Abstract
Association between let-7-KRAS rs712 polymorphism and cancer risk was inconsistent. We therefore conducted this meta-analysis to clarify the association between let-7-KRAS rs712 polymorphism and cancer risk with STATA 14.0 software. A systemic literature search in online databases (PubMed, Embase, CNKI and Wanfang database) was preformed to obtain relevant articles. A total of 13 case-control studies involving 3,453 patients and 4,470 controls were identified up to May 16, 2015. The pooled results indicated that significantly increased risk were observed in Chinese population in T vs. G (OR = 1.21, 95% CI = 1.03–1.42) and TT vs. GG + GT genetic models (OR = 1.69, 95% CI = 1.17–2.42). Sensitivity analysis was conducted and the result without heterogeneity showed significant associations in all five genetic models. Subgroup analyses of cancer type indicated a similar result in digestive cancer (for T vs. G: OR = 1.41, 95% CI = 1.26–1.57; GT vs. GG: OR = 1.24, 95% CI = 1.07–1.43; TT vs. GG: OR = 2.53, 95% CI = 1.86–3.44; GT + TT vs. GG: OR = 1.36, 95% CI = 1.19–1.56; TT vs. GG + GT: OR = 2.35, 95% CI = 1.73–3.19). In summary, these evidences demonstrate that let-7-KRAS rs712 G > T polymorphism might be associated with digestive system cancer risk in the Chinese population.
Keywords: let-7, KRAS, polymorphism, cancer
INTRODUCTION
Cancer, one of the major common malignant diseases contributes to death worldwide, which has become an important healthy problem [1]. In China, accompanied with the accelerated deterioration of the environment and the population aging, the incidence of cancer has been rising in recent ten year. Today, cancer has become the leading cause of death in China, more than 4292,000 new cancer patents and 2814,000 deaths would occur in 2015 [2]. Dysfunction, deformity, and mental stress have seriously reduced the quality of life of cancer patients. Furthermore, the increasing medical costs have become a heavy economic burden on families and society [3, 4]. Unfortunately, the development mechanism of cancer has not been clearly explained, although numerous epidemiological and molecular biology researches has shown that live habits, nutritional intake, mental state, and chronic inflammation are contributed to cancer risk [5].
To date, a large numbers of studies have indicated that genetic abnormity maybe result in tumorigenesis [6, 7]. MicroRNA always consist of short, single-stranded, noncoding RNAs with 20–22 nucleotides long, which could take part in the genetic post-transcriptional regulation and influenced the cell procedures of differentiation, proliferation, apoptosis [8]. Lethal-7 (let-7) is the earliest discovered microRNA family, which is an important genetic regulators through controlling cancer oncogene expression by binding to the complementary elements in the 3′ untranslated regions (UTRs) of their target messenger RNAs (mRNAs) [9]. Let-7 could decrease KRAS expression through a let-7-KRAS binding located at specific sites of the 3′ UTRs of KRAS, which has been proved one of the most frequently activated oncogenes [10].
Gene mutation, including single-nucleotide polymorphisms (SNPs), such as interleukin gene family polymorphisms and microRNA polymorphisms has been proved to be associated with cancer risk. Regarding KARS gene, several common SNPs located at the 3′-UTR region have been identified, such as rs712 G > T polymorphism. A recent study with luciferase vector reporter system demonstrated that the let-7 would decrease the activity of KRAS, but the rs712 minor allele would compromise the interaction between let-7g and KRAS 3′-UTR [11].
From 2014, two meta-analyses were conducted, only 6 case-control studies were included in both meta-analyses [12, 13]. Today, more than ten studies that assessed the association between rs712 G > T polymorphism and cancer risk published. Until now, most of the studies focused on Chinese population without consistent conclusion. Therefore, we performed this updated meta-analysis to further investigate an accurate association between rs712 G > T polymorphism and cancer risk in the Chinese population.
RESULTS
Study characteristics
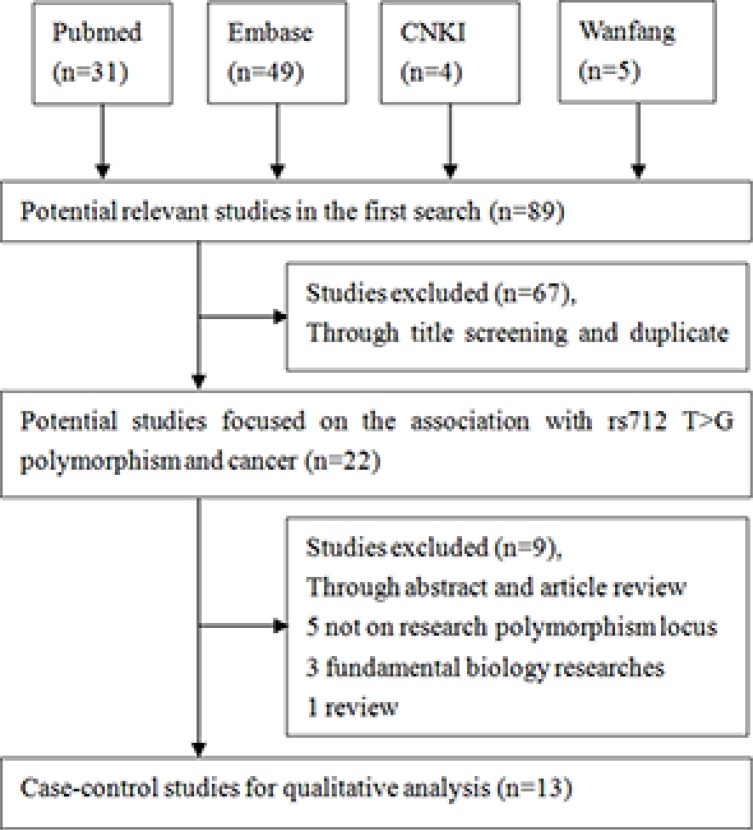
A total of 89 studies were identified initially. Figure 1 showed the selecting procession of studies step by step. After reviewed the titles and abstracts, 67 articles were excluded. Through reading full texts, we deleted another 9 articles. Finally, 13 articles involving 3,453 patients and 4,470 controls were selected in our meta-analysis based on the inclusion criteria [11, 14–25]. Among them, 5 articles on digestive system cancer with 1,798 patients and 2,145 controls [15, 17, 22–24], which included three studies on Colorectal cancers, one study on gastric cancer and the other study on Hepatocellular cancer. 3 articles on head and neck cancer with 593 patients and 850 controls [16, 18, 19], 2 articles on lung cancer with 215 patients and 502 controls [11, 14], 2 articles on cervical cancer with 619 patients and 722 controls [20, 21, 25], one article breast cancer with 228 patients and 251 controls [25]. In term of genotyping method, 11 studies used polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), one study adopted Real-time PCR [11] and another study used iMLDR [22] method. The genotype distributions in controls are all satisfy with HWE. All included characteristics of each study were summarized in Table 1.
Figure 1. Flow diagram of the study selection process.
Table 1. Characteristics of case-control studies on Let-7-KRAS rs712 G > T polymorphism and cancer risk included in the meta-analysis.
| First author | Year | Genotype method | Control design | Case | Control | Genotype distribution | P for HWE |
MAF | Location | NOS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||||||||||
| GG | GT | TT | GG | GT | TT | Case | Control | ||||||||||
| Peng | 2010 | PCR-RFLP | HB | 83 | 80 | 49 | 31 | 3 | 51 | 25 | 4 | 0.68 | 0.22 | 0.21 | Lung | 7 | |
| Li | 2013 | PCR-RFLP | HB | 181 | 674 | 105 | 60 | 16 | 442 | 211 | 21 | 0.49 | 0.25 | 0.19 | Gastric | 8 | |
| Yan | 2013 | PCR-RFLP | HB | 153 | 204 | 83 | 56 | 14 | 137 | 61 | 6 | 0.80 | 0.27 | 0.18 | Glioma | 6 | |
| Pan1 | 2014 | PCR-RFLP | HB | 339 | 313 | 188 | 125 | 26 | 203 | 100 | 10 | 0.58 | 0.26 | 0.19 | Colorectal | 8 | |
| Pan2 | 2014 | PCR-RFLP | HB | 188 | 356 | 112 | 64 | 12 | 201 | 138 | 17 | 0.34 | 0.23 | 0.24 | Nasopharyngeal | 7 | |
| Jin | 2014 | PCR-RFLP | HB | 252 | 290 | 154 | 84 | 14 | 183 | 92 | 15 | 0.44 | 0.22 | 0.21 | Thyroid | 7 | |
| Ni | 2015 | PCR-RFLP | HB | 204 | 218 | 112 | 73 | 19 | 145 | 67 | 6 | 0.60 | 0.27 | 0.18 | Cervical | 6 | |
| Liang | 2015 | PCR-RFLP | HB | 415 | 504 | 257 | 144 | 14 | 327 | 163 | 14 | 0.23 | 0.21 | 0.19 | Cervical | 8 | |
| Hu | 2015 | Real-time PCR | HB | 132 | 422 | 22 | 38 | 72 | 12 | 132 | 278 | 0.43 | 0.69 | 0.82 | Lung | 7 | |
| Dai | 2015 | iMLDR | HB | 430 | 430 | 253 | 145 | 32 | 283 | 130 | 17 | 0.67 | 0.24 | 0.19 | Colorectal | 8 | |
| Xiong | 2015 | PCR-RFLP | PB | 262 | 252 | 150 | 92 | 20 | 162 | 79 | 11 | 0.73 | 0.25 | 0.20 | Hepatocellular | 9 | |
| Jiang | 2015 | PCR-RFLP | HB | 586 | 476 | 372 | 176 | 38 | 331 | 133 | 12 | 0.75 | 0.22 | 0.16 | Colorectal | 8 | |
| Huang | 2015 | PCR-RFLP | HB | 228 | 251 | 155 | 65 | 8 | 173 | 71 | 7 | 0.93 | 0.18 | 0.17 | Breast | 7 | |
aHWE in control
Abbreviation: MAF: Minor allele frequency in control group.
HB: Hospital-based; PB: Population-based.
Quantitative analysis
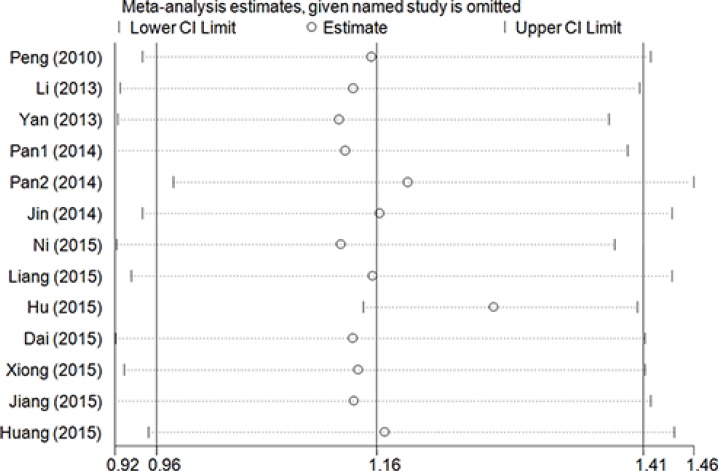
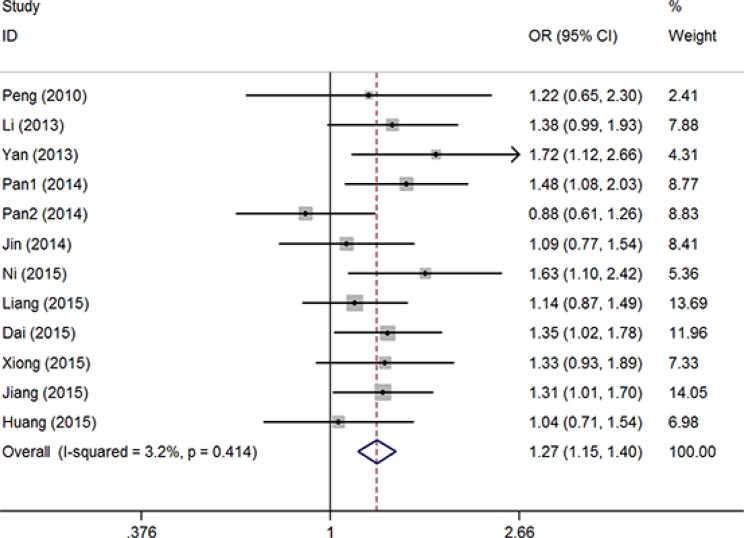
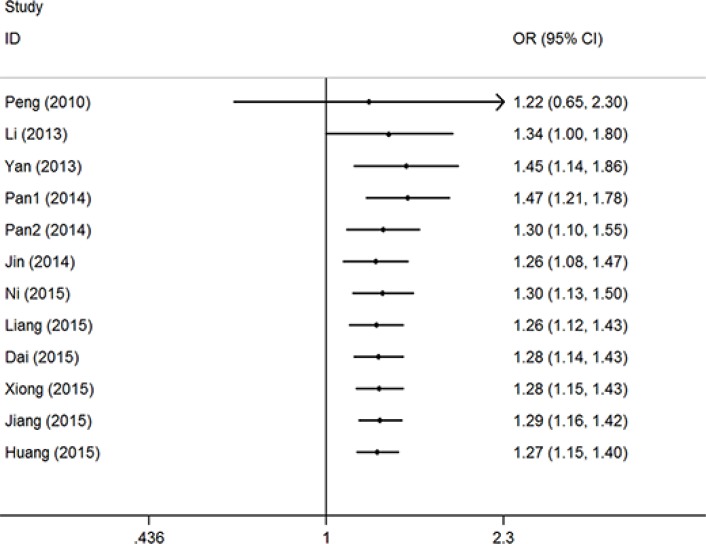
Overall, significantly elevated risks were observed with combined studies in allele contrast model (T vs. G: OR = 1.21, 95% CI = 1.03–1.42, P = 0.03, I2 = 75.8%) and recessive model (TT vs. GG + GT: OR = 1.69, 95% CI = 1.17–2.42, P = 0.01, I2 = 67.4%). Obviously heterogeneities were found in all analyzed genetic models. Sensitive analysis were conducted though deleting each study one by one, and the results indicated that the report by Hu et al. [11] maybe the critical important factor which result in these heterogeneities (Figure 2). Then, significant associations presented in all five genetic models with obviously reduced heterogeneities without the study of Hu et al. [11] (T vs. G: OR = 1.30, 95% CI = 1.20–1.41, P < 0.01, I2 = 33.7%; GT vs. GG: OR = 1.18, 95% CI = 1.07–1.31, P < 0.01, I2 = 0%; TT vs. GG: OR = 2.07, 95% CI = 1.65–2.59, P < 0.01, I2 = 24.8%; GT + TT vs. GG: OR = 1.27, 95% CI = 1.15–1.41, P < 0.01, I2 = 3.2%, (Figure 3); TT vs. GG + GT: OR = 1.96, 95% CI = 1.57–2.44, P < 0.01, I2 = 14.8%) (Supplementary Figure 1). Accumulative analysis indicated that the increased risk with rs712 T > G polymorphism could be found in 2013, and the result was further confirmed with added researches (Figure 4 for GT + TT vs. GG model) (Supplementary Figure 2).
Figure 2. Sensitivity analysis through deleting each study to reflect the influence of the individual dataset to the pooled ORs in GT+TT vs. GG model of rs712 G > T polymorphism.
Figure 3. OR and 95% CIs of the associations between rs712 G > T polymorphism and cancer risk in GT + TT vs. GG model.
Figure 4. Cumulative meta-analyses according to publication year in GT + TT vs. GG model of rs712 G > T polymorphism.
Subgroup analysis based on cancer location, control resource, and genotype methods were conducted. Significant cancer risk were also found without heterogeneity in the subgroup of digestive system cancer (T vs. G: OR = 1.41, 95% CI = 1.26–1.57, P < 0.01, I2 = 0%; GT vs. GG: OR = 1.24, 95% CI = 1.07–1.43, P < 0.01, I2 = 0%; TT vs. GG: OR = 2.53, 95% CI = 1.86–3.44, P < 0.01, I2 = 0%; GT + TT vs. GG: OR = 1.36, 95% CI = 1.19–1.56, P < 0.01, I2 = 0%; TT vs. GG + GT: OR = 2.35, 95% CI = 1.73–3.19, P < 0.01, I2 = 0%). Furthermore, other significant increased associations were also found in the subgroup analysis by control resource, and genotype methods (Table 2).
Table 2. Summary ORs and 95% CI of Let-7-KRAS rs712 G > T polymorphisms and cancer risk.
| N* | T vs. G | GT vs. GG | TT vs. GG | GT+TT vs. GG | TT vs. GG+GT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unadjusted | OR | 95% CI | P | I2(%) | OR | 95% CI | P | I2(%) | OR | 95% CI | P | I2(%) | OR | 95% CI | P | I2(%) | OR | 95% CI | P | I2(%) | |||||
| Total | 13 | 1.21 | 1.03-1.42 | 0.03 | 75.8 | 1.11 | 0.93-1.31 | 0.24 | 62.0 | 1.61 | 0.98-2.65 | 0.06 | 79.8 | 1.16 | 0.96-1.41 | 0.12 | 72.8 | 1.69 | 1.17-2.42 | 0.01 | 67.4 | ||||
| Sensitive analysis# | 12 | 1.30 | 1.20-1.41 | <0.01 | 33.7 | 1.18 | 1.07-1.31 | <0.01 | 0 | 2.07 | 1.65-2.59 | <0.01 | 24.8 | 1.27 | 1.15-1.41 | <0.01 | 3.2 | 1.96 | 1.57-2.44 | <0.01 | 14.8 | ||||
| Location | |||||||||||||||||||||||||
| Digestive system | 5 | 1.41 | 1.26-1.57 | <0.01 | 0 | 1.24 | 1.07-1.43 | <0.01 | 0 | 2.53 | 1.86-3.44 | <0.01 | 0 | 1.36 | 1.19-1.56 | <0.01 | 0 | 2.35 | 1.73-3.19 | <0.01 | 0 | ||||
| Head and neck | 3 | 1.20 | 0.86-1.67 | 0.29 | 70.6 | 1.09 | 0.79-1.50 | 0.61 | 49.8 | 1.65 | 0.82-3.32 | 0.16 | 52.7 | 1.16 | 0.81-1.67 | 0.42 | 64.2 | 1.58 | 0.86-2.92 | 0.14 | 40.1 | ||||
| Lung | 2 | 0.72 | 0.34-1.56 | 0.41 | 84.1 | 0.46 | 0.06-3.59 | 0.46 | 93. | 0.29 | 0.06-1.52 | 0.14 | 73.9 | 0.43 | 0.05-3.41 | 0.42 | 94.6 | 0.63 | 0.43-0.92 | 0.02 | 0 | ||||
| Cervical | 2 | 1.35 | 0.90-2.02 | 0.14 | 75.5 | 1.21 | 0.96-1.52 | 0.11 | 0 | 2.20 | 0.70-6.94 | 0.18 | 72.0 | 1.32 | 0.93-1.88 | 0.12 | 54.8 | 2.03 | 0.70-5.90 | 0.19 | 68.4 | ||||
| Breast | 1 | 1.06 | 0.76-1.48 | 0.73 | NA | 1.02 | 0.68-1.52 | 0.92 | NA | 1.28 | 0.45-3.60 | 0.65 | NA | 1.04 | 0.71-1.54 | 0.83 | NA | 1.27 | 0.45-3.55 | 0.65 | NA | ||||
| Design | |||||||||||||||||||||||||
| HB | 12 | 1.20 | 1.00-1.43 | 0.05 | 77.6 | 1.09 | 0.91-1.31 | 0.34 | 65.0 | 1.58 | 0.92-2.72 | 0.10 | 81.4 | 1.15 | 0.93-1.41 | 0.20 | 74.9 | 1.68 | 1.14-2.49 | 0.01 | 69.9 | ||||
| PB | 1 | 1.33 | 0.99-1.78 | 0.06 | NA | 1.24 | 0.85-1.80 | 0.26 | NA | 1.94 | 0.90-4.18 | 0.09 | NA | 1.33 | 0.93-1.89 | 0.12 | NA | 1.80 | 0.84-3.83 | 0.13 | NA | ||||
| Genotype method | |||||||||||||||||||||||||
| PCR-RFLP | 11 | 1.29 | 1.18-1.41 | <0.01 | 39.0 | 1.17 | 1.05-1.31 | <0.01 | 0 | 2.06 | 1.62-2.63 | <0.01 | 31.6 | 1.26 | 1.14-1.40 | <0.01 | 10.5 | 1.96 | 1.54-2.49 | <0.01 | 22.5 | ||||
| Others | 2 | 0.83 | 0.31-2.21- | 0.71 | 96.0 | 0.46 | 0.06-3.49 | 0.45 | 95.7 | 0.55 | 0.04-7.79 | 0.66 | 96.7 | 0.46 | 0.05-4.01 | 0.48 | 96.7 | 1.08 | 0.35-3.31 | 0.90 | 89.6 | ||||
| Adjusted | |||||||||||||||||||||||||
| Total | 12 | 1.32 | 1.18-1.79 | <0.01 | 45.6 | 1.20 | 1.08-1.34 | <0.01 | 0 | 1.58 | 0.93-2.70 | 0.09 | 82.0 | 1.12 | 0.73-1.70 | 0.61 | 87.0 | 1.94 | 1.26-3.00 | <0.01 | 16.6 | ||||
| Sensitive analysis# | 10 | 1.32 | 1.18-1.79 | <0.01 | 45.6 | 1.20 | 1.08-1.34 | <0.01 | 0 | 1.99 | 1.49-2.66 | <0.01 | 32.4 | 1.39 | 1.21-1.61 | <0.01 | 21.8 | 1.94 | 1.26-3.00 | <0.01 | 16.6 | ||||
| Location | |||||||||||||||||||||||||
| Digestive system | 5 | 1.41 | 1.27-1.55 | <0.01 | 0 | 1.30 | 1.12-1.51 | <0.01 | 0 | 2.42 | 1.76-3.33 | <0.01 | 0 | 1.44 | 1.16-1.79 | <0.01 | 14.0 | 2.45 | 1.44-4.15 | <0.01 | 0 | ||||
| Head and neck | 3 | 1.20 | 0.86-1.67 | 0.29 | 71.2 | 1.09 | 0.79-1.51 | 0.60 | 50.4 | 1.65 | 0.82-3.33 | 0.16 | 52.7 | 1.73 | 1.12-2.67 | 0.01 | NA | NA | NA | NA | NA | ||||
| Lung | 1 | NA | NA | NA | NA | NA | NA | NA | NA | 0.14 | 0.07-0.29 | <0.01 | NA | 0.15 | 0.07-0.32 | <0.01 | NA | NA | NA | NA | NA | ||||
| Cervical | 2 | 1.69 | 1.22-2.34 | <0.01 | NA | 1.20 | 0.96-1.51 | 0.12 | 0 | 2.19 | 0.69-6.96 | 0.18 | 72.3 | 1.32 | 0.92-1.88 | 0.13 | 55.6 | 1.22 | 0.57-2.60 | <0.01 | NA | ||||
| Breast | 1 | 0.94 | 0.65-1.35 | 0.74 | NA | 0.98 | 0.66-1.46 | 0.92 | NA | 0.78 | 0.28-2.19 | 0.64 | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||||
| Design | |||||||||||||||||||||||||
| HB | 11 | 1.32 | 1.16-1.50 | <0.01 | 51.6 | 1.18 | 1.05-1.31 | <0.01 | 0 | 1.52 | 0.86-2.69 | 0.15 | 83.3 | 1.01 | 0.62-1.65 | 0.97 | 88.9 | 1.85 | 0.85-4.01 | 0.12 | 57.7 | ||||
| PB | 1 | 1.35 | 1.02-1.79 | 0.04 | NA | 1.64 | 1.08-2.50 | 0.02 | NA | 12.56 | 1.05-6.25 | 0.04 | NA | 1.75 | 1.16-2.65 | <0.01 | NA | 2.08 | 0.87-4.96 | 0.10 | NA | ||||
| Genotype method | |||||||||||||||||||||||||
| PCR-RFLP | 10 | 1.31 | 1.14-1.50 | <0.01 | 49.3 | 1.19 | 1.06-1.33 | <0.01 | 3.7 | 2.00 | 1.55-2.59 | <0.01 | 39.1 | 1.39 | 1.21-1.61 | <0.01 | 21.8 | 1.94 | 1.26-3.00 | <0.01 | 16.6 | ||||
| Others | 2 | 1.42 | 1.21-1.66 | <0.01 | NA | 1.31 | 0.93-1.84 | 0.12 | NA | 0.52 | 0.04-6.70 | 0.62 | 96.6 | 0.15 | 0.07-0.32 | <0.01 | NA | NA | NA | NA | NA | ||||
* Numbers of comparisons
a Test for heterogeneity
# Sensitive analysis without the report of Hu et al.
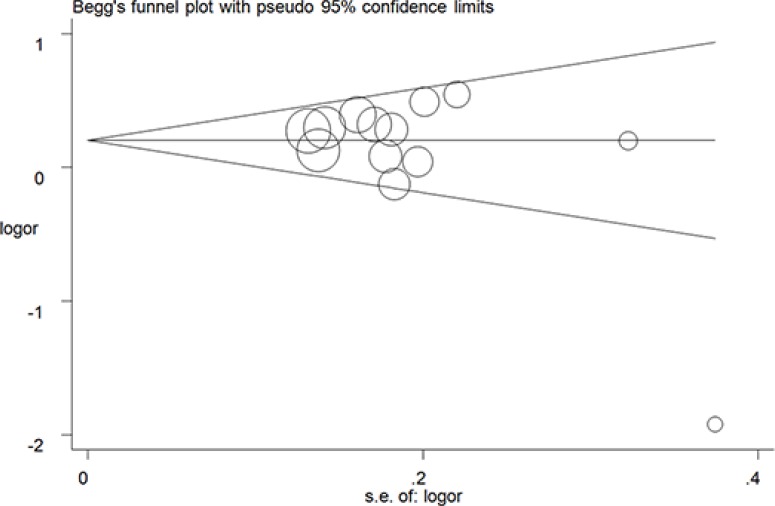
Begg's tests were performed with funnel plot to assess publication bias. No apparently asymmetry was found (Figure 5 for GT + TT vs. GG model) (Supplementary Figure 3), and there results were further guaranteed by Egger's test (T vs. G, P = 0.62; GT vs. GG: P = 0.08; TT vs. GG, P = 0.82; GT + TT vs. GG, P = 0.08; TT vs. GG + GT, P = 0.08).
Figure 5. Funnel plot analysis to detect publication bias for GT + TT vs. GG model of rs712 G > T polymorphism. Circles represent the weight of the studies.
DISCUSSION
In 2012, there were more than 14.1 million new cancer patient and 8.2 million deaths worldwide [26]. Today, cancer is still the most common malignant disease due to death and disability. The incidence of cancer in the developing countries is gradually increasing, with the aging of the population and the deterioration of environmental factors. China has the largest population in the world, and the incidence of cancer in China has been high, leading to a decline in quality of the living standards [2]. The occurrence of cancer is the result of interaction of various factors. Diet, living habits, cell abnormalities, gene mutations are one of the factors that lead to the development of tumor. Different regions, racial diversity, may be the cause of the changes in cancer susceptibility.
Let-7 family has several members, recent studies have found that the Let-7 could function as a tumor suppressor during the development of solid tumors through inhibiting cancer proliferation by targeting some oncogenes/antioncogenes, including KRAS gene Chun-Yan Deng [27, 28]. KRAS is one of the critical oncogene, which locates at 12p12.1. Some single nucleotide polymorphisms (SNPs) residing in 3′ UTRs of KRAS gene have been found effected cancer risk through altering the activation of KRAS gene. Such as the KRAS-LCS (rs61764370) polymorphism had been proved to influence the KRAS transcription, resulting in an increased KRAS expression in non-small cell lung cancer [29]. Today, the interaction mechanism of many polymorphism locus located in different genes sites is more and more attracting our attentions. A large number studies had suggested that, many gene mutations, such as BRAF, E-Cadherin, and TP53 genes play an important role during the process of cancer development. The synergistic effects of these polymorphisms maybe initiate the procedure of abnormal changes of normal cells, and to accelerate the formation of the tumor solid [30–32].
Recently, the SNP of rs712 G > T polymorphism in the let-7-KRAS binding site has been reported and drawn more attentions. Kim et al. reported that the rs712 G allele would downgrade more than 15% activity compared with rs712 T allele with luciferase reporter in vitro [33]. In 2010, Peng et al. conducted the first case-control study and didn't found any significant association between the patients with lung cancer and healthy controls in a Chinese population [14]. Since then, a lot of case-control studies have been conducted, but the conclusions were not consistent. In 2014, Ying et al. conducted the first meta-analysis including only six case-controls studies. The results demonstrated that no significant association was found between rs712 polymorphism and cancer susceptibility in the overall population, but the subgroup analysis found that the allele T (T vs. G: OR = 1.33, 95% CI = 1.08–1.64, P = 0.01) and dominant genotype (GT + TT vs. GG: OR = 1.30, 95% CI = 1.11–1.55, P < 0.01) would increase the risk of cancer in Chinese population. Moreover, Zhao et al conducted another meta-analysis and obtained the similar results with the same six studied. However, there were only five case-controls of Chinese population were included in the two meta-analysis. No further subgroup analysis of cancer location, control resource, and genotype methods was conducted owe to the limited number of researches and small sample size. To our knowledge, meta-analysis is a scientific statistical method to draw more precise results through expanding sample with as much as possible homogeneity studies. Today, there are seven new researcher articles on the association between the rs712 G > T polymorphism and cancer risk had been published, and all these seven articles focused on Chinese population. So, we conducted the updated systematic meta-analysis to answer this question “Does this mutation of rs712 G > T increase the susceptibility of cancer in Today's China population?” fatherly. Our results demonstrate that the rs712 G > T polymorphism might be increase the digestive system cancer risk in the Chinese population. Furthermore, there were three studies focused on colorectal cancer and the results of meta-analysis indicated that the rs712 G > T might be associate with increased colorectal cancer risk (T vs. G: OR = 1.41, 95% CI = 1.23–1.61, P < 0.01, I2 = 0%; GT vs. GG: OR = 1.25, 95% CI = 1.05–1.48, P = 0.01, I2 = 0%; TT vs. GG: OR = 2.52, 95% CI = 1.71–3.71, P < 0.01, I2 = 0%; GT + TT vs. GG: OR = 1.37, 95% CI = 1.16–1.61, P < 0.01, I2 = 0%; TT vs. GG + GT: OR = 2.34, 95% CI = 1.59–3.42, P < 0.01, I2 = 0%). More researches, larger sample size and further subgroup analysis were included in our meta-analysis, in order to enhance statistical efficiency and increase the accuracy of the effect. Furthermore, we extracted the adjusted data (OR and 95% CI) and pooled them for investigating the interactions between genetic polymorphisms and environmental risk factors. These results were almost consistent with the former results that we conducted with unadjusted data.
In this meta-analysis, our result demonstrated that the G allele would increase the cancer susceptibility in Chinese population with some apparently heterogeneities. Through the sensitivity analysis, we found the data of Hu et al. maybe the main reason leading to heterogeneity through reviewing of the minor allele frequency of controls [11]. The results indicated that the frequency distribution of minor allele frequency in his article was higher than other studies. And the heterogeneity was alleviated through deleting the data of Hu et al. In the next subgroup analysis, the pooled results indicated that the rs712 G > T polymorphism was associated with the development of digestive system cancer. This difference suggested that gene mutations may be associated with a specific susceptibility to some cancer, which may provide us an important approach to early screening and prevention in the future.
Moreover, some limitations of this meta-analysis should to be raised. Firstly, only one SNP locus (rs712 G > T) was analysis in this article, and the statistical calculations were conducted without other risk factors, such as lifestyle, bad habits (smoking and drinking), environmental deterioration, and other gene mutations. Secondly, the racial bias could not be eliminated due to only Chinese population was included in this research. And the conclusion should be tested before applying to other populations again. Third, although we have included thirteen studies, the sample size is still insufficient, which could made some deviations from the truly results.
In conclusion, our meta-analysis suggest that the rs712 G > T polymorphism is associated with an increased cancer risk in Chinese population, especially in digestive system cancer. Additional studies with large sample sizes in other ethnic populations are needed to guarantee our findings further.
MATERIALS AND METHODS
This meta-analysis was conducted following Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [34]. All included data were collected from published studies, and no ethical issues were involved.
Search strategy
Four electronic databases (PubMed, Embase, CNKI and Wanfang database) were searched with following terms: “let”, “KRAS”, “polymorphism”, and “cancer” through the review of “let-7[All Fields] AND (“proto-oncogene proteins p21(ras)”[MeSH Terms] OR (“proto-oncogene”[All Fields] AND “proteins” [All Fields] AND “p21(ras)”[All Fields]) OR “proto-oncogene proteins p21(ras)”[All Fields] OR “kras”[All Fields]) AND (“polymorphism, genetic”[MeSH Terms] OR (“polymorphism”[All Fields] AND “genetic”[All Fields]) OR “genetic polymorphism”[All Fields] OR “polymorphism”[All Fields]) AND (“neoplasms”[MeSH Terms] OR “neoplasms”[All Fields] OR “cancer”[All Fields])”, up to 10 June, 2016. Only studies written with English and Chinese were selected.
Study selection
All included studies according to these inclusion criteria: (1) case-control studies; (2) research focus on rs712 G > T polymorphism and cancer risk; and (3) the publication with adequate information to calculate the odds ratio (OR) and 95% confidence interval (CI). The exclusion criteria: (1) review studies; (2) molecular fundamental studies; (3) data not about the related locus or with in sufficient outcome; and (4) duplicated or overlapping data of the same author or issue.
Data extraction
Two independent reviewers (Du and Xie) review and extracted the relevant the data from all included studies: the first author's name, study published date, sources of controls, genotyping method, cancer location, adjusted OR and its 95% CI, MAF (Minor allele frequency) in cases and controls, and the distributed number of genotypes in cases and controls. Quality assessment of included studies was evaluated with modified Newcastle-Ottawa scale (NOS) by two authors, and the scores ranged from 0 points (worst) to 9 points (best) (Table 3) [35].
Table 3. Scale for quality assessment.
| Criteria | Score |
|---|---|
| Representativeness of cases | |
| Consecutive/randomly selected form case population with clearly defined sampling frame | 2 |
| Consecutive/randomly selected form case population without clearly defined sampling frame or with extensive | 1 |
| Not described | 0 |
| Source of controls | |
| Population- or Healthy-based | 2 |
| Hospital-bases | 1 |
| Not described | 0 |
| Hardy-Weinberg equilibrium in controls | |
| Hardy-Weinberg equilibrium | 2 |
| Hardy-Weinberg disequilibrium | 1 |
| Genotyping examination | |
| Genotyping done under “blinded” condition | 1 |
| Unblinded done or not mentioned | 0 |
| Association assessment | |
| Assess association between genotypes and cancer with appropriate statistics and adjustment for confounders | 2 |
| Assess association between genotypes and cancer with appropriate statistics and without adjustment for confounders | 1 |
| Inappropriate statistics used | 0 |
Statistical analysis
First, Hardy-Weinberg equilibrium (HWE) in controls of every included study was calculated by Chi-square test. Second, Crude ORs with 95% CIs were used to assess the association between rs712 G > T polymorphism and cancer risk. Five genetic models were analyses, involving allele contrast (T vs. G), co-dominant (GT vs. GG and TT vs. GG), dominant (GT + TT vs. GG) and recessive (TT vs. GG + GT) models. Stratified assessments were calculated based on cancer location, control resource, and genotype methods. Heterogeneity between studies was calculated with the I2 value and Cochran's Q test. The fixed-effect model (the Mantel-Haenszel method) was applied when the I2 value less than 50% and P > 0.10 for the Q test; otherwise, a random effects model (the DerSimonian and Laird method) was adopted. Third, cumulative meta-analyses were conducted to identify a possible trend of the pooled results with new studies added. Sensitivity analyses were also conducted to examine the stability of the results through deleting each study one by one. Finally, publication bias was assessed with Begg's funnel plot and Egger's test. All statistical analyses were performed using STATA version 14.0 (Stata Corporation, College Station, TX, USA). A P value < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIALS FIGURES
ACKNOWLEDGMENTS AND FUNDING
This study was supported by grants from the Foundation of Ministry of Education of Hubei Province (D20142102), Natural Science Foundation of Hubei province (2016CFB567), and Taihe Hospital (2013PY02, EBM2013006). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Kinoshita H, Maeda I, Morita T, Miyashita M, Yamagishi A, Shirahige Y, Takebayashi T, Yamaguchi T, Igarashi A, Eguchi K. Place of death and the differences in patient quality of death and dying and caregiver burden. J Clin Oncol. 2015;33:357–363. doi: 10.1200/JCO.2014.55.7355. [DOI] [PubMed] [Google Scholar]
- 4.Mallath MK, Taylor DG, Badwe RA, Rath GK, Shanta V, Pramesh CS, Digumarti R, Sebastian P, Borthakur BB, Kalwar A, Kapoor S, Kumar S, Gill JL, et al. The growing burden of cancer in India: epidemiology and social context. Lancet Oncol. 2014;15:e205–212. doi: 10.1016/S1470-2045(14)70115-9. [DOI] [PubMed] [Google Scholar]
- 5.Tsilidis KK, Papadimitriou N, Capothanassi D, Bamia C, Benetou V, Jenab M, Freisling H, Kee F, Nelen A, O'Doherty MG, Scott A, Soerjomataram I, Tjonneland A, et al. Burden of Cancer in a Large Consortium of Prospective Cohorts in Europe. J Natl Cancer Inst. 2016:108. doi: 10.1093/jnci/djw127. [DOI] [PubMed] [Google Scholar]
- 6.van der Weyden L, Arends MJ, Rust AG, Poulogiannis G, McIntyre RE, Adams DJ. Increased tumorigenesis associated with loss of the tumor suppressor gene Cadm1. Mol Cancer. 2012;11:29. doi: 10.1186/1476-4598-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall H. Genetic and epigenetic factors in development of lung cancer. Lancet Oncol. 2012;13:1188. doi: 10.1016/s1470-2045(12)70523-5. [DOI] [PubMed] [Google Scholar]
- 8.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Kranenburg O. The KRAS oncogene: past, present, and future. Biochim Biophys Acta. 2005;1756:81–82. doi: 10.1016/j.bbcan.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Hu H, Zhang L, Teng G, Wu Y, Chen Y. A variant in 3′-untranslated region of KRAS compromises its interaction with hsa-let-7g and contributes to the development of lung cancer in patients with COPD. Int J Chron Obstruct Pulmon Dis. 2015;10:1641–1649. doi: 10.2147/COPD.S83596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying HQ, Wang F, He BS, Pan YQ, Gao TY, Xu YQ, Li R, Deng QW, Sun HL, Wang SK. The involvement of Kras gene 3′-UTR polymorphisms in risk of cancer and influence on patient response to anti-EGFR therapy in metastatic colorectal cancer: a meta-analysis. Onco Targets Ther. 2014;7:1487–1496. doi: 10.2147/OTT.S65496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao WH, Qu XF, Xing ZG, Zhao LQ, Qin L, Lv C. Association of rs712 polymorphism in Kras gene 3′-luntranslated region and cancer risk: a meta-analysis. J BUON. 2015;20:309–316. [PubMed] [Google Scholar]
- 14.Peng X, Zhao J, Lei Z, Liu R, Liu Z, Jiang X, Zhang H. Association of A SNP in MicroRNA Let-7 Complementary Region in KRAS 3′UTR with Non-small Cell Lung Cancer (Chinese) Suzhou University Journal of Medical Science. 2010:30. [Google Scholar]
- 15.Li ZH, Pan XM, Han BW, Guo XM, Zhang Z, Jia J, Gao LB. A let-7 binding site polymorphism rs712 in the KRAS 3′ UTR is associated with an increased risk of gastric cancer. Tumour Biol. 2013;34:3159–3163. doi: 10.1007/s13277-013-0885-x. [DOI] [PubMed] [Google Scholar]
- 16.Yan L, Wang Q, Tian K. Association between the single nucleotide polymorphisms of the let-7 target gene KRAS-binding site rs712 and risk of glioma (Chinese) Chinese Journal of Cancer Prevention and Treatment. 2013;20:811–814. [Google Scholar]
- 17.Pan XM, Sun RF, Li ZH, Guo XM, Zhang Z, Qin HJ, Xu GH, Gao LB. A let-7 KRAS rs712 polymorphism increases colorectal cancer risk. Tumour Biol. 2014;35:831–835. doi: 10.1007/s13277-013-1114-3. [DOI] [PubMed] [Google Scholar]
- 18.Pan XM, Jia J, Guo XM, Li ZH, Zhang Z, Qin HJ, Xu GH, Gao LB. Lack of association between let-7 binding site polymorphism rs712 and risk of nasopharyngeal carcinoma. Fam Cancer. 2014;13:93–97. doi: 10.1007/s10689-013-9681-4. [DOI] [PubMed] [Google Scholar]
- 19.Jin H, Liang Y, Wang X, Zhu J, Sun R, Chen P, Nie X, Gao L, Zhang L. Association between a functional polymorphism rs712 within let-7-binding site and risk of papillary thyroid cancer. Med Oncol. 2014;31:221. doi: 10.1007/s12032-014-0221-3. [DOI] [PubMed] [Google Scholar]
- 20.Ni S, Chen X, Sun L. Association between a single nucleotide polymorphism within let-7 target gene KRAS and risk of cervical cancer (Chinese) Chinese Journal of Birth Health & Heredity. 2015;23:24–25. [Google Scholar]
- 21.Liang Y, Sun R, Li L, Yuan F, Liang W, Wang L, Nie X, Chen P, Zhang L, Gao L. A Functional Polymorphism in the Promoter of MiR-143/145 Is Associated With the Risk of Cervical Squamous Cell Carcinoma in Chinese Women: A Case-Control Study. Medicine (Baltimore) 2015;94:e1289. doi: 10.1097/MD.0000000000001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai Q, Wei HL, Huang J, Zhou TJ, Chai L, Yang ZH. KRAS polymorphisms are associated with survival of CRC in Chinese population. Tumour Biol. 2016;37:4727–4734. doi: 10.1007/s13277-015-4314-1. [DOI] [PubMed] [Google Scholar]
- 23.Xiong D, Song YP, Xiong W, Liang YD. An let-7 KRAS rs712 polymorphism increases hepatocellular carcinoma risk. Genet Mol Res. 2015;14:14050–14055. doi: 10.4238/2015.October.29.24. [DOI] [PubMed] [Google Scholar]
- 24.Jiang QH, Peng HX, Zhang Y, Tian P, Xi ZL, Chen H. rs712 polymorphism within let-7 microRNA-binding site might be involved in the initiation and progression of colorectal cancer in Chinese population. Onco Targets Ther. 2015;8:3041–3045. doi: 10.2147/OTT.S89746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X, Yang Y, Guo Y, Cao ZL, Cui ZW, Hu TC, Gao LB. Association of a let-7 KRAS rs712 polymorphism with the risk of breast cancer. Genet Mol Res. 2015;14:16913–16920. doi: 10.4238/2015.December.14.19. [DOI] [PubMed] [Google Scholar]
- 26.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 27.Barh D, Malhotra R, Ravi B, Sindhurani P. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr Oncol. 2010;17:70–80. doi: 10.3747/co.v17i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 29.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palacio-Rua KA, Isaza-Jimenez LF, Ahumada-Rodriguez E, Muneton-Pena CM. Genetic analysis in APC, KRAS, and TP53 in patients with stomach and colon cancer. Rev Gastroenterol Mex. 2014;79:79–89. doi: 10.1016/j.rgmx.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Phua LC, Ng HW, Yeo AH, Chen E, Lo MS, Cheah PY, Chan EC, Koh PK, Ho HK. Prevalence of KRAS, BRAF, PI3K and EGFR mutations among Asian patients with metastatic colorectal cancer. Oncol Lett. 2015;10:2519–2526. doi: 10.3892/ol.2015.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan P, Manna A, Saha S, Mohanty S, Mukherjee S, Mazumdar M, Guha D, Das T. Aspirin inhibits epithelial-to-mesenchymal transition and migration of oncogenic K-ras-expressing non-small cell lung carcinoma cells by down-regulating E-cadherin repressor Slug. BMC Cancer. 2016;16:39. doi: 10.1186/s12885-016-2078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim M, Chen X, Chin LJ, Paranjape T, Speed WC, Kidd KK, Zhao H, Weidhaas JB, Slack FJ. Extensive sequence variation in the 3′ untranslated region of the KRAS gene in lung and ovarian cancer cases. Cell Cycle. 2014;13:1030–1040. doi: 10.4161/cc.27941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niu YM, Du XY, Cai HX, Zhang C, Yuan RX, Zeng XT, Luo J. Increased risks between Interleukin-10 gene polymorphisms and haplotype and head and neck cancer: a meta-analysis. Sci Rep. 2015;5:17149. doi: 10.1038/srep17149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.