Abstract
Two cellular transcription factors, nuclear factor I (NFI) and octamer binding protein (Oct-1), bind simultaneously to their recognition sequences in the Ad5 origin of replication thereby enhancing initiation. Using scanning force microscopy we have previously shown that NFI induces a 60° bend in the origin DNA. Here we demonstrate that Oct-1 induces a 42° bend in the origin DNA. Simultaneous binding of NFI and Oct-1 induces an 82° collective bend suggesting that both bends are oriented towards each other. In functional replication assays we further demonstrate that this extensive DNA bending leads to a synergistic enhancement of DNA replication. We propose that collective DNA bending induced by NFI and Oct-1 facilitates the optimal assembly of the preinitiation complex and plays an important role in the stimulatory mechanism of NFI and Oct-1 in replication.
INTRODUCTION
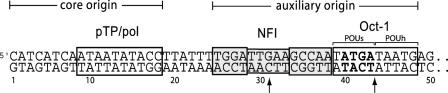
Adenovirus serotype 5 (Ad5) contains a 36 kb double-stranded linear DNA genome with terminal proteins (TP) covalently attached at the 5′ ends. The origins of DNA replication are located at each end of the DNA in 103 bp long inverted terminal repeats (ITR). Three viral proteins are involved in Ad5 DNA replication, precursor terminal protein (pTP) that serves as a primer, adenovirus DNA polymerase (pol) and DNA binding protein (DBP) [reviewed in (1–5)]. pTP and pol form a stable heterodimer in solution and bind as a complex in the conserved core origin at position 9–18 (Figure 1). The core origin is followed by the auxiliary origin that is recognised by two cellular transcription factors, Nuclear Factor I (NFI) and Octamer binding protein (Oct-1) (6) (Figure 1). NFI and Oct-1 considerably enhance the replication process with the highest effect at low (physiological) pol concentrations (7,8).
Figure 1.
The origin of Ad5 DNA replication. The Ad5 origin consists of a highly conserved core origin with the pTP/pol binding site and an auxiliary origin containing the consensus sequences recognised by the NFI and Oct-1 (boxed). The arrows indicate the middle of the NFI consensus sequence at position 31 and the middle of the Oct-1 recognition sequence between position 43 and 44. The binding half-sites for the POUs and POUh are indicated. The ATGA sequence (in bold) within the Oct-1 binding site represents the apparent centre of DNA bending induced by Oct-1 as demonstrated by biochemical assays (37).
NFI binds as a dimer to the consensus sequence 5′-TGGA(N5)GCCAA-3′ located in the Ad5 origin at position 25–38 (9–12) (Figure 1). This sequence is essential for optimal replication (13,14). Contact point analysis demonstrates that almost all contacts of the NFI dimer are clustered at one side of the DNA helix, mainly in the major groove (15,16). The conserved N-terminal domain of NFI is required for DNA binding and protein dimerization and is sufficient for the stimulation of Ad DNA replication (17,18). The C-terminal domain is involved in transcription regulation (18).
Oct-1 binds as a monomer to the recognition sequence 5′-GATAATG-3′ located in the Ad5 origin at position 39–48 (Figure 1) (19,20) and the presence of this sequence in the origin stimulates viral replication (21). The Oct-1 interaction with DNA involves many contact points, mainly in the major groove, that are located on different sides of the DNA helix (22,23). Oct-1 consists of a centrally located bipartite DNA binding domain (POU domain) flanked by two transcription activation domains. The POU domain is sufficient for the stimulatory function of Oct-1 in Ad5 DNA replication (24). It consists of a POU specific domain (POUs) that binds DNA with high specificity but low affinity, and a POU homeodomain (POUh) that binds DNA with low specificity but high affinity (25–27).
There are strict spacing requirements for the position of the NFI and Oct-1 recognition sequences with respect to the core origin bound by the pTP/pol complex. Insertion or deletion of one or two nucleotides between the core and the auxiliary origin severely inhibits the stimulatory function of NFI or Oct-1 (28–31). NFI and Oct-1 have a similar mode of action resulting in stimulation of replication. They stabilise binding of the pTP/pol complex to the origin (7,32). They also influence the kinetics of replication by increasing the Vmax of initiation, without affecting the Km, suggesting that they do not change the activity of the initiating pTP/pol complex, but rather increase the number of active initiation complexes (7). In solution, NFI interacts with pol and Oct-1 interacts with pTP suggesting that NFI and Oct-1 recruit the pTP/pol complex to the origin (17,28,32–34).
In a previous study using scanning force microscopy (SFM), we demonstrated that NFI induces a 60° bend in the Ad5 origin DNA that is important for the stimulation of replication (35). In this study we show that Oct-1 induces a 42° DNA bend in the origin. Moreover, we demonstrate that simultaneous binding of NFI and Oct-1 resulted in an 82° collective bend, indicating that the two bends are in the same direction. Also, stimulation of replication is greatly enhanced when both proteins bind simultaneously compared with the individual stimulation of replication by NFI or Oct-1 alone. We propose that the assembly of the preinitiation complex involves additive bending by NFI and Oct-1 of the origin DNA, thereby facilitating protein–protein and protein–DNA interactions resulting in a maximised stimulation of replication.
MATERIALS AND METHODS
Proteins
For all experiments, the DNA binding domain of rat NFI type A1 (NFI-BD consisting of 5–242 amino acids out of 499 amino acids) and the DNA binding domain of Oct-1 (POU domain consisting of 280–439 amino acids out of 743 amino acids) were used. The NFI-BD will be further referred to in the text as NFI and the Oct-1 POU domain will be referred to as Oct-1. NFI, Ad5 DNA pol, Ad5 pTP and Ad5 DBP were expressed using the baculovirus expression system and the proteins were purified to near homogeneity as described previously (8,34,36,37). Oct-1 was expressed in bacteria as a glutathione S-transferase (GST)-tagged (GST–Oct-1, 47 kDa) or 6×His-tagged (his–Oct-1, 20 kDa) fusion protein and purified as described previously (32,34). The protein dilution buffer contained 25 mM HEPES–KOH, pH 7.5, 150 mM NaCl, 15% glycerol, 5 mM DTT and 0.5 μg/μl BSA.
Scanning force microscopy
For the SFM studies of Oct-1–DNA or NFI–Oct-1–DNA complexes a 717 bp DNA fragment was used. This fragment contains the first 103 bp of the Ad5 ITR from the left end of the viral genome with the Ad5 origin of replication starting from position 209. The Oct-1 binding site (Figure 1) is located between 39 and 48 bp downstream from the beginning of the origin, at position 247 and 256 of the fragment (35% of DNA fragment length). The middle of the auxiliary origin (Figure 1) is located at 34% of the fragment length. The DNA fragment was obtained by PCR using the PHRI plasmid (14) with the following primers: forward 5′-GTGAAATACCGCACAGATGCGTAAGGAG-3′, reverse 5′-CTTTTGCTCACATGTTCTTTCCTGCGTTATC-3′. The PCR fragment was purified from a 1% agarose gel using the QIAquick Gel Extraction Kit (Qiagen) and resuspended in 10 mM HEPES–KOH, pH 8.0. For analysis of a DNA bend induced by Oct-1 alone, GST–Oct-1 of 47 kDa was used to facilitate detection in SFM images, since his–Oct-1 (20 kDa) was difficult to detect (data not shown). However, for the study of the DNA bend induced by NFI and Oct-1 together we did use his–Oct-1 to avoid possible steric hindrance from the GST tag and also because the GST tag apparently interferes with the stimulatory effect of Oct-1 on replication (34). We assume that the bend angles induced by his-Oct-1 and GST–Oct-1 are the same. The SFM depositions were performed as follows. NFI (4.5 pmol) and/or Oct-1 (17 pmol his–Oct-1 or 10 pmol GST–Oct-1) were incubated with the DNA fragment (0.19 pmol) for 20 min on ice in replication buffer (see below). The final mixture was diluted 20–30 times in deposition buffer (5 mM HEPES–KOH, pH 7.8, 5 mM MgCl2) and deposited onto freshly cleaved mica. After 1 min the surface of the mica was washed with 3 ml of high-performance liquid chromatography (HPLC) grade water and dried in a stream of air. The complexes were imaged in the tapping mode using a Nanoscope IIIa (Digital Instruments, Santa Barbara, CA). The DNA contour length and the DNA bend angle were measured using the Image SXM v 1.69 software, an NIH Image version modified for use with SXM images by Steve Barrett, Surface Science Research Centre, University of Liverpool, UK. For each measurement we analysed 70–120 molecules. The data obtained were analysed in Sigma Plot.
Protein–DNA interactions
For the protein–DNA binding studies TD50, a double-stranded oligonucleotide consisting of the first 50 nt of the template (T50) and displaced (D50) strand of the Ad5 origin was used.T50: 5′-CTCATTATCATATTGGCTTCAATCCAAAATAAGGTATATTATTGATGATG-3′, D50: 5′-CATCATCAATAATATACCTTATTTTGGATTGAAGCCAATATGATAATGAG-3′. For the preparation of TD50, the D50 oligonucleotide was end-labelled using T4 polynucleotide kinase and [γ-32P] ATP (4500 Ci/mmol) in a standard kinase buffer. D50 was subsequently hybridized with T50 and the labelled TD50 was purified from a 12% polyacrylamide gel. DNA binding was determined by band shift assays (electrophoretic mobility shift assays) as follows: 0.86 pmol of NFI or/and 2 pmol of his–Oct-1 was incubated with DNA for 60 min on ice in binding buffer (25 mM HEPES–KOH, pH 7.5, 4 mM MgCl2, 0.4 mM DTT, 4% Ficoll, 80 mM NaCl) including 1 μg poly (dI-dC)–(dI-dC) as a non-specific DNA competitor. TD50–protein complexes were separated on a 5% polyacrylamide gel in TBE buffer at 4°C and the intensity of the bands was quantified using a Storm 820 phosphorimager at the linear range of the signal.
In vitro DNA replication
In vitro DNA replication was performed using 9 ng of Ad5 DNA polymerase, 9 ng of Ad5 pTP, 1 μg of Ad5 DBP and the indicated amounts of NFI and his–Oct-1 in a 15 μl reaction mixture containing replication buffer (25 mM HEPES–KOH, pH 7.5, 50 mM NaCl, 1.5 mM MgCl2, 1 mM DTT), 40 μM of dATP, dTTP, dGTP, 0.7 μM of dCTP and 3 μCi of [α-32P] dCTP (3000 Ci/mmol). As a template, 60 ng of XhoI digested viral TP-DNA was used. TP-DNA is the 36 kb linear Ad5 genome with the TP covalently attached to each DNA end that was isolated from Ad5 virions as described previously (38). XhoI digestion generated seven fragments, two of which (6.2 kb fragment B and 5.8 kb fragment C) contain the origin and are replicated. The replication mixtures were incubated for 45 min at 37°C and stopped by the addition of 2 μl stop mix (40% sucrose, 1% SDS, 0.1% bromophenol blue and 0.1% xylene cyanol). The replication products were separated on a 1% agarose gel containing 0.1% SDS in 0.5× TBE/0.1% SDS buffer. The replicated bands were quantified using a Storm 820 phosphorimager at the linear range of the signal. The level of stimulation by NFI and/or Oct-1 was calculated as the ratio between the replication signal (the sum of the ds and the ss B and C bands) in the presence of NFI and/or Oct-1, and the average basal signal.
RESULTS
Oct-1 induces a 42° DNA bend in the Ad5 origin of replication
In order to study Oct-1-induced changes in the origin DNA structure by SFM, a 717 bp double-stranded DNA (dsDNA) fragment containing the Ad5 origin of replication was used. The middle of the Oct-1 binding site (Figure 1) is located at 35% of the fragment length from one end. The remaining part of the DNA contains no obvious Oct-1 recognition sequences. Oct-1 and the 717 bp DNA fragment were incubated at a 53-fold molar excess of Oct-1 over DNA in conditions similar to those used for the functional replication assays.
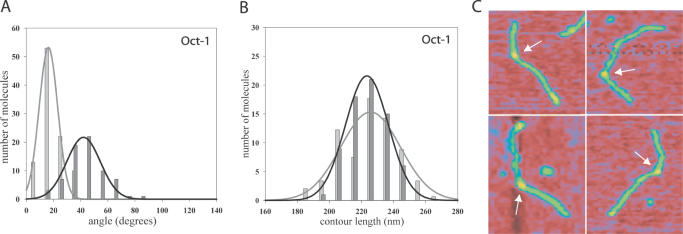
SFM images showed that Oct-1 bound specifically to DNA at the position of the origin (Figure 2C). Very few Oct-1 molecules were bound to other sites on the DNA. The position of Oct-1 bound to DNA was determined by measuring the length of the DNA from the centre of the protein in a protein–DNA complex, to each end of the DNA. The binding position was then expressed as the ratio r of the length of the shorter DNA arm divided by the total DNA length (contour length) with a theoretically calculated value of r = 0.35 for the middle of the Oct-1 consensus sequence in the origin. The protein position was on average 0.35 ± 0.05 (70 molecules measured), which confirms that Oct-1 is specifically bound to the origin (Table 1). Additionally, the DNA contour length of the 717 bp DNA fragment was analysed by tracing the DNA molecules from one end to the other in the absence and in the presence of Oct-1. There were no obvious differences in DNA contour length observed upon Oct-1 binding (Figure 2B, Table 1), showing that Oct-1 does not induce DNA shortening by DNA compaction upon binding or DNA wrapping around Oct-1.
Figure 2.
Oct-1 induces a 42° bend when bound to the Ad5 origin DNA. Two histograms represent the DNA bend angle distributions (A) and contour length distributions (B) of the protein-free DNA molecules [light grey bars, data taken from Mysiak et al. (35)] and the Oct-1–DNA complexes (dark grey bars) measured by SFM. The grey and black lines represent the Gaussian fitting of the distribution as defined in equation 1 of Schulz et al. (39). A 717 bp DNA fragment was used that contains the Ad5 origin of replication with the middle of the Oct-1 recognition sequence at 35% of the DNA fragment. The mean values with standard deviations are presented in Table 1. (C) SFM images of the representative Oct-1–DNA complexes. The arrow indicates Oct-1 bound to DNA. Colour indicates height from 0 nm (red) to 1 nm (yellow).
Table 1. Summary of the SFM data.
| Protein position (r) | Angle (degrees) | Contour length (nm) | |
|---|---|---|---|
| Bare origina | – | 17 ± 7 | 226 ± 19 |
| Origin + NFIa | 0.36 ± 0.09 | 60 ± 19 | 217 ± 26 |
| Origin + Oct-1 | 0.35 ± 0.05 | 42 ± 12 | 223 ± 13 |
| Origin + NFI + Oct-1 | 0.36 ± 0.03 | 73 ± 20, 82b ± 12 | 223 ± 11 |
The DNA bend angle is defined as the angle by which a DNA segment departs from linearity. The bend angle induced at the Ad5 origin by Oct-1 was determined by measuring the angle between two 20 nm long straight segments of DNA on each side of Oct-1 followed by subsequent calculation of the angle by which DNA deviates from its linearity. Gaussian curve fitting according to equation 1 in Schulz et al. (39) was used to determine the average values of bend angles with standard deviations. The intrinsic bending of the origin was determined in the absence of Oct-1 as described in Mysiak et al. (35). The Ad5 origin DNA showed an intrinsic 17° ± 7 DNA bend and after addition of Oct-1 a clear shift in DNA bend angle distribution was observed resulting in the average bend of 42° ± 12 (Figure 2A, Table 1).
Simultaneous binding of NFI and Oct-1 to the Ad5 origin increases DNA bending
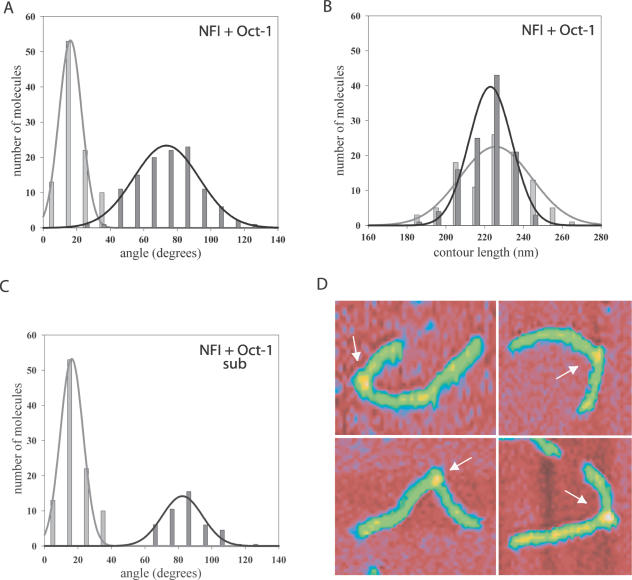
Previously we determined that NFI induces a 60° bend in the Ad5 origin DNA (35). In order to study the collective DNA bending induced by NFI and Oct-1 bound simultaneously to the origin we incubated the 717 bp DNA fragment with NFI and Oct-1 under conditions used for the replication assays. Since NFI binds to the origin with a higher affinity than Oct-1 (25,40,41), we used a 12-fold molar excess of NFI dimers over DNA and a 90-fold excess of Oct-1 over DNA. The protein position on the 113 protein–DNA complexes analysed, corresponded well with the auxiliary origin (experimental value r = 0.36 ± 0.03, theoretical value r = 0.34 calculated for the middle of the auxiliary origin, since we now look at the binding of NFI and Oct-1) (Figure 3D, Table 1). Also, the protein binding was specific since very few molecules were bound outside the origin region. We also analysed the DNA contour length of the 717 bp DNA fragment in the presence of NFI and Oct-1 and no noticeable differences were observed compared with the contour length of the protein-free DNA (Figure 3B, Table 1). As presented in Figure 3A the average DNA bend induced in the origin in the presence of NFI and Oct-1 together was 73° ± 20, which is more that the individual bends induced by NFI or Oct-1 alone, indicating that NF1 and Oct-1 together induce additional bending in the origin DNA.
Figure 3.
The DNA bend angle increases when both NFI and Oct-1 are bound to the Ad5 origin DNA. The SFM data were collected in the presence of both NFI and Oct-1. The histograms represent DNA bend angle distributions (A) and contour length distributions (B) of the protein-free DNA molecules [light grey bars, data from Mysiak et al. (35)] and protein–DNA complexes (dark grey bars). The grey and black lines represent the Gaussian fitting of the distribution. The mean values with standard deviations are presented in Table 1. (C) The distribution of DNA bends induced by NFI alone [based on (35)] was subtracted from the NFI–Oct-1 distribution. (D) SFM images of the representative protein–DNA complexes. The arrow indicates proteins bound to DNA. Colour indicates height from 0 nm (red) to 1 nm (yellow).
One should consider the fact that among the protein–DNA complexes analysed some DNA molecules are bound by NFI alone or Oct-1 alone. In these SFM images it was not possible to distinguish between the different protein–DNA complexes. Due to the small size of Oct-1 used in this experiment (see Materials and Methods), we assumed that Oct-1–DNA complexes are not clearly distinguishable from free DNA and therefore were not selected for analysis. Indeed, the distribution of bend angles induced by Oct-1 alone does not appear to contribute to the bend angle distribution of NFI and Oct-1 together, since in Figure 2A the distribution is centred at 42°, whereas in Figure 3A the distribution is almost empty <50°. Therefore, the distribution measured in the presence of NFI and Oct-1 is made up of two overlapping distributions. One is the distribution of bend angles induced by binding of NFI alone, and the other is the distribution of bend angles induced by NFI and Oct-1 together. Based on the DNA binding data presented below, we assumed that 68% of the complexes are bound by NFI alone under these conditions. In order to eliminate the contribution of DNA bending angles induced by binding of NFI alone, this distribution [based on (35)] of 77 molecules was subtracted from the distribution obtained in the presence of both NFI and Oct-1 (113 molecules, Figure 3C). The remaining distribution represents the DNA bend angle induced by NFI and Oct-1 simultaneously bound to DNA. Similar analysis was performed previously in the study of the photolyase–DNA complexes (42,43). The average angle obtained from this corrected distribution was 82° ± 12 (Figure 3C, Table 1), indicating that the bends are additive.
NFI and Oct-1 bind the Ad5 origin DNA simultaneously
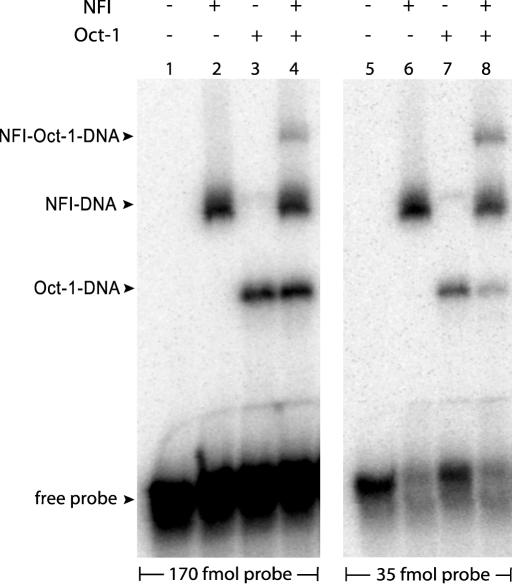
The Ad5 origin DNA binding sites recognized by NFI and Oct-1 are located next to each other (Figure 1). In order to confirm independently that NFI and Oct-1 can bind the origin simultaneously and analyse if their binding is cooperative we performed band shift assays using a double-stranded TD50 DNA fragment representing the first 50 bp of the origin (Figure 4). Two concentrations of the DNA probe were used, 170 fmol (Figure 4, lanes 1–4) and 35 fmol (Figure 4, lanes 5–8). Addition of NFI resulted in the formation of a single NFI–DNA complex (Figure 4, lanes 2, 6) and addition of Oct-1 gave rise to a single Oct-1–DNA complex (Figure 4, lanes 3, 7). When both proteins were incubated with the DNA probe, in addition to single NFI–DNA and Oct-1–DNA complexes, a third supershifted band appeared on the gel (Figure 4, lanes 4, 8). This complex represents both proteins simultaneously bound to a single DNA molecule, demonstrating that NFI and Oct-1 do not exclude each other from binding to the Ad5 origin, despite the close proximity. When 170 fmol of DNA probe was used the simultaneous binding of NFI and Oct-1 to the origin did not seem to be very cooperative, since the supershifted complexes represented 7% of all protein–DNA complexes (Figure 4, lane 4). When 35 fmol of DNA probe was used, which is similar to conditions used for the SFM analysis, the amount of supershifted complexes increased to 22% (Figure 4, lane 8), showing that an increase of the protein–DNA ratio promotes simultaneous binding of NFI and Oct-1. This validates the formation of the NFI–Oct-1–DNA complexes and allows estimation of the proportion of NFI–DNA complexes (68%) in the SFM experiments that included both proteins.
Figure 4.
NFI and Oct-1 bind the Ad5 origin of replication simultaneously. Binding of NFI and Oct-1 to the Ad5 origin was studied by band shift assays using the TD50 probe containing the first 50 nt of the Ad5 origin. In lanes 1–4 and 5–8, respectively, 0.17 pmol and 35 fmol of TD50 probe were used. The position of the protein–DNA complexes and free probe are marked with arrowheads. Lanes 1 and 5 represent free probe.
Simultaneous stimulation of replication by NFI and Oct-1 is cooperative
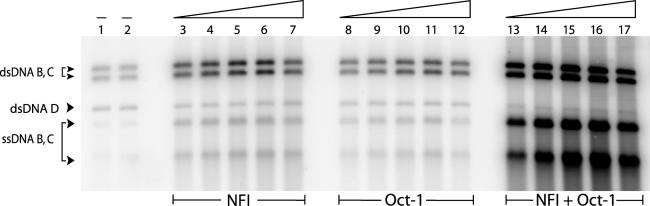
To analyse if simultaneous binding and presumably additive DNA bending of the Ad5 origin by NFI and Oct-1 enhances their stimulatory effect on replication we performed in vitro replication assays. The natural XhoI-digested TP-DNA isolated from Ad5 virions was used as a template. TP-DNA is the 36 kb long linear double stranded viral genome containing TPs covalently attached to each 5′ end. XhoI digestion generated seven fragments, but only two of them contain the origin and are replicated (DNA fragments B and C). First we analysed the ability of NFI or Oct-1 alone to stimulate replication. In order to obtain the maximal stimulatory effect, low pTP and pol concentrations (9 ng pTP and 9 ng pol) were used (6–8). Addition of NFI (Figure 5, lanes 3–7) stimulated replication maximally (9-fold) at 165 ng and addition of Oct-1 (Figure 5, lanes 8–12) stimulated replication maximally (5-fold) at 120 ng.
Figure 5.
NFI and Oct-1 stimulate Ad5 DNA replication. The in vitro replication assays were performed using Ad5 XhoI-digested TP-DNA as a template. As a result of replication two dsDNA fragments containing the origin (dsDNA B and C) and single-strands originated from the second and subsequent rounds of replication (ssDNA B and C) are generated. The D fragment (dsDNA D) is lacking an origin, but is nonspecifically labelled by polymerase. The first two lanes represent the basal level of replication in the absence of NFI and Oct-1. To stimulate replication 50, 75, 110, 165 and 250 ng of NFI (lanes 3–7 and 13–17) or 35, 53, 80, 120 and 180 ng of Oct-1 (lanes 8–12 and 13–17) were added. Lanes 13–17 represent replication in the presence of both NFI and Oct-1. The level of stimulation was determined by comparing the replication signal (the sum of the ds and ss B and C bands) in the presence of NFI and/or Oct-1 with the average basal signal. A second independent experiment gave similar results.
The presence of NFI and Oct-1 resulted in a large stimulation of replication, up to 83-fold at 165 ng of NFI and 120 ng of Oct-1 (Figure 5, lanes 13–17). The reaction was very efficient as evidenced by the large amount of single-stranded DNA (ssDNA) fragments displaced during the replication process, indicative of second and subsequent rounds of replication. Theoretically, the level of combined stimulation by NFI and Oct-1 can be calculated from the multiplication of the stimulation levels of NFI and Oct-1 alone and thus would be 9 × 5 = 45. The 83-fold stimulation observed indicates that NFI and Oct-1 act synergistically in this reaction. Similar levels of synergistic stimulation of replication were observed for other concentrations of NFI and Oct-1 (Figure 5). The molar excess of NFI and Oct-1 over DNA in these reactions was similar to SFM and protein–DNA interaction experiments favouring simultaneous binding of both transcription factors and additive DNA bending.
DISCUSSION
Protein induced DNA bending of the Ad5 origin of replication
In this study, using SFM, we demonstrate that the two DNA bends induced in the Ad5 origin of replication by NFI and Oct-1 are additive, since the collective angle induced by both proteins simultaneously bound to the origin increased up to 82°, compared to the individual angles of 60 and 42° induced by NFI and Oct-1, respectively. Based on biochemical assays and on the co-crystal structure, the Oct-1-induced DNA bend is estimated to be 30–37° with the apparent centre of DNA bending located in the left part of the recognition sequence contacted by POUs (Figure 1) (23,37). However, if both NFI and Oct-1 bends were in the same plane, a calculated theoretical value of the collective bend would be 102°. Since NFI and Oct-1 bind very close to each other (Figure 1), it is possible that they influence each other's ability to bend DNA resulting in a lower value of the collective angle. Alternatively, both DNA bends may not be exactly in the same plane and projection of a 3D arrangement of DNA bends onto a 2D surface may influence the value measured by this method.
Contact point analysis of NFI and Oct-1 binding to the Ad5 origin reveals that both proteins bind to the DNA major groove and that NFI binds on the opposite side of the Ad5 origin DNA than the POUs domain of Oct-1 (15,16,22,23). It is also suggested that the POUs domain of Oct-1 bends DNA by compression of a major groove (37). Up till now the mechanism of DNA bending by NFI is not known. Given the distance between the binding sites of NFI and Oct-1, we propose that in order to gain an additive effect of the collective DNA bending, NFI will most likely bend DNA by broadening of the major groove.
NFI and Oct-1 interaction with the origin DNA
NFI and Oct-1 bind the Ad5 origin in close proximity to each other (Figure 1) (15,20,22). Using band shift assays we showed that despite the close spacing, NFI and Oct-1 can bind to the origin together and they do not exclude each other (Figure 4). Since both proteins have the ability to stimulate binding of the pTP/pol complex to DNA (7,32), possibly by inducing a DNA bend, they could also enhance their simultaneous binding to DNA. However, we did not observe an increased binding affinity of NFI or Oct-1 when both proteins were incubated together with the origin DNA (Figure 4), suggesting that these transcription factors do not bind the origin cooperatively.
In the early stage of infection the amount of a viral DNA in infected cells is minimal, whereas viral DNA replication needs to be enhanced rapidly (6). Since both transcription factors, NFI and Oct-1 are ubiquitously expressed, the concentration ratio between these proteins and viral DNA in the early stage of infection would be very high. We demonstrated that under such conditions NFI and Oct-1 can simultaneously bind to the origin (Figure 4), collectively inducing an 82° bend in the origin DNA and thereby contributing to optimal stimulation of DNA replication (Figure 5). Moreover, at the early stages of infection in vivo pTP/pol concentration is low, which is similar to the condition in which NFI and Oct-1 have the highest stimulatory effect in vitro (6–8,44).
Arrangement of the preinitiation complex
This study reveals that two DNA bends induced in the Ad5 origin by NFI and Oct-1 are oriented towards each other resulting in an increased DNA curvature of 82°. Assembly of the preinitiation complex involves several protein–protein interactions among the proteins involved in Ad5 DNA replication (17,28,34,45–47). Since all these proteins interact with an origin consisting of only 50 bp (Figure 1), extensive DNA bending by NFI and Oct-1 would facilitate the formation of an optimal nucleoprotein structure of the preinitiation complex. The resulting specific arrangement of protein–protein and protein–DNA interactions is presumably required for optimal stimulation of replication. We have previously shown that the ability of NFI to stimulate replication depends specifically on the degree of DNA bending, since the introduction of mutations in the origin leading to a DNA bend angle <60° reduced this activity (35). Protein-induced DNA bending is an important architectural feature of specific nucleoprotein complexes involved in several different DNA replication processes as well as in the regulation of transcription and DNA repair (48–54).
In addition to DNA bending by NFI and Oct-1, there are other changes induced in the structure of the origin DNA. DBP changes the DNA structure in a way that apparently facilitates binding of NFI and the pTP/pol complex to the origin (55–58). During initiation the core origin has to be unwound since pol uses ssDNA as a template for initiation while the displaced strand is protected by DBP (59,60). Another distortion is the intrinsic 17° bend of the origin (16,35). Moreover, the high content of A and T bases is likely to facilitate DNA destabilisation, unwinding or even bending (61). Indeed, the A/T-rich stretch located between the core origin and auxiliary origin is essential for an optimal bend angle induced by NFI (35), although it is not involved in DNA bending and stimulation of replication by Oct-1 (data not shown).
The precise position of the NFI and Oct-1 recognition sequences with respect to the core origin (Figure 1) is very important, since insertion or deletion of only one or two nucleotides in front of the auxiliary origin abolishes the stimulatory effect of NFI or Oct-1 (28–31). We propose that such insertions or deletions would change the direction of the DNA bend induced by NFI or Oct-1 in relation to the core origin DNA bound by the pTP/pol complex. Consequently this change in bend direction could disrupt the optimal architecture of the replication complex and reduce the stimulatory effect of NFI or Oct-1 on replication. The direction of protein-induced DNA bending was shown to play an important role in other examples of replication and transcription (62,63).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Lars Meijer, Richard Heideman and Marjoleine Bleijenberg for helpful discussions.
REFERENCES
- 1.Hay R.T. (1996) Adenovirus DNA replication. In DePamphilis,M. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 699–719. [Google Scholar]
- 2.Hay R.T., Freeman,A., Leith,I., Monaghan,A. and Webster,A. (1995) Molecular interactions during adenovirus DNA replication. Curr. Top. Microbiol. Immunol., 199, 31–48. [DOI] [PubMed] [Google Scholar]
- 3.Liu H., Naismith,J.H. and Hay,R.T. (2003) Adenovirus DNA replication. Curr. Top. Microbiol. Immunol., 272, 131–164. [DOI] [PubMed] [Google Scholar]
- 4.van der Vliet P.C. (1995) Adenovirus DNA replication. Curr. Top. Microbiol. Immunol., 199, 1–30. [DOI] [PubMed] [Google Scholar]
- 5.de Jong R.N., van der Vliet,P.C. and Brenkman,A.B. (2003) Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP. Curr. Top. Microbiol. Immunol., 272, 187–211. [DOI] [PubMed] [Google Scholar]
- 6.de Jong R.N. and van der Vliet,P.C. (1999) Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene, 236, 1–12. [DOI] [PubMed] [Google Scholar]
- 7.Mul Y.M. and van der Vliet,P.C. (1992) Nuclear factor I enhances adenovirus DNA replication by increasing the stability of a preinitiation complex. EMBO J., 11, 751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mul Y.M., Verrijzer,C.P. and van der Vliet,P.C. (1990) Transcription factors NFI and NFIII/Oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J. Virol., 64, 5510–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gronostajski R.M., Adhya,S., Nagata,K., Guggenheimer,R.A. and Hurwitz,J. (1985) Site-specific DNA binding of nuclear factor I: analyses of cellular binding sites. Mol. Cell. Biol., 5, 964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gronostajski R.M., Knox,J., Berry,D. and Miyamoto,N.G. (1988) Stimulation of transcription in vitro by binding sites for nuclear factor I. Nucleic Acids Res., 16, 2087–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leegwater P.A., Van Driel,W. and van der Vliet,P.C. (1985) Recognition site of nuclear factor I, a sequence-specific DNA-binding protein from HeLa cells that stimulates adenovirus DNA replication. EMBO J., 4, 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagata K., Guggenheimer,R.A. and Hurwitz,J. (1983) Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc. Natl Acad. Sci. USA, 80, 6177–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay R.T. (1985) Origin of adenovirus DNA replication. Role of the nuclear factor I binding site in vivo. J. Mol. Biol., 186, 129–136. [DOI] [PubMed] [Google Scholar]
- 14.Hay R.T. (1985) The origin of adenovirus DNA replication: minimal DNA sequence requirement in vivo. EMBO J., 4, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Vries E., Van Driel,W., van den Heuvel,S.J. and van der Vliet,P.C. (1987) Contactpoint analysis of the HeLa nuclear factor I recognition site reveals symmetrical binding at one side of the DNA helix. EMBO J., 6, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zorbas H., Rogge,L., Meisterernst,M. and Winnacker,E.L. (1989) Hydroxyl radical footprints reveal novel structural features around the NF I binding site in adenovirus DNA. Nucleic Acids Res., 17, 7735–7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M., Mermod,N. and Horwitz,M.S. (1990) Protein–protein interactions between adenovirus DNA polymerase and nuclear factor I mediate formation of the DNA replication preinitiation complex. J. Biol. Chem., 265, 18634–18642. [PubMed] [Google Scholar]
- 18.Mermod N., O'Neill,E.A., Kelly,T.J. and Tjian,R. (1989) The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell, 58, 741–753. [DOI] [PubMed] [Google Scholar]
- 19.Verrijzer C.P., van Oosterhout,J.A. and van der Vliet,P.C. (1992) The Oct-1 POU domain mediates interactions between Oct-1 and other POU proteins. Mol. Cell. Biol., 12, 542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruijn G.J., Van Driel,W. and van der Vliet,P.C. (1986) Nuclear factor III, a novel sequence-specific DNA-binding protein from HeLa cells stimulating adenovirus DNA replication. Nature, 322, 656–659. [DOI] [PubMed] [Google Scholar]
- 21.Hatfield L. and Hearing,P. (1993) The NFIII/OCT-1 binding site stimulates adenovirus DNA replication in vivo and is functionally redundant with adjacent sequences. J. Virol., 67, 3931–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruijn G.J., van Miltenburg,R.T., Claessens,J.A. and van der Vliet,P.C. (1988) Interaction between the octamer-binding protein nuclear factor III and the adenovirus origin of DNA replication. J. Virol., 62, 3092–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klemm J.D., Rould,M.A., Aurora,R., Herr,W. and Pabo,C.O. (1994) Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell, 77, 21–32. [DOI] [PubMed] [Google Scholar]
- 24.Verrijzer C.P., Kal,A.J. and van der Vliet,P.C. (1990) The DNA binding domain (POU domain) of transcription factor oct-1 suffices for stimulation of DNA replication. EMBO J., 9, 1883–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verrijzer C.P., Alkema,M.J., van Weperen,W.W., van Leeuwen,H.C., Strating,M.J. and van der Vliet,P.C. (1992) The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J., 11, 4993–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dekker N., Cox,M., Boelens,R., Verrijzer,C.P., van der Vliet,P.C. and Kaptein,R. (1993) Solution structure of the POU-specific DNA-binding domain of Oct-1. Nature, 362, 852–855. [DOI] [PubMed] [Google Scholar]
- 27.Sturm R.A. and Herr,W. (1988) The POU domain is a bipartite DNA-binding structure. Nature, 336, 601–604. [DOI] [PubMed] [Google Scholar]
- 28.Bosher J., Robinson,E.C. and Hay,R.T. (1990) Interactions between the adenovirus type 2 DNA polymerase and the DNA binding domain of nuclear factor I. New Biol., 2, 1083–1090. [PubMed] [Google Scholar]
- 29.Adhya S., Shneidman,P.S. and Hurwitz,J. (1986) Reconstruction of adenovirus replication origins with a human nuclear factor I binding site. J. Biol. Chem., 261, 3339–3346. [PubMed] [Google Scholar]
- 30.Wides R.J., Challberg,M.D., Rawlins,D.R. and Kelly,T.J. (1987) Adenovirus origin of DNA replication: sequence requirements for replication in vitro. Mol. Cell. Biol., 7, 864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coenjaerts F.E., De Vries,E., Pruijn,G.J., Van Driel,W., Bloemers,S.M., Van der Lugt,N.M. and van der Vliet,P.C. (1991) Enhancement of DNA replication by transcription factors NFI and NFIII/Oct-1 depends critically on the positions of their binding sites in the adenovirus origin of replication. Biochim. Biophys. Acta, 1090, 61–69. [DOI] [PubMed] [Google Scholar]
- 32.van Leeuwen H.C., Rensen,M. and van der Vliet,P.C. (1997) The Oct-1 POU homeodomain stabilizes the adenovirus preinitiation complex via a direct interaction with the priming protein and is displaced when the replication fork passes. J. Biol. Chem., 272, 3398–3405. [DOI] [PubMed] [Google Scholar]
- 33.Botting C.H. and Hay,R.T. (1999) Characterisation of the adenovirus preterminal protein and its interaction with the POU homeodomain of NFIII (Oct-1). Nucleic Acids Res., 27, 2799–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Jong R.N., Mysiak,M.E., Meijer,L.A., van der Linden,M. and van der Vliet,P.C. (2002) Recruitment of the priming protein pTP and DNA binding occur by overlapping Oct-1 POU homeodomain surfaces. EMBO J., 21, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mysiak M.E., Bleijenberg,M.H., Wyman,C., Holthuizen,P.E. and van der Vliet,P.C. (2004) Bending of adenovirus origin DNA by nuclear factor I as shown by scanning force microscopy is required for optimal DNA replication. J. Virol., 78, 1928–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brenkman A.B., Heideman,M.R., Truniger,V., Salas,M. and van der Vliet,P.C. (2001) The (I/Y)XGG motif of adenovirus DNA polymerase affects template DNA binding and the transition from initiation to elongation. J. Biol. Chem., 276, 29846–29853. [DOI] [PubMed] [Google Scholar]
- 37.Verrijzer C.P., van Oosterhout,J.A., van Weperen,W.W. and van der Vliet,P.C. (1991) POU proteins bend DNA via the POU-specific domain. EMBO J., 10, 3007–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coenjaerts F.E. and van der Vliet,P.C. (1995) Adenovirus DNA replication in a reconstituted system. Meth. Enzymol., 262, 548–560. [DOI] [PubMed] [Google Scholar]
- 39.Schulz A., Mucke,N., Langowski,J. and Rippe,K. (1998) Scanning force microscopy of Escherichia coli RNA polymerase sigma 54 holoenzyme complexes with DNA in buffer and in air. J. Mol. Biol., 283, 821–836. [DOI] [PubMed] [Google Scholar]
- 40.Rosenfeld P.J. and Kelly,T.J. (1986) Purification of nuclear factor I by DNA recognition site affinity chromatography. J. Biol. Chem., 261, 1398–1408. [PubMed] [Google Scholar]
- 41.Verrijzer C.P., Kal,A.J. and van der Vliet,P.C. (1990) The oct-1 homeo domain contacts only part of the octamer sequence and full oct-1 DNA-binding activity requires the POU-specific domain. Genes Dev., 4, 1964–1974. [DOI] [PubMed] [Google Scholar]
- 42.van Noort J., Orsini,F., Eker,A., Wyman,C., de Grooth,B. and Greve,J. (1999) DNA bending by photolyase in specific and non-specific complexes studied by atomic force microscopy. Nucleic Acids Res., 27, 3875–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bustamante C. and Rivetti,C. (1996) Visualizing protein–nucleic acid interactions on a large scale with the scanning force microscope. Annu. Rev. Biophys. Biomol. Struct., 25, 395–429. [DOI] [PubMed] [Google Scholar]
- 44.Ramachandra M. and Padmanabhan,R. (1995) Expression, nuclear transport, and phosphorylation of adenovirus DNA replication proteins. Curr. Top. Microbiol. Immunol., 199, 50–88. [PubMed] [Google Scholar]
- 45.de Jong R.N., Meijer,L.A.T. and van der Vliet,P.C. (2003) DNA binding properties of the adenovirus DNA replication priming protein pTP. Nucleic Acids Res., 31, 3274–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enomoto T., Lichy,J.H., Ikeda,J.E. and Hurwitz,J. (1981) Adenovirus DNA replication in vitro: purification of the terminal protein in a functional form. Proc. Natl Acad. Sci. USA, 78, 6779–6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coenjaerts F.E., van Oosterhout,J.A. and van der Vliet,P.C. (1994) The Oct-1 POU domain stimulates adenovirus DNA replication by a direct interaction between the viral precursor terminal protein–DNA polymerase complex and the POU homeodomain. EMBO J., 13, 5401–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grosschedl R. (1995) Higher-order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr. Opin. Cell Biol., 7, 362–370. [DOI] [PubMed] [Google Scholar]
- 49.Guo F., Gopaul,D.N. and Van Duyne,G.D. (1999) Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proc. Natl Acad. Sci. USA, 96, 7143–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janicijevic A., Sugasawa,K., Shimizu,Y., Hanaoka,F., Wijgers,N., Djurica,M., Hoeijmakers,J.H. and Wyman,C. (2003) DNA bending by the human damage recognition complex XPC-HR23B. DNA Repair (Amst), 2, 325–336. [DOI] [PubMed] [Google Scholar]
- 51.Carr E.A., Mead,J. and Vershon,A.K. (2004) Alpha1-induced DNA bending is required for transcriptional activation by the Mcm1–alpha1 complex. Nucleic Acids Res., 32, 2298–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scaffidi P. and Bianchi,M.E. (2001) Spatially precise DNA bending is an essential activity of the sox2 transcription factor. J. Biol. Chem., 276, 47296–47302. [DOI] [PubMed] [Google Scholar]
- 53.Schultz J.R., Loven,M.A., Melvin,V.M., Edwards,D.P. and Nardulli,A.M. (2002) Differential modulation of DNA conformation by estrogen receptors alpha and beta. J. Biol. Chem., 277, 8702–8707. [DOI] [PubMed] [Google Scholar]
- 54.Gillitzer E., Chen,G. and Stenlund,A. (2000) Separate domains in E1 and E2 proteins serve architectural and productive roles for cooperative DNA binding. EMBO J., 19, 3069–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cleat P.H. and Hay,R.T. (1989) Co-operative interactions between NFI and the adenovirus DNA binding protein at the adenovirus origin of replication. EMBO J., 8, 1841–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Breukelen B., Holthuizen,P. and van der Vliet,P.C. (2002) Adenovirus type 5 DNA binding protein stimulates binding of DNA polymerase to the replication origin. J. Virol., 77, 915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stuiver M.H., Bergsma,W.G., Arnberg,A.C., van Amerongen,H., van Grondelle,R. and van der Vliet,P.C. (1992) Structural alterations of double-stranded DNA in complex with the adenovirus DNA-binding protein. Implications for its function in DNA replication. J. Mol. Biol., 225, 999–1011. [DOI] [PubMed] [Google Scholar]
- 58.Stuiver M.H. and van der Vliet,P.C. (1990) Adenovirus DNA-binding protein forms a multimeric protein complex with double-stranded DNA and enhances binding of nuclear factor I. J. Virol., 64, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Breukelen B., Kanellopoulos,P.N., Tucker,P.A. and van der Vliet,P.C. (2000) The formation of a flexible DNA-binding protein chain is required for efficient DNA unwinding and adenovirus DNA chain elongation. J. Biol. Chem., 275, 40897–40903. [DOI] [PubMed] [Google Scholar]
- 60.Franklin M.C., Wang,J. and Steitz,T.A. (2001) Structure of the replicating complex of a pol alpha family DNA polymerase. Cell, 105, 657–667. [DOI] [PubMed] [Google Scholar]
- 61.Ramstein J. and Lavery,R. (1988) Energetic coupling between DNA bending and base pair opening. Proc. Natl Acad. Sci. USA, 85, 7231–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bashaw J.M. and Yates,J.L. (2001) Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J. Virol., 75, 10603–10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perez-Martin J. and Espinosa,M. (1991) The RepA repressor can act as a transcriptional activator by inducing DNA bends. EMBO J., 10, 1375–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]