Abstract
The Escherichia coli AlkB protein repairs 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) lesions in DNA and RNA by oxidative demethylation, a reaction requiring ferrous iron and 2-oxoglutarate as cofactor and co-substrate, respectively. Here, we have studied the activity of AlkB proteins on 3-methylthymine (3-meT) and 1-methylguanine (1-meG), two minor lesions which are structurally analogous to 1-meA and 3-meC. AlkB as well as the human AlkB homologues, hABH2 and hABH3, were all able to demethylate 3-meT in a DNA oligonucleotide containing a single 3-meT residue. Also, 1-meG lesions introduced by chemical methylation of tRNA were efficiently removed by AlkB. Unlike 1-meA and 3-meC, nucleosides or bases corresponding to 1-meG or 3-meT did not stimulate the uncoupled, AlkB-mediated decarboxylation of 2-oxoglutarate. Our data show that 3-meT and 1-meG are repaired by AlkB, but indicate that the recognition of these substrates is different from that in the case of 1-meA and 3-meC.
INTRODUCTION
Alkylating agents are abundant in the environment and are also generated inside cells. Such agents introduce a number of premutagenic and cytotoxic lesions in DNA and are often highly carcinogenic in mammals. To counteract the deleterious effects of the lesions, most organisms possess several mechanisms for repairing alkylation damage in DNA. In the bacterium Escherichia coli, the adaptive response to alkylation damage regulates the expression of several alkylation repair proteins through the Ada regulon (1,2), and three different mechanisms for repairing alkylation damage have been described. First, AlkA and Tag are alkylbase glycosylases which excise purines alkylated in the 3- or 7-position, such as 3-methyladenine and 7-methylguanine (7-meG) from DNA (3,4). Second, the alkyltransferases Ada and Ogt directly reverse the damage at O-alkylated bases, such as O6-methylguanine, by an irreversible reaction where the methyl group is transferred to a cysteine residue of the transferase, which is then consumed in the reaction (5). Third, the AlkB protein was recently found to be an oxidative demethylase which directly reverses 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) lesions (6,7).
AlkB belongs to the superfamily of 2-oxoglutarate- and iron(II)-dependent oxygenases (8). These enzymes perform various oxidation reactions, where molecular oxygen is the oxidizing agent, and ferrous iron and 2-oxoglutarate are required as a cofactor and co-substrate, respectively (9). AlkB catalyses the oxidation of the harmful methyl group in 1-meA or 3-meC to a hydroxymethyl moiety, which is spontaneously released as formaldehyde, thereby regenerating the normal base (6,7). The co-substrate 2-oxoglutarate is decarboxylated, yielding succinate and CO2. It was recently shown that 1-ethyladenine lesions in DNA (10), as well as 5′ phosphorylated 1-methyldeoxyadenosine mono- and triphos-phates (11), can also be dealkylated by AlkB, albeit at reduced efficiency compared with 1-meA present in DNA. AlkB catalyses the decarboxylation of 2-oxoglutarate to succinate and CO2 at a significant rate even in the absence of a DNA substrate (‘uncoupled reaction’), and the addition of methylated DNA substrate causes stoichiometric increase in 2-oxoglutarate turnover (6,7). Also, it was demonstrated that 2-oxoglutarate turnover was stimulated by some nucleosides and bases containing AlkB relevant lesions, but these compounds were not demethylated by AlkB (12). Thus, it appears that the proper geometry for optimal AlkB action is only acquired in the context of a DNA oligomer.
All three mechanisms for repairing alkylated DNA in E.coli are also present in human cells, and sequence homology searches indicate that a number of AlkB homologues exist in the human proteome (8,13). However, only two of these, denoted hABH2 and hABH3, have been reported to share the ability of E.coli AlkB to demethylate 1-meA and 3-meC (10,14). Interestingly, E.coli AlkB and hABH3 are also able to efficiently remove 1-meA and 3-meC lesions from RNA (14), and, in the case of tRNA and mRNA, such repair is also associated with the recovery of RNA function (15).
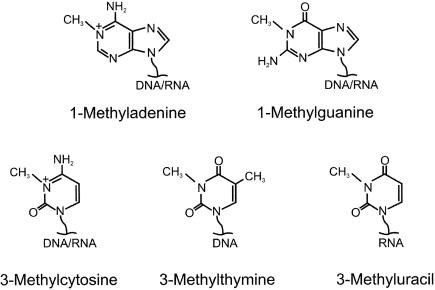
The lesions 1-methylguanine (1-meG) and 3-methylthymine (3-meT) are structural analogues of 1-meA and 3-meC, respectively (Figure 1). 1-meA and 3-meC are methylated at N-atoms that are involved in Watson–Crick base pairing of the corresponding normal bases, and this is also the case for 1-meG and 3-meT. Although less prevalent than 1-meA and 3-meC, both 3-meT and 1-meG have been reported to be introduced upon treatment of DNA or RNA with alkylating agents, both in vitro (16–20) and in vivo (21,22). In the present study, we report that both 1-meG and 3-meT can be repaired by AlkB proteins from humans and bacteria.
Figure 1.
Nucleobases obtained by N1-methylation of purines and N3-methylation of pyrimidines.
MATERIALS AND METHODS
Enzymes and reagents
The DNA oligonucleotide containing a single 3-meT lesion was obtained from Synthegen (Houston, TX). Other DNA oligonucleotides were from Invitrogen (Glasgow, Scotland). E.coli total RNA was from Roche (Germany). The RNA oligonucleotide was from Dharmacon (Lafayette, CO). AlkB, hABH2 and hABH3 proteins were expressed and purified as described previously (6,14). Adenosine, guanosine, 1-methyladenosine and deoxythymidine were from Sigma. 3-Methyldeoxythymidine (3-mdT) was from ChemGenes (Ashland, MA). 3-meT was from Specs (Delft, The Netherlands), 1-meG was from Fluka (Milwaukee, WI) and 1-methylguanosine was from MP Biomedicals (Irvine, CA). 2-oxo[5-14C]glutarate was from Moravek Biochemicals (Brea, CA)
Reaction conditions for AlkB-mediated repair
Reactions were performed for 30 min at 37°C in 10 μl (Figure 2), 100 μl (Figure 3) or 50 μl (Figure 4) reaction mixtures containing 50 mM Tris–HCl, pH 8.0 (Figure 2) or 50 mM HEPES/KOH, pH 7.0 (Figures 3 and 4), as well as 2 mM ascorbic acid, 40 μM Fe2SO4, 100 μM 2-oxoglutarate, repair enzyme as indicated, and DNA or RNA substrate. The concentration of DNA oligonucleotide substrate (Figures 2 and 3) was 1 μM. In the case of the RNA substrates, 2 μg (5000 c.p.m.) of N-[3H]methyl-N-nitrosourea ([3H]MNU)-treated RNA oligonucleotide (Figure 4A) or 25 μg (5000 c.p.m.) of [14C]methyliodide (MeI)-treated tRNA (Figure 4B) was used in each reaction.
Figure 2.
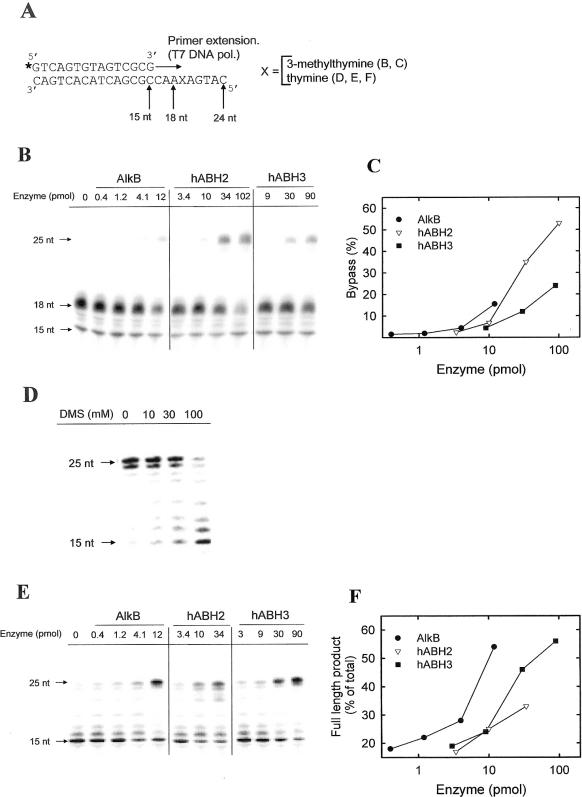
Repair of replication blocking lesions by AlkB proteins. (A) Schematic outline of the experiments. 32P-end-labelled (indicated by asterisk) 15mer oligonucleotide was extended by T7 DNA polymerase on a 24mer oligonucleotide template. (B) Release of 3-meT-induced extension block by AlkB proteins. The 3-meT containing template was subjected to repair by the indicated amounts of human or bacterial AlkB proteins, and subsequently used as template in a primer extension experiment. The primer extension products were analysed by denaturing polyacrylamide analysis and phophorimaging. (C) Quantification of the data shown in (B). The degree of bypass was calculated as the intensity of the 25 nt band, relative to the sum of the 18 and 25 nt bands. (D) Induction of extension block by DMS treatment of template. The 24mer oligonucleotide (without 3-meT) was incubated with the indicated concentrations of DMS, and, subsequently, used as template in a primer extension experiment. (E) Release of DMS-induced extension block by AlkB proteins. The 24mer oligonucleotide (without 3-meT) was treated with 100 mM DMS, then incubated with the indicated amounts of AlkB proteins, and, finally, used as template for primer extension. (F) Quantification of the data shown in (E). The amount of radioactivity present in the 25 nt band has been expressed relative to the sum of the radioactivity contained in all the bands in each lane.
Figure 3.
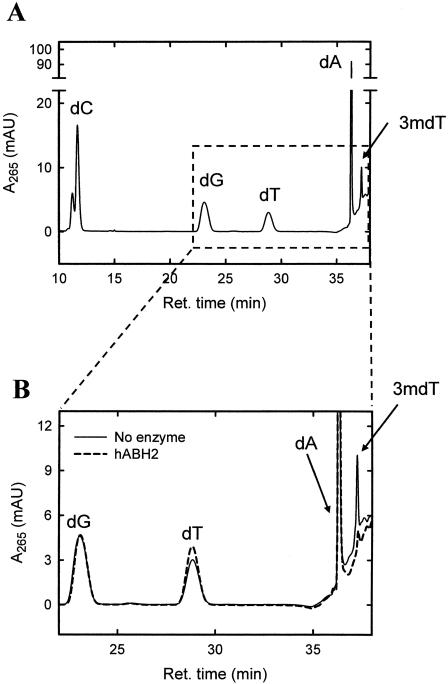
Repair of 3-meT lesions by hABH2 catalysed oxidative demethylation. The 3-meT containing oligonucleotide was incubated in the absence (solid line) or presence (dashed line) of hABH2, then degraded enzymatically to its constituent nucleosides by treatment with nuclease P1 and calf intestine alkaline phosphatase, and, finally, subjected to reverse-phase HPLC. The identity of the peaks corresponding to 2′-deoxycytidine (dC), 2′-deoxyguanosine (dG), 2′-deoxythymidine (dT), 2′-deoxyadenosine (dA), and 3-methyl-2′-deoxythymidine (3mdT) was inferred from comparison of their elution times with those of the corresponding reference standards.
Figure 4.
AlkB-mediated repair of 1-meG in chemically methylated tRNA. [3H]MNU-treated 24mer RNA oligonucleotide (A) or [14C]MeI-treated E.coli tRNA from (B) was incubated in the absence (filled circles) or presence (open circles) of AlkB, then enzymatically degraded to nucleosides, which were subjected by reverse-phase HPLC. Fractions were collected, and the radioactivity present in the fractions determined by scintillation counting. Arrows indicate the elution times of internal nucleoside reference standards corresponding to the modified bases.
Methylation of DNA and RNA in vitro
The 24mer DNA oligonucleotide (0.6 nmol) was treated with dimethylsulfate (DMS) for 30 min at 30°C in a reaction volume of 50 μl. The DMS was removed by ethanol precipitation of the oligonucleotide, which was redissolved in 20 μl of H2O. [3H]MNU treatment of the RNA oligonucleotide (23) and [14C]MeI treatment of tRNA (15) were performed as described previously.
Primer extension assay
The 15mer primer (50 pmol) was 32P-end-labelled by incubation for 30 min at 37°C in a 10 μl reaction mixture containing 5 pmol [γ-32P]ATP (3000 Ci/mmol; Amersham), 100 pmol unlabelled ATP and 10 U T4 polynucleotide kinase (MBI Fermentas, Germany). The oligonucleotide was recovered by ethanol precipitation. Primer extension was performed with Sequenase (Amersham Biosciences), a genetically modified T7 DNA polymerase. Annealing of the 32P-labelled 15mer primer (0.2 pmol) to the unlabelled 24mer template (2 pmol; corresponding to 2 μl of the repair reaction) was performed by heating to 55°C, followed by slow cooling to 25°C, in a 5 μl reaction mixture, which contained 20 mM Tris–HCl, pH 7.5, 10 mM MgCl2, and 25 mM NaCl (as well as the components from the repair reaction mixture). To the annealing reaction was added DTT (6.5 mM), dNTPs (16 μM of each) and 0.8 U Sequenase Version 2.0 DNA polymerase, and primer extension (7.75 μl reaction volume) was performed for 10 min at 37°C. The reaction was then stopped by the addition of 6 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF). The primer extension products were electrophoresed in a 15% denaturing polyacrylamide gel. The gel was dried, and then subjected to phosphorimaging and analysis by the software ImageQuant.
High-pressure liquid chromatography (HPLC) analysis of nucleosides from DNA and RNA
Methylated DNA or RNA was subjected to repair as indicated, and then degraded to nucleosides by incubation with 125 μg/ml nuclease p1 (Sigma) in 100 μl reaction buffer (50 mM sodium acetate, pH 5.3, 0.2 mM ZnCl2) for 60 min at 37°C, followed by the addition of 10 μl of 1 M Tris–HCl, pH 8.0 and 1 U calf intestine alkaline phosphatase (Roche, Germany), and continued the incubation further for 60 min at 37°C. The resulting hydrolysate was subjected to reverse-phase HPLC which was performed using two Chrompack Inertsil 5 ODS-2 150 × 4 mm columns in series at a flow rate of 0.5 ml/min. For the RNA samples, the mobile phase was 50 mM Na-acetate, pH 5.0, 3% methanol (solution B), and a gradient was used where the concentration of methanol (solution A) was gradually increased, according to a profile (0–25 min: 0% A; 25–30 min: linear increase to 10% A; 30–40 min: 10% A; 40–55 min: linear increase to 100% A). In the case of the DNA samples, the mobile phase was 50 mM Na-acetate, pH 5.0, 10% methanol (solution B), and the concentration of methanol was gradually increased (0–20 min: 100% B; 20–40 min: linear increase to 100% A; 40–50 min: 100% A). Non-labelled nucleosides were measured by ultraviolet detection, and radiolabelled nucleosides were measured by scintillation counting of the radioactivity in collected fractions.
Measurement of AlkB-mediated decarboxylation of 2-oxoglutarate
The reaction conditions were similar to those described by Welford et al. (12), but instead of using 2-oxo[1-14C]glutarate and measuring the release of 14CO2, 2-oxo[5-14C]glutarate was used and the generation of [1-14C]succinate was measured. The reaction mixtures (50 μl) contained 50 mM Tris–HCl (pH 7.5), 4 mM ascorbic acid, 80 μM FeSO4, 160 μM 2-oxo[5-14C]glutarate (50 000 c.p.m. per reaction, obtained by mixing [5-14C]radiolabelled and unlabelled 2-oxoglutarate) and 12.5 μM AlkB. Nucleosides and bases were added to a final concentration of 400 μM from 10 mM stock solutions in dimethyl sulfoxide (DMSO). The reactions were incubated for 10 min at 37°C. The amount of succinate formed during the reaction was determined by precipitating remaining 2-oxo[5-14C]glutarate with 2,4-dinitrophenyl hydrazine, and then measuring the radioactivity in the supernatant, as described by Kaule and Gunzler (24).
RESULTS
Release of a 3-meT-induced replication block by treatment with AlkB proteins
3-meT lesions in DNA have previously been shown to block primer extension by DNA polymerases (25), demonstrating that 3-meT is a replication blocking lesion. To test whether AlkB proteins can repair 3-meT lesions in DNA, we studied their ability to rescue a 3-meT-induced replication block. We performed T7 DNA polymerase mediated extension of a 32P-end-labelled 15mer primer on a 24mer oligonucleotide template containing a single 3-meT residue in position 19 from the 3′ end (Figure 2A). Primer extension yielded exclusively an 18mer extension product (Figure 2B, lane 1), indicating that the T7 DNA polymerase is not able to incorporate a nucleoside opposite the 3-meT residue. Extension on an otherwise identical template, where the 3-meT residue had been replaced by thymine, yielded a mixture of 24 and 25 nt products (Figure 2D, lane 1). (The 25mer product results from the ability of T7 DNA polymerase to add an additional residue after completing template-directed synthesis). However, when the 3-meT containing template had been incubated with human or bacterial AlkB proteins as well as the cofactors Fe2+ and 2-oxoglutarate prior to primer extension, a partial rescue of the primer extension block was observed (Figure 2B), i.e. an additional 25mer primer extension product was observed. However, even at the highest enzyme concentrations, we were not able to observe complete repair, as indicated by the presence of the 18mer extension product. Further increasing the enzyme concentrations did not increase the repair efficiency but rather caused adverse effects, such as substrate degradation and/or inhibition of primer extension (data not shown). Normalized for the amounts of enzyme used, the relative repair efficiency of the three enzymes can be summarized as follows: AlkB ≈ hABH2 > hABH3 (Figure 2C). In contrast, when we previously introduced 1-meA and 3-meC residues by [3H]MNU treatment of the corresponding 3-meT-free oligonucleotide, the relative repair efficiencies were AlkB > hABH2 ≈ hABH3 (23). To more directly compare the activity of these enzymes on 1-meA and 3-meC with that observed for 3-meT, the 3-meT-free oligonucleotide was treated with DMS to introduce 1-meA and 3-meC lesions, and similar primer extension experiments were performed. Clearly, the DMS treatment inhibited the primer extension, leading to a substantial reduction in the amount of the 24mer/25mer extension products (Figure 2D). The pattern of observed stop sites corresponded reasonably well to the positions of A and C bases in the template, indicating that the extension block is caused by the introduction of 1-meA and 3-meC. As observed with the 3-meT containing oligonucleotide, incubation of the DMS-treated oligonucleotide with AlkB, hABH2 or hABH3 partially or completely reversed the primer extension block, thus demonstrating repair (Figure 2E). The relative repair efficiencies of these three enzymes were similar to those previously obtained when using the [3H]MNU-treated substrate (23), i.e. AlkB > hABH2 ≈ hABH3 (Figure 2F), but somewhat different from that obtained when using the 3-meT containing oligonucleotide (AlkB ≈ hABH2 > hABH3). In summary, these experiments indicate that 3-meT lesions can be reversed by AlkB, hABH2 and hABH3, but that the relative activity of the enzymes on this lesion is different from that observed with 1-meA and 3-meC.
Reversal of 3-meT measured by HPLC analysis
We considered it highly likely that the observed AlkB-mediated release of a 3-meT-induced replication block was caused by an actual oxidative demethylation of 3-meT to thymine. However, it could not be formally excluded that the result of the oxidative action of the human and bacterial AlkB proteins on 3-meT is not thymine, but rather another product which allows bypass by T7 DNA polymerase. To study this, the 3-meT containing oligonucleotide was digested to nucleosides by enzymatic hydrolysis with nuclease P1 and alkaline phosphatase, and the resulting nucleosides analysed by reverse-phase HPLC. Five peaks were detected in the chromatogram; four major peaks corresponding to the unmodified nucleosides adenosine, cytidine, guanosine and thymidine, and a minor peak corresponding to the single 3-mdT residue (Figure 3A). However, when the oligonucleotide was treated with hABH2 prior to HPLC analysis of its constituent nucleosides, the peak corresponding to 3-mdT was barely visible, while the thymidine peak increased slightly, indicating reversal of the 3-meT lesion to the normal base thymine (Figure 3B). In accordance with the results of the primer extension experiments, the highest degree of repair was observed in the case of hABH2, and we were only able to see partial reversal of the 3-meT lesion with AlkB or hABH3 (data not shown). Our HPLC analysis confirms the major finding from the primer extension experiments, i.e. that 3-meT is a substrate for AlkB, and also demonstrates that the lesion is repaired by demethylation.
AlkB-mediated repair of 1-meG lesions
When previously studying the ability of AlkB to repair RNA, we used [3H]MNU-treated RNA oligonucleotide (23) and [14C]MeI-treated tRNA as substrates (15), and AlkB repaired these two substrates at similar efficiencies. Here, we have analysed the contents of methylated bases in these substrates by subjecting the constituent nucleosides to HPLC analysis, and some important differences were noticed. In the case of the [3H]MNU-treated oligonucleotide substrate, the major lesions introduced were 1-meA, 3-meC and 7-meG, and, as expected, AlkB-mediated removal was only observed in the case of 1-meA and 3-meC (Figure 4A). However, when the nucleosides from the [14C]MeI-treated tRNA were analysed by HPLC, two additional peaks were observed (Figure 4B). One of these peaks coeluted with a nucleoside standard corresponding to 1-meG and was removed by the AlkB treatment. We have not established the identity of the second peak, which was not affected by the AlkB treatment. Our data indicate that 1-meG is a major lesion introduced by MeI treatment of tRNA, and that this lesion is efficiently removed by AlkB treatment.
1-meG and 3-meT do not stimulate AlkB-mediated decarboxylation of 2-oxoglutarate
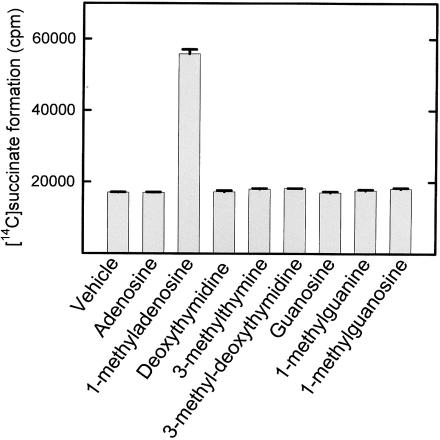
Many members of the superfamily of iron- and 2-oxoglutarate-dependent oxygenases are able to catalyse the decarboxylation of 2-oxoglutarate to succinate at a low rate even in the absence of their primary substrate (‘uncoupled reaction’), and this is also the case for AlkB (6,7). Moreover, the uncoupled AlkB reaction can be stimulated by various nucleosides and bases representing the AlkB relevant lesions, i.e. the nucleosides 1-methyladenosine, 1-methyl-2′-deoxyadenosine, 3-methylcytidine, 3-methyl-2′-deoxycytidine and the free base 1-meA (12). Therefore, we also tested the ability of 1-meG and 3-meT, as well as corresponding nucleosides, to stimulate the uncoupled AlkB reaction. However, while 1-methyladenosine, as previously reported, caused a substantial increase in the decarboxylation of 2-oxoglutarate, no such effect was observed with 1-meG, 3-meT, 3-methyl-2′-deoxythymidine or 1-methylguanosine (Figure 5).
Figure 5.
Inability of 1-meG and 3-meT, and corresponding nucleosides, to stimulate AlkB-mediated uncoupled decarboxylation of 2-oxoglutarate. Reaction mixtures containing AlkB and [5-14C]2-oxoglutarate, and 400 μM of the indicated compounds were incubated for 10 min at 37°C. Remaining [5-14C]2-oxoglutarate was precipitated with 2,4,dinitrophenylhydrazine, and the [1-14C]succinate in the supernatant was measured by scintillation counting. Error bars represent the range between duplicate measurements. ‘Vehicle’ indicates the addition of an amount of DMSO equivalent to that added with the bases and nucleosides.
DISCUSSION
The results presented here further extend the substrate specificity of the AlkB proteins, by demonstrating that also 1-meG lesions in RNA and 3-meT lesions in DNA are efficiently demethylated by AlkB. Thus, it now appears likely that all N1-methylated purine bases and N3-methylated pyrimidine bases in nucleic acids (shown in Figure 1) are AlkB substrates (it has not yet been tested whether 3-methyluracil is demethylated by AlkB).
3-meT has previously been shown to block DNA polymerases when present in the DNA template or in the incoming deoxynucleotide (25), and may therefore be a cause of both genomic instability and mutation. The introduction of 3-meT in DNA upon exposure to methylating agents is quite inefficient, compared with the level of 1-meA and 3-meC (26). However, numerous studies have reported the introduction of 3-meT in DNA in vitro (16,18,19,27), and also in vivo (21), so it appears likely that this lesion poses a threat to genomic integrity in living cells. Compared with the 1-meA and 3-meC containing substrate (obtained by DMS treatment), the 3-meT-containing substrate was a poorer substrate for AlkB and hABH3 (hABH2 was approximately equally efficient on both substrates). Thus, although the frequency of introduction of 3-meT lesions is quite low, this lesion may be as deleterious as the more frequent 1-meA and 3-meC lesions, due to its inefficient repair.
Here, we have shown that [14C]MeI treatment of tRNA introduces 1-meG lesions, and that these lesions are repaired by AlkB. Previous studies have shown that 1-meG is the major methylation product obtained by MeI treatment of guanosine under basic conditions (28), and that other methylating agents can induce 1-meG in RNA and DNA in vitro (16–18) and in vivo (22). We also attempted to introduce 1-meG residues in DNA by treatment of a G-rich oligonucleotide with [14C]MeI, but were unsuccessful (P. Ø. Falnes and R. F. Johansen, unpublished data). In our previous studies, we have titrated the amounts of AlkB and hABH3 required to demethylate the [14C]MeI-treated tRNA substrate used in Figure 4B (15), and we have also performed similar titrations for the [3H]methylated RNA oligonucleotide used in Figure 4A (23). Several conclusions can be drawn by combining our previous data with those presented here. When treating the [14C]methylated tRNA substrate with hABH3, the maximal extent of demethylation corresponded to the plateau level obtained with E.coli AlkB (Figure 5 in ref. 15), indicating that 1-meG is also a substrate for hABH3. Furthermore, the titration curve of the activity of AlkB on the [14C]methylated tRNA substrate containing the AlkB substrates 1-meG, 1-meA and 3-meC was not biphasic (Figure 5 in ref. 15), and very similar to that obtained when using the 1-meA and 3-meC containing [3H]methylated RNA oligonucleotide (Figure 4A in ref. 23). This may suggest that 1-meG, 1-meA and 3-meC in RNA are demethylated at similar efficiencies by AlkB. Kroger and Singer (29) studied the transcription of homopolymeric DNA templates and found that the introduction of 1-meG lesions completely abrogated transcription, while 1-meA or 3-meC only decreased its rate. These data demonstrate that methylation of guanosine in the N1-position severely compromises the ability of DNA to act as template for RNA polymerases, and suggest to us that 1-meG may also block DNA replication and mRNA translation, as is the case for 1-meA and 3-meC (6,15).
Similarly to 1-meA and 3-meC, 1-meG exists as a naturally occurring modified base in tRNA. The presence of 1-meG residues in some tRNAs is important for maintaining the proper reading frame in translation (30), and bacterial mutants deficient in incorporating 1-meG in tRNA display decreased growth rates (31). Thus, the ability of AlkB to demethylate 1-meG appears counterproductive in this context, and it will be interesting to study whether naturally occurring bases in tRNA are protected from AlkB-mediated demethylation, or whether a low level of such demethylation is actually tolerated.
A recent article reported that the bases and nucleosides corresponding to the lesions 1-meA and 3-meC strongly stimulated the AlkB-mediated uncoupled decarboxylation of 2-oxoglutarate to succinate (12). Here, we have measured the effect of 1-meG and 3-meT on this reaction but were not able to detect any stimulation. These data indicate that the interaction of the neutral bases 1-meG and 3-meT with the AlkB active site may be weaker than in the case of 1-meA and 3-meC, which are positively charged at physiological pH (Figure 1). This is also supported by the observation that AlkB was more efficient at relieving a replication block at the DMS-treated substrate (containing 1-mA and 3-meC), than at the 3-meT-containing oligo (c.f. Figure 2C and F). Mechanistic insight on several enzymes that bind methylated bases in DNA and RNA has been obtained by studying the 3D structure of the protein in complex with a free methylated base or nucleoside (32,33). This approach may also be useful in the case of AlkB, particularly, since bases and nucleosides corresponding to 1-meA and 3-meC apparently can interact with the AlkB active site, but are not AlkB substrates. However, 1-meG and 3-meT may not be equally useful for this purpose; their inability to stimulate the uncoupled, AlkB-mediated decarboxylation of 2-oxoglutarate may suggest that their interaction with the AlkB active site is weaker than in the case of 1-meA and 3-meC.
Since the initial discovery that 1-meA and 3-meC in DNA are demethylated by AlkB (6,7), the number of known AlkB substrates has increased considerably (10,11). Similarly, the family of putative human AlkB homologues has grown. For many years, only one such homologue was known (34), but recent searches in protein databases have identified eight putative human AlkB proteins, denoted hABH1–hABH8 (8,10,13,14). Of these, an enzymatic activity has only been demonstrated in the case of hABH2 and hABH3, which, like E.coli AlkB, are able to demethylate 1-meA and 3-meC lesions in nucleic acids (10,14). A wide range of roles can be imagined for the remaining proteins, such as repair of other lesions in nucleic acids or reversal of nucleic acid or protein methylations that regulate gene activity. These unsolved questions represent important challenges for future research.
Note: After these data were obtained, two studies have reported AlkB-mediated repair of 3-meT (35,36) and 1-meG (35) lesions, using experimental strategies rather different from those used here. The reported data are consistent with those obtained in the present study. Koivisto et al. (36) reported oxidative demethylation by AlkB, hABH2 and hABH3 of 3-meT lesions induced by treatment of poly(dT) with methylating agents. Delaney and Essigmann (35) used single-stranded DNA substrates containing a single 1-meG or 3-meT lesion to study the mutagenicity and cytotoxicity of these lesions in wild-type and alkB mutant E.coli, and their results indicated that both 1-meG and 3-meT are AlkB substrates in vivo.
Acknowledgments
ACKNOWLEDGEMENTS
The technical assistance of Rune F. Johansen is greatly appreciated. I thank Per Arne Aas and Hans Krokan for providing purified hABH2 and hABH3, and Arne Klungland for a critical reading of the manuscript. The experimental part of this work was performed in the laboratory of Erling Seeberg, and I am most grateful for his help and support. This work was supported by the Norwegian Cancer Society and the Research Council of Norway.
REFERENCES
- 1.Landini P. and Volkert,M.R. (2000) Regulatory responses of the adaptive response to alkylation damage: a simple regulon with complex regulatory features. J. Bacteriol., 182, 6543–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sedgwick B. and Lindahl,T. (2002) Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene, 21, 8886–8894. [DOI] [PubMed] [Google Scholar]
- 3.Evensen G. and Seeberg,E. (1982) Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature, 296, 773–775. [DOI] [PubMed] [Google Scholar]
- 4.Karran P., Hjelmgren,T. and Lindahl,T. (1982) Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature, 296, 770–773. [DOI] [PubMed] [Google Scholar]
- 5.Karran P., Lindahl,T. and Griffin,B. (1979) Adaptive response to alkylating agents involves alternation in situ of O6-methylguanine residues in DNA. Nature, 280, 76–77. [DOI] [PubMed] [Google Scholar]
- 6.Falnes P.O., Johansen,R.F. and Seeberg,E. (2002) AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature, 419, 178–182. [DOI] [PubMed] [Google Scholar]
- 7.Trewick S.C., Henshaw,T.F., Hausinger,R.P., Lindahl,T. and Sedgwick,B. (2002) Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature, 419, 174–178. [DOI] [PubMed] [Google Scholar]
- 8.Aravind L. and Koonin,E.V. (2001) The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol., 2, RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schofield C.J. and Zhang,Z. (1999) Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr. Opin. Struct. Biol., 9, 722–731. [DOI] [PubMed] [Google Scholar]
- 10.Duncan T., Trewick,S.C., Koivisto,P., Bates,P.A., Lindahl,T. and Sedgwick,B. (2002) Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA, 99, 16660–16665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koivisto P., Duncan,T., Lindahl,T. and Sedgwick,B. (2003) Minimal methylated substrate and extended substrate range of Escherichia coli AlkB protein, a 1-methyladenine-DNA dioxygenase. J. Biol. Chem., 278, 44348–44354. [DOI] [PubMed] [Google Scholar]
- 12.Welford R.W., Schlemminger,I., McNeill,L.A., Hewitson,K.S. and Schofield,C.J. (2003) The selectivity and inhibition of AlkB. J. Biol. Chem., 278, 10157–10161. [DOI] [PubMed] [Google Scholar]
- 13.Kurowski M.A., Bhagwat,A.S., Papaj,G. and Bujnicki,J.M. (2003) Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics, 4, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aas P.A., Otterlei,M., Falnes,P.O., Vagbo,C.B., Skorpen,F., Akbari,M., Sundheim,O., Bjoras,M., Slupphaug,G., Seeberg,E. et al. (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature, 421, 859–863. [DOI] [PubMed] [Google Scholar]
- 15.Ougland R., Zhang,C.M., Liiv,A., Johansen,R.F., Seeberg,E., Hou,Y.M., Remme,J. and Falnes,P.O. (2004) AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell, 16, 107–116. [DOI] [PubMed] [Google Scholar]
- 16.Ashworth D.J., Baird,W.M., Chang,C.J., Ciupek,J.D., Busch,K.L. and Cooks,R.G. (1985) Chemical modification of nucleic acids. Methylation of calf thymus DNA investigated by mass spectrometry and liquid chromatography. Biomed. Mass Spectrom., 12, 309–318. [DOI] [PubMed] [Google Scholar]
- 17.Chang C. and Lee,C.G. (1981) Chemical modification of ribonucleic acid. A direct study by carbon-13 nuclear magnetic resonance spectroscopy. Biochemistry, 20, 2657–2661. [DOI] [PubMed] [Google Scholar]
- 18.Chang C.J., Gomes,J.D. and Byrn,S.R. (1983) Chemical modification of deoxyribonucleic acids: a direct study by C-13 nuclear magnetic-resonance spectroscopy. J. Org. Chem., 48, 5151–5160. [Google Scholar]
- 19.Saffhill R. and Abbott,P.J. (1978) Formation of O2-methylthymine in poly(dA-dT) on methylation with N-methyl-N-nitrosourea and dimethyl sulphate. Evidence that O2-methylthymine does not miscode during DNA synthesis. Nucleic Acids Res., 5, 1971–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singer B. (1975) Methylation and ethylation of uridylic acid and thymidylic acid. Reactivity of the ring and phosphate as a function of pH and alkyl group. Biochemistry, 14, 4353–4357. [DOI] [PubMed] [Google Scholar]
- 21.Daoud A.H. and Irving,C.C. (1977) Methylation of DNA in rat liver and intestine by dimethylnitrosamine and N-methylnitrosourea. Chem. Biol. Interact., 16, 135–143. [DOI] [PubMed] [Google Scholar]
- 22.Kang J.O. (1994) Methylated purine bases in hepatic and colonic RNA of rats treated with 1,2-dimethylhydrazine. Biochem. Med. Metab. Biol., 53, 52–57. [DOI] [PubMed] [Google Scholar]
- 23.Falnes P.O., Bjoras,M., Aas,P.A., Sundheim,O. and Seeberg,E. (2004) Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res., 32, 3456–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaule G. and Gunzler,V. (1990) Assay for 2-oxoglutarate decarboxylating enzymes based on the determination of [1–14C]succinate: application to prolyl 4-hydroxylase. Anal. Biochem., 184, 291–297. [DOI] [PubMed] [Google Scholar]
- 25.Huff A.C. and Topal,M.D. (1987) DNA damage at thymine N-3 abolishes base-pairing capacity during DNA synthesis. J. Biol. Chem., 262, 12843–12850. [PubMed] [Google Scholar]
- 26.Singer B. and Grunberger,D. (1983) Molecular Biology of Mutagens and Carcinogens. Plenum Press, NY. [Google Scholar]
- 27.Ahmmed Z. and Laval,J. (1984) Enzymatic repair of O-alkylated thymidine residues in DNA: involvement of a O4-methylthymine-DNA methyltransferase and a O2-methylthymine DNA glycosylase. Biochem. Biophys. Res. Commun., 120, 1–8. [DOI] [PubMed] [Google Scholar]
- 28.Singer B. (1975) The chemical effects of nucleic acid alkylation and their relation to mutagenesis and carcinogenesis. Prog. Nucleic Acid Res. Mol. Biol., 15, 219–284. [PubMed] [Google Scholar]
- 29.Kroger M. and Singer,B. (1979) Ambiguity and transcriptional errors as a result of methylation of N-1 of purines and N-3 of pyrimidines. Biochemistry, 18, 3493–3500. [DOI] [PubMed] [Google Scholar]
- 30.Bjork G.R., Wikstrom,P.M. and Bystrom,A.S. (1989) Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science, 244, 986–989. [DOI] [PubMed] [Google Scholar]
- 31.Hagervall T.G., Ericson,J.U., Esberg,K.B., Li,J.N. and Bjork,G.R. (1990) Role of tRNA modification in translational fidelity. Biochim. Biophys. Acta, 1050, 263–266. [DOI] [PubMed] [Google Scholar]
- 32.Drohat A.C., Kwon,K., Krosky,D.J. and Stivers,J.T. (2002) 3-Methyladenine DNA glycosylase I is an unexpected helix–hairpin–helix superfamily member. Nature Struct. Biol., 9, 659–664. [DOI] [PubMed] [Google Scholar]
- 33.Hu G., Gershon,P.D., Hodel,A.E. and Quiocho,F.A. (1999) mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc. Natl Acad. Sci. USA, 96, 7149–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei Y.F., Carter,K.C., Wang,R.P. and Shell,B.K. (1996) Molecular cloning and functional analysis of a human cDNA encoding an Escherichia coli AlkB homolog, a protein involved in DNA alkylation damage repair. Nucleic Acids Res., 24, 931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaney J.C. and Essigmann,J.M. (2004) Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl Acad. Sci. USA, 101, 14051–14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koivisto P., Robins,P., Lindahl,T. and Sedgwick,B. (2004) Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J. Biol. Chem., 279, 40470–40474. [DOI] [PubMed] [Google Scholar]