Abstract
Helicobacter pylori is a genetically diverse bacterial species that chronically infects human stomachs and sometimes causes severe gastroduodenal disease. Studies of polymorphic DNA sequences can suggest geographic origins of individual strains. Here, we describe a 180-bp insertion (ins180), which is just after the translation stop of a gene of unknown function, near the promoter of jhp0152-jhp0151 two-component signal transduction genes in strain J99, and absent from this site in strain 26695. This ins180 insertion was found in 9 of 9 Gambian (West African), 9 of 20 (45%) South African, and 9 of 40 (23%) Spanish strains but in only 2 of 20 (10%) North American strains and none of 20 Lithuanian, 20 Indian, and 20 Japanese strains. Four South African isolates that lacked ins180 and that belonged to an unusual outlier group contained a 480-bp insertion at this site (ins480), whereas none of 181 other strains screened contained ins480. In further tests 56% (10 of 18) of strains from African Americans but only 17% (3 of 18) of strains from Caucasian Americans carried ins180 (P < 0.05). Thus, the H. pylori strains of modern African Americans seem to retain traces of African roots, despite the multiple generations since their ancestors were taken from West Africa. Fragmentary ins180-like sequences were found at numerous sites in H. pylori genomes, always between genes. Such sequences might affect regulation of transcription and could facilitate genome rearrangement by homologous recombination. Apparent differences between African-American and Caucasian-American H. pylori gene pools may bear on our understanding of H. pylori transmission and disease outcome.
Helicobacter pylori is a gram-negative microaerophilic bacterium that chronically infects the human gastric mucosa, typically for years or decades (8, 13, 19). It can cause a spectrum of gastroduodenal diseases including peptic ulcer and gastric cancer (8, 13) and has been designated a class I carcinogen by the World Health Organization (13). H. pylori acquisition occurs mostly in childhood (10, 36), often via maternal transmission (12, 24, 36).
Extensive geographic differences have been found in DNA sequences from H. pylori strains from different human populations (11, 14, 20, 24, 41). This led to a proposal that historical patterns of human migration and of racial admixture can be predicted from DNA sequences of H. pylori housekeeping genes (14, 41). An ability to discern H. pylori lineages from distinct human populations should also have medical relevance, because disease outcomes associated with H. pylori infection vary geographically (22, 27, 28, 38, 39), and some of this might stem from genetic differences between H. pylori populations (25, 26, 37).
More than 80% of Africans are infected with H. pylori, but their gastric cancer rates are lower than might be predicted based on this high prevalence (6, 22, 25-28, 38, 39). Infection rates in North America and Europe are often less than half of those in Africa, but yet distal gastric carcinoma is more common in these regions (22, 38, 39), a discordance referred to as the “African Enigma” (22). Gastric cancer rates also vary by country in Asia (29). Those in Japan and Korea are remarkably high (male, 82.7/100,000, and female, 32.8/100,000), some 10- to 20-fold higher than in Thailand and 100-fold higher than in India (29), whereas H. pylori seroprevalences in these south Asian countries are higher than those of Japan and Korea. This discordance is referred to as the “Asian Enigma” (29). These discordances have been ascribed variously to societal, human, bacterial genetic, and environmental factors, including concurrent helminth infections (the frequency of which varies among societies) (18, 25, 26, 27).
Although overall rates of gastric adenocarcinoma have decreased in the United States over the last 60 years (2), those in African-American, Hispanic, and Native American populations remain two to three times those in the Caucasian population (15). Again, much of this difference may stem from environmental, societal, or human genetic factors, but we infer that H. pylori genetic differences could also be important, i.e., disease outcomes might be affected by population differences in gene content or gene regulation in predominant H. pylori strains. Here we characterize 180-bp and 480-bp sequences (ins180 and ins480, respectively) found just downstream of a highly conserved but function-unknown gene and near a promoter of two-component signal transduction genes in a subset of H. pylori strains, positions that might allow it to affect expression of either the upstream or downstream genes. The 180-bp insert, in particular, was more common in strains from Africans and African Americans than in those from other human populations. ins480 was found only in an unusual outlier subset of South African strains.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. pylori strains were cultured on Trypticase soy agar II plates with 5% sheep blood (BBL) at 37°C in humidified ambient air containing 5% CO2. All strains used here were from our lab collections (M.H.F., R.M.P., and D.E.B.), and most have been described previously (7, 23, 31-34).
Among Lithuanian isolates (n = 20), 18 contained the cag pathogenicity island (PAI) and two did not. Twelve of the Lithuanian source patients were diagnosed with gastritis, six were diagnosed with duodenal ulcer, and two were diagnosed with gastric ulcer. Isolates from India (n = 20) each contained the cag PAI and were each from patients diagnosed with duodenal ulcer. All Japanese isolates (n = 20) contained the cag PAI and included strains from patients with gastric carcinoma (n = 4) and gastritis only (n = 4). All 29 African strains (9 Gambian and 20 South African) were from black Africans with gastric complaints. Four of nine Gambian patients had duodenal ulcer, and 2 of 20 South African patients had gastric or prepyloric ulcers. In addition, seven of nine Gambian and 14 of 20 South African strains contained the cag PAI and the other two Gambian and six South African strains did not. North American H. pylori isolates used in this study included 30 that contained and 24 that lacked the cag PAI. Included were 18 strains from African Americans. Clinical diagnoses of source patients included gastritis, gastric ulcer, gastric erosions, duodenal ulcer, esophagitis, and Barrett's esophagus. Spanish isolates (n = 40) were from patients with gastric complaints, and 24 strains contained the cag PAI and 16 did not.
Molecular biology techniques.
Genomic DNA was extracted using cetyltrimethylammonium bromide (CTAB), a cationic detergent, essentially as described previously (5). Briefly, 24- to 48-h H. pylori cultures were harvested from solid medium into 0.9% NaCl solution, and cell lysis was accomplished by addition of sodium dodecyl sulfate to a final volume of 0.5% and incubation at 37°C for 30 min. A 5 M concentration of NaCl was added to a final concentration of 725 mM, followed by 0.144 volume of 10% CTAB-0.7 M NaCl detergent. Bacterial lysates were phenol-chloroform and chloroform extracted, and genomic DNA was isopropanol precipitated using 0.86 volume of isopropanol, washed with 70% ethanol, and resuspended in 10 mM Tris, pH 8.5. PCR was performed using Taq DNA polymerase (Promega) and manufacturer-provided reaction buffer. Reaction mixtures consisted of 1× Mg2+-free polymerase reaction buffer, 2 mM MgCl2, 0.8 mM deoxynucleoside triphosphates (200 nM dATP, 200 nM dTTP, 200 nM dGTP, and 200 nM dCTP), 400 ng of each oligonucleotide, 200 ng of template DNA, and 1.25 U of Taq DNA polymerase in a final reaction volume of 50 μl. Amplification conditions consisted of a 5-min hot start and 25 to 30 cycles as follows: 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min. The oligonucleotide primers used to amplify ins180-ins480 were insFwd, 5′-GTGGCGCGTTTCTTGCAATACC-3′, and insRev, 5′-AACTCGCTCAAAAACTCGGC-3′. Cloning of PCR amplicons was accomplished using pGEM-T Easy and T4 DNA ligase (Promega). Plasmids were prepared using Qiagen spin columns, and restriction enzyme digestions were performed according to standard protocols (4). PCR analysis of the intergenic region of jhp0153 to jhp0152 was performed on 164 different H. pylori clinical isolates to determine the prevalence of the 180-bp and 480-bp insertions. One hundred forty-nine of these strains were from a global collection, and the others were from age- and gender-matched African-American (n = 18) and Caucasian (n = 18) patients in Tennessee (Table 1). Before running these tests, the researcher carrying them out was unaware of the origin of each of these 36 isolates.
TABLE 1.
Prevalence of ins180 and ins480 in the jhp0153-jhp0152 intergenic region among a panel of H. pylori clinical isolates from geographically and ethnically diverse source patients
| Parameter | Lithuanian | Indian | Japanese | Gambian | South African | North Americana | Spanishb | African Americanc | Caucasian Americanc |
|---|---|---|---|---|---|---|---|---|---|
| No. of strains tested | 20 | 20 | 20 | 9 | 20 | 20 | 40 | 18 | 18 |
| No. ins180 positive | 0 | 0 | 0 | 9 (100%) (P < 0.001) | 9 (45%) (P < 0.001) | 2 (10%) | 9 (23%) (P < 0.001) | 10 (56%) (P < 0.05) | 3 (17%) |
| No. ins480 positive | 0 | 0 | 0 | 0 | 4 (20%) (P < 0.001) | 0 | 0 | 0 | 0 |
Patient cohort largely Caucasian from central Tennessee; however, patient data are incomplete.
Five ins180-positive strains possessed the cag PAI, and four did not.
Isolates from age- and gender-matched patients living in central Tennessee.
DNA sequencing.
PCR products destined for sequencing were purified using a Qiagen PCR purification kit. DNA sequencing reactions performed on the purified amplicons were accomplished using Big Dye Terminator version 3.1 (Applied Biosystems, Inc.) and the sequencing reaction mixture described by the manufacturer. The cycling conditions were 94°C for 5 min, followed by 26 cycles of 94°C for 45 s, 50°C for 30 s, and 60°C for 4 min. Excess dye terminators were removed with DTF gel filtration cartridges (Edge Biosystems) according to the manufacturer's protocol. Sequencing was performed at the College of William and Mary on an ABI Prism 3100 genetic analyzer.
Nucleotide sequence accession number.
The sequences of ins180 and ins480 elements, determined here, have been deposited in GenBank (accession numbers AY731183, AY731182, and AY669071).
RESULTS
ins180 and ins480 in the jhp0153-jhp0152 intergenic region: inserted DNAs that are most common in African H. pylori strains.
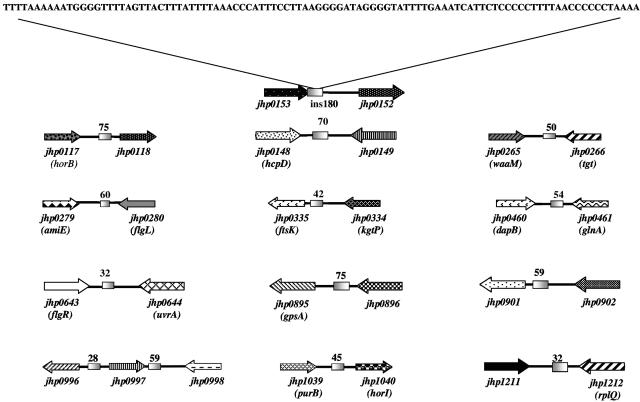
Comparison of the large intergenic region between two-component signal transduction genes jhp0152-jhp0151 and the conserved function-unknown gene jhp0153 of H. pylori strain J99 with the corresponding region of strain 26695 (between genes hp0166-hp0165 and hp0167) revealed a 180-bp segment (ins180) in J99 that was absent from the corresponding site in 26695. A BLAST search of this ins180 sequence against the complete H. pylori genome sequences revealed several smaller sequences of about 32 to 75 bp that were well matched to ins180 (>78% match; E value, ≤1 × 10−4) at 13 sites in the J99 genome (Fig. 1) and five sites in the 26695 genome (two of them at sites also identified in J99, downstream of jhp0643 and jhp1211 in Fig. 1). Every one of the 16 sequences so identified was between genes, not within them. Seven of the 13 in strain J99 were between 3′ ends of convergently transcribed genes, five were between the 3′ and 5′ ends of neighboring genes transcribed in the same direction (as with canonical ins180 in the jhp0153-jhp0152 site), and one was between the 5′ ends of divergently transcribed genes.
FIG. 1.
ins180 contains a highly repetitive element. A 99-bp subsequence of ins180 is shown. This sequence, internal to ins180, contains all subsequences found in 13 other intergenic regions of H. pylori J99 and which along with ins180 are represented as shaded boxes between adjacent open reading frames. The numbers above each box represent the length (in base pairs) of ins180 subsequence found at each location. Each ins180 subsequence region was identified by BLAST search, and each had an E value of ≤1 × 10−4. Typical percent homologies between these subsequences and ins180 were from 78 to 100. All genes with predicted functions are designated with three-letter gene designations within parentheses. Those genes without such designations were predicted to encode hypothetical proteins (1).
To better assess if canonical ins180 is unique to strain J99 or widespread in H. pylori populations (or if the corresponding deletion is unique to strain 26695 or common), we surveyed the jhp0152-jhp0153 (hp0166-hp0167) intergenic regions of 149 strains from several geographic regions, using primers for sites flanking the ins180 insertion site (Fig. 2) and scoring either the presence or absence of ins180 based on formation of 530-bp or 350-bp amplicons, respectively (Fig. 3A, lanes 2 and 3). Of the H. pylori strains tested, all those from Lithuania (n = 20), India (n = 20), and Japan (n = 20) lacked ins180, whereas all nine Gambian strains and 45% (9 of 20) of South African strains carried ins180 at this locus (P < 0.001), as did 23% of Spanish H. pylori strains (9 of 40). Among Spanish strains, 5 of 24 that carried the virulence-associated cag PAI and 4 of 16 that lacked the cag PAI carried ins180. Two of 20 U.S. strains also carried this element (Table 1).
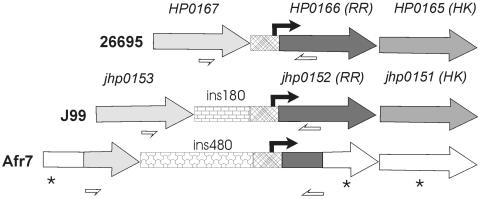
FIG. 2.
Sequence comparison of response regulator promoter regions from H. pylori strains 26695, J99, and Afr7. Matched shadings denote sequence similarities of predicted coding regions, and patterned regions illustrate extragenic sequence homology. Asterisks indicate unsequenced regions where nucleotide homology is unknown. Black arrows indicate transcription start points for the response regulator (RR)-histidine kinase (HK) operon that was previously determined in H. pylori strain 26695 (10, 17). Predicted transcription start points in H. pylori strains Afr7 and J99 are based upon sequence homology with the orthologous region in strain 26695. White half-arrows represent binding sites for primers insFwd and insRev. The amplicon without ins180 is 350 bp, and amplicons with ins180 and ins480 are ∼530 and ∼830 bp long, respectively.
FIG. 3.
ins180 is more frequent among H. pylori isolates from African Americans than from Caucasians. A PCR survey for ins180 in the response regulator jhp0152 promoter region from 36 H. pylori strains from both African Americans (n = 18) and Caucasians (n = 18) was performed. The jhp0153-jhp0152 intergenic region amplicon without ins180 is ∼350 bp, while the amplicons possessing ins180 are 530 bp. Intergenic regions possessing ins480 yield amplicons of ∼830 bp. (A) Lane 1, H. pylori strain Afr7 (containing ins480); lane 2, strain 26695 (no ins180 or ins480); lane 3, strain J99 (containing ins180); lane 4, 100-bp ladder. (B and C) H. pylori strains derived from18 African Americans and 18 U.S. Caucasians, respectively.
Four of the South African strains that lacked ins180 contained an even larger (480-bp) segment in this region (Fig. 3A, lane 1). Separate multilocus sequence typing (G. Dailide and D. E. Berg, unpublished data) has shown that these four strains belong to an outlier H. pylori type, which has been postulated variously to have evolved in an animal host and to have jumped to humans recently (9) or, when seen by others (14), to be an ancient lineage that evolved in a remote human population (e.g., possibly the Khoisan) (14). This ins480 sequence was not found in any of the nine Gambian strains (none of which are outliers). Taken together, our PCR tests indicated that African (especially West African) H. pylori strains are unusual in often containing the ins180 sequence. This sequence was also more common in Spanish than in Lithuanian or other Eurasian strains tested (P < 0.001).
Sequence analysis of ins180 and ins480.
Amplicons that putatively contained ins180 from one North American strain (B166) and one South African strain (Afr82) were sequenced. The jhp0152-jhp0153 intergenic regions of these strains were 90 to 95% identical to that of H. pylori strain J99 (GenBank accession numbers AY731182 and AY731183), indicating that the larger (530-bp) amplicons from North American and African strains do indeed reflect presence of the same 180-bp insertion. These ins180 elements seemed to be inserted at the same site as that occupied by ins180 in strain J99 (just downstream of the translation stop of hp0167) and contained a potential 41-codon open reading frame, which may not, however, be an authentic coding region, because it lacks a consensus Shine-Dalgarno sequence and would start translation with a UUG codon.
As noted above, 4 of 20 South African strains (each of an unusual outlier group) carried a 0.5-kb insertion between jhp0153 and jhp0152 that was not found in any of the strains from other regions that we tested. Sequence analysis of this region from one such strain, Afr7, revealed an insertion of a unique 480-bp segment in this strain's intergenic region, at the same site as ins180 (Fig. 2) (GenBank accession number AY669071). This ins480 segment is not found in either of the fully sequenced genomes (strain 26695 or J99), although an ∼80-bp internal sequence exhibits 70% identity with a segment of ins180 (data not shown). A search for open reading frames in ins480 revealed several small (160- to 190-bp) possible open reading frames with no significant DNA or protein-level homology to other current database entries.
ins180 is more frequent in H. pylori isolates from African Americans than in those from Caucasian Americans.
Given the presence of ins180 in each of our nine West African strains and also its presence in nearly half of the South African strains, we tested for this or other characteristic segments in this region in H. pylori from infected African-American patients, relative to isolates from age- and gender-matched Caucasian-American patients (n = 18 in each group). Thirteen of 36 (36%) strains yielded amplicons of approximately 530 bp, indicating the presence of ins180, whereas the other 23 (64%) yielded amplicons of 350 bp, indicating a simple empty site (Fig. 3 and Table 1). According to sorting of these data by patient ethnicity, 10 of 18 (56%) strains from African Americans versus 3 of 18 (17%) strains from other patients carried ins180 (P < 0.05) (Fig. 3B and C and Table 1). There was no correlation between the presence or absence of ins180 and carriage of the cag PAI or patient disease outcome (data not shown).
DISCUSSION
We identified a 180-bp segment (ins180) in the promoter region of jhp0152 in H. pylori strain J99 that is absent from the corresponding locus of strain 26695. These two strains, both from Caucasian males, are closely matched in DNA sequences flanking the ins180 site, which suggested that this element might be a useful marker for distinguishing different types of strains. In addition, jhp0152 and the downstream jhp0151 genes are coexpressed from a single promoter (10, 17), and ins180 is approximately 125 bp upstream of the transcriptional start site of jhp0152. We hypothesize that ins180 may introduce or disrupt a cis-acting sequence that affects downstream gene transcription. Since the external stimulus to which jhp0152 and jhp0151 (which encode components of a two-component signal transduction system) respond is likely to be a pH-dependent signal (35), it is interesting that ins180 might affect H. pylori colonization or persistence in its special gastric environment. Alternatively, the placement of these elements immediately downstream of the stop codon of jhp0153, which encodes a conserved H. pylori-specific protein of unknown function, raises the possibility of effects on posttranscriptional regulation of this gene. Sequences in 3′ untranslated regions can have profound effects on expression of the upstream gene (“retroregulation”) by affecting access to RNases and thereby mRNA stability (21, 30).
A BLAST search with the ins180 sequence identified conserved subsequences at 5 and 13 other sites in the genomes of the two sequenced H. pylori strains. Others have reported that ∼78% of the H. pylori genome was well conserved in a set of 15 strains (37). This represents ∼1,300 genes, most specifying metabolic and cellular functions. Many of the other nonconserved genes encoded DNA restriction and modification enzymes, transposases, and cell surface proteins (37). H. pylori strains also exhibit marked differences in gene arrangement (mostly due to inversion) and extensive point mutation (1, 19). Given the repetitive nature of ins180 and related sequences, we suggest that these elements constitute substrates for homologous recombination, which occurs readily in short repetitive sequences in H. pylori (4).
The ins180 sequence was nonrandomly distributed geographically (Table 1). Each of nine strains from The Gambia in West Africa, nearly half of South African strains, and one-fourth of Spanish strains contained ins180, whereas only a few of the other isolates that we first surveyed (all North American) carried this element. Also of note was the larger ins480 element, which was inserted at the same site and was found in an additional 4 of 29 South African strains (14%). Each of these four was of an unusual outgroup that is distinct from mainstream H. pylori strains of Africa or other continents (9; Dailide and Berg, unpublished). Reinforcing the view that such polymorphisms can be useful in identifying geographic origins of different H. pylori strains, 10 of 18 H. pylori isolates from African-American patients but only 3 of another 18 isolates from a matched Caucasian-American patient cohort carried this ins180 sequence. This is interpreted as reflecting H. pylori′s predominately intrafamilial transmission (3, 24, 36, 40) but is still most remarkable, given the multiple generations since the ancestors of most African Americans were taken from their West African homelands. In this context, it is tempting to imagine that ins180 in nearly one-fourth of Spanish strains might stem from the Moorish conquest and other movements of peoples between northwest Africa and the Iberian Peninsula (16).
The population correlations seen with ins180, in particular, make it attractive to consider that phylogenetic analyses of other loci in H. pylori strains from African Americans will also show evidence of West African origins. This may allow us to more clearly focus on bacterial versus other factors that contribute to the higher risk of gastric cancer in African Americans than in Africans. More generally, we suggest that easily scored markers such as ins180 and ins480 will be particularly useful in deciphering bacterial genetic versus other causes of particular disease associations, as epitomized by the African Enigma.
Acknowledgments
We thank Giedrius Dailide for valuable discussion and permission to cite unpublished data.
This work was supported by grants from the National Institutes of Health (AI 53062) and the Thomas F. and Kate Miller Jeffress Memorial Trust (J-602) to M.H.F. as well as grants DK 58587 and CA 77955 from the National Institutes of Health to R.M.P. and grants AI 38166, DK 53727, and DK 63041 from the National Institutes of Health to D.E.B. O.S. was the recipient of a UNESCO-ASM Travel Award. This research was also supported in part by a Howard Hughes Medical Institute grant through the Undergraduate Biological Sciences Education Program to the College of William and Mary.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Ming, B. L. King, E. D. Brown, and P. C. Doig. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society, Inc. 2004. Cancer facts and figures 2004. Online presentation. http://www.cancer.org/downloads/STT/CAFF_finalPWSecured.pdf. Accessed 19 February 2004.
- 3.Ando, T., R. M. Peek, D. Pride, S. M. Levine, T. Takata, Y. C. Lee, K. Kusugami, A. van der Ende, E. J. Kuipers, J. G. Kusters, and M. J. Blaser. 2002. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J. Clin. Microbiol. 40:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aras, R. A., J. Kang, A. I. Tschumi, Y. Harasaki, and M. J. Blaser. 2003. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc. Natl. Acad. Sci. USA 100:13579-13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
- 6.Campbell, D. I., B. F. Warren, J. E. Thomas, N. Figura, J. L. Telford, and P. B. Sullivan. 2001. The African enigma: low prevalence of gastric atrophy, high prevalence of chronic inflammation in west African adults and children. Helicobacter 6:263-267. [DOI] [PubMed] [Google Scholar]
- 7.Chalkauskas, H., D. Kersulyte, I. Cepuliene, V. Urbonas, D. Ruzeviciene, A. Barakauskiene, A. Raudonikiene, and D. E. Berg. 1998. Genotypes of Helicobacter pylori in Lithuanian families. Helicobacter 3:296-302. [PubMed] [Google Scholar]
- 8.Cover, T. L., D. E. Berg, M. J. Blaser, and H. L. T. Mobley. 2001. H. pylori pathogenesis, p. 509-558. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 9.Dailidiene, D., G. Dailide, K. Ogura, M. Zhang, A. K. Mukhopadhyay, K. A. Eaton, G. Cattoli, J. G. Kusters, and D. E. Berg. 2004. Helicobacter acinonychis: genetic and rodent infection studies of a Helicobacter pylori-like gastric pathogen of cheetahs and other big cats. J. Bacteriol. 186:356-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184:350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Disotell, T. R. 2003. Discovering human history from stomach bacteria. Genome Biol. 4:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drumm, B., G. I. Perez-Perez, M. J. Blaser, and P. M. Sherman. 1990. Intrafamilial clustering of Helicobacter pylori infection. N. Engl. J. Med. 322:359-363. [DOI] [PubMed] [Google Scholar]
- 13.Ernst, P. B., and B. D. Gold. 2000. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu. Rev. Microbiol. 54:615-640. [DOI] [PubMed] [Google Scholar]
- 14.Falush, D., et al. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582-1585. [DOI] [PubMed] [Google Scholar]
- 15.Fennerty, M. B., J. C. Emerson, R. E. Sampliner, D. L. McGee, L. J. Hixson, and H. S. Garewal. 1992. Gastric intestinal metaplasia in ethnic groups in the southwestern United States. Cancer Epidemiol. Biomark. Prev. 1:293-296. [PubMed] [Google Scholar]
- 16.Flores, C., N. Maca-Meyer, A. M. Gonzalez, P. J. Oefner, P. Shen, J. A. Perez, A. Rojas, J. M. Larruga, and P. A. Underhill. 2004. Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography. Eur. J. Hum. Genet. 12:855-863. [DOI] [PubMed] [Google Scholar]
- 17.Forsyth, M. H., P. Cao, P. P. Garcia, J. D. Hall, and T. L. Cover. 2002. Genome-wide transcriptional profiling in a histidine kinase mutant of Helicobacter pylori identifies members of a regulon. J. Bacteriol. 184:4630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox, J. G., P. Beck, C. A. Dangler, M. T. Whary, T. C. Wang, H. N. Shi, and C. Nagler-Anderson. 2000. Concurrent enteric helminth infection modulates inflammation and gastric immune responses and reduces helicobacter-induced gastric atrophy. Nat. Med. 6:535-542. [DOI] [PubMed] [Google Scholar]
- 19.Ge, Z., and D. E. Taylor. 1999. Contributions of genome sequencing to understanding the biology of Helicobacter pylori. Annu. Rev. Microbiol. 53:353-387. [DOI] [PubMed] [Google Scholar]
- 20.Ghose, C., G. I. Perez-Perez, M.-G. Dominguez-Bello, D. T. Pride, C. M. Bravi, and M. J. Blaser. 2002. East Asian genotypes of Helicobacter pylori strains in Amerindians provide evidence for its ancient human carriage. Proc. Natl. Acad. Sci. USA 99:15107-15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins, C. F., R. S. McLaren, and S. F. Newbury. 1988. Repetitive extragenic palindromic sequences, mRNA stability and gene expression: evolution by gene conversion? A review. Gene 72:3-14. [DOI] [PubMed] [Google Scholar]
- 22.Holcombe, C. 1992. Helicobacter pylori: the African enigma. Gut 33:429-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatino, W. W. Su, Z. J. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcon, M. Lopez-Brea, G. Balakrish Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Wong, S. K. Lam, F. O. Olfat, T. Boren, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kivi, M., Y. Tindberg, M. Sorberg, T. H. Casswall, R. Befrits, P. M. Hellstrom, C. Bengtsson, L. Engstrand, and M. Granstrom. 2003. Concordance of Helicobacter pylori strains among families. J. Clin. Microbiol. 41:5604-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuipers, E. J., and G. A. Meijer. 2000. Helicobacter pylori gastritis in Africa. Eur. J. Gastroenterol. Hepatol. 12:601-603. [DOI] [PubMed] [Google Scholar]
- 26.Louw, J. A., M. S. G. Kidd, A. F. Kummer, K. Taylor, U. Kotze, and D. Hanslo. 2001. The relationship between Helicobacter pylori infection, the virulence genotypes of the infecting strain and gastric cancer in the African setting. Helicobacter 6:268-273. [DOI] [PubMed] [Google Scholar]
- 27.McFarlane, G., D. Forman, and G. Lachlan. 2001. A minimum estimate for the incidence of gastric cancer in eastern Kenya. Br. J. Cancer 85:1322-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McFarlane, G., J. Wyatt, D. Forman, and G. W. Lachlan. 2000. Trends over time in Helicobacter pylori gastritis in Kenya. Eur. J. Gastroenterol. Hepatol. 12:617-621. [DOI] [PubMed] [Google Scholar]
- 29.Miwa, H., M. F. Go, and N. Sato. 2002. H. pylori and gastric cancer: the Asian enigma. Am. J. Gastroenterol. 97:1106-1112. [DOI] [PubMed] [Google Scholar]
- 30.Montanez, C., J. Bueno, U. Schmeissner, D. L. Court, and G. Guarneros. 1986. Mutations of bacteriophage lambda that define independent but overlapping RNA processing and transcription termination sites. J. Mol. Biol. 191:29-37. [DOI] [PubMed] [Google Scholar]
- 31.Peek, R. M., L. J. van Doorn, J. P. Donahue, K. T. Tham, C. Figueiredo, M. J. Blaser, and G. G. Miller. 2000. Quantitative detection of Helicobacter pylori gene expression in vivo and relationship to gastric pathology. Infect. Immun. 68:5488-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peek, R. M., Jr., H. P. Wirth, S. F. Moss, M. Yang, A. M. Abdalla, K. T. Tham, T. Zhang, L. H. Tang, I. M. Modlin, and M. J. Blaser. 2000. Helicobacter pylori alters gastric epithelial cell cycle events and gastrin secretion in Mongolian gerbils. Gastroenterology 118:48-59. [DOI] [PubMed] [Google Scholar]
- 33.Peek, R. M., Jr., M. J. Blaser, D. J. Mays, M. H. Forsyth, T. L. Cover, S. Y. Song, U. Krishna, and J. A. Pietenpol. 1999. Helicobacter pylori strain-specific genotypes and modulation of the gastric epithelial cell cycle. Cancer Res. 59:6124-6131. [PubMed] [Google Scholar]
- 34.Peek, R. M., Jr., S. A. Thompson, J. P. Donahue, K. T. Tham, J. C. Atherton, M. J. Blaser, and G. G. Miller. 1998. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Assoc. Am. Physicians 110:531-544. [PubMed] [Google Scholar]
- 35.Pflock, M., P. Dietz, J. Schar, and D. Beier. 2004. Genetic evidence for histidine kinase HP0165 being an acid sensor of Helicobacter pylori. FEMS Microbiol. Lett. 234:51-61. [DOI] [PubMed] [Google Scholar]
- 36.Rothenbacher, D., G. Bode, G. Berg, U. Knayer, T. Gonser, G. Adler, and H. Brenner. 1999. Helicobacter pylori among pre-school children and their parents: evidence of parent-child transmission. J. Infect. Dis. 179:398-402. [DOI] [PubMed] [Google Scholar]
- 37.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segal, I., R. Ally, and H. Mitchell. 2001. Helicobacter pylori: an African perspective. Q. J. Med. 94:561-565. [DOI] [PubMed] [Google Scholar]
- 39.Segal, I., R. Ally, and H. Mitchell. 2001. Gastric cancer in sub-Saharan Africa. Eur. J. Cancer Prev. 10:479-482. [DOI] [PubMed] [Google Scholar]
- 40.Tindberg, Y., C. Bengtsson, F. Grannath, M. Blennow, O. Nyren, and M. Granstrom. 2001. Helicobacter pylori infection in Swedish school children: lack of evidence of child-to-child transmission outside the family. Gastroenterology 121:310-316. [DOI] [PubMed] [Google Scholar]
- 41.Wirth, T., X. Wang, B. Linz, R. P. Novick, J. K. Lum, M. Blaser, G. Morelli, D. Falush, and M. Achtman. 2004. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proc. Natl. Acad. Sci. USA 101:4746-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]