Abstract
Summer-type hypersensitivity pneumonitis (SHP) is type III or IV allergies developed by repeated inhalation of arthroconidia of Trichosporon species. We identified 105 strains obtained from the homes of 36 SHP patients by analysis of the intergenic spacer (IGS) 1 region, which is located between the 26S and 5S rRNA genes; in addition, we analyzed the IGS genotypes of the strains. Serologically, Trichosporon species are classified as serotype I, II, III, or I-III. Of the 105 strains, 43 (41.1%), 53 (50.5%), and 9 (8.6%) strains were isolated as serotypes I, II, and III, respectively. Serotype I, II, and III strains were recovered from 19 (52.8%), 29 (80.6%), and 4 (11.1%) of the 36 houses of SHP patients, respectively. No serotype I-III strains were isolated from the houses. Of 43 serotype I strains, 42 (97.7%) were identified as Trichosporon dermatis, and the remaining one was T. terricola. Of 53 serotype II strains, 37 (69.8%) were identified as T. asahii, and the remaining serotype II isolates were T. aquatile (1.9%), T. coremiiforme (7.5%), T. faecale (1.9%), T. japonicum (15.1%), and T. ovoides (3.8%). There were nine serotype III strains comprised of T. montevideense (77.8%) and T. domesticum (22.2%). Intraspecies diversity was found only in T. asahii. This microorganism also causes opportunistic infections (trichosporonosis); seven genotypes of its IGS 1 region have been identified. While the strains of T. asahii obtained from Japanese patients with trichosporonosis were genotype I, the strains from the houses of SHP patients were genotype III. Based on our analysis, we conclude that the strains that play the most significant roles in the development of SHP are T. dermatis, T. asahii genotype 3, and T. montevideense, representing serotypes I, II, and III, respectively.
Trichosporon Behrend is a medically important genus that includes the causative agents of systemic, mucosa-associated, and superficial infections, including white piedra. Since 1970, when Watson and Kallichurum (22) first reported disseminated infection (trichosporonosis) caused by Trichosporon species, there have been an increasing number of reports (8) of this rare disease in immunocompromised hosts. The majority of leukemia and lymphoma patients with fatal disseminated fungemia are profoundly neutropenic when the infection develops (20). Systemic trichosporonosis has a high mortality rate, and the prognosis for patients is very poor. The principal etiologic agents of trichosporonosis differ in each type of infection: Trichosporon asahii and T. mucoides are involved in systemic infections (7), T. asteroides and T. cutaneum are associated with superficial infections, and T. ovoides and T. inkin are involved in white piedra of the head and the genital area, respectively.
In addition to infections, Trichosporon species are also responsible for the development of summer-type hypersensitivity pneumonitis (SHP) (1, 15). SHP follows the development of type III or IV allergies by repeated inhalation of Trichosporon arthroconidia, which often contaminate home environments during the summer season. In western and southern Japan, the summer is hot, humid, and rainy. Such conditions favor the growth of Trichosporon species, and most patients initially experience symptoms during the summer. Although SHP is considered peculiar to Japan, a case was recently reported in Korea (2). Since 1984, when Shimazu et al. (15) first reported that SHP was induced by Trichosporon, the number of patients reported to suffer from SHP has been increasing steadily. When the role of Trichosporon in SHP was first ascertained, T. cutaneum was considered to be the causative antigen, and four serotypes were identified, namely I, II, III, and I-III (9, 12). Patients with SHP had positive reactions to inhalation challenge with antigens of the same serotype of T. cutaneum isolated from their houses, indicating that patients are sensitized by exposure to the dominant Trichosporon species in their home environment (3). Based on this evidence, it is clear that the Trichosporon serotype plays a significant role in the development of SHP.
However, the taxonomy of T. cutaneum was problematic (5), leading to a revision of the taxonomy of the genus Trichosporon by Guého et al. (6) based on rRNA gene sequence analysis. Therefore, T. cutaneum was divided into >10 distinct species. Consequently, each serotype corresponds to a group of species and to a phylogenetic clade (12). For instance, serotype II species comprise T. aquatile, T. asahii, T. asteroides, T. coremiiforme, T. faecale, T. inkin, T. japonicum, and T. ovoides, which group together in the Ovoides clade. Nishiura et al. (12) revealed that the causative antigens of SHP were assigned to T. mucoides (serotype I), T. asahii (serotype II), and T. montevideense (serotype III) after an environmental investigation of the houses of SHP patients and an analysis of serum antibodies of SHP patients. Twenty years have passed since Trichosporon was proposed as the causative antigen of SHP.
In the present study, we reidentified the Trichosporon strains obtained from SHP patients' houses using current taxonomic tools and further reassessed the causative agents of SHP.
MATERIALS AND METHODS
Trichosporon isolates.
One hundred and five Trichosporon strains were isolated from 36 SHP patients' houses in Japan, as shown in Table 1. The organisms were obtained from tatami mats, pillars, and beds in the houses of SHP patients by swabbing them with a cotton applicator (Transwab Medical Wire and Equipment, Corsham City, Wiltshire, United Kingdom). Each cotton applicator was then spread onto Sabouraud dextrose agar plates containing 50 μg of chloramphenicol (Sankyo, Tokyo, Japan)/ml, 400 IU of penicillin (Meijiseika, Tokyo, Japan)/ml, and 400 IU of streptomycin (Meijiseika)/ml. The plates were incubated at 27°C until yeast colonies were recovered.
TABLE 1.
Trichosporon species isolated from the houses of patients with SHP
| Species | Strain
|
House
|
||
|---|---|---|---|---|
| No. | % | No. | % | |
| Serotype I | 43 | 41.0 | 19 | 52.8 |
| T. dermatis | 42 | 40.0 | 18 | 50.0 |
| T. terricola | 1 | 1.0 | 1 | 2.8 |
| Serotype II | 53 | 50.5 | 29 | 80.6 |
| T. aquatile | 1 | 1.0 | 1 | 2.8 |
| T. asahii | 37 | 35.2 | 24 | 66.7 |
| Genotype 1 | 2 | 1.9 | 2 | 5.6 |
| Genotype 3 | 30 | 28.6 | 18 | 50.0 |
| Genotype 4 | 1 | 1.0 | 1 | 2.8 |
| Genotype 5 | 4 | 3.8 | 3 | 8.3 |
| T. coremiiforem | 4 | 3.8 | 4 | 11.1 |
| T. faecale | 1 | 1.0 | 1 | 2.8 |
| T. japonicum | 8 | 7.6 | 1 | 2.8 |
| T. ovoides | 2 | 1.9 | 2 | 5.6 |
| Serotype III | 9 | 8.6 | 4 | 11.1 |
| T. domesticum | 2 | 1.9 | 2 | 5.6 |
| T. montevideense | 7 | 6.7 | 2 | 5.6 |
Serotyping.
The serotypes of the fungal isolates were determined using the cell slide agglutination test based on agglutination of heat-killed cells from 3-day cultures (27°C on Sabouraud dextrose agar) by specific-factor sera for Trichosporon serotypes, as described by Ikeda et al. (9).
Identification of Trichosporon isolates.
The Trichosporon isolates were identified by DNA sequence analysis of the intergenic spacer (IGS) 1 region, which is located between the 26S and 5S rRNA genes. Genomic DNA was extracted by the method of Makimura et al. (11). The IGS 1 region was sequenced directly from PCR products using the primer pairs 26SBF (5′-AGCTGCTGCCAATGCTAGCTC) and 5SR (5′-AGCTTGACTTCGCAGATCGG) (19) and an ABI 310 DNA-sequencing apparatus with the BigDye Terminator Cycle Sequencing Ready Reaction kit version 3.1 (Perkin-Elmer Applied Biosystems, Foster City, Calif.), according to the manufacturer's instructions.
Diversity of IGS 1 DNA sequences in T. asahii.
The IGS 1 sequences of the T. asahii strains obtained from SHP patients' houses were compared with those of strains isolated from 30 Japanese patients with trichosporonosis from a previous report (GenBank accession numbers AB066375 to AB066393, AB066402, AB072599 to AB072606, and AB072612) (19). The IGS 1 sequences of six additional clinical isolates obtained from the blood of Japanese patients with systemic trichosporonosis were also analyzed for the present study.
Nucleotide sequence accession numbers.
The GenBank accession numbers for new genotypes are AB180192 for genotype 6 and AB180193 and AB180194 for genotype 7.
RESULTS
Serotyping of the isolates.
Of 105 isolates, 43 (41.1%), 53 (50.5%), and 9 (8.6%) were serotype I, II, and III, respectively. Strains of serotype I, II, and III were recovered from 19 (52.8%), 29 (80.6%), and 4 (11.1%) houses of SHP patients, respectively. No serotype I-III strains were isolated from the houses (Table 1).
Identification of the isolates by IGS 1 sequence analysis.
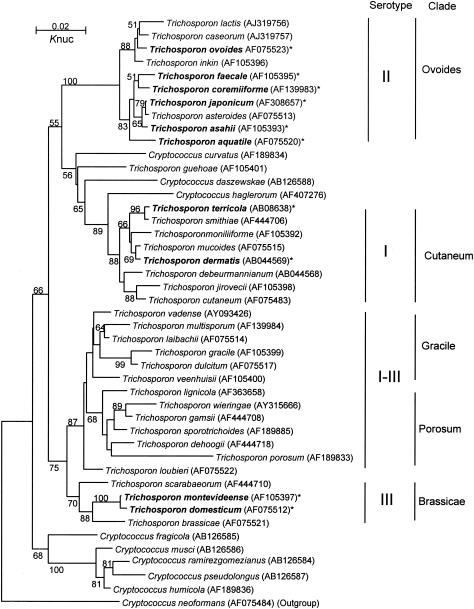
The phylogenetic relationships of the Trichosporon isolates and other Trichosporon species are shown in Fig. 1.
FIG. 1.
Molecular phylogenetic tree of Trichosporon isolates and other Trichosporon members. A tree was constructed from D1-D2 26S ribosomal DNA sequences using neighbor-joining analysis (13) after the sequences were aligned using Clustal W (21). The distances between sequences were calculated using Kimura's two-parameter model (10). The numbers indicate the confidence levels from 100 replicate bootstrap samplings (frequencies of <50% are not shown) (4). *, species isolated from SHP patients' houses.
(i) Serotype I species.
Of 43 serotype I isolates, 42 (97.7%) were identified as T. dermatis and the remaining strain was T. terricola (Table 1). No DNA sequence diversity was found in the IGS 1 region of either species. The IGS 1 sequence of each isolate was identical to that of the type strain for each species (i.e., GenBank accession numbers AB066412 for T. dermatis and AB086383 for T. terricola).
(ii) Serotype II species.
Of 53 serotype II strains, 37 (69.8%) were T. asahii strains and the remaining isolates were identified as T. aquatile (1.9%), T. coremiiforme (7.5%), T. faecale (1.9%), T. japonicum (15.1%), and T. ovoides (3.8%) (Table 1). With the exception of T. asahii, no DNA sequence diversity of the IGS 1 region was found in the serotype II species. The IGS 1 sequence of each non-asahii isolate was identical to that of the type strain of the respective species (i.e., GenBank accession numbers AB066403 for T. aquatile, AB066406 for T. coremiiforme, AB066413 for T. faecale, AB066426 for T. japonicum, and AB066434 for T. ovoides). While serotype II species were recovered from 29 (80.6%) of 36 patients' houses, T. aquatile, T. faecale, and T. japonicum were isolated from only 1 house each.
(iii) Serotype III species.
Serotype III species were less frequently isolated than were the other serotypes. Only 9 (8.6%) of the 105 isolates were serotype III, and only 4 (11.1%) of 36 patients' houses harbored this serotype. Among the serotype III isolates, T. montevideense (77.8%) and T. domesticum (22.2%) were identified (Table 1). Neither species had variations in the IGS 1 DNA sequence. The IGS 1 sequence of each isolate was identical to that of the type strain of each species (i.e., GenBank accession numbers AB066431 for T. montevideense and AB066416 for T. domesticum).
Diversity of Trichosporon isolated from the houses of SHP patients.
The numbers of Trichosporon species isolated from each SHP patient's house are shown in Table 2. On average, 1.6 ± 0.7 species were isolated per house. One or two species were obtained from most houses (88.9%), while only four houses harbored three species simultaneously (11.1%).
TABLE 2.
Variety of species isolated from houses of SHP patients
| Species (serotype) | No. of houses (%) |
|---|---|
| Single species (19 of 36) | 19 (52.8) |
| T. dermatis (I) | 3 (8.3) |
| T. asahii (II) | 11 (30.6) |
| T. coremiiforme (II) | 1 (2.8) |
| T. faecale (II) | 1 (2.8) |
| T. ovoides (II) | 2 (5.6) |
| T. domesticum (III) | 1 (2.8) |
| Two species (13 of 36) | 13 (36.1) |
| T. dermatis (I) + T. terricola (I) | 1 (2.8) |
| T. dermatis (I) + T. asahii (II) | 9 (25.0) |
| T. dermatis (I) + T. montevideense (III) | 2 (5.6) |
| T. asahii (II) + T. coremiiforme (II) | 1 (2.8) |
| Three species (4 of 36) | 4 (11.1) |
| T. dermatis (I) + T. asahii (II) + T. coremiiforme (II) | 1 (2.8) |
| T. dermatis (I) + T. asahii (II) + T. japonicum (II) | 1 (2.8) |
| T. dermatis (I) + T. aquatile (II) + T. coremiiforme (II) | 1 (2.8) |
| T. dermatis (I) + T. asahii (II) + T. domesticum (III) | 1 (2.8) |
| Total | 36 |
IGS 1 genotypes in the T. asahii isolates.
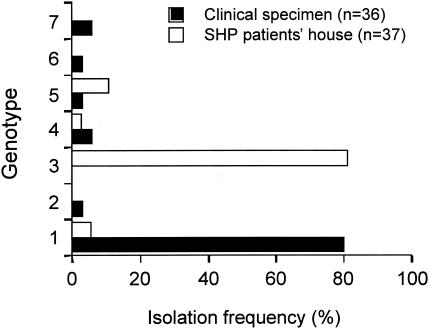
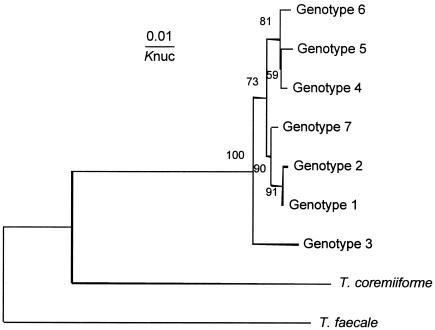
Our previous study showed that T. asahii had five genotypes of the IGS 1 region (19). Since that report, six additional strains of T. asahii isolated from the blood of patients with systemic trichosporonosis have been genotyped, permitting the identification of two new genotypes, 6 and 7. In strains obtained from SHP patients' houses, genotypes 1, 3, 4, and 5 were identified. Of 36 T. asahii strains, 30 (81.1%) were genotype 3 (Fig. 2). In contrast, of 37 clinical isolates from Japanese cases of systemic trichosporonosis, most strains were genotype 1 (80.6%) and no genotype 3 isolates were found. A molecular phylogenetic tree of the seven genotypic strains of T. asahii is shown in Fig. 3.
FIG. 2.
Genotypes of T. asahii isolated from the houses of Japanese patients with SHP and from clinical specimens from trichosporonosis patients.
FIG. 3.
Molecular phylogenetic tree of the seven IGS genotypes of T. asahii. The tree was constructed using the same method described in the legend to Fig. 1. Knuc, Kimura’s parameter (10).
DISCUSSION
Although the identification and taxonomy of microorganisms are progressing rapidly, these processes require continual reassessment to remain current with the most recent scientific findings. The sequences of the 18S and 26S rRNA genes and of the internal transcribed spacer (ITS) region have been determined for almost all Trichosporon species (14, 16, 18). As Trichosporon species are phylogenetically very closely related to each other, there are more than a few cases in which analyses of the 18S, 26S, and ITS sequences fail to distinguish between species. We previously found that the IGS sequence is most suitable for differentiating between phylogenetically close species, because this region, which is located between the 26S and 18S rRNA genes, showed more sequence diversity than did the 26S rRNA gene or ITS sequences (19).
The causative antigens of SHP have previously been reported to be T. mucoides for serotype I, T. asahii for serotype II, and T. montevideense for serotype III (12). Unfortunately, that study identified only one representative strain for each serotype. Our results differ from that report only for serotype I. Although it is true that T. dermatis had not yet been described at the time of the previous report, the fact remains that, for reasons not yet known, T. mucoides was not isolated from the houses of SHP patients in the present study. Based on the very high frequency of the isolation of T. dermatis, we consider this species, not T. mucoides, to play a significant role in SHP induction in this study population. Although T. asahii was the most frequently isolated species among the serotype II species found in the patients' houses, a greater diversity of serotype II species were isolated than in the other serotypes; an additional five serotype II species were recovered (T. aquatile, T. coremiiforme, T. faecale, T. japonica, and T. ovoides).
Patients with SHP had positive reactions to inhalation challenge with antigens of the same serotype of T. cutaneum that had been isolated from their houses, indicating that patients are typically sensitized by exposure to the dominant Trichosporon species in their home environments (3). Based on this evidence, it appears that the Trichosporon serotype plays a significant role in the induction of SHP. While T. dermatis (serotype I), T. asahii (serotype II), and T. montevideense (serotype III) were the principal causative antigens identified in our mycological investigation, other species have also been considered to cause SHP. Sugita et al. (17) isolated T. ovoides (serotype II) from an SHP patient's house, and the crude antigen extracted from cultures of that isolate provoked a positive reaction in the patient when used in an inhalation challenge (personal communication). Although serotype I-III species are widely distributed in the environment, they were not isolated from the houses of patients in the present study. Previously, we identified a serotype I-III species, T. laibachii, isolated from the house of a Korean patient (unpublished data), as having the potential to cause SHP.
Our findings concerning the genotypes of T. asahii are of particular interest. While T. asahii isolates from the SHP patients' houses were mostly IGS genotype 3, T. asahii isolates recovered from clinical specimens were mostly genotype 1 (19). Our genotypic analysis of the cases of Japanese SHP and trichosporonosis caused by T. asahii revealed that each was dominated by a specific genotype. Therefore, the genotype should be considered when studying the relationship between T. asahii and SHP or trichosporonosis.
Acknowledgments
This study was supported in part by a Grant for the Promotion of the Advancement of Education and Research in Graduate Schools from the Ministry of Education, Culture, Sports, Science, and Technology of Japan
REFERENCES
- 1.Ando, M., K. Arima, R. Yoneda, and M. Tamura. 1991. Japanese summer-type hypersensitivity pneumonitis. Geographic distribution, home environment, and clinical characteristics of 621 cases. Am. Rev. Respir. Dis. 144:765-769. [DOI] [PubMed] [Google Scholar]
- 2.Ando, M., M. Suga, Y. Nishiura, and M. Miyajima. 1995. Summer-type hypersensitivity pneumonitis. Intern. Med. 34:707-712. [DOI] [PubMed] [Google Scholar]
- 3.Ando, M., T. Sakata, K. Yoshida, H. Yamasaki, S. Araki, K. Onoue, and T. Shinoda. 1998. Serotype-related antigen of Trichosporon cutaneum in the induction of summer-type hypersensitivity pneumonitis: correlation between serotype of inhalation challenge-positive antigen and that of the isolates from patients' homes. J. Allergy Clin. Immunol. 85:36-44. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 5.Guého, E., J. Tredick, and H. J. Phaff. 1984. DNA base composition and DNA relatedness among species of Trichosporon Behrend. Antonie Leeuwenhoek 50:17-32. [DOI] [PubMed] [Google Scholar]
- 6.Guého, E., M. T. Smith, G. S. de Hoog, G. Billon-Grand, R. Christen, and W. H. Batenburg-van der Vegte. 1992. Contributions to a revision of the genus Trichosporon. Antonie Leeuwenhoek 61:289-316. [DOI] [PubMed] [Google Scholar]
- 7.Guého, E., L. Improvisi, G. S. de Hoog, and B. Dupont. 1994. Trichosporon on humans: a practical account. Mycoses 37:3-10. [DOI] [PubMed] [Google Scholar]
- 8.Hazen, K. C. 1995. New and emerging yeast pathogens. Clin. Microbiol. Rev. 8:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda, R., M. Yokota, and T. Shinoda. 1996. Serological characterization of Trichosporon cutaneum and related species. Microbiol. Immunol. 40:813-819. [DOI] [PubMed] [Google Scholar]
- 10.Kimura, M. 1980. A simple method for estimation of evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 11.Makimura, K., S. Y. Murayama, and H. Yamaguchi. 1994. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 40:358-364. [DOI] [PubMed] [Google Scholar]
- 12.Nishiura, Y., K. Nakagawa-Yoshida, M. Suga, T. Shinoda, E. Guého, and M. Ando. 1997. Assignment and serotyping of Trichosporon species: the causative agents of summer-type hypersensitivity pneumonitis. J. Med. Vet. Mycol. 35:45-52. [DOI] [PubMed] [Google Scholar]
- 13.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 14.Scorzetti, G., J. W. Fell, A. Fonseca, and A. Statzell-Tallman. 2002. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2:495-517. [DOI] [PubMed] [Google Scholar]
- 15.Shimazu, K., M. Ando, T. Sakata, K. Yoshida, and S. Araki. 1984. Hypersensitivity pneumonitis induced by Trichosporon cutaneum. Am. Rev. Respir. Dis. 130:407-411. [DOI] [PubMed] [Google Scholar]
- 16.Sugita, T., and T. Nakase. 1998. Molecular phylogenetic study of the basidiomycetous anamorphic yeast genus Trichosporon and related taxa based on small subunit ribosomal DNA sequences. Mycoscience 39:7-13. [Google Scholar]
- 17.Sugita, T., A. Nishikawa, R. Ikeda, T. Shinoda, H. Sakashita, Y. Sakai, and Y. Yoshizawa. 1998. First report of Trichosporon ovoides isolated from the home of a summer type hypersensitivity pneumonitis patient. Microbiol. Immunol. 42:475-478. [DOI] [PubMed] [Google Scholar]
- 18.Sugita, T., A. Nishikawa, R. Ikeda, and T. Shinoda. 1999. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J. Clin. Microbiol. 37:1985-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugita, T., M. Nakajima, R. Ikeda, T. Matsushima, and T. Shinoda. 2002. Sequence analysis of the ribosomal DNA intergenic spacer 1 regions of Trichosporon species. J. Clin. Microbiol. 40:1826-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tashiro, T., H. Nagai, P. Kamberi, Y. Goto, H. Kikuchi, M. Nasu, and S. Akizuki. 1994. Disseminated Trichosporon beigelii infection in patients with malignant diseases: immunohistochemical study and review. Eur. J. Clin. Microbiol. Infect. Dis. 13:218-224. [DOI] [PubMed] [Google Scholar]
- 21.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson, K. C., and S. Kallichurum. 1970. Brain abscess due to Trichosporon cutaneum. J. Med. Microbiol. 3:191-193. [DOI] [PubMed] [Google Scholar]