An autophagy process termed chlorophagy is induced by photodamage and serves to eliminate entire damaged chloroplasts via transport to the vacuole in Arabidopsis leaves.
Abstract
Turnover of dysfunctional organelles is vital to maintain homeostasis in eukaryotic cells. As photosynthetic organelles, plant chloroplasts can suffer sunlight-induced damage. However, the process for turnover of entire damaged chloroplasts remains unclear. Here, we demonstrate that autophagy is responsible for the elimination of sunlight-damaged, collapsed chloroplasts in Arabidopsis thaliana. We found that vacuolar transport of entire chloroplasts, termed chlorophagy, was induced by UV-B damage to the chloroplast apparatus. This transport did not occur in autophagy-defective atg mutants, which exhibited UV-B-sensitive phenotypes and accumulated collapsed chloroplasts. Use of a fluorescent protein marker of the autophagosomal membrane allowed us to image autophagosome-mediated transport of entire chloroplasts to the central vacuole. In contrast to sugar starvation, which preferentially induced distinct type of chloroplast-targeted autophagy that transports a part of stroma via the Rubisco-containing body (RCB) pathway, photooxidative damage induced chlorophagy without prior activation of RCB production. We further showed that chlorophagy is induced by chloroplast damage caused by either artificial visible light or natural sunlight. Thus, this report establishes that an autophagic process eliminates entire chloroplasts in response to light-induced damage.
INTRODUCTION
Chloroplasts are the photosynthetic organelles in autotrophic eukaryotes such as plants and algae. As they convert solar energy into chemical energy to produce most of the energy for plant growth, chloroplasts also suffer sunlight-induced damage. UV-B (wavelengths between 280 and 315 nm) is the most energetic and shortest wavelength light reaching the surface of the Earth (x-rays and gamma rays are blocked by the Earth’s atmosphere). Although UV-B is ineffective for photosynthesis, it is directly absorbed by intracellular macromolecules such as proteins, lipids and DNA, causing them damage and ultimately inhibiting photosynthesis (Kataria et al., 2014). In addition, excess PAR (wavelengths between 400 and 700 nm) causes photodamage to the chloroplast apparatus and decreases photosynthetic capacity, a phenomenon termed photoinhibition (Sonoike, 1998; Li et al., 2009; Takahashi and Badger, 2011; Tikkanen et al., 2014). Photodamaged chloroplasts and chloroplast proteins must be eliminated or repaired both to maintain optimal chloroplast function and to avoid further cellular damage. Plants have diverse and highly controlled mechanisms to avoid, relieve, or repair chloroplast photodamage (Sonoike, 1998; Asada, 2006; Li et al., 2009; Kato and Sakamoto, 2010; Takahashi and Badger, 2011; Kataria et al., 2014; Kong and Wada, 2014; Nishimura and van Wijk, 2015). However, to date, the mechanism by which entire damaged chloroplasts are removed has remained poorly understood.
Organelle turnover in eukaryotic cells is typically achieved via autophagy, a process during which cytoplasm and damaged organelles are sequestered by a double membrane-bound vesicle called an autophagosome and then transported either to the vacuole in yeast and plants or to the lysosome in animals. The outer membrane of the autophagosome then fuses to the vacuolar/lysosomal membrane, thereby releasing an inner membrane-bound structure called the autophagic body into the vacuolar/lysosomal lumen for subsequent degradation (Nakatogawa et al., 2009; Mizushima and Komatsu, 2011). To date, 41 autophagy-related genes (ATGs) have been identified in yeast (Yao et al., 2015), among which 15 core ATGs (ATG1-10, 12-14, 16, and 18) are involved in the nucleation and elongation of the autophagosomal membrane and are essential for any type of autophagy (Nakatogawa et al., 2009). Orthologs of core ATGs are found in many plant species and have been demonstrated to serve similar functions in studies using Arabidopsis thaliana mutants (Yoshimoto, 2012).
Autophagy was initially recognized as a process for bulk degradation of cytoplasmic components to allow nutrient recycling, especially under starvation conditions (Takeshige et al., 1992). However, recent advances have demonstrated vital roles for autophagy in the selective elimination of damaged organelles. A well characterized example is mitophagy, the selective elimination of dysfunctional mitochondria in yeast and mammals (Green et al., 2011; Youle and Narendra, 2011; Kanki et al., 2015). Mitochondria produce most of the energy used by heterotrophs, but concomitantly cause and accumulate oxidative damage. In yeast, mitochondria with oxidative damage are selectively degraded by mitophagy during the stationary phase of growth under nonfermentative conditions (Kanki and Klionsky, 2008). Impaired mitophagy leads to the accumulation of reactive oxygen species (ROS) and mutations in mitochondrial DNA (Kurihara et al., 2012). In mammals, a selective mitophagy process is mediated by phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1) and an E3 ubiquitin ligase known as Parkin (Matsuda et al., 2010; Narendra et al., 2010; Vives-Bauza et al., 2010). Loss-of-function mutations in PINK1 and Parkin can cause a type of Parkinson’s disease in humans (Kitada et al., 1998; Valente et al., 2004), indicating a possible link between impaired mitophagic turnover and human neurodegenerative disorders. These studies highlight the crucial role of mitophagy in the maintenance of cellular homeostasis in yeast and mammals.
Our previous studies revealed autophagic recycling of chloroplast proteins during leaf senescence or energy starvation. In this process, part of the stroma is actively degraded via a type of autophagosome known as the Rubisco-containing body (RCB) to allow efficient amino acid recycling (Ishida et al., 2014). It has also been reported that, during starvation, a distinct type of autophagy-related vesicles decorated by AUTOPHAGY8 (ATG8)-INTERACTING PROTEIN1 delivers some plastid proteins, including certain stroma, envelope, and thylakoid proteins to the vacuole (Michaeli et al., 2014). In addition, electron microscopy observations suggest that entire chloroplasts can be transported to the vacuole during leaf senescence (Minamikawa et al., 2001). Similarly, our study in Arabidopsis plants revealed that chloroplasts that have been shrunken due to the activation of the RCB pathway are degraded as entire organelles via an autophagy process termed chlorophagy in the late stage of starvation-induced senescence (Wada et al., 2009).
However, it is not clear whether there is a relationship between chloroplast-targeted autophagy and sunlight-induced damage. Recent studies have indicated that other pathways facilitate the vacuolar degradation of chloroplast proteins under photodamaging conditions. The CHLOROPLAST VESICULATION (CV) transcript is upregulated by abiotic stresses such photooxidative stress, which can induce the release of CV-containing vesicles from chloroplasts (Wang and Blumwald, 2014). Such vesicles contain stroma, envelope, and thylakoid proteins, which are degraded in the vacuole independent of autophagy. In addition, the accumulation of singlet oxygen (1O2), a type of ROS, induces a chloroplast degradation process that is induced by ubiquitination via a cytoplasmic ubiquitin E3 ligase PLANT U-BOX 4 (Woodson et al., 2015).
In this work, we found that photodamage induces vacuolar transport of entire chloroplasts in mature, nonstarved Arabidopsis leaves. Photodamage resulted in collapsed chloroplasts, which were transported into the vacuole via chlorophagy in wild-type plants but accumulated in the cytoplasm in autophagy-deficient atg mutants. This study thus establishes that entire photodamaged chloroplasts are turned over by an autophagy-dependent pathway.
RESULTS
Autophagy Is Important in Plant Responses to UV-B Damage
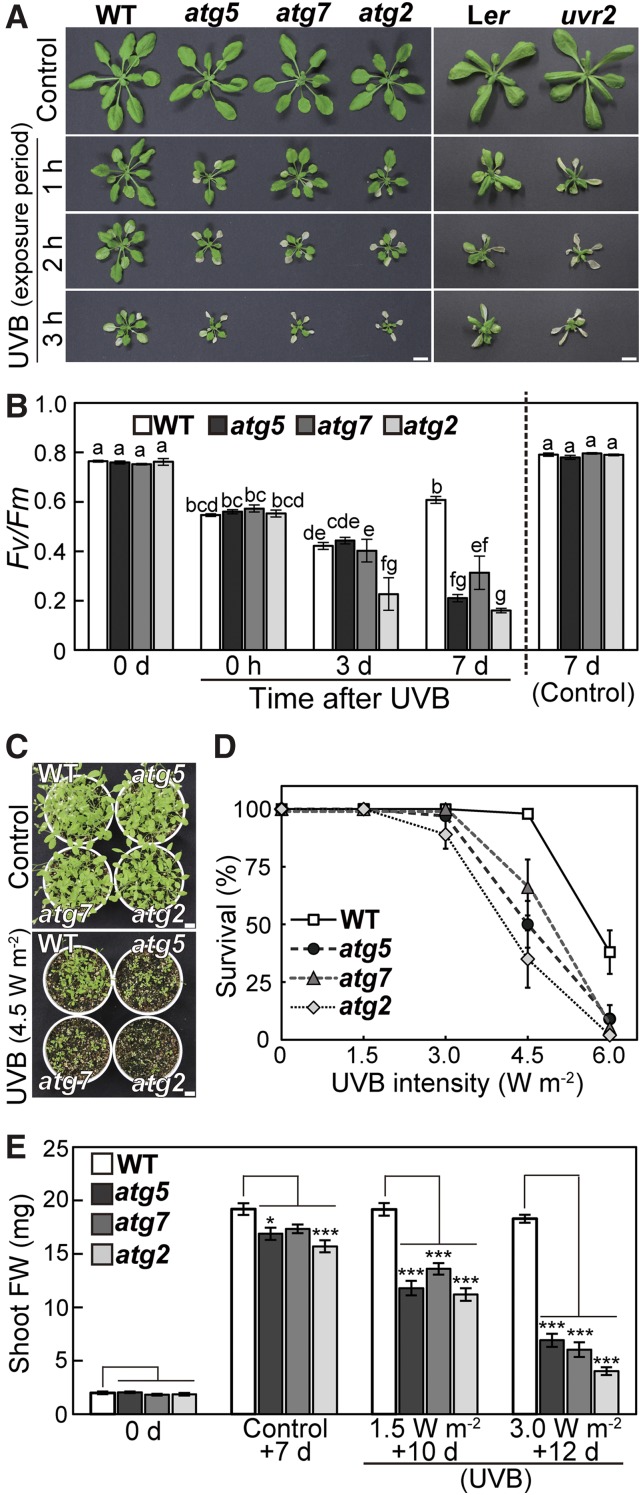
We first examined the relationship between plant responses to UV-B damage and autophagy. Wild-type Arabidopsis ecotype Columbia and three lines of knockout mutants in core ATGs, namely, atg5, atg7, and atg2, were subjected to UV-B exposure (1.5 W m−2) for 1 to 3 h, and their visible phenotypes were observed 7 d later (Figure 1A, left). Shoot size in wild-type plants was decreased by UV-B exposure, and the amount of decrease was dependent on the exposure period. The reductions in shoot size were more severe in atg mutants, which additionally exhibited leaf death. This visible UV-B sensitivity of atg mutants was similar to that of a previously characterized UV-B-sensitive mutant, uv resistance locus2 (uvr2) (Figure 1A, right), which is defective in an enzyme that repairs UV-B-induced DNA damage (Landry et al., 1997). The maximum quantum yield of photosystem II (Fv/Fm) was measured as an indicator of photosynthetic performance following a 2-h exposure to UV-B (Figure 1B). Although the declines of Fv/Fm just after the period of UV-B exposure were the same regardless of genotype, Fv/Fm in atg mutants significantly decreased during the 7 d following treatment.
Figure 1.
Sensitivity of Autophagy-Defective atg Mutants to UV-B-Induced Damage.
(A) Phenotypes of Arabidopsis plants 7 d after UV-B treatment. Wild-type ecotype Columbia (WT), three atg mutants in the Columbia background, atg5, atg7, and atg2, wild-type ecotype Landsberg erecta (Ler), and a previously identified UV-B-sensitive mutant uvr2 in the Ler background were exposed to UV-B (wavelength between 280 and 315 nm) at 1.5 W m−2 for 1, 2, or 3 h. Bars = 10 mm.
(B) The Fv/Fm ratio in leaves of wild-type Columbia, atg5, atg7, and atg2 plants during a 7-d period following a 2-h UV-B treatment at 1.5 W m−2 (±se, n = 4). Different letters denote significant differences from each other based on Tukey’s test (P < 0.05).
(C) Phenotypes of seedlings of wild-type Columbia, atg5, atg7, and atg2 plants 10 d after a 1-h UV-B exposure of 11-d-old seedlings at 4.5 W m−2. Control indicates untreated, 21 d-old plants. Bars = 10 mm.
(D) and (E) The survival (D) and shoot fresh weight of indicated seedlings (E) after a 1-h UV-B treatment at 1.5 to 6.0 W m−2, based on experiments described in (C) ([D], ±se, n = 3 [0 and 1.5 W m−2 conditions] or 5 [all other conditions]; [E], ±se, n = 8). Asterisks denote significant differences from wild-type Columbia based on Dunnett’s test (*P < 0.05, **P < 0.01, and ***P < 0.001).
We next characterized the dose dependence of UV-B sensitivity in atg mutants (Figures 1C to 1E). When 11-d-old seedlings were exposed to different intensities of UV-B for 1 h, the survival of atg mutants was reduced compared with that of the wild type at 4.5 and 6.0 W m−2 UV-B (Figures 1C and 1D). Although shoot fresh weight in atg mutants was 82 to 90% of that in the wild type in control conditions, it declined to 58 to 71% or 22 to 38% of that in the wild type after 1.5 or 3.0 W m−2 UV-B, respectively (Figure 1E). Analogous dose-dependent declines occurred in the uvr2 mutant, although they were more severe than those in atg mutants (Supplemental Figure 1). Therefore, we concluded that atg mutants are UV-B sensitive, indicating that impaired autophagy reduces the tolerance to UV-B damage in Arabidopsis plants. These results suggest that autophagy plays a role in the plant response to UV-B-induced damage.
Chlorophagy Is Induced in UV-B-Exposed Leaves
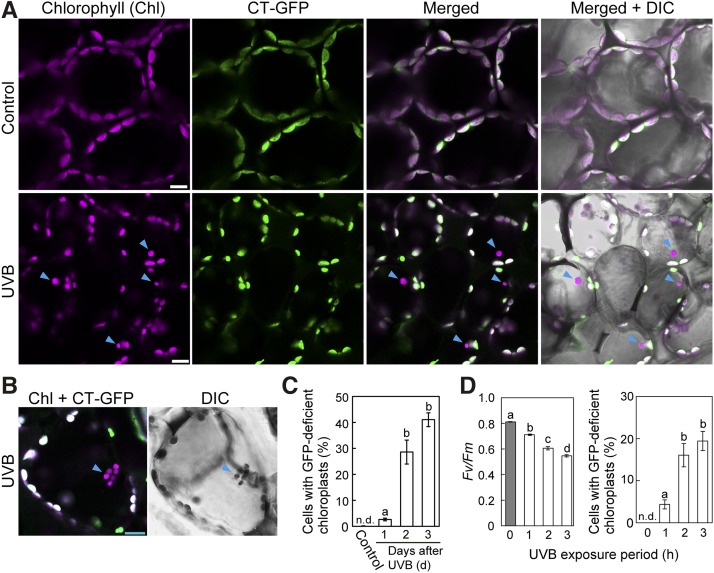
The decrease in Fv/Fm immediately following UV-B exposure indicated damage to the chloroplast apparatus (Figure 1B). We explored whether chloroplast-targeted autophagy was induced by such damage using transgenic plants expressing a chloroplast stroma-targeted GFP, in which the plastid-targeting transit peptide of a DNA recombination enzyme, RECA1, was fused to GFP (Pro35S:CT-GFP; Köhler et al., 1997). In these plants, chlorophyll autofluorescence is a marker of the thylakoid membrane and GFP fluorescence is a marker of soluble stromal fractions (Ishida et al., 2008). When untreated leaves of Pro35S:CT-GFP were observed via laser scanning confocal microscopy (LSCM), all chloroplasts exhibiting chlorophyll autofluorescence had CT-GFP signals (Figure 2A, top). By contrast, chloroplasts lacking CT-GFP were frequently observed in UV-B-exposed leaves 3 d after treatment (Figure 2A, arrowheads). These spherical bodies were visible in differential interference contrast (DIC) images, with more than one found in some mesophyll cells (Figure 2B, arrowheads).
Figure 2.
Induction of Stromal Fluorescent Protein-Deficient Chloroplasts in UV-B-Exposed Leaves.
(A) and (B) Confocal images of leaf mesophyll cells expressing stromal CT-GFP from untreated control plants or plants exposed to UV-B (1.5 W m−2) for 2 h. Images were taken 3 d after treatment. Chlorophyll (magenta), stromal CT-GFP (green), and DIC images are shown. Arrowheads indicate CT-GFP-deficient chloroplasts in the central area of mesophyll cells in UV-B-exposed leaves. In merged images, overlapping signals appear white. Bars = 10 μm in all images.
(C) The proportion of cells containing CT-GFP-deficient chloroplasts in a fixed region (212 × 212 × 40 μm each) 1, 2, and 3 d after 2-h UV-B treatment at 1.5 W m−2 (±se, n = 3).
(D) The Fv/Fm ratio immediately following UV-B exposure at 1.5 W m−2 for 1, 2, or 3 h and the proportion of cells containing CT-GFP-deficient chloroplasts in a fixed region (212 × 212 × 40 μm each) 2 d afterward (±se, n = 4). CT-GFP-deficient chloroplasts in the central area of mesophyll cells were not detected (n.d.) in untreated control plants. Different letters in each graph denote significant differences from each other based on Tukey’s test (P < 0.05).
To evaluate the frequency of this phenomenon, we performed a three-dimensional observation starting from the top of epidermal cells (Supplemental Figure 2) and measured the proportion of cells containing CT-GFP-deficient chloroplasts among all mesophyll cells within a fixed area (four regions of 212 × 212 × 40 μm each per plant). Dead cells in the UV-B-exposed leaves (Supplemental Figures 2A and 2B, asterisks) were excluded from measurement. Plastids exhibiting strong CT-GFP signals and faint chlorophyll autofluorescence (for a representative one, see arrows in Supplemental Figure 2A) were visible in both control and UV-B-exposed leaves (Supplemental Figures 2A and 2B). Such chloroplasts, which correspond to chloroplasts in epidermal cells (Caplan et al., 2015; Supplemental Figures 2C and 3) were sometimes observed in the images focusing on mesophyll cells because of an unequal boundary between the epidermal cell layers and mesophyll cell layers (such as in Figures 2A and 2B).
Quantification of CT-GFP-deficient chloroplasts 1 to 3 d after UV-B exposure indicated that the appearance of such chloroplasts was actively induced from 2 d after treatment (Figure 2C). This phenomenon is likely a response to damage because its frequency increased with increased chloroplast damage, represented by larger decline of Fv/Fm, upon longer exposures to UV-B (Figure 2D). CT-GFP-deficient chloroplasts were located in the central area of mesophyll cells and appeared to move randomly (Figure 2A; Supplemental Movie 1), similarly to vacuolar-accumulated autophagic bodies (Ishida et al., 2008), suggesting that such chloroplasts are located in and drift within the vacuole.
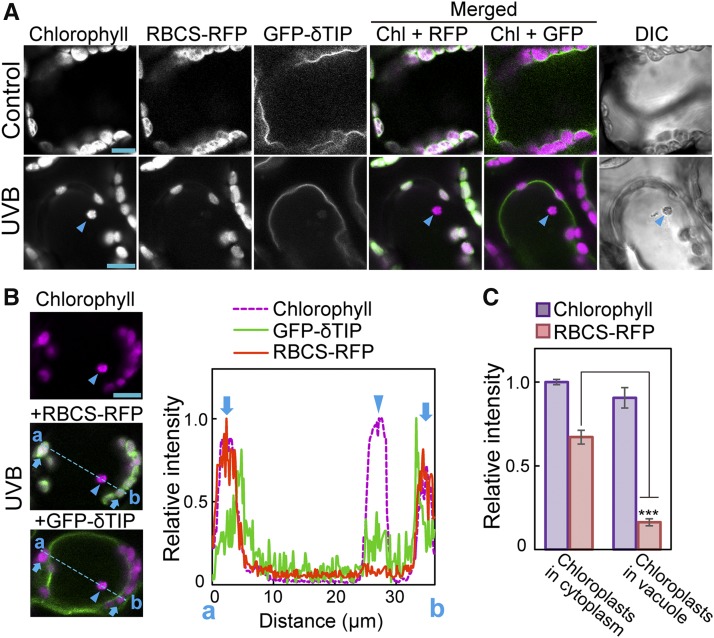
To further define the localization of these chloroplasts, we generated transgenic plants expressing the tonoplast marker delta tonoplast intrinsic protein (δTIP)-GFP, Pro35S:δTIP-GFP (Cutler et al., 2000), along with a different stromal marker (Rubisco small subunit [RBCS]-monomeric RFP, ProRBCS:RBCS-mRFP [Ono et al., 2013]). When these plants were exposed to UV-B, RBCS-RFP-deficient chloroplasts were observed inside the tonoplast (Figure 3A) and appeared to move randomly (Supplemental Movie 2). Measurement of fluorescence intensity (along the blue dotted line in Figure 3B) indicated that chloroplasts containing chlorophyll but not stromal RBCS-RFP were located inside the tonoplast (Figure 3B). This result was consistently observed in individual plants such that the intensity of the stromal RBCS-RFP signal in vacuolar chloroplasts decreased to 24% of that in cytosolic chloroplasts, whereas the chlorophyll signal in vacuolar chloroplasts was 90% of that in cytosolic chloroplasts (Figure 3C). These results indicated that chloroplasts lacking the stromal fluorescent marker in the central area of cells were incorporated into the vacuolar lumen.
Figure 3.
Chloroplasts Lacking Stromal Fluorescent Protein in the Central Area of Mesophyll Cells Are Incorporated into the Vacuole.
(A) to (C) Confocal images of leaf mesophyll cells expressing tonoplast δTIP-GFP and stromal RBCS-RFP from untreated control plants or plants exposed to UV-B (1.5 W m−2) for 2 h. Images were taken 2 d following exposure. Arrowheads indicate chloroplasts lacking stromal RBCS-RFP. In merged images, chlorophyll appears magenta, and GFP and RFP appear green. Bars = 10 μm in all images.
(B) Fluorescence intensity along the blue dotted line (a and b) in the images. The intensity is shown relative to the maximum intensity in each type of fluorescence, which is represented as 1. Cytosolic chloroplasts along the blue dotted line are indicated by arrows.
(C) The intensity of chlorophyll and RBCS-RFP fluorescence in the vacuolar or cytosolic chloroplasts from 13 individual cells. The intensity is shown relative to the chlorophyll fluorescence level in cytosolic chloroplast, which is represented as 1 (±se, n = 13). Asterisks denote significant difference between cytosolic and vacuolar chloroplasts based on a t test (***P < 0.001).
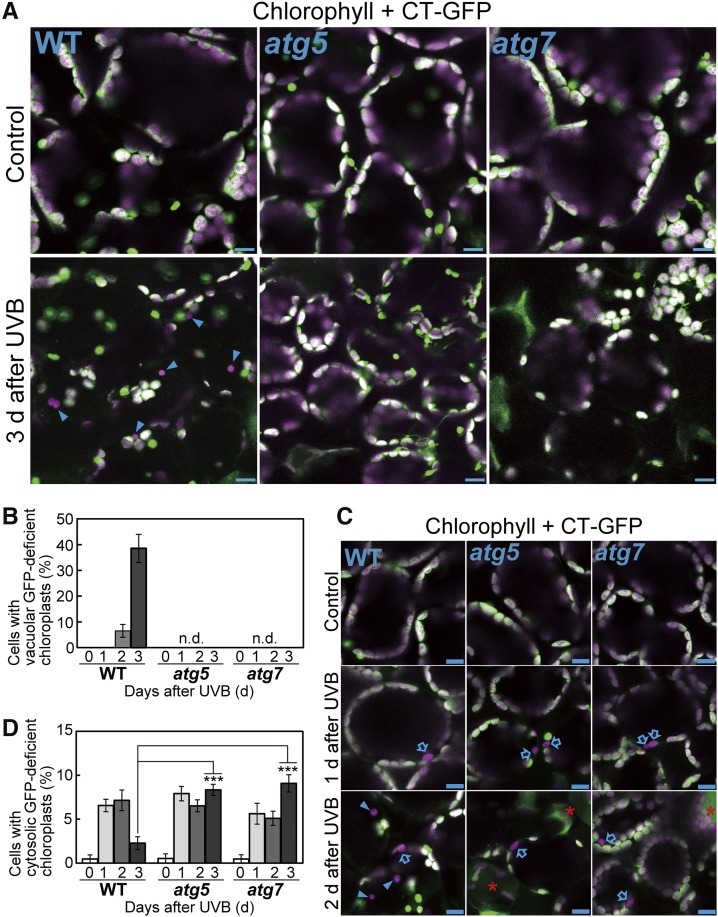
To check whether this phenomenon corresponds to activation of an autophagic pathway, plants expressing Pro35S:CT-GFP in the atg5 or atg7 background were exposed to UV-B and their leaves were observed 3 d afterward. Vacuole-localized chloroplasts that lack CT-GFP and appear to move randomly were not observed in atg5 and atg7 during the 3 d following UV-B treatment (Figures 4A and 4B). Therefore, we concluded that the presence of such chloroplasts reflects autophagic transport of entire chloroplasts, i.e., chlorophagy. The vacuolar chloroplasts in Figure 2A and Supplemental Movie 1 appeared similar in size to cytosolic chloroplasts. The largest were ∼14.3 μm2 and 15.7 μm2 in the vacuole and in the cytoplasm, respectively, supporting our conclusion that chloroplasts are transported in their entirety via chlorophagy in UV-B-exposed leaves.
Figure 4.
Vacuolar Chloroplasts in UV-B-Damaged Leaves Reflect the Autophagic Transport of Entire Chloroplasts, i.e., Chlorophagy.
(A) and (C) Confocal images of mesophyll cells from untreated control leaves or leaves exposed to UV-B (1.5 W m−2) for 2 h. Images were taken 1, 2, and 3 d after treatment of wild-type, atg5, and atg7 plants. Chlorophyll appears magenta and stromal CT-GFP appears green; merged images are shown. Arrowheads indicate vacuolar CT-GFP-deficient chloroplasts, which were located in the central area of mesophyll cells and appeared to move randomly. Arrows in (C) indicate cytosolic vacuolar chloroplasts, which were located at the periphery of cells and did not appear to move randomly. Representative movement for both types of chloroplasts is shown in Supplemental Movie 3. Bars = 10 μm. Red asterisks in (C) indicate dead cells.
(B) and (D) The proportion of cells containing vacuolar CT-GFP-deficient chloroplasts (B) or cytosolic CT-GFP-deficient chloroplasts (D), which are described in (A) and (C), in a fixed region (212 × 212 × 40 μm each) 0 (before treatment), 1, 2, and 3 d following a 2-h UV-B treatment at 1.5 W m−2 (±se, n = 3–5). Vacuolar CT-GFP-deficient chloroplasts were not detected (n.d.) in atg5 and atg7. Asterisks denote significant differences compared with the wild type among the treatments 3 d after UV-B treatment based on Dunnett’s test (***P < 0.001).
Damaged Chloroplasts Accumulate in Autophagy-Defective Mutants
We found that chloroplasts lacking stromal GFP were present at the periphery of the cells before the active induction of chlorophagy in the wild type (1–2 d after UV-B exposure; Figure 4C). Such chloroplasts did not appear to move randomly (Supplemental Movie 3) and were also observed in atg5 and atg7 (Figure 4C). In ProRBCS:RBCS-mRFP; Pro35S:δTIP-GFP plants, chloroplasts that lacked stromal RBCS-RFP but did not appear to move randomly were located outside the GFP-δTIP-labeled tonoplast (Supplemental Figure 4A, arrowheads, and Supplemental Movie 4), indicating their localization in the cytoplasm.
We hypothesized that stromal marker-deficient chloroplasts in the cytoplasm are caused by defects of the envelope. To observe the envelope structure via confocal microscopy, we generated plants expressing an OUTER ENVELOPE PROTEIN7 (OEP7)-GFP fusion protein (Pro35S:OEP7-GFP; Lee et al., 2001). In ProRBCS:RBCS-mRFP; Pro35S:OEP7-GFP plants, RBCS-RFP-deficient chloroplasts located in the cytoplasm consistently had a partly disconnected envelope as visualized by OEP7-GFP (Supplemental Figure 4B, arrows), suggesting that UV-B damage leads to cytosolic collapsed chloroplasts with ruptured envelopes.
We sequentially evaluated the appearance of cytosolic CT-GFP-deficient chloroplasts in Pro35S:CT-GFP plants during the 3 d following exposure to UV-B (1.5 W m−2) for 2 h. Such chloroplasts decreased 3 d after UV-B treatment in wild-type plants (Figure 4D), and this agreed with the increase of vacuolar chloroplasts via chlorophagy 3 d after treatment during this experiment (Figure 4B). In atg5 and atg7, the decreases in cytosolic CT-GFP-deficient chloroplasts were attenuated compared with the wild type (Figure 4D).
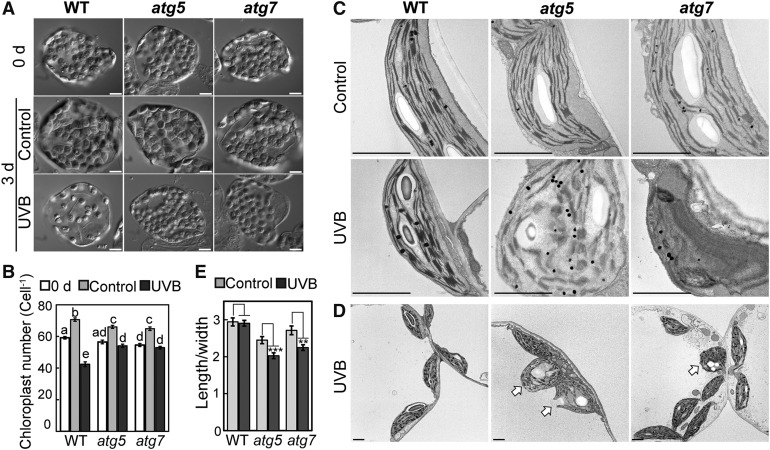
Observations of the accumulation of collapsed chloroplasts in the cytoplasm of atg mutants via confocal microscopy suggested that chlorophagy is involved in the degradation of photodamaged chloroplasts in wild-type plants. To further investigate this possibility, we examined the number and morphology of chloroplasts in UV-B-damaged leaves. We assessed the number of chloroplasts using chemically fixed leaves (Pyke and Leech, 1991). In fixed mesophyll cells, the chloroplast population was markedly sparse 3 d after UV-B exposure in some cells of the wild type compared with cells before treatment or control cells (Figure 5A). In atg5 and atg7, the chloroplast population after UV-B treatment appeared to be similar to that in cells before treatment or in untreated plants (Figure 5A). Consistent with this observation, the chloroplast number per cell decreased significantly, to 72% of that before treatment, in UV-B-exposed wild-type leaves, but did not decrease in those of atg5 and atg7 (Figure 5B). The decrease in chloroplast number in wild-type leaves was not attributable to reduced cell size, as the area of mesophyll cells did not decrease after UV-B damage (Supplemental Figure 5A), and the resulting ratio of chloroplast number to cell area in UV-B-exposed leaves was lower in the wild type than in atg5 and atg7 (Supplemental Figure 5B). These results indicate that the number of chloroplasts decrease after UV-B damage due to the induction of chlorophagy.
Figure 5.
The Accumulation of Abnormal Chloroplasts in the Cytoplasm in UV-B-Exposed Leaves of atg Mutant Plants.
(A) DIC images of fixed mesophyll cells from leaves before treatment (0 d), from untreated leaves as control or from leaves exposed to UV-B (1.5 W m−2) for 2 h. Images were taken 3 d following treatment of wild-type, atg5, and atg7 plants. Bars = 10 μm.
(B) Quantification of the number of chloroplasts per cell from experiments described in (A) (±se, n = 50). Different letters denote significant differences from each other based on Tukey’s test (P < 0.05).
(C) and (D) Transmission electron micrographs of ultrathin sections from control leaves and leaves exposed to UV-B (1.5 W m−2) for 2 h. Images were taken 3 d following treatment of wild-type, atg5, and atg7 plants. Chloroplasts exhibiting a round shape and disorganized thylakoid membranes in UV-B-exposed leaves of atg5 or atg7 are indicated by arrows in (D). Bars = 2 μm.
(E) The ratio of chloroplast length to width from observations described in (C) and (D) (±se, n = 66–72). Asterisks denote significant differences between control leaves and UV-B-exposed leaves based on a t test (**P <0.01 and ***P < 0.001).
We monitored subcellular morphological changes of chloroplasts using transmission electron microscopy (TEM) 3 d after treatment (Figures 5C to 5E). In the wild type, chloroplasts in UV-B-exposed leaves were smaller and had more abundant plastoglobules than those in control leaves. However, their ultrastructure was similar in both conditions. By contrast, chloroplasts in UV-B-exposed leaves of atg5 and atg7 had an abnormal shape with disorganized thylakoid membranes or collapsed envelopes (Figures 5C and 5D). We observed such collapsed chloroplasts along with visibly normal chloroplasts exhibiting an elliptical shape within the same cells (Figure 5D, arrows). To evaluate the abnormality of chloroplast shape, we measured the ratio of chloroplast length to width (Figure 5E). This ratio was reduced 3 d after UV-B treatment in atg5 and atg7, but was not reduced in the wild type. These observations indicated that damaged chloroplasts with an abnormal shape are eliminated by chlorophagy in the wild type but remain in the cytoplasm of atg mutants. Overall, both confocal and TEM observations support the notion that chlorophagy plays an important role in the elimination of cytosolic collapsed chloroplasts that are a result of photodamage.
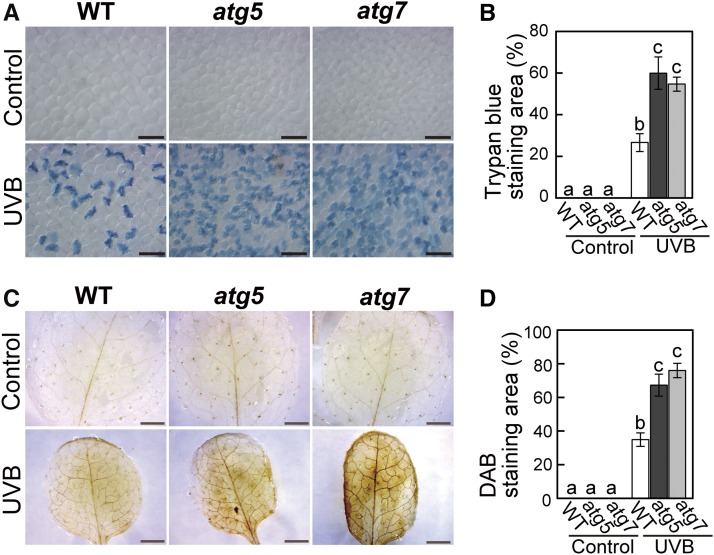
Impaired Autophagy Enhances UV-B-Induced Cell Death and ROS Accumulation
During the quantification of cytosolic GFP-deficient chloroplasts, we found that the appearance of collapsed cells after UV-B treatment increased in atg5 and atg7 plants compared with the wild type (Figure 4C, asterisks). To directly assess the frequency of dead cells, we stained leaves with Trypan blue, which is permeable only in dead cells. In UV-B-exposed leaves, we observed enhanced Trypan blue staining relative to the wild type in atg5 and atg7 (Figures 6A and 6B). In addition, the accumulation of H2O2, a ROS, was enhanced in atg mutants compared with the wild type (Figures 6C and 6D). These results suggest that impaired turnover of UV-B-damaged organelles in atg mutants might enhance ROS accumulation and cell death, which eventually results in the UV-B-sensitive phonotype (Figure 1).
Figure 6.
Cell Death and ROS Accumulation Is Enhanced in UV-B-Exposed Leaves of atg Mutant Plants.
(A) and (B) Dead cells stained by trypan blue in untreated control leaves and leaves exposed to UV-B (1.5 W m−2) for 2 h. Images were taken 3 d following treatment in the wild type, atg5, and atg7. The graph in (B) shows the quantification of Trypan blue-stained areas (±se, n = 4). Bars = 0.1 mm.
(C) and (D) H2O2 stained by 3,3′-diaminobenzidine (DAB) in untreated control leaves and leaves exposed to UV-B (1.5 W m−2) for 2 h. Images were taken 5 d following treatment of wild-type, atg5, and atg7 plants. The graph in (D) shows the quantification of the DAB-stained area (±se, n = 4). Different letters in each graph denote significant differences from each other based on Tukey’s test (P < 0.05). Bars = 2 mm.
Induction of Chlorophagy Involves the Accumulation of ROS Resulting from UV-B Damage
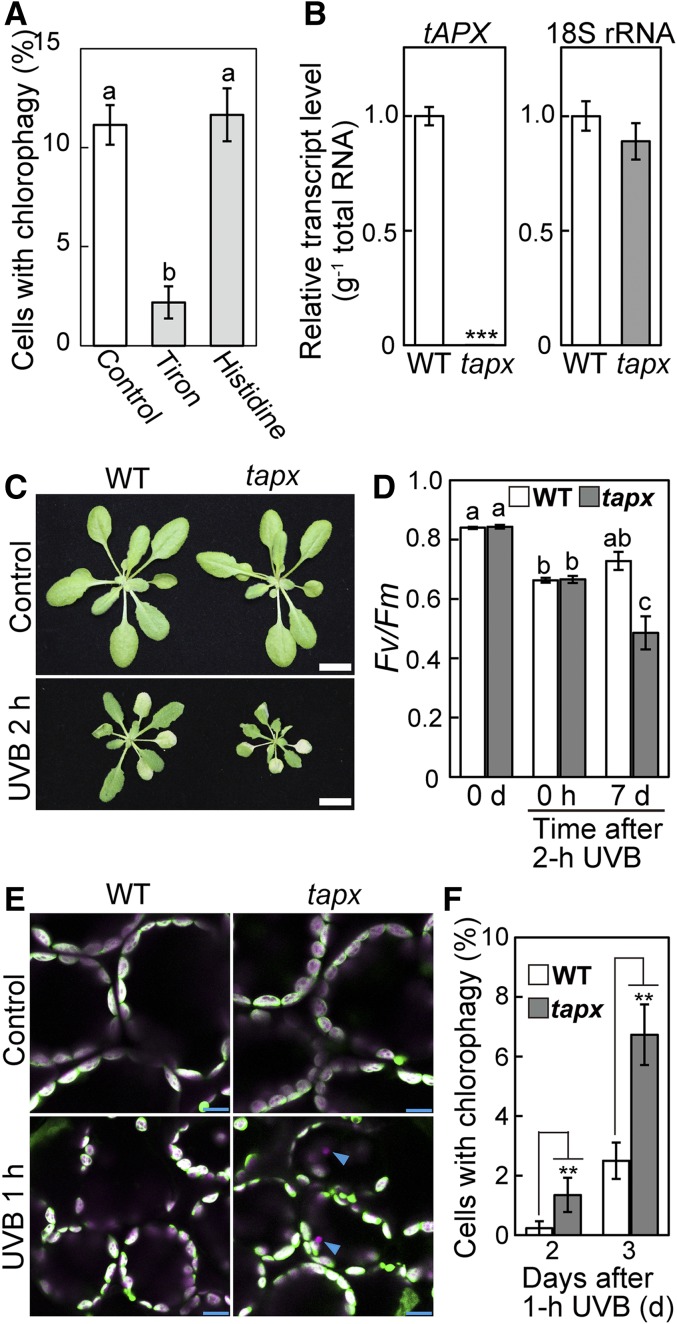
It is widely recognized that UV-B exposure causes the accumulation of ROS in plant leaves (Hideg et al., 2002; Kataria et al., 2014). The enhanced accumulation of H2O2 in atg mutants after UV-B damage further suggested that ROS accumulation is involved in the regulation of chlorophagy. We therefore assessed the effects of 1,2-dihydroxybenzene-3,5-disulfonic acid (Tiron) and histidine, which are scavengers of superoxide (O2−) and singlet oxygen (1O2), respectively. Our previous study confirmed the effects of both reagents as ROS scavengers in chloroplasts of living leaves (Nakano et al., 2006). When leaves were incubated with ROS scavengers 1 d after UV-B exposure, Tiron suppressed the induction of chlorophagy, but histidine did not (Figure 7A). These results suggested that O2− is involved in the induction of chlorophagy.
Figure 7.
The Involvement of a Type of ROS in the Induction of Chlorophagy in UV-B-Exposed Leaves.
(A) The effects of the presence of Tiron, a scavenger of O2−, or histidine, a scavenger of 1O2, on the induction of chlorophagy. Leaves were excised 1 d after a 2-h UV-B exposure (1.5 W m−2) and were then incubated in 10 mM MES-NaOH (pH 5.5) with or without 50 mM Tiron or 10 mM histidine for 20 h. The proportions of cells containing vacuolar CT-GFP-deficient chloroplasts in a fixed region (212 × 212 × 40 μm each) were measured (±se, n = 5).
(B) Transcript levels of tAPX and 18s rRNA (an internal standard) in wild-type and tapx mutants (±se, n = 4). Asterisks denote significant differences between wild-type and tapx plants based on a t test (***P < 0.001).
(C) Phenotypes of wild-type and tapx mutant plants that were either untreated (control) or exposed to after 2-h UV-B treatment (1.5 W m−2). Images were taken 7 d following UV-B exposure. Bars = 10 mm.
(D) The Fv/Fm ratio in leaves before treatment (0 d), immediately following a 2-h UV-B treatment (0 h) or 7 d after treatment (±se, n = 4). Different letters denote significant differences from each other based on Tukey’s test (P < 0.05).
(E) Confocal images of mesophyll cells from untreated control leaves or leaves from wild-type and tapx plants that were exposed to UV-B (1.5 W m−2) for 1 h. Images were taken 2 d after treatment. Chlorophyll appears magenta and stromal CT-GFP appears green; merged images are shown. Arrowheads indicate vacuolar CT-GFP-deficient chloroplasts. Bars = 10 μm.
(F) The proportion of cells containing vacuolar CT-GFP-deficient chloroplasts in a fixed region (212 × 212 × 40 μm each) 1 or 2 d following a 1-h UV-B treatment (1.5 W m−2) of wild-type and tapx plants (±se, n = 6). Asterisks denote significant differences between wild-type and tapx plants based on a t test (**P < 0.01).
O2− is enzymatically detoxified in chloroplasts by two enzymes; O2− is converted to H2O2 by superoxide dismutase and thereafter reduced to H2O by ascorbate peroxidase (APX) (Asada, 2006). In Arabidopsis, stromal APX (sAPX) and thylakoid APX (tAPX) are localized in chloroplasts, and knocking out tAPX leads to increased oxidative damage under photooxidative conditions (Maruta et al., 2012). We generated a knockout mutant of tAPX (tapx) expressing stroma-targeted GFP (Pro35S:CT-GFP; Figures 7B and 7C). Although tapx plants were indistinguishable from wild-type plants under untreated, control conditions, they exhibited more severe growth defects and declines of the Fv/Fm ratio 7 d after UV-B treatment at 1.5 W m−2 for 2 h compared with the wild type (Figures 7C and 7D). This finding indicates the substantial role of tAPX in chloroplasts during the response to UV-B damage. The induction of chlorophagy 2 or 3 d after UV-B exposure for 1 h increased in the tapx mutant compared with the wild type (Figures 7E and 7F). These results support the notion that the accumulation of O2− or subsequent damage is involved in the induction of chlorophagy in UV-B-exposed leaves.
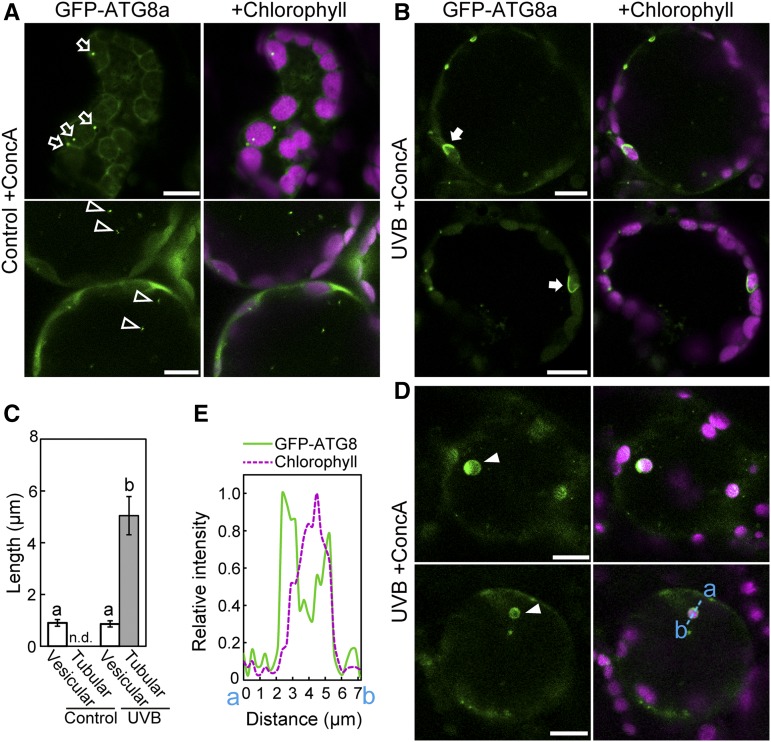
UV-B-Induced Chlorophagy Uses a Transport Process Mediated by an Autophagosomal Membrane
GFP- ATG8 is a marker for the autophagosomal membrane, and the use of concanamycin A (ConcA), an inhibitor of vacuolar ATPase activity, allows the stabilization of GFP-ATG8-labeled autophagic structures through the inhibition of lytic degradation (Ishida et al., 2008; Nakayama et al., 2012). To observe the transport process related to UV-B-induced chlorophagy, Pro35S:GFP-ATG8a plants were exposed to UV-B and then treated with ConcA. In leaves of control plants, autophagosome structures were observed as vesicles of ∼1 μm diameter in the cytoplasm (Figure 8A, top) or in the vacuole (Figure 8A, bottom). By contrast, in UV-B-exposed leaves, we observed large autophagosome structures with a tubular shape surrounding individual cytosolic chloroplasts (Figure 8B, arrows). The size of these tubular autophagosome structures were 5.6 times that of vesicular autophagosomes (Figure 8C). Similar chloroplasts were also observed in the vacuole (Figures 8D, arrowheads, and 8E). These results indicate that vacuolar chloroplasts are transported via sequestering by autophagosomal membrane.
Figure 8.
Tubular Autophagosomal Membranes Are Localized Around Chloroplasts in UV-B-Exposed Leaves.
(A), (B), and (D) Confocal images of mesophyll cells from ConcA-treated leaves expressing GFP-ATG8a. GFP-ATG8a appears green and chlorophyll appears magenta. Leaves of untreated control plants (A) or leaves 1 d following a 2-h UV-B exposure at 1.5 W m−2 ([B] and [D]) were treated with ConcA (1 μM) for 20 h. GFP-ATG8a-labeled vesicles in the cytoplasm and in the vacuole in (A) are indicated by open arrows and open arrowheads, respectively. GFP-ATG8a-labeled tubular structures engulfing chloroplasts in the cytoplasm in (B) are indicated by closed arrows. GFP-ATG8a-labeled chloroplasts in the vacuole in (D) are indicated by closed arrowheads. In (B) and (D), two independent representative images are shown.
(C) Length of GFP-ATG8a-labeled structures exhibiting vesicular or tubular structures from observations described in (A) and (B) (±se, n = 6). GFP-ATG8a-labeled tubular structures engulfing chloroplasts were not detected (n.d.) in control leaves. Different letters denote significant differences from each other based on Tukey’s test (P < 0.05).
(E) Fluorescence intensity of GFP-ATG8a and chlorophyll along the blue dotted line (a and b) in the bottom panels in (D) is shown relative to the maximum intensity of each type of fluorescence, which is represented as 1. Bars = 10 μm in all images; left panels show GFP-ATG8a and right panels show merged images of GFP-ATG8a and chlorophyll.
Photooxidative Damage Activates Chlorophagy without Prior Activation of the RCB Pathway
Our model describing chloroplast-targeted autophagy for recycling during senescence or starvation suggested that the activation of partial degradation via the RCB pathway leads to shrinkage of chloroplasts, which then become targets for degradation as entire organelles (Ishida et al., 2014). Chloroplast autophagy for recycling is particularly activated in individually darkened leaves (IDLs), in which energy starvation due to impaired photosynthesis accelerates senescence (Wada et al., 2009). Therefore, we tested whether chloroplast autophagy under UV-B damage occurs in the same manner as during starvation-induced senescence in IDLs.
We characterized the phenotypic differences between IDLs and UV-B-exposed leaves (Supplemental Figure 6). In IDLs, starch was consumed and transcript levels of dark-inducible genes were highly elevated (Supplemental Figures 6A and 6C). By contrast, starch was detected and H2O2 accumulated in UV-B-exposed leaves (Supplemental Figures 6A and 6B). In addition, transcripts of photooxidative stress-inducible genes were upregulated in UV-B-exposed leaves (Supplemental Figure 6C). These differences suggest that UV-B-exposed leaves suffer photooxidative stress but not sugar starvation.
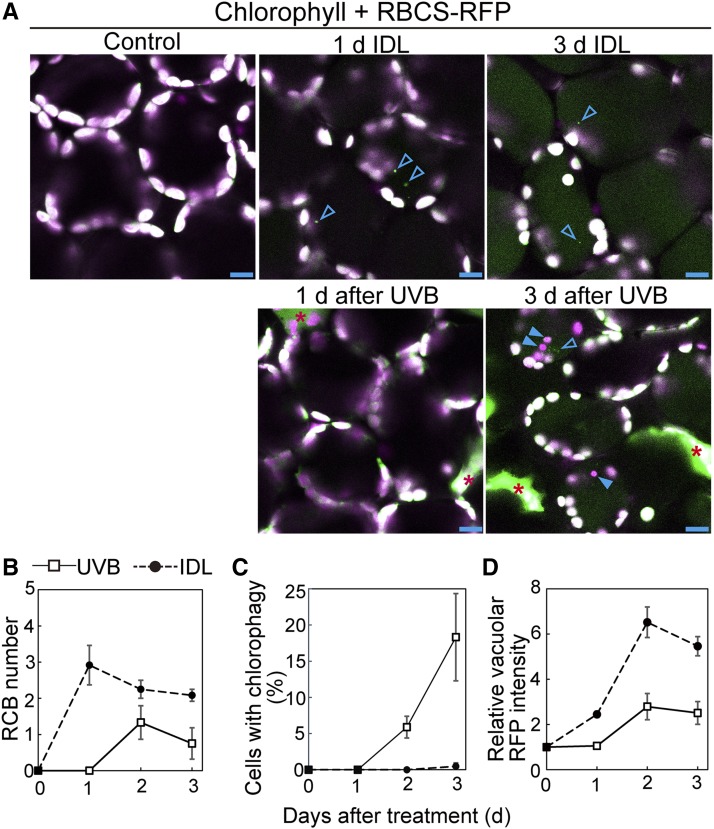
Next, we directly compared the induction levels of the RCB pathway and chlorophagy in IDLs with those of UV-B-exposed leaves using ProRBCS:RBCS-mRFP plants (Figure 9). RCBs are vesicles of ∼1 μm diameter that contain stromal RBCS-RFP but not thylakoid components, including chlorophyll (Figure 9A, arrowheads). The accumulation of RCBs was observed from 1 d of dark treatment (Figure 9B). Chlorophagy was rarely observed during 3 d of dark treatment (Figure 9C). The accumulation of the vacuolar RFP signal began at 1 d of treatment (Figure 9D), which corresponded with the beginning of RCB accumulation (Figure 9B). These observations support our model of starvation-induced chloroplast autophagy. In UV-B-exposed leaves, both chlorophagy and RCB structures were observed during the 3 d after treatment (Figure 9A). In contrast to IDLs, chlorophagy occurred from 2 d after treatment without prior activation of the RCB pathway after UV-B damage (Figures 9B and 9C), and this agreed with the increase of vacuolar RFP intensity (Figure 9D). Accumulation of vacuolar RFP signal in UV-B-exposed leaves of ProRBCS:RBCS-mRFP plants was indeed an autophagy-dependent phenomenon, as it did not occur in the atg5 mutant background (Supplemental Figure 7). In addition, the presence of Tiron, a ROS scavenger, or the knockout mutation of tAPX did not affect the RCB production during incubation of leaves in darkness (Supplemental Figure 8), unlike the induction of chlorophagy in UV-B-exposed leaves (Figure 7). These results indicate that photooxidative stress-induced chloroplast autophagy occurs in a different manner from starvation-induced chloroplast autophagy.
Figure 9.
Chlorophagy Occurs without Prior Activation of Piecemeal Chloroplast Autophagy in UV-B-Exposed Leaves.
(A) Confocal images of mesophyll cells expressing stromal RBCS-mRFP from IDLs or leaves exposed to UV-B (1.5 W m−2) for 2 h. Chlorophyll appears magenta and RBCS-RFP appears green; merged images are shown. Open arrowheads indicate RCBs, which are small, 1-μm-diameter vesicles exhibiting the RBCS-RFP signal. Closed arrowheads indicate chloroplasts in the vacuole. IDLs and UV-B-exposed leaves were observed before treatment and 1, 2, and 3 d after treatment. Representative images taken before treatment and 1 and 3 d following treatment are shown. Red asterisks denote dead cells. Note the spread, faint signal of RFP in the vacuole 3 d after treatment. Bars = 10 μm.
(B) to (D) Quantification of RCB appearance (B), chlorophagy appearance (C), and vacuolar accumulation of RFP signals (D) from observations described in (A) (±se, n = 3–4). RCB number in a fixed area (212 × 212 μm each) was measured. As the indicator of chlorophagy, the proportion of the cells containing vacuolar RBCS-RFP-deficient chloroplasts in a fixed region (212 × 212 × 40 μm each) was measured. Vacuolar RFP intensity is shown relative to the data before treatment, which is represented as 1.
Our previous studies showed that the degradation of stromal proteins in IDLs progresses in atg mutants similarly to in the wild type, whereas the RCB production does not occur in atg mutants (Wada et al., 2009; Ono et al., 2013). We measured the changes of chloroplast protein content in untreated and UV-B-exposed leaves of equal fresh weight (Supplemental Figure 9A). The contents of Rubisco (as a representative stromal protein), chloroplast ATPase COUPLING FACTOR1 (CF1), and cytochrome f (Cyt f) (as representative thylakoid proteins) similarly decreased 3 d after UV-B treatment among the wild type, atg5, and atg7. Thus, impaired autophagy did not compromise the UV-B-induced decrease of some chloroplast proteins, similarly to the decrease during starvation in IDLs. UV-B-exposed leaves also showed upregulation of CV transcript accumulation (Supplemental Figure 9B), which can activate an autophagy-independent degradation of a portion of the chloroplast containing stroma, thylakoid, and envelope proteins via CV-containing vesicles (Wang and Blumwald, 2014). To estimate the contribution of vacuolar lytic activity to UV-B-induced degradation of chloroplast proteins in living cells of the wild type, we examined the effects of ConcA on the decrease of stroma and thylakoid proteins in UV-B-exposed leaves (Supplemental Figure 10). The abundance of both stromal and thylakoid proteins decreased even in the presence of ConcA similarly to the situation in the absence of ConcA 2 d after UV-B exposure. This further supports the notion that the decrease of chloroplast proteins in UV-B-damaged leaves is independent of the vacuolar degradation of chloroplasts by autophagy. UV-B exposure caused cell death in leaves, which also leads to the decrease of cytosolic components including chloroplasts due to the collapse of vacuolar membrane (Hara-Nishimura and Hatsugai, 2011). UV-B-induced cell death was enhanced in atg5 and atg7 (Figures 6A and 6B). These results indicate that the degradation of chloroplast proteins after UV-B damage progressed by different processes that facilitate their degradation.
Chlorophagy Is Activated by Visible Light- or Natural Sunlight-Induced Chloroplast Damage
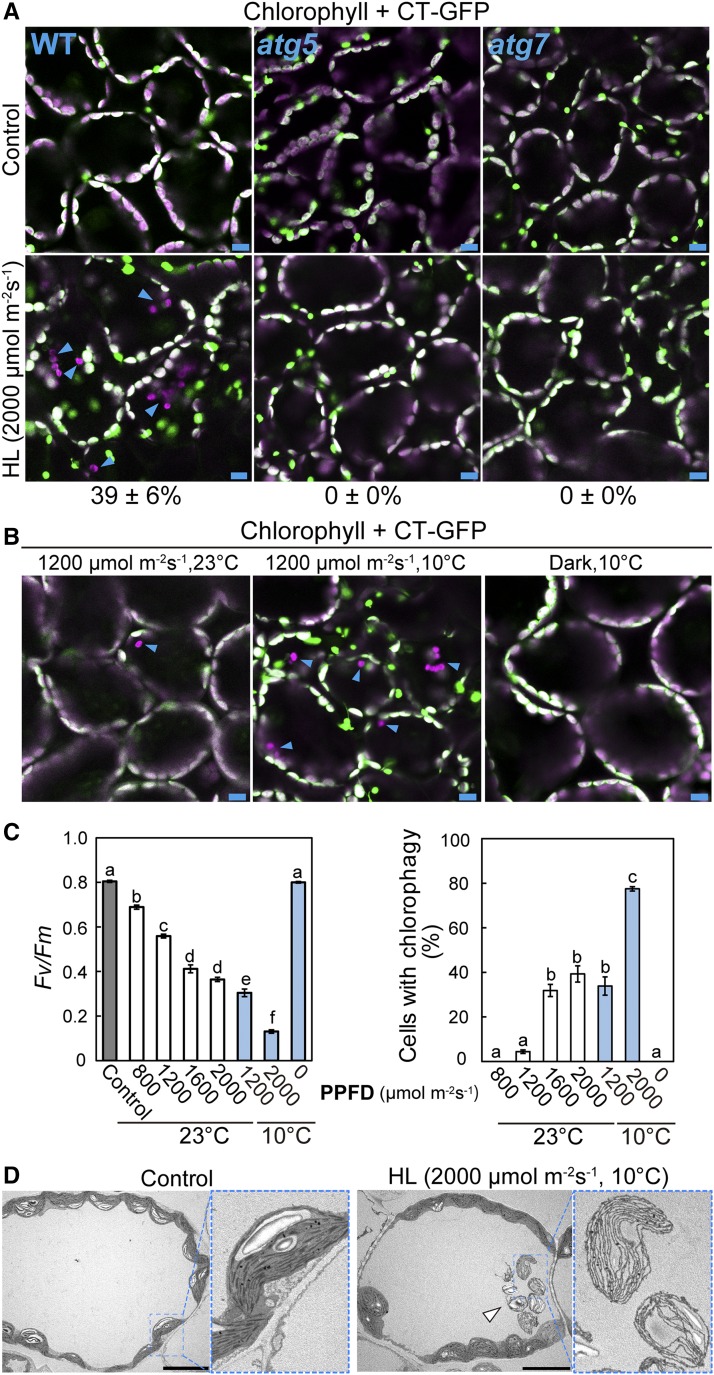
Excess energy from visible light causes damage mainly to the photosynthetic apparatus within chloroplasts, in contrast to the UV-B-related damage that directly accumulates in various macromolecules. These differences prompted us to examine whether chlorophagy is induced by exposure to high visible light. When Pro35S:CT-GFP plants were exposed to visible light at 2000 μmol m−2 s−1, many chloroplasts lacking CT-GFP and appearing to move randomly were observed 2 d later (Figure 10A, arrowheads). This phenomenon was suppressed in leaves of atg5 and atg7 (Figure 10A). Thus, chlorophagy is activated by visible light-induced damage.
Figure 10.
The Induction of Chlorophagy in Response to Chloroplast Damage Due to High Visible Light.
(A) and (B) Confocal images of leaf mesophyll cells expressing stromal CT-GFP from untreated control plants or plants exposed to high visible light for 3 h. Leaves from wild-type, atg5, and atg7 plants 2 d after 3-h visible light irradiation at 2000 μmol m−2 s−1 were observed in (A). Leaves from wild-type plants 2 d after a 3-h visible light irradiation at 0 (dark) or 1200 μmol m−2 s−1 with or without additional low-temperature treatment (10°C) were observed in (B). GFP appears green and chlorophyll appears magenta; merged images are shown. Arrowheads indicate CT-GFP-deficient chloroplasts in the vacuole. Values in (A) represent the proportion of cells containing vacuolar CT-GFP-deficient chloroplasts in a fixed region (212 × 212 × 40 μm each) in wild-type, atg5, and atg7 plants (±se, n = 4).
(C) The Fv/Fm ratio immediately following a 3-h visible light exposure at various photosynthetic photon flux densities between 800 and 2000 μmol m−2 s−1 and the frequency of chlorophagy as the proportion of cells containing vacuolar CT-GFP-deficient chloroplasts 2 d later (±se, n = 4). Different letters denote significant differences from each other based on Tukey’s test (P < 0.05).
(D) Transmission electron micrographs from wild-type control leaves and leaves exposed to 2000 μmol m−2 s−1 visible light at 10°C. An arrowhead indicates vacuolar chloroplasts containing thylakoid membranes but not stroma.
Bars = 10 μm in all images.
Low temperature enhances visible light-induced damage in the thylakoid apparatus (Sonoike, 1998). Combined low temperature treatment (10°C) and exposure to visible light exposure at 1200 μmol m−2 s−1 strongly enhanced the frequency of chlorophagy (Figure 10B). We examined the relationship between the extent of chloroplast damage immediately following visible light exposure and the induction levels of chlorophagy 2 d later and found that more severe declines in Fv/Fm due to higher PPFDs or additional 10°C treatment led to increased activation of chlorophagy (Figure 10C). This agreement between the declines in Fv/Fm and the frequency of chlorophagy clearly indicates that chlorophagy occurs in response to visible light-induced chloroplast damage.
In TEM images of high visible light-exposed leaves (Figure 10D, arrow), we observed that vacuolar chloroplasts retained thylakoid structures, but the electron density in their stromal region was similar to that in the vacuole, indicating that vacuolar chloroplasts had lost their stroma. This observation by TEM was consistent with the observations of vacuolar chloroplasts with fluorescent stromal markers by confocal microscopy (Figures 2 and 10).
In the wild, plants accumulate damage due to combined effects of UV and visible light. We examined whether chlorophagy is induced in response to natural sunlight-induced damage using nontransgenic wild-type plants (Supplemental Figure 11), taking advantage of the fact that we could distinguish vacuolar chloroplasts from cytoplasmic chloroplasts without any fluorescent markers due to their apparently random movement upon observation by DIC and chlorophyll autofluorescence (Supplemental Movie 1). Irradiation in natural sunlight for 3 h at a wavelength greater than 300 nm (Supplemental Figure 11A) caused significant declines of Fv/Fm in leaves (Supplemental Figure 11B), and it also stimulated chlorophagy (Supplemental Figure 11C, arrow, and Supplemental Movie 5). These findings indicate that chlorophagic elimination can function in response to natural sunlight-induced damage.
DISCUSSION
In this study, we observed autophagosome-mediated transport of entire chloroplasts to the vacuole in leaves that had been photodamaged by exposure to UV-B. This transport did not occur in autophagy-defective mutants, which accumulated collapsed chloroplasts. This chlorophagy process was induced by visible light- or natural sunlight-induced chloroplast damage. Therefore, our results establish that chlorophagy leads to the elimination of entire photodamaged chloroplasts. There has been great progress in our understanding of selective autophagy of damaged organelles in recent years. The quality and quantity of mitochondria in several organisms including yeast and mammals are maintained via selective mitophagy (Green et al., 2011; Youle and Narendra, 2011; Kanki et al., 2015). In plants, recent numerous studies showed the involvement of autophagy in the turnover of oxidized peroxisomes or stressed endoplasmic reticulum (Liu et al., 2012; Farmer et al., 2013; Kim et al., 2013; Shibata et al., 2013; Yoshimoto et al., 2014). Identification of photodamage-induced chlorophagy, which is a previously uncharacterized type of autophagy, enhances our understanding of the roles of autophagy in eukaryotes.
Our previous studies showed that nutrient recycling during senescence or starvation is achieved mainly by piecemeal autophagy of chloroplasts via the RCB pathway (Ishida et al., 2008; Ono et al., 2013; Izumi et al., 2015). RCB pathways are specifically activated during energy limitation due to impaired photosynthetic conditions such as darkness (Izumi et al., 2010, 2013). This suggests that energy availability is an important factor regulating the RCB pathway in leaves. In accordance with this hypothesis, vacuolar transport of stromal fluorescent proteins via the RCB pathway was preferentially induced during dark starvation-induced senescence (Figure 9). By contrast, UV-B exposure led to photooxidative stress but did not lead to energy starvation stress (Supplemental Figure 6), in which chlorophagy occurred without prior activation of the RCB pathway (Figure 9). In addition, the frequency of chlorophagy increased in parallel with the amount of chloroplast damage, as represented by declines in Fv/Fm (Figures 2 and 10). Thus, chlorophagy is more likely to be mainly responsible for the elimination of entire collapsed chloroplasts by oxidative damage, in contrast to the larger role of RCBs in the chloroplast-targeted autophagy for nutrient recycling. We propose that two distinct forms of autophagy, the RCB pathway and chlorophagy, enable the optimal chloroplast turnover in response to environmental or developmental circumstances.
Recent progress in our understanding of degradation mechanisms suggests that chloroplasts are degraded via diverse pathways under photodamaging conditions. One recent report demonstrated that a portion of chloroplasts including some stroma, thylakoid, and envelope proteins are degraded in the vacuole via autophagy-independent vesicles known as CV-containing vesicles, which are around 1 μm diameter under photooxidative conditions (Wang and Blumwald, 2014). This process may be activated following UV-B damage (Supplemental Figure 9B). The involvement of ubiquitination in the removal of damaged chloroplasts accumulating 1O2 has also been indicated (Woodson et al., 2015). In these studies, there were no observations of transport of entire chloroplasts containing thylakoid structure such as that seen in this current study (Figures 2 and 10). In addition, we observed that the decrease of chloroplast number in mesophyll cells occurred in the wild type but not in atg5 and atg7 (Figure 5; Supplemental Figure 5). Therefore, our results suggest that the degradation of photodamaged chloroplasts in their entirety is accomplished by autophagy. O2− is the dominant ROS caused by UV exposure (Hideg et al., 2002), and our data suggest that photodamage-induced chlorophagy is linked to the accumulation of O2− (Figure 7). O2− is converted to H2O2 by superoxide dismutase, and H2O2 and 1O2 distinctly influence nuclear gene expression (Laloi et al., 2007). When considered together, it is conceivable that the diverse mechanisms for the elimination of chloroplasts are differentially regulated in response to the type of chloroplast damage, perhaps via the type of dominantly accumulated ROS.
The activation of autophagosome production by treatment with H2O2 or methylviologen, a ROS-producing reagent, has been observed in Arabidopsis root tips (Xiong et al., 2007). Autophagosome production in roots during starvation or salt stress was suppressed by inhibitors of plasma membrane NADPH-dependent oxidase, an enzyme involved in production of ROS as signaling molecules (Liu et al., 2009). ROS is likely involved in the induction of several types of plant autophagy including UV-B-induced chlorophagy, whereas its molecular mechanism is largely unknown. Although ROS can act as signaling molecules that regulate gene expression in plants, the accumulation of ROS subsequently causes damage to organelles and proteins. Thus, it is also possible that subsequent damage within chloroplasts due to the accumulation of O2− induces chlorophagy in UV-B-damaged leaves. The activation of autophagy by the presence of H2O2 or methylviologen was similarly observed in the green alga Chlamydomonas reinhardtii, and this autophagy was enhanced by impaired biosynthesis of carotenoids (Pérez-Pérez et al., 2012). Because carotenoids are pigments that quench excess light energy and ROS in chloroplasts, the induction of autophagy is suggested to be linked to chloroplast photooxidative damage in Chlamydomonas. One recent study indicated that the activity of Chlamydomonas ATG4 protease, a core ATG protein required for the elongation of autophagosome membrane, is regulated by cellular redox states (Pérez-Pérez et al., 2016). However, in algal cells, the process of chloroplast-targeted autophagy is not clearly established. Overall, the mechanism of ROS-mediated regulation of chloroplast-targeted autophagy in photosynthetic eukaryotes has not been elucidated.
To selectively eliminate dysfunctional organelles, the autophagy machinery must distinguish damaged organelles from healthy organelles, as in the selective mitophagy found in yeast or mammals (Green et al., 2011; Youle and Narendra, 2011; Kanki et al., 2015). Our data indicated that UV-B-damaged, abnormal chloroplasts remain in the cytoplasm in atg mutants (Figures 4 and 5). In addition, apparently specialized tubular forms of autophagosomal membrane were associated with chloroplasts in mesophyll cells of UV-B-exposed leaves (Figure 8). These observations support the notion that chlorophagy is regulated in a selective manner. Further studies are needed to clarify the selectivity of this process.
Our observations via confocal microscopy indicated that UV-B damage leads to cytosolic collapsed chloroplasts exhibiting a ruptured envelope (Supplemental Figure 4). The number of such chloroplasts decreased in the wild type during 3 d following treatment, and this decrease was attenuated in atg mutants (Figure 4D). These results suggest that chloroplasts with damaged envelopes are selectively transported to the vacuole. However, the proportion of cytosolic collapsed chloroplasts was less than that of vacuolar chloroplasts that are results of chlorophagy (Figures 4B and 4D). CT-GFP-deficient chloroplasts in the cytoplasm were rarely observed 2 d following high visible light exposure even in atg5 and atg7 (Figure 10A). In ProRBCS:RBCS-mRFP plants, stromal RBCS-RFP was transported to the vacuole 2 d after UV-B treatment, in agreement with the occurrence of chlorophagy (Figure 9), suggesting that stromal components are also transported by chlorophagy. Therefore, it is also possible that cytosolic stroma-deficient chloroplasts are one of the targets of chlorophagy or that damaged chloroplasts remain in the cytoplasm in atg mutants, thereafter collapse, and eventually are observed as cytosolic stroma-deficient chloroplasts. Although our data suggested that the accumulation of O2− or the subsequent related damage is a possible signal inducing chlorophagy in UV-B-exposed leaves, it is currently unclear what type of chloroplast damage directly induces chlorophagy. It is widely recognized that local damage of chloroplasts such as in the reaction center protein of photosystem II is quickly repaired via the protease-dependent degradation inside the chloroplasts (Kato and Sakamoto, 2010; Takahashi and Badger, 2011; Tikkanen et al., 2014; Nishimura and van Wijk, 2015). Thus, it is anticipated that other damage representing wholesale defects of chloroplast function could be the signal for chlorophagy.
Chloroplasts are much larger than typical autophagosomes, which are 1 μm diameter (Figure 8C; Thompson et al., 2005; Yoshimoto, 2012). In UV-B-exposed leaves, large autophagosome structures exhibiting a tubular shape surrounded individual chloroplasts in the cytoplasm or in the vacuole (Figures 8B and 8D). We conclude that the autophagosome structure was elongated to sequester individual chloroplasts for chlorophagic transport because autophagic cargo needs to be sequestered for transport and degradation. However, even on the vacuolar chloroplasts in the presence of ConcA, the intensity of GFP-ATG8a-labeled structures was stronger on one side than on the other side (Figures 8D and 8E). It is also anticipated that unknown structures labeled by GFP-ATG8a are associated with the sequestering of these large organelles in plant cells. During micropexophagy, a type of autophagic degradation of peroxisomes in the methylotrophic yeast Pichia pastoris, an ATG8-associated structure called the micropexophagy-specific apparatus fuses with the vacuolar membrane to sequester a peroxisome cluster (Oku and Sakai, 2016). Similarly, membrane dynamics involved in the sequestering of chloroplasts need to be examined to understand the mechanism of chlorophagy.
In summary, based on the findings presented here, we propose that chlorophagy serves to eliminate chloroplasts that have collapsed due to sunlight-induced damage. This process would complement the multiple well known photoprotective mechanisms in plants, so that severely damaged, dysfunctional chloroplasts would be degraded in their entirety in the vacuole via autophagy. Our findings open up new biological questions relating to both molecular mechanisms of autophagy and plant responses to photodamage, including what type of chloroplast damage induces chlorophagy and how collapsed chloroplasts are recognized and recruited for autophagy.
METHODS
Plant Materials
Arabidopsis thaliana plants were grown in soil in chambers at 23°C under a 12-h-light/12-h-dark photoperiod using fluorescent lamps (140 μmol m−2 s−1). For analyses of dose dependence of UV-B sensitivity, plants were grown under a 16-h-light/8-h-dark photoperiod using fluorescent lamps (60 μmol m−2 s−1). The Arabidopsis T-DNA insertion lines atg5 (atg5-1), atg7 (atg7-2), and atg2 (atg2-1) in the ecotype Columbia background were previously described (Thompson et al., 2005; Hofius et al., 2009; Yoshimoto et al., 2009). The T-DNA insertion line of tapx (tapx-2) was obtained from the Arabidopsis Biological Resource Center. Transgenic Arabidopsis plants expressing chloroplast stroma-targeted GFP driven by the CaMV35S promoter (Pro35S:CT-GFP), RBCS2B-RFP driven by the RBCS2B promoter (ProRBCS:RBCS-mRFP), GFP-ATG8a driven by the CaMV35S promoter (Pro35S:GFP-ATG8), and GFP-δTIP driven by the CaMV35S promoter (Pro35S:GFP-δTIP) were previously described (Köhler et al., 1997; Cutler et al., 2000; Nakayama et al., 2012; Ono et al., 2013). Plants expressing CaMV35S promoter-driven OEP7-GFP (Pro35S:OEP7-GFP) were generated as follows. The protein coding region of OEP7 was amplified from Arabidopsis cDNA by RT-PCR using the primers OEP7-F and OEP7-R (Supplemental Table 1). The amplicon was cloned into pENTR/D/TOPO (Invitrogen), transferred to the pGWB505 vector (Nakagawa et al., 2007), and transformed into Arabidopsis plants using the floral dip method (Clough and Bent, 1998), ecotype Columbia. The atg mutants or tapx mutants containing Pro35S:CT-GFP were obtained by sexual crossing. The expression of Pro35S:CT-GFP was drastically reduced in atg2-1, likely due to transgene silencing (Daxinger et al., 2008). ProRBCS:RBCS-mRFP plants in the atg5 background (atg5-4) were previously described (Ono et al., 2013). Transgenic plants expressing two types of fluorescent marker proteins were generated by sexual crossing. The Arabidopsis point mutation line uvr2 (uvr2-1) is in the Landsberg erecta ecotype and is defective in cyclobutane pyrimidine dimer-photolyase, as previously described (Landry et al., 1997).
Light Treatments
UV-B exposure was provided with UV-B-fluorescent tubes (FL20SE; Toshiba). Visible light exposure was provided by a Xenon light source (MAX-303; Asahi Spectra) equipped with a mirror module (MAX-VIS; Asahi Spectra) to extract visible light (wavelengths between 385 and 740 nm) and a rod lens (RLQL80-1; Asahi Spectra) to emit light with uniform intensity. Light treatments were done in a chamber at 23°C or 10°C. The intensity of UV-B or PAR was measured with a data logger (LI-1400; Li-Cor) equipped with a UV-B sensor (SD204B; Li-Cor) or a photosynthetic photon flux density sensor (LI-190SA; Li-Cor), respectively. Exposure to natural sunlight was performed on a sunny day in June 2015 in the field at Tohoku University (Sendai, Japan), during which control conditions were generated using shade nets. The spectra between 250 and 800 nm of natural sunlight, Xenon lamps, and UV-B lamps were obtained at 1-nm resolution with a spectroradiometer (USR-45DA; Ushio) and are shown in Supplemental Figure 11 along with a control spectrum obtained in the shade. For IDL treatments, third rosette leaves were covered with aluminum foil.
Chlorophyll Fluorescence Measurements
The maximum quantum yield of PSII (Fv/Fm) was measured with a pulse-modulated fluorometer (Junior-PAM; Walz). After 20 min of dark incubation of each plant at room temperature (23 to 25°C), F0 and Fm were measured with a measuring light and a saturating pulse.
Detection of H2O2, Starch, and Dead Cells
H2O2 staining was performed as previously described (Orozco-Cardenas and Ryan, 1999), with slight modification. Shoots or leaves were collected and infiltrated with an H2O2 staining solution (1 mg/mL 3,3′-diaminobenzidine, pH 3.8) using a 50 mL-syringe and incubated for 7 h in darkness with gentle shaking. The samples were boiled in a bleaching solution (60% ethanol, 20% acetic acid, and 20% glycerol) until the tissues were completely decolorized due to chlorophyll loss and transferred to water for photography.
Starch staining was performed as previously described (Caspar et al., 1985), with slight modifications. The shoots were collected and boiled in the bleaching solution until completely decolorized. Then, the samples were incubated for 5 min in the starch staining solution (5.7 mM iodine and 43.4 mM potassium iodide in 0.2 n HCl) and transferred to water for photography.
Staining of dead cells was performed as previously described (Koch and Slusarenko, 1990), with slight modification. The leaves were collected and stained by boiling for 1 min in alcoholic lactophenol trypan blue solution followed by destaining in a chloral hydrate solution (250 g chloral hydrate dissolved in 100 mL distilled water) for 30 min. The images were observed using a microscope (Axio Imager D1; Carl Zeiss) with an EC Plan-Neofluar 20× objective (numerical aperture = 0.5; Carl Zeiss).
qRT-PCR
Total RNA was isolated from rosette leaves using the RNeasy kit (Qiagen) and used for cDNA synthesis using random hexamer and oligo(dT) primers with the PrimeScript RT reagent kit and gDNA Eraser (Takara). An aliquot of the synthesized cDNA derived from 4.0 ng total RNA was subjected to qRT-PCR analysis using the KAPA SYBR FAST qPCR Kit (KAPA Biosystems) using a real-time PCR detection system (CFX96, Bio-Rad). Analyses of dark-inducible genes and photooxidative stress-marker genes were described in previous studies (Baena-González et al., 2007; Ramel et al., 2012; Shao et al., 2013). The level of 18S rRNA was measured as an internal standard (Izumi et al., 2012). The gene-specific primers were used in previous studies (Baena-González et al., 2007; Carbonell-Bejerano et al., 2010; Izumi et al., 2012; Maruta et al., 2012; Ramel et al., 2012; Shao et al., 2013; Wang and Blumwald, 2014). The sequences of primers for qRT-PCR analysis are shown in Supplemental Table 1.
Imaging Using LSCM
Confocal microscopy was performed using an inverted Carl Zeiss LSM710 or LSM800 system equipped with an EC Plan-Neofluar 40× oil-immersion objective (numerical aperture = 1.30; Carl Zeiss) or a C-Apochromat LD63× water-immersion objective (numerical aperture = 1.15; Carl Zeiss). GFP was excited with a 488-nm laser, chlorophyll autofluorescence was excited with a 488-nm and a 633-nm laser, and RFP was excited with a 561-nm laser. For UV-B treatment, 18 d-old plants were exposed to UV-B and the third rosette leaves were observed. For high visible light treatment, 14 d-old plants were exposed to visible light and the second rosette leaves were observed. Cross sections of leaves were observed in 100-μm slices of chemically fixed leaves in fixation buffer (50 mM PBS, 10 mM EGTA, and 5 mM MgSO4, pH 7.0) containing 2% formaldehyde and 0.3% glutaraldehyde (Supplemental Figure 3) as previously described (Iwabuchi et al., 2016).
For quantitative evaluation of the frequency of chlorophagy, four different regions (212 × 212 × 40 μm each) per plant were monitored by LSCM with changing focus to calculate the percentage of cells containing vacuolar chloroplasts lacking signal for stromal fluorescent proteins and appearing to move randomly in the central area of mesophyll cells or for cytosolic collapsed chloroplasts that lacked signal for stromal fluorescent proteins and were located on the verge of mesophyll cells. For evaluation of the frequency of chlorophagy in nontransgenic plants (Supplemental Figure 11), the percentage of cells containing randomly moving chloroplasts in their central area was calculated. For the inhibition of vacuolar degradation activity, leaves of control plants or UV-B-exposed plants were excised 1 d after treatment and infiltrated with 10 mM MES-NaOH (pH 5.5) containing 1 μM ConcA using a 1-mL syringe and were then incubated for 20 h in the growth condition described. For the treatment with ROS scavengers, leaves of control plants or UV-B-exposed plants were excised 1 d after treatment and infiltrated with 10 mM MES-NaOH (pH 5.5) containing 50 mM Tiron or 10 mM histidine with or without 1 μM ConcA using a 1-mL syringe. Leaves were then incubated for 20 h in the growth condition described or in darkness.
For quantitative evaluation of the accumulations of RCBs or vacuolar RFP, images of four different areas (212 × 212 μm each) per plant were obtained by LSCM and either the number of RCBs were counted or the RFP intensity in the central area of mesophyll cells was measured.
Transmission Electron Microscopy
Leaves were fixed with 50 mM cacodylate buffer (pH 7.4) containing 2% glutaraldehyde and 2% paraformaldehyde overnight at 4°C. Next, samples were rinsed with 50 mM cacodylate buffer and postfixed with 2% osmium tetroxide for 3 h at 4°C. After dehydration in an ethanol gradient (50, 70, 90, and 100%), samples were embedded and polymerized in resin at 60°C for 2 d. Resin blocks were ultra-thin sectioned (70 or 80 nm) with a diamond knife attached to an ultramicrotome (Ultracut UCT; Leica). These sections were placed on copper grids and stained with 2% uranyl acetate. The grids were observed using a transmission electron microscope (JEM-1400Plus; JEOL), and images were taken by a CCD camera (VELETA; Olympus).
Measurement of the Number of Chloroplasts
Leaves were fixed in 3.5% glutaraldehyde and observed as previously described (Pyke and Leech, 1991). DIC images were observed using a microscope (Axio Imager D1; Carl Zeiss) with a αPlan-Apochromat 100× oil immersion objective (numerical aperture = 1.46; Carl Zeiss), and the chloroplast number in 50 individual cells from five independent plants was counted with changing focus to avoid duplicate and uncounted chloroplasts.
Immunoblotting
Immunoblotting was performed as previously described (Ishida et al., 2008; Izumi et al., 2015), with slight modification. Leaves were homogenized in HEPES-NaOH (pH 7.5) containing 14 mM 2-mercaptoethanol, 10% (v/v) glycerol, 2% (w/v) SDS, and protease inhibitor cocktail (Roche) and then centrifuged at 10,000g for 5 min. The supernatants were incubated for 5 min at 95°C, and the total protein from leaves of the equal fresh weight or the equal amount of total protein was analyzed by SDS-PAGE using 4% stacking gels and 12% separation gels. The content of total protein in the supernatants was measured by Lowry assay (Bio-Rad). Immunoblotting was performed with an anti-Rubisco large subunit antibody (Ishida et al., 1997), an anti-chloroplast ATPase CF1 antibody (Hidema et al., 1991), an anti-Cyt f antibody (Hidema et al., 1991), and an anti-GFP antibody (MBL). Signals were visualized using horseradish peroxidase-conjugated secondary antibodies (Pierce) and a chemiluminescent reagent (Pierce) and then detected using a ChemiDoc system (Bio-Rad).
Statistical Analysis
Statistical analysis in this study was performed with JMP software (SAS Institute). A Student's t test was used for the comparison of paired samples, a Tukey’s test, or a Dunnett’s test was used for the comparison of multiple samples, as indicated in figure legends.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database or the Arabidopsis Genome Initiative database under the following accession numbers: APX1, At1g07890; ATG2, At3g19190; ATG5, At5g17290; ATG7, At5g45900; ATG8a, At4g21980; CV, At2G25625; DIN1, At4g35770; DIN6, At3g47340; DIN10, At5g20250; OEP7, At3G52420; PUB4, At2g23140; RBCS2B, At5g38420; RECA1, At1g79050; tAPX, At1g77490; TH8, At1g69880; UVR2, At1g12370; and δTIP, At3g16240. Arabidopsis T-DNA insertion lines in the Columbia background were as follows: atg5-1, SAIL_129_B07; atg7-2, GABI_655B06; atg2-1, SALK_076727; tapx-2, GABI_070G07; and atg5-4, SALK_151148.
Supplemental Data
Supplemental Figure 1. Sensitivity of uvr2 mutant plants to UV-B-induced damage.
Supplemental Figure 2. 3D observation of the appearance of CT-GFP-deficient chloroplasts in leaves of Pro35S:CT-GFP plants.
Supplemental Figure 3. Chloroplasts exhibiting strong CT-GFP and faint chlorophyll are located in epidermal cells.
Supplemental Figure 4. Cytosolic collapsed chloroplasts exhibit damaged envelopes.
Supplemental Figure 5. Mesophyll cell area does not decrease due to UV-B exposure.
Supplemental Figure 6. Comparison of phenotypes between individually darkened leaves and UV-B-exposed leaves.
Supplemental Figure 7. The accumulation of vacuolar RFP in UV-B-exposed leaves of ProRBCS:RBCS-mRFP plants is an autophagy-dependent phenomenon.
Supplemental Figure 8. RCB production is not affected by the presence of a scavenger of O2− or the knockout mutation of tAPX.
Supplemental Figure 9. The changes of chloroplast protein content in total leaf protein extracts after UV-B damage does not reflect the impaired chlorophagy in atg5 and atg7.
Supplemental Figure 10. Chloroplast protein abundance in UV-B-damaged leaves after incubation with or without ConcA, an inhibitor of vacuolar lytic activity.
Supplemental Figure 11. Activation of chlorophagy by natural sunlight-induced damage.
Supplemental Table 1. The sequences of primers for qRT-PCR analysis.
Supplemental Movie 1. Chloroplasts lacking CT-GFP in the central area of mesophyll cells appear to move randomly in UV-B-exposed leaves.
Supplemental Movie 2. Chloroplasts lacking RBCS-RFP induced by UV-B damage appears to move randomly inside the GFP-δTIP-labeled tonoplast.
Supplemental Movie 3. Chloroplasts lacking CT-GFP on the verge of mesophyll cells do not appear to move randomly, unlike those in the central area.
Supplemental Movie 4. Chloroplasts lacking RBCS-RFP outside the GFP-δTIP-labeled tonoplast do not appear to move randomly.
Supplemental Movie 5. Vacuolar chloroplasts induced by the irradiation of natural sunlight appeared to move randomly.
Supplementary Material
Acknowledgments
We thank Tsuyoshi Nakagawa for providing us with the pGWB vector, Kohki Yoshimoto and Yoshinori Ohsumi for the use of atg mutant plants, Maureen R. Hanson for the use of Pro35S:CT-GFP and critical reading of the manuscript, and Ling Qihua and Paul Jarvis for critical reading of the manuscript. We thank the Nottingham Arabidopsis Resource Centre for providing uvr2 mutant plants, the Arabidopsis Biological Resource Center for providing tapx mutants and Pro35S:GFP-δTIP, and the Katahira Technical Support Center, Technology Center for Research and Education Activities (Tohoku University), for providing the LSM 710 analytical instrument. This work was supported, in part, by JSPS KAKENHI (Grants 26506001 to M.I., 25119703 and 15H04626 to H.I., 16J03408 to S.N., and 25120702 and 15H05945 to J.H.), Building of Consortia for the Development of Human Resources in Science and Technology (to M.I.), JST PRESTO (to M.I.), Program for Creation of Interdisciplinary Research (to M.I.) in Frontier Research Institute for Interdisciplinary Sciences (Tohoku University), and a JSPS Research Fellowships for Young Scientists (to S.N.)
AUTHOR CONTRIBUTIONS
M.I. conceived the study. M.I., H.I., and J.H. designed the experiments. M.I., H.I., S.N., and J.H. performed the experiments. M.I. and S.N. analyzed the data. M.I. wrote the article with advice from the other authors.
Glossary
- ROS
reactive oxygen species
- RCB
Rubisco-containing body
- LSCM
laser scanning confocal microscopy
- DIC
differential interference contrast
- TEM
transmission electron microscopy
- Tiron
1,2-dihydroxybenzene-3,5-disulfonic acid
- APX
ascorbate peroxidase
- ConcA
concanamycin A
- IDL
individually darkened leaf
Footnotes
Articles can be viewed without a subscription.
References
- Asada K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J.M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942. [DOI] [PubMed] [Google Scholar]
- Caplan J.L., Kumar A.S., Park E., Padmanabhan M.S., Hoban K., Modla S., Czymmek K., Dinesh-Kumar S.P. (2015). Chloroplast stromules function during innate immunity. Dev. Cell 34: 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Bejerano P., Urbez C., Carbonell J., Granell A., Perez-Amador M.A. (2010). A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol. 154: 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T., Huber S.C., Somerville C. (1985). Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 79: 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L., Hunter B., Sheikh M., Jauvion V., Gasciolli V., Vaucheret H., Matzke M., Furner I. (2008). Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends Plant Sci. 13: 4–6. [DOI] [PubMed] [Google Scholar]
- Farmer L.M., Rinaldi M.A., Young P.G., Danan C.H., Burkhart S.E., Bartel B. (2013). Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25: 4085–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D.R., Galluzzi L., Kroemer G. (2011). Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science 333: 1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I., Hatsugai N. (2011). The role of vacuole in plant cell death. Cell Death Differ. 18: 1298–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideg E., Barta C., Kálai T., Vass I., Hideg K., Asada K. (2002). Detection of singlet oxygen and superoxide with fluorescent sensors in leaves under stress by photoinhibition or UV radiation. Plant Cell Physiol. 43: 1154–1164. [DOI] [PubMed] [Google Scholar]
- Hidema J., Makino A., Mae T., Ojima K. (1991). Photosynthetic characteristics of rice leaves aged under different irradiances from full expansion through senescence. Plant Physiol. 97: 1287–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H.T., Mattsson O., Jørgensen L.B., Jones J.D.G., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783. [DOI] [PubMed] [Google Scholar]
- Ishida H., Izumi M., Wada S., Makino A. (2014). Roles of autophagy in chloroplast recycling. Biochim. Biophys. Acta 1837: 512–521. [DOI] [PubMed] [Google Scholar]
- Ishida H., Nishimori Y., Sugisawa M., Makino A., Mae T. (1997). The large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is fragmented into 37-kDa and 16-kDa polypeptides by active oxygen in the lysates of chloroplasts from primary leaves of wheat. Plant Cell Physiol. 38: 471–479. [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., Ohsumi Y., Hanson M.R., Mae T. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148: 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi K., Hidema J., Tamura K., Takagi S., Hara-Nishimura I. (2016). Plant nuclei move to escape ultraviolet-induced DNA damage and cell death. Plant Physiol. 170: 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Hidema J., Makino A., Ishida H. (2013). Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol. 161: 1682–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Hidema J., Wada S., Kondo E., Kurusu T., Kuchitsu K., Makino A., Ishida H. (2015). Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol. 167: 1307–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Tsunoda H., Suzuki Y., Makino A., Ishida H. (2012). RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. J. Exp. Bot. 63: 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Wada S., Makino A., Ishida H. (2010). The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol. 154: 1196–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Klionsky D.J. (2008). Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 283: 32386–32393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Furukawa K., Yamashita S. (2015). Mitophagy in yeast: Molecular mechanisms and physiological role. Biochim. Biophys. Acta 1853: 2756–2765. [DOI] [PubMed] [Google Scholar]
- Kataria S., Jajoo A., Guruprasad K.N. (2014). Impact of increasing ultraviolet-B (UV-B) radiation on photosynthetic processes. J. Photochem. Photobiol. B 137: 55–66. [DOI] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. (2010). New insights into the types and function of proteases in plastids. Int. Rev. Cell Mol. Biol. 280: 185–218. [DOI] [PubMed] [Google Scholar]
- Kim J., Lee H., Lee H.N., Kim S.H., Shin K.D., Chung T. (2013). Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. (1998). Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605–608. [DOI] [PubMed] [Google Scholar]
- Koch E., Slusarenko A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler R.H., Cao J., Zipfel W.R., Webb W.W., Hanson M.R. (1997). Exchange of protein molecules through connections between higher plant plastids. Science 276: 2039–2042. [DOI] [PubMed] [Google Scholar]
- Kong S.G., Wada M. (2014). Recent advances in understanding the molecular mechanism of chloroplast photorelocation movement. Biochim. Biophys. Acta 1837: 522–530. [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Kanki T., Aoki Y., Hirota Y., Saigusa T., Uchiumi T., Kang D. (2012). Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 287: 3265–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C., Stachowiak M., Pers-Kamczyc E., Warzych E., Murgia I., Apel K. (2007). Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry L.G., Stapleton A.E., Lim J., Hoffman P., Hays J.B., Walbot V., Last R.L. (1997). An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc. Natl. Acad. Sci. USA 94: 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Kim D.H., Kim Y.W., Hwang I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260. [DOI] [PubMed] [Google Scholar]
- Liu Y., Xiong Y., Bassham D.C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5: 954–963. [DOI] [PubMed] [Google Scholar]
- Liu Y., Burgos J.S., Deng Y., Srivastava R., Howell S.H., Bassham D.C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T., Noshi M., Tanouchi A., Tamoi M., Yabuta Y., Yoshimura K., Ishikawa T., Shigeoka S. (2012). H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J. Biol. Chem. 287: 11717–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., et al. (2010). PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S., Honig A., Levanony H., Peled-Zehavi H., Galili G. (2014). Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 26: 4084–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamikawa T., Toyooka K., Okamoto T., Hara-Nishimura I., Nishimura M. (2001). Degradation of ribulose-bisphosphate carboxylase by vacuolar enzymes of senescing French bean leaves: immunocytochemical and ultrastructural observations. Protoplasma 218: 144–153. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Komatsu M. (2011). Autophagy: renovation of cells and tissues. Cell 147: 728–741. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007).. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nakano R., Ishida H., Makino A., Mae T. (2006). In vivo fragmentation of the large subunit of ribulose-1,5-bisphosphate carboxylase by reactive oxygen species in an intact leaf of cucumber under chilling-light conditions. Plant Cell Physiol. 47: 270–276. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009). Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10: 458–467. [DOI] [PubMed] [Google Scholar]
- Nakayama M., Kaneko Y., Miyazawa Y., Fujii N., Higashitani N., Wada S., Ishida H., Yoshimoto K., Shirasu K., Yamada K., Nishimura M., Takahashi H. (2012). A possible involvement of autophagy in amyloplast degradation in columella cells during hydrotropic response of Arabidopsis roots. Planta 236: 999–1012. [DOI] [PubMed] [Google Scholar]
- Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. (2010). PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., van Wijk K.J. (2015). Organization, function and substrates of the essential Clp protease system in plastids. Biochim. Biophys. Acta 1847: 915–930. [DOI] [PubMed] [Google Scholar]
- Oku M., Sakai Y. (2016). Pexophagy in yeasts. Biochim. Biophys. Acta 1863: 992–998. [DOI] [PubMed] [Google Scholar]
- Ono Y., Wada S., Izumi M., Makino A., Ishida H. (2013). Evidence for contribution of autophagy to rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ. 36: 1147–1159. [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas M., Ryan C.A. (1999). Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 96: 6553–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez M.E., Couso I., Crespo J.L. (2012). Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 8: 376–388. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez M.E., Lemaire S.D., Crespo J.L. (2016). Control of autophagy in Chlamydomonas is mediated through redox-dependent inactivation of the ATG4 protease. Plant Physiol. 172: 2219–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. (1991). Rapid image-analysis screening-procedure for identifying chloroplast number mutants in mesophyll-cells of Arabidopsis thaliana (L) Heynh. Plant Physiol. 96: 1193–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M. (2012). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA 109: 5535–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N., Duan G.Y., Bock R. (2013). A mediator of singlet oxygen responses in Chlamydomonas reinhardtii and Arabidopsis identified by a luciferase-based genetic screen in algal cells. Plant Cell 25: 4209–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M., Oikawa K., Yoshimoto K., Kondo M., Mano S., Yamada K., Hayashi M., Sakamoto W., Ohsumi Y., Nishimura M. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25: 4967–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoike K. (1998). Various aspects of inhibition of photosynthesis under light/chilling stress: “Photoinhibition at chilling temperatures” versus “Chilling damage in the light”. J. Plant Res. 111: 121–129. [Google Scholar]
- Takahashi S., Badger M.R. (2011). Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci. 16: 53–60. [DOI] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. (1992). Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119: 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M., Mekala N.R., Aro E.M. (2014). Photosystem II photoinhibition-repair cycle protects photosystem I from irreversible damage. Biochim. Biophys. Acta 1837: 210–215. [DOI] [PubMed] [Google Scholar]
- Valente E.M., et al. (2004). Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304: 1158–1160. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C., et al. (2010). PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA 107: 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., Makino A. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149: 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Blumwald E. (2014). Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles. Plant Cell 26: 4875–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.D., Joens M.S., Sinson A.B., Gilkerson J., Salomé P.A., Weigel D., Fitzpatrick J.A., Chory J. (2015). Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350: 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Nguyen P.Q., Bassham D.C. (2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Delorme-Axford E., Backues S.K., Klionsky D.J. (2015). Atg41/Icy2 regulates autophagosome formation. Autophagy 11: 2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K. (2012). Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol. 53: 1355–1365. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Shibata M., Kondo M., Oikawa K., Sato M., Toyooka K., Shirasu K., Nishimura M., Ohsumi Y. (2014). Organ-specific quality control of plant peroxisomes is mediated by autophagy. J. Cell Sci. 127: 1161–1168. [DOI] [PubMed] [Google Scholar]
- Youle R.J., Narendra D.P. (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.