gogos JA, Mannisto PT. Site-specific role of catechol-O-methyltransferase in dopamine overflow within prefrontal cortex and dorsal striatum. J Neurosci. 2007;27:10196–10209. doi: 10.1523/JNEUROSCI.0665-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Bergstrom R, Klemm M, Lederman B, Nelson H, Ticknor C, Jaenisch R, Schimenti J. Chromosomal deletion complexes in mice by radiation of embryonic stem cells. Nat Genet. 1997;15:285–288. doi: 10.1038/ng0397-285. [DOI] [PubMed] [Google Scholar]
- Yu T, Clapcote SJ, Li Z, Liu C, Pao A, Bechard AR, Carattini-Rivera S, Matsui S, Roder JC, Baldini A, Mobley WC, Bradley A, Yu YE. Deficiencies in the region syntenic to human 21q22.3 cause cognitive deficits in mice. Mamm Genome. 2010;21:258–267. doi: 10.1007/s00335-010-9262-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Bradley A. Engineering chromosomal rearrangements in mice. Nat Rev Genet. 2001;2:780–790. doi: 10.1038/35093564. [DOI] [PubMed] [Google Scholar]
- Yu YE, Morishima M, Pao A, Wang DY, Wen XY, Baldini A, Bradley A. A deficiency in the region homologous to human 17q21.33-q23.2 causes heart defects in mice. Genetics. 2006;173:297–307. doi: 10.1534/genetics.105.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Chattarji S, Barbarosie M, Rondi-Reig L, Philpot BD, Miyakawa T, Bear MF, Tonegawa S. Forebrain-specific calcineurin knockout selectively impairs bidirectional synaptic plasticity and working/episodic-like memory. Cell. 2001;107:617–629. doi: 10.1016/s0092-8674(01)00585-2. [DOI] [PubMed] [Google Scholar]
- Zheng B, Mills AA, Bradley A. Introducing defined chromosomal rearrangements into the mouse genome. Methods. 2001;24:81–94. doi: 10.1006/meth.2001.1160. [DOI] [PubMed] [Google Scholar]
- Ziats MN, Goin-Kochel RP, Berry LN, Ali M, Ge J, Guffey D, Rosenfeld JA, Bader P, Gambello MJ, Wolf V, Penney LS, Miller R, Lebel RR, Kane J, Bachman K, Troxell R, Clark G, Minard CG, Stankiewicz P, Beaudet A, Schaaf CP. The complex behavioral phenotype of 15q13.3 microdeletion syndrome. Genet Med. 2016 doi: 10.1038/gim.2016.9. [DOI] [PubMed] [Google Scholar]