Abstract
G-protein coupled receptors (GPCRs), which activate heterotrimeric G proteins, are an essential class of transmembrane receptors that are responsible for a myriad of signaling events in normal and pathologic conditions. Two members of the G protein family, Gαq and Gα11, activate one of the main GPCR pathways and function as oncogenes by integrating mitogen-stimulated signaling cascades that are active under malignant conditions. Recently, it has been shown that targeted deletion of Gα11 and Gαq from endothelial cells impairs the Rho-mediated formation of focal adherens junctions, suggesting that Gα11/q signaling may also play a significant role in cytoskeletal-mediated cellular responses in epithelial cells. Indeed, combined deletion of Gα11 and Gαq confers a significant migratory defect in keratinocytes that delays cutaneous wound healing in an in vivo setting. This delay can be attributed to a defect during the reepithelialization phase due to significantly attenuated migratory capacity of Gαq-null keratinocytes under combined Gα11 deficiency. In fact, cells lacking Gα11/q demonstrate a severely reduced ability to respond to mitogenic and migratory signals in the microenvironment, leading to inappropriate and premature terminal differentiation. These results suggest that Gα11/q signaling pathways may be critical for integrating mitogenic signals and cytoskeletal function to achieve normal physiological responses. Emergence of a malignant phenotype may therefore arise from both under- and overexpression of Gα11/q signaling, implicating its upstream regulation as a potential therapeutic target in a host of pathologic conditions.
Introduction
The signal transduction mechanisms in the cell that mediate gene expression, cell survival, and growth rely heavily on the function of the largest family of cell-surface molecules, the G-protein coupled receptors (GPCRs) [1]. The GPCR family transduces signals from the cellular microenvironment via receptor coupling and subsequently activating heterotrimeric G proteins. This activation in turn leads to the second-messenger systems that influence transcription, translation, viability, and normal and malignant cell growth. Within the heterotrimeric G proteins, activating mutations in multiple members of the Gα subunit family have been shown to function as oncogenes. Among the heterotrimeric G proteins, Gαq and Gα11 share around 90% amino acid homology and are the main components of the Gq family of alpha proteins [2]. Gain of function in Gαq and Gα11 leads oncogenic transformation [3, 4] and is particularly prevalent in the development and progression of uveal melanoma, where Gαq and Gα11 share activating mutations and are currently being evaluated for both diagnostic and therapeutic purposes [5–8]. However, relatively little is known about the pathological consequences of Gα11 and Gαq loss.
GNAQ (Gαq) and GNA11 (Gα11) comprise the Gq family of alpha proteins that transduces many of the mitogen-stimulated GPCR signaling cascades. Gαq stimulates Phospholipase C (PLC) to achieve downstream activation of Protein Kinase C (PKC) and extracellular signal-related kinase (ERK) [9]. Combined deletion of Gα11 and Gαq leads to embryonic lethality at day 11 due to myocardial hypertrophy, while global deletion of GNA11 alone causes no apparent behavioral and morphological defects and global deletion of GNAQ alone leads to ataxia and impaired motor control [10, 11]. Thus, due to their compensatory functions, investigation into the role of Gnaq and Gna11 requires conditional deletion of one of these genes in the context of total loss of its compensatory family member. Using this approach, it has been shown that conditional deletion of Gαq in the context of Gna11 abolishes the embryonic lethality and hypertrophic response in cardiomyocytes observed previously [12]. This approach has been also used to demonstrate that loss of Gαq from endothelial cells under combined Gα11 deficiency impairs the Rho-mediated formation of focal adherens junctions after histamine stimulation [13]. Gα11 and Gαq have also been found to play a large role in the genetics of the skin [14, 15]. However, it is unclear if cutaneous targeted deletion of Gα11 and Gαq may also have an impact on epidermal cytoskeletal integrity and function.
Here, we show that targeted deletion of Gα11/Gαq in the skin does not result in cancer formation, but rather underlies an acute migratory defect in keratinocytes. While not deleterious under physiologic conditions, this defect manifests as a severe attenuation in the rate of cutaneous wound healing. We further demonstrate that this migratory defect may be partly attributed to an inability of Gα11/q-null keratinocytes to respond to the mitogenic signals that mediate the transition between a differentiated, static cell and a more stem-like phenotype capable of engaging in highly migratory reepithelialization programs. These results suggest that the Gα11/q signaling pathway may be an important mediator of pathologic stress, where low or inactivated Gαq results in a chronic wound phenotype that may contribute to malignant transformation. Therefore, understanding the factors that contribute to Gαq expression and regulate its function may have a broad impact in a variety of pathologic disease conditions and open new therapeutic avenues in the future.
Materials and methods
Mice
All animal studies were carried out according to NIH-approved protocols, in compliance with the Guide for the Care and Use of Laboratory Animals. All mice used were in C57BL/6 background, 8 weeks old or older and both males and females were used for experiments. The generation of null alleles of the genes encoding Gα11 (Gna11) and of the floxed alleles of the genes encoding Gαq (Gnaq) have been described previously [16]. For the experiments probing Gα11 deficiency only, Gna11-/- mice and littermate controls were used. Pups were obtained from a heterozygote mating where Gna11+/- x Gna11+/- animals were crossed, and upon genotyping the Gna11+/+ and Gna11-/- littermates were selected for experiments. Epidermal-specific Gαq knockouts were obtained by crossing the aforementioned mice with mice carrying a constitutive Cre-mediated recombination system driven by the K14 promoter (K14-Cre, [17]), where Gna11-/-Gnaqf/f K14-Cre+/- mice were crossed with Gna11-/-Gnaqf/f K14-Cre-/- such that all pups were deficient for Gα11 and double floxed for Gαq. Expression of Gaq was therefore driven by inheritance of the K14-Cre promoter, where 50% of the pups expressed the K14-Cre promoter (Gna11-/-Gnaqf/f K14-Cre+/-, referred to as Gα11KO/Gαq-eKO) and were the experimental group, while the other 50% did not expressing the K14 promoter (Gna11-/-Gnaqf/f K14-Cre-/-, referred to as Gα11KO/Gαq-WT) and were used as controls. Both males and female mice were used for the experiments.
Reagents and antibodies
Cell culture surfaces were coated with 10 μg/ml of poly-L-lysine (Sigma), type I rat tail collagen (BD Biosciences) or fibronectin (Invitrogen). Recombinant human TGF-β1 from Peprotech. Deep space black and warp red chromogen kits were from Biocare medical. Antibodies for western blotting and immunohistochemistry were used as described in Table 1.
Table 1. Antibodies and experimental conditions.
This table details the antibodies, their formulation, experimental application, dilution, and source for all experiments in this manuscript.
| Antibody | Conjugation | Primary/Secondary | Application | Dilution | Company |
|---|---|---|---|---|---|
| Actin | None | Primary | Western | 1:5000 | Cell Signaling Technology |
| Goat a-rabbit | Biotinylated | Secondary | IHC | 1:400 | Vector Laboratories |
| Goat a-rabbit | HRP | Secondary | Western | 1:40,000 | Southern Biotech |
| Goat a-rat | Biotinylated | Secondary | IHC | 1:400 | Vector Laboratories |
| Gαq | None | Primary | Western | 1:1000 | Santa Cruz Biotechnology |
| K10 | None | Primary | IHC | 1:2000 | Covance |
| K13 | None | Primary | IHC | 1:5000 | Abcam |
| K14 | None | Primary | IHC | 1:1000 | Covance |
| Ki67 | None | Primary | IHC | 1:50 | Dako |
| Phospho-SMAD2 | None | Primary | Western | 1:1000 | Cell Signaling Technology |
| SMAD2 | None | Primary | Western | 1:1000 | Cell Signaling Technology |
Cell lines, keratinocyte cell culture, and transfections
All cells were cultured at 37°C in the presence of 5% CO2. HEK293ΔGαq cells, which lack functional Gαq and Gα11, were described previously [18]. Cells were cultured in DMEM (Invitrogen) containing 10% fetal bovine serum (FBS; Sigma-Aldrich) and antibiotic/antimycotic solution (Sigma-Aldrich). Cells were transfected with Turbofect (Fermentas) according to the manufacturer’s instructions. Fibroblasts and keratinocytes from newborn mouse skin were isolated as previously described [19]. Keratinocytes were cultured in defined keratinocyte serum-free medium (KSFM; Invitrogen) and subdermal fibroblasts were cultured in DMEM 10% FBS. Cells were transfected with 10ng/cm2 of either pCAGGS-Gαq or vector control and/or 200ng/cm2 LifeAct-GFP and assessed 48 hours post transfection.
Immunoblot analysis
Cells were lysed in RIPA buffer [50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate] supplemented with protease and phosphatase inhibitors (Thermo Fisher) for 15 minutes on ice, scraped, and then sonicated for 3x10s on 30% power. Lysates were cleared by centrifugation at 10,000xg for 15 minutes at 4°C and concentration was determined using Bio-Rad DC protein assay. Ten micrograms total protein was separated by SDS-PAGE and transferred to PVDF membrane overnight at 4°C. Membranes were blocked for 1 hour at room temperature in 5% milk in TBST and then probed with primary antibodies overnight at 4°C. Membranes were washed four times in TBST, probed with HRP-conjugated secondary antibodies for 1h at RT in 5% milk, washed four times in TBST, and detected using chemiluminescent substrate (Millipore) with the FluorChem E system image analyzer (Cell Biosciences, Santa Clara, CA)
Histopathologic and immunohistologic analysis
Formalin-fixed paraffin-embedded (FFPE) tissue slides were deparaffinized in 100% xylene, hydrated through a series of graded alcohols (100%, 95%, 80%, and 70%), and incubated in sodium citrate buffer (pH6.0) to unmask the antigen. Endogenous peroxidase activity was blocked by incubate with 3% H2O2, blocked in PBS with 3% BSA, and then incubated with primary antibody in blocking buffer overnight at 4°C. The slides were washed in PBS three times, incubated with a biotin-conjugated secondary antibody at room temperature for 30 minutes followed by the avidin-biotin complex (Vector Stain Elite Standard, ABC kit, Vector Laboratories) for 30 minutes at room temperature. For single stained slides, the slides were washed and developed in 3,3′-diaminobenzidine (Sigma FASTDAB tablet, Sigma Chemical) under microscopic observation. The reaction was stopped in tap water and the tissues were counterstained with hematoxylin, dehydrated, and mounted. For double stained images, slides were developed with Deep Space Black chromogen kit according to manufacturer’s instructions (Biocare Medical). The reaction was stopped in tap water, washed with TBS, blocked in TBS with 3% BSA, and then incubated with primary antibody in blocking buffer overnight at 4°C. The slides were washed in TBS three times, incubated with alkaline phosphatase-conjugated secondary antibody at room temperature for 30 minutes followed by the AP complex (Vector Stain Elite AP, ABC kit, Vector Laboratories) for 30 minutes at room temperature. Slides were washed and developed using Warp Red chromogen kit according to manufacturer’s instructions (Biocare Medical) under microscopic observation. Images were taken using Scanscope (Aperio). Quantification of slides stained for Ki67 was performed using Aperio Scanscope software and the Nuclear v9 algorithm, while quantification of K10 and K13 was performed using IHC Membrane v9 algorithm. H-scores were determined as the product of the staining intensity (0, absent; 1, weak staining; 2, moderate staining; and 3, strong staining) multiplied by the percentage of positive cells quantified.
Proliferation assay (EdU incorporation assay)
To visualize individual cells synthesizing DNA we used the Click–It kit (Invitrogen) according to manufacturer’s instructions. This kit allows for robust statistical analysis in small populations of cells while utilizing the highly-specific labeling methodology of BrdU or [3H]-thymidine incorporation. Briefly, subconfluent cells were grown in 96-well plates in quadruplicate, starved, and transferred to complete media for 16 h. Then EdU (10 μM) was incorporated for 4 h and cells were fixed in a 4% paraformaldehide/PBS solution (Electron Microscopy Sciences, PA USA). Samples were counterstained with Hoescht 33342 and visualized under Axio Imager Z1 microscope equipped with ApoTome system controlled by AxioVision software (Carl Zeiss).
Colony formation assay
For the colony formation analysis, cells were plated in 6-well plates at 100 live cells/well (counted with trypan blue) and cultured until visible colonies around 1–2 mm in diameter appeared in control wells. The cells were then stained with crystal violet fixing solution (0.05% crystal violet in PBS with 4% paraformaldehyde) for 30 min RT, washed with water, and the resulting colonies were quantified by size and number per plate with ImageJ.
Scratch wound closure
Scratch assays were performed with keratinocytes or fibroblasts grown to confluence on fibronectin-coated wells and starved overnight. Scratches were made with a plastic pipette tip across the diameter of each well. Phase contrast images of the wound were obtained every 6 h for fibroblasts and every 24 h for the keratinocytes, Quantitative analysis of uncovered wound area was performed using the Axiovision Rel. 4.7 (Carl Zeiss, Thornwood, NY). Calculations were based on three replicates per experiment from three independent experiments.
Data mining and GEO analysis
Data regarding the expression levels of migratory genes was downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/, accessed June 30, 2015) for GSE8531. Significant genes, defined as those with a p-value less than 0.05 and logFC value >2 or <-2, were inputted into the ENRICHR algorithm (http://amp.pharm.mssm.edu/Enrichr/, accessed June 30, 2015). The GO Molecular Function ontology was downloaded and represented according to the internally calculated p-value.
Migration assay
Cells were serum starved for six hours and placed in the top well of a Boyden chamber while serum-free media or serum-free media supplemented with 5% FBS, 1 ng/ml TGF-β1, or 10 ng/ml TGF-β1 was placed in the bottom well. The two chambers were separated by an 8 μm pore polyvinylpropylene-free membrane coated with 10 μg/ml collagen. Cells were incubated at 37°C in a humidified chamber overnight. The next day, the apparatus was disassembled and the membrane was fixed for 30 minutes in methanol at room temperature. The membrane was then stained with hematoxylin for 20 minutes at RT, washed three times with distilled water, mounted face down on glass slides, and nonmigrated cells were scraped off with cotton swabs. Membranes were scanned and quantified using ImageJ. Calculations were based on 6 imaging fields each from three independent experiments. Statistical significance was determined using Student’s t-test.
Immunofluorescence
Cells were transfected with LifeAct-GFP and either empty vector or Gαq and grown on cover slips coated with collagen I. Cells were starved for 6 hours and then treated for 3 hours with either 0, 1, or 10 ng/ml TGF-β1. Cells were fixed with 4% paraformaldehyde, washed in PBS, and mounted on uncharged slides. The images were taken using an Axio Imager Z1 microscope equipped with an ApoTome system.
Wound healing assay and experimental mice
This study was approved by the Animal Care and User Committee, according to National Institutes of Health (NIH) animal study protocols approved by the Animal Care and Use Committee protocol 12–667, National Institute of Dental and Craniofacial Research, in compliance with the “Guide for the Care and Use of Laboratory Animals”. Animals were housed on 12-h light/dark cycles and received food, standard rodent chow, and water ad libitum in compliance with Association for Assessment and Accreditation of Laboratory Animal Care International guidelines. The animals were observed daily by the investigators and animal care staff. Any animals displaying signs of discomfort, wasting, ruffled hair coat, hunching, or other signs indicative of distress were treated appropriately to alleviate discomfort or euthanized if recommended by animal care staff or the facility veterinary. Mice were anesthetized and dorsal surfaces were shaved and cleaned with betadine topical solution. Fifteen millimeters full-thickness incisional skin wounds were made in the mid-dorsal area. The wound sites were monitored and measured daily utilizing wound closure as endpoint as previously [20]. At indicated time points, the wound field was excised and fixed in aqueous buffered zinc formalin for 24 h, transferred to 70% ethanol, paraffin embedded and sectioned. Wound size was determined as the factor of wound width and length, normalized to the wound area at Day 0. Wound half-life was determined from the nonlinear regression of wound closure over time per animal.
Statistical analysis
All analyses were performed with a minimum of three technical replicates per experiment from three independent experiments and the means obtained were used for ANOVA or independent t-tests. Additional technical and biological replicates were used as indicated. Statistical analyses, variation estimation and validation of test assumptions were carried out using the Prism 6 statistical analysis program (GraphPad). Asterisks denote statistical significance (nonsignificant or NS, P > 0.05; *P < 0.05; **P < 0.01; and ***P < 0.001). All data are reported as mean ± standard error of the mean (s.e.m.).
Results
Epidermal integrity is maintained under homeostasis despite targeted deletion of Gα proteins
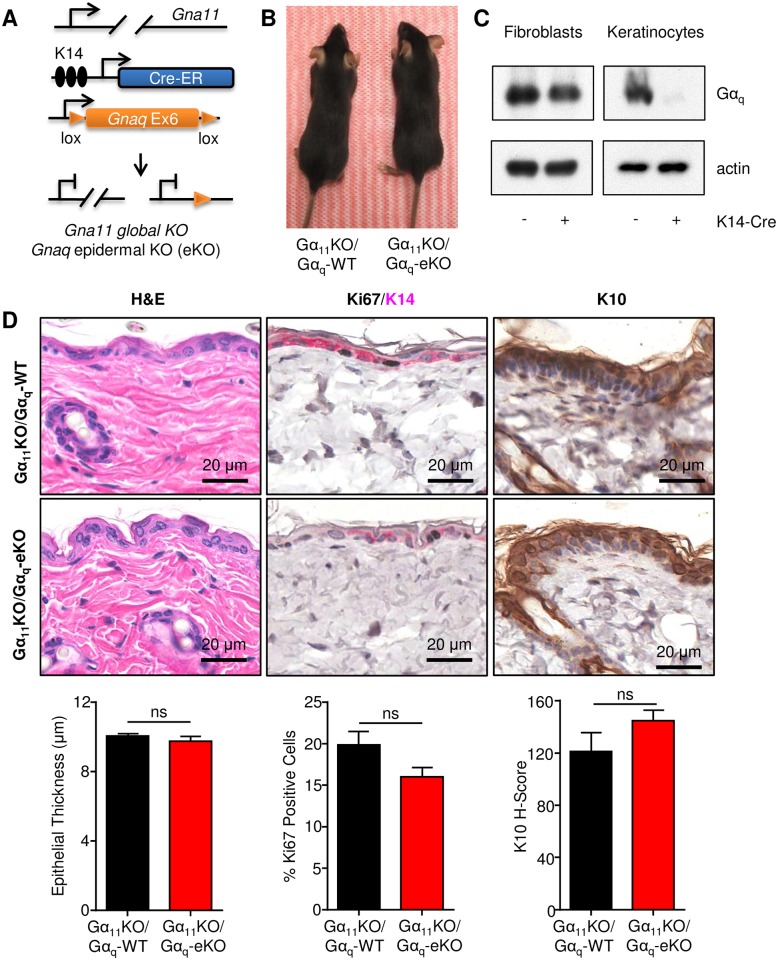
We have recently shown that activating mutations in Gαq function as drivers of oncogenesis in uveal melanoma and are also found in a variety of other malignancies [21, 22], while deletion of Gnaq in the context of Gna11 deficiency appears to have significant defects in developmental and Rho-mediated endothelial cell functions [11, 13]. Deletion of Gnaq alone results in ataxia, impaired motor control, and skin pigmentation [10, 11, 14, 23]. However, a specific functional role for Gαq proteins in the skin has not been identified partially due to the compensatory roles that Gαq and Gα11 play in vivo. To determine if Gαq proteins might have a specific effect on epidermal function, we used a mouse model harboring global deletion of Gα11 (Gα11KO) and targeted deletion of Gαq in the skin (Gαq-eKO) driven by the K14 promoter (Fig 1A). Gα11KO/Gαq-eKO animals displayed no gross abnormalities (Fig 1B) despite complete deletion of Gαq proteins from the skin (Fig 1C). Histologically, both littermate controls bearing global Gα11 deletion but expressing WT Gαq and Gα11KO/Gαq-eKO animals demonstrated no differences in epidermal thickness (Fig 1D). There was a slight decrease in Ki67 proliferating cells that corresponded with a slight increase in K10+ differentiated cells in Gα11KO/Gαq-eKO animals, suggesting that there might be a stronger tendency towards terminal differentiation within these cells (Fig 1D). However, these animals were largely phenotypically normal under homeostatic conditions and did not present the endogenous tumorigenic susceptibility of the Gαs model.
Fig 1. Mice with epidermal loss of Gαq do not demonstrate phenotypic defects under normal conditions.
A) Schematic representation of the animal model used to derive a delete Gnaq exon 6 (Ex6) from the basal epidermal compartment concomitant with global deletion of Gna11. B) Representative pictures of Gα11KO/Gαq-WT control and Gα11KO/Gαq-eKO mice. C) Western blot demonstrating Gαq deletion in the epidermal keratinocytes but not fibroblasts after expression of K14-Cre. D) Normal skin samples from Gα11KO/Gαq-WT control and Gα11KO/Gαq-eKO animals (N = 4 each) were examined for histology (H&E), proliferation (Ki67), and differentiation (K10).
Loss of Gαq confers a strong migratory defect in keratinocytes
To determine if any defect might be present within Gα11/q-null keratinocytes, we isolated cells from mice and analyzed them according to the key hallmarks of carcinomas, namely increased proliferation, increased colony formation, and increased migration. Similar to what we observed in histologic sections, Gα11/q-null keratinocytes did not demonstrate any differences in proliferation (Fig 2A). Similarly, no statistical differences could be detected in the size or number of colonies (Fig 2B). However, a remarkable phenotype was observed upon induction of migration, where Gα11/q-null keratinocytes had a profound defect in their ability to migrate after scratch wounding (Fig 2C). Over a 96-hour time course, control keratinocytes migrated to close the gap formed after a scratch wound assay, while Gα11/q-null keratinocytes were able to close less than half of the gap in the same time frame (Fig 2C). This was due specifically to loss of Gα11/q and not due to loss of Gα11 alone, as Gα11-null fibroblasts isolated from the same mice showed no such defect and were identical to fibroblasts from control mice (Fig 2C). Together, this suggested that rather than promoting a carcinogenic phenotype, deletion of Gαq from the epidermis contributed to a migratory defect. To determine if a similar phenotype had been observed in other studies, we searched for gene expression data related to alteration of a migration phenotype in keratinocytes. By annotating the GEO dataset from a study that used TGF ligands to assess migration-specific genes [24], we found that among the ten most statistically significantly altered ontologies amongst regulated genes, nine were related to Gα protein signaling events and specifically to events associated with Gαq activity, including phospholipase C activity (S1 Fig).
Fig 2. Gα11/q-null keratinocytes have normal proliferative and stem-like phenotypes but demonstrate a significant migratory defect.
Keratinocytes from Gα11KO/Gαq-WT control and Gα11KO/Gαq-eKO animals were assayed for proliferation (A), colony formation (B) and scratch wound closure (C). To confirm the migratory defect, keratinocyte and fibroblast wound closure from Gα11KO/Gαq-WT control and Gα11KO/Gαq-eKO were quantified. Statistical significance was determined using Student’s t-test, *p<0.05, **p<0.01, ***p<0.001.
TGF-β has been largely characterized in cell migration due to its pleiotropic effects in a wide variety of cell types and in particular its contribution to proliferation and migration in cutaneous injury [25–27]. Taking advantage of the CRISPR targeted deletion system, we tested the possibility that loss of Gαq might be necessary for integrating migration stimulatory signals. First, we compared parental HEK293 cells with Gα11/Gαq CRISPR-deleted HEK293 migration and found that consistent with our previous observations, the Gα11/q-null cells migrated significantly less under serum-starved and FBS-stimulated conditions (Fig 3A). To further investigate whether this was a migration-specific defect due to Gαq expression and not due to competing mitogen signaling or clonality issues from the CRISPR cell cloning, we reintroduced Gαq with low levels of transfection plasmid or used empty vector as a control and then assessed their TGF-β-dependent migration. Gα11/q-null cells showed a modest, dose-dependent response to TGF-β, indicating that they are capable of responding to TGF-β signaling (Fig 3B). However, reintroduction of Gαq resulted in a far more robust response to TGF-β and these cells migrated significantly more than empty vector controls (Fig 3B). To confirm that this was not due to a defect in TGF-β signaling, we stimulated the cells for 30 minutes and then performed western blots for downstream targets of TGF-β, namely SMAD2. Both the Gαq-expressing and vector control cells responded to TGF-β signaling to approximately the same degree, as indicated by accumulation of p-SMAD2 (Fig 3C). This suggests that TGF-β does not signal directly through Gαq, but rather that Gαq participates at some level in orchestrating the signaling cascade that is initiated upon TGF-β in order to facilitate migration. Migration requires reorganization of the actin cytoskeleton, so we imaged vector control and Gαq-restored cells after treatment with TGF-β via fluorescently-labelled actin. Here, vector control cells acquired a ruffled cytoskeletal appearance at high concentrations of TGF-β, but Gαq-restored cells were able to achieve a more efficient reorganization of their cytoskeleton at low doses of TGF-β (Fig 3D). Together, these results demonstrate that loss of Gαq confers a strong migratory defect in epithelial cells that is specific to migration-related signaling events and independent of other growth related signaling cues.
Fig 3. Gαq enhances TGF-β stimulated migration and cytoskeletal remodeling.
A) Parental and Gα11/q-null HEK293 cells were assessed for migration towards serum free media (SFM) or 5% fetal bovine serum (FBS). B) Gα11/q-null or Gα11/q-null with transfected, reconstituted expression of Gαq were allowed to migrate towards 0, 1, or 10 ng/ml TGF-β1. C) Western blot demonstrating efficacy of TGF-β signaling in Gα11/q-null and reconstituted cells after 30 minutes treatment with 0, 1, or 10 ng/ml TGF-β1. D) Gα11/q-null and reconstituted cells co-expressing LifeActGFP were treated with 0, 1, or 10 ng/ml TGF-β1 for 3 hours and assessed for cytoskeletal changes. Statistical significance was determined by Student’s t-test, *p<0.05, **p<0.01, ***p<0.001.
Gαq-induced migratory defects in keratinocytes leads to significant delay in cutaneous wound healing
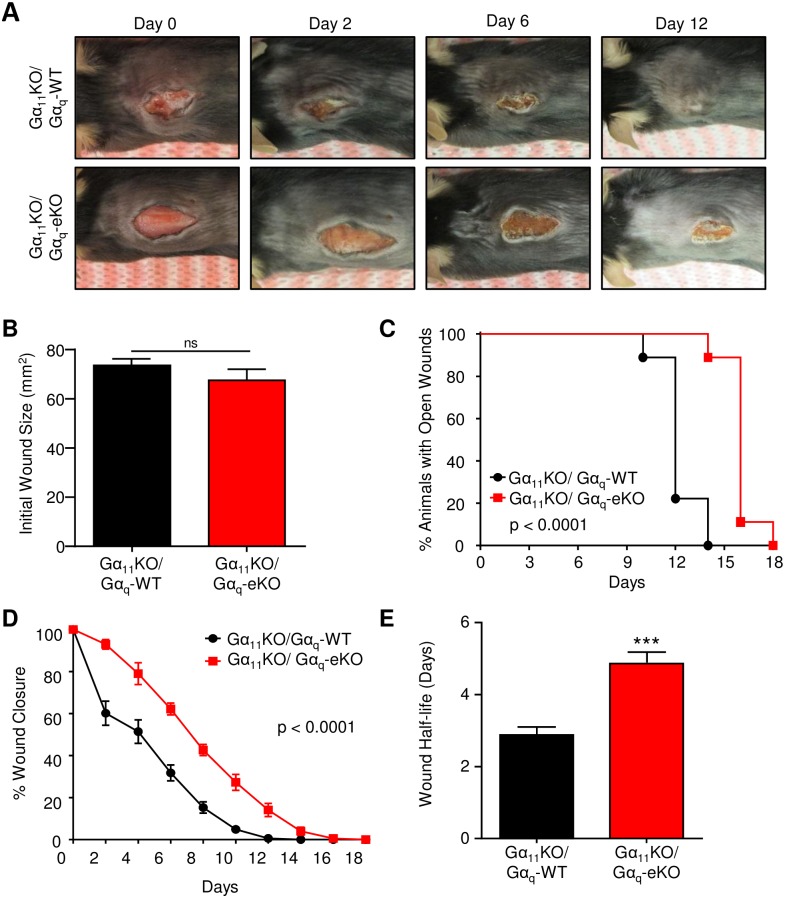
The ability of keratinocytes to migrate into the wound bed is an essential step in cutaneous wound healing and essential for reepithelialization. As such, we reasoned that the pronounced migratory defect in Gα11/q-null animals might attenuate wound healing and lead to a delay in wound closure. Gα11KO/Gαq-WT control and Gα11KO/Gαq-eKO littermates were subjected to 15 mm dorsal cutaneous wounds and observed continuously during the wound healing process (Fig 4A). Initial wound sizes were nearly identical between Gα11KO/Gαq-WT control and Gα11KO/Gαq-eKO animals (Fig 4B). Despite this, Gα11KO/Gαq-eKO mice healed significantly later than their control littermates (Fig 4C). This delay in wound healing was directly attributed to loss of both epidermal Gαq proteins, Gαq and Gα11 as Gα11-null control animals alone healed similarly to wild-type animals (S2 Fig). Analysis of the rate of wound healing between Gα11KO/Gαq-WT control and Gα11KO/Gαq-eKO animals demonstrated a significant shift at all time points, leading to a highly significant difference in the wound healing curves (Fig 4D). Nonlinear regression of these curves revealed that the Gα11KO/Gαq-eKO animals had a wound half-life nearly twice that of Gα11KO/Gαq-WT control mice (Fig 4E). Together, this data indicates that the migratory defect incurred upon loss of Gαq has significant impact on the rate of cutaneous wound healing and loss of epidermal Gαq may lead to the development of chronic wounds.
Fig 4. Gα11KO/Gαq-eKO mice have delayed wound healing.
Gα11KO/Gαq-WT control and Gα11KO/Gαq-eKO mice received 15 mm incisional wound and closure was monitored over 18 days. A) Gross examination of the wounds in Gα11KO/Gαq-WT control (N = 15) and Gα11KO/Gαq-eKO (N = 10) mice. B) Initial wound size. Statistical significance determined by Student’s t-test. C) Kaplan-Meier survival curve of wound closure. Statistical significance was determined by log-rank test. D) Wound closure over time. Statistical significance was determined by two-way ANOVA. E) Wound half-life for Gα11KO/Gαq-WT control and Gα11KO/Gαq-eKO animals, as determined from the rate of wound closure over time. Statistical significance was determined by Student’s t-test, ***p<0.001.
Gαq-mediated wound healing defects correlate with insensitivity to the regulation of differentiation signals
Defects in the keratinocyte migration impair subsequent reepithelialization phase of wound healing and can be seen by a failure to form an epithelial tongue adjacent to the wound bed. This was confirmed by H&E analysis, where the length of the migrating epithelial tongue in Gα11KO/Gαq-eKO cells was significantly smaller than their Gα11KO/Gαq-WT control counterparts (Fig 5, top row). While migrating cells closest to the wound bed do not demonstrate significant levels of proliferation, the trailing end of the epithelial tongue engages in robust proliferation to facilitate reepithelialization [28]. Compared to normal adjacent epithelium, WT cells demonstrate the spectrum of proliferation associated with this phase of wound healing, where the proximal segment is less proliferative than normal keratinocytes but a sharp doubling in proliferation can be seen at the supporting edge (Fig 5, second row). In contrast, KO cells demonstrate a relatively stable level of proliferation throughout the epithelial tongue that is not overtly different from the adjacent normal cells (Fig 5, second row). In the same manner, Gα11KO/Gαq-eKO animals demonstrate a profound inability to downregulate markers of differentiation at the leading edge of the epithelial tongue. While control littermates show a steady and significant decrease in the early differentiation marker K10 as the epithelial tongue approaches the wound bed, a nearly uniform expression of K10 is observed in all segments of the epithelial tongue of Gα11KO/Gαq-eKO animals (Fig 5, third row). This correlated to a nearly 13-fold reduction in slope for the K10 H-score across the epithelial tongue (Fig 5, third row). Defects in reepithelialization are associated with chronic wounds and malignant transformation in which reciprocal expression of K10 and K13 are often observed [29, 30]. Again, we observed that Gα11KO/Gαq-WT control animals had a steady, nearly linear increase in K13 H-score across the epithelial tongue that was abrogated in the Gα11KO/Gαq-eKO animals. While a slight increase was detected, it was still nearly 4 times less than that of the control animals. Together, this data indicates that the migratory defect that results from epidermal loss of Gαq manifests in a failure of the epithelial tongue to migrate during wound healing. Further, it suggests that the deficiency in reepithelialization is due to an inability to engage the signaling pathways that originate within the wound bed.
Fig 5. Loss of epidermal Gαq correlates with defects in reepithelialization of cutaneous wounds.
Top panel: the epithelial tongue as evidenced by H&E staining (yellow dashed line) is diagrammed and the length quantified. Representative stains and quantification of Ki67 (second panel), K10 (third panel), and K13 (bottom panel) are shown. All quantifications are based on an N = 4 mice per group and proliferation is shown relative to normal adjacent epithelium. Slope was determined by linear regression and statistical significance was determined by Student’s t-test, p<0.05, **p<0.01, ***p<0.001.
Discussion
Migration is a complex and highly orchestrated process that is essential for maintaining many cell functions under normal and pathologic conditions. Here, we demonstrate a novel role for Gαq in facilitating acquisition of a stress-induced migratory phenotype in epidermal cells in the context of Gα11 loss. Under normal conditions, loss of Gα11/q signaling does not demonstrate any overt effects but upon wounding, Gα11/q-null cells exhibit a remarkable inability to initiate the standard migratory response both in vitro and in vivo. This migratory defect correlates strongly with an inability to engage in the normal differentiation and proliferative programs that are initiated upon wounding and consequently lead to significantly delayed wound closure. These results have strong implications for our current understanding for the management of chronic wounds and aggressive malignancies whose pathologic hallmarks include the acquisition of a migratory phenotype.
Migration of keratinocytes, either collectively or as single cells, depends on the integration of internal and external cues to achieve directed movement [31]. These extracellular cues involve growth factors including members of the EGF, FGF, insulin, and VEGF families [32]. Gα proteins have been shown to be critical in the transactivation of many of these growth factor receptors and in some cases this transactivation has been shown to have pathologic consequences including tumor development and metastasis [33–35]. Despite the significant contribution of other Gα family members with migration, remarkably little is known regarding the specific role of Gαq in epidermal motility. LPA has been shown to activate Gα11/q signaling pathways and drive cell proliferation, differentiation, and migration in keratinocytes, similar to the effect observed upon TGF-β stimulation [36]. Conversely, transactivation of the insulin-like growth factor I receptor (IGF1R) by UTP-stimulated P2Y purinergic receptors inhibits PI3K activation in a Gαq-dependent manner, blocking subsequent keratinocyte migration [37, 38]. However, in other contexts, Gαq is critical for activation of PI3K, MAPK, and other motility-associated pathways [39], suggesting that the role of Gαq in migration may be to integrate multiple and sometimes conflicting extracellular signals to achieve appropriate cellular responses.
Wound healing is a clear example of a process that involves such an incorporation of multiple complex signaling networks. In order to restore barrier function, keratinocytes must coordinate migration, proliferation, and differentiation within a dynamic extracellular space. After acute injury, keratinocytes at the margin of wounds begin to migrate towards the wound bed. Proximity to the proinflammatory cytokines secreted at the wound bed leads to a number of phenotypical changes, including downregulation of proliferative and differentiation markers such as Ki67 and K10 [28]. At the same time, epidermal cells trailing the migrating keratinocytes engage in a hyperproliferative program to support the wound healing process [28]. Indeed, WT keratinocytes demonstrate a burst of proliferation at the trailing edge of the epithelial tongue that is nearly twice that of normal epithelium while simultaneously downregulating proliferation at the leading edge. Conversely, Gα11/q-null keratinocytes show a remarkably dampened response to stimuli, with an only slightly increased level of proliferation in the distal tongue section and a relatively unchanged level of proliferation at the leading edge. A similar trend is observed with respect to differentiation, as Gα11/q-null cells show little to no downregulation of K10, while WT cells actively engage in dedifferentiation at the wound bed.
Failure to enact the reepithelialization phase of wound healing due to diminished keratinocyte migration is clinically associated with chronic wounds. Cutaneous squamous cell carcinomas (SCCs) in particular have been shown to arise from precancerous lesions associated with chronic wounds and impaired wound healing paradigms [40]. Activation of the epithelial stem cell compartment is a critical element of wound healing, and epidermal loss of another Gα family member, Gαs, was associated with subsequent formation of basal cell carcinomas [41]. Therefore, chronic wounds demonstrating loss of Gαq function may be more prone to development of precancerous and cancerous lesions in the skin.
In addition to activation of the stem cell compartment, chronic wounds and tumors share many other similar mechanistic and molecular manifestations, including irreversible loss of keratinocyte differentiation, uncontrolled proliferation, and enhanced migration [42]. Impaired signaling, proliferation, and migration of fibroblasts in chronic wounds have been observed and are in part attributable to decreased sensitivity to TGF-β1 [43]. Given the decreased responsiveness of Gα11/q-null cells to TGF-β1, this suggests that loss of Gαq-mediated signaling may contribute to the development of chronic wounds that could progress to SCCs. K13, a common marker of malignant progression, typically replaces expression of K10 during tumor progression and is also seen at higher levels at the leading edge of the epithelial tongue in wound healing [29, 30]. Only cells expressing Gαq were able to differentially regulate expression of K13 (Fig 5), indicating that Gαq is essential for transmitting the multilayered cues within the wound microenvironment. Taken together, it is tempting then to speculate then that under conditions of chronic inflammation and pathologic injury, loss of Gαq may contribute to chronic wound formation and subsequent malignant transformation in the epithelial compartment.
Gαq and Gα11 are integral members of the Gα family of proteins and play multifaceted roles in the regulation of gene expression, viability, and growth. Here, we propose that Gαq proteins may have pleiotropic effects in the epithelial compartment under pathologic conditions to coordinate reepithelialization. Loss of epithelial Gα11/q during the wound healing process manifests in a severe migratory phenotype punctuated by an inability of Gα11/q-null cells to differentially regulate proliferation and differentiation proximal to the wound bed. As such, Gαq proteins are essential for the integration of the complex signaling networks in pathologic conditions and may provide a unique opportunity for clinical intervention.
Supporting information
Expression levels of statistically significant differentially regulated genes involved in migration were downloaded and analyzed for ontologic similarities using the ENRICHR program. The GO Molecular Function ontology is represented according to the internally calculated p-value.
(TIF)
Wild-type C57B/L6 mice and Gα11KO/Gαq-WT mice were given 15 mm incisional wounds and closure was monitored over 18 days. A) Initial wound size. Statistical significance determined by Student’s t-test. B) Kaplan-Meier survival curve of wound closure. Statistical significance was determined by log-rank test. C) Wound closure over time. Statistical significance was determined by two-way ANOVA. D) Wound half-life for WT and Gα11KO as determined from the rate of wound closure over time. Statistical significance was determined by Student’s t-test, ns = not significant.
(TIF)
Acknowledgments
We thank Dr. Roberto Weigert (NIDCR, National Institutes of Health) for providing the LifeAct plasmid.
This research was supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, and in part by Marian University Indianapolis. A.I. was funded by JST, PRESTO Grant #13408007.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, and in part by Marian University Indianapolis. AI received Grant #13408007 from PRESTO, Japan Science and Technology Agency. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
References
- 1.O'Hayre M, Degese MS, Gutkind JS. Novel insights into G protein and G protein-coupled receptor signaling in cancer. Curr Opin Cell Biol. 2014;27:126–35. Epub 2014/02/11. 10.1016/j.ceb.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griewank K V S; Bastian BC. GNA11 (guanine nucleotide binding protein (G protein), alpha 11 (Gq class)). Atlas Genet Cytogenet Oncol Haematol. 2011;15(10):828–30. [Google Scholar]
- 3.Kalinec G, Nazarali AJ, Hermouet S, Xu N, Gutkind JS. Mutated alpha subunit of the Gq protein induces malignant transformation in NIH 3T3 cells. Mol Cell Biol. 1992;12(10):4687–93. Epub 1992/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Hayre M, Vazquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, et al. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer. 2013;13(6):412–24. Epub 2013/05/04. 10.1038/nrc3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363(23):2191–9. 10.1056/NEJMoa1000584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griewank KG, Schilling B, Scholz SL, Metz CH, Livingstone E, Sucker A, et al. Oncogene status as a diagnostic tool in ocular and cutaneous melanoma. Eur J Cancer. 2016;57:112–7. 10.1016/j.ejca.2016.01.010 [DOI] [PubMed] [Google Scholar]
- 7.Chattopadhyay C, Kim DW, Gombos DS, Oba J, Qin Y, Williams MD, et al. Uveal melanoma: From diagnosis to treatment and the science in between. Cancer. 2016;122(15):2299–312. 10.1002/cncr.29727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decatur CL, Ong E, Garg N, Anbunathan H, Bowcock AM, Field MG, et al. Driver Mutations in Uveal Melanoma: Associations With Gene Expression Profile and Patient Outcomes. JAMA Ophthalmol. 2016;134(7):728–33. 10.1001/jamaophthalmol.2016.0903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol Sci. 2001;22(7):368–76. Epub 2001/06/30. [DOI] [PubMed] [Google Scholar]
- 10.Offermanns S, Zhao LP, Gohla A, Sarosi I, Simon MI, Wilkie TM. Embryonic cardiomyocyte hypoplasia and craniofacial defects in G alpha q/G alpha 11-mutant mice. EMBO J. 1998;17(15):4304–12. 10.1093/emboj/17.15.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Offermanns S, Hashimoto K, Watanabe M, Sun W, Kurihara H, Thompson RF, et al. Impaired motor coordination and persistent multiple climbing fiber innervation of cerebellar Purkinje cells in mice lacking Galphaq. Proc Natl Acad Sci U S A. 1997;94(25):14089–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wettschureck N, Rutten H, Zywietz A, Gehring D, Wilkie TM, Chen J, et al. Absence of pressure overload induced myocardial hypertrophy after conditional inactivation of Galphaq/Galpha11 in cardiomyocytes. Nat Med. 2001;7(11):1236–40. 10.1038/nm1101-1236 [DOI] [PubMed] [Google Scholar]
- 13.Mikelis CM, Simaan M, Ando K, Fukuhara S, Sakurai A, Amornphimoltham P, et al. RhoA and ROCK mediate histamine-induced vascular leakage and anaphylactic shock. Nat Commun. 2015;6:6725 Epub 2015/04/11. 10.1038/ncomms7725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Raamsdonk CD, Fitch KR, Fuchs H, de Angelis MH, Barsh GS. Effects of G-protein mutations on skin color. Nat Genet. 2004;36(9):961–8. 10.1038/ng1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitch KR, McGowan KA, van Raamsdonk CD, Fuchs H, Lee D, Puech A, et al. Genetics of dark skin in mice. Genes Dev. 2003;17(2):214–28. 10.1101/gad.1023703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korhonen H, Fisslthaler B, Moers A, Wirth A, Habermehl D, Wieland T, et al. Anaphylactic shock depends on endothelial Gq/G11. J Exp Med. 2009;206(2):411–20. Epub 2009/01/28. 10.1084/jem.20082150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131(10):2257–68. Epub 2004/04/23. 10.1242/dev.01125 [DOI] [PubMed] [Google Scholar]
- 18.Schrage R, Schmitz AL, Gaffal E, Annala S, Kehraus S, Wenzel D, et al. The experimental power of FR900359 to study Gq-regulated biological processes. Nat Commun. 2015;6:10156 10.1038/ncomms10156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dlugosz AA, Glick AB, Tennenbaum T, Weinberg WC, Yuspa SH. Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 1995;254:3–20. Epub 1995/01/01. [DOI] [PubMed] [Google Scholar]
- 20.Romer J, Bugge TH, Pyke C, Lund LR, Flick MJ, Degen JL, et al. Impaired wound healing in mice with a disrupted plasminogen gene. Nat Med. 1996;2(3):287–92. Epub 1996/03/01. [DOI] [PubMed] [Google Scholar]
- 21.Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25(6):831–45. Epub 2014/06/03. 10.1016/j.ccr.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaque JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q, et al. A genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol Cell. 2013;49(1):94–108. Epub 2012/11/28. 10.1016/j.molcel.2012.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Offermanns S, Toombs CF, Hu YH, Simon MI. Defective platelet activation in G alpha(q)-deficient mice. Nature. 1997;389(6647):183–6. 10.1038/38284 [DOI] [PubMed] [Google Scholar]
- 24.Cheng CF, Fan J, Bandyopahdhay B, Mock D, Guan S, Chen M, et al. Profiling motility signal-specific genes in primary human keratinocytes. J Invest Dermatol. 2008;128(8):1981–90. Epub 2008/03/08. 10.1038/jid.2008.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hata A, Chen YG. TGF-beta Signaling from Receptors to Smads. Cold Spring Harb Perspect Biol. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koivisto L, Heino J, Hakkinen L, Larjava H. Integrins in Wound Healing. Adv Wound Care (New Rochelle). 2014;3(12):762–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hameedaldeen A, Liu J, Batres A, Graves GS, Graves DT. FOXO1, TGF-beta regulation and wound healing. Int J Mol Sci. 2014;15(9):16257–69. 10.3390/ijms150916257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Usui ML, Mansbridge JN, Carter WG, Fujita M, Olerud JE. Keratinocyte migration, proliferation, and differentiation in chronic ulcers from patients with diabetes and normal wounds. J Histochem Cytochem. 2008;56(7):687–96. Epub 2008/04/17. 10.1369/jhc.2008.951194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betz P, Nerlich A, Tubel J, Penning R, Eisenmenger W. The time-dependent expression of keratins 5 and 13 during the reepithelialization of human skin wounds. Int J Legal Med. 1993;105(4):229–32. Epub 1993/01/01. [DOI] [PubMed] [Google Scholar]
- 30.Glick AB, Kulkarni AB, Tennenbaum T, Hennings H, Flanders KC, O'Reilly M, et al. Loss of expression of transforming growth factor beta in skin and skin tumors is associated with hyperproliferation and a high risk for malignant conversion. Proc Natl Acad Sci U S A. 1993;90(13):6076–80. Epub 1993/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188(1):11–9. Epub 2009/12/03. 10.1083/jcb.200909003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seeger MA, Paller AS. The Roles of Growth Factors in Keratinocyte Migration. Adv Wound Care (New Rochelle). 2015;4(4):213–24. Epub 2015/05/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filardo EJ, Quinn JA, Bland KI, Frackelton AR Jr. Estrogen-induced activation of Erk-1 and Erk-2 requires the G protein-coupled receptor homolog, GPR30, and occurs via trans-activation of the epidermal growth factor receptor through release of HB-EGF. Mol Endocrinol. 2000;14(10):1649–60. Epub 2000/10/24. 10.1210/mend.14.10.0532 [DOI] [PubMed] [Google Scholar]
- 34.Hart S, Fischer OM, Prenzel N, Zwick-Wallasch E, Schneider M, Hennighausen L, et al. GPCR-induced migration of breast carcinoma cells depends on both EGFR signal transactivation and EGFR-independent pathways. Biol Chem. 2005;386(9):845–55. Epub 2005/09/17. 10.1515/BC.2005.099 [DOI] [PubMed] [Google Scholar]
- 35.Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, et al. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402(6764):884–8. Epub 2000/01/06. 10.1038/47260 [DOI] [PubMed] [Google Scholar]
- 36.Sauer B, Vogler R, Zimmermann K, Fujii M, Anzano MB, Schafer-Korting M, et al. Lysophosphatidic acid interacts with transforming growth factor-beta signaling to mediate keratinocyte growth arrest and chemotaxis. J Invest Dermatol. 2004;123(5):840–9. Epub 2004/10/16. 10.1111/j.0022-202X.2004.23458.x [DOI] [PubMed] [Google Scholar]
- 37.Taboubi S, Milanini J, Delamarre E, Parat F, Garrouste F, Pommier G, et al. G alpha(q/11)-coupled P2Y2 nucleotide receptor inhibits human keratinocyte spreading and migration. Faseb J. 2007;21(14):4047–58. Epub 2007/07/05. 10.1096/fj.06-7476com [DOI] [PubMed] [Google Scholar]
- 38.Taboubi S, Garrouste F, Parat F, Pommier G, Faure E, Monferran S, et al. Gq-coupled purinergic receptors inhibit insulin-like growth factor-I/phosphoinositide 3-kinase pathway-dependent keratinocyte migration. Mol Biol Cell. 2010;21(6):946–55. Epub 2010/01/22. 10.1091/mbc.E09-06-0497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7(2):79–94. Epub 2007/01/26. 10.1038/nrc2069 [DOI] [PubMed] [Google Scholar]
- 40.Leiter U, Eigentler T, Garbe C. Epidemiology of skin cancer. Adv Exp Med Biol. 2014;810:120–40. [DOI] [PubMed] [Google Scholar]
- 41.Iglesias-Bartolome R, Torres D, Marone R, Feng X, Martin D, Simaan M, et al. Inactivation of a Galphas-PKA tumour suppressor pathway in skin stem cells initiates basal-cell carcinogenesis. Nat Cell Biol. 2015;17(6):793–803. Epub 2015/05/12. 10.1038/ncb3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–9. Epub 1986/12/25. 10.1056/NEJM198612253152606 [DOI] [PubMed] [Google Scholar]
- 43.Kim BC, Kim HT, Park SH, Cha JS, Yufit T, Kim SJ, et al. Fibroblasts from chronic wounds show altered TGF-beta-signaling and decreased TGF-beta Type II receptor expression. J Cell Physiol. 2003;195(3):331–6. 10.1002/jcp.10301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression levels of statistically significant differentially regulated genes involved in migration were downloaded and analyzed for ontologic similarities using the ENRICHR program. The GO Molecular Function ontology is represented according to the internally calculated p-value.
(TIF)
Wild-type C57B/L6 mice and Gα11KO/Gαq-WT mice were given 15 mm incisional wounds and closure was monitored over 18 days. A) Initial wound size. Statistical significance determined by Student’s t-test. B) Kaplan-Meier survival curve of wound closure. Statistical significance was determined by log-rank test. C) Wound closure over time. Statistical significance was determined by two-way ANOVA. D) Wound half-life for WT and Gα11KO as determined from the rate of wound closure over time. Statistical significance was determined by Student’s t-test, ns = not significant.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.