CIPK23, a kinase that regulates the most abundant nitrate transporter (NPF6;3) and activates potassium channel AKT1, inhibits the high-affinity plant ammonium transporters, AMT1s, by phosphorylation.
Abstract
Ion transport in plants is not only strictly regulated on a transcriptional level, but it is also regulated posttranslationally. Enzyme modifications such as phosphorylation provide rapid regulation of many plant ion transporters and channels. Upon exposure to high ammonium concentrations in the rhizosphere, the high-affinity ammonium transporters (AMTs) in Arabidopsis thaliana are efficiently inactivated by phosphorylation to avoid toxic accumulation of cytoplasmic ammonium. External ammonium stimulates the phosphorylation of a conserved threonine in the cytosolic AMT1 C terminus, which allosterically inactivates AMT1 trimers. Using a genetic screen, we found that CALCINEURIN B-LIKE INTERACTING PROTEIN KINASE23 (CIPK23), a kinase that also regulates the most abundant NO3− transporter, NPF6;3, and activates the K+ channel AKT1, inhibits ammonium transport and modulates growth sensitivity to ammonium. Loss of CIPK23 increased root NH4+ uptake after ammonium shock and conferred hypersensitivity to ammonium and to the transport analog methylammonium. CIPK23 interacts with AMT1;1 and AMT1;2 in yeast, oocytes, and in planta. Inactivation of AMT1;2 by direct interaction with CIPK23 requires kinase activity and the calcineurin B-like binding protein CBL1. Since K+, NO3−, and NH4+ are major ions taken up by plants, CIPK23 appears to occupy a key position in controlling ion balance and ion homeostasis in the plant cell.
INTRODUCTION
The transport of nitrogen across membranes is vital for plants. Having a sufficient nitrogen supply is a prerequisite for optimal plant growth. Ion balance and homeostasis require that the uptake of the two major nitrogen ions, NO3− and NH4+, is adjusted with that of other major ions, notably potassium, the most abundant cation (Haynes, 1990). All transporter proteins that are involved in the passage of these ions across the plasma membrane are regulated by phosphorylation (Lanquar et al., 2009; Neuhäuser et al., 2007; Ho et al., 2009; Cheong et al., 2007; Lan et al., 2011; Xu et al., 2006). The major Arabidopsis thaliana root K+ transporter, AKT1, is phosphorylated and activated by the kinase CALCINEURIN B-LIKE INTERACTING PROTEIN KINASE23 (CIPK23). This regulation requires the activation of CIPK23 by one of the two calcineurin B-like binding proteins, CBL1 or CBL9 (Cheong et al., 2007; Lan et al., 2011). CBL1 binding recruits CIPK23 to the plasma membrane and enables autophosphorylation of the kinase (Cheong et al., 2007). Autophosphorylation and CBL1 binding lead to complete activity of the kinase (Hashimoto et al., 2012). The phosphorylation of AKT1 is essential for its activation at low external potassium concentrations. Interestingly, the nitrate transporter NPF6;3 is also phosphorylated by CIPK23. Low external nitrate concentrations are the trigger for the phosphorylation of Thr-101 in NPF6;3 (Ho et al., 2009). Phosphorylation of the transporter leads to a decoupling of the two NPF6;3 monomers and a dissociation of the dimerization. The monomers show a higher flexibility and a higher nitrate affinity than the dimer (Sun et al., 2014).
Phosphorylation of the C-terminal protein tail from AMTs was first identified in a screen for phosphorylated membrane proteins (Nühse et al., 2004). In this screen, Thr-460 from AtAMT1;1 was found to be phosphorylated, even under normal nitrogen supply. A threonine at this position is highly conserved in AMT1 proteins, with the exception of AMT1;5 (Neuhäuser et al., 2007). Mutational analysis of the conserved threonine to mimic phosphorylation showed an inactivation of AtAMT1;1, AtAMT1;2, and other plant AMT1s. Subunit contacts in the trimer might be involved in the inactivation, as the phosphorylation site is adjacent to the neighboring subunits. Coexpression experiments showed that the phosphorylation of one monomer is sufficient to inactivate the complete AMT1 trimer (Loqué et al., 2007; Neuhäuser et al., 2007). This yields a fast and efficient shutdown mechanism for plant NH4+ uptake to avoid ammonium toxicity. Indeed, the phosphorylation of AtAMT1;1 is rapidly triggered by high external ammonium concentrations (Lanquar et al., 2009).
Which kinase might mediate this regulation of plant AMT1s has been a long-standing question. In the yeasts Saccharomyces cerevisiae and Candida albicans, the kinase NPR1 regulates ammonium transport (Boeckstaens et al., 2007; Neuhäuser et al., 2011), but the kinase that regulates plant AMT1s is currently unclear. Plant kinases related to S. cerevisiae NPR1 are members of the CIPK family, and some CIPK genes in plants are upregulated during the ammonium response (Canales et al., 2014; Patterson et al., 2010; Engelsberger and Schulze, 2012). However, whether CIPKs are involved in ammonium transport in plants is currently unclear. Here, we report that CIPK23 inhibits AMT1 activity. This regulation is dependent on CIPK23 activation by CBL1. CIPK23 and CBL1 are therefore major regulators of NO3−, K+, and NH4+ homeostasis in Arabidopsis.
RESULTS
Genetic Screen for the Ammonium Transport Regulating Kinases
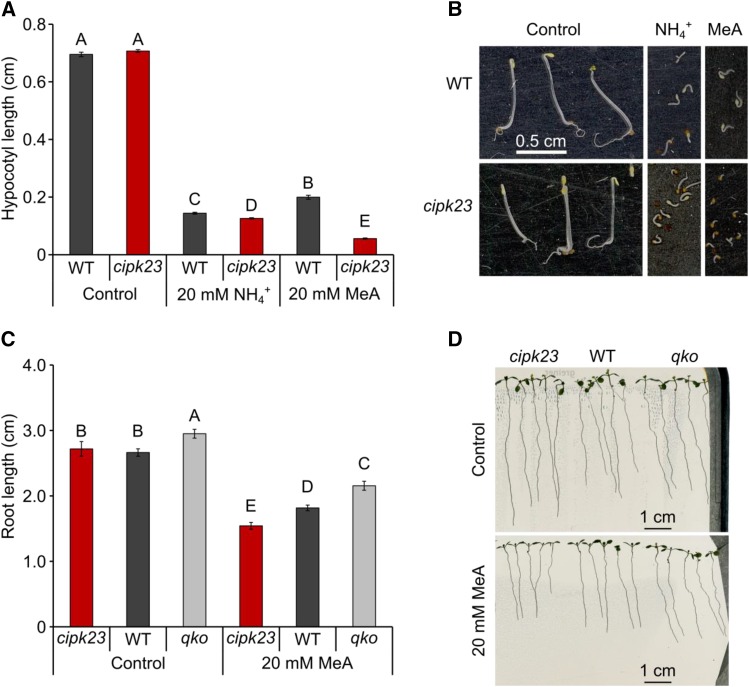
The Arabidopsis genome encodes more than 950 kinases, including more than 850 Ser/Thr kinases. As ammonium is taken up primarily via the root, we selected potential candidate kinases for ammonium transport regulation with expression in the root (and in the hypocotyl) from public databases. Fifty-two hetero- and homozygous mutant lines with T-DNA insertions in or close to the candidate genes were ordered and amplified (Supplemental Table 1). To test these candidate kinase knockout lines, we implemented a hypocotyl elongation screen in the dark. Ammonium as well as the transport analog methylammonium (MeA) impaired primary root growth and elongation (Yang et al., 2015), but root development of individual isogenic seedlings on agar plates was occasionally rather heterogeneous. Hypocotyl elongation showed greater homogeneity and was sensitive to NH4+ and strongly impaired by MeA (Supplemental Figure 1). This allowed reliable screening of a larger number of mutants and identified a mutant that was hypersensitive to toxic MeA and NH4+ concentrations (Figures 1A and 1B). This mutant line (SALK_036154C; lks1-3; Xu et al., 2006) carried a homozygous T-DNA insertion in the 7th intron of AT1G30270, leading to a complete loss of the CIPK23 transcript (Supplemental Figure 2). In light-grown seedlings, high concentrations of MeA in the growth medium weakly impaired primary root growth of the wild type but dramatically reduced primary root length in cipk23 seedlings (Figures 1C and 1D). By contrast, a mutant lacking four AMTs (qko) (Yuan et al., 2007) was less affected by MeA. In addition to the reduced primary root length, the mean lateral root number was strongly reduced in cipk23 in a MeA-dependent manner (Supplemental Figure 3). A second T-DNA insertion line in the 10th intron of CIPK23 (SALK_ 112091C) was isolated, but this line still had residual CIPK23 transcript and weaker phenotypes.
Figure 1.
Ammonium Hypersensitivity of cipk23 Plants.
(A) Hypocotyl length in wild-type and cipk23 plants in the presence of 20 mM ammonium/MeA and under control conditions (0 mM MeA/NH4Cl). Hypocotyl length is given in centimeters. Values are means ± se; n ≥ 150. Different letters (A–C) indicate significant differences by Student’s t test, P value < 0.01. WT, wild-type plants.
(B) Representative photographs of wild-type and cipk23 plants grown in control or 20 mM ammonium/MeA conditions.
(C) Primary root length in wild-type, cipk23, and qko plants after 10 d of growth on control plates (Hoagland [HL] medium containing 2 mM KNO3) or on selective plates (HL medium containing 20 mM MeA and 2 mM KNO3). Values are means ± sd; n ≥ 9. Different letters (A–C) indicate significant differences by Student’s t test, P value < 0.01.
(D) Representative photographs of plants grown for 10 d on control plates or on selective plates.
Phenotype of the cipk23 T-DNA Insertion Line
Since CIPK23 is a positive regulator of potassium uptake, loss of this gene might indirectly lead to ammonium hypersensitivity via altered cation homeostasis. However, experimental variation and elevated K+ showed that hypocotyl hypersensitivity to MeA and NH4+ was not dependent on the external potassium supply (Supplemental Figure 4).
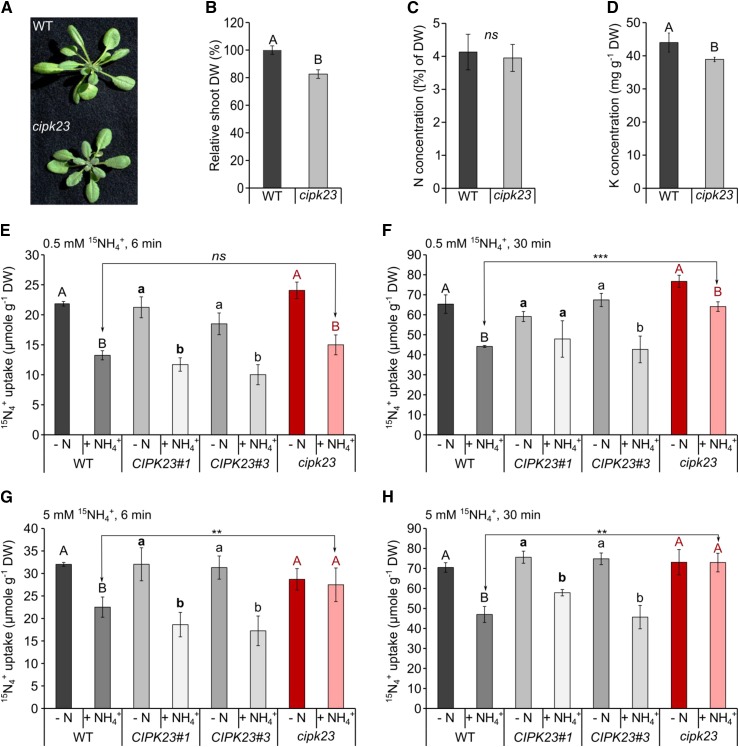
Adult short-day-grown cipk23 plants only accumulated roughly half the biomass of the wild type (Col-0) in standard hydroponics solution (Supplemental Figure 5), but mutant growth was partially rescued by the addition of 5 mM potassium to the nutrient medium (Figures 2A and 2B). Nitrogen and carbon concentrations, by contrast, were not different in mutant plants grown in ammonium-nitrate solution compared with the wild type (Figure 2C; Supplemental Figure 6). Under high K+ conditions, cipk23 had ∼15% less root and shoot biomass and only ∼10% less potassium in the shoot compared with the wild type, which was sufficient for growth and well above the deficiency range (Figure 2D; Supplemental Figure 6). Since no second T-DNA insertion line was available, we complemented the SALK_036154C line with a CIPK23 cDNA construct under its endogenous promoter to test whether the increased susceptibility to high NH4+ and MeA were caused by the loss of function of CIPK23. Two independent complementation lines exhibited fully restored CIPK23 expression to the wild type level (Supplemental Figure 7A) and complemented toxicity phenotypes, and the growth reduction of the cipk23 plants was also complemented (Supplemental Figures 7B and 7C).
Figure 2.
Phenotypic Characterization of Adult cipk23 Plants and Short-Term (6 min/30 min) High (0.5 mM) and Low (5 mM) Affinity Uptake of 15N-Labeled Ammonium.
(A) Rosette size of wild-type and cipk23 plants cultivated hydroponically for 6 weeks in HL with 1 mM NH4NO3 under elevated potassium conditions (5 mM potassium).
(B) Shoot dry weight of 6-week-old plants. Data are shown as means ± sd; n ≥ 60.
(C) Nitrogen concentration in the shoots of wild-type (WT) and cipk23 plants. Data are shown as means ± sd; n ≥ 30. DW, dry weight.
(D) Potassium content in the shoots of wild-type and cipk23 plants under elevated potassium conditions. Significance tested with t test and indicated by A and B. P < 0.001; n = 15.
(E) to (H) Six-week-old plants were grown hydroponically under continuous supply of 1 mM ammonium nitrate and then deprived of nitrogen for 4 d (−N). After 4 d, half of the plants were resupplied with 2 mM NH4+ [1 mM (NH4)2SO4] for 30 min (+NH4+). N-starved plants and plants after ammonium resupply were moved to the HL solution containing 0.5/5 mM 15N [0.25/2.5 mM (15NH4)2SO4] for (E) and (G) 6 min or (F) and (H) 30 min. cipk23, CIPK23 knockout plant; CIPK23#1 and CIPK23#3, complementation lines 1 and 3, respectively. Data are shown as means ± sd. Different uppercase and lowercase letters indicate significant differences between conditions within individual lines at the P value < 0.05 level, ***P value < 0.001, **P value < 0.01; ns, not significant; n = 6.
Ammonium Transport Capacity in cipk23 Mutant and Complementation Lines
In order to test whether the ammonium hypersensitivity of cipk23 plants was caused by altered (higher) ammonium transporter activity in the roots, a short-term 15NH4+ uptake experiment was performed. Two different ammonium concentrations, 0.5 and 5 mM, were chosen that reflect high-affinity ammonium transport via AMTs and low-affinity uptake via a combination of AMTs and the molecularly unknown low-affinity transport system or the apoplastic ammonium flow. Hydroponically grown adult plants were nitrogen-starved for 4 d before the uptake experiment to maximize transcriptional upregulation of high-affinity ammonium transporter transcripts (Yuan et al., 2007) and uptake capacity. It was demonstrated that 30 min of ammonium shock leads to complete phosphorylation of ammonium transporters (inactive AMTs), while under nitrogen starvation, AMTs remain nonphosphorylated and active (Lanquar et al., 2009). Plant lines with a disrupted kinase that regulates ammonium transport activity should therefore exhibit similar rates of ammonium uptake after nitrogen starvation and ammonium shock.
Directly before the uptake experiment, the roots were exposed to a 30-min ammonium shock with 2 mM ammonium to trigger phosphorylation of the AMTs in the wild type and therefore create an AMT phosphorylation/activity gradient between wild-type and cipk23 plants.
Surprisingly, all plant lines showed no major differences in root ammonium uptake activity when treated with 0.5 mM ammonium for 6 min (Figure 2E), although the previous seedling growth experiments with MeA suggested that high-affinity uptake via AMTs was targeted in the cipk23 line. Wild-type and CIPK23 complementation lines had very similar rates of ammonium uptake, but in the cipk23 line, a trend of less transport reduction after the shock was observed. However, in all lines, high affinity root uptake was still massively reduced by previous exposure to unlabeled 2 mM NH4+ (ammonium shock). Interestingly, cipk23 accumulated significantly (P value < 0.01) more ammonium after ammonium shock than wild-type plants when the uptake assay was performed for 30 min (wild type = 27%; cipk23 = 13% reduction in uptake after shock) (Figure 2F; Supplemental Table 2). Contrary to our initial expectations, a more pronounced effect was observed at high ammonium (5 mM). For both uptake durations, the ammonium shock strongly reduced uptake at 5 mM in all lines, except for the cipk23 line (Figures 2G and 2H), indicating that the CIPK23 kinase impaired the ammonium transport activity of AMTs after ammonium shock.
Rescue of cipk23 by Reduced AMT1 Expression
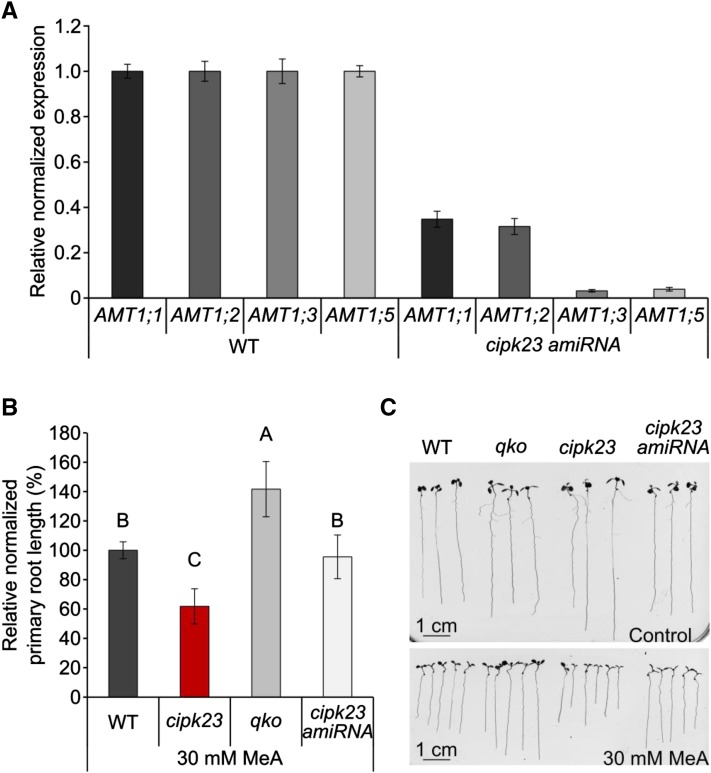
The higher ammonium uptake after shock and the increased sensitivity of cipk23 were not explained by elevated AMT expression in cipk23 plants (Supplemental Figure 8) or altered localization of AMT1s (Supplemental Figure 9), as AMT expression and cellular and tissue AMT1 localization were unchanged in the cipk23 background. As a consequence, higher AMT1 transport activity after the shock is likely causal for the larger NH4+ influx. If AMT1 transport activity is targeted by CIPK23, the absence or reduction of these ammonium transporters is expected to rescue cipk23 plants from elevated MeA (and NH4+) toxicity. In order to suppress all relevant AMT1 genes simultaneously, we used an artificial microRNA (amiRNA) approach to generate plants with reduced AMT1 expression in the cipk23 background. The reduced expression of individual AMTs was checked and verified by qRT-PCR. In the chosen cipk23-amiRNA line, the expression of AMT1;3 and AMT1;5 was strongly reduced, while AMT1;1 and AMT1;2 transcripts were still detectable at around 30% of the wild-type expression level (Figure 3A). When cipk23-amiRNA plants were grown on sterile agar medium supplemented with 30 mM MeA, phenotypic analysis revealed that MeA hypersensitivity was lost in cipk23-amiRNA plants, bringing MeA sensitivity back to the wild-type level (Figures 3B and 3C). This is in agreement with the hypothesis that CIPK23 targets high-affinity AMT1 proteins.
Figure 3.
Knockdown of AMT1 Transcripts by amiRNA Rescues the cipk23 Growth Deficiency.
(A) The knockdown was confirmed by determining the relative normalized expression of ammonium transporters by quantitative RT-PCR: AMT1;1, AMT1;2, AMT1;3, and AMT1;5 in wild-type (WT) and cipk23-amiRNA plants. Representative data from three replicates are shown with values expressed as means ± sd; n = 3 with each sample consisting of 1 g pooled plant material from 14-d-old plants.
(B) Relative normalized primary root length in wild-type, cipk23, qko, and cipk23-amiRNA plants after 10 d of growth on agar containing 30 mM MeA. Values are means ± sd; n ≥ 14. Different letters (A–C) indicate significant differences by Student’s t test, P value < 0.001.
(C) Representative photographs of plants grown for 10 d on control plates (HL medium containing 2 mM KNO3) or on selective plates (HL medium containing 30 mM MeA and 2 mM KNO3).
Regulation of CIPK23
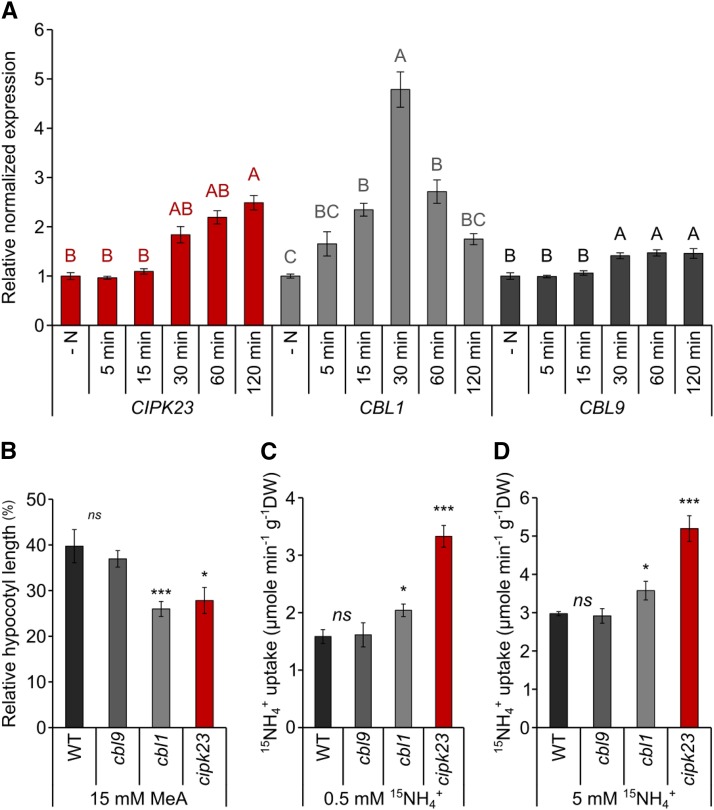
To efficiently regulate ammonium transport activity, the responsible kinase may be transcriptionally regulated by ammonium availability. In line with this notion, CIPK23 expression increased with time after ammonium resupply. Thirty minutes after an ammonium shock, CIPK23 transcripts reached twice the initial levels of N-starved plants. Even after 1 or 2 h, the CIPK23 mRNA levels were still increasing (Figure 4A).
Figure 4.
The Interplay of CIPK23 and CBL1 Is Important in Ammonium Uptake Regulation.
(A) Relative normalized expression of CIPK23, CBL1, and CBL9 in 6-week-old wild-type plants after 4 d of nitrogen starvation (−N) and 5, 15, 30, 60, and 120 min after a 2 mM NH4+ resupply. Data are shown as the mean ± sd of three biological replicates (each two technical replicates and at least 10 plants pooled each). Significance calculated with ANOVA, Turkey test, P < 0.05.
(B) Hypocotyl length in wild-type, cbl9, cbl1, and cipk23 plants in the presence of 15 mM MeA. Hypocotyl length is given in percentage relative to control conditions (0 mM MeA/NH4Cl). Error bars are se; n ≥ 100.
(C) Short term (6 min, 0.5 mM) 15N-ammonium uptake after an ammonium shock in wild-type, cbl9, cbl1, and cipk23 mutants.
(D) Short term (6 min., 5 mM) 15N-ammonium uptake after ammonium shock in wild-type, cbl9, cbl1, and cipk23 mutants. Plants were grown for 6 weeks in standard HL medium with low (1 mM) potassium. Significance calculated with t test; ns, not significant; *P < 0.05; **P < 0.01; n = 8.
As stated before, the kinase activity of CIPK23 is dependent on CBL calcium sensors. Previous analysis showed that CIPK23 regulates AKT1 (Xu et al., 2006; Cheong et al., 2007) and NPF6;3 (Ho et al., 2009) activity in a complex with CBL1 and/or CBL9. Therefore, we investigated the response of CBL1 and CBL9 expression to ammonium. The expression of CBL9 did not change, but CBL1 was transiently stimulated 5-fold by ammonium, with its expression peak after 30 min. A similar expression pattern was found for CBL1 and CBL9 in the cipk23 mutant background (Supplemental Figure 10). Homozygous knockout lines (cbl1, SALK_110426C; cbl9, SALK_142774C) (Supplemental Figure 11) were used to test the effect of CBL1 and CBL9 on the regulation of CIPK23 by ammonium. Hypocotyl elongation of cbl1 plants completely resembled the cipk23-hypersensitive phenotype (Figure 4B), and the rate of ammonium uptake after ammonium shock was significantly higher (P value < 0.05) in cbl1 plants than in Col-0 plants grown in 1 mM potassium. By contrast, cbl9 failed to show differences in the hypocotyl or 15N uptake tests compared with wild-type plants (Figures 4C and 4D).
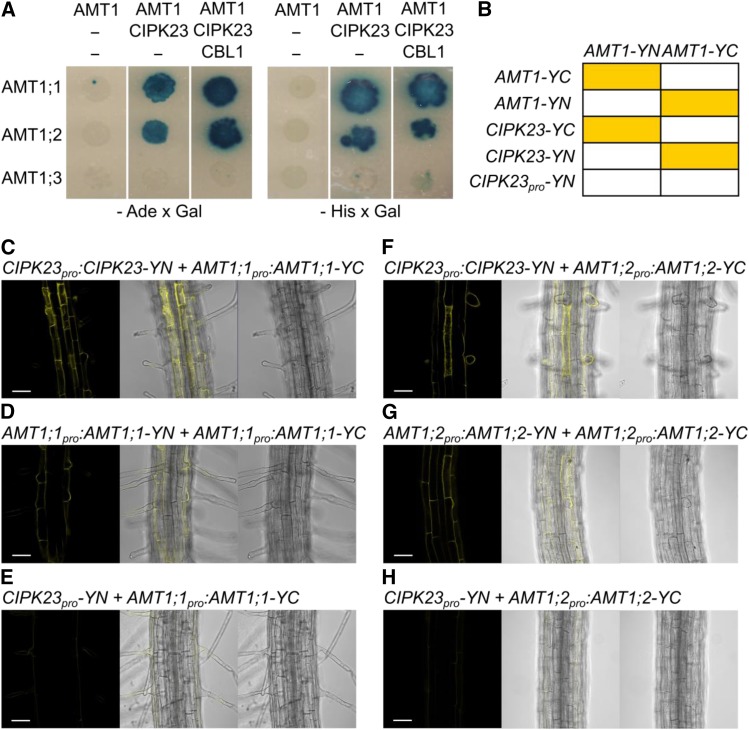
AMT1-CIPK23 Interaction
We investigated whether CIPK23 is directly or indirectly involved in the regulation of ammonium uptake in yeast and in planta. For phosphor-regulation of AMT1 by CIPK23, a direct physical interaction between AMTs and the kinase must occur. To examine potential protein-protein interaction, a yeast two-hybrid assay using Ade2 (enables growth without adenine) and His3 (enables growth without histidine), as well as β-galactosidase as reporter genes was performed and the results were verified using a stable bimolecular fluorescence complementation assay (BiFC) in plants. Using all three reporters, the yeast two-hybrid assay indicated interactions of AMT1;1 and AMT1;2 with CIPK23, but no interaction of CIPK23 with AMT1;3 (Figure 5A). These interactions in yeast were independent of CBL1. For BiFC analysis in plants, translational fusions of AMT1;1, AMT1;2, and CIPK23, driven by their endogenous promoters, were generated in the BiFC vector system. A chart showing which combinations yielded interaction and thereby showed fluorescence can be seen in Figure 5B.
Figure 5.
Protein Interactions of CIPK23 with AMT1;1 and AMT1;2.
(A) Representative photographs of yeast growth after 5 d in 28°C without adenine (left) or without histidine (right) and stained with X-Gal.
(B) Chart showing which combinations yielded interaction and thereby fluorescence in planta.
(C) and (F) Representative photographs of stable BiFC coexpression in roots of CIPK23pro:CIPK23-YN with AMT1;1pro:AMT1;1-YC (C) and CIPK23pro:CIPK23-YN with AMT1;2pro:AMT1;2-YC (F) in a wild-type background.
(D) Interactions of ammonium transporters with different subunits in the trimer AMT1;1pro:AMT1;1-YN coexpressed with AMT1;1pro:AMT1;1-YC.
(E) and (H) Coexpression of the N fragment of YFP under the CIPK23 promoter with AMT1;1pro:AMT1;1-YC (E) and AMT1;2pro:AMT1;2-YC (H). From left to right, fluorescence BiFC-YFP signal, overlay, and bright-field image.
(G) AMT1;2pro:AMT1;2-YN coexpressed with AMT1;2pro:AMT1;2-YC. Bars = 50 µm.
The coexpression of CIPK23 together with AMT1;1 or AMT1;2 (fused to the complemental halves of YFP) generated fluorescence, indicating functional YFP molecules (Figures 5C and 5F). This not only indicated that the AMTs and CIPK23 have overlapping tissue expression patterns, but it also strongly suggested that these CIPK23-AMT pairs directly interact. Coexpression of ammonium transporters fused with N- and C-fragments of YFP was used as a positive control (Figures 5D and 5G). Whether fluorescence resulted from coincidental protein-protein proximity, as the genes were expressed in the same root cell types, was excluded by coexpressing AtAMT1;1pro:AtAMT1;1-N-YFP and AtAMT1;2pro:AtAMT1;2-N-YFP with the C-terminal fragment of YFP under the control of the CIPK23 promoter region (1500 bp upstream of the gene) as a negative control (Figures 5E and 5H). Reconstitution (fluorescence) was not observed for AtAMT1;1 nor AtAMT1;2, suggesting direct protein-protein interactions between CIPK23 and AMT1;1 / AMT1;2 transporters, not only in yeast, but in planta as well.
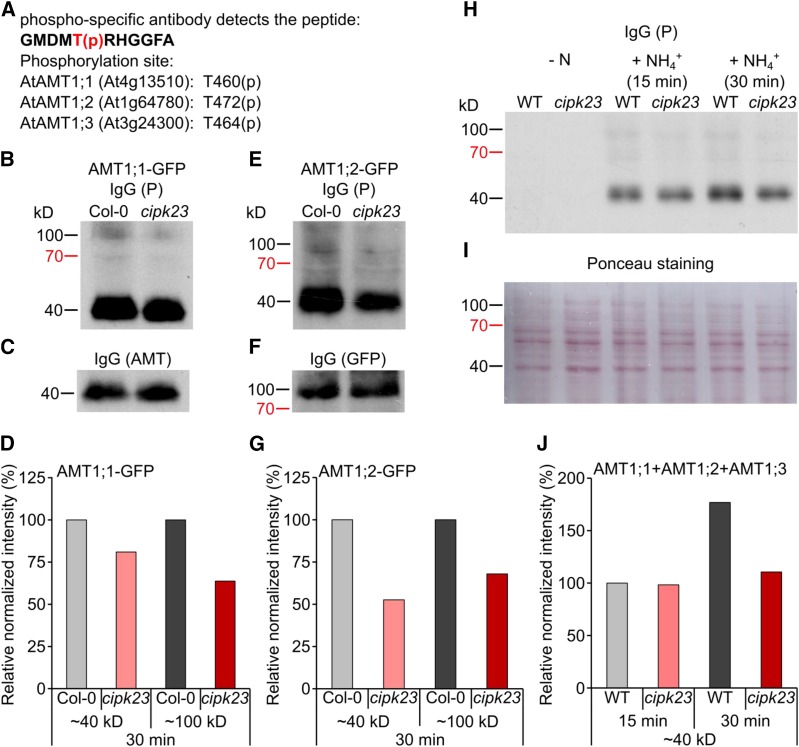
Reduced AMT1 Phosphorylation upon Ammonium Shock in cipk23
Whether CIPK23 is capable of phosphorylating AMT1s at the crucial C-terminal threonine was assayed by protein gel blot analysis. Ammonium resupply to N-starved seedlings has been shown to induce phosphorylation of several residues in the conserved C terminus of ammonium transporters. A single residue, Thr-460 in AMT1;1, was shown to be responsible for transporter inactivation (Lanquar et al., 2009); however, the corresponding residue also exists in the same peptide moiety in AMT1;2 and AMT1;3, but not in AMT1;5. Mutations in this residue have similar effects in the other AMT1s (Neuhäuser et al., 2007; Yuan et al., 2013). The phospho-specific antibody (anti-P) raised against the phospho-peptide GMDMT(p)RHGGFA (Figure 6A) was therefore expected to collectively detect phosphorylated AMT1;1, AMT1;2, and AMT1;3 proteins at the expected molecular mass of monomeric AMT proteins between 33 and 55 kD (Blakey et al., 2002; Ludewig et al., 2003; Lanquar et al., 2009; Yuan et al., 2013). Using protein gel blot analysis with this phosphorylation-specific, but AMT-unselective, antibody, phosphorylation of AMT1s after 15 and 30 min ammonium shock was verified. In contrast to the wild-type background, the phosphorylation of AMT1s in cipk23 plants was slightly reduced after 15 min and more clearly after 30 min (Figures 6H to 6J). However, as AMT1;3 does not interact with CIPK23, it may be phosphorylated in both wild-type and cipk23 plants, thus leading to background phosphorylation in the mixed AMT1 band. AMT1;1 and AMT1;2 together comprise ∼50 to 70% of all root-localized AMT1 proteins, but residual phosphorylation of these two transporters is still manifested in cipk23 plants. As a consequence, the maximal expected reduction in total AMT1 phosphorylation is only expected in the range of 30 to 40%. Normalizing the 45-kD band intensity to the control band, a reduction of this intensity by ∼30 to 40% was indeed observed and corresponds to a reduction of total AMT1 phosphorylation by 30 to 40%. To test the phosphorylation of individual AMT1s, Col-0 and cipk23 plants expressing AMT1;1-GFP or AMT1;2-GFP under their endogenous promoters were used. In these plants, two copies of the corresponding AMT1 are expressed, but only one of these is tagged as a translational fusion with GFP. As a consequence, its monomer size is ∼30 to 40 kD larger than the endogenous AMTs. The AMT1-GFP fusion proteins were quantified with an anti-GFP antibody at similar levels in the wild-type and cipk23 background, with a molecular mass of around 80 to 100 kD. In the cipk23 background, a mild reduction in phosphorylation of the monomeric AMT1 bands (∼45 kD) was again apparent after ammonium shock. Furthermore, the weaker bands at 80 to 100 kD indicated that AMT1;1-GFP and AMT1;2-GFP proteins were detected and also phosphorylated after shock (Figures 6B to 6G), but phosphorylated AMT1-GFP levels were strongly reduced in the cipk23 background. Since the 45-kD band represents all three root-expressed AMT1 transporters, it is much more strongly visible than the AMT-GFP fusion protein band. This band represents at maximum one quarter of the complete AMT1 protein, but in all blots its visibility was relatively low, potentially indicating some silencing of the additional introduced AMT-GFP. As AMT1;2 only represents a maximum of 20 to 30% of AMT1 present in the root, the amount of AMT1;2-GFP is even lower. In summary, the ammonium-dependent phosphorylation of the regulatory threonines in the AMT1;1 and AMT1;2 C termini was substantially reduced in cipk23.
Figure 6.
Ammonium Transporter Phosphorylation in Wild-Type and cipk23 Roots.
(A) Peptide recognized by the anti-AMT1-P antibody GMDMT(pH)RHGGFA.
(B) to (F) Protein gel blot analysis using the anti-AMT1-P antibody of plants expressing AtAMT1;1-GFP (B) and AtAMT1;2-GFP (E) under the endogenous promoter and the respective control with anti-AMT or anti-GFP antibody ([C] and [F]).
(D) and (G) The intensities of the AMT1 and the AMT1-GFP-P bands were measured by ImageJ and normalized to the control.
(H) and (I) Protein gel blot analysis of wild-type (WT) and cipk23 plants (H) using anti-AMT1-P antibody and Ponceau staining of the nitrocellulose membrane (I).
(J) The intensities of the AMT1-P bands were measured by ImageJ and normalized to the Ponceau staining.
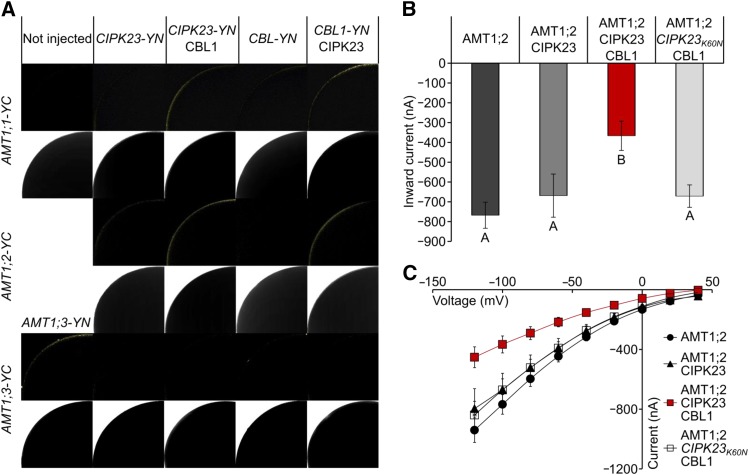
Functional Interaction and Reconstitution in Xenopus laevis Oocytes
We used the split YFP system to confirm the interaction of CIPK23, CBL1, and the single AMT1s at the plasma membrane of African clawed frog (Xenopus laevis) oocytes (Figure 7A). These experiments again showed that CIPK23-YN interacts with AMT1;1-YC and AMT1;2-YC in a CBL-dependent way. Interestingly, we could not detect an interaction of CIPK23-YN with AMT1;3-YC, even though AMT1;3 was expressed at the plasma membrane in the oocytes, as shown by the positive interaction of AMT1;3-YN and AMT1;3-YC. The coexpression of the tagged proteins further showed a CIPK23-dependent interaction of CBL-YN with AMT1;1-YC and AMT1;3-YC. This indicates that the three proteins build a trimeric regulatory complex, since YFP reconstitution could only be seen in oocytes expressing all three interaction partners.
Figure 7.
Functional Interaction of AMT1;2 and CIPK23 Depends on CBL1.
(A) Protein-protein interaction of AMT1;1 (rows 1 and 2), AMT1;2 (rows 3 and 4), and AMT1;3 (rows 5 and 6) with CIPK23 and CBL1 in oocytes. First row shows YFP fluorescence by reconstitution of C- and N-terminal YFP; lower row shows the corresponding bright-field photographs.
(B) Coexpression of AMT1;2, CIPK23, and CBL in oocytes. Inward currents by 1 mM NH4Cl at −100 mV from oocytes injected with mixtures of cRNAs always containing an equal amount of AMT1;2 cRNA supplemented with CIPK23 cRNA, CIPK23, and CBL1 cRNA or inactive CIPK23K60N and CBL1 cRNA are shown. Data are from six to 21 oocytes and three independent experiments. Values are means ± se. Different letters (A and B) indicate significant differences by Student’s t test, P value ≤ 0.05.
(C) Current-voltage plot of currents by AMT1;2 (black circles), AMT1;2 + CIPK23 (black triangles), AMT1;2 + CIPK23 + CBL1 (red squares), or AMT1;2 + CIPK23K60N + CBL1 (white squares) induced by 1 mM ammonium. Data are from six to 21 oocytes and three independent experiments.
To finally prove a direct functional interaction of the proteins without tags, CIPK23, CBL1, and AMT1;2 were coexpressed in X. laevis oocytes. Unfortunately, the low amount of AMT1;1 cRNA, which needed to be injected for titratable coexpression, was not producing sufficient AMT1;1-mediated ammonium currents, impairing the possibility of directly testing the reduction in these currents by CBL1 and CIPK23.
Oocytes exclusively expressing AMT1;2 showed large NH4+-specific currents resulting from transport of the charged NH4+ substrate into the oocyte. Coexpression of AMT1;2 with CIPK23 did not reduce these currents, but coexpression of AMT1;2 with CBL1 and CIPK23 substantially reduced the magnitude of the ammonium-induced currents by roughly 50%. Interestingly, when AMT1;2 was coexpressed with CBL1 and the inactive CIPK23K60N mutant (Li et al., 2006), no reduction in the ammonium currents was apparent. These results clearly indicate that only the active kinase, in combination with CBL1, inactivated transport by AMT1;2 (Figures 7B and 7C).
DISCUSSION
Although ammonium is the preferred inorganic nitrogen source in Arabidopsis and other plants (Gazzarrini et al., 1999), ammonium uptake via the roots is tightly controlled, since elevated ammonium concentrations are toxic. The ammonium-dependent inhibition of the rhizodermal plasma membrane ammonium transporter AMT1;1 by phosphorylation of Thr-460 is best understood, but likely applies to other AMT1s (Neuhäuser et al., 2007; Lanquar et al., 2009; Yuan et al., 2013). As a consequence, we hypothesized that the lack of the functional kinase that phosphorylates AMT1s and inhibits ammonium uptake would lead to persistently active ammonium transporters. Plants devoid of this kinase are expected to lose the ability to control ammonium transport by phosphorylation and thus to accumulate more ammonium over time compared with the wild type. This would lead to a higher susceptibility to toxic concentrations of ammonium or MeA.
Indeed, a screen for hypocotyl length identified a plant line with a T-DNA insertion in AT1G30270 (CIPK23). Interestingly, a genomic region harboring the CIPK23 gene is responsible for oversensitivity to ammonium in the ammonium oversensitive2 mutant (Li et al., 2012). The identified T-DNA insertion line SALK_036154C showed hypersensitivity to ammonium and MeA (Figure 1). A similar line, SAIL_402_F05 (cipk23-2) with a T-DNA insertion in the 7th intron, was previously used to study the impact of CIPK23 on potassium transport (Cheong et al., 2007). Comparable to this study, cipk23 plants were smaller when grown under low potassium conditions. Addition of potassium to a total concentration of 5 mM only partially rescued the growth reduction but increased the potassium concentration to the wild-type level. The still reduced biomass of the cipk23 plants at elevated potassium concentrations may imply that plants experience another imbalance in ion uptake, possibly a higher ammonium/nitrate ratio.
By investigating short-term uptake of 15N-labeled ammonium, it was directly confirmed that the ammonium-hypersensitive phenotype of cipk23 plants is related to a deregulation of ammonium uptake and a consequentially higher accumulation of ammonium in the plant. The restriction in ammonium uptake observed in the wild type after an ammonium shock was impaired in cipk23, but the functional defects of the mutant were fully complemented by expression of the gene under its endogenous promoter (Figure 2). Interestingly, the increased 15N-ammonium uptake in cipk23 roots compared with the wild type was most significant at higher concentrations of ammonium and longer incubation times, suggesting that the lower-affinity AMT1;2, which is expressed in cortical and in endodermal cells, or the molecularly unknown low-affinity transport system, may be the prime targets of CIPK23 (Figure 2).
The potassium transporter AKT1 had been suggested as a candidate low-affinity transporter of ammonium into plant roots (ten Hoopen et al., 2010) and requires activation by CIPK23 (Xu et al., 2006; Lee et al., 2007). As the loss of CIPK23 abolished K+ channel activity, hypothetical low-affinity NH4+ transport by AKT1 must be lost in the absence of CIPK23. However, low-affinity ammonium influx was even increased in cipk23, excluding the notion that CIPK23 action on AKT1 was responsible for the observed effects.
Analysis of knockdown mutants of several AMTs produced by amiRNA further implied that AMT1s are likely the physiological targets of CIPK23. Disruption of CIPK23 affected root morphology on ammonium and MeA, which was rescued by amiRNA-mediated suppression of AMT1s (Figure 3). As MeA is primarily transported into roots via AMT1 transporters, but not the ammonium low-affinity transport system, these data strongly suggest that CIPK23 regulates AMT1s.
The activity as well as membrane localization of CIPK23 was shown to be CBL1 or CBL9 dependent. The role of CBLs in AKT1 regulation is redundant for CBL1 and CBL9 (Xu et al., 2006; Cheong et al., 2007). With respect to AMT1 function, the loss of CBL1 in cbl1, but not of CBL9, which also interacts with CIPK23 in two-hybrid assays (Xu et al., 2006), was equivalent to cipk23 (Figure 4). This may imply a CBL-mediated specificity, since NPF6;3 was shown to be mainly phosphorylated in a CBL9-dependent manner (Ho et al., 2009).
AMT1;1 and AMT1;2 physically interacted with CIPK23 in the yeast two-hybrid assay, but this was not CBL1 dependent. However, the same assay showed a surprising specificity of CIPK23 for these two transporters, since there was no interaction of CIPK23 with AMT1;3 detected in yeast and oocytes, even though its conserved phosphorylation target site is indistinguishable by sequence. The interaction of these two AMT1s with CIPK23 was confirmed by BiFC experiments in planta (Figure 5).
Protein gel blot analysis with a phosphorylation-specific antibody showed a clear time- and ammonium-dependent phosphorylation of the complete AMT1 pool (Figure 6). The phosphorylation was substantially reduced in cipk23 plants. Nevertheless, it was still prominent in cipk23 and was triggered by ammonium shock. This indicates that other, partially redundant, kinases are further involved in high-affinity AMT1 phosphorylation. The interaction experiments imply that the residual phosphorylated AMT1 is an ensemble composed of AMT1;3, as well as AMT1;1 and AMT1;2, since residual phosphorylation of the two transporter-GFP protein fusions was detected, although this was reduced in the cipk23 background.
CIPK23-independent phosphorylation of AMT1;3 might explain the lower impact of cipk23 under high affinity conditions. Due to hetero-oligomerization of AMT1;1 and AMT1;3, the phosphorylation of AMT1;3 should almost completely inactivate high-affinity ammonium transport at the epidermis. Furthermore, rhizodermal AMT1s might be internalized upon high external ammonium concentrations, as shown for AtAMT1;3 (Wang et al., 2013). The high background uptake therefore likely consists of an apoplastic flow into the inner root cell layers. AMT1;2 is the only ammonium transporter located in the plasma membrane of the endodermis. The apoplastic, radial flow of nutrients across this layer into the vasculature is blocked by the Casparian strip; thus, inhibition of transport of toxic compounds in the endodermis will effectively reduce further long-distance transport to the shoot. Therefore, blockage of the ammonium flux into the vasculature exclusively depends on AMT1;2 regulation, which is impaired in cipk23 plants. Coexpression experiments in oocytes showed that a complex of AMT1;2 with CBL1 and the active CIPK23 kinase is required for AMT1;2 regulation, while a noncatalytic CIPK23 is not sufficient to inactivate the transport of NH4+ (Figure 7).
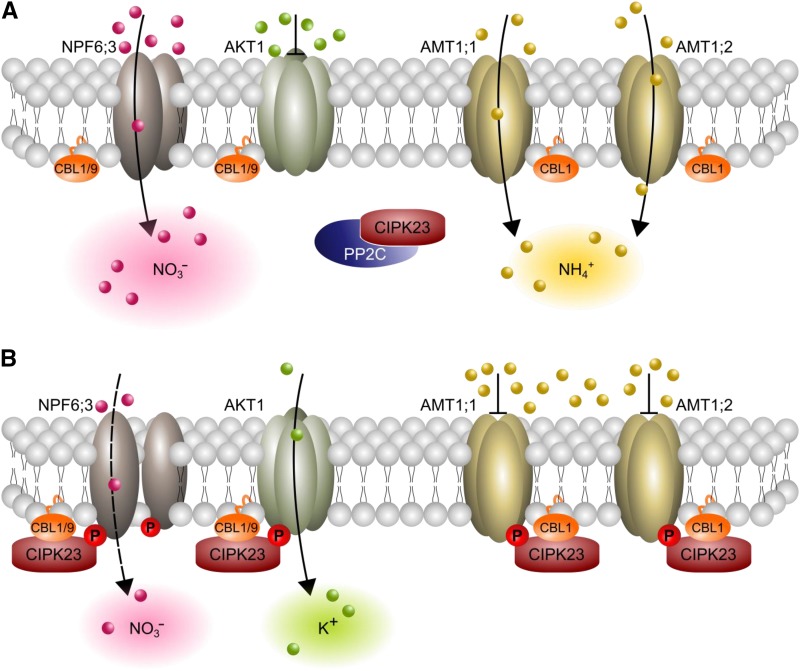
While the identification of a kinase that directly interacts with ammonium transporters is interesting per se, the fact that this kinase is encoded by CIPK23, a major regulator of root NO3− and K+ uptake (Ho et al., 2009; Xu et al., 2006; Tsay et al., 2011; Ho and Tsay, 2010), is intriguing. The uptake of major cations and the two inorganic nitrogen forms is known to be well balanced (Haynes, 1990), but integrators of ion transport to maintain charge balance had not been molecularly identified. Here, we postulate that CIPK23 and CBL1 might regulate the balance between NO3− and its counterion by antipodal regulation of AKT1, AMT1;1, and AMT1;2 (Figure 8). Low availability of nitrate leads to the phosphorylation of Thr-101 in NPF6;3 by the CIPK23/CBL complex and a corresponding change in affinity (Ho et al., 2009). Recently, it was shown that nitrogen starvation after exclusive nitrate supply reduces AMT1;1 Thr-461 phosphorylation (Menz et al., 2016). This contradicts the idea of nitrate as a common signal for phosphorylation of N-ion and K+ transporters. Lacking phospho-proteomic approaches for comparing potassium sufficient and deficient plants, we can only hypothesize whether potassium deficiency might not only induce the phosphorylation of AKT1, but nitrogen ion transporters as well. CIPK23 expression increases due to low external potassium conditions (Cheong et al., 2007). Combined with the observation that CIPK23 expression increases due to high external ammonium concentrations, this indicates that CIPK23 regulates the K+/NH4+ uptake ratio according to the external nutrient concentrations. Future research is needed to clarify the role of single ions in this regulation. In our postulated model, Arabidopsis balances NO3− uptake with AMT1 mediated NH4+ uptake under high nitrogen demand and high nitrate conditions (Figure 8A). By contrast, under low nitrogen demand or potential NH4+ toxicity, NO3− uptake is balanced by K+ uptake through the activation of AKT1 and simultaneous inactivation of AMT1s (Figure 8B). Potential NH4+ toxicity might occur due to high external NH4+ concentrations or low nitrate concentrations, which potentiate the toxic effects of NH4+. CIPK23 and CBL1 therefore appear to occupy a key position in cellular ion homeostasis, which likely affects metabolic pathways that depend on ion balance. Whether CIPK23 directly senses all three ions, NH4+, NO3−, and K+, to exert its effects, or whether other proteins and signaling cascades are involved, is an interesting question for future research.
Figure 8.
Model Showing How CIPK23 Regulates Ion Homeostasis.
(A) Under moderate ammonium conditions and/or high nitrogen demand, CIPK23 is bound by PP2Cs in the cytoplasm and is inactive. NO3− uptake is balanced by NH4+ uptake for ion charge balance by nonphosphorylated and active AMT1s.
(B) Under toxic ammonium conditions (or possibly low nitrogen demand), CIPK23 inactivates AMT1;1 and AMT1;2 by phosphorylation. The nitrogen demand is then met by high-affinity NO3− uptake by NPF6;3, with K+ serving as a balancing counter ion. Therefore, AKT1 is phosphorylated and consequently activated under low potassium conditions.
METHODS
Hypocotyl Test
Arabidopsis thaliana seeds (Col-0 as well as T-DNA insertion lines in the Col-0 background) were surface sterilized with ethanol + 0.05% Triton X-100. Approximately 100 seeds were placed on sterile filter paper that was moistened with 1 mL water or water containing variable ammonium/MeA concentrations as chloride salt. The plates were wrapped with Parafilm and dormancy was broken by incubation in the dark for 2 d. Germination was initiated by incubation in a Percival AR-66L incubator with 150 µmol m−2 s−1 light (bulb type: Philips F17T8/TL841, 17 W) and 22°C for 6 h. The plates were then incubated in the dark at a temperature rhythm of 22°C from 6 am to 10 pm and 20°C from 10 pm to 6 am.
Hypocotyls were then taken from the filter and scanned on plastic foil. The length of the hypocotyls was measured using the freeware program ImageJ (https://imagej.nih.gov/ij/index.html).
Toxicity Test on Agar Medium
Seeds were surface-sterilized with 70% ethanol, containing 0.001% Triton X-100, germinated, and grown for 4 d on quarter-strength Hoagland medium (HL + 0.8% agar, pH 6.0) containing 2 mM KNO3 as a single nitrogen source. Afterwards, the seedlings were either transferred to control medium (HL with 2 mM KNO3) or to medium containing 20 or 30 mM MeA. The plates were placed vertically in a plant growth chamber (Percival Scientific) for 10 d (short day: 8 h/16 h day/night; 22°C/18°C). Root systems were scanned and analyzed using ImageJ software.
Plant Growth and Analysis
Plants were grown hydroponically for 6 weeks in quarter-strength Hoagland solution, pH 6.0 (HL) (Supplemental Table 3), containing 1 mM NH4NO3 and 2 mM K2SO4 under controlled environment conditions (8 h/16 h day/night; light intensity 100–120 μmol m−2 s−1 [bulb type: HQI-T2000/D Osram Powerstar]; temperature 22°C/18°C, and humidity range 50 to 60%).
Short-Term Uptake Assay
After 6 weeks of hydroponic growth as described above, the medium was changed to HL without nitrogen for 4 d. Plants were divided into (1) plants immediately analyzed and (2) plants transferred into HL + 1 mM (NH4)2SO4 for 30 min (ammonium shock). For 15N uptake tests, roots were washed twice in 1 mM CaSO4 and incubated for 6 min or 30 min in HL solution containing 0.5 or 5 mM 15NH4+, followed by another two washing steps in 1 mM CaSO4. Afterwards, the roots were briefly dried, separated from shoots, immediately frozen in liquid nitrogen, and cold-dried. Samples were ground, and 1.0 mg (0.5 mM 15NH4+) or 0.5 mg (5 mM 15NH4+) powder was used for 15N, total N, and total C determination by isotope ratio mass spectrometry.
Gene Expression Analysis
To extract total RNA from plants, an innuPREP Plant RNA extraction kit from Analytik Jena was used. The concentration and purity of the extracted RNA were determined using a NanoDrop2000 spectrophotometer. Ten plants were pooled into one sample, three biological replicates (each from a separate experiment) were used, and for each replicate, two technical replications were performed. Reverse transcription was performed according to the manufacturer’s instructions (reverse transcription kit; Qiagen). qPCR was performed using a KAPA SYBR FAST qPCR kit (Peqlab) and C1000 Thermo Cycler combined with a CFX 384 real-time system (Bio-Rad). Each RT-PCR contained 0.05 µg of total RNA in a total volume of 15 µL. Primers used are shown in Supplemental Table 4. PCR cycling parameters were set as follows: 95°C for 3 min, 45 cycles of 3 s at 95°C, 20 s at 60°C, and a final melting curve of 65 to 95°C with an increment of 0.2°C.
Yeast Two-Hybrid Assay
AMT1s were cloned into the pBT3-C vector and CIPK23 into the pPR3-N vector from the DUALmembrane starter kit (Dualsystems Biotech) as described in the user manual. CBL1 was clone into the multiple cloning site (MCS) of pOO2 by PstI and SalI digestion and then subcloned using PstI and SmaI into the MCS of pDR199. Different combinations were transformed into NMY51 yeast cells provided in the kit, and positive transformed colonies were selected on SD-Trp-Leu-Ura medium. For the assay, positive colonies were inoculated with shaking in liquid SD-Trp-Leu-Ura and grown overnight at 28°C. The overnight culture was washed tree times in water and OD600 was determined. Ten microliters of OD600 = 2 dilutions were spotted onto SD plates -Trp-Leu-Ura-His, -Trp-Leu-Ura-Ade, or -Trp-Leu-Ura-His-Ade. Growth was assayed after 5 d in 28°C. The X-Gal overlay assay was performed as described by Barral et al. (1995).
In Planta Interactions
The YFP molecule was split between amino acids 153 and 154, producing the N-terminal fragment (N-eYFP, 1–153 amino acids) and the C-terminal fragment (C-eYFP, 154–239 amino acids). PCR of the N-terminal fragment was performed with primers including MluI-XhoI restriction sites and of the C-terminal fragment with primers including SalI-BcuI restriction sites. The reverse primer of N-eYFP was designed to establish translation termination after amino acid 152 to produce the N-eYFP fragment.
The eYFP fragments were individually inserted into the MCS of the pTkan+ and pTbar vector, thereby constructing the pTkan+YN 153stop and pTbar YC stop vectors. Respective AMT and CIPK23 were cloned into these vectors using appropriate single cutting enzymes in the MCS. Different plasmid combinations were transformed into Arabidopsis plants using the floral-dip method (Clough and Bent, 1998) with the Agrobacterium tumefaciens strain GV3101 (Supplemental Table 5). Transgenic plants were analyzed under an LSM700_ZEN_2010 microscope (Zeiss).
Protein Gel Blot Analysis
To prepare microsomes, plants were grown hydroponically as described before. After ammonium shock, roots were immediately harvested in liquid nitrogen. Samples were ground, and 1.0 g powder was resuspended in 5 mL ice-cold extraction buffer (250 mM Tris-HCl, pH 8.5, 290 mM sucrose, 25 mM EDTA, 2 mM DTT and 5 mM β-Me) containing protease inhibitors, aprotinin, leupeptin, pepstatin, PMSF, EGTA, and phosphatase inhibitor cocktail 2 (Sigma-Aldrich); phosphatase inhibitor cocktail 3 (Sigma-Aldrich), and phosphatase inhibitor cocktail (Roche PhosSTOP) and incubated with constant agitation for 15 min at 4°C. The samples were centrifuged 4500g for 15 min at 4°C. The supernatants were filtered through four layers of Miracloth (Calbiochem) and centrifuged at 41,000g for 2 h at 4°C. The pellets were resuspended in 100 μL of ice-cold storage buffer (400 mM mannitol, 10% [v/v] glycerol, 6 mM MES-Tris, pH 8.0, and protease/phosphatase inhibitors as described above). Protein concentration was measured and standard SDS gel electrophoresis and protein gel blot analysis were performed using the following antibodies: primary anti-GFP (rabbit, polyclonal IgG; Rockland; code 600-401-215, lot 28839; dilution 1:20,000), anti-phospho-pep40227 (anti-P) raised against p-peptide, CG-Nle-D-Nle-pT-RHGGFA-amide (rabbit, monoclonal IgG; BioGenes; dilution 1:1000), and secondary anti-rabbit (goat, polyclonal IgG, conjugated to horseradish peroxidase; Roth; charge 304217180; dilution 1:5000).
Electrophysiological Measurements and Injection of Oocytes
Split YFP constructs were obtained by amplifying AMT1;1, AMT1;2, and CIPK23 with split YFP tags attached from the respective plant expression vectors using gene-specific primers containing suitable restriction sites for cloning into the pOO2 plasmid. AMT1;3 and CBL1 were also amplified by PCR and cloned into pOO2-C-YFP and pOO2-N-YFP by adding suitable restriction sites into the primers. The electrophysiological methods are described in more detail elsewhere (Mayer and Ludewig, 2006). Briefly, oocytes were ordered at Ecocyte Bioscience (Castrop-Rauxel), presorted again, and injected with 50 nL of cRNA (5–50 ng/nL). Oocytes were kept in ND96 for 4 d at 18°C and then placed in a small recording chamber. The recording solution was 110 mM choline chloride, 2 mM CaCl2, 2 mM MgCl2, and 5 mM MES, pH adjusted to 5.5 with Tris. Variable ammonium concentrations were added as NH4Cl salt. Currents without added ammonium were subtracted at each voltage.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers AT4G13510 (AMT1;1), AT1G64780 (AMT1;2), AT3G24300 (AMT1;3), AT3G24290 (AMT1;5), AT1G30270 (CIPK23), AT4G17615 (CBL1), and AT5G47100 (CBL9).
Supplemental Data
Supplemental Figure 1. Susceptibility of Col-0 seedlings grown in the dark to toxic concentrations of ammonium and methylammonium.
Supplemental Figure 2. Test for homozygosity of cipk23 and CIPK23 expression in cipk23.
Supplemental Figure 3. Root phenotypes of the cipk23 and qko mutants.
Supplemental Figure 4. Influence of potassium on hypocotyl elongation.
Supplemental Figure 5. Comparative growth under normal (1 mM) and elevated potassium conditions (5 mM potassium).
Supplemental Figure 6. Nutrient concentrations in 6-week-old wild-type and cipk23 plants grown under normal (1 mM) and elevated potassium conditions (5 mM potassium).
Supplemental Figure 7. Complementation of the cipk23 line.
Supplemental Figure 8. Relative normalized expression of ammonium transporter genes in wild-type and cipk23 plants after nitrogen starvation and after subsequent ammonium shock.
Supplemental Figure 9. AMT localization in the Arabidopsis thaliana Col-0 (upper panel) and cipk23 (lower panel) backgrounds.
Supplemental Figure 10. Relative normalized expression of CBL1 and CBL9 in 6-week-old cipk23 plants.
Supplemental Figure 11. CBL1 and CBL9 expression in cbl1 and cbl9 T-DNA insertion lines.
Supplemental Table 1. T-DNA insertion lines used in the hypocotyl screen.
Supplemental Table 2. Combined 15N ammonium uptake rates from Figure 2.
Supplemental Table 3. Modified Hoagland medium used for plant growth.
Supplemental Table 4. Primers used in qRT PCR.
Supplemental Table 5. Newly generated plant lines in this study.
Supplementary Material
Acknowledgments
We thank D. Schnell and E. Dachtler for their excellent technical assistance, M. Brünnel for help with the yeast two-hybrid experiments, and the Deutsche Forschungsgemeinschaft for funding (DFG NE 1727/2-1).
AUTHOR CONTRIBUTIONS
T.S. designed and performed most of the experiments, except the hypocotyl growth, the oocyte measurements, and the yeast two-hybrid experiments, which were performed by B.N. B.N. and U.L. designed the project, obtained funding by the Deutsche Forschungsgemeinschaft, supervised the project, and wrote the manuscript.
Glossary
- MeA
methylammonium
- BiFC
bimolecular fluorescence complementation assay
- MCS
multiple cloning site
References
- Barral Y., Jentsch S., Mann C. (1995). G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 9: 399–409. [DOI] [PubMed] [Google Scholar]
- Blakey D., Leech A., Thomas G.H., Coutts G., Findlay K., Merrick M. (2002). Purification of the Escherichia coli ammonium transporter AmtB reveals a trimeric stoichiometry. Biochem. J. 364: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckstaens M., André B., Marini A.M. (2007). The yeast ammonium transport protein Mep2 and its positive regulator, the Npr1 kinase, play an important role in normal and pseudohyphal growth on various nitrogen media through retrieval of excreted ammonium. Mol. Microbiol. 64: 534–546. [DOI] [PubMed] [Google Scholar]
- Canales J., Moyano T.C., Villarroel E., Gutiérrez R.A. (2014). Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front. Plant Sci. 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong Y.H., Pandey G.K., Grant J.J., Batistic O., Li L., Kim B.-G., Lee S.-C., Kudla J., Luan S. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52: 223–239. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Engelsberger W.R., Schulze W.X. (2012). Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J. 69: 978–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S., Lejay L., Gojon A., Ninnemann O., Frommer W.B., von Wirén N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K., Eckert C., Anschütz U., Scholz M., Held K., Waadt R., Reyer A., Hippler M., Becker D., Kudla J. (2012). Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J. Biol. Chem. 287: 7956–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes R.J. (1990). Active ion uptake and maintenance of cation-anion balance: A critical examination of their role in regulating rhizosphere pH. Plant Soil 120: 247–264. [Google Scholar]
- Ho C.-H., Lin S.-H., Hu H.-C., Tsay Y.-F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194. [DOI] [PubMed] [Google Scholar]
- Ho C.-H., Tsay Y.-F. (2010). Nitrate, ammonium, and potassium sensing and signaling. Curr. Opin. Plant Biol. 13: 604–610. [DOI] [PubMed] [Google Scholar]
- Lan W.-Z., Lee S.-C., Che Y.-F., Jiang Y.-Q., Luan S. (2011). Mechanistic analysis of AKT1 regulation by the CBL-CIPK-PP2CA interactions. Mol. Plant 4: 527–536. [DOI] [PubMed] [Google Scholar]
- Lanquar V., Loqué D., Hörmann F., Yuan L., Bohner A., Engelsberger W.R., Lalonde S., Schulze W.X., von Wirén N., Frommer W.B. (2009a). Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell 21: 3610–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Lan W.-Z., Kim B.-G., Li L., Cheong Y.H., Pandey G.K., Lu G., Buchanan B.B., Luan S. (2007). A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. USA 104: 15959–15964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Dong G., Li B., Li Q., Kronzucker H.J., Shi W. (2012). Isolation and characterization of a novel ammonium overly sensitive mutant, amos2, in Arabidopsis thaliana. Planta 235: 239–252. [DOI] [PubMed] [Google Scholar]
- Li L., Kim B.-G., Cheong Y.H., Pandey G.K., Luan S. (2006). A Ca(2)+ signaling pathway regulates a K(+) channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 12625–12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D., Lalonde S., Looger L.L., von Wirén N., Frommer W.B. (2007). A cytosolic trans-activation domain essential for ammonium uptake. Nature 446: 195–198. [DOI] [PubMed] [Google Scholar]
- Ludewig U., Wilken S., Wu B., Jost W., Obrdlik P., El Bakkoury M., Marini A.-M., André B., Hamacher T., Boles E., von Wirén N., Frommer W.B. (2003). Homo- and hetero-oligomerization of ammonium transporter-1 NH4 uniporters. J. Biol. Chem. 278: 45603–45610. [DOI] [PubMed] [Google Scholar]
- Mayer M., Ludewig U. (2006). Role of AMT1;1 in NH4+ acquisition in Arabidopsis thaliana. Plant Biol. (Stuttg.) 8: 522–528. [DOI] [PubMed] [Google Scholar]
- Menz J., Li Z., Schulze W.X., Ludewig U. (2016). Early nitrogen-deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and (phospho-) proteome levels between nitrate and ammonium nutrition. Plant J. 88: 717–734. [DOI] [PubMed] [Google Scholar]
- Neuhäuser B., Dunkel N., Satheesh S.V., Morschhäuser J. (2011). Role of the Npr1 kinase in ammonium transport and signaling by the ammonium permease Mep2 in Candida albicans. Eukaryot. Cell 10: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhäuser B., Dynowski M., Mayer M., Ludewig U. (2007). Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol. 143: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse T.S., Stensballe A., Jensen O.N., Peck S.C. (2004). Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K., Cakmak T., Cooper A., Lager I., Rasmusson A.G., Escobar M.A. (2010). Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ. 33: 1486–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Bankston J.R., Payandeh J., Hinds T.R., Zagotta W.N., Zheng N. (2014). Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507: 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hoopen F., Cuin T.A., Pedas P., Hegelund J.N., Shabala S., Schjoerring J.K., Jahn T.P. (2010). Competition between uptake of ammonium and potassium in barley and Arabidopsis roots: molecular mechanisms and physiological consequences. J. Exp. Bot. 61: 2303–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay Y.-F., Ho C.-H., Chen H.-Y., Lin S.-H. (2011). Integration of nitrogen and potassium signaling. Annu. Rev. Plant Biol. 62: 207–226. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhao Y., Luo W., Li R., He Q., Fang X., Michele R.D., Ast C., von Wirén N., Lin J. (2013). Single-particle analysis reveals shutoff control of the Arabidopsis ammonium transporter AMT1;3 by clustering and internalization. Proc. Natl. Acad. Sci. USA 110: 13204–13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li H.-D., Chen L.-Q., Wang Y., Liu L.-L., He L., Wu W.-H. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360. [DOI] [PubMed] [Google Scholar]
- Yang H., von der Fecht-Bartenbach J., Friml J., Lohmann J.U., Neuhäuser B., Ludewig U. (2015). Auxin-modulated root growth inhibition in Arabidopsis thaliana seedlings with ammonium as the sole nitrogen source. Funct. Plant Biol. 42: 239–251. [DOI] [PubMed] [Google Scholar]
- Yuan L., Gu R., Xuan Y., Smith-Valle E., Loqué D., Frommer W.B., von Wirén N. (2013). Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. Plant Cell 25: 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Loqué D., Kojima S., Rauch S., Ishiyama K., Inoue E., Takahashi H., von Wirén N. (2007). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19: 2636–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.