Abstract
Cryptococcus gattii causes life-threatening infection of the pulmonary and central nervous systems in hosts with normal immunity and traditionally has been considered to be restricted geographically to tropical and subtropical climates. The recent outbreak of C. gattii in the temperate climate of Vancouver Island, BC, Canada, led to a collaborative investigation. The objectives of the current study were to ascertain the environmental source of the outbreak infections, survey the molecular types of the outbreak and environmental cryptococcal isolates, and determine the extent of genetic diversity among the isolates. PCR-fingerprinting and amplified fragment length polymorphism (AFLP) were used to examine the genotypes, and mating assays were performed to determine the mating type of the isolates. All outbreak and environmental isolates belonged to C. gattii. Concordant results were obtained by using PCR-fingerprinting and AFLP analysis. The vast majority of clinical and veterinary infections were caused by isolates of the molecular type VGII/AFLP6, but two were caused by molecular type VGI/AFLP4. All environmental isolates belonged to molecular type VGII/AFLP6. Two or three subtypes were observed within VGII/AFLP6 among outbreak and environmental isolates. All mating-competent isolates were of the α-mating type. The emergence of this usually tropical pathogen on Vancouver Island highlights the changing distribution of this genotype and emphasizes the importance of an ongoing collaborative effort to monitor the global epidemiology of this yeast.
Keywords: PCR fingerprinting, amplified fragment length polymorphism (AFLP), restriction fragment length polymorphism (RFLP), mating type
The Cryptococcus neoformans species complex comprises basidiomycetous yeasts causing life-threatening disease of the central nervous system, lung, and skin in humans and animals. C. neoformans var. grubii and C. neoformans var. neoformans have been isolated worldwide, typically causing disease in hosts with impaired immunity. C. neoformans var. gattii, recently raised to species level as Cryptococcus gattii (1), was until now considered to be restricted to tropical and subtropical climates and is most often the cause of infection in hosts with normal immunity (2, 3).
A recent outbreak of C. gattii infection in the temperate climate of Vancouver Island, BC, Canada (4, 5), led to the initiation of a collaborative effort to investigate the extent of this outbreak, to describe the temporal, geographic, demographic, microbiological, and clinical characteristics of cryptococcosis cases, and to identify possible environmental reservoirs responsible for the outbreak.
Vancouver Island lies on the west coast of Canada, with an area of 32,284 km2. The island has a population of ≈697,000, two-thirds of which live in the southern part (http://geodepot2.statcan.ca/Diss/Highlights). Vancouver Island has a mean temperature of 2.7°C in winter and 17.6°C in summer, and the mean precipitation is 136 mm/month in winter and 40 mm/month in summer (www.islands.bc.ca/general/frclimate.html).
The incidence of C. gattii infection among the people of Vancouver Island from 1999 to 2003 was between 8.5 and 37 cases per million residents per year (5). This incidence is significantly greater than that of C. gattii infection typically observed in Australia (0.94 cases per million residents per year), where C. gattii is endemic (3, 6). A public advisory was issued by the British Columbia Centre for Disease Control on June 6, 2002 (www.bccdc.org) advising the public, medical practitioners, and veterinarians of the potential health risk and telling them to be alert for symptoms and to seek early diagnosis. Because the majority of patients were immunocompetent, cryptococcal infection was listed as a notifiable disease in BC.
A preliminary study of the clinical aspects of the outbreak revealed that 38 human cryptococcosis cases diagnosed between January 1999 and December 2001 were caused by C. gattii.Ofthose cases, 22 (58%) were male and 36 (94.7%) were Caucasian, whereas 2 (5.3%) were of Asian descent. Of the cases, 72% had lung involvement and 26% had CNS involvement (ref. 5 and M.F., L.M., K.H.B., M. G. Romney, S.E.K., C. Stephen, M. Starr, M. Pearce, S. Mak, T.B., W.M., and P. Kibsey, unpublished data). The mean age at the point of diagnosis was 59.7 years (range, 11–87 years). However a case-control study found that infection status was not age-dependent (M.F., L.M., K.H.B., M. G. Romney, S.E.K., C. Stephen, M. Starr, M. Pearce, S. Mak, T.B., W.M., and P. Kibsey, unpublished data).
The molecular epidemiological aspects of the outbreak form the basis for this study. The aims were to obtain cryptococcal isolates from environmental sources on Vancouver Island to ascertain the source of outbreak infections, survey the molecular types of outbreak and environmental cryptococcal isolates, and determine the extent of genetic diversity among those isolates.
Materials and Methods
Selection of Outbreak Cases. The criteria for human cryptococcal infection to qualify as an outbreak case included observing symptoms consistent with cryptococcal infection accompanied with isolation of C. gattii from normally sterile sites or bronchoalveolar lavage, or HIV-negative status accompanied by cryptococcal isolation. For this study, 21 isolates from immunocompetent humans were available. Six isolates from HIV-positive hosts from BC, regarded as non-outbreak-related, were included for the purposes of quality control.
Cryptococcal infection was diagnosed in 35 animals during 2000 and 2001, including 13 dogs (Canis familiaris), 17 cats (Felis catus), 2 ferrets (Mustela putorious furo), and 3 wild Dall's porpoises (Phocoenidae dalli) (4).
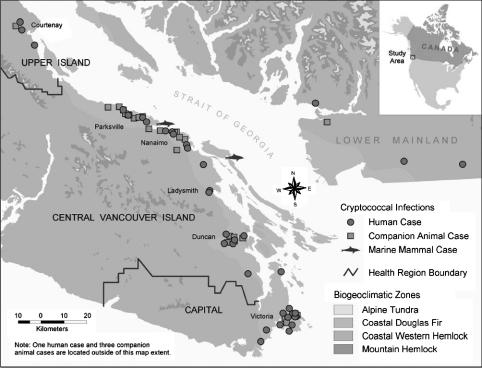
Thirty-five of the 38 (92.1%) human cases and 28 terrestrial animals of the 35 (80.0%) animal cases resided on the east coast of Vancouver Island, designated the Coastal Douglas Fir biogeoclimatic zone (CDF). The remaining three human cases and four terrestrial animal cases resided on the BC mainland but had visited Vancouver Island within a year before diagnosis. The corpses of three porpoises were found on the shores of some Gulf Islands in the Strait of Georgia, located between Vancouver Island and the BC mainland. The geographical distribution of the human and animal cases is illustrated in Fig. 1.
Fig. 1.
Geographical distribution of human and animal cases of cryptococcosis diagnosed between January 1999 and January 2002. All Vancouver Island cases occurred in patients and animals that lived on the east coast of Vancouver Island in the CDF biogeoclimatic zone. All mainland-based cases occurred in people who had traveled to the CDF zone on Vancouver Island within 1 year prior to the onset of disease. Environmental sampling primarily was concentrated on areas within the CDF, particularly around residences and recreational areas of patients and animals.
Clinical and veterinary isolates were sent to the BC Centre for Disease Control laboratories for culturing and identification. Samples were cultured directly on Sabouraud's dextrose agar containing 20 mg/liter gentamycin and grown at 37°C for 48 h.
Environmental Cryptococcal Isolation. Environmental sampling on Vancouver Island was conducted near patient or companion animal residences, including backyards, walking trails, parks, and recreational areas. Soil, woody debris, bark, and leaves were sampled as described in ref. 7. Between October 2001 and June 2002, 732 samples were taken from environmental sources on Vancouver Island (13 different sites), the BC lower mainland (1 site), and two of the Gulf islands. These samples contained 634 swabs from 347 different trees, including eucalypts (Eucalyptus spp.), arbutus (Arbutus menziesii), alder (Alnus spp.), cedar (Cedrus spp.), Douglas fir (Pseudotsuga spp.), and others. Sterile transport swabs containing Amies medium without charcoal (Starplex Scientific, Etobicoke, ON, Canada) were immersed in saline and used to swab probable areas of cryptococcal habitation, such as hollows (hole, niche, or scar), areas of woody debris, shallow cavities, and under bark. For some samples, a single defined hollow in the tree was swabbed, whereas for other samples, one swab was used to sample from several different areas of a single tree (composite swab). In some cases, repeated samples (retests) from a single tree were taken at different time points to determine whether the tree was truly colonized. Replicate samples were taken, where multiple swabs were used in a single hollow at a single time point, to assess the predictive value of having an individual positive or negative swab. Swabs were transferred to birdseed agar (8) and incubated at 30°C for up to 10 days for detection of the melanized colonies characteristic of cryptococcal isolates.
Eleven air samples were taken to test for the existence of airborne cryptococcal propagules in the environs of the trees where swabs had been taken. The Andersen six-stage air sampling head (Andersen Instruments, Smyrna, GA) was connected to an Air-Con2 pump (Gilian Instruments, West Caldwell, NJ). The six stages of the Andersen sampling head correspond to different sampling efficiencies, with each stage having a progressively narrowing air inlet, designed to mimic the narrowing of human bronchial passages: stage 1, >7.0 μm; stage 2, 4.7–7.0 μm; stage 3, 3.3–4.7 μm; stage 4, 2.1–3.3 μm; stage 5, 1.1–2.1 μm; and stage 6, 0.65–1.1 μm. Each stage of the sampling head contained a plate of birdseed agar. The airflow rate was controlled at 28.3 liters/min for 10 min. Birdseed agar plates were incubated as described above.
We collected 87 samples of soil or tree debris in individual ziplock bags. In the laboratory, ≈2 g of soil or debris was suspended in 10 ml of sterile distilled water and homogenized by vigorous vortexing, and 0.5-ml aliquots of the soil/debris suspension were transferred to birdseed agar plates, followed by incubation as described above.
Identification of Yeast Isolates. Cryptococcal isolates from tree swabs and air samples, detected on birdseed agar, were further subcultured on Sabouraud's dextrose agar to obtain a pure culture. Cryptococcal strains were identified by using the API 20C AUX identification system (BioMerieux, Charbonnier les Bains, France). The yeasts were determined to be C. gattii by culturing the isolates on l-canavanine-glycine-bromothymol blue medium (9). Serotyping was performed with the Crypto Check Kit (Iatron Laboratories, Tokyo). Information for all studied isolates is given in Table 1, which is published as supporting information on the PNAS web site, including strain number, serotype, molecular type [determined by PCR-fingerprinting, URA5-restriction fragment length polymorphism (RFLP) and AFLP analysis], mating type, host factors, and geographical source.
Genotyping. High molecular weight genomic DNA was extracted from cryptococcal cells as described in ref. 10. PCR-fingerprinting using the minisatellite specific primer M13 (5′-GAGGGTGGCGGTTCT-3′) was performed as described in ref. 10. The software gelcomparii (Version 2.5, Applied Maths, Kortrijk, Belgium) was used to calculate the PCR-fingerprint profile similarity of the 94 outbreak and environmental isolates from Vancouver Island. Normalization of the gels was conducted by using the 1-kb size markers (GIBCO/BRL), as external standards and common bands within the profiles as internal standards. The DNA bands for each fingerprint profile were defined manually with a band position tolerance of 0.8% and an optimization setting of 0.2%. Pairwise similarity was calculated by using the Dice coefficient, and a phenogram was generated with the unweighted pairgroup method using arithmetic means (UPGMA). URA5-RFLP analysis was performed as described in ref. 11. AFLP analysis was performed as described in ref. 12 by using both UPGMA and single linkage. A set of eight standard laboratory strains, representing each of the eight previously defined molecular types (WM148 = serotype A, VNI/AFLP1; WM626 = serotype A, VNII/AFLP1A; WM628 = serotype AD, VNIII/AFLP3; WM629 = serotype D, VNIV/AFLP2; WM179 = serotype B, VGI/AFLP4; WM178 = serotype B, VGII/AFLP6; WM161 = serotype B, VGIII/AFLP5; and WM779 = serotype C, VGIV/AFLP7) (Table 1 and refs. 10, 11, and 13) were included to assign the molecular type and ensure reproducibility. Determination of Mating Type. The serotype B clinical isolates NIH 198 (MATa) and NIH 112 (MATα) and the environmental isolate CBS 7750 (MATα) (Table 1) were used as tester strains in matings to determine the mating types. Cells from each mating partner were mixed on V8 juice agar adjusted to pH 5 (14) and incubated in darkness at 25°C for up to 4 weeks. Isolates also were incubated alone on V8 juice agar to detect haploid fruiting (15). Plates were examined regularly for evidence of hyphae and basidiospore chains indicative of mating.
Results
Outbreak Isolates. We analyzed 21 clinical and 6 veterinary isolates of C. gattii originating from Vancouver Island or the closely surrounding lower BC mainland with PCR-fingerprinting by using the primer M13, URA5-RFLP and AFLP analysis, to discern molecular/genotypes and genetic variation. Six non-outbreak-related serotype A (A1M R266, A1M R312, and A1M R449) and D (A1M R270, A1M R314, and A1M R461) isolates from BC HIV-positive hosts were included for quality control.
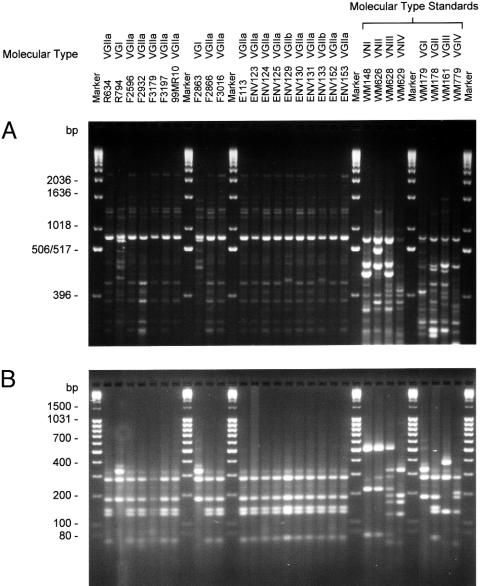
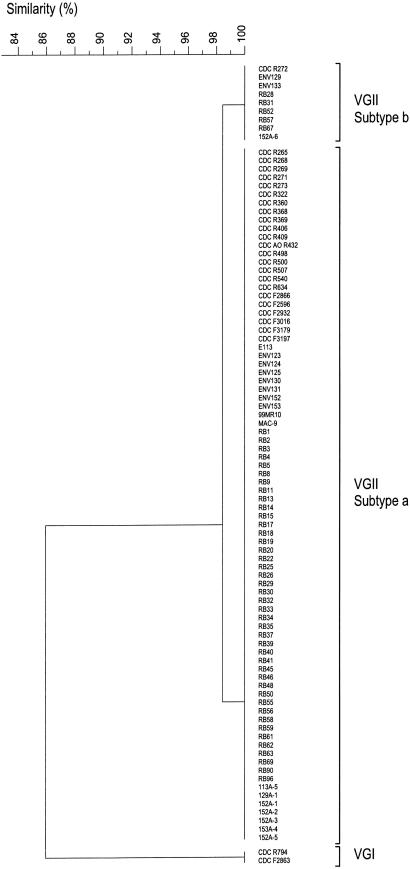
Selected PCR-fingerprint patterns and URA5-RFLP profiles are shown in Fig. 2 A and B, respectively. The molecular-type classification for each isolate was the same with both typing methods (Table 1). Of the 21 clinical isolates, 20 (95.2%) were VGII, whereas 1 was VGI. Of the 6 veterinary isolates, 5 (83.3%) were VGII, whereas 1 was VGI. PCR-fingerprinting (10) divided all outbreak and environmental VGII isolates into two subgroups, designated VGIIa and VGIIb. The PCR-fingerprint profiles of the two subgroups were 98% similar, distinguishable by a single band of ≈1,218 bp. PCR-fingerprints of the two VGI, all VGIIa, and all VGIIb isolates were found to be identical within each group (Fig. 3). AFLP analysis (12) identified the majority of outbreak isolates as AFLP6/VGII, whereas two isolates were AFLP4/VGI (Fig. 6, which is published as supporting information on the PNAS web site). The overall similarity of the AFLP profiles of the clinical isolates was 84.6% using UPGMA and 94.9% using single linkage. Most of the isolates formed a major cluster, designated AFLP6A/VGIIa. Four clinical isolates belonging to VGIIa differed by one additional AFLP band and were subgrouped as AFLP6C (Fig. 6). One clinical isolate belonged to the AFLP6B/VGIIb subgroup, differing by the presence of three additional bands and the absence of one band in the AFLP patterns (Figs. 4 and 6). PCR-fingerprint data are consistent with the AFLP data, but the latter method discerned more polymorphisms between the clinical isolates (Fig. 6).
Fig. 2.
Examples of molecular profiles obtained from isolates from the Vancouver Island cryptococcosis outbreak. (A) PCR-fingerprint profiles amplified by using the M13 primer (5′-GAGGGTGGCGGTTCT-3′). Profiles were separated on a 1.4% agarose gel with 1× Tris-borate-EDTA (TBE) buffer [10.8 g/liter Tris base/5.5 g/liter boric acid/4 mM EDTA (pH 8.0)] to 14 cm, with a 1-kb size marker (GIBCO/BRL). (B) URA5-RFLP used enzymes HhaI and Sau96I. Profiles separated on a 3.0% agarose-TBE gel with a 100-bp mass ruler (Fermentas, Hanover, MD).
Fig. 3.
Phenogram of PCR-fingerprint profiles obtained from Vancouver Island outbreak and environmental isolates with the single primer M13, created with the program gelcompar II (Applied Maths), with Dice coefficient and UPGMA.
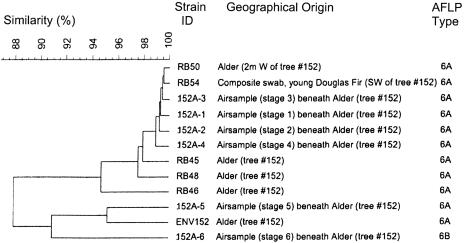
Fig. 4.
Clustering of AFLP patterns of isolates from a single Douglas fir tree (no. 152) by using UPGMA and band matching. Both AFLP genotypes, AFLP6A (11 isolates) and AFLP6B (1 isolate), occurred on this tree.
Environmental Isolates. Of the 732 environmental samples taken until June 2002 across 16 different sites in BC, 58 C. gattii isolates were obtained from 25 trees in two different locations on Vancouver Island. These included 57 isolates from 24 trees at Rathtrevor Beach Provincial Park (RBPP), located in Parksville, and 1 isolate from MacMillan Park, located in Cathedral Grove, ≈31 km west of Parksville. Both parks are located in the CDF of Vancouver Island. C. gattii was isolated from the following five native tree species: alder (n = 5), cedar (n = 1), Douglas fir (n = 16), grand fir (n = 1), and Garry oak (n = 2). There were no positive samples from eucalypts.
Three of the 11 air samples taken were positive, yielding a total of eight isolates, recovered from stage 1 (129A-1), stage 5 (113A-5), and all six stages (152A-1 to 152A-6) of the air sampler. The 11 air samplings yielded a range of 0–1,081 cryptococcal colony-forming units (cfu) per m3, with a geometric mean of 7 cfu/m3 air and a geometric SD of 10.72. The positive air samples were obtained near trees that had C. gattii positive swabs.
We analyzed 87 samples of soil and tree debris, with only one soil sample being positive for C. gattii (115 cfu/g soil).
All 67 isolates belonged to molecular type VGII/AFLP6 as determined by PCR-fingerprinting and URA5-RFLP and AFLP analysis, with 59 belonging to the subtype VGIIa/AFLP6A and 8 to VGIIb/AFLP6B (Figs. 3 and 6). Some polymorphisms were apparent among the AFLP banding patterns from the environmental isolates (Fig. 6).
Genetic Variation. There was no genetic variation observed between the two isolates of VGI/AFLP4, which were obtained from a human female (A1M R794) and a wild Dall's porpoise (A1M F2863). There were two subtypes observed, designated VGIIa/AFLP6A (n = 83; 90.2%) and VGIIb/AFLP6B (n = 9; 9.8%), differing by one band in their PCR-fingerprint profiles and four bands in the AFLP patterns (see above). One of the strains belonging to the VGIIb subtype was isolated from a clinical source (A1M R272), whereas the remaining eight isolates were environmental sources (ENV129, ENV133, RB28, RB31, RB52, RB57, RB67, and 152A-6), including one from an air sample (152A-6). In addition, two clinical (A1M F2932 and A1M F3179) and two veterinary (A1M F2866 and A1M F2596) VGIIa isolates possessed an additional AFLP band (Fig. 6, arrowhead) and formed a subgroup, AFLP6C.
Of the eight isolates from air samples, six were obtained from a single collection beneath alder tree no. 152, one from each of the six stages of the air sampling head; isolates from the first five stages were VGIIa (152A-1, 152A-2, 152A-3, 152A-4, and 152A-5), whereas the isolate from the sixth stage was VGIIb (152A-6). AFLP analysis revealed three subclusters for those isolates: ENV152 and 152A-5 (95.2% similarity using UPGMA, ALFP6A/VGIIa), isolates 152A-1 to 152A-4 showing 99.0% similarity using UPGMA (AFLP6A/VGIIa), and isolate 152A-6 belonging to AFLP 6B/VGIIb (Figs. 4 and 6). Analysis of the isolates from tree no. 152 clearly demonstrate that this tree was colonized by isolates of two molecular types, VGIIa/AFLP6A and VGIIb/AFLP6B.
The eight environmental VGIIb/AFLP6B isolates were obtained from eight different tree environs in RBPP, the same park from which the VGIIa/AFLP6A isolates were obtained. Six were obtained from swabs of Douglas fir trees nos. 126, 129, 130, 133, 193, and 196, one from grand fir tree no. 169, and one by air sampling close to alder tree no. 152. In the case of two Douglas fir trees, replicate samples from the same hollow yielded VGIIb/AFLP6B isolates (tree no. 126, RB57; tree no. 130, RB67) as well as VGIIa/AFLP6A isolates (tree no. 126, RB58; tree no. 130, RB69). The only available isolate (MAC-9) in the study obtained outside of RBPP was isolated from a cedar tree in MacMillan Park, in Cathedral Grove, Vancouver Island, and was identified as VGIIa/AFLP6A.
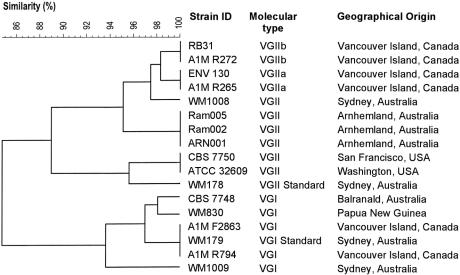
To determine the degree of similarity between the outbreak isolates and globally isolated C. gattii strains, we compared selected outbreak strains and other VGI and VGII strains randomly chosen from a PCR-fingerprinting database of ≈1,000 cryptococcal strains (S.E.K. and W.M., unpublished data) (Fig. 5). PCR-fingerprint profiles of the two VGII subtypes characteristic for the outbreak isolates have a high similarity of 97.5 or 95.1% with the Australian VGII isolates WM1008, or Ram2, Ram5, and ARN1, respectively. The similarity to the VGII isolates CBS 7750 and ATCC 32609 from the U.S. was 89.0%. PCR-fingerprints from the two VGI outbreak isolates were found to be identical to that of the Australian VGI strain WM179 and highly similar to the Australian VGI isolate CBS 7748 and the Papua New Guinea isolate WM830 (97.0%). However, because there has been no identification of VGI from environmental sources on Vancouver Island, the possibility remains that these infections may have been acquired in a different location.
Fig. 5.
Comparison of PCR-fingerprint similarity between selected outbreak isolates and previously studied global VGI and VGII isolates. PCR-fingerprints were generated by using the M13 primer and analyzed with gelcompar II software (Applied Maths) by using Dice coefficient and UPGMA.
Mating Type. Mating studies revealed that 83 of 92 (90%) isolates were able to mate. The ability to mate was independent of the clinical or environmental source of the isolates, because 23 of 26 (88%) outbreak and 60 of 66 (91%) of the environmental isolates were mating-competent (Table 1). All mating-competent strains had the MATα mating type, because they mated with the MATa tester strain (NIH 198) but not with either of the two MATα tester strains (NIH 112 and CBS 7750). Self-fertility of the isolates was ruled out, because all isolates grew vegetatively when incubated alone on V8 juice agar. Nine isolates failed to mate with the tester strains. An association between mating-competency and molecular subtype was observed. Among the isolates that did not mate, only one belonged to VGIIa (A1M F2866, from a porpoise), seven belonged to VGIIb [one clinical (A1M R272) and six environmental (ENV133, RB31, RB52, RB57, RB67, and 152A-6)], and one belonged to VGI (A1M R794, a human isolate). Thus, seven of the nine VGIIb isolates (78%) and one of the two VGI isolates (50%) were not mating-competent.
Discussion
Until the recent emergence of cryptococcal infection on Vancouver Island, C. gattii had been considered to be restricted to areas with tropical and subtropical climates (2). The identification of large-scale colonization of C. gattii in the environment occurring in a temperate climate zone indicates a striking change in the distribution of this species. Furthermore, the identification of the C. gattii genotype VGII/AFLP6 as the causative organism is of great significance. It represents the largest environmental reservoir of this genotype reported to date. Previously, environmental VGII/AFLP6 isolations have been sporadic, with sources including Eucalyptus camaldulensis bark debris in California (12, 16), eucalypt species in Australia (3), a variety of tree species in Brazil (11, 17, 18), a wasp nest in Uruguay (12, 19), and a single isolate from insect frass associated with Eucalyptus tereticornis in Western Sydney, Australia (13). Isolates belonging to the VGII/AFLP6 genotype have been documented from clinical and veterinary sources, including Aborigines from Arnhemland in the Northern Territory of Australia (3, 20), a sick goat in Aruba (12), and clinical isolates from Brazil (17). This genotype had never been reported previously from clinical or environmental sources in Canada. However, a single isolate (ATCC 32609, also known as CBS6956 or NIH444) cultured from sputum of a patient in Seattle, circa 1971, was retrospectively found to belong to VGII/AFLP6 (12), indicating that this genotype may have been present in the area for >30 years rather than being a recent introduction. This finding also may suggest that a more sufficient generation of infectious propagules has occurred because of a recent unknown event, resulting in a widespread airborne proliferation of these propagules into the environment and a corresponding increase in the prevalence of C. gattii infections on Vancouver Island.
Reports of cryptococcosis and cryptococcal isolation within Canada are sparse. A study of cryptococcosis in Canada did not examine the variety and serotype of the causative agents, but at least 23% of the patients were HIV positive (21). The same study found that the provinces of Quebec, Ontario, and Alberta have the highest incidence of diagnosed cryptococcal infection (21). A single-case study of canine cryptococcosis, probably originating from Vancouver Island, also did not examine the variety or serotype of the causative organism (22). In addition, C. neoformans was inadvertently isolated from a patient in Montréal, Quebec, during a routine sampling of dermatophytes between 1963 and 1973, but this isolate was not investigated further (23). A 1984 global epidemiology study of C. neoformans (2) found that only 7.7% of 78 isolates from Canada belonged to C. gattii (one was serotype C; the remainder were serotype B); the majority, 79.5%, were C. neoformans var. grubii (serotype A), 6.4% were C. neoformans var. neoformans (serotype D), and 6.4% belonged to the hybrid serotype AD. These serotype frequencies were very similar to those among U.S. isolates, with the exception of Southern California, where a relatively high occurrence of C. gattii serotype B isolates was observed (2).
At the time that the environmental samples were obtained, the concentration of C. gattii appeared to be highest in RBPP, with several trees yielding positive swabs with replicate samplings and repeated sampling at different time points. Tree no. 113 in particular appeared to be associated with a high concentration of cryptococcal propagules. C. gattii isolates were consistently obtained from replicate and retest swabs taken over a 4-month period (April-June 2002); one isolate was obtained from soil collected from the base of the tree, and another isolate was obtained from stage 5 of an air sample taken beneath the canopy of the tree. Another positive air sample, taken from beneath the canopy of tree no. 152, yielded cryptococcal isolates from each of the six stages of the air-sampling head. This result indicates not only a high concentration of airborne cryptococcal particles in that area but that the size of the propagule was sufficiently small (<1.1 μm) to reach the deepest levels of the air sampler and, potentially, the deeper areas of the human lung. Although it is clear that RBPP and MacMillan Park, the sources of the environmental isolates in this study, are a reservoir of potentially infectious cryptococcal isolates, it is not conclusive whether these parks were the specific source of individual infections, because subsequent sampling in different areas of Vancouver Island and BC has yielded additional isolates.‡‡ However, it is known that 46% of all patients had a history of visiting RBPP within 12 months before their diagnosis.
In this study, we have demonstrated an overall concordance between PCR-fingerprinting and AFLP genotypes. Both methods found the outbreak VGIIa isolates to be identical to the environmental VGIIa isolates. However, AFLP analysis revealed some polymorphic bands among all groups of isolates. There were no VGI/AFLP4 isolates obtained from environmental sources in this study, leaving the potential source of VGI infections in the human and porpoise cases unknown.
PCR-fingerprinting and AFLP analysis support the presence of different subtypes among the VGII/AFLP6 Vancouver Island isolates. These results may suggest that distinct C. gattii strains were introduced to Vancouver Island and then colonized the island for an extended period. The presence of polymorphisms within each of the subtypes, detected by AFLP analysis, may indicate that some microevolution or recombination between different subtypes has occurred, leading to minor genetic differences.
Mating experiments showed that all typeable isolates in this study are MATα. However, this finding does not preclude the existence of MATa isolates on Vancouver Island. The observed association between mating-incompetence and the VGIIb/AFLP6B subtype suggests that recombination between subtypes is unlikely to have occurred and, thus, supports the scenario that multiple strains that were slightly distinct genetically were introduced.
Cryptococcal infection has been diagnosed in a wide variety of animal species from BC. The incidence of cryptococcal infection in porpoises and other sea mammals is relatively rare and a surprising finding in the investigation of this outbreak. There also has been a recent report of C. gattii infection in a male Atlantic bottlenose dolphin (Tursiops truncatus) held in captivity at the U.S. Navy Marine Mammal Facility in San Diego (25). Molecular typing revealed that this isolate belongs to the VGI molecular type (S.E.K., unpublished data). Additional reports of cryptococcal infection in a striped dolphin (Stenella coeruleoalba) and a bottlenose dolphin from Australia and the USA speculated that seabirds might be the source of infection (26, 27). Cryptococcal cells have been demonstrated to be airborne and to be able to colonize trees in RBPP, as well as several other areas of Vancouver Island. The close proximity of RBPP to seawater (Strait of Georgia) may explain how the Dall's porpoises came to acquire infection with C. gattii VGII/AFLP6. In addition, although not a true halophile, C. gattii recently was shown to be able to survive for long periods of time in 3.5% sodium chloride solution, a concentration equivalent to that of seawater.‡‡ The presence of cryptococcal cells in the seawater and airborne cells close to the surface of the Strait of Georgia has not yet been examined, but it would be a useful analysis to provide clues as to the route of cryptococcal infection in sea mammals.
Since 1999, the incidence of C. gattii infection among the people of Vancouver Island has risen to 37 cases per million in 2002 and 2003. This is a significantly higher rate of infection than that typically observed in Australia, an area in which C. gattii is endemic (≈0.94 cases per million residents annually) (6). The reasons for this increase in C. gattii infection and environmental colonization are unclear. However, speculation might allow suggestions of importation and/or changing climatic conditions to allow distribution and thriving of C. gattii propagules into new ecological niches. The comparison of selected VGII/AFLP6 strains from different geographical locations with those from Vancouver Island revealed high similarity (Figs. 5 and 6), suggesting that this genotype has the potential to cause infection in temperate climates around the world. It is well documented that fungi can be dispersed on trees, vehicles, and other vectors, after which they might colonize native plants or trees in new surroundings (28). No importations of established C. gattii host plants to Vancouver Island are known, and all sampled eucalypts were negative for both C. neoformans and C. gattii. Regardless of the source of C. gattii, a warming trend on the island may have supported its recent emergence. The CDF on Vancouver Island's east coast is characterized by warm, dry summers and mild, wet winters (29). Global warming has caused an increase in the mean global surface temperature of 0.55°C in the last century, with accelerated warming in the last two decades (http://yosemite.epa.gov/oar/globalwarming.nsf/content/climate.html). Environment Canada reported six consecutive seasons of above average temperatures since 1998, with an increase of >3°C in some seasons, due to global warming (www.msc-smc.ec.gc.ca/ccrm/bulletin). These data suggest that the climate in some parts of Canada could become favorable to allow colonization with C. gattii. However, areas of similar climatic conditions, such as Washington state, which shares a border and coastline with BC, have not reported an increased incidence of cryptococcal infection among its residents, so the microclimate on Vancouver Island may have played an important role in the outbreak. The possibility of a link between the outbreak of C. gattii on Vancouver Island and climatic changes is also the subject of ongoing research.
The emergence of C. gattii in a temperate area, with a relatively high incidence of infection, is uncharacteristic, particularly in marine mammals, and emphasizes the changing distribution of the C. neoformans species complex as observed in an ongoing global epidemiological study (refs. 10, 11, and 24, and S.E.K. and W.M., unpublished data). This unprecedented outbreak highlights the importance of continued surveillance of this primary pathogen.
Supplementary Material
Acknowledgments
We thank Kristin Tangen and Jim Kronstad (Biotechnology Laboratory, University of British Columbia) for organization of isolates and collaborations; Louise Stein and Sultana Mithani (British Columbia Centre for Disease Control) and Krystyna Maszewska (Molecular Mycology Laboratory, Westmead Hospital) for their assistance with cultures; and Vincent Robert, Bart Theelen, and Gijs Dingemans (Centraalbureau voor Schimmelcultures) for their assistance in the interpretation of the AFLP data. This work was supported by National Health and Medical Research Council Grant 99738 (to W.M.).
Author contributions: T.B. and W.M. designed research; S.E.K., F.H., R.L.T., M.H., and W.M. performed research; S.E.K., F.H., R.L.T., K.H.B., T.B., K.J.K.-C., and W.M. analyzed data; S.E.K., T.B., and W.M. wrote the paper; K.H.B. collected environmental strains; and M.F. and L.M. collected human and veterinary strains.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AFLP, amplified fragment length polymorphism; CDF, Coastal Douglas Fir biogeoclimatic zone; cfu, colony-forming units; RBPP, Rathtrevor Beach Provincial Park; RFLP, restriction fragment length polymorphism; UPGMA, unweighted pairgroup method using arithmetic means.
Footnotes
Bartlett, K. H., Fyfe, M. W. & MacDougall, L. A. (2003) Am. J. Respir. Crit. Care Med. 167, A499 (abstr.).
References
- 1.Kwon-Chung, K. J., Boekhout, T., Fell, J. W. & Diaz, M. (2002) Taxon 51, 804–806. [Google Scholar]
- 2.Kwon-Chung, K .J. & Bennett, J.E. (1984) Am. J. Epidemiol. 120, 123–130. [DOI] [PubMed] [Google Scholar]
- 3.Sorrell, T. C. (2001) Med. Mycol. 39, 155–168. [PubMed] [Google Scholar]
- 4.Stephen, C., Lester, S., Black, W., Fyfe, M. & Raverty, S. (2002) Can. Vet. J. 43, 792–794. [PMC free article] [PubMed] [Google Scholar]
- 5.Hoang, L. M. N., Maguire, J. A., Doyle, P., Fyfe, M. & Roscoe, D. L. (2004) J. Med. Microbiol. 53, 935–940. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S., Sorrell, T., Nimmo, G., Speed, B., Currie, B., Ellis. D., Marriott, D., Pfeiffer, T., Parr, D. & Byth, K. (2000) Clin. Infect. Dis. 31, 499–508. [DOI] [PubMed] [Google Scholar]
- 7.Ellis, D. H. & Pfeiffer, T. J. (1990) J. Clin. Microbiol. 28, 1642–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staib, F., Seibold, M., Artweiler, E. & Frohlich, B. (1987) Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. A 266, 167–177. [DOI] [PubMed] [Google Scholar]
- 9.Kwon-Chung, K. J., Polachek, I. & Bennett, J. E. (1982) J. Clin. Microbiol. 15, 535–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer, W., Marszewska, K., Amirmostofian, M., Igreja, R. P., Hardtke, C., Methling, K., Viviani, M. A., Chindamporn, A., Sukroongreung, S., John, M. A., et al. (1999) Electrophoresis 20, 1790–1799. [DOI] [PubMed] [Google Scholar]
- 11.Meyer, W., Castañeda, A., Jackson, S., Huynh, M., Castañeda, E. & the IberoAmerican Cryptococcal Study Group (2003) Emerg. Infect. Dis. 9, 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boekhout, T., Theelen, B., Diaz, M., Fell, J. W., Hop, W. C. J., Abeln, E. C. A., Dromer, F. & Meyer, W. (2001) Microbiology 147, 891–907. [DOI] [PubMed] [Google Scholar]
- 13.Kidd, S. E., Sorrell, T. C. & Meyer, W. (2003) Med. Mycol. 41, 171–176. [DOI] [PubMed] [Google Scholar]
- 14.Kwon-Chung, K. J., Bennett, J. E. & Rhodes, J. C. (1982) Antonie van Leeuwenhoek 48, 25–38. [DOI] [PubMed] [Google Scholar]
- 15.Wickes, B. L., Mayorga, M. E., Edman, U. & Edman, J. C. (1996) Proc. Natl. Acad. Sci. USA 93, 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorrell, T. C., Brownlee, A. G., Ruma, P., Malik, R., Pfeiffer, T. J. & Ellis, D. H. (1996) J. Clin. Microbiol. 34, 1261–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barreto de Oliveira, M. T., Boekhout, T., Theelen, B., Hagen, F., Baroni, F. C., Lazera, M., Lengeler, K. B., Heitman, J, Rivera, I. N. G. & Paula, C. R. (2003) J. Clin. Microbiol. 42, 1356–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trilles, L., Lazéra, M., Wanke, B., Theelen, B. & Boekhout, T. (2003) Med. Mycol. 41, 383–390. [DOI] [PubMed] [Google Scholar]
- 19.Gezuele, E., Calegari, L., Sanabria, D., Davel, G. & Civilia, E. (1993) Rev. Iberoamericana Micol. 10, 5–6. [Google Scholar]
- 20.Chen, S. C. A., Currie, B. J., Campbell, H. M., Fisher, D. A., Pfeiffer, T. J., Ellis, D. H. & Sorrell, T. C. (1997) Trans. R. Soc. Trop. Med. Hyg. 91, 547–550. [DOI] [PubMed] [Google Scholar]
- 21.Sekhon, A. S., Bennerjee, S. N., Mielke, B. M., Idikio, H., Wood, G. & Dixon, J. M. (1990) Mycoses 33, 73–80. [DOI] [PubMed] [Google Scholar]
- 22.Brocklebank, J. (1997) Can. Vet. J. 38, 724. [PMC free article] [PubMed] [Google Scholar]
- 23.Dion, W.M. & Kapica, L. (1975) Can. Med. Assoc. J. 112, 712–716. [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis, D., Marriott, D., Hajjeh, R. A., Warnock, D., Meyer, W. & Barton, R. (2000) Med. Mycol. 38, Suppl. 1, 173–182. [PubMed] [Google Scholar]
- 25.Miller, W. G., Padhye, A. A., van Bonn, W., Jensen, E., Brandt, M. E. & Ridgway, S. H. (2002) J. Clin. Microbiol. 40, 721–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gales, N., Wallace, G. & Dickson, J. (1985) J. Wildl. Dis. 21, 443–446. [DOI] [PubMed] [Google Scholar]
- 27.Migaki, G., Gunnels, R. D. & Casey, H. W. (1978) Lab. Anim. Sci. 28, 603–606. [PubMed] [Google Scholar]
- 28.Allen, E. A. (2002) Can. J. Plant. Pathol. 24, 103–110. [Google Scholar]
- 29.Meidinger, D. & Pojar, J., eds. (1991) Ecosystems of British Columbia, Special Report Series (Research Branch, Ministry of Forests, Victoria, BC, Canada) Vol. 6.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.