Abstract
Two‐wing flyingfish (Exocoetus spp.) are widely distributed, epipelagic, mid‐trophic organisms that feed on zooplankton and are preyed upon by numerous predators (e.g., tunas, dolphinfish, tropical seabirds), yet an understanding of their speciation and systematics is lacking. As a model of epipelagic fish speciation and to investigate mechanisms that increase biodiversity, we studied the phylogeny and biogeography of Exocoetus, a highly abundant holoepipelagic fish taxon of the tropical open ocean. Morphological and molecular data were used to evaluate the phylogenetic relationships, species boundaries, and biogeographic patterns of the five putative Exocoetus species. We show that the most widespread species (E. volitans) is sister to all other species, and we find no evidence for cryptic species in this taxon. Sister relationship between E. monocirrhus (Indo‐Pacific) and E. obtusirostris (Atlantic) indicates the Isthmus of Panama and/or Benguela Barrier may have played a role in their divergence via allopatric speciation. The sister species E. peruvianus and E. gibbosus are found in different regions of the Pacific Ocean; however, our molecular results do not show a clear distinction between these species, indicating recent divergence or ongoing gene flow. Overall, our phylogeny reveals that the most spatially restricted species are more recently derived, suggesting that allopatric barriers may drive speciation, but subsequent dispersal and range expansion may affect the distributions of species.
Keywords: cryptic speciation, epipelagic, species delimitation
1. Introduction
Marine fish habitats are typically large, continuous, and lack definitive boundaries. Fishes that inhabit the epipelagic zone are generally less taxonomically diverse than species found in other habitats (benthic, coastal, reef‐associated, estuarine), possibly because the overall homogeneity of epipelagic habitats may reduce rates of speciation (Hamner, 1995). Nevertheless, some widespread and diverse fish families such as scombrids, belonids, hemiramphids, and exocoetids have circumtropical distributions that include a diversity of habitats (Gaither et al., 2015). The underlying mechanisms responsible for diversification in these fishes remain unclear, at least in part because their phylogenetic relationships are poorly resolved and life history characteristics little known. Phylogenetic characterizations are necessary to understand speciation because they define the sequence of lineage and species diversification. Also, phylogenies can clarify species identity when taxa are morphologically very similar (cryptic species), thereby improving understanding of species geographic distributions (Bass et al., 2005; Colborn et al., 2001; Quattro et al., 2005).
Comprehensive species phylogenies can provide key insights regarding speciation in marine lineages with high dispersal potential, wide ranges, and overlapping distributions. Exocoetus (two‐wing flyingfish) is a monophyletic genus of five species found in the epipelagic waters of tropical and subtropical oceans worldwide (Lewallen et al., 2011; Parin & Shakhovskoy, 2000). Gliding on elongated pectoral fins (Figure 1) separates Exocoetus from most other members of the family Exocoetidae that can use elongated pectoral, pelvic, and sometimes dorsal fins to achieve prolonged aerial glides. As with many widely distributed fishes, Exocoetus has buoyant, pelagic eggs, and larvae that persist in the epipelagic zone during maturation, which occurs at lengths of 130–155 mm (SL) (Grudtsev et al., 1987). Exocoetus individuals live for approximately 1 year, are small [max SL ≤ 207 mm (Grudtsev et al., 1987)], slow swimming, and incapable of long‐distance migrations (Parin, 1968). Curiously, the distribution of each Exocoetus species overlaps with at least one other species, suggesting they may have evolved in parapatry or sympatry. Species ranges vary from circumtropical (e.g., E. volitans) to single oceanographic regions (e.g., E. peruvianus), indicating differences in habitat specialization. Although three species of Exocoetus were traditionally recognized [E. volitans, E. monocirrhus, E. obtusirostris (Kovalevskaya, 1982; Parin, 1961)], two cryptic species previously grouped within E. obtusirostris have been described more recently (E. peruvianus and E. gibbosus) (Lewallen et al., 2011; Parin & Shakhovskoy, 2000). A phylogenetic hypothesis for Exocoetus (Parin & Shakhovskoy, 2000) that was previously proposed based on 11 morphological characters (Figure 2a) has not been tested using strict inference methods or molecular data.
Figure 1.

An Exocoetus fish gliding along the surface of epipelagic water in the eastern tropical Pacific. Photo credit: EAL (first author)
Figure 2.

Morphology‐based phylogenetic hypotheses for Exocoetus. (a) Phylogenetic hypothesis presented by Parin and Shakhovskoy (2000). Illustrations of adults and juveniles were compiled from the following publications: Exocoetus volitans (Parin, 2002), Exocoetus obtusirostris (Parin, 2002), Exocoetus monocirrhus adult (Parin, 1984); Exocoetus monocirrhus juvenile (Heemstra & Parin, 1986), Exocoetus peruvianus (Parin & Shakhovskoy, 2000), Exocoetus gibbosus (Parin & Shakhovskoy, 2000). (b) Phylogenetic hypothesis using the same 11 morphological characters as Parin and Shakhovskoy (2000), with added morphological data from outgroups Fodiator acutus and Fodiator rostratus; strict consensus of 4 equally parsimonious trees of 12 steps each
A thorough examination of genetic diversity in Exocoetus is greatly needed, considering the potential for uncovering cryptic species (especially within the globally distributed E. volitans). Here, through extensive sampling and phylogenetic analysis, we improve the resolution of evolutionary lineages within Exocoetus, thereby providing new data on how speciation occurs in the epipelagic zone. We specifically focused on the following questions: (1) What are the phylogenetic relationships within Exocoetus based on molecular data, and how do they compare to the most recent morphological hypothesis? (2) Do the currently recognized Exocoetus species represent distinct monophyletic lineages, and are there cryptic species? (3) What biogeographic patterns of speciation are revealed by phylogenetic arrangements within this genus?
2. Materials and Methods
2.1. Taxon sampling
A total of 429 flyingfish specimens (422 Exocoetus and seven outgroup specimens) were collected at night using long‐handled dipnets and/or donated by collaborators (Appendix S1). Animals were euthanized in an ice‐water bath. Post‐mortem handling included shipboard freezing in seawater, removal of lateral muscle tissue for DNA analysis (95% ethanol), whole‐specimen fixation (10% formalin), and long‐term museum archiving (70% ethanol). Each specimen was identified using key diagnostic characters (e.g., gill raker counts and body depth measurements) as presented in (Parin & Shakhovskoy, 2000). All voucher specimens are archived with catalogue numbers at the Royal Ontario Museum or Scripps Institution of Oceanography (Appendix S1). We note that 15 specimens used in the current study were included in a previous study (Lewallen et al., 2011). Also, 266 E. volitans specimens were sequenced (Cytb) for a previous population genetic analysis (Lewallen et al., 2016). Details regarding which specimens are common among studies are provided in Appendix S1.
2.2. Morphological data
Parin and Shakhovskoy (2000) presented a series of morphological characters for Exocoetus, and a phylogenetic hypothesis for the genus (Figure 2a). However, their study used dichotomous morphological character analyses to discern species rather than explicit phylogenetic analyses. Importantly, only 11 characters in Parin & Shakhovskoy's study were informative for distinguishing species and could be clearly coded for phylogenetic analysis. To test the morphology‐based phylogeny for this genus, we tabulated the characters presented in Parin and Shakhovskoy (2000) into a data matrix (Table 1). Data for the 11 characters for two outgroup taxa (Fodiator acutus and F. rostratus) were obtained from the literature (Parin & Belyanina, 2002; Parin & Shakhovskoy, 2000; Table 1).
Table 1.
Morphological character matrix for the 11 characters described by Parin and Shakhovskoy (2000). Characters were coded as binary (1 or 0). Outgroup taxa (Fodiator acutus and F. rostratus) were added to this matrix using morphological data presented by Parin and Belyanina (2002) and Parin and Shakhovskoy (2000)
| Morphological characters | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| E. volitans | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| E. monocirrhus | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| E. obtusirostris | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 |
| E. peruvianus | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| E. gibbosus | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| F. acutus | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| F. rostratus | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
Character Coding: 1, Supraoccipital with one posterior process; 2, Presence of posteromedian process on cleithrum; 3, Humpbacked juveniles; 4, Ventral fins are anteriorly shifted; 5, Increased number of scales in transverse row; 6, Shortened ventral fins; 7, Jaw teeth much reduced; 8, Absence of barbel in juveniles; 9, High body depth in juveniles; 10, Maximum development of posterolateral process on cleithrum; 11, High number of rays in pectoral fin.
2.3. Molecular data
For phylogenetic analysis of Exocoetus, mitochondrial encoded cytochrome b gene (Cytb; 1,082 bps) and nuclear recombination activating gene 2 (Rag2; 882 bps) sequence data were obtained for 14 individuals including 2 representatives of each Exocoetus species and 4 outgroup specimens (Cheilopogon xenopterus, F. rostratus, Hirundichthys marginatus, Parexocoetus brachypterus; Table 2). To test for cryptic speciation, we generated an expanded molecular dataset by collecting mitochondrial sequence data (Cytb) for 422 Exocoetus specimens (266 E. volitans, 9 E. peruvianus, 2 E. gibbosus, 9 E. obtusirostris, and 136 E. monocirrhus), and 4 outgroup specimens (2 P. hillianus and 2 P. brachypterus). The globally distributed E. volitans was collected from the Atlantic (n = 150), Pacific (n = 111), and Indian (n = 5) Oceans. For E. monocirrhus, we collected individuals from the eastern and central Pacific (n = 131), as well as Indian (n = 5) Ocean. E. obtusirostris specimens (n = 9) were obtained from the Atlantic Ocean and Gulf of Mexico. Because they have restricted distributions, only small numbers of E. peruvianus (n = 9) and E. gibbosus (n = 2) were obtained from waters of the Peruvian Upwelling Current and South Pacific Subtropical Gyre, respectively.
Table 2.
Exocoetus specimens with cytochrome b (Cytb) and recombination activating gene 2 (Rag2) sequence data. Ingroup and outgroup taxa are specified and voucher catalogue numbers, collection localities, Genbank accession numbers, and citations are listed
| Specimen data | Genbank accession number | ||||||
|---|---|---|---|---|---|---|---|
| Genus | Species | Specimen No. | Voucher No. | Locality | Cytb | Rag2 | Citation |
| Exocoetus | gibbosus | 3717 | ROM‐79289 | ETP | KY382508 | KY385897 | This study |
| Exocoetus | gibbosus | 4568 | ROM‐92584 | ETP | KY382509 | KY385898 | This study |
| Exocoetus | monocirrhus | 1572 | SIO‐07‐129 | ETP | HQ325628 | HQ325695 | Lewallen et al. (2011) |
| Exocoetus | monocirrhus | 5801 | ROM‐79270 | ETP | HQ325629 | HQ325696 | Lewallen et al. (2011) |
| Exocoetus | obtusirostris | 1851 | NMNH380590 | Atlantic | HQ325630 | HQ325697 | Lewallen et al. (2011) |
| Exocoetus | obtusirostris | 1854 | NMNH380574 | Atlantic | HQ325631 | HQ325698 | Lewallen et al. (2011) |
| Exocoetus | peruvianus | 1611 | SIO‐07‐125 | ETP | HQ325632 | HQ325699 | Lewallen et al. (2011) |
| Exocoetus | peruvianus | 1612 | SIO‐07‐125 | ETP | HQ323633 | HQ325700 | Lewallen et al. (2011) |
| Exocoetus | volitans | 1585 | SIO‐07‐132 | ETP | HQ325634 | HQ325701 | Lewallen et al. (2011) |
| Exocoetus | volitans | 1586 | SIO‐07‐132 | ETP | HQ325635 | HQ325702 | Lewallen et al. (2011) |
| Cheilopogon | xenopterus | 3785 | ROM‐79248 | ETP | HQ325621 | HQ325688 | Lewallen et al. (2011) |
| Fodiator | rostratus | 1570 | SIO‐07‐128 | ETP | HQ325638 | HQ325705 | Lewallen et al. (2011) |
| Hirundichthys | marginatus | 3181 | ROM‐79330 | ETP | HQ325644 | HQ325711 | Lewallen et al. (2011) |
| Parexocoetus | brachypterus | 4148 | ROM‐79331 | ETP | HQ325656 | HQ325723 | Lewallen et al. (2011) |
ETP, Eastern Tropical Pacific.
Genomic DNA was extracted using DNeasy kits (Qiagen, Valencia, CA, USA). A portion of both the Cytb and Rag2 genes were amplified using previously published primers ExoCBFwd, ExoCBRev, and Ffly‐Ch, Rfly‐Ch, respectively (Lewallen et al., 2011). One advantage of using Rag2 over some other nuclear genes is that it does not contain introns in the coding region (Peixoto, Mikawa, & Brenner, 2000). PCR conditions, internal sequencing primers (ExoFwd1 and ExoRev1 for Cytb; F16‐Ch and R17‐Ch for Rag2), and sequence alignment methods followed Lewallen et al. (2011).
2.4. Maximum Parsimony
Maximum parsimony (MP) analyses were conducted on the following five datasets using PAUP* 4.10b (Swofford, 2000). Morphological and genetic data were concatenated using MacClade 4.07 (Maddison & Maddison, 2005): Set 1: morphological data, 7 taxa, 11 characters (Parin & Belyanina, 2002; Parin & Shakhovskoy, 2000; Table 1); Set 2: Cytb data, 14 specimens, 1,137 bps each (Table 2); Set 3: Rag2 data, 14 specimens, 882 bps each (Table 2); Set 4: All data combined, 14 specimens, morphology, Cytb, Rag2 (Tables 1 and 2); Set 5: An expanded Cytb dataset, 427 specimens, 1,082 bps each (Appendix S1). Morphological data (Set 1) were analyzed using the exhaustive search algorithm (MP) within PAUP* 4.10b (Swofford, 2000). Fodiator acutus and Fodiator rostratus were defined as outgroup taxa and a strict consensus of the four most parsimonious trees was generated.
For MP analysis of Cytb (Set 2) and Rag2 (Set 3) data, we used heuristic searches (10,000 random addition sequence replicates and TBR branch swapping). Cheilopogon xenopterus, Hirundichthys marginatus, Parexocoetus brachypterus, and Fodiator rostratus were defined as outgroups, and strict consensus trees were calculated. Support for nodes was measured by performing 100 bootstrap replicates (BS), with 10,000 random addition sequence replicates per bootstrap iteration. For the combined analysis (Set 4), C. xenopterus, H. marginatus, P. brachypterus, and F. rostratus were defined as outgroup taxa. A heuristic search using 10,000 random addition sequence replicates and TBR branch swapping was performed. Bootstrap support was calculated with 100 bootstrap replicates and 10,000 random addition sequence replicates per bootstrap iteration. The expanded Cytb dataset (Set 5) was analyzed using a heuristic search of 1,000 random addition sequence replicates, and TBR branch swapping. For this analysis, four non‐Exocoetus sequences were included (2 P. hillianus and 2 P. brachypterus) and designated as outgroups (Appendix S2).
2.5. Bayesian inference
We analyzed Sets 2 through 5 (see above) using Bayesian inference (BI) implemented by BEAST 1.6.1 (Drummond & Rambaut, 2007). Outgroup taxa for each dataset were the same as in MP analyses above. As in previous phylogenetic analyses of these taxa and molecular markers (Lewallen et al., 2011), a general time reversible model with invariant sites and gamma distribution (GTR + I + Γ) was determined as the best model of evolution, and was used for this study. Using a random starting tree, 10 million MCMC generations were run, saving one of every 1,000 trees, and the first 10% of saved trees were discarded as burn‐in. TRACER 1.4 (Rambaut & Drummond, 2007a) was used to view the posterior distribution of sampled trees and assess convergence, and TreeAnnotator 1.4 (Rambaut & Drummond, 2007b) was used to calculate a maximum clade credibility tree. Phylograms were generated using TreeView (Page, 1996), with branch lengths corresponding to substitutions per site and Bayesian posterior probabilities (BPP) presented at each node.
2.6. Genetic distance
To estimate genetic distances among sampled individuals, mean Kimura two‐parameter (K2P) values (Kimura, 1980) were calculated using MEGA 5 (Tamura et al., 2011). All possible pairwise comparisons were calculated among individuals within each species, and also between each species. Between‐species genetic distance estimates were then used to obtain an overall mean for the genus.
3. Results
3.1. Maximum parsimony
Our MP analysis of the 11 morphological characters (Set 1) used by Parin and Shakhovskoy (Parin & Shakhovskoy, 2000) provided limited phylogenetic resolution (Figure 2b). The tree contains a polytomy of four Exocoetus species (E. gibbosus, E. peruvianus, E. obtusirostris, and E. monocirrhus), and this lineage is placed in an unresolved polytomy with E. volitans and F. rostratus (Figure 2b). Our MP analyses that included molecular data for 14 individuals (Sets 2–4) support the monophyly of Exocoetus (Figures 3a,c, 4a). These analyses supported E. volitans as the sister taxon to all other Exocoetus species with BS ≥ 98. The arrangement of E. monocirrhus as sister to an E. obtusirostris – E. peruvianus – E. gibbosus clade proposed by Parin and Shakhovskoy (Parin & Shakhovskoy, 2000) (Figure 2a) is not supported by any of the MP analyses conducted here, including analysis involving morphological characters (Figures 2b, 3a,c, 4a). Similarly, the grouping of E. obtusirostris as sister to E. gibbosus – E. peruvianus (Figure 2a) is also not supported (Figures 2b, 3a,c, 4a). Instead, our analyses show sister clades of E. monocirrhus – E. obtusirostris and E. peruvianus – E. gibbosus with high support (BS ≥ 98) in the combined analysis (Figure 4a), and independent Cytb (Figure 3a) and Rag2 (Figure 3c) phylogenies.
Figure 3.

Phylogenetic analyses of Exocoetus specimens using each gene (Cytb = cytochrome b, Rag2 = recombination activating gene 2) and inference method (MP = maximum parsimony using PAUP* 4.10b (Swofford, 2000), BI = Bayesian inference using BEAST 1.6.1 (Drummond & Rambaut, 2007)). Bootstrap proportions (BS) and Bayesian posterior probabilities (BPP) are listed above nodes. Cheilopogon xenopterus, Hirundichthys marginatus, Parexocoetus brachypterus, and Fodiator rostratus were used as outgroup taxa. (a) MP analysis of Cytb data, (b) BI analysis of Cytb data, (c) MP analysis of Rag2 data, (d) BI analysis of Rag2 data
Figure 4.
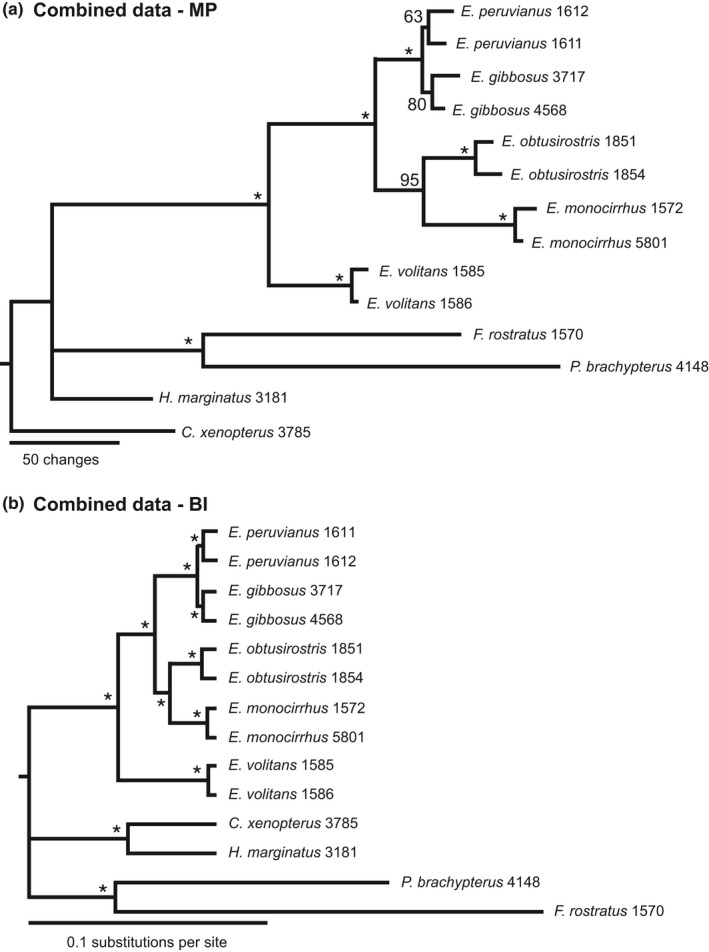
Combined evidence phylogenies analyzed using maximum parsimony (MP) and Bayesian inference (BI) methods and 2,030 total characters (Cytb = 1,137, Rag2 = 882, morphology = 11). Cheilopogon xenopterus, Hirundichthys marginatus, Parexocoetus brachypterus and Fodiator rostratus were used as outgroup taxa. (a) MP phylogeny with bootstrap proportions (BS) listed next to each node (*100). (b) BI phylogeny with branch lengths corresponding to the number of base pair differences between sequences. *indicates Bayesian posterior probabilities ≥0.96
The monophyletic grouping of multiple representatives of each species was generally observed in the Cytb (Figure 3a, BS ≥ 79) and combined analyses (Figure 4a, BS ≥ 63). However, analysis of Rag2 data did not resolve E. gibbosus and E. peruvianus individuals as distinct species (Figure 3c), and E. gibbosus individuals were not monophyletic in the Cytb analysis (Figure 3a).
Maximum parsimony analysis of the expanded Cytb dataset (Set 5) including 426 individuals (1,082 total bps, 319 parsimony informative) grouped multiple individuals of E. volitans, E. obtusirostris, and E. monocirrhus into monophyletic clades corresponding to species, but did not clearly segregate E. peruvianus from E. gibbosus (Appendix S2). Rather, E. gibbosus individuals were nested within E. peruvianus. This topology shows E. volitans as sister to all other Exocoetus species, and E. monocirrhus and E. obtusirostris as sister taxa.
3.2. Bayesian Inference
Bayesian inference analyses of Sets 2, 3, and 4 produced topologies similar to MP analyses, and consistently supported E. volitans as sister to all other Exocoetus species (BPP ≥ 0.96, Figures 3b,d, 4b). As in MP results, E. monocirrhus and E. obtusirostris are sister taxa in every analysis, with strong support (BPP ≥ 0.96). The arrangement of E. peruvianus and E. gibbosus individuals in a monophyletic clade was well supported across analyses, but individuals from each of these species did not form monophyletic groups in the Rag2 analysis (Figure 3d). We did not find evidence for the phylogenetic arrangement of E. monocirrhus as sister to an E. obtusirostris–E. peruvianus–E. gibbosus clade in any of the BI analyses.
The BI analysis of the expanded Cytb dataset (Set 5) resulted in well‐supported clades (BPP ≥ 0.97) with Exocoetus monophyletic and E. volitans as the sister to all other species. A clade comprised of E. peruvianus–E. gibbosus, was sister to a clade comprised of E. obtusirostris–E. monocirrhus. Multiple representatives of E. volitans, E. monocirrhus, and E. obtusirostris grouped together in every analysis, whereas E. gibbosus and E. peruvianus were not clearly distinguished.
3.3. Genetic distance
Pairwise estimates of K2P genetic distances for Cytb between individuals within each Exocoetus species were 0.012 (±0.002) for E. peruvianus, 0.011 (±0.002) for E. obtusirostris, 0.010 (±0.002) for E. monocirrhus, and 0.007 (±0.001) for E. volitans. For E. gibbosus, the K2P genetic distance between the two individuals was 0.009. Genetic distance comparisons between species ranged from 0.011 (E. gibbosus vs. E. peruvianus) to 0.087 (E. monocirrhus vs. E. volitans), with an overall mean K2P value of 0.060 (Figure 5).
Figure 5.

Summary of genetic distance estimates (Kimura two‐Parameter; K2P) within and between Exocoetus species based on the expanded Cytb dataset (n = 1,082 bps). (a) Mean Cytb genetic distances (K2P ± standard error; S.E.) within each species of Exocoetus. (b) Mean Cytb genetic distance estimates (K2P ± standard error; S.E.) between Exocoetus species. Species are abbreviated using the following labels: EXVO = E. volitans, EXMO = E. monocirrhus, EXOB = E. obtusirostris, EXPE = E. peruvianus, EXGI = E. gibbosus
4. Discussion
4.1. Phylogeny of exocoetus
Phylogenetic analyses of Exocoetus species yielded consistent, well‐supported evidence for the arrangement of four monophyletic groups irrespective of the method used. First, Exocoetus is monophyletic, which corroborates the findings of other authors (Collette et al., 1984; Lewallen et al., 2011; Parin, 1961; Parin & Shakhovskoy, 2000). Second, E. volitans is sister to all other Exocoetus species, with E. monocirrhus + E. obtusirostris + E. peruvianus + E. gibbosus forming a monophyletic clade. Third, E. monocirrhus is sister to E. obtusirostris. Fourth, a monophyletic group containing E. peruvianus and E. gibbosus is well supported. Our results agree with the hypothesis of Parin and Shakhovskoy (2000), with the exception of several key relationships. We did not find evidence for an E. obtusirostris + E. gibbosus + E. peruvianus clade sister to E. monocirrhus. Additionally, none of our analyses yielded support for the arrangement of E. obtusirostris as sister to an E. gibbosus–E. peruvianus clade. Furthermore, in all analyses (except using morphological data only), we found support for a sister species relationship between E. monocirrhus and E. obtusirostris, which was not included in the Parin and Shakhovskoy (2000) hypothesis. Although there is support for a clade containing E. peruvianus and E. gibbosus, sequences from these species were not always reciprocally monophyletic, suggesting that they are not genetically distinct species, or that speciation has been rapid, recent and/or ongoing (see below).
4.2. Species distinctions
To assess the genetic distinctiveness of Exocoetus species, we performed a genetic survey of specimens collected worldwide and found that E. volitans, E. monocirrhus, and E. obtusirostris are distinct species that clearly form monophyletic groups. These three species can easily be distinguished using morphological or molecular characters. E. gibbosus and E. peruvianus comprise a well‐supported monophyletic group, yet the evolutionary separation of these species is less clear. These species are distributed allopatrically, with E. peruvianus found offshore of Peru, and E. gibbosus found in the South Pacific. The two species are therefore separated by the Eastern Pacific Barrier, a 4,000–7,000 km wide stretch of deep ocean without islands. However, despite this apparent allopatric distribution, phylogenetic and genetic distinctiveness of these taxa is lacking. Individuals were not reciprocally monophyletic by species, and genetic divergence was low (K2P = 1.1%). Morphological characters distinguishing E. peruvianus from E. gibbosus are subtle, involving proportional body form measurements. For example, in adults (>150 mm SL), the body depth at the pectoral fin base is 19–22% SL in E. gibbosus and 15–18.5% SL in E. peruvianus. Additionally, in juveniles (<80 mm SL), the head depth is 22.5–28.5% SL in E. gibbosus, and 19.5–23% in E. peruvianus (Parin & Shakhovskoy, 2000).
Two main situations could result in the lack of evidence for the taxonomic distinctiveness of E. peruvianus and E. gibbosus. One possibility is very recent speciation, accompanied by incomplete lineage sorting. Gene trees may not be congruent with a species tree when the rate of speciation exceeds the rate at which allelic polymorphisms achieve reciprocal monophyly in separated gene pools (Harrison, 1991). Although we have not calibrated a molecular clock for this study, the very low amounts of divergence between individuals of the two putative species are indicative of very recent divergence. A second possibility is that E. peruvianus and E. gibbosus represent a single species, with regular gene flow between two distant allopatric populations, sufficient to prevent them from becoming reproductively isolated. Observed morphological differences might be due to phenotypic plasticity associated with the occupation of slightly different habitats. If this is the case, these species would be better classified as regional morphotypes of the same species, a pattern that has been observed in other flyingfishes (Parin & Belyanina, 1998). Our results point to the need for additional sampling and genetic analyses to confirm whether E. peruvianus and E. gibbosus represent distinct species. Increasing the number of sampled individuals, or using higher‐resolution genetic markers would likely improve our ability to differentiate between the two scenarios described above. We also note that adding samples for lineages with lower numbers of individuals sequenced would reduce any possible biases caused by differences in sample number across species analyzed. For example, we are likely to have incompletely sampled the total Cytb variation of E. peruvianus and E. gibbosus, and further sequencing may provide clearer indication of whether these putative species are genetically isolated.
4.3. Cryptic species
Cryptic species (Bickford et al., 2007) have long posed taxonomic challenges and may be identified using anatomical, ecological, behavioral, biogeographic, and/or molecular characteristics. DNA comparisons can provide particularly useful information about species distinctiveness. Hebert et al. (2003) suggested that genetic distance estimates above 3% for the DNA “barcode” gene cytochrome oxidase I should be used as a threshold for defining species, and genetic distance estimates above 2% have been proposed for distinguishing vertebrate species using Cytb data (Avise & Walker, 1999). However, other studies have shown that model selection for genetic distance calculations can also affect species delimitation (Barley & Thomson, 2016). In our study, very low mean Cytb genetic distance estimates (K2P = 0.7–1.2%) within each species suggests an absence of cryptic species. In addition, in the case of E. volitans, an analysis across the range of the species found minimal population genetic structure at a global scale (Lewallen et al., 2016).
Single widely distributed marine taxa are sometimes found to consist of morphologically cryptic, but genetically distinct, independent evolutionary lineages (species) segregated by ocean basin (Briggs, 1960), or oceanographic factors (Gaither et al., 2015). Examples include bristlemouths (Miya & Nishida, 1997), goliath groupers (Craig et al., 2009), bonefish (Bowen, Karl, & Pfeiler, 2007), ocean sunfish (Bass et al., 2005), and hammerhead sharks (Quattro et al., 2005). In contrast to these documented cases of cryptic species, we find no evidence of this phenomenon in Exocoetus, despite the multi‐ocean distributions of E. volitans and E. monocirrhus. At least some globally connected species maintain global population connectivity by dispersal (e.g., pelagic wahoo; Theisen et al., 2008). For Exocoetus, buoyant pelagic eggs likely provide an adequate mechanism for dispersal across large distances. The exact pelagic larval duration of Exocoetus is not known, but Mora et al. (2012) recently demonstrated that the larvae of many tropical reef fishes persist in the water column long enough for regular breaching of the Eastern Pacific Barrier. Thus, long‐distance dispersal of pelagic eggs could explain the lack of genetic differentiation of Exocoetus populations from different Oceans.
4.4. Exocoetus biogeography
At the base of the Exocoetus tree, E. volitans is distributed throughout all tropical oceans and sympatric with every other species in the genus (Figure 6) although granular patterns of habitat preference might preclude contact among individuals from different species (e.g., seasonally sympatric/parapatric). As sister to all other species in this genus, we conclude that the ancestor of Exocoetus fishes may have been similar to E. volitans, both in terms of morphology and distribution. The distribution of the sister lineage to E. volitans may also have had an expansive distribution, so inferring the geographic context of divergence within Exocoetus is difficult. Sympatric diversification between two globally distributed lineages is a possibility, but equally realistic is allopatric diversification followed by significant dispersal and range expansion.
Figure 6.

Phylogeny of Exocoetus with distribution maps derived from collection localities presented in Parin & Shakhovskoy (Parin & Shakhovskoy, 2000). Polygons were produced using ArcMap 9.3.1 (ESRI, Redlands, CA, USA)
Because E. volitans individuals from the Atlantic and Indo‐Pacific are not genetically diverged (Lewallen et al., 2016), we suggest that the species either dispersed between these regions very recently, or that there is regular gene flow between Oceans, presumably across the Benguela Barrier. The Benguela Barrier results from the upwelling of cold waters near the tip of South Africa and can prevent dispersal between the tropical Atlantic and tropical Indian Oceans for some marine fishes (Briggs, 1995; Rocha, Craig, & Bowen, 2007). However, Rocha et al. (2005) provided compelling evidence to suggest that at least some tropical marine fishes (Gnatholepis gobies) have breached the Benguela Barrier to invade the Atlantic Ocean from the Indian Ocean. Additionally, Craig, Hastings and Pondella (2004) showed support for a sister species relationship between Caribbean and Western Indian Ocean species of the grouper genus Dermatolepis, demonstrating trans‐Atlantic dispersal and crossing of the Benguela Barrier by reef fishes (Craig et al., 2004). The distribution of E. volitans, and the minimal genetic divergence between individuals from the Atlantic and Indian Oceans suggest that Exocoetus may be capable of similar dispersals.
The two most distal nodes of the Exocoetus tree (E. obtusirostris + E. monocirrhus and E. peruvianus + E. gibbosus) provide better opportunities for determining the biogeographic context of diversification (Figure 6). In particular, previously identified marine biogeographic barriers (Rocha et al., 2007) are relevant to the phylogenetic and geographic patterns we observe in Exocoetus. In the case of the sister relationship between E. monocirrhus and E. obtusirostris, their respective distributions in the Indian and Pacific Oceans versus the Atlantic Ocean suggest that the Isthmus of Panama and Benguela Barriers may have provided effective boundaries to limit gene flow, resulting in speciation. On the west side of the Atlantic Ocean, the Isthmus of Panama is a well‐known land barrier that has resulted in the speciation of many Atlantic and Pacific sister lineages of marine fishes (Banford, Bermingham, & Collette, 2004). According to Banford et al. (2004), at least four periods in the last 10 million years provided marine fishes with opportunities for allopatric speciation on opposite sides of the Isthmus of Panama as it gradually formed. Perhaps a result of species‐specific thermal tolerances, the cold waters of South Africa (Benguela Barrier) seem to effectively confine E. obtusirostris and E. monocirrhus to their respective tropical Atlantic and tropical Indo‐Pacific distributions.
The sister relationship between E. peruvianus and E. gibbosus, in combination with their respective distributions in the Peruvian Upwelling Current and South Pacific Subtropical Gyre (Parin & Shakhovskoy, 2000), suggests that the Eastern Pacific Barrier (Lessios & Robertson, 2006) may segregate these putative species. The Eastern Pacific Barrier is an expanse of deep water (4,000–7,000 km wide) that separates coastally distributed fishes, although Lessios and Robertson (2006) showed examples of species that can cross this barrier. The lack of islands in the eastern Pacific makes it a particularly effective barrier for some reef‐inhabiting organisms. However, this barrier should, in principle, only influence coastal or neritic flyingfish species (e.g., Fodiator, Parexocoetus), or species that have island‐associated life stages (e.g., Cheilopogon atrisignis, Cypselurus angusticeps), and would be expected irrelevant to holoepipelagic species, such as Exocoetus. Thus, E. peruvianus and E. gibbosus may be separated by some other oceanographic barrier. Additional samples are required to first determine whether these taxa are distinct, and then reveal if and how gene flow occurs across this putative barrier. Given the low amount of sequence divergence between E. peruvianus and E. gibbosus, we favor the hypothesis that this species pair is the result of recent divergence and provides a rare example of incipient speciation in an epipelagic fish lineage. As such, these taxa are good candidates for addressing the mechanisms by which speciation occurs in the epipelagic zone.
Conflict of Interest
None declared.
Supporting information
Acknowledgments
For assistance with collecting specimens, we thank the crews and researchers aboard the following ships: Endeavor (R. McMunn, R. Chase, B. Collins, P. Roussell, J. Montminy, P. Quigley, K. Walsh, G. Maltby, T. Varney, A. Wright, M. Brennan, B. Wilson, B. Fanning, A. Tucker, K. Pohl, V. Pascal, R. Cooper, L. Koren, H. Hamner, and R. Lohmann); David Starr Jordan; McArthur; McArthur II; Kahana; Gunter; Oscar Elton Sette; Oregon II; Malcolm Baldrige; and the Protected Resources Division of the NOAA Southwest Fisheries Science Center (J. Cotton, L. Ballance, K. Forney, E. Archer, J. Redfern, A. Henry, C. Hall, T. Gerrodette, A. Ü, E. V. Morquecho, J. C. Salinas, L. Zele, M. Force, R. Rowlett, R. Driscoll, S. Rankin, S. Webb, S. Yin, J. Barlow). Tissues were graciously donated by the National Marine Fisheries Service (B. Collette) and Florida Museum of Natural History (C. Obordo). D. Xiao, E. Holm, F. Pardo, D. Stacey, and H.J. Walker assisted specimen handling and preservation. Molecular data collection and data management were aided by A. Shah and B. Shah. This manuscript was improved by helpful comments from D. Lewallen, R. Winterbottom, C. Healy, A. Mason, and P. Hastings. Funding for the study was provided by an NSERC Discovery Grant (to NRL) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (F32 AR68154 to EAL; R01 AR049069 to AJVW). The scientific results and conclusions, as well as any views or opinions expressed herein, are those of the author(s) and do not necessarily reflect those of NOAA or the United States Department of Commerce.
Lewallen EA, Bohonak AJ, Bonin CA, van Wijnen AJ, Pitman RL, Lovejoy NR. Phylogenetics and biogeography of the two‐wing flyingfish (Exocoetidae: Exocoetus). Ecol Evol. 2017;7:1751–1761. https://doi.org/10.1002/ece3.2786
Contributor Information
Eric A. Lewallen, Email: lewallen.eric@mayo.edu.
Nathan R. Lovejoy, Email: lovejoy@utsc.utoronto.ca
References
- Avise, J. C. , & Walker, D. (1999). Species realities and numbers in sexual vertebrates: Perspectives from an asexually transmitted genome. Proceedings of the National Academy of Sciences, 96(3), 992–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banford, H. M. , Bermingham, E. , & Collette, B. B. (2004). Molecular phylogenetics and biogeography of transisthmian and amphi‐Atlantic needlefishes (Belonidae: Strongylura and Tylosurus): Perspectives on New World marine speciation. Molecular Phylogenetics and Evolution, 31(3), 833–851. [DOI] [PubMed] [Google Scholar]
- Barley, A. J. , & Thomson, R. C. (2016). Assessing the performance of DNA barcoding using posterior predictive simulations. Molecular Ecology, 25 (9), 1944–1957. [DOI] [PubMed] [Google Scholar]
- Bass, A. L. , Dewar, H. , Thys, T. , Streelman, J. T. , & Karl, S. A. (2005). Evolutionary divergence among lineages of the ocean sunfish family, Molidae (Tetraodontiformes). Marine Biology, 148(2), 405–414. [Google Scholar]
- Bickford, D. , Lohman, D. J. , Sodhi, N. S. , Ng, P. K. L. , Meier, R. , Winker, K. , … Das, I. (2007). Cryptic species as a window on diversity and conservation. Trends in Ecology and Evolution, 22(3), 148–155. [DOI] [PubMed] [Google Scholar]
- Bowen, B. W. , Karl, S. A. , & Pfeiler, E. (2007). Resolving evolutionary lineages and taxonomy of bonefishes (Albula spp.) In Ault J. S. (Ed.), Biology and Management of the World Tarpon and Bonefish Fisheries, (pp. 147–154) . Boca Raton: CRC Press. [Google Scholar]
- Briggs, J. C. (1960). Fishes of worldwide (circumtropical) distribution. Copeia, 1960, 171–180. [Google Scholar]
- Briggs, J. C. (1995). Global biogeography. Amsterdam: Elsevier. [Google Scholar]
- Colborn, J. , Crabtree, R. E. , Shaklee, J. B. , Pfeiler, E. , & Bowen, B. W. (2001). The evolutionary enigma of bonefishes (Albula spp.): Cryptic species and ancient separations in a globally distributed shorefish. Evolution, 55(4), 807–820. [DOI] [PubMed] [Google Scholar]
- Collette, B. , McGowen, G. E. , Parin, N. V. , & Mito, S. (1984). Beloniformes: Development and relationships In Moser H. G. (Ed.), Ontogeny and systematics of fishes (pp. 335–354). American Society of Ichthyologists and Herpetologists Special Publication 1, 1–760. [Google Scholar]
- Craig, M. T. , Hastings, P. A. , & Pondella, D. J. (2004). Speciation in the Central American Seaway: The importance of taxon sampling in the identification of trans‐isthmian geminate pairs. Journal of Biogeography, 31(7), 1085–1091. [Google Scholar]
- Craig, M. T. , Graham, R. T. , Torres, R. A. , Hyde, J. R. , Freitas, M. O. , Ferreira, B. P. , … Robertson, D. R. (2009). How many species of goliath grouper are there? Cryptic genetic divergence in a threatened marine fish and the resurrection of a geopolitical species. Endangered Species Research, 7(3), 167–174. [Google Scholar]
- Drummond, A. J. , & Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7(1), 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaither, M. R. , Bowen, B. W. , Rocha, L. A. , & Briggs, J. C. (2015). Fishes that rule the world: Circumtropical distributions revisited. Fish and Fisheries, 17(3), 664–679. [Google Scholar]
- Grudtsev, M. , Salekhova, L. P. , & Lushchina, V. G. , & SSR (1987). Distribution, ecology and intraspecific variability of flyingfishes of the genus Exocoetus of the Atlantic Ocean. Journal of Ichthyology, 27, 39–50. [Google Scholar]
- Hamner, W. M. (1995). Predation, cover, and convergent evolution in epipelagic oceans. Marine and Freshwater Behaviour and Physiology, 26(2–4), 71–89. [Google Scholar]
- Harrison, R. G. (1991). Molecular changes at speciation. Annual Review of Ecology and Systematics, 22, 281–308. [Google Scholar]
- Hebert, P. D. , Cywinska, A. , & Ball, S. L. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society – Biological Sciences, 270(1512), 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemstra, P. C. , & Parin, N. V. (1986). Exocoetidae In Smith M. M. & Heemstra P. C. (Eds.), Smith's sea fishes (pp. 391–396). Berlin: Springer‐Verlag. [Google Scholar]
- Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16(2), 111–120. [DOI] [PubMed] [Google Scholar]
- Kovalevskaya, N. (1982). Superfluous reproduction and development of flying fishes of the family Exocoetidae. Journal of Ichthyology, 22(4), 48–54. [Google Scholar]
- Lessios, H. A. , & Robertson, D. R. (2006). Crossing the impassable: Genetic connections in 20 reef fishes across the eastern Pacific barrier. Proceedings of the Royal Society – Biological Sciences, 273(1598), 2201–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewallen, E. A. , Pitman, R. L. , Kjartanson, S. L. , & Lovejoy, N. R. (2011). Molecular systematics of flyingfishes (Teleostei: Exocoetidae): Evolution in the epipelagic zone. Biological Journal of the Linnean Society, 102(1), 161–174. [Google Scholar]
- Lewallen, E. A. , Bohonak, A. J. , Bonin, C. A. , van Wijnen, A. J. , Pitman, R. L. , & Lovejoy, N. R. (2016). Population genetic structure of the tropical two‐wing Flyingfish (Exocoetus volitans). PLoS One, 11(10), e0163198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison, D. , & Maddison, W. (2005). MacClade 4 release version 4.07 for OSX. Sunderland, MA: Sinauer. [Google Scholar]
- Miya, M. , & Nishida, M. (1997). Speciation in the open ocean. Nature, 389(6653), 803–804. [Google Scholar]
- Mora, C. , Treml, E. A. , Roberts, J. , Crosby, K. , Roy, D. , & Tittensor, D. P. (2012). High connectivity among habitats precludes the relationship between dispersal and range size in tropical reef fishes. Ecography, 35(1), 89–96. [Google Scholar]
- Page, R. (1996). Treeview program version 161. Computer Applications in the Biosciences, 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Parin, N. (1961). Principles of classification of flying fishes (Oxyporhamphidae and Exocoetidae). Trudy Institute of Oceanology, 43, 92–183. [Google Scholar]
- Parin, N. (1968). Ichthyofauna of oceanic epipelagic zone. Moscow: Nauka Press. [Google Scholar]
- Parin, N. (1984). Exocoetidae In Fischer W. & Bianchi G. (Eds.), FAO species identification sheets for fishery purposes. Western Indian Ocean (Fishing Area 51), Volume 2, (pp. 1–8). Rome: FAO. [Google Scholar]
- Parin, N. (2002). Exocoetidae (Flyingfishes) In Carpenter K. (Ed.), The living marine resources of the western central Atlantic. Vol. 2: Bony fishes part 1 (Acipenseridae to Grammatidae). FAO Species Identification Guide for Fishery Purposes. American Society of Ichthyologists and Herpetologists, Special Publication 5. (pp. 1116–1134). Rome: FAO. [Google Scholar]
- Parin, N. , & Belyanina, T. (1998). Age and geographic variability and distribution of the flying fish Cheilopogon furcatus (Exocoetidae, beloniformes), with a description of two new sub species. Journal of Ichthyology, 38(8), 557–573. [Google Scholar]
- Parin, N. , & Belyanina, T. (2002). Flying fishes of the genus Fodiator (Exocoetidae): Systematics and distribution. Journal of Ichthyology, 42(5), 357–367. [Google Scholar]
- Parin, N. , & Shakhovskoy, I. (2000). A review of the flying fish genus Exocoetus (Exocoetidae) with descriptions of two new species from the southern Pacific Ocean. Journal of Ichthyology, 40(1), S31. [Google Scholar]
- Peixoto, B. R. , Mikawa, Y. , & Brenner, S. (2000). Characterization of the recombinase activating gene‐1 and 2 locus in the Japanese pufferfish, Fugu rubripes . Gene, 246(1), 275–283. [DOI] [PubMed] [Google Scholar]
- Quattro, J. M. , Stoner, D. S. , Driggers, W. B. , Anderson, C. A. , Priede, K. A. , Hoppmann, E. C. , … Grady, J. M. (2005). Genetic evidence of cryptic speciation within hammerhead sharks (Genus Sphyrna). Marine Biology, 148(5), 1143–1155. [Google Scholar]
- Rambaut, A. , & Drummond, A. J. (2007a). Tracer v1. 4. http://www.beast.bio.ed.ac.uk/Tracer. [Google Scholar]
- Rambaut, A. , & Drummond, A. J. (2007b). TreeAnnotator v1.4.8. http://beast.bio.ed.ac.uk/TreeAnnotator. [Google Scholar]
- Rocha, L. , Craig, M. , & Bowen, B. (2007). Phylogeography and the conservation of coral reef fishes. Coral Reefs, 26(3), 501–512. [Google Scholar]
- Rocha, L. A. , Robertson, D. , Rocha, C. R. , Tassell, J. L. , Craig, M. T. , & Bowen, B. W. (2005). Recent invasion of the tropical Atlantic by an Indo‐Pacific coral reef fish. Molecular Ecology, 14(13), 3921–3928. [DOI] [PubMed] [Google Scholar]
- Swofford, D. (2000). PAUP and other methods. Phylogenetic analysis using parsimony. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. , & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen, T. C. , Bowen, B. W. , Lanier, W. , & Baldwin, J. D. (2008). High connectivity on a global scale in the pelagic wahoo, Acanthocybium solandri (tuna family Scombridae). Molecular Ecology, 17(19), 4233–4247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


