Abstract
Jumonji domain-containing protein 1A (JMJD1A) plays a key role in the development and progression of several cancers. Here, we showed that the expression of JMJD1A is increased in cervical cancer cells and tissues, and that suppression of JMJD1A inhibits proliferation, migration, and invasion of cervical cancer cells. JMJD1A induced transcription of c-Myc, which is essential for cervical cancer growth and progression. Clinical data showed that JMJD1A expression correlated with lymph node metastasis (P=0.031) and FIGO stage (P=0.007). Increased c-Myc levels were associated with tumor differentiation (P=0.007) and FIGO stage (P<0.001). JMJD1A protein levels correlated with c-Myc expression (P<0.001), and high co-expression of the two proteins correlated with a poor prognosis. Survival analysis showed that JMJD1A and c-Myc levels are independent prognostic factors for cervical cancer patients. These results suggest that JMJD1A is a promising therapeutic target in cervical cancer.
Keywords: cervical cancer, JMJD1A, c-Myc, prognosis, immunohistochemistry
INTRODUCTION
Cervical cancer continues to be a leading cause of death among women in less developed countries including China [1, 2]. Many recent studies have focused on cervical cancer, leading to reduced mortality rates; however, the underlying molecular mechanisms remain unclear [3-6]. The diversity of pathological characteristics complicates the study of cervical cancer molecular mechanisms. Precision medicine may help identify effective treatments on an individual basis in the future [7-9].
Jumonji domain containing 1A (JMJD1A), also known as JHDM2A and KDM3A, is a histone demethylase that removes mono- and di-methyl groups from histone H3K9 (specifically, from H3K9me1 or H3K9me2) [10], thereby mediating transcriptional activation [11, 12]. JMJD1A acts on various target genes to regulate such processes as metabolism, spermatogenesis, stem cell self-renewal, sex determination, and differentiation [13-15]. Recent studies have also indicated that increased JMJD1A expression is associated with the development of different types of cancer, including renal cell carcinoma [16], prostate cancer [17], and liver cancer [18]. In addition, elevated JMJD1A levels have been associated with poor cancer prognosis [18-20]. However, the expression patterns and the potential prognostic role of JMJD1A in the pathogenesis of cervical cancer remain unclear. In the present study, we investigated the expression and function of JMJD1A in cervical cancer cells and assessed its prognostic significance in cervical cancer.
RESULTS
JMJD1A expression is increased in cervical cancer
We initially analyzed JMJD1A expression in samples of tumor and adjacent normal tissues from ten patients with cervical cancer. The results showed that in eight of the tumor samples, JMJD1A expression was increased as compared to matched paracancerous controls (P<0.05, Figure 1A). JMJD1A mRNA and protein levels in the cervical cancer HeLa, SiHa, ME-180, and C-33A cells were higher than in the non-tumorigenic human epithelial cell line HaCaT (P<0.05, Figure 1B, 1C). These data indicate that the JMJD1A expression is increased in cervical cancer.
Figure 1. JMJD1A expression level is significantly upregulated in cervical cancer.
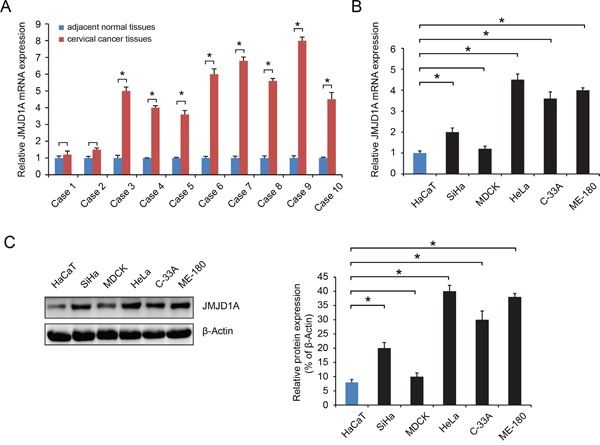
A. RT-qPCR was conducted to examine the expression levels of JMJD1A in 10 cases of cervical cancer tissues and their matched adjacent non-tumor tissues. JMJD1A expression was significantly higher in eight cases of tumor tissues than in matched paracancerous controls. B and C. RT-qPCR (B) and western blot (C) was employed to examine the expression levels of JMJD1A in five common human cervical cell lines, HeLa, SiHa, ME-180 and C-33A, as well as in a non-tumorigenic human epithelial cell line HaCaT. JMJD1A mRNA and protein expression was higher in cervical cell lines. We have set the mRNA and protein control levels to 1.0, relative to internal standards GAPDH. *P < 0.05.
Downregulation of JMJD1A inhibits cervical cancer cell proliferation, migration and invasion in vitro
We used a lentiviral shRNA vector to stably knock down the expression of JMJD1A in HeLa cells, which have relatively high levels of JMJD1A (P<0.05, Figure 1B, 1C). Compared to negative control, silencing with siJMJD1A suppressed more than 80% of the JMJD1A protein expression (P<0.05, Figure 2A). Thus, stably transfected cells HeLa-conJMJD1A and HeLa-siJMJD1A were used in the following experiments.
Figure 2. Stable downregulation of JMJD1A expression inhibits cervical cancer cell proliferation, migration and invasion in vitro.
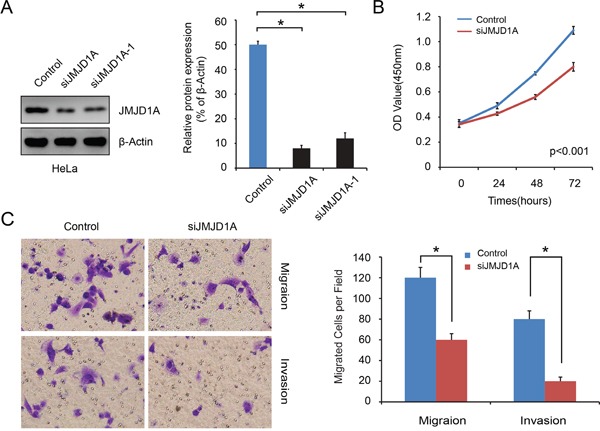
A. Western blot analysis was performed to detect the knockdown effect on JMJD1A expression in HeLa cell. It was found that siJMJD1A achieved a high efficacy in silencing JMJD1A expression compared to the negative control. B. Effect of JMJD1A knockdown on proliferation was evaluated by Cell Counting Kit-8 assays. Suppressing JMJD1A significantly reduced cell proliferation in comparison with control. C. Transwell and Boyden chamber assay was conducted to examine cell migration and invasion ability. Knockdown of JMJD1A clearly inhibits cell migration and invasion of pancreatic cancer in vitro.*P < 0.05.
We used CCK-8 assays to examine the effect JMJD1A expression on cervical cancer cell growth. The growth curves showed that suppression of JMJD1A significantly reduced cell proliferation in comparison with control (P<0.05, Figure 2B). Furthermore, Transwell and Boyden chamber assays showed that there were significantly fewer migrated cells in the siJMJD1A group than in the control group (P<0.05, Figure 2C). To eliminate the possibility that the decrease in cell migration and invasion in JMJD1A-suppressed cells was a secondary consequence of reduced cell proliferation, we placed serum-free medium in the upper chambers. These in vitro results indicate that JMJD1A knockdown inhibits cervical cancer cell proliferation, migration and invasion.
JMJD1A regulates c-Myc transcriptional activity
An earlier study showed that JMJD1A stimulates prostate cancer cell proliferation by increasing c-Myc expression [21]. Our results showed that JMJD1A suppression decreased c-Myc expression in cervical cancer cells (P<0.05, Figure 3A, 3B). Using luciferase assays with transfected cancer cells (HeLa-conJMJD1A or HeLa-siJMJD1A), we showed that JMJD1A knockdown significantly inhibited c-Myc expression (P<0.05, Figure 3C). To further provide evidence that JMJD1A binds directly to the c-Myc promoter, we conducted a Chip assay in HeLa-conJMJD1A and HeLa-siJMJD1A cells. Compared to HeLa-conJMJD1A cells, HeLa-siJMJD1A cells exhibited a reduced JMJD1A recruitment to c-Myc promoter (P<0.05, Figure 3D). In contrast, knockdown of JMJD1A increased H3K9me2 levels at the c-Myc promoter (P<0.05, Figure 3E).
Figure 3. JMJD1A regulates c-Myc transcriptional activity.
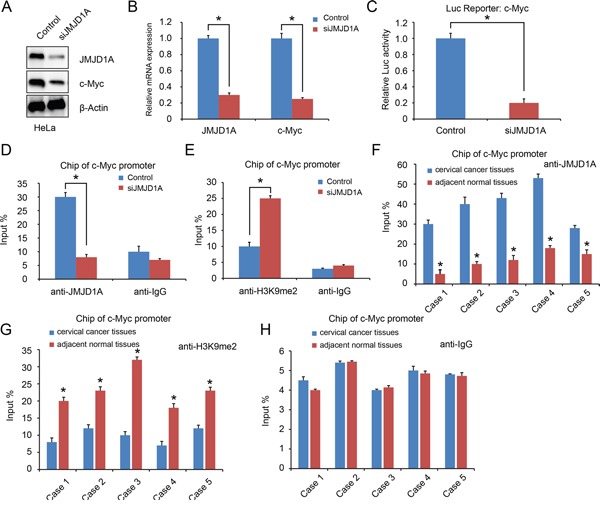
A and B. JMJD1A knockdown dramatically decreased c-Myc expression at protein (A) and mRNA (B) level. C. HeLa cells were transfected with the indicated c-Myc promoter reporter containing JMJD1A-binding sites. The c-Myc promoter activity was normalized via co-transfection of a β-actin/Renilla luciferase reporter containing a full-length Renilla luciferase gene. The luciferase activity in the HeLa cells was quantified using a dual luciferase assay system (Promega) 24 hours after transfection. Renilla expression plasmid was used as an internal control. Knockdown of JMJD1A significantly inhibited the c-Myc promoter activity. D and E. Chip assay using chromatins prepared from HeLa-conJMJD1A and HeLa-siJMJD1A. Chip analysis was conducted on c-Myc promoter regions using the indicated antibodies. Enrichment was determined relative to input controls. (D). Compare to the HeLa-conJMJD1A group, reduced JMJD1A recruitment to the c-Myc promoter was found in HeLa-siJMJD1A. (E). Knockdown of JMJD1A increased H3K9me2 levels at the c-Myc promoter significantly. F, G and H. Chip assay using chromatins prepared from cervical cancer tissues and adjacent normal tissues. (F). Compare to the adjacent normal tissues, increased JMJD1A recruitment to the c-Myc promoter was found in cervical cancer tissues. (G). Enhanced H3K9me2 levels at the c-Myc promoter in cervical cancer tissues significantly. (H). Normal human IgG was used as a negative control. *P < 0.05.
Next, we analyzed JMJD1A recruitment to c-Myc promoter in five paired cervical cancer tissues and adjacent normal tissues. Compared with normal tissues, in cervical cancer tissues, JMJD1A recruitment to c-Myc promoter was increased, while H3K9me2 recruitment was decreased (P<0.05, Figure 3F-3H). Taken together, these data suggest that JMJD1A binds to c-Myc promoter to mediate c-Myc transcription in cervical cancer cells.
JMJD1A-induced cell proliferation, migration, and invasion is dependent on c-Myc in cervical cancer cells
When we overexpressed c-Myc in HeLa-siJMJD1A and HeLa-conJMJD1A cells, expression of c-Myc was increased in HeLa-siJMJD1A-c-Myc cells as compared to HeLa-conJMJD1A-pcDNA3.0 cells (P<0.05, Figure 4A, 4B). HeLa-siJMJD1A-c-Myc cells exhibited increased proliferation, migration, and invasive ability compared to HeLa-conJMJD1A-pcDNA3.0 cells (P<0.05, Figure 4C-4E). Thus JMJD1A-induced cervical cancer cell proliferation, migration, and invasion are dependent on c-Myc.
Figure 4. Overexpression of c-Myc rescues the loss of JMJD1A-mediated repression activity in cervical cancer.
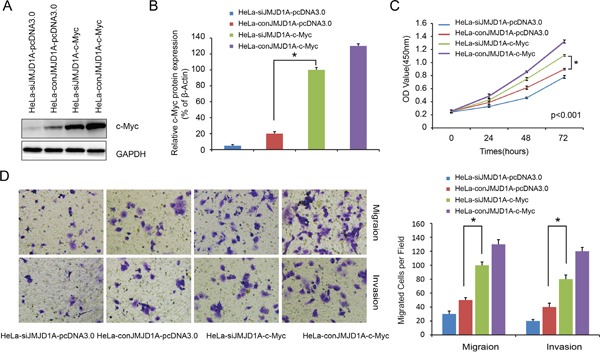
A. Western blot analysis was performed to detect the c-Myc expression. We found that c-Myc expression was upregulated in HeLa-siJMJD1A-c-Myc compared to HeLa-conJMJD1A-pcDNA3.0 cell. B. Effect of c-Myc overexpression on proliferation was evaluated by Cell Counting Kit-8 assays. The cell proliferation ability was increased in HeLa-siJMJD1A-c-Myc compared to HeLa-conJMJD1A-pcDNA3.0 cell. C. Transwell and Boyden chamber assay was performed to examine cell migration and invasion ability. The cell migration and invasion ability was increased in HeLa-siJMJD1A-c-Myc compared to HeLa-conJMJD1A-pcDNA3.0 cell. *P < 0.05.
JMJD1A and c-Myc protein levels correlate with pathologic features of cervical cancer
To further confirm this hypothesis that JMJD1A regulates c-Myc transcription, we investigated the expression of JMJD1A and c-Myc in 80 cervical cancer specimens by immunohistochemical analysis. Both JMJD1A (Figure 5A, low; Figure 5B, High) and c-Myc (Figure 5C, low; Figure 5D, High) were localized in the nucleus. Association of JMJD1A and c-Myc expression with clinicopathological factors is summarized in Table 1. Increased JMJD1A expression correlated with lymph node metastasis (P=0.031) and FIGO stage (P=0.007), while increased c-Myc expression correlated with tumor differentiation (P=0.007) and FIGO stage (P<0.001). Furthermore, we found a positive correlation between JMJD1A and c-Myc expression (P<0.001, Table 2), with Spearman correlation coefficients of 0.397.
Figure 5. Immunohistochemical expression levels and localization of JMJD1A and c-Myc in cervical cancer tissues.
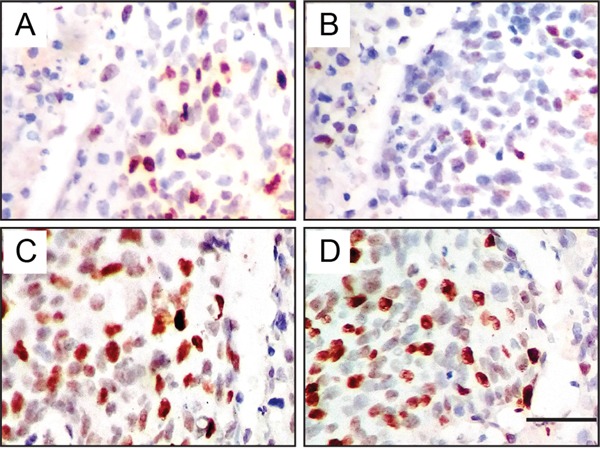
Both JMJD1A (A, low; B, High) and c-Myc (C, low; D, High) are localized in the nucleus. Representative pictures show positive correlation of concomitant expression of JMJD1A and c-Myc. A, C. Low JMJD1A and c-Myc expression from one patient with FIGO stage I. B, D. High JMJD1A and c-Myc expression from another patient with FIGO stage II.
Table 1. Correlation between the clinicopathologic characteristics and JMJD1A and c-Myc expression in cervical cancer (n = 80).
| Variable | Cases | JMJD1A expression | P-value | c-Myc expression | P-value | ||
|---|---|---|---|---|---|---|---|
| Low(n=34) | High(n=46) | Low(n=38) | High(n=42) | ||||
| Age(years) | |||||||
| <45 | 32 | 13 | 19 | 0.782 | 11 | 21 | 0.055 |
| ≥45 | 48 | 21 | 27 | 27 | 21 | ||
| Tumor size(cm) | |||||||
| <4 | 42 | 16 | 26 | 0.402 | 20 | 22 | 0.982 |
| ≥4 | 38 | 18 | 20 | 18 | 20 | ||
| Histological type | |||||||
| SCC | 62 | 27 | 35 | 0.725 | 33 | 29 | 0.057 |
| NSCC | 18 | 7 | 11 | 5 | 13 | ||
| Parametrial infiltration | |||||||
| No | 36 | 12 | 24 | 0.134 | 14 | 22 | 0.163 |
| Yes | 44 | 22 | 22 | 24 | 20 | ||
| Differentiation | |||||||
| Grade 1+2 | 51 | 21 | 30 | 0.751 | 30 | 21 | 0.007* |
| Grade 3 | 29 | 13 | 16 | 8 | 21 | ||
| Lymph node metastasis | |||||||
| No | 67 | 32 | 35 | 0.031* | 34 | 33 | 0.187 |
| Yes | 13 | 2 | 11 | 4 | 9 | ||
| FIGO stage | |||||||
| <II | 31 | 19 | 12 | 0.007* | 23 | 8 | <0.001* |
| ≥II | 49 | 15 | 34 | 15 | 34 | ||
SCC, squamous cell carcinoma; NSC, non-squamous cell cancer; FIGO, International Federation of Gynecology and Obstetrics
P<0.05 indicates a significant association among the variables(2-tailed).
Table 2. Association between JMJD1A and c-Myc expression in cervical cancer.
| Tissue sample | c-Myc expression | r | P-value | |
|---|---|---|---|---|
| Low(n=38) | High(n=42) | |||
| JMJD1A Low(n=34) | 24 | 10 | 0.397 | <0.001* |
| JMJD1A High(n=46) | 14 | 32 | ||
P<0.05 indicates that correlation is significant at the 0.05 level(2-tailed).
JMJD1A and c-Myc protein levels correlate with survival of cervical cancer patients
Kaplan-Meier analysis and log-rank test were used to investigate the prognostic value of JMJD1A and c-Myc protein levels and classic clinicopathological characteristics of patients' survival. Using univariate analysis, both JMJD1A and c-Myc protein levels were closely associated with overall survival (OS) of cervical cancer patients (P=0.006 and P<0.001, Table 3). The log-rank test results showed that increased levels of JMJD1A and c-Myc correlated strongly with poor survival (P=0.005 and P < 0.001, Figure 6A, 6B).
Table 3. Summary of univariate and multivariate Cox regression analysis of overall survival duration in all cervical cancer patients (n = 80).
| Clinicopathological parameters | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| JMJD1A(High/Low) | 2.229 | 1.258-3.948 | 0.006* | 1.899 | 1.025-3.519 | 0.042* |
| c-Myc(High/Low) | 3.489 | 1.960-6.211 | <0.001* | 2.502 | 1.323-4.731 | 0.005* |
| Age(years)(≥45/<45) | 0.587 | 0.338-1.019 | 0.058 | |||
| Tumor size(cm)(≥4/<4) | 1.292 | 0.749-2.228 | 0.357 | |||
| Histological type(NSCC/SCC) | 2.402 | 1.256-4.593 | 0.008* | |||
| Parametrial infiltration(Yes/No) | 0.839 | 0.484-1.454 | 0.532 | |||
| Differentiation(Grade 3/Grade 1+2) | 2.650 | 1.451-4.839 | 0.002* | 2.317 | 1.217-4.414 | 0.011* |
| Lymph node metastasis(Yes/No) | 2.426 | 1.169-5.035 | 0.017* | |||
| FIGO stage(≥II/<II) | 2.314 | 1.289-4.153 | 0.005* | |||
HR hazard ratio, 95% CI 95% confidence interval
indicates P <0.05.
Figure 6. Cumulative Kaplan-Meier overall survival curves of 80 cervical cancer patients segmented by JMJD1A (A), c-Myc (B), and high-risk combination group (JMJD1A and c-Myc combinations) (C).
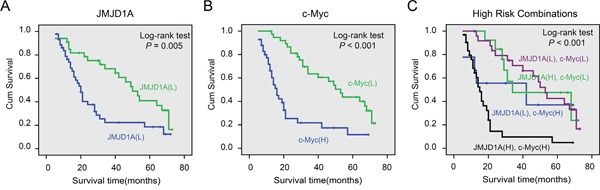
A. Compared to JMJD1A low expression group, patients with high levels of JMJD1A had poor survival. B. Patients with increased expression of c-Myc had a worse prognosis. C. We compared the prognosis with different groups including JMJD1A/c-Myc high/high, low/low, high/low and low/high samples. The results showed that the group with worst prognosis is the high/high group. P-values were calculated by the log-rank test. *P < 0.05.
Univariate analysis indicated that histological type, tumor differentiation, lymph node metastasis, and FIGO stage correlated with patient survival (P=0.008, P=0.002, P=0.017, and P= 0.005, respectively). Multivariate analysis showed that JMJD1A and c-Myc expression, and tumor differentiation were independent prognostic factors for cervical cancer patients (Table 3). Histological type, tumor differentiation, lymph node metastasis, and FIGO stage were not associated with survival (Table 3). In addition, the log-rank test showed that strong co-expression of JMJD1A and c-Myc correlated with shorter OS (P<0.001, Figure 6C). The 5-year survival rate was only 5% (95% CI, -0.048–0.148) in the high co-expression group and 42% (95% CI, 0.263–0.577) in other groups.
DISCUSSION
Our study reveals the important role played by JMJD1A in cervical cancer. JMJD1A is upregulated in cervical cancer compared with para-carcinoma. JMJD1A knockdown inhibits cervical cancer cell proliferation, migration, and invasion. Furthermore, JMJD1A induces transcription of c-Myc, which is crucial for cervical cancer growth and progression. Survival analysis reveals that both JMJD1A and c-Myc protein levels are independent prognostic factors for cervical cancer patients. High co-expression of these two proteins correlates with poor prognosis. Our results indicate JMJD1A is a promising therapeutic target in cervical cancer.
JMJD1A is an H3K9me1/2 histone demethylase [10]. Because H3K9 methylation in promoter regions is inhibitory to transcription [22, 23], gene expression tends to be enhanced by JMJD1A activity [24-26]. Moreover, JMJD1A appears to be an important regulator affecting the development and progression of various malignancies. Yamada et al [18] reported that JMJD1A is a useful prognostic marker and may enhance malignant transformation in hepatocellular carcinoma. Uemura et al [20] showed that JMJD1A is a useful biomarker for hypoxic tumor cells and may be a promising therapeutic target in colorectal cancer. Yang et al [19] have shown that JMJD1A suppression inhibits gastric cancer cell proliferation, and suppresses MAPK pathway via transcriptional downregulation of long noncoding RNA MALAT1. JMJD1A-MALAT1-MAPK signaling might participate in the JMJD1A-induced proliferation of gastric cancer cells. Zhan et al [27] reported that JMJD1A promotes tumorigenesis and forms a feedback loop with EZH2/let-7c in non-small cell lung cancer.
We observed that JMJD1A levels are increased in cervical cancer tissue as compared to matched non-tumor tissue. In addition, JMJD1A suppression inhibited cervical cancer cell proliferation, migration, and invasion. c-Myc is reportedly upregulated in cervical cancer, suggesting the possibility that c-Myc overexpression drives cervical cancer progression [28, 29]. Here, we showed that JMJD1A binds to the c-Myc promoter and induces c-Myc transcription, which is consistent with earlier findings [21]. In addition, c-Myc overexpression rescues the loss of JMJD1A-mediated repression activity in cervical cancer. More importantly, our observation that c-Myc levels correlated with those of JMJD1A in cervical cancer tissue samples, suggest JMJD1A contributes to c-Myc overexpression in at least some human cervical cancers. Furthermore, elevated JMJD1A and c-Myc levels correlate with poor patient prognosis and survival.
In sum, our findings reveal the oncogenic effects of JMJD1A in cervical cancer and elucidate a possible mechanism by which JMJD1A and c-Myc act to enhance cervical cancer growth and progression. These findings suggest JMJD1A is a potential therapeutic target for the treatment cervical cancer.
MATERIALS AND METHODS
Patients and specimens
The study was conducted according to the Declaration of Helsinki and approved by the Ethics Committee of the Second Affiliated Hospital of University of South China. Written informed consent was obtained from all patients. Human fresh cervical cancer tissue samples and adjacent noncancerous control tissues were obtained by surgical resection from ten patients at the Department of Obstetrics and Gynecology, the Second Affiliated Hospital of University of South China. All samples were derived from patients who had not received adjuvant treatment including radiotherapy or chemotherapy prior to surgery. All samples were snap-frozen and stored in liquid nitrogen after collection.
Additionally, a total of 80 paraffin embedded cervical cancer tissue samples were collected from the Department of Obstetrics and Gynecology, the Second Affiliated Hospital of University of South China. The construction of tissue microarray (TMA) was performed by ShGnghGi Outdo Biotech Company (China). The median age of patients was 50 years (ranging from 35 to 75 years old). The overall survival time ranged from 5 to 73 months, with a median of 21 months. Detailed information can be found in Table 1.
Immunohistochemistry
After deparaffinization and rehydration, TMA sections were subjected 5 minutes to high pressure for antigen retrieval. Endogenous peroxidase activity was blocked using 100 μL of peroxidase block solution for 10 min. The slides were subsequently incubated overnight at 4°C with primary antibodies as follows: JMJD1A (dilution 1:100; Sigma-Aldrich, USA), c-Myc (dilution 1:100, Abcam, Cambridge, MA). Sections were then incubated with biotinylated secondary antibodies (Zymed, San Francisco, CA) for 30 min at room temperature, followed by incubation with streptavidin horseradish peroxidase complex. Finally, sections were incubated with DAB for 2 min. Positive controls were used in each experiment following supplier's instructions. Negative controls using appropriate IgG to replace primary antibody were also run in each experiment (Supplementary Figure 1).
The staining was scored according to the staining intensity as previously described [30]. The percentage of positive cells was scored as follows: 0% (0), 1%–10% (1), 11%–50% (2) and 51%–100% (3). Staining intensity was scored as follows: no staining (0), weak staining (1), moderate staining (2), and strong staining (3). Comprehensive score = staining percentage × intensity. JMJD1A or c-Myc expression was classified as follows: <6 low expression, ≥6 high expression. Immunostaining was independently scored by two pathologists blinded to the clinicopathological characteristics.
Cell culture
Cervical cancer-derived cell lines SiHa, MDCK, HeLa, C-33A and ME-180, and the non-tumorigenic human epithelial cell line HaCaT were obtained from Cell Bank of Chinese Academy of Science. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Gibco, Carslbad, CA, USA), at 37°C with an atmosphere of 5% CO2 and 90% relative humidity.
RNA isolation and RT-qPCR analysis
Total RNA was isolated by using miniBEST universal RNA extraction kit (Takara Bio, Inc.) according to the manufacturer's instructions. Reverse transcription and RT-qPCR kits (Takara Bio, Inc.) were used to evaluate mRNA expression of JMJD1A and c-Myc. β-Actin expression was used to normalize for variance. The PCR primer pairs were as follows:
JMJD1A: 5’-CAGGAGCTCCACATCAGGTT-3’ (F), 5’-TGCATCTTTCACTGCATGGT-3’(R). c-Myc: 5’-GGCTCCTGGCAAAAGGTCA-3’ (F), 5’-AGTTGTG CTGATGTGTGGAGA-3’ (R). β-Actin:5’-CGGGAAAT CGTCCGTGACATTAAG-3’(F), 5’-TGATCTCCTTCT GCATCCTGTCGG-3’ (R).
Western blotting analysis
Cells were lysed in RIPA lysis buffer and the protein concentration was determined by a standard Bradford assay (Beyotime Institute of Biotechnology, Haimen, China). Whole cell extracts were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes by Pharmacia Phast gel electrophoresis system (Roche Diagnostics, Indianapolis, IN, USA). The PVDF membrane was blocked with 3% bovine serum albumin for 1 h. The immunoblots were incubated overnight at 4°C with the following primary antibodies: anti-JMJD1A (Sigma-Aldrich, USA), anti-c-Myc (Abcam, Cambridge, MA), and β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). The membranes were washed, incubated with appropriate secondary antibodies (room temperature, 1 h), and detected using the enhanced chemiluminescence detection system. The data were normalized against β-actin, used as a loading control.
Lentiviral vector packaging and infection of HeLa cells
Lentiviral vector encoding shRNA JMJD1A was packaged in 293T cells by the calcium phosphate transfection. The supernatants containing lentiviral particles were collected 48 h after transfection (GeneChem, Shanghai, China). HeLa cells were transduced with the supernatant of lentiviral particles in the presence of polybrene (8μg/ml) for 24 h, before replacement with fresh growth media. Polyclonal cells with puromycin resistance were selected for further experiments.
Plasmid construction and transfection of HeLa cells
Recombinant plasmid pcDNA3.0/c-Myc was constructed by the insertion of human c-Myc cDNA into pcDNA3.0 vector (GeneChem, Shanghai, China). HeLa-conJMJD1A or HeLa-siJMJD1A cells were transfected with pcDNA3.0/c-Myc or pcDNA3.0 vector control. All transfections were carried out using lipofectamine RNAi-MAX (Invitrogen, USA). Transfected cells were analyzed by Western blotting.
Luciferase reporter assay
Cervical cancer cells were transfected with the indicated c-Myc promoter reporter containing JMJD1A-binding sites (constructed by GeneChem, Shanghai, China). The c-Myc promoter activity was normalized via co-transfection of a β-actin/Renilla luciferase reporter containing a full-length Renilla luciferase gene [31]. The luciferase activity in the cells was quantified using a dual luciferase assay system (Promega) 24 hours after transfection.
Cell proliferation assay
The effect of JMJD1A knockdown on cell proliferation was determined by measuring absorbance at 450 nm. Briefly, cervical cancer cells in 96-well plates (1×103 cells/well) in triplicate were added (10 μl) to cell counting kit-8 (CCK-8, Dojindo Molecular Technologies, Kumamoto, Japan) and incubated for 24, 48, and 72 h. The absorbance at 450 nm was measured 1 h later.
Migration and invasion assays
The cell invasive and migratory potential was evaluated using Boyden chamber and Transwell assays, respectively [32]. Briefly, the Boyden chamber assay was conducted using specialized MilliCell chambers, which included a 24-well tissue culture plate with 12 cell culture inserts (Millipore, Bedford, MA, USA). The inserts contained an 8 μm pore size polycarbonate membrane with a pre-coated thin layer of Matrigel (BD Biosciences). Ten percent fetal bovine serum-containing medium was placed in the lower chambers to act as a chemo-attractant. Then, 1×105 cells in a 100 μl volume of serum-free medium were placed in the upper chambers and incubated at 37°C for 36 h. Invasive cells on the bottom surface of the membrane, which had invaded the Matrigel and had migrated through the polycarbonate membrane, were stained by the staining solution, and counted under a microscope in ten randomly selected fields at a magnification of ×200. Transwell assay was the same as the Boyden chamber with the exception that no Matrigel was used and the permeating time for cells was 24 h.
Chromatin immunoprecipitation (Chip) assay
Chip assays were performed using Imprint Chromatin Immunoprecipitation Kit as described [33], according to the manufacturer's instructions (Millipore, USA). Briefly, cells (5×106) were cross-linked with 1% formaldehyde for 10 min, quenched by addition of 0.125 M glycine, and collected by centrifugation at 800 g for 5 min at 4°C. Cross-linked cells were resuspended in sodium dodecyl sulfonate lysis buffer containing a protease inhibitor cocktail and the soluble chromatin was sheared to fragment DNA. The fragmented chromatin samples were aliquoted as genomic input DNA or immunoprecipitated with JMJD1A antibodies (Sigma-Aldrich, USA) or IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), and incubated at 4°C with rotation overnight. Immunocomplexes were collected by magnetic separator washed, and eluted with Chip elution buffer. DNA was purified on spin columns. The Chip products and genomic input DNA were analyzed by real-time PCR with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). The primer pairs for c-Myc were: 5’-CCAGCGAATTATTCAGAA-3’, 5’-AATTACCATTGACTTCCTC-3’.
Statistical analysis
All data are presented as the mean ± standard deviation. Means of two groups were compared by Student's t-test; means of three or more groups were compared with one-way analysis of variance using SPSS for Windows version 13.0 (SPSS, Inc., Chicago, IL, USA). The χ2 test was used to analyze the correlation between the clinicopathological characteristics and JMJD1A and c-Myc expression. Overall survival (OS) was defined as the interval from date of diagnosis until death from any cause. Data were analyzed for living patients and patients lost between follow-ups. The OS was estimated using the Kaplan-Meier method and compared using the log-rank test. Significant variables were further analyzed by multivariate analysis to test for independent prognosis. Bivariate correlations between variable factors were calculated by Spearman rank correlation coefficients. P<0.05 was considered to indicate a statistically significant difference.
SUPPLEMENTARY MATERIALS FIGURE
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Schmitt M, Dalstein V, Waterboer T, Clavel C, Gissmann L, Pawlita M. Diagnosing cervical cancer and high-grade precursors by HPV16 transcription patterns. Cancer Res. 2010;70:249–256. doi: 10.1158/0008-5472.CAN-09-2514. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Wu X, Cheng X. Advances in diagnosis and treatment of metastatic cervical cancer. J Gynecol Oncol. 2016;27:e43. doi: 10.3802/jgo.2016.27.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgioti C, Chatoupis K, Moulopoulos LA. Current imaging strategies for the evaluation of uterine cervical cancer. World J Radiol. 2016;8:342–354. doi: 10.4329/wjr.v8.i4.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crafton SM, Salani R. Beyond Chemotherapy: An Overview and Review of Targeted Therapy in Cervical Cancer. Clin Ther. 2016;38:449–458. doi: 10.1016/j.clinthera.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Shrager J, Tenenbaum JM. Rapid learning for precision oncology. Nat Rev Clin Oncol. 2014;11:109–118. doi: 10.1038/nrclinonc.2013.244. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt KT, Chau CH, Price DK, Figg WD. Precision Oncology Medicine: The Clinical Relevance of Patient Specific Biomarkers Used to Optimize Cancer Treatment. J Clin Pharmacol 2016. [DOI] [PMC free article] [PubMed]
- 9.Egatz-Gomez A, Wang C, Klacsmann F, Pan Z, Marczak S, Wang Y, Sun G, Senapati S, Chang HC. Future microfluidic and nanofluidic modular platforms for nucleic acid liquid biopsy in precision medicine. Biomicrofluidics. 2016;10:32902. doi: 10.1063/1.4948525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 11.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goda S, Isagawa T, Chikaoka Y, Kawamura T, Aburatani H. Control of histone H3 lysine 9 (H3K9) methylation state via cooperative two-step demethylation by Jumonji domain containing 1A (JMJD1A) homodimer. J Biol Chem. 2013;288:36948–36956. doi: 10.1074/jbc.M113.492595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 14.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroki S, Matoba S, Akiyoshi M, Matsumura Y, Miyachi H, Mise N, Abe K, Ogura A, Wilhelm D, Koopman P, Nozaki M, Kanai Y, Shinkai Y, Tachibana M. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;341:1106–1109. doi: 10.1126/science.1239864. [DOI] [PubMed] [Google Scholar]
- 16.Guo X, Shi M, Sun L, Wang Y, Gui Y, Cai Z, Duan X. The expression of histone demethylase JMJD1A in renal cell carcinoma. Neoplasma. 2011;58:153–157. doi: 10.4149/neo_2011_02_153. [DOI] [PubMed] [Google Scholar]
- 17.Suikki HE, Kujala PM, Tammela TL, van Weerden WM, Vessella RL, Visakorpi T. Genetic alterations and changes in expression of histone demethylases in prostate cancer. Prostate. 2010;70:889–898. doi: 10.1002/pros.21123. [DOI] [PubMed] [Google Scholar]
- 18.Yamada D, Kobayashi S, Yamamoto H, Tomimaru Y, Noda T, Uemura M, Wada H, Marubashi S, Eguchi H, Tanemura M, Doki Y, Mori M, Nagano H. Role of the hypoxia-related gene, JMJD1A, in hepatocellular carcinoma: clinical impact on recurrence after hepatic resection. Ann Surg Oncol. 2012;19(Suppl 3):S355–S364. doi: 10.1245/s10434-011-1797-x. [DOI] [PubMed] [Google Scholar]
- 19.Yang H, Liu Z, Yuan C, Zhao Y, Wang L, Hu J, Xie D, Wang L, Chen D. Elevated JMJD1A is a novel predictor for prognosis and a potential therapeutic target for gastric cancer. Int J Clin Exp Pathol. 2015;8:11092–11099. [PMC free article] [PubMed] [Google Scholar]
- 20.Uemura M, Yamamoto H, Takemasa I, Mimori K, Hemmi H, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Doki Y, Mori M. Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer: in vivo identification from hypoxic tumor cells. Clin Cancer Res. 2010;16:4636–4646. doi: 10.1158/1078-0432.CCR-10-0407. [DOI] [PubMed] [Google Scholar]
- 21.Fan L, Peng G, Sahgal N, Fazli L, Gleave M, Zhang Y, Hussain A, Qi J. Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene. 2016;35:2441–2452. doi: 10.1038/onc.2015.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi J, Nakayama K, Cardiff RD, Borowsky AD, Kaul K, Williams R, Krajewski S, Mercola D, Carpenter PM, Bowtell D, Ronai ZA. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell. 2010;18:23–38. doi: 10.1016/j.ccr.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wade MA, Jones D, Wilson L, Stockley J, Coffey K, Robson CN, Gaughan L. The histone demethylase enzyme KDM3A is a key estrogen receptor regulator in breast cancer. Nucleic Acids Res. 2015;43:196–207. doi: 10.1093/nar/gku1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrish JK, Sechler M, Winn RA, Jedlicka P. The histone demethylase KDM3A is a microRNA-22-regulated tumor promoter in Ewing Sarcoma. Oncogene. 2015;34:257–262. doi: 10.1038/onc.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tee AE, Ling D, Nelson C, Atmadibrata B, Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, Pasquier E, Haber M, Norris MD, et al. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget. 2014;5:1793–1804. doi: 10.18632/oncotarget.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhan M, Wen F, Liu L, Chen Z, Wei H, Zhou H. JMJD1A promotes tumorigenesis and forms a feedback loop with EZH2/let-7c in NSCLC cells. Tumour Biol. 2016;37:11237–11247. doi: 10.1007/s13277-016-4999-9. [DOI] [PubMed] [Google Scholar]
- 28.Kubler K, Heinenberg S, Rudlowski C, Keyver-Paik MD, Abramian A, Merkelbach-Bruse S, Buttner R, Kuhn W, Schildhaus HU. c-myc copy number gain is a powerful prognosticator of disease outcome in cervical dysplasia. Oncotarget. 2015;6:825–835. doi: 10.18632/oncotarget.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao LM, Sun XY, Liu AW, Wu JB, Cheng XL, Lin JX, Zheng M, Huang L. Low expression of long noncoding XLOC_010588 indicates a poor prognosis and promotes proliferation through upregulation of c-Myc in cervical cancer. Gynecol Oncol. 2014;133:616–623. doi: 10.1016/j.ygyno.2014.03.555. [DOI] [PubMed] [Google Scholar]
- 30.Jiao F, Hu H, Han T, Zhuo M, Yuan C, Yang H, Wang L, Wang L. Aberrant expression of nuclear HDAC3 and cytoplasmic CDH1 predict a poor prognosis for patients with pancreatic cancer. Oncotarget. 2016;7:16505–16516. doi: 10.18632/oncotarget.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K, Huang S. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008;68:8733–8742. doi: 10.1158/0008-5472.CAN-08-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–2492. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 33.Han T, Jiao F, Hu H, Yuan C, Wang L, Jin ZL, Song WF, Wang LW. EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer. Oncotarget. 2016;7:11194–11207. doi: 10.18632/oncotarget.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


