Abstract
Massive cancer genomics data have facilitated the rapid revolution of a novel oncology drug discovery paradigm through targeting clinically relevant driver genes or mutations for the development of precision oncology. Natural products with polypharmacological profiles have been demonstrated as promising agents for the development of novel cancer therapies. In this study, we developed an integrated systems pharmacology framework that facilitated identifying potential natural products that target mutated genes across 15 cancer types or subtypes in the realm of precision medicine. High performance was achieved for our systems pharmacology framework. In case studies, we computationally identified novel anticancer indications for several US Food and Drug Administration‐approved or clinically investigational natural products (e.g., resveratrol, quercetin, genistein, and fisetin) through targeting significantly mutated genes in multiple cancer types. In summary, this study provides a powerful tool for the development of molecularly targeted cancer therapies through targeting the clinically actionable alterations by exploiting the systems pharmacology of natural products.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Massive cancer genomic data has facilitated the revolution of a novel oncology drug discovery paradigm through targeting clinically relevant driver genes or mutations for the development of precision oncology.
WHAT QUESTION DOES THIS STUDY ADDRESS?
☑ Natural products with polypharmacological space have been recognized as important agents in the development of novel therapies for various complex diseases, including cancer. It is urgently needed to develop novel approach, such as a systems pharmacology approach, for the development of targeted cancer therapeutics that target clinically relevant alterations by exploiting the polypharmacology of natural products.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
☑ This study demonstrates that a systems pharmacology framework that integrates drug–target interaction network of natural products and cancer mutant genes from the cancer genome projects would be useful for the development of novel targeted cancer therapies.
HOW MIGHT THIS CHANGE DRUG DISCOVERY, DEVELOPMENT, AND/OR THERAPEUTICS?
☑ This study will provide useful systems pharmacology approaches for the development of precision oncology through targeting the clinically relevant driver alterations by exploiting the polypharmacology of natural products.
Despite rapid technological advancements and massive research and development (R&D) investments, oncology drug development remains a great challenge. In the past several decades, traditional oncology drug discovery that focused on synthesized compounds has shown high risk in clinical trials.1 A recent study revealed that ∼7.5% of the oncology drugs were able to enter phase I trials and only 33.2% of the drugs that entered phase III trials were eventually approved.1 Compared with traditional oncology drugs, natural products are better templates with ideal pharmacokinetics/pharmacodynamics (PK/PD) properties, of which scaffolds are repeatedly considered “privileged” in drug discovery.2 So far, more than half of the new drugs introduced after 1990 could be traced to naturally derived products or their analogs.3, 4 Since natural products possess enormous structural and chemical diversity and are abundant, they have served as great inspiration for the next generation of cancer therapeutics.5, 6
With rapidly growing, revolutionary next‐generation sequencing technologies, several large‐scale cancer genome projects, such as The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC), have generated massive amounts of somatic mutation profiles.7 Such massive amounts of cancer genomic data have helped us better understand cancer biology and improve cancer prognosis, diagnosis, and treatment.8, 9 For most solid tumors, their genomes harbor hundreds of DNA‐level genetic alterations, which are composed of massive bystander mutations (“passenger mutations”) with no oncogenic potential and few cancer‐driving genomic aberrations (“driver mutations”).10 Driver mutations represent mutations that have a selective growth advantage in tumor cells, further contributing to tumor initiation, progression, and drug resistance via activating oncogenes or inactivating tumor suppressor genes.11 Therefore, cancer drugs that target clinically relevant driver mutations, such as kinase inhibitors, have shown high selectivity on tumor cells and demonstrated valuable opportunities for precision oncology.12 For example, BCR‐ABL mutations have been found to predict clinical responses to imatinib, the first US Food and Drug Administration (FDA)‐approved targeted agent, in chronic myelogenous leukemia (CML). Epidermal growth factor receptor (EGFR) mutations (e.g., T790M) are used to predict clinical response to EGFR tyrosine kinase inhibitors in non‐small cell lung cancer (NSCLC).13 Although molecularly targeted agents (e.g., kinase inhibitors) greatly improved the clinical benefit and life quality of some cancer types, the benefit has remained disappointing for many solid tumors due to drug resistance or other issues.14, 15, 16 Natural products, which have the unprecedented scaffolds with good pharmacokinetic/pharmacodynamic (PK/PD) profiles, provide alternative opportunities for overcoming cross‐resistance to many of the known cancer drugs. Nowadays, an urgent need is to develop more efficiently therapeutic agents derived from natural products via targeting the mutated cancer genes for precision oncology.
In this study we systematically examined natural products for precision oncology by targeting cancer mutated genes through an integrated systems pharmacology framework (Figure 1). Specifically, we constructed a comprehensive drug–target interaction network for natural products by integrating data from about 10 chemoinformatics/bioinformatics sources currently available. We then collected cancer Significantly Mutated Genes (SMGs) for 15 cancer types or subtypes from over 20 large‐scale cancer genome sequencing projects. Finally, we developed an integrated statistical approach under systems pharmacology framework by incorporating drug–target interaction network of natural products into our curated cancer SMGs to prioritize new anticancer indications of natural products for each individual cancer type. Systematic validations revealed the high performance of our approach. Moreover, we repurposed multiple potential anticancer indications or off‐label cancer treatment for several FDA‐approved or clinically investigational natural products with new molecular mechanisms in case studies.
Figure 1.
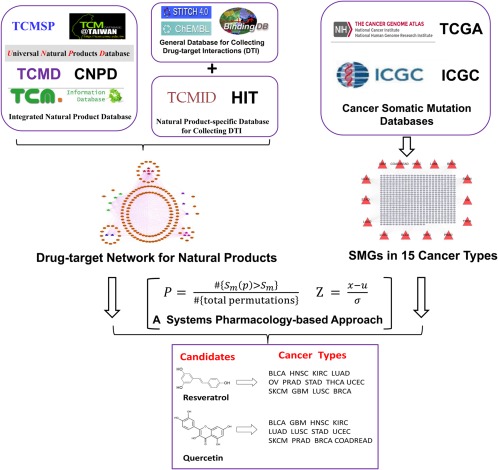
Diagram of an integrated systems pharmacology framework for prioritizing new anticancer indications by mapping the polypharmacology of natural products into significantly mutated genes (SMGs) in cancers across 15 cancer types or subtypes. The abbreviations of 15 major cancer types/subtypes are: acute myeloid leukemia (LAML), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon and rectal adenocarcinoma (COADREAD), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), prostate adenocarcinoma (PRAD), skin cutaneous melanoma (SKCM), stomach adeno‐carcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC). The SMGs are derived from various large‐scale cancer genome projects as described in a previous study.28 The drug–target interaction network for natural products were built via integration data from several commonly used chemoinformatics and bioinformatics databases (see Methods).
METHODS
Integration of drug data for natural products
We collected drug information for natural products by integrating data from six data sources, including a traditional Chinese medicine database,17 Chinese natural product database,18 traditional Chinese medicine integrated database (TCMID),19 traditional Chinese medicine systems pharmacology (TCMSP),20 traditional Chinese medicine database@Taiwan (TCM@Taiwan),21 and universal natural product database (UNPD).22 For each database, drug structures for natural products were initially stored in ISIS Base 2.5 (MDL Information Systems, San Ramon, CA), and converted to SDF format. Subsequently, six SDF files were merged to a single SDF, which contained all drug structures from six data sources. InChIKey, a fixed‐length (25 character) condensed digital representation of the InChI, was generated by Open Babel23 for each drug. After removing the duplicates according to InChiKey, 259,547 unique natural products were obtained. Finally, InChIKey for each drug was retrieved to obtain the corresponding PubChem CID information with the python script “PubChemPy” (https://pypi.python.org/pypi/PubChemPy) and mapped to the DrugBank database to link the corresponding drug information.
Construction of drug–target interaction network for natural products
Drug–target interaction mappings were performed for two types of data sources: 1) general chemoinformatics databases (including ChEMBL,24 BindingDB,25 and STITCH26), and 2) natural products‐specific chemoinformatics databases (including HIT27 and TCMID19). We downloaded the latest data as: ChEMBL (v. 21), BindingDB (accessed in June 2016), and STITCH (accessed in June 2016). For ChEMBL and BindingDB, only data items that met the following criteria were retained: i) Ki, Kd, IC50, or EC50 ≤ 10 μM; ii) the target is a human protein; iii) the target can be represented in a unique UniProt accession number; and iv) the drug can be successfully represented in the canonical SMILES format. For the STITCH source, the thickness of each interaction pair represents the confidence score of the association. Only drug–target interactions from Homo sapiens were downloaded, and high‐confidence interactions (score ≥0.7) were kept in this study. Finally, the duplicated drug–target pairs were removed.
Traditional Chinese Medicine (TCM) provides an important source for natural products. As representative TCM databases, HIT and TCMID manually curated thousands of known herb ingredients' targets information. To enlarge the scope of the known drug–target interactions for natural products, we extracted drug–target interactions from HIT and TCMID using a web crawler approach. For TCMID, we excluded the computationally predicted drug–target interactions from STITCH, and only kept the known drug–target interactions identified by experimental data in human. Then we merged the known drug–target interactions from HIT and TCMID. Subsequently, we mapped 259,547 unique natural products into the five drug–target databases mentioned above to extract the experimentally reported drug–target interactions using the “InChIKey.”
Manual curation of mutated genes for 15 cancer types/subtypes
We manually collected SMGs for 15 cancer types/subtypes from over 20 cancer genome analysis projects. The details were provided in our previous study.28 These 15 major cancer types consist of acute myeloid leukemia (LAML), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon and rectal adenocarcinoma (COADREAD), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), prostate adenocarcinoma (PRAD), skin cutaneous melanoma (SKCM), stomach adeno‐carcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC). The details are available in online Supplementary Figure 1 and Supplementary Table 1. We annotated all SMGs using gene Entrez ID, chromosome location, and the official gene symbols from the National Center for Biotechnology Information (NCBI) database (accessed on June 12, 2015).29
Table 1.
Statistics of drug‐target interaction (DTI) network for natural products
| Data resource | # of drug targets | # of natural products | # of DTIs |
|---|---|---|---|
| ChEMBL | 550 | 2,004 | 5,905 |
| BindingDB | 614 | 1,360 | 3,245 |
| STITCH | 2,648 | 840 | 6,594 |
| HIT & TCMID | 1,276 | 527 | 5,079 |
| Total | 3,546 | 2,988 | 18,008 |
Prioritizing new anticancer indications for natural products
In this study we developed an integrated statistical framework to prioritize new anticancer indications or off‐label cancer treatment for FDA‐approved or clinically investigational natural products by incorporating drug–target interaction network of natural products into SMGs identified by several cancer genome projects. The hypothesis of our integrated statistical framework asserts that a natural product with polypharmacological profiles shows a high potential for a particular anticancer indication if its targets are more likely to be SMGs in this specific cancer. We proposed a permutation testing to calculate the statistical significance of a natural product to be prioritized for treating a specific cancer type or subtype. The null hypothesis posits that drug target proteins of natural products equally distribute at protein products of SMGs against other human genome‐wide gene products (proteins). The alternative hypothesis asserts that cancer drug target proteins are more likely to be protein products of SMGs than other proteins for a natural product that has a potential anticancer indication. We performed the permutation test as below:
| (1) |
Here we performed 100,000 permutations (#[total permutations]) by randomly selecting the same number of SMGs in a specific cancer type from protein products at the genome‐wide scale (20,462 human protein‐coding genes from the NCBI database; Supplementary Table 2). In each permutation, we repeated the calculation of Sm (the number of SMGs targeted by a given natural product in a specific cancer type) and denoted it as Sm(p). A nominal P was the computed for each natural product by counting the number of permutations that have Sm(p) greater than the real case, divided by the total number of permutations. Then the resulting P‐values generated from the permutation tests were corrected as adjusted P‐values (q values) using the Benjamini–Hochberg multiple test correction method.30
Finally, we further calculated a Z‐score for each natural product in a specific cancer during permutations:
| (2) |
where x is the real number of SMGs targeted by a given natural product in a specific cancer type, μ is the mean number of SMGs targeted by a given natural product during 100,000 permutations in a specific cancer type, and σ is the standard deviation.
Statistical analysis and network visualization
The statistical analysis was performed using the Python (v. 3.2, https://www.python.org/) and R platforms (v. 3.01, http://www.r-project.org/). Network visualization and analysis (such as degree calculation) were conducted using Cytoscape (v. 3.2.0, http://www.cytoscape.org/).
RESULTS
Construction of drug–target interaction network for natural products
The final drug–target interaction network contains 18,008 known natural product‐specific drug–target interaction pairs, which connects 2,988 unique natural products and 3,546 target proteins (Table 1). The 18,008 interaction pairs (Supplementary Table 3) are composed of five data sources, including ChEMBL (5,905 pairs), BindingDB (3,245 pairs), STITCH (6,594 pairs), and the combined HIT and TCMID (HIT&TCMID, 5,079 pairs).
Figure 2 shows a bipartite drug–target interaction network for 5,681 interactions connecting 409 FDA‐approved or clinically investigational natural products annotated in DrugBank database and 2,210 target proteins. The detailed data are provided in Supplementary Table 4. The average degree (i.e., connectivity) of a natural product is 6.03 (18,008/2988), which is stronger than 2.22 of nonnatural product drugs in DrugBank (Supplementary Table 5). The Wilcoxon test shows that there is a significant difference (P‐value < 2.2 × 10−16) for the number of known targets between natural products and non‐natural product drugs in DrugBank, suggesting a significant polypharmacology for natural products. Network analysis suggests that the average number of known target proteins for the FDA‐approved and clinically investigational natural products in DrugBank is 13.89 and the average degree for each target protein is 2.57. Among the 409 FDA‐approved or clinically investigational natural products, eight are linked by over 100 target proteins: DB04077 (glycerol), DB04216 (quercetin), DB02709 (resveratrol), DB00396 (progesterone), DB04272 (citric acid), DB07352 (apigenin), DB01645 (genistein), and DB00131 (adenosine monophosphate). Among 2,210 known target proteins, 154 are products of SMGs, while the rest of 2,056 are products of non‐SMGs. For example, eight SMGs have drug degrees (K) greater than 10: TP53 (K = 27), BLM (K = 22), ESR1 (K = 16), ABCB1 (K = 16), AKT1 (K = 13), CDKN1A (K = 12), MMP2 (K = 11), and PTGS1 (K = 11). Taken together, the observed polypharmacological profiles of natural products motivated us to develop a new computational approach for prioritizing potential anticancer indications to FDA‐approved or clinically investigational natural products via targeting SMGs in cancers.
Figure 2.
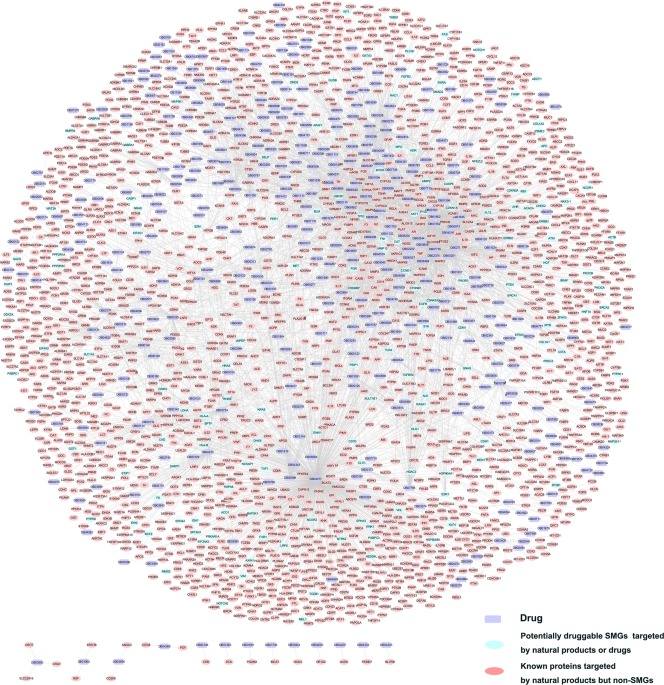
A global bipartite drug–target interaction network for natural products. This network connects 409 FDA‐approved or clinically investigational natural products annotated in the DrugBank database and 2,210 known drug target proteins, including proteins encoded by 154 significantly mutated genes (SMGs) and 2,056 non‐SMGs in cancers.
Systems pharmacology‐based prediction of new anticancer indications for natural products
Here, we proposed a statistical systems pharmacology framework (Figure 1, see Methods) to prioritize new anticancer indications for natural products. By applying the threshold of q < 0.05, we identified 848 anticancer indication pairs connecting 224 natural products and 15 cancer types/subtypes (Supplementary Table 6). Table 2 summarizes 154 significant anticancer indication pairs for 45 FDA‐approved or clinically investigational natural products (including known anticancer (off‐label use) and non‐anticancer (drug repurposing) natural products) across 15 cancer types/subtypes. Interestingly, several natural products were predicted to have anticancer indications for multiple cancers, such as resveratrol, glycerol, quercetin, fisetin, and genistein. Heat maps in Figure 3 shows Z‐scores and q‐values of the predicted indications for 45 FDA‐approved or clinically investigational natural products against 15 cancer types/subtypes.
Table 2.
List of discovered anticancer indications for 45 FDA‐approved or clinically investigational selected natural products
| Name | Pubchem CID | Cancer Indications (Z‐score) |
|---|---|---|
| Resveratrol | CID445154 | **BLCA (7.4), **HNSC (7.5), **KIRC (7. 6), **LUAD (6.1), **OV (6.94), **PRAD (6.44), **STAD (5.57), **THCA (6.79), **UCEC (7.21), **SKCM (7.03), *GBM (4.98), *LUSC (4.80), *BRCA (4.77) |
| Glycerol | CID753 | **GBM (5.47), **HNSC (6.24), **KIRC (6.44), **LUAD (8.66), **LUSC (7.50), **STAD (6.95), **UCEC (7.03), **SKCM (7.31), *PRAD (4.76), *BLCA (4.90), *BRCA (4.34), *OV (4.07) |
| Quercetin | CID5280343 | **BLCA (9.03), **GBM (6.16), **HNSC (9.09), **KIRC (10.58), **LUAD (11.89), **LUSC (7.57), **STAD (8.64), **UCEC (9.40), **SKCM (8.27), *PRAD (5.94), *BRCA (5.60), *COAD/READ (5.25) |
| Fisetin | CID5281614 | **HNSC (6.72), **LUAD (9.02), **LUSC (7.29), **PRAD (7.41), *COAD/READ (8.38), *SKCM (6.51), *THCA (6.59), *BLCA (6.65), *UCEC (5.95), *LAML (6.11), *STAD (4.98) |
| Genistein | CID5280961 | **BLCA (6.24), **BRCA (7.03), **HNSC (7.34), **KIRC (7.93), **LUAD (9.74), **LUSC (8.05), **PRAD (8.96), **SKCM (7.42), *COAD/READ (5.56), *UCEC (6.48), *GBM (4.86) |
| Estradiol | CID5757 | **BLCA (8.21), **HNSC (8.37), **LUAD (9.92), **LUSC (9.07), **PRAD (7.06), **UCEC (6.41), **SKCM (7.54), *COAD/READ (4.87) |
| Ethanol | CID702 | **LUAD (6.63), **THCA (13.05), *UCEC (6.31), *LAML (8.10), *LUSC (5.71), *SKCM (5.68) |
| Isopropyl Alcohol | CID3776 | **THCA (9.65), *LUSC (5.53), *HNSC (5.00), *PRAD (5.23), *STAD (4.76) |
| 1,3‐Diphenylurea | CID7595 | *LAML (11.61), *THCA (12.28), *OV (7.75), *PRAD (7.09) |
| Citric Acid | CID88113319 | **LAML (8.37), **LUSC (6.73), *SKCM (4.71), *GBM (4.69) |
| Ellagic acid | CID5281855 | **LUAD (10.40), *UCEC (5.15), *GBM (5.14), *LUSC (4.99) |
| Paclitaxel | CID36314 | *BRCA (6.54), *BLCA (5.97), *KIRC (5.50), *GBM (4.91) |
| Deoxycholic Acid | CID222528 | **LAML (10.61), *PRAD (6.27), *KIRC (5.62) |
| Apigenin | CID5280443 | *BLCA (5.11), *HNSC (5.17), *SKCM (4.75) |
| Dimethyl sulfoxide | CID679 | *LUAD (6.40), *GBM (6.04), *THCA (6.67) |
| Kaempherol | CID5280863 | *KIRC (5.85), *UCEC (5.26), *LAML (5.50), |
| L‐Citrulline | CID9750 | *HNSC (8.01), *STAD (7.63), *SKCM (6.59) |
| Progesterone | CID5994 | **GBM (6.64), *LUSC (5.38), *LUAD (5.35) |
| Trometamol | CID6503 | **HNSC (6.95), *STAD (5.34), *THCA (5.69) |
| 8‐azaguanine | CID8646 | *THCA (10.33), *LAML (9.71) |
| Acetylsalicylic acid | CID2244 | *LUAD (7.52), *SKCM (6.61) |
| Caffeine | CID2519 | **PRAD (7.76), *THCA (6.91) |
| Cisplatin | CID24191118 | *LAML (10.55), *PRAD (6.38) |
| Dicoumarol | CID54676038 | *LAML (8.05), *THCA (8.24) |
| Digitoxin | CID441207 | *THCA (9.02), *LAML (8.29) |
| Digoxin | CID2724385 | *THCA (8.23), *LAML (7.83) |
| Emodin | CID3220 | *LUAD (5.97), *LUSC (5.13) |
| Flavone | CID10680 | *LUSC (6.08), *BLCA (5.45) |
| Hydralazine | CID3637 | *LAML (7.84), *THCA (8.42) |
| Mercaptopurine | CID667490 | *LAML (8.96), *THCA (9.48) |
| Theophylline | CID2153 | **COAD/READ (8.19), *HNSC (6.59) |
| Xylometazoline | CID5709 | *THCA (7.57), *LAML (7.01) |
| Acetaminophen | CID1983 | *COAD/READ (6.78) |
| Butanoic Acid | CID264 | *OV (7.52) |
| Capsaicin | CID1548943 | *SKCM (4.89) |
| Chenodeoxycholic acid | CID10133 | *PRAD (7.46) |
| D‐Glutamine | CID5961 | *BLCA (6.51), |
| Dimethylformamide | CID6228 | *THCA (10.20) |
| Morphine | CID5288826 | *STAD (5.20) |
| Nicotine | CID942 | *PRAD (5.63) |
| Pyruvic acid | CID1060 | *THCA (6.23) |
| Reserpine | CID5770 | **GBM (7.99) |
| Riboflavin | CID493570 | *OV (7.44) |
| Rofecoxib | CID5090 | *COAD/READ (6.56) |
| Staurosporinone | CID3815 | *SKCM (6.10) |
*1.0 × 10−5 < q < 0.05; **q < 1.0 × 10−5.
Figure 3.
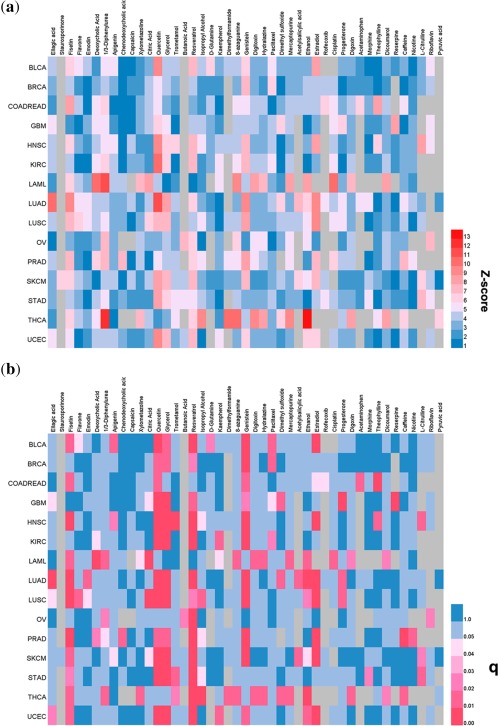
The heat maps show the predicted indications for 45 FDA‐approved or clinically investigational natural products against 15 cancer types/subtypes. The predicted Z‐scores (a) and q‐values (b) for 45 natural products against 15 cancer types/subtypes. The area in gray represents the nonavailable value since no significantly mutated genes are overlapped with the known targets of a specific natural product. The area in red represents the natural product having the high Z‐score and the low q value across specific cancer indications. The abbreviations of 15 major cancer types/subtypes are: acute myeloid leukemia (LAML), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon and rectal adenocarcinoma (COADREAD), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), prostate adenocarcinoma (PRAD), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC).
To evaluate the accuracy of our systems pharmacology‐based model, we further systematically searched the literature in PubMed for the predicted indications on these 45 FDA‐approved or clinically investigational natural products. Among the 45 natural products, 31 (31/45, 68.9% success rate) were previously reported to have anticancer indications in the previously published literature (Supplementary Table 7). This success rate of 68.9% suggests a reasonable accuracy for our systems pharmacology framework.
DISCUSSION
Natural products with polypharmacological profiles have been demonstrated as promising agents for the development of oncology drugs. In this study we constructed a comprehensive drug–target interaction network for 2,988 natural products through integration of various chemoinformatics and bioinformatics resources (Table 1 and Figure 1). We further developed an integrated statistical systems pharmacology framework with the permutation test for prioritizing new anticancer indications of natural products via targeting cancer relevant mutations and mutant genes. By applying our integrated systems pharmacology approach to 15 cancer types/subtypes, we found 848 significant anticancer indications for 224 natural products (q < 0.05). We systematically examined the predictions for 45 FDA‐approved or clinically investigational natural products annotated in the DrugBank database via searching various literature data. High performance (31/45, 68.9% successful rate) was yielded in the evaluation of our systems pharmacology framework based on currently available literature evidence.
To further demonstrate the accuracy or usefulness of our approach, we selected four typical FDA‐approved or clinically investigational natural products (resveratrol, quercetin, fisetin, and genistein) which have abundant experimentally validated data in the literature as example drugs to illustrate their anticancer profiles and mechanism‐of‐action (MOA).
Resveratrol is a non‐flavonoid polyphenol in grape skin and has been reported to have antioxidative and proapoptotic effects in several cancer cell lines.31 Several ongoing or completed clinical trials (http://clinicaltrials.gov/) for resveratrol are being conducted to treat various cancers, such as colon cancer (NCT00256334), neuroendocrine tumor (NCT01476592), and liver cancer (NCT02261844). Figure 4 a shows that resveratrol is predicted to have potential indications for 13 cancer types in our study: BLCA (Z = 7.36, q < 1.0 × 10−5), HNSC (Z = 7.48, q < 1.0 × 10−5), KIRC (Z = 7.56, q < 1.0 × 10−5), LUAD (Z = 6.12, q < 1.0 × 10−5), OV (Z = 6.94, q < 1.0 × 10−5), PRAD (Z = 6.44, q < 1.0 × 10−5), STAD (Z = 5.57, q < 1.0 × 10−5), THCA (Z = 6.79, q < 1.0 × 10−5), UCEC (Z = 7.21, q < 1.0 × 10−5), SKCM (Z = 7.03, q < 1.0 × 10−5), GBM (Z = 4.98, q = 0.0125), LUSC (Z = 4.80, q = 0.0206), and BRCA (Z = 4.77, q = 0.0277). Among the 13 cancer indications, six (HNSC, KIRC, STAD, SKCM, LUSC, and BRCA) were also predicted by our previous study.28 Surprisingly, the effect of resveratrol was well studied in breast cancer, and the results showed that patients with high total intake of resveratrol had a lower risk of breast cancer compared to a group with a low level of ingestion.32 Specifically, protein kinase B and tumor suppressor p53 (resveratrol targets) are encoded by two key SMGs in various cancer types: AKT1 and TP53. AKT1 regulates multiple critical biological processes in cancer, such as metabolism, proliferation, cell survival, growth, and angiogenesis, while p53 plays a key role in apoptosis, genomic stability, and inhibition of angiogenesis.33 A recent study reported that resveratrol potently inhibited glioblastoma multiforme (GBM) and glioblastoma stem‐like cells (GSC) growth and infiltration, acting partially via AKT deactivation and p53 induction, and suppressed glioblastoma growth in vivo.34
Figure 4.
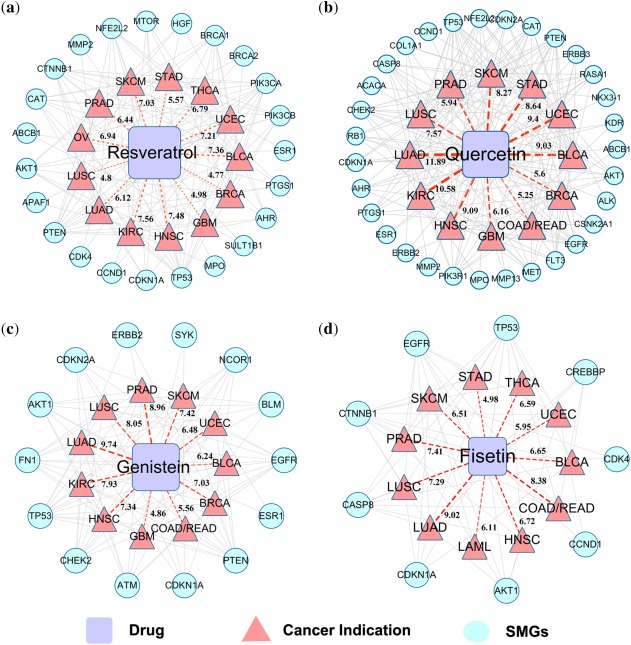
The reconstructed networks for four typical natural products. The networks display the predicted indications for four typical natural products, resveratrol (a), quercetin (b), genistein (c), and fisetin (d), against 15 cancer types/subtypes and their corresponding targets of the significantly mutated genes (SMGs) in multiple cancers. The gray lines denote SMGs in a specific cancer. The dotted red lines denote the predicted indications. The thickness (value) of a dotted red line is proportional to the Z‐score (see Methods). The abbreviations of 15 major cancer types/subtypes are: acute myeloid leukemia (LAML), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon and rectal adenocarcinoma (COADREAD), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), prostate adenocarcinoma (PRAD), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC).
Quercetin, a bioactive flavonol, is mainly found in plant foods such as onions, apples, and berries.35 It governs various intracellular targets, including the proteins involved in apoptosis (e.g., RELA and PTGS2), cell cycle (e.g., CDKs), antioxidant replication, and metastasis and angiogenesis (e.g., MMPs). Figure 4 b shows that quercetin is predicted to have potential anticancer indications for 12 cancer types, including BLCA (Z = 9.03, q < 1.0 × 10−5), GBM (Z = 6.16, q < 1.0 × 10−5), HNSC (Z = 9.09, q < 1.0 × 10−5), KIRC (Z = 10.58, q < 1.0 × 10−5), LUAD (Z = 11.89, q < 1.0 × 10−5), LUSC (Z = 7.57, q < 1.0 × 10−5), STAD (Z = 8.64, q < 1.0 × 10−5), UCEC (Z = 9.40, q < 1.0 × 10−5), SKCM (Z = 8.27, q < 1.0 × 10−5), PRAD (Z = 5.94, q = 0.0023), BRCA (Z = 5.60, q = 0.0055), and COAD/READ (Z = 5.25, q = 0.0093). Recently, several clinical trials for quercetin to treat or prevent various cancers are ongoing or completed, including prostate cancer (NCT01912820), colorectal cancer (NCT00003365), renal cell carcinoma (NCT02446795), and advanced pancreatic cancer (NCT01879878). Our prediction is consistent with several previous in vitro or in vivo studies for quercetin.36, 37, 38 For example, quercetin targets, such as epidermal growth factor receptor (EGFR) and matrix metalloprotease 2 (MMP2), are significantly mutated in multiple cancer types (Figure 4 b). Both EGFR and MMP2 play important roles in tumor metastasis and angiogenesis.36 Previous studies showed that quercetin acted as a chemopreventive agent against prostate cancer in an in vivo model by inhibiting the EGFR signaling pathway.37 In addition, a previous pharmacological study suggested that quercetin could directly inhibit the activities of MMP2 (IC50 = 6.68 μM) and MMP13 (IC50 = 8.46 μM).38 Hence, targeting MMP2 and EGFR pathways are potential anticancer mechanisms for quercetin.
Genistein belongs to a multifunctional natural isoflavonoid class of flavonoids. Figure 4 c shows that genistein is predicted to have potential anticancer indications for 11 cancer types, such as BLCA (Z = 6.24, q < 1.0 × 10−5), BRCA (Z = 7.03, q < 1.0 × 10−5), and LUAD (Z = 9.74, q < 1.0 × 10−5). A recent study has reported that genistein acted as a chemotherapeutic agent against different types of cancers via altering apoptosis, cell cycle, and angiogenesis or inhibiting metastasis.39 In addition, genistein has been widely used and tested in several clinical cancer studies, such as breast cancer (NCT00244933), bladder cancer (NCT00118040), prostate cancer (NCT01325311), and lung cancer (NCT01628471). Estrogen receptors (ERs) play an important role in the development and progression of breast cancer, acting via two subtypes: ERα and ERβ encoded by ESR1 and ESR2, respectively. Genistein was reported to have strong inhibitory activities (IC50) on human ERα (395 nM) and ERβ (10 nM).40 A recent study showed that genistein inhibited the proliferation and differentiation of MCF‐7 human breast cancer cells via the regulation of ERα expression and induction of apoptosis.41
Similar to quercetin and genistein, the flavonol fisetin is also mostly present in fruits and vegetables. A previous study has suggested that fisetin inhibited cancer growth through alteration of the cell cycle, inducing apoptosis, angiogenesis, invasion, and metastasis.42, 43 Herein, fisetin was predicted to exert anticancer activity for 11 cancer types (Table 2), such as PRAD (Z = 7.41, q < 1.0 × 10−5), LUAD (Z = 9.02, q < 1.0 × 10−5), and LUSC (Z = 7.29, q < 1.0 × 10−5). Figure 4 d shows that fisetin targets protein products of nine SMGs in various cancer types. For example, several fisetin targets, such as Cyclin‐dependent kinases (e.g., CDK2 and CDK4) and AKT, play critical roles in the cell cycle.42, 43 Specifically, fisetin arrests HT29 colon cancer cells from G1 to S phase by inhibiting the activities of CDK2 and CDK4,42 and also arrests A375 melanoma cell growth at the G2 phase through dephosphorylation of AKT and inhibition of its downstream molecules.43
In summary, we computationally identified several potential anticancer indications for resveratrol, quercetin, fisetin, and genistein by targeting various key cancer signaling pathways (Figure 4). These high‐confidence predictions provide potential candidates for future experimental investigations or clinical validations in order to develop new molecularly targeted cancer therapies. However, further in vitro and in vivo experimental assays are needed to validate these predicted anticancer effects and potential MOA before moving to preclinical or clinical studies.
There are several limitations in the current systems pharmacology‐based framework. First, the incomplete drug–target interaction network for natural products curated from publicly available databases may influence the predicted results. Recently, several network‐based approaches provide useful tools for prediction of drug–target interactions with high accuracy.44, 45, 46 Hence, integration of the predicted drug–target interactions for natural products via previously reported network‐based approaches may improve the performance of the systems pharmacology framework. Second, only significantly mutated genes reported by the previous cancer genome projects were used in this study. Data quality and data incompleteness of the cancer mutant genes may be influenced by the limited cohorts of cancer genome projects and tumor heterogeneity. Big cancer samples and types (e.g., rare cancers) are sequenced and their SMGs have been reported or will be released soon. We will integrate the incoming large‐scale cancer genomics data in our future analysis. Third, our validation of the approach is only limited to literature evidence. We will expand the validation by using other approaches or data in the future, like cross‐validation. Fourth, it is commonly known that most cancer significantly mutated genes are not targetable by current small molecular drugs, owing to the “undruggable” properties of certain cancer mutant genes (e.g., KRAS). In addition, tumor suppressor genes (e.g., p53) with loss‐of‐functions cannot be considered as drug targets. One alternative way for development of new therapies for loss‐of‐function genes is to target the pathways or subnetworks (e.g., neighbors in the protein interaction network) perturbed by the inactivation mutations in tumor suppressor genes.28 For example, integrating systems biology resources, such as the human protein–protein interaction network, may assist in targeting the “undruggable” mutant cancer genes for natural products by indirectly targeting their neighbors in the human protein–protein interaction network, regulatory network, or biological pathways.47, 48, 49 Finally, integration of drug‐induced transcriptome data in cancer cells, such as LINCLOUD (http://www.lincscloud.org/), for natural products, may help identify novel anticancer indications of natural products by the systems pharmacology framework in the future.50
In conclusion, this study presents an integrative systems pharmacology framework for identifying potential natural products that may target cancer mutant genes derived from the large‐scale cancer genome sequencing projects. Our approach would provide a useful tool for the development of targeted cancer therapies by exploiting the systems pharmacology of natural products in the post era of cancer genomics studies.
Supporting information
Supporting Information
Supporting Information
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81603318 and 81573020). Dr. Zhao was supported by National Institutes of Health grant R01LM011177. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the article.
Author Contributions
Z.Z., J.F., and F.C. wrote the article; Z.Z., J.F., and F.C. designed the research; J.F. and C.C. performed the research; J.F., C.C., Q.W., P.L., and F.C. analyzed the data; F.C. contributed new reagents/analytical tools.
Conflict of Interest
The authors declare that no conflicts of interest exist.
Contributor Information
Z Zhao, Email: zhongming.zhao@uth.tmc.edu.
F Cheng, Email: fxcheng1985@gmail.com.
References
- 1. Toniatti, C. , Jones, P. , Graham, H. , Pagliara, B. & Draetta, G. Oncology drug discovery: planning a turnaround. Cancer Discov. 4, 397–404 (2014). [DOI] [PubMed] [Google Scholar]
- 2. Rodrigues, T. , Reker, D. , Schneider, P. & Schneider, G. Counting on natural products for drug design. Nat. Chem. 8, 531–541 (2016). [DOI] [PubMed] [Google Scholar]
- 3. Li, J.W. & Vederas, J.C. Drug discovery and natural products: end of an era or an endless frontier? Science 325, 161–165 (2009). [DOI] [PubMed] [Google Scholar]
- 4. DeCorte, B.L. Underexplored opportunities for natural products in drug discovery. J Med. Chem. 59, 9295–9304 (2016). [DOI] [PubMed] [Google Scholar]
- 5. Shen, B. A new golden age of natural products drug discovery. Cell. 163, 1297–1300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee, K.W. , Bode, A.M. & Dong, Z. Molecular targets of phytochemicals for cancer prevention. Nat. Rev. Cancer 11, 211–218 (2011). [DOI] [PubMed] [Google Scholar]
- 7. Cheng, F. , Zhao, J. & Zhao, Z. Advances in computational approaches for prioritizing driver mutations and significantly mutated genes in cancer genomes. Brief. Bioinform. 17, 642–656 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitra, R. & Zhao, Z. The oncogenic and prognostic potential of eight microRNAs identified by a synergetic regulatory network approach in lung cancer. Int. J. Comput. Biol. Drug Des. 7, 384–393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eifert, C. & Powers, R.S. From cancer genomes to oncogenic drivers, tumour dependencies and therapeutic targets. Nat. Rev. Cancer 12, 572–578 (2012). [DOI] [PubMed] [Google Scholar]
- 10. Chin, L. , Andersen, J.N. & Futreal, P.A. Cancer genomics: from discovery science to personalized medicine. Nat. Med. 17, 297–303 (2011). [DOI] [PubMed] [Google Scholar]
- 11. Croce, C.M. Oncogenes and cancer. N. Engl. J. Med. 358, 502–511 (2008). [DOI] [PubMed] [Google Scholar]
- 12. Zhao, J. , Cheng, F. , Wang Y., Arteaga, C.L. & Zhao, Z. Systematic prioritization of druggable mutations in approximately 5000 genomes across 16 cancer types using a structural genomics‐based approach. Mol. Cell. Proteomics 15, 642–656 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gazdar, A.F. & Minna, J.D. Precision medicine for cancer patients: lessons learned and the path forward. J Natl. Cancer Inst. 105, 1262–1263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang, M. , Shen, A. , Ding, J. & Geng, M. Molecularly targeted cancer therapy: some lessons from the past decade. Trends Pharmacol. Sci. 35, 41–50 (2014). [DOI] [PubMed] [Google Scholar]
- 15. Tannock, I.F. & Hickman, J.A. Limits to personalized cancer medicine. N. Engl. J. Med. 375, 1289–1294 (2016). [DOI] [PubMed] [Google Scholar]
- 16. Prasad, V. Perspective: The precision‐oncology illusion. Nature 537, S63 (2016). [DOI] [PubMed] [Google Scholar]
- 17. He, M. , Yan, X. , Zhou, J. & Xie, G. Traditional Chinese medicine database and application on the Web. J. Chem. Inf. Comput. Sci. 41, 273–277 (2001). [DOI] [PubMed] [Google Scholar]
- 18. Shen, J. et al Virtual screening on natural products for discovering active compounds and target information. Curr. Med. Chem. 10, 2327–2342 (2003). [DOI] [PubMed] [Google Scholar]
- 19. Xue, R. , Fang, Z. , Zhang, M. , Yi, Z. , Wen, C. & Shi, T. TCMID: Traditional Chinese Medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res. 41, D1089–1095 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ru, J. et al TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 6, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen, C.Y. TCM Database@Taiwan: the world's largest traditional Chinese medicine database for drug screening in silico. PLoS One 6, e15939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gu, J. , Gui, Y. , Chen, L. , Yuan, G. , Lu, H.Z. & Xu, X. Use of natural products as chemical library for drug discovery and network pharmacology. PLoS One 8, e62839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O'Boyle, N.M. , Banck, M. , James, C.A. , Morley, C. , Vandermeersch, T. & Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 3, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bento, A.P. et al The ChEMBL bioactivity database: an update. Nucleic Acids Res. 42, D1083–1090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gilson, M.K. , Liu, T. , Baitaluk, M. , Nicola, G. , Wang, L. & Chong, J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44, D1045–1053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuhn, M. et al STITCH 4: integration of protein‐chemical interactions with user data. Nucleic Acids Res. 42, D401–407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ye, H. et al HIT: linking herbal active ingredients to targets. Nucleic Acids Res. 39, D1055–1059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng, F. , Zhao, J. , Fooksa, M. & Zhao, Z. A network‐based drug repositioning infrastructure for precision cancer medicine through targeting significantly mutated genes in the human cancer genomes. J. Am. Med. Inform. Assoc. 23, 681–691 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Agarwala, R. et al Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44, D7–19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B (Methodological). 289–300 (1995). [Google Scholar]
- 31. Jang, M. et al Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 275, 218–220 (1997). [DOI] [PubMed] [Google Scholar]
- 32. Levi, F. et al Resveratrol and breast cancer risk. Eur. J. Cancer Prev. 14, 139–142 (2005). [DOI] [PubMed] [Google Scholar]
- 33. Mantovani, F. , Walerych, D. & Del, S.G. Targeting mutant p53 in cancer: a long road to precision therapy. FEBS J.; e-pub ahead of print 2016. doi: 10.1111/febs.13948. [DOI] [PubMed] [Google Scholar]
- 34. Clark, P.A. et al Resveratrol targeting of AKT and p53 in glioblastoma and glioblastoma stem‐like cells to suppress growth and infiltration. J. Neurosurg. 15, 1–13. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khan, F. et al Molecular targets underlying the anticancer effects of quercetin: an update. Nutrients 8, 529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kashyap, D. et al Molecular mechanisms of action of quercetin in cancer: recent advances. Tumour Biol. 37,12927–12939 (2016). [DOI] [PubMed] [Google Scholar]
- 37. Firdous, A.B. et al Quercetin, a natural dietary flavonoid, acts as a chemopreventive agent against prostate cancer in an in vivo model by inhibiting the EGFR signaling pathway. Food Funct. 5, 2632–2645 (2014). [DOI] [PubMed] [Google Scholar]
- 38. Wang, L. et al Natural products as a gold mine for selective matrix metalloproteinases inhibitors. Bioorg. Med. Chem. 20, 4164–4171 (2012). [DOI] [PubMed] [Google Scholar]
- 39. Spagnuolo, C. et al Genistein and cancer: current status, challenges, and future directions. Adv. Nutr. 6, 408–419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Collini, M. D. et al 7‐Substituted 2‐phenyl‐benzofurans as ER beta selective ligands. Bioorg. Med. Chem. Lett. 14, 4925–4929 (2004). [DOI] [PubMed] [Google Scholar]
- 41. Choi, E.J. , Jung, J.Y. & Kim, G.H. Genistein inhibits the proliferation and differentiation of MCF‐7 and 3T3‐L1 cells via the regulation of ERα expression and induction of apoptosis. Exp. Ther. Med. 8, 454–458 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu, X. et al Fisetin inhibits the activities of cyclin‐dependent kinases leading to cell cycle arrest in HT‐29 human colon cancer cells. J. Nutr. 135, 2884–2890 (2005). [DOI] [PubMed] [Google Scholar]
- 43. Syed, D.N. et al Fisetin inhibits human melanoma cell growth through direct binding to p70S6K and mTOR: findings from 3‐D melanoma skin equivalents and computational modeling. Biochem. Pharmacol. 89, 349–360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheng, F. et al Prediction of drug–target interactions and drug repositioning via network‐based inference. PLoS Comput. Biol. 8, e1002503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu, Z. , Cheng, F. , Li, J. , Li, W. , Liu, G. & Tang, Y. SDTNBI: an integrated network and chemoinformatics tool for systematic prediction of drug–target interactions and drug repositioning. Brief. Bioinform.; e-pub ahead of print 2016. doi: 10.1093/bib/bbw012. [DOI] [PubMed] [Google Scholar]
- 46. Wu, Z. et al In silico prediction of chemical mechanism‐of‐action via an improved network‐based inference method. Br. J. Pharmacol. 173, 3372–3385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cheng, F. , Jia, P. , Wang, Q. , Lin, C.C. , Li, W.H. & Zhao, Z. Studying tumorigenesis through network evolution and somatic mutational perturbations in the cancer interactome. Mol. Biol. Evol. 31, 2156–2169 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng, F. , Jia, P. , Wang Q. & Zhao, Z. Quantitative network mapping of the human kinome interactome reveals new clues for rational kinase inhibitor discovery and individualized cancer therapy. Oncotarget 5, 3697–3710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cheng, F. , Murray, J.L. , Zhao, J. , Sheng, J. , Zhao, Z. & Rubin, D.H. Systems biology‐based investigation of cellular antiviral drug targets identified by gene‐trap insertional mutagenesis. PLoS Comput. Biol. 12, e1005074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheng, F. , Hong, H. , Yang, S. & Wei, Y. Individualized network‐based drug repositioning infrastructure for precision oncology in the panomics era. Brief. Bioinform.; e-pub ahead of print 2016. doi: 10.1093/bib/bbw051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information


