Phosphorylation of transcription elongation factor SPT5 determines the trimethylated lysine 4 on histone 3 (H3K4me3) status in Arabidopsis.
Abstract
The elongation factor suppressor of Ty 5 homolog (Spt5) is a regulator of transcription and histone methylation. In humans, phosphorylation of SPT5 by P-TEFb, a protein kinase composed of Cyclin-dependent kinase 9 (CDK9) and cyclin T, interacts with the RNA polymerase II-associated factor1 (PAF1) complex. However, the mechanism of SPT5 phosphorylation is not well understood in plants. Here, we examine the function of SPT5 in Arabidopsis thaliana and find that spt5 mutant flowers early under long-day and short-day conditions. SPT5 interacts with the CDK-activating kinase 4 (CAK4; CDKD;2) and is specifically phosphorylated by CDKD;2 at threonines. The phosphorylated SPT5 binds VERNALIZATION INDEPENDENCE5 (VIP5), a subunit of the PAF1 complex. Genetic analysis showed that VIP5 acts downstream of SPT5 and CDKD;2. Loss of SPT5 or CDKD;2 function results in early flowering because of decreased amounts of FLOWERING LOCUS C (FLC) transcript. Importantly, CDKD;2 and SPT5 are required for the deposition of VIP5 and the enhancement of trimethylation of histone 3 lysine 4 in the chromatin of the FLC locus. Together, our results provide insight into the mechanism by which the Arabidopsis elongation factor SPT5 recruits the PAF1 complex via the posttranslational modification of proteins and suggest that the phosphorylation of SPT5 by CDKD;2 enables it to recruit VIP5 to regulate chromatin and transcription in Arabidopsis.
INTRODUCTION
Transcriptional elongation is a critical and tightly regulated step in the control of gene expression (Orphanides and Reinberg, 2002). The entry of RNA polymerase II (Pol II) into the elongation stage promotes the recruitment of factors involved in mRNA maturation, mRNA export, and chromatin remodeling and modification. Numerous associated factors, including transcription elongation factor S-II, TFIIF, DRB sensitivity inducing factor (DSIF), Elongin, RNA polymerase II elongation factor ELL, negative elongation factor (NELF), the RNA polymerase II-associated factor1 (PAF1) complex, and the histone chaperone FACT (Conaway and Conaway, 1993; Aso et al., 1995; Shilatifard et al., 1997), help Pol II to overcome pausing at specific DNA sequences (Li et al., 2007).
The DSIF subunits Spt4 and Spt5 are evolutionarily conserved in organisms ranging from yeast to mammals. In mammalian cells, SPT5 contains a conserved repetitive pentapeptide motif (consensus = G-S-[R/Q]-T-P) in its C-terminal repeat (CTR) region, which is similar to the C-terminal domain (CTD) of Pol II (Yamada et al., 2006). SPT4 and SPT5 form a heterodimer that both positively and negatively regulate transcriptional elongation. NELF interacts with DSIF to repress transcriptional elongation in the promoter-proximal region, and the relief of this repression involves the phosphorylation of DSIF by P-TEFb, a protein kinase composed of Cyclin-dependent kinase 9 (CDK9) and cyclin T. P-TEFb phosphorylates the threonine residues of the SPT5 CTR in human DSIF as well as serine 2 of the Pol II CTD (Wada et al., 1998; Yamada et al., 2006). Phosphorylation of the SPT5 CTR promotes the efficient progression of Pol II along the gene, while phosphorylation of the Pol II CTD leads to the dissociation of NELF, enabling DSIF to activate elongation (Yamada et al., 2006). In Saccharomyces cerevisiae, Spt5 contains 15 repeats of a hexapeptide with the consensus sequence S[T/A]WGG[A/Q] and is a substrate of the BUR kinase. Spt5 acts as a platform for the association of proteins that promote both transcriptional elongation and histone modification in transcribed regions (Liu et al., 2009; Zhou et al., 2009). A loss of BUR2 or Spt5 function leads to the attenuation of methylation of lysine 4 and lysine 36 of histone H3 (Zhou et al., 2009).
The PAF1 complex influences gene expression through its ability to modify chromatin structure and regulate RNA processing (Krogan et al., 2003). The Paf1 complex consists of Rtf1, Paf1, Cdc73, Leo1, and Ctr9 subunits and was originally characterized in yeast based on its association with Pol II during elongation (Krogan et al., 2002; Mueller and Jaehning, 2002). The PAF1 complex is required for H2B ubiquitination and the downstream H3K4 and H3K79 methylation in yeast (Krogan et al., 2003; Ng et al., 2003) and humans (Zhu et al., 2005; Kim et al., 2009). In yeast, recruitment of the SET1/COMPASS (Complex Proteins Associated with SET1) complex and Dot1p, which catalyze the methylation of lysine 4 and lysine 79 of histone 3, respectively, requires the Paf1 complex (Krogan et al., 2003).
In Arabidopsis thaliana, the VERNALIZATION INDEPENDENCE (VIP) genes VIP2/ELF7, VIP4, VIP5, and VIP6/ELF8 encode proteins closely related to Paf1, Leo1, Rtf1, and Ctr9, respectively. Loss-of-function vip mutants flower earlier; this phenotype is accompanied by a reduction of trimethylated lysine 4 on histone 3 (H3K4me3) and silencing of FLOWERING LOCUS C (FLC) (Zhang and van Nocker, 2002; He et al., 2004; Oh et al., 2004). FLC encodes a MADS box transcriptional regulator that inhibits the floral transition largely by reducing the expression of flowering time integrators such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) and FLOWERING LOCUS T (Michaels and Amasino, 1999; Sheldon et al., 1999; Hepworth et al., 2002). FLC is repressed by the autonomous flowering pathway and upregulated by FRIGIDA (FRI) (Michaels and Amasino, 1999, 2001). The delayed flowering caused by FRI activity or autonomous pathway mutations can be suppressed by the loss-of-function mutant flc (Michaels and Amasino, 2001), suggesting that FLC acts downstream of the autonomous pathway genes and FRI. FLC activity early in development also requires the SET domain group (SDG) proteins SDG8/EFS, ARABIDOPSIS TRITHORAX-RELATED1 (ATX1), and SDG25/ATXR7, which are associated with the methylation of H3K4 and H3K36 at the FLC chromatin (Alvarez-Venegas et al., 2003; Kim et al., 2005; Zhao et al., 2005; Pien et al., 2008; Berr et al., 2009; Tamada et al., 2009). Silencing of FLC in response to growth in cold temperatures (vernalization) is associated with a loss of H3 acetylation and H3K4me3 and concurrent accumulation of H3K27me2/me3 (Bastow et al., 2004; Sung et al., 2006; Angel et al., 2011; Yang et al., 2014).
Three CDKs, CDK7, CDK8, and CDK9, phosphorylate the heptapeptide sequence repeats (Y1S2P3T4S5P6S7) of the CTD of Pol II. CTD phosphorylation occurs mostly at Ser-5 and Ser-2. CDK7 and its S. cerevisiae homolog Kin28 are responsible for most of the Ser5 phosphorylation, whereas CDK9 (and its yeast homologs Ctk1 and BUR1) is required for Ser2 phosphorylation (Egloff and Murphy, 2008)
The Arabidopsis genome encodes two CDK9-like proteins and four predicted CDK-activating kinases (CAKs). Arabidopsis CAK1 (CDKF;1) phosphorylates the T-loop of CAK2 (CDKD;3) and CAK4 (CDKD;2) to activate their kinase activity (Shimotohno et al., 2004). CDKD;3 and CDKD;2 have strong sequence similarity to CDK7/p40MO15, a vertebrate CAK that phosphorylates CDKs and the CTD of a Pol II subunit (Shimotohno et al., 2003). In contrast to CDKF;1, CDKD;2, and CDKD;3, Arabidopsis CDKD;1 lacks kinase activity on CDKs and CTDs. CDKC;1 and CDKC;2 are closely related to human CDK9 and interact with cyclin T partners, CYCT1;4 and CYCT1;5. Both CDKC;1 and CDKC;2 exhibit CTD kinase activity, and a loss of CDKC function results in delayed flowering time and partial resistance to CaMV (Cui et al., 2007).
In yeast, the recruitment of the Paf1 complex to target genes requires BUR2 and Spt5 CTR, which enable the binding of Rtf1; furthermore, in yeast, phosphorylated Spt5 affects H2B ubiquitination, H3K4me3, and H3K36me3 modifications. The mechanism by which proteins, or complexes, affect the accumulation of the PAF1 complex in plants is less clear. Here, we report that CDKD;2 specifically phosphorylates threonines in the SPT5 CTR, enabling its interaction with VIP5 and ability to regulate histone modification and FLC expression in Arabidopsis.
RESULTS
SPT5 Is Involved in the FLC-Dependent Flowering Pathway
The Arabidopsis proteins SPT5 and global transcription factor group A2 (GTA2) (originally named SPT5-1 and SPT5-2, respectively) are homologs of Spt5 in yeast and humans (Dürr et al., 2014). SPT5 has 19 exons with encoding 989 amino acids in length; GTA2 has 22 exons with encoding 1041 amino acids (Figure 1A; Supplemental Figure 1A). Arabidopsis SPT5 and GTA2 contain a eukaryotic N-utilization substance G N-terminal (NGN) domain, a CTR, and multiple KOW domains, which are conserved in yeast, flies, and humans. Phylogenetic analyses indicated that the Arabidopsis SPT5 and GTA2 were highly related to SPT5 homologs of other species (Supplemental Figure 2 and Supplemental Data Set 1), suggesting that their roles as regulators of chromatin and transcription in plants might be evolutionarily conserved as well.
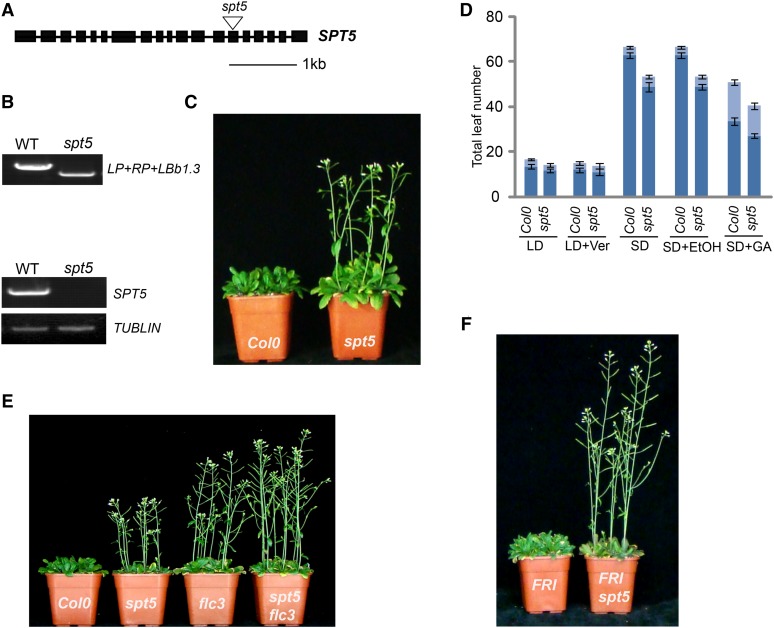
Figure 1.
Mutation in SPT5 Results in Early Flowering.
(A) Gene structure of SPT5. Exons are indicated by boxes and introns are indicated by lines. The T-DNA insertion is indicated by a triangle.
(B) Genotypic and RT-PCR analysis of SPT5. The genotype was analyzed with the left genomic primer (LP), right genomic primer (RP), and a primer in the vector (LBb1.3) (top panel). The transcripts of SPT5 are shown in the bottom panel. TUBLIN was used as internal control.
(C) The spt5 mutant exhibits an early-flowering phenotype.
(D) Flowering time of Col-0 and spt5 mutants under different conditions or treatments. Flowering time was assessed by counting the number of rosette leaves and cauline leaves at bolting under long-day photoperiod (LD), vernalization (LD+Ver), short-day photoperiod (SD), and GA treatment (SD+GA). SD+EtOH (ethanol) treatment was used as a control for GA treatment. The rosette leaves are indicated in dark blue, and the cauline leaves are indicated in light blue. Values shown are mean ± sd of rosette and cauline leaves; 40 plants were scored for each line.
(E) and (F) Phenotypic analysis of different mutants. Flowering time of spt5 and flc3 (E) and spt5 with FRI (F).
To functionally characterize the Arabidopsis SPT5 homologs, we identified T-DNA insertion mutants for one spt5 and two gta2 alleles. Genotypic analyses confirmed a T-DNA insertion in exon 14 in the spt5 mutant (Figures 1A and 1B) and in exon 1 and 20 in the gta2 mutants (Supplemental Figures 1A to 1C). Transcriptional analyses revealed that spt5 is a knockout mutant (Figure 1B), gta2-1 is a knockdown allele, and gta2-2 is a knockout mutant (Supplemental Figure 1D). However, as the gta2 mutant plants did not display a significant phenotype in our hands (Supplemental Figure 1E), they were not included in our further studies here. By contrast, the loss of SPT5 resulted in earlier flowering in both long- and short-day conditions, indicating that SPT5 is dispensable for the photoperiod response (Figure 1C). The spt5 mutant exhibited a normal response to vernalization and gibberellin (GA) treatment, suggesting that SPT5 might be involved in the FLC-dependent flowering pathway (Figure 1D). To confirm that the early-flowering phenotype is caused by the spt5 mutation, we generated an endogenous SPT5 promoter-driven SPT5 fused with a flag epitope tag (FLAG; ProSPT5:FLAG-SPT5) and introduced it into spt5. The early-flowering phenotype of spt5 was completely rescued by SPT5-FLAG (Supplemental Figure 3).
To evaluate the relationship between SPT5 and FLC, we generated spt5 flc-3 double mutants, which showed early flowering with a similar phenotype to flc-3 mutants (Figure 1E, Table 1), suggesting that SPT5 affected flowering time via FLC. To confirm that the early-flowering phenotype of spt5 is dependent on FLC, we introduced the spt5 mutant into lines carrying a dominant FRI allele. The late-flowering phenotype of plants containing FRI (Hu et al., 2014) was suppressed by the loss of SPT5 function (Figure 1F). Additional evidence for the relationship between SPT5 and FLC was provided by the similar flowering phenotypes displayed by the FRI spt5 flc-3 and FRI flc-3 plants (Table 1).
Table 1. Primary Leaf Number at Bolting of cdkd;2 and spt5 Mutants.
| Lines | Rosette Leaves | Cauline Leaves |
|---|---|---|
| Col-0 | 13.4 ± 0.74 | 3.4 ± 0.42 |
| spt5 | 11.7 ± 0.9 | 2.9 ± 0.5 |
| cdkd2-1 | 10.7 ± 1.0 | 2.8 ± 0.5 |
| cdkd2-2 | 11.1 ± 0.9 | 2.8 ± 0.4 |
| spt5/cdkd2-1 | 10.5 ± 0.9 | 2.9 ± 0.6 |
| spt5/cdkd2-2 | 10.7 ± 0.9 | 2.9 ± 0.5 |
| flc3 | 9.6 ± 0.5 | 2.8 ± 0.4 |
| spt5/flc3 | 9.7 ± 0.7 | 2.6 ± 0.6 |
| cdkd2-1/flc3 | 9.5 ± 0.8 | 2.7 ± 0.5 |
| cdkd2-2/flc3 | 10.1 ± 1.0 | 2.8 ± 0.4 |
| vip5 | 7.6 ± 0.5 | 2.3 ± 0.4 |
| spt5/vip5 | 7.4 ± 0.8 | 2.9 ± 0.7 |
| cdkd2-1/vip5 | 7.5 ± 0.8 | 2.9 ± 0.6 |
| cdkd2-2/vip5 | 7.5 ± 0.4 | 2.7 ± 0.6 |
| FRI | 27.2 ± 2.2 | 6.5 ± 1.1 |
| FRI/spt5 | 15.8 ± 1.3 | 3.9 ± 0.9 |
| FRI/cdkd2-1 | 12.7 ± 1.1 | 3.5 ± 0.5 |
| FRI/cdkd2-2 | 14.4 ± 0.9 | 3.4 ± 0.5 |
| FRI/spt5/cdkd2-1 | 12.8 ± 1.4 | 3.3 ± 0.6 |
| FRI/spt5/cdkd2-2 | 14.3 ± 1.2 | 3.4 ± 0.8 |
| FRI/flc3 | 12.1 ± 0.9 | 3.0 ± 0.6 |
| FRI/spt5/flc3 | 12.6 ± 0.6 | 3.1 ± 0.6 |
| FRI/cdkd2-1/flc3 | 12.2 ± 1.1 | 2.8 ± 0.7 |
| FRI/cdkd2-1/flc3 | 12.4 ± 1.3 | 2.9 ± 0.9 |
| FRI/vip5 | 8.5 ± 0.6 | 2.8 ± 0.7 |
| FRI/spt5/vip5 | 8.3 ± 0.7 | 2.7 ± 0.5 |
| FRI/cdkd2-1/vip5 | 8.7 ± 0.5 | 2.9 ± 0.3 |
Values shown are mean number ± sd of rosette and cauline leaves; 40 plants were scored for each line.
CDKD;2 Phosphorylates the C-Terminal Repeat of SPT5 at Threonine and Interacts with SPT5
In yeast, phosphorylation of Spt5 has important regulatory effects on Paf1 complex accumulation (Liu et al., 2009; Zhou et al., 2009). Here, we performed in vitro phosphorylation assays using GST-fused Arabidopsis CDK proteins. SPT5 was divided into the SPT5 N terminus (SPT5N), which included the NGN domain and five KOW domains, and the SPT5 C terminus (SPT5C), which incorporated the CTR and a KOW domain (Figure 2A). Neither CDKC;1 nor CDKC;2 exhibited phosphorylation activity on SPT5N or SPT5C (Supplemental Figures 4A and 4B). CDKD;1, CDKD;2, CDKD;3, and CDKF;1 also did not phosphorylate SPT5N (Supplemental Figure 4C). CDKD;2 strongly phosphorylated SPT5C (Figure 2B), although CDKD;1, CDKD;3, and CKF;1 did not. Within SPT5C, CDKD;2 phosphorylated the CTR, but not the C-terminal KOW domain (Figure 2C). We therefore concluded that CDKD;2 kinase phosphorylates the SPT5 CTR in vitro.
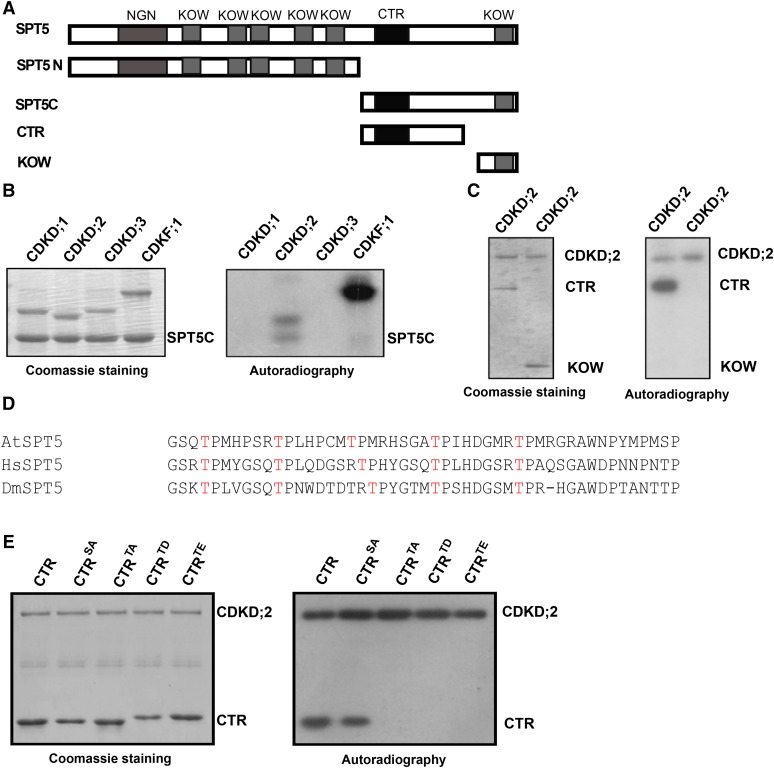
Figure 2.
CDKD;2 Phosphorylates SPT5.
(A) Different regions of SPT5 containing the indicated domains (NGN, KOW, and CTR) tested in the phosphorylation and yeast two-hybrid analyses are shown.
(B) CDKD;2 phosphorylates the C terminus of SPT5. The ability of CDKD;1, CDKD;2, CDKD;3, and CDKF;1 to phosphorylate the C terminus of SPT5 was assessed.
(C) CDKD;2 phosphorylates the CTR of SPT5. The positions of the CTR and KOW domains are indicated on the right.
(D) Alignment of the CTR domain with other closely related SPT5s using ClustalW. The amino acid sequences of Arabidopsis SPT5 (744–792 amino acids), human SPT5 (772–829 amino acids), and Drosophila Spt5 (822–869 amino acids). The conserved threonines are indicated in red.
(E) The activity and specificity of CDKD;2 kinase assessed using different substrates. Substrates contain the SPT5 CTR fused with His and are either wild-type (CTR) or have all serines converted into alanine (CTRSA) or all threonines converted into alanine (CTRTA), aspartic acid (CTRTD), or glutamic acid (CTRTE).
For (B), (C), and (E), the left panel shows the Coomassie blue-stained gel. The positions of the different CDKs and SPT5 fragments are indicated on the right. Autoradiography (right panel) indicates any kinase activity and its specificity.
The CTR of Arabidopsis SPT5 possesses several threonine residues that are also conserved in human and Drosophila melanogaster SPT5 (Figure 2D). We investigated the specific activity of CDKD;2 on the SPT5 CTR using His-tagged CTRs with amino acid substitutions, including constructs where all serine residues were replaced with alanines (total S-to-A replacement, CTRSA), all threonines were replaced by alanines (CTRTA), all threonine by aspartic acid (CTRTD), and all threonines by glutamic acid (CTRTE). We found that CDKD;2 phosphorylated the wild-type forms of the CTR and CTRSA, but not CTRTA, CTRTD, or CTRTE, suggesting that CDKD;2 is specific for the threonines of the CTR (Figure 2E).
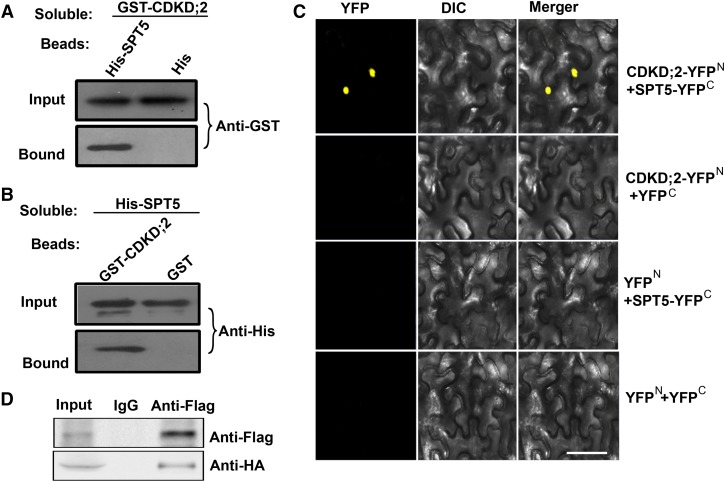
We also assessed whether CDKD;2 interacts with SPT5 directly. GST-CDKD;2 was observed to bind beads containing a His-tagged SPT5, but not His alone (Figure 3A). In complementary experiments, soluble His-tagged SPT5 bound to beads attached to GST-CDKD;2, but not to the GST control (Figure 3B). This pull-down interaction was confirmed by bimolecular fluorescence complementation (BiFC). Functional YFP in the nucleus was observed following the coexpression of CDKD;2 fused to the N terminus of YFP (CDKD;2-YFPN) and SPT5 fused to the C terminus of YFP (SPT5-YFPC), but not in the controls, thereby providing further evidence that SPT5 binds directly to CDKD;2 (Figure 3C). This interaction was further validated by coimmunoprecipitation; FLAG-tagged SPT5 and HA-tagged CDKD;2 were cotransformed into Arabidopsis protoplasts and immunoprecipitated using an anti-FLAG antibody. SPT5, but not the antibody alone, bound to CDKD;2 (Figure 3D). These results suggest that SPT5 interacts with CDKD;2 in vitro and in vivo.
Figure 3.
CDKD;2 Interacts with SPT5.
(A) Beads containing a His tag or a His tag fused to SPT5 were assayed for their ability to bind a soluble GST fused to CDKD;2. Input or bound protein was detected with an antibody to GST (Anti-GST).
(B) Beads containing a GST tag or a GST tag fused to CDKD;2 were assessed for their ability to bind a soluble SPT5 fused to a His tag (His-SPT5) and detected with antibody to His (Anti-His). All experiments were repeated three times.
(C) CDKD;2 fused to the N terminus of YFP was tested for its ability to bind to the C terminus of YFP or the C terminus of YFP fused to SPT5. Yellow fluorescence (YFP) and a bright-field image were recorded and the resulting images were merged. DIC, differential interference contrast. Bar = 100 µm.
(D) Coimmunoprecipitation of CDKD;2 and SPT5. FLAG-SPT5 and HA-CDKD;2 were cotransformed into Arabidopsis protoplasts, followed by immunoprecipitation using an anti-FLAG antibody.
Threonine Phosphorylation of the CTR Is Critical for SPT5 Function
We sought to investigate whether the threonine phosphorylation of the CTR by CDKD;2 is critical for the proper functioning of SPT5. The in vivo function of threonine phosphorylation was evaluated using a complementation assay. For this assay, we generated constructs containing FLAG-SPT5, in which the CTR nucleotides encoding threonine were substituted with nucleotides encoding alanine (SPT5TA) or aspartic acid (SPT5TD). These constructs were expressed under the control of the endogenous SPT5 promoter (ProSPT5) and were transformed into the spt5 mutant. Eight representative individual lines for each transformation, containing ProSPT5:FLAG-SPT5TA (A9, A13, A15, and A41) and ProSPT5:FLAG-SPT5TD (D4, D11, D21, and D43), were used in the phenotypic analysis (Supplemental Figure 5). The early flowering phenotype of spt5 was complemented by the wild-type form of SPT5, but not by the nonphosphorylatable (SPT5TA) or the phosphomimic (SPTTD) forms of SPT5 (Supplemental Figures 3 and 5). Thus, threonine phosphorylation of the CTR is essential for SPT5 function.
Mutations in CDKD;2 Result in Early Flowering
To investigate the biological roles of CDKD;2, we obtained two T-DNA insertion mutant cdkd;2 lines. Genotypic analyses demonstrated that T-DNA insertions were presented in intron 3 and exon 4 of the cdkd;2-1 and cdkd;2-2 alleles, respectively, and RT-PCR confirmed that full-length CDKD;2 mRNA was not transcribed in the two mutants (Figures 4A and 4B). The cdkd;2-1 and cdkd;2-2 plants displayed early-flowering phenotypes in both long- and short-day growth conditions and had a normal response to vernalization and GA, similar to spt5 (Figures 4C and 4D), suggesting that CDKD;2 participates in the FLC-dependent flowering pathway.
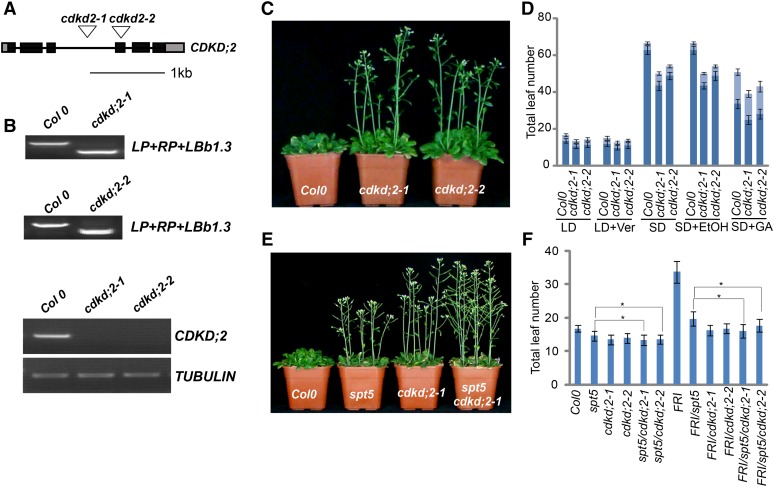
Figure 4.
Mutations in CDKD;2 Result in Early Flowering.
(A) Gene structure of CDKD;2. Boxes represent exons and lines represent introns. T-DNA insertions are indicated with a triangle.
(B) Genotypic and RT-PCR analysis of cdkd;2 mutants. The genotype was analyzed with the left genomic primer (LP), right genomic primer (RP), and primer in the vector (LBb1.3). Genotype analysis is shown in the top and middle panels; transcription is shown in the bottom panel. TUBULIN was used as internal control.
(C) The cdkd;2 mutants exhibit an early-flowering phenotype.
(D) Flowering time of Col-0 and the cdkd;2 mutants under different conditions or treatments. Flowering time was assessed by counting the number of rosette leaves and cauline leaves in bolting plants under a long-day photoperiod (LD), vernalization (LD+Ver), short-day photoperiod (SD), and GA treatment (SD+GA). SD+EtOH (ethanol) treatment was used as a GA treatment control. The rosette leaves are indicated in dark blue and the cauline leaves are indicated in light blue. Values shown are mean number ± sd of rosette and cauline leaves; 40 plants were scored for each line.
(E) Flowering time of Col-0, spt5, cdkd;2-1, and the spt5 cdkd;2-1 double mutant under LD photoperiod.
(F) The leaf number of Col-0, spt5, cdkd;2-1, and spt5 cdkd;2-1 with or without FRI under LD photoperiod. Total leaf number at flowering is shown for each indicated strain; asterisk indicates statistically significant by t test. Values shown are mean number ± sd of total leaves; 40 plants were scored for each line.
The possibility of a genetic interaction between CDKD;2 and FLC was tested by introducing the cdkd;2 alleles into the flc-3 mutant background. The cdkd;2 flc-3 double mutants showed early flowering and were phenotypically similar to flc-3 (Table 1). To confirm that the early-flowering phenotype of cdkd;2 is dependent on FLC, we introduced the two cdkd;2 mutant alleles independently into the FRI background and found that these alleles suppressed the late-flowering phenotype of FRI. The genetic interaction of CDKD;2 and FLC was further validated by independently introducing the cdkd;2 flc-3 double mutants into the FRI background (Table 1). These results indicate that the loss of CDKD;2 function affected the early-flowering phenotype mediated by FLC.
Next, we tested the genetic relationship between CDKD;2 and SPT5 by introducing cdkd;2 into the spt5 mutant. The cdkd;2 spt5 double mutants exhibited a similar flowering time to cdkd;2 single mutants (Figures 4E and 4F, Table 1), indicating that CDKD;2 and SPT5 might work in the same pathway. The genetic relationship between CDKD;2 and SPT5 was further evaluated by examining the FRI spt5 cdkd;2 mutant lines. Both FRI spt5 cdkd;2 and FRI cdkd;2 plants flowered at similar times (Figure 4F, Table 1).
SPT5 Physically Interacts with VIP5
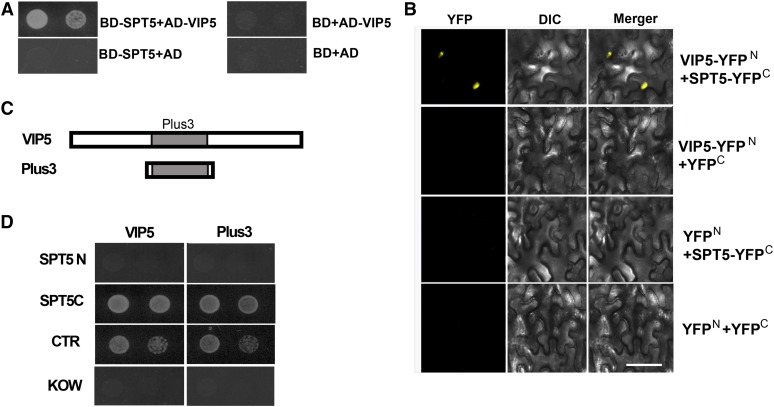
Yeast Spt5 interacts with Rtf1 in vivo and phosphorylated human SPT5 binds RTF1 in vitro (Mayekar et al., 2013; Wier et al., 2013). Given that SPT5 and the PAF1 complex are conserved in different species, we asked whether Arabidopsis SPT5 might also interact with partners involved in the regulation of histone modification. To investigate whether SPT5 and VIP5 interact with each other, we generated a binding domain fusion of full-length SPT5 (BD-SPT5) for yeast two-hybrid analysis with activation domain-tagged VIP5 (AD-VIP5). The intact SPT5 protein bound strongly to VIP5 (Figure 5A). The yeast two-hybrid interactions were verified by BiFC assays. Coexpression of VIP5-YFPN and SPT5-YFPC reconstituted a functional YFP in the nucleus, whereas the controls did not (Figure 5B), providing further evidence that SPT5 binds directly to VIP5.
Figure 5.
SPT5 Interacts with VIP5.
(A) DNA binding domain (BD) or a BD-SPT5 fusion was tested for its ability to bind to an activation domain (AD) fused to VIP5. The growth of two dilutions (1 × 10−1and 2 × 10−2) of the yeast culture on SD medium lacking Trp, Leu, His, and adenine is shown.
(B) SPT5 fused to the N terminus of YFP or the N terminus of YFP alone were tested for their ability to bind to the C terminus of YFP alone or the C terminus of YFP fused to VIP5. Yellow fluorescence (from YFP) and a bright-field image were recorded and the resulting images were merged. DIC, differential interference contrast. Bar = 100 µm.
(C) VIP5 containing the Plus3 domain tested in the yeast two-hybrid analyses is shown.
(D) BD fused with various SPT5 domains shown in (Figure 2A) were tested for their interaction with either AD-VIP5 or AD-Plus3 (C). The growth of two dilutions (1 × 10−1 and 2 × 10−2) of the yeast culture on synthetic defined medium lacking Trp, Leu, His, and adenine is shown.
Next, we examined which domain is critical for the interaction of SPT5 and VIP5. SPT5C, but not SPT5N, bound VIP5. Within SPT5C, the CTR, but not the KOW domain, bound VIP5 and the individual Plus3 domain of VIP5 was sufficient to bind SPT5 (Figures 5C and 5D). We conclude that the interaction between SPT5 and VIP5 occurs at their respective CTR and Plus3 domains.
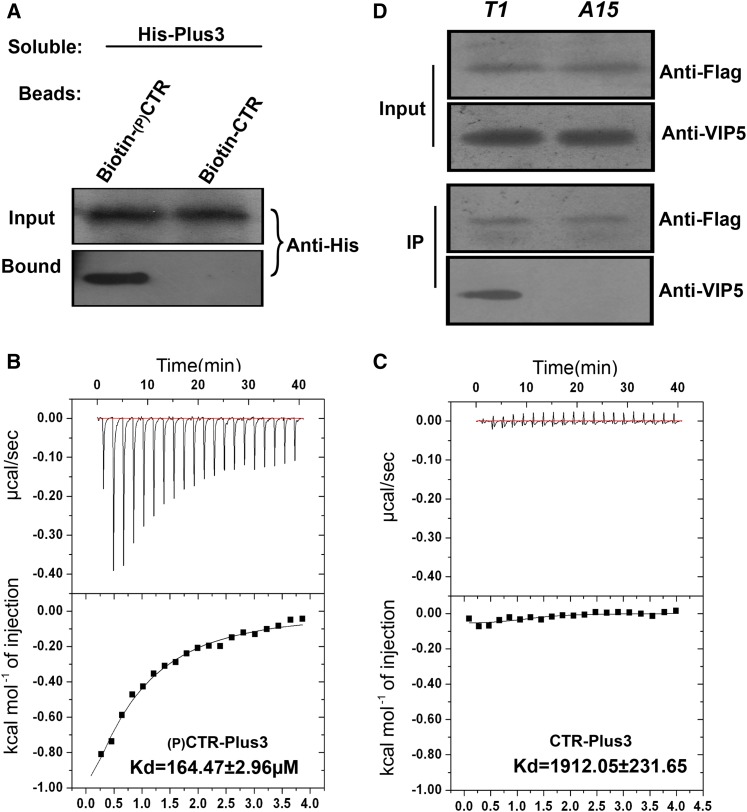
VIP5 Preferentially Binds the Phosphorylated Form of the CTR of SPT5
The interaction between human SPT5 and RTF1 depends on the phosphorylation of the SPT5 CTR. We sought to characterize the Arabidopsis VIP5-SPT5 interaction and determine whether this interaction was also phosphorylation specific. To clarify the interpretation of the biochemical results, we synthesized two CTR peptides corresponding to residues 744 to 762 of Arabidopsis SPT5. These peptides, containing three complete CTR repeats including its spacer, were constructed either with or without a phosphothreonine at position T747. A protein containing His fused to the VIP5 Plus3 domain (His-Plus3) was observed to bind to beads fused to the phosphorylated form of the CTR peptide (pCTR) but not to beads fused to the unphosphorylated form (Figure 6A). To confirm the pull-down interaction, we quantitatively measured the biomolecular interactions using isothermal titration calorimetry. The Plus3 domain bound to the pCTR with a calculated equilibrium dissociation constant (Kd) of 164.47 ± 2.96 μM. Binding to the unphosphorylated peptide resulted in a negligible change in anisotropy, with an estimated Kd of >1 mM (Figure 6C). These data indicate that interactions between phosphorylated SPT5 and the PAF1 complex were likely conserved between mammals and plants.
Figure 6.
Interaction between SPT5 and VIP5 Depends on CTR Phosphorylation.
(A) Binding of soluble His-Plus to bead-bound peptides containing three complete CTR repeats was measured. The 18-mer peptides were either nonphosphorylated (Non-P) or phosphorylated at threonine. The amount of His-VIP5 protein bound to the peptides on the beads was determined by an immunoblot analysis with antibody to His (Anti-His).
(B) and (C) Measurement of phosphorylated or nonphosphorylated peptides described as binding to the Plus3 domain of VIP5 (A) using isothermal titration calorimetry at 25°C. A 200-μL sample of 150 μM Plus3 was titrated with 2 μL injections of 3 mM phosphorylated peptides (B) or nonphosphorylated peptide (C) at 120-s intervals. Curves are the best fits to the data that enable the acquisition of the stoichiometry and thermodynamic parameters. Experiments were repeated at least three times, and a representative experiment is shown.
(D) Coimmunoprecipitation of the wild-type or nonphosphorylatable form of SPT5 with VIP5. The cell extractions from the SPT5-complemented (T1) or nonphosphorylatable SPT5-complemented spt5 seedling (A15) were immunoprecipitated with an anti-FLAG antibody and then detected with anti-VIP5 antibody.
Next, we investigated whether this interaction depends on the phosphorylation of SPT5 in vivo. The cell extracts from FLAG-tagged wild-type ProSPT5:FLAG-SPT5 and nonphosphorylatable ProSPT5:FLAG-SPT5TA seedlings were immunoprecipitated using an anti-FLAG antibody, followed by blotting with a VIP5-specific antibody (Supplemental Figure 6). The wild-type form of SPT5, but not SPT5TA, bound VIP5 (Figure 6D), indicating that threonine phosphorylation of SPT5 is required for the interaction between SPT5 and VIP5 in vivo.
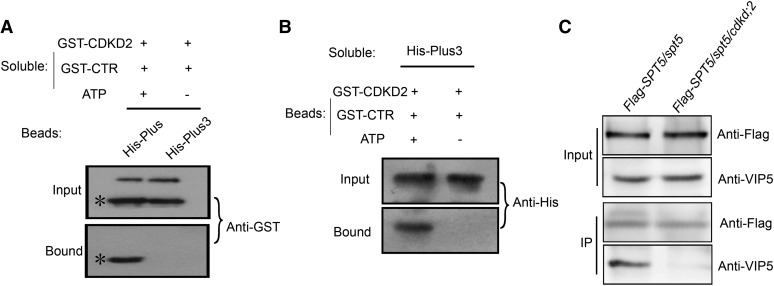
CDKD;2 Is Required for the Interaction between VIP5 and SPT5 in Vitro and in Vivo
To test whether the interaction between the phosphorylated SPT5 CTR and VIP5 is caused by CDKD;2, we developed a phosphorylation reaction with or without ATP and then isolated CDKD;2 and SPT5 CTR. A protein containing the VIP5 Plus3 domain fused with His (His-Plus3) was observed to bind the CTR in the presence, but not the absence, of ATP (Figure 7A). In the complementary experiment, GST-CTR bound to His-Plus3 after the phosphorylation reaction with ATP, whereas this binding was not observed in the absence of ATP (Figure 7B). These results indicated that the interaction between SPT5 and VIP5 depends on the threonine phosphorylation of CTR, which is mediated by CDKD;2.
Figure 7.
Interaction between SPT5 and VIP5 Depends on CDKD;2.
(A) Binding of soluble phosphorylated SPT5 CTR to bead-bound His-Plus3. A phosphorylation reaction between CDKD;2 and SPT5 CTR was performed with or without ATP. The purified proteins were assessed to determine their binding ability to the Plus3 domain. The amount of GST-CTR protein was determined by an immunoblot analysis with antibody to GST (Anti-GST). Asterisk denotes CTR bands.
(B) Binding of soluble His-Plus3 to phosphorylated CTR. The amount of His-CTR protein was determined by an immunoblot analysis with an antibody to His (Anti-His).
(C) Coimmunoprecipitation of SPT5 with VIP5 in the wild type and cdkd;2 mutants.
Seedlings containing FLAG-SPT5 in spt5 or in the spt5 cdkd;2 double mutant was used for immunoprecipitation with an anti-FLAG antibody, followed by blotting with anti-VIP5.
To confirm that this interaction depends on CDKD;2, we crossed ProSPT5:FLAG-SPT5/spt5 into the cdkd;2-1 background to generate the ProSPT5:FLAG-SPT5/spt5 cdkd;2-1 mutant. Cell extracts from these plants were immunoprecipitated with an anti-FLAG antibody and then detected with a VIP5-specific antibody. As expected, VIP5 was observed to interact with SPT5 in ProSPT5:FLAG-SPT5/spt5 seedlings, but not in the ProSPT5:FLAG-SPT5/spt5 cdkd;2-1 mutant seedlings (Figure 7C), indicating that CDKD;2 is required for the interaction between SPT5 and VIP5 in vivo.
VIP5 Acts in the Same Pathway as SPT5 and CDKD;2
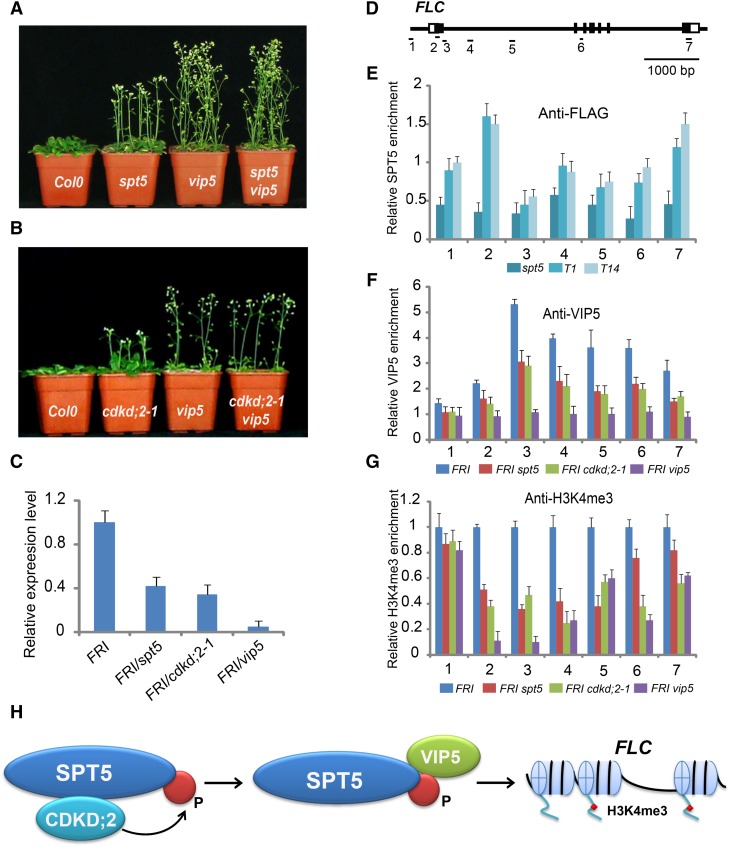
The vip5 mutant containing a T-DNA insertion flowers early, as previously described (Oh et al., 2004). To study the functional relationship between SPT5 and VIP5, we constructed a spt5 vip5 double mutant. The flowering time of spt5 vip5 was similar to that of vip5 (Figure 8A, Table 1), suggesting that VIP5 acts in the same pathway of SPT5 with regard to flowering time.
Figure 8.
Distribution of SPT5, VIP5, and H3K4me3 on FLC.
(A) Flowering time of Col-0, spt5, vip5, and the spt5 vip5 double mutant in long-day photoperiod.
(B) Flowering time of Col-0, cdkd;2-1, vip5, and the cdkd;2-1 vip5 double mutant in long-day photoperiod.
(C) Transcripts of FLC were assessed in FRI, FRI spt5, FRI cdkd;2-1, and FRI vip5. RNA from 10-d-old seedlings was used for RT-PCR. Experiments were repeated at least three times, each experiment included three replicates, and the representative experiments shown indicate the mean ± se, n = 3 replicates.
(D) Gene structure of FLC. Exons are indicated as boxes and introns are indicated as lines. The locations of the gene regions analyzed by ChIP-PCR are marked.
(E) to (G) The amounts of SPT5, VIP5, and H3K4me3 at different regions of FLC were determined by ChIP-PCR. The y axis denotes enrichment relative to ubiquitin. Experiments were repeated at least three times, each experiment included three replicates, and the representative experiments shown indicate the mean ± se, n = 3 replicates. T1 and T14 are indicated in Supplemental Figure 3.
(H) A model showing that phosphorylation of SPT5 by CDKD;2 is required for VIP5 recruitment and H3K4me3 of FLC.
Next, we inspected the genetic relationship between CDKD;2 and VIP5 by introducing cdkd;2 into the vip5 background. The early flowering phenotype of the cdkd;2 vip5 double mutant was similar to that of the vip5 single mutant (Figure 8B, Table 1), indicating that VIP5 acts in the same pathway as CDKD;2 in flowering time. The genetic results were validated by introducing FRI into the cdkd;2 vip5 and spt5 vip5 double mutants. In summary, these observations indicated that CDKD;2 and SPT5 are required for early flowering via VIP5.
SPT5 and CDKD;2 Are Required for the Deposition of VIP5 and Methylation of Lysine 4 of Histone 3 at FLC
VIP5 is a homolog of yeast and human Rtf1, which is critical for the methylation of lysine 4 and lysine 36 of histone 3 in yeast. The loss of VIP5 function in Arabidopsis leads to the downregulation of FLC and other members of the FLC/MAF MADS box gene family (Oh et al., 2004).
The interaction between VIP5 and phosphorylated SPT5 is mediated by CDKD;2 and VIP5 acts downstream of CDKD;2 and SPT5, suggesting that, in the loss-of-function mutant cdkd;2 or spt5, VIP5 may be misregulated, affecting the transcription of downstream genes. We first asked whether the transcript level of FLC was affected in either cdkd;2 or spt5 mutants. A quantitative PCR analysis revealed relatively lower levels of FLC transcripts in cdkd;2 and spt5 than in FRI, with a larger reduction in vip5 (Figure 8C), suggesting that FLC expression is impaired in the three mutants and that CDKD;2, SPT5, and VIP5 are required for flowering time via FLC. The presence of residual transcripts of FLC in spt5 or cdkd;2 mutants suggests that SPT5 and CDKD;2 function to promote FLC transcription.
Since FLC transcripts were reduced in spt5 mutants, we investigated whether SPT5 binds FLC directly. The profiles of SPT5 were investigated using two lines carrying the ProSPT5:FLAG -SPT5 transgene in the spt5 background, where it complemented the mutant phenotype. The binding of SPT5 was measured by chromatin immunoprecipitation (ChIP) with a FLAG-specific antibody, followed by quantitative PCR analysis of the amount of DNA enrichment. The amount of DNA precipitated by anti-FLAG was highly enriched across the FLC locus in the ProSPT5:FLAG-SPT5 lines, but not in the spt5 mutant that lacked the transgene (Figures 8D and 8E), indicating that SPT5 binds FLC directly.
Next, we tested if the distribution of VIP5 binding to FLC depends on CDKD;2 and SPT5. The distribution of VIP5 was measured in a ChIP-PCR assay with a VIP5-specific antibody. The levels of VIP5 binding to FLC were strongly affected by the vip5 mutation, as well as the cdkd;2 and spt5 mutations. In addition to a main peak at region 3, the amount of VIP5 was somewhat higher across the gene in the wild-type line, but not in the spt5, cdkd;2, and vip5 mutants. These results indicate that both SPT5 and CDKD;2 are required for the binding of VIP5 across the FLC gene (Figures 8D and 8F).
The loss of PAF1 subunits has been shown to lead to the deposition of H3K4me3 (He et al., 2004; Yu and Michaels, 2010). Therefore, we next investigated the distribution of H3K4me3 marks in different genotypic backgrounds. The H3K4me3 mark is enriched in the wild type, whereas it was greatly reduced in the spt5, cdkd;2, and vip5 mutants (Figure 8G), indicating that SPT5 and CDKD;2, like VIP5, are involved in deposition of H3K4me3 at FLC.
DISCUSSION
Our study demonstrated that Arabidopsis SPT5 has a conserved CTR, with a consensus sequence that contains the repetitive pentapeptide motif (G/P)S(Q/R)TP. Although this SPT5 CTR sequence is not identical to the corresponding human sequence, our data show that, like human SPT5, the Arabidopsis SPT5 CTR is phosphorylated at threonine by CDKD;2 (Figures 2D and 2E). The yeast Spt5 CTR, which contains 15 repeats of a hexapeptide with a S[T/A]WGG[Q/A] consensus sequence, differs from the CTR of mammals and plants and can be phosphorylated at both serine and threonine by the BUR kinase (Zhou et al., 2009).
Threonine phosphorylation of the SPT5 CTR is necessary for protein interactions between SPT5 and VIP5 and for SPT5 function (Figures 6 and 7). These findings were supported by complementation experiments in which the wild-type form of SPT5 was observed to rescue the early-flowering phenotype of spt5, but neither the nonphosphorylatable (SPT5TA) nor phosphomimic (SPT5TD) forms of SPT5 could (Supplemental Figures 3 and 5). Nonphosphorylatable SPT5 results in the reduced expression and reduced H3K4me3 level of FLC because the unphosphorylated CTR of SPT5 failed to bind VIP5 in vitro and in vivo (Figures 6 to 8). Although a CTR deletion in yeast resulted in the reduction of global H3K4me3 levels and transcriptional regulation, it is still not known whether the interaction of Spt5 and Rtf1 was dependent on CTR phosphorylation. Collectively, our data illustrated that the posttranslational modification of the SPT5 CTR threonine mediated by CDKD;2 is critical for the functions of SPT5 in regulating flowering time, transcription, and chromatin modification in Arabidopsis (Figure 8H).
Human CDK9, the catalytic subunit of P-TEFb, phosphorylates the threonine of SPT5 CTR; by contrast, yeast BUR2, a homolog of CDK9, phosphorylates the CTR at threonine and serine (Yamada et al., 2006; Zhou et al., 2009). Here, we have shown that the phosphorylation of the Arabidopsis SPT5 CTR at threonine requires CDKD;2, not CDKC;1, CDKC;2, or other CDKD;2 homologs (Figure 2; Supplemental Figure 4). Arabidopsis CDKC;1 and CDKC;2 are closely related to CDK9 and both interact with cyclin T to perform Ser-2 phosphorylation of Pol II CTD in vitro (Cui et al., 2007). There are four proteins closely related to CDK7 in Arabidopsis, namely, CDKD;1, CDKD;2, CDKD;3, and CDKF;1. Both CDKD;1 and CDKD;3 have the C-terminal extensions along with significant sequence similarity to rice (Oryza sativa) CDKD;1 (Umeda et al., 2005). Although CDKD;2, CDKD;3, and CDKF;1 had CAK2 and CTD kinase activity, only the CDKF;1 was able to completely rescue the temperature sensitivity of the cak1 mutant in budding yeast (Shimotohno et al., 2003, 2004), indicating that CDKD;2 is not orthologous to CAK1, which is itself an ortholog of human CDK7. The difference between CDKD;2 and CAK1 (or CDK7) was further shown by the discovery that CDKD;2 phosphorylates Ser-5 and Ser-2 of the Pol II CTD (Hajheidari et al., 2012). In this study, we found that cdkd;2 displayed an early-flowering phenotype (Figures 4C and 4D), whereas cdkc;1 and cdkc;2 exhibited a later-flowering phenotype (Cui et al., 2007). Therefore, CDKD;2 reflects the evolutionary divergence of the members of the Arabidopsis CDK7 subgroup and may have evolved as a functional kinase for the CTR of SPT5.
H3K4me3 is a hallmark of transcriptional initiation in yeast, mammalian cells, and plants (Pokholok et al., 2005; Guenther et al., 2007; Saleh et al., 2007). SPT5 is a transcriptional elongation factor required for VIP5 recruitment. The loss-of-function spt5 mutant had reduced levels of VIP5 recruitment and H3K4me3, indicating that VIP5 recruitment and H3K4me3 modification are determined by transcriptional elongation. ATX1-mediated H3K4me3 modification has previously been reported to be involved in transcription elongation (Ding et al., 2012), and in this study, we provide insight into the relationship between these processes. Both SPT5 and VIP5 were observed to bind across FLC and were required for the transcription and chromatin modification of FLC (Figures 8C to 8G), suggesting that SPT5 and VIP5 regulate FLC directly. Unlike in the vip5 mutant, residual transcripts and H3K4me3 of FLC were observed in the spt5 mutant. One possible reason for this is that the SPT5 protein in spt5 is still partially functional as the T-DNA insertion was located in the middle of the gene. The SPT5 homolog GTA2 may also play a partially redundant function with SPT5. In contrast to an earlier study (Dürr et al., 2014), the gta2 mutants tested here did not display visible phenotypes (Supplemental Figure 1). It is unclear what caused this discrepancy and further studies would be necessary to resolve this contradiction; it is noted, however, that the alleles tested here were not the same as those in the earlier report.
The Arabidopsis PAF1 complex contains the VIP proteins VIP2/ELF7, VIP4, VIP5, and VIP6/ELF8. The disruption of a subunit of PAF1 leads to early flowering, with a reduction of FLC transcription and H3K4me3 modification. Our data demonstrated that mutations that affect SPT5 or CDKD;2 result in the deposition of VIP5, leading to the repression of transcription and H3K4me3 levels of FLC, suggesting that these two factors could be involved in both transcriptional elongation and histone modification. Moreover, these results are also supported by the genetic results that show that VIP5 acts downstream of SPT5 and CDKD;2 (Figures 8A and 8B). Therefore, the reduced H3K4me3 in spt5 and cdkd;2 is caused by a disordered SPT5-VIP5 interaction, which depends on CTR phosphorylation mediated by CDKD;2 in vitro and in vivo (Figure 7).
The yeast Paf1 complex is involved in recruiting Set1 to the Pol II elongation complex (Krogan et al., 2003; Ng et al., 2003). Set1 is the only methyltransferase that catalyzes the methylation of histone 3 on Lys-4 (K4) to produce H3K4me3. Although the relationship between the phosphorylated Spt5 CTR and the Paf1 complex is unclear in yeast, a recent study demonstrated that Rtf1 is necessary and sufficient for the physical interaction between the Paf1 complex and Spt5 (Mayekar et al., 2013). Deletion of the CTR of yeast Spt5 disrupts the interaction between Rtf1 and Spt5 and releases the Paf1 complex from the chromatin. In addition, both H3K4me3 and H3K36me3 are eliminated when either BUR2 or the CTR of yeast Spt5 is deleted (Zhou et al., 2009). Although CDK9 phosphorylates the CTR of human SPT5, enabling it to bind the Plus3 domain of RTF1 in vitro, little is known of the consequence of histone modification with a loss-of-function of human SPT5. ATX1, a histone H3K4 methyltransferase, regulates transcription and H3K4me3 via a direct interaction with the CTD of Pol II, indicating that ATX1 has conserved functions similar to that of yeast Set1 (Ding et al., 2011, 2012; Fromm and Avramova, 2014). However, there is no evidence that ATX1 is recruited by the PAF1 complex in Arabidopsis. Collectively, our study provides an elegant example of molecular regulation involving the interaction of transcriptional elongation and histone methylation in Arabidopsis.
METHODS
Plant Materials
The Arabidopsis thaliana ecotype Col-0 was grown at 22°C under a long-day photoperiod with a 16-h-light/8-h-dark cycle and light intensity of 180 µmol m−2 s−1, or short-day photoperiod with an 8-h-light/16-h-dark cycle. For the GA treatment, 10 µmol GA was sprayed weekly, starting with 2-week-old seedlings. The vernalization treatment followed the method described previously (Gendall et al., 2001; Michaels and Amasino, 2001; De Lucia et al., 2008). Briefly, the seedlings were grown in a growth chamber at 4°C for 5 weeks and then transferred to 22°C under long-day conditions. The mutant lines, obtained from the SALK collection, were as follows: spt5, SALKseq_9711; cdkd;2-1, SALK_065163; cdkd;2-2, SALK_063929; gta2-1, SALK_126891; gta2-2, CS813305; and vip5, SALK_062223.
Plasmid Constructs
The plasmids were constructed with the DNA primers and protocols described in Supplemental Table 1. All cloned DNAs were confirmed by DNA sequencing.
Protein and Peptide Pull-Down Assays, Immunoprecipitation, and Immunoblot Assays
Protein expression and purification were performed as previously described (Ding et al., 2007), and the protocols for protein isolation from the leaves and protein immunoprecipitations were previously described by Ding et al. (2011). Briefly, 3 to 5 g of leaf material was ground in buffer (0.4 M sucrose, 10 mM Tris, pH 8.0, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail [P9599; Sigma-Aldrich]) and filtered through Miracloth. After centrifugation, the pellet was suspended in buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1 mM PMSF, and protease inhibitor cocktail) and resuspended using a Dounce homogenizer. After centrifugation, the supernatant was precleared with protein A or protein G magnetic beads, and specific antibodies or control IgG serum was added for overnight incubation at 4°C. Antibody complexes were precipitated with protein A or protein G magnetic beads. The beads were washed with buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% Triton X-100, and protease inhibitor cocktail) and then boiled for 5 min in SDS loading buffer, after which the proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (Bio-Rad). Immunoblots were analyzed with antibodies to FLAG (Sigma-Aldrich; F1804, lot SLBQ6349V), VIP5 (Abcam; ab84564, lot GR265927-2), HA (Sigma-Aldrich; H9658, lot 095M4778V), H3K4me3 (Abcam; ab8580, lot GR288375-1), and H3 (Abcam; ab1791, lot GR293151-1).
For the pull-down assay, beads were incubated with 3 µg of fusion protein and then incubated with 3 µg of soluble protein overnight at 4°C. Mock controls included extracts prepared from either the His tag or GST vectors. The beads were washed five times (20 mM Tris, pH 7.4, 150 mM NaCl, and 0.05% Tween 20), separated on an SDS-PAGE gel, and analyzed by immunoblot using an anti-GST antibody (Abmart; M20007, lot 252311) or His (Abmart; M20001, lot 273884).
For the peptide pull-down assay, 1 to 2 µg of peptides were bound to 0.5 mg of streptavidin-coated beads (Dynabeads M280; Invitrogen) by incubating them in 50 mL of high-salt binding buffer (25 mM Tris-HCl, pH 8.0, 1 M NaCl, 1 mM DTT, 5% glycerol, and 0.03% Nonidet P-40) at 4°C for 2 h. The bead-bound peptides were washed once with high-salt binding buffer and twice with binding buffer (25 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM DTT, 5% glycerol, and 0.03% Nonidet P-40) and finally resuspended in 50 mL of binding buffer. The beads were incubated with 3 µg of His-Plus3 protein overnight at 4°C. The beads were washed five times with buffer (20 mM Tris, pH 7.4, 150 mM NaCl, and 0.05% Tween 20), and the remaining proteins were eluted from the washed beads in SDS loading buffer. The samples were separated by SDS-PAGE and detected by immunoblot analysis with a His antibody. The sequence of the peptides was GSQTPMHPSRTPLHPCMTP and the phosphorylated form of the peptide had the same sequence, with a phosphate group on the first threonine.
Isothermal Titration Calorimetry Assay
An isothermal titration calorimetry assay was performed as previously described (Leavitt and Freire, 2001), using the proteins and peptides in buffer (100 mM Tris-HCl, pH 7.4, and 50 mM NaCl) and a MicroCal iTC200 titration calorimeter (GE Healthcare) at 25°C. Briefly, an initial 1.0-μL injection was followed by 19 injections of 2.0 μL of sample in solution (peptides with or without phosphorylated threonine) into a 200-μL protein solution of His-Plus3 constantly stirred at 1000 rpm, and the data were recorded for 120 s between injections. The generation of heat due to dilution was determined in a separate experiment by diluting the protein into buffer and subtracting these as blank values for each injection. The corrected heat values were assessed using a nonlinear least squares curve-fitting algorithm (Microcal Origin 7.0) to obtain the stoichiometry (n), binding constants (Ka and Kd), and change in enthalpy for each enzyme-ligand interaction (ΔH).
Phosphorylation Reaction in Vitro
The phosphorylation reaction assay was performed according to the method described by Demidov et al. (2005). Briefly, 2 µg of protein in reaction buffer (50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 2 mM EDTA, and 50 µM ATP) was incubated with 2.5 µCi [ɣ-32P]ATP at 30°C for 1 h. The reaction products were separated by SDS-PAGE and autoradiographed with X-film.
Complementation Assay
For the ProSPT5:SPT5-FLAG construct, full-length genomic SPT5 fused with FLAG was inserted into pCambia1300 containing the 2500-bp SPT5 promoter region. All cloned DNAs were confirmed by DNA sequencing. The primers used to generate the constructs are described in Supplemental Table 1.
Transient Expression and BiFC
SPT5, CDKD;2, and VIP5 were cloned into the pUC-SPYCE (amino acids 156–239) or pUC-SPYNE (amino acids 1–155) vectors. Transient expression assays were performed as described previously (Ndamukong et al., 2010). Briefly, colonies of Agrobacterium tumefaciens were grown overnight The cells were collected and resuspended in an equal volume of induction medium [60 mM K2HPO4, 33 mM KH2PO4, 7.5 mM (NH4)2SO4, 1.7 mM NaCitrate·2H2O, 10 mM MES pH5.6, 1 mM MgSO4, 0.2% glucose, 0.5% glycerol, antibiotics, and 50 µg/mL of acetosyringone]. After vigorous shaking at 28°C for 6 h, the cells were resuspended to an optical density of 0.6 in infiltration medium (10 mM MgCl2, 10 mM MES, and 150 μM acetosyringone) and injected into the abaxial surface of Nicotiana benthamiana leaves. The YFP was visualized 24 to 48 h after inoculation, using a confocal laser scanning microscope (Zeiss LSM700).
Transient Expression in Arabidopsis Protoplasts
Arabidopsis mesophyll protoplast isolation and transformation were performed as described previously (Yoo et al., 2007). Briefly, the leaves of 3-week-old Arabidopsis plants were detached onto double-sided tape, digested with enzymes, and then washed with MMG buffer (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES, pH 5.7). The protoplasts were cotransformed with FLAG-SPT5 and HA-CDKD;2, followed by immunoprecipitation. The primers used to generate the constructs are displayed in Supplemental Table 1.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed according to the manufacturer’s protocol (Clontech; user manual 630489). Briefly, the Saccharomyces cerevisiae strain Y190 was transformed with the bait constructs pGBKT-SPT5, pGBKT-SPT5N, pGBKT-SPT5C, pGBKT-CTR, or pGBKT-KOW and then transformed with pGADT7-VIP5 or pGADT7-Plus3. Vectors lacking coding region insertions were used as negative controls. The yeast was scored for protein interaction based on their ability to grow on synthetic defined medium lacking Trp, Leu, His, and adenine. The primers used to generate the constructs are displayed in Supplemental Table 1.
Reverse Transcription and Real-Time PCR
Total RNA isolation and reverse transcription with oligo(dT) primers (Promega) were performed as described previously (Ding et al., 2007), and the amounts of individual transcripts were measured with gene-specific primers. Real-time PCR analysis was performed using a CFX real-time PCR instrument (Bio-Rad) and SYBR Green mixture (Roche). The relative expression of genes was quantitated with the 2−ΔΔCT Ct calculation, using UBIQUITIN as the reference housekeeping gene for the expression analyses or relative to the input DNA for chromatin immunoprecipitation assays. See Supplemental Table 1 for primers.
ChIP Assay
The ChIP assay was performed using a previously described method (Ding et al., 2011) with slight modifications. Briefly, 3 g of 10-d-old seedlings was fixed with 1% formaldehyde for 10 min and quenched in 0.125 M glycine. The leaves were ground in a mortar and pestle in buffer I (0.4 M sucrose, 10 mM Tris, pH 8.0, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail) and filtered through Miracloth. After centrifugation, the pellet was extracted using buffer II (0.25 M sucrose, 10 mM Tris, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail) and then buffer III (1.7 M sucrose, 10 mM Tris, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail). The nuclei were then lysed in lysis buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 1% SDS, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail), and the extract was sonicated to fragment the DNA to a size range of 300 to 500 bp. After centrifugation, the supernatant was diluted using dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris, pH 8.0, 167 mM NaCl, 0.1 mM PMSF, and protease inhibitor cocktail) and then precleared with protein A or protein G magnetic beads. Specific antibodies (anti-FLAG [Sigma-Aldrich; F1804, lot SLBQ6349V], anti-VIP5 [Abcam; ab84564, lot GR265927-2], and anti-H3K4me3 [Abcam; ab8580, lot GR288375-1]) or control IgG serum were added to the precleared supernatants for an overnight incubation at 4°C. The antibody protein complexes were isolated by binding to protein A or protein G beads. The washed beads were heated at 65°C for 8 h with proteinase K to reverse the formaldehyde cross-linking and digest proteins. The sample was then extracted with phenol/chloroform and the DNA was precipitated in ethanol and resuspended in water. The purified DNA was analyzed by real-time PCR using gene-specific primers presented in Supplemental Table 1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SPT5, AT2G34210; GTA2, AT4G08350; CDKD;2, AT1G66750; VIP5, AT1G61040; AtSPT5, NP_180968.1; AtGTA2, NP_192575.3; HsSPT5.1, NP_001104490; DmSPT5, NP_652610.1; and ScSPT5, NP_013703.1
Supplemental Data
Supplemental Figure 1. Phenotypic analysis of gta2 mutants.
Supplemental Figure 2. Phylogenetic analysis of SPT5 proteins from Arabidopsis, humans, Drosophila, and yeast.
Supplemental Figure 3. Complementation of spt5 with ProSPT5::FLAG-SPT5.
Supplemental Figure 4. The phosphorylation activity of CDKC and CDK proteins.
Supplemental Figure 5. Complementation of spt5 with mutated SPT5.
Supplemental Figure 6. Commercial antibody to mammalian RTF1 recognizes Arabidopsis VIP5.
Supplemental Data Set 1. Alignments used to generate the phylogeny presented in Supplemental Figure 2B.
Supplementary Material
Acknowledgments
We thank Xiangyang Hu from Shanghai University for kindly providing the transgenic FRI seeds and Zoya Avramova from University of Nebraska-Lincoln for critical reading and editing. We thank all members of the Ding group for the discussions during this study. This work was supported by the National Natural Science Foundation of China (Grants 31571315, 31371306, and 91435101 to Y.D.).
AUTHOR CONTRIBUTIONS
C.L. and Y.D. conceived the study and designed the experiments. All authors performed the experiments and took part in interpreting the results and preparing the manuscript. Y.D. wrote the manuscript.
Glossary
- Pol II
polymerase II
- CTR
C-terminal repeat
- CTD
C-terminal domain
- BiFC
bimolecular fluorescence complementation
- ChIP
chromatin immunoprecipitation
- GA
gibberellin
Footnotes
Articles can be viewed without a subscription.
References
- Alvarez-Venegas R., Pien S., Sadder M., Witmer X., Grossniklaus U., Avramova Z. (2003). ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 13: 627–637. [DOI] [PubMed] [Google Scholar]
- Angel A., Song J., Dean C., Howard M. (2011). A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476: 105–108. [DOI] [PubMed] [Google Scholar]
- Aso T., Lane W.S., Conaway J.W., Conaway R.C. (1995). Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science 269: 1439–1443. [DOI] [PubMed] [Google Scholar]
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167. [DOI] [PubMed] [Google Scholar]
- Berr A., Xu L., Gao J., Cognat V., Steinmetz A., Dong A., Shen W.-H. (2009). SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 151: 1476–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R.C., Conaway J.W. (1993). General initiation factors for RNA polymerase II. Annu. Rev. Biochem. 62: 161–190. [DOI] [PubMed] [Google Scholar]
- Cui X., Fan B., Scholz J., Chen Z. (2007). Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell 19: 1388–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F., Crevillen P., Jones A.M.E., Greb T., Dean C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. USA 105: 16831–16836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov D., Van Damme D., Geelen D., Blattner F.R., Houben A. (2005). Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 17: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Avramova Z., Fromm M. (2011). Two distinct roles of ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1) at promoters and within transcribed regions of ATX1-regulated genes. Plant Cell 23: 350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Ndamukong I., Xu Z., Lapko H., Fromm M., Avramova Z. (2012). ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 8: e1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Wang X., Su L., Zhai J., Cao S., Zhang D., Liu C., Bi Y., Qian Q., Cheng Z., Chu C., Cao X. (2007). SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr J., Lolas I.B., Sørensen B.B., Schubert V., Houben A., Melzer M., Deutzmann R., Grasser M., Grasser K.D. (2014). The transcript elongation factor SPT4/SPT5 is involved in auxin-related gene expression in Arabidopsis. Nucleic Acids Res. 42: 4332–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., Murphy S. (2008). Cracking the RNA polymerase II CTD code. Trends Genet. 24: 280–288. [DOI] [PubMed] [Google Scholar]
- Fromm M., Avramova Z. (2014). ATX1/AtCOMPASS and the H3K4me3 marks: how do they activate Arabidopsis genes? Curr. Opin. Plant Biol. 21: 75–82. [DOI] [PubMed] [Google Scholar]
- Gendall A.R., Levy Y.Y., Wilson A., Dean C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. (2007). A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M., Farrona S., Huettel B., Koncz Z., Koncz C. (2012). CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA polymerase II. Plant Cell 24: 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Doyle M.R., Amasino R.M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18: 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S.R., Valverde F., Ravenscroft D., Mouradov A., Coupland G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21: 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Kong X., Wang C., Ma L., Zhao J., Wei J., Zhang X., Loake G.J., Zhang T., Huang J., Yang Y. (2014). Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during vernalization. Plant Cell 26: 4763–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Guermah M., McGinty R.K., Lee J.S., Tang Z., Milne T.A., Shilatifard A., Muir T.W., Roeder R.G. (2009). RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., He Y., Jacob Y., Noh Y.-S., Michaels S., Amasino R. (2005). Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17: 3301–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Dover J., Wood A., Schneider J., Heidt J., Boateng M.A., Dean K., Ryan O.W., Golshani A., Johnston M., Greenblatt J.F., Shilatifard A. (2003). The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11: 721–729. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Kim M., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F. (2002). RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22: 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt S., Freire E. (2001). Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr. Opin. Struct. Biol. 11: 560–566. [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. (2007). The role of chromatin during transcription. Cell 128: 707–719. [DOI] [PubMed] [Google Scholar]
- Liu Y., Warfield L., Zhang C., Luo J., Allen J., Lang W.H., Ranish J., Shokat K.M., Hahn S. (2009). Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29: 4852–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayekar M.K., Gardner R.G., Arndt K.M. (2013). The recruitment of the Saccharomyces cerevisiae Paf1 complex to active genes requires a domain of Rtf1 that directly interacts with the Spt4-Spt5 complex. Mol. Cell. Biol. 33: 3259–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C.L., Jaehning J.A. (2002). Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22: 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I., Jones D.R., Lapko H., Divecha N., Avramova Z. (2010). Phosphatidylinositol 5-phosphate links dehydration stress to the activity of ARABIDOPSIS TRITHORAX-LIKE factor ATX1. PLoS One 5: e13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Dole S., Struhl K. (2003). The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278: 33625–33628. [DOI] [PubMed] [Google Scholar]
- Oh S., Zhang H., Ludwig P., van Nocker S. (2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16: 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., Reinberg D. (2002). A unified theory of gene expression. Cell 108: 439–451. [DOI] [PubMed] [Google Scholar]
- Pien S., Fleury D., Mylne J.S., Crevillen P., Inzé D., Avramova Z., Dean C., Grossniklaus U. (2008). ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D.K., et al. (2005). Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527. [DOI] [PubMed] [Google Scholar]
- Saleh A., Al-Abdallat A., Ndamukong I., Alvarez-Venegas R., Avramova Z. (2007). The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 35: 6290–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Burn J.E., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A., Duan D.R., Haque D., Florence C., Schubach W.H., Conaway J.W., Conaway R.C. (1997). ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc. Natl. Acad. Sci. USA 94: 3639–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno A., Matsubayashi S., Yamaguchi M., Uchimiya H., Umeda M. (2003). Differential phosphorylation activities of CDK-activating kinases in Arabidopsis thaliana. FEBS Lett. 534: 69–74. [DOI] [PubMed] [Google Scholar]
- Shimotohno A., Umeda-Hara C., Bisova K., Uchimiya H., Umeda M. (2004). The plant-specific kinase CDKF;1 is involved in activating phosphorylation of cyclin-dependent kinase-activating kinases in Arabidopsis. Plant Cell 16: 2954–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., He Y., Eshoo T.W., Tamada Y., Johnson L., Nakahigashi K., Goto K., Jacobsen S.E., Amasino R.M. (2006). Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 38: 706–710. [DOI] [PubMed] [Google Scholar]
- Tamada Y., Yun J.-Y., Woo S.C., Amasino R.M. (2009). ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21: 3257–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M., Shimotohno A., Yamaguchi M. (2005). Control of cell division and transcription by cyclin-dependent kinase-activating kinases in plants. Plant Cell Physiol. 46: 1437–1442. [DOI] [PubMed] [Google Scholar]
- Wada T., Takagi T., Yamaguchi Y., Watanabe D., Handa H. (1998). Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17: 7395–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier A.D., Mayekar M.K., Héroux A., Arndt K.M., VanDemark A.P. (2013). Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc. Natl. Acad. Sci. USA 110: 17290–17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., Handa H. (2006). P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21: 227–237. [DOI] [PubMed] [Google Scholar]
- Yang H., Howard M., Dean C. (2014). Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr. Biol. 24: 1793–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu X., Michaels S.D. (2010). The Arabidopsis Paf1c complex component CDC73 participates in the modification of FLOWERING LOCUS C chromatin. Plant Physiol. 153: 1074–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., van Nocker S. (2002). The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J. 31: 663–673. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Yu Y., Meyer D., Wu C., Shen W.-H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 7: 1256–1260. [DOI] [PubMed] [Google Scholar]
- Zhou K., Kuo W.H.W., Fillingham J., Greenblatt J.F. (2009). Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. USA 106: 6956–6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Mandal S.S., Pham A.D., Zheng Y., Erdjument-Bromage H., Batra S.K., Tempst P., Reinberg D. (2005). The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 19: 1668–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.