Abstract
The genomic landscape of adenomas and polyps may help define disease pathways. Expression of miRNAs in adenomas and polyps may importantly contribute to these pathways. We evaluated miRNA expression in 293 polyp-normal colorectal mucosa pairs. Polyps were classified as either adenomatous polyp (AD), hyperplastic polyp (HP), or sessile serrated polyp (SSP). We compared these miRNA expression profiles in polyps to miRNA expression in microsatellite unstable (MSI) and stable (MSS) tumors. A False Discovery Rate (FDR) of 0.05 based on Benjamini and Hochberg was used to adjust for multiple comparisons. There were 70 miRNAs with differential expression by polyp type with a fold change <0.75 or >1.34 after adjusting for multiple comparisons. The major differences in miRNA expression were observed between AD and SSP and AD and HP, with few differences in expression noted for SSP and HP. AD polyps were more likely to be up-regulated from normal colonic mucosa, while SSP and HP were more likely to be down-regulated from normal colonic mucosa. MiRNA expression in the SSP and HP tumors almost uniformly go in opposite directions from the MSS tumor miRNA expression and was mixed with MSI tumors. We conclude that different types of polyps have unique miRNA expression profiles.
Keywords: miRNA, adenoma, adenomatous polyp, microsatellite instability, sessile serrated polyp, hyperplastic polyp
Introduction
Certain types of colorectal polyps are thought to be precursor lesions for colorectal carcinomas (CRC). The histological characteristics of polyps are thought to be related to unique pathways to CRC [1, 2]. For example, sessile serrated polyps (SSP) have been linked to CIMP high, MSI high and BRAF-mutated tumors, while the more common adenoma carcinoma sequence has been linked to microsatellite stable tumors [2]. Hyperplastic polyps (HP), usually considered as a non-carcinoma precursor, have some overlapping characteristics with SSP’s. The molecular signature of polyps may be important in identifying clinical and molecular features of subsequent carcinomas.
MiRNAs are small, non-protein-coding RNA molecules that regulate gene expression [3, 4]. We have previously shown that miRNAs are commonly dysregulated in CRC and that some miRNAs are dysregulated as the tumor progresses, i.e. some are expressed differently in adenoma than in normal mucosa but have similar levels of expression in cancers, while others are similar in expression to normal mucosa but different from carcinoma expression[5]. Additionally, we have shown that differences in miRNA expression by tumor molecular phenotype exist [6].
In this study we examined miRNA expression profiles in polyp subtypes, evaluating adenomatous polyps (AD), SSP, and HP. We did this to determine similarities and differences in polyp types as well as their alignment with MSI tumors. Thus, we compared miRNA expression between polyp subtype and MSI and MSS miRNA expression profiles. Our goal was to identify miRNAs that can help characterize MSI or MSS molecular pathways of CRC and help describe the genomic landscape of polyps.
Materials and Methods
Study Participants
The study was approved by the Institutional Review Board of the University of Utah; study participants signed informed consent. Study participants came from two population-based case-control studies that included all incident colon and rectal cancers between 30 to 79 years of age who resided along the Wasatch Front in Utah or were members of the Kaiser Permanente Medical Care Program in Northern California [7, 8]. Cases had to have tumor registry verification of a first primary adenocarcinoma of the colon or rectum and were diagnosed between October 1991 and September 1994 for the colon cancer and between June 1997 and May 2001 for the rectal cancer. Individuals with familial adenomatous polyposis coli, inflammatory bowel disease, Crohn’s disease, and known Lynch Syndrome were excluded. Tumor tissue was obtained for 97% of all Utah cases diagnosed and for 85% of all Kaiser Permanente Medical Care Program in Northern California study participants [9]. Detailed study methods have been described [5]. The study population available to examine miRNA profiles in synchronous adenomas included 293 samples with either an AD, SSP, or HP. Of the 293 individual pathologies reviewed, 265 individuals only had AD polyps, eight individuals had SSP only, seven individuals had an HP only, five individuals had both an AD and SSP, seven individuals had both an AD and HP, and one person had both an SSP and HP.
miRNA processing
RNA was extracted from formalin-fixed paraffin embedded tissue using Ambion’s RecoverAll Total Nucleic Acid isolation kit. Normal mucosa adjacent to the carcinoma tissue was used. The Agilent Human miRNA Microarray V19.0 was used given the number of miRNAs, its high level of reliability (repeatability coefficient was 0.98 in our data) and the amount of RNA needed to run the platform. The microarray contains probes for 2006 unique human miRNAs. Data were required to pass stringent quality control parameters established by Agilent that included tests for excessive background fluorescence, excessive variation among probe sequence replicates on the array, and measures of the total gene signal on the array to assess low signal. The Agilent platform was shown to have high repeatability (r=0.98) and good correlation with other platforms such as Nanostring [5]. Other analysis of a subset of 22 important miRNAs showed 100% agreement in terms of directionality of differential expression and fold change when comparing results from the Agilent platform to those obtained from qPCR [10]. While the results were identical in terms of directionality and Agilent platform often slightly underestimated the fold change detected in qPCR analysis.
Tumor molecular phenotype
We have previously evaluated microsatellite instability by the mononucleotide repeats BAT26 and TGFβR2 and a panel of 10 tetranucleotide repeats that were correlated highly with the Bethesda Panel [11].
Statistical Methods
To normalize differences in miRNA expression that could be attributed to the array, amount of RNA, location on array, or other factors that could erroneously influence expression, total gene signal was normalized by multiplying each sample by a scaling factor [12] (http://genespring-support.com/files/gs_12_6/GeneSpring-manual.pdf), which was the median of the 75th percentiles of all the samples divided by the individual 75th percentile of each sample. Data were log2 transformed prior to analysis and were analyzed using the significance analysis of microarrays technique implemented in the R package siggenes [13]; p-values were based upon 1000 permutations. Only miRNAs with a large percentage of samples expressing above the background were included. Comparisons were made between polyp type (AD, SSP, and HP) to assess miRNA expression differences. We compared differences based on both polyp miRNA expression as well as differential expression between polyp and normal colonic mucosa expression. To adjust for multiple comparisons we applied a False Discovery Rate (FDR) level of significance of 0.05 based on Benjamini and Hochberg [14]. Additionally, we evaluated miRNA expression by polyp type with both MSI and MSS tumors.
To visualize differences in expression by polyp subtype, first we compared differential expression between the three different polyp types and normal colonic mucosa using the two class test for paired polyp/normal mucosa data in siggenes, applied an FDR level of 0.05, and fold change less than log2(0.75) or greater than log2(1.5). Using these results we created heatmaps of the fold changes of the different polyp types in SAS 9.4. We recalculated the fold changes of the significant results comparing the MSI or MSS carcinoma samples and polyp samples to paired normal colonic mucosa results and created heatmaps of the fold changes of the different polyp types and MSI or MSS.
Results
The majority of the study population was male (Table 1). AD were somewhat equally distributed across colorectal tumor sites, while SSP was predominately in the proximal colon and HP was predominately from the rectum. The majority of AD polyps were associated with TP53-mutated and KRAS-mutated tumors, SSP were associated with MSI and CIMP tumors, while HP were associated mainly with either a TP53-mutated or CIMP high tumor.
Table 1.
Description of Study Population
| Adenomatous | Adenoma type Sessile Serrated | Hyperplastic | ||||
|---|---|---|---|---|---|---|
| Mean | (SD) | Mean | (SD) | Mean | (SD) | |
| Age | 65.9 | (9.2) | 67.4 | (9.3) | 61.5 | (10.3) |
| N | (%) | N | (%) | N | (%) | |
| Sex | ||||||
| Men | 164 | (59.2) | 8 | (57.1) | 8 | (53.3) |
| Women | 113 | (40.8) | 6 | (42.9) | 7 | (46.7) |
| Location | ||||||
| Proximal Colon | 84 | (30.3) | 11 | (78.6) | 3 | (20.0) |
| Distal Colon | 84 | (30.3) | 2 | (14.3) | 2 | (13.3) |
| Rectal | 109 | (39.4) | 1 | (7.1) | 10 | (66.7) |
| Related Tumor Molecular Phenotype | ||||||
| MSI + | 15 | (5.7) | 5 | (41.7) | 2 | (13.3) |
| CIMP high | 45 | (19.1) | 7 | (63.6) | 5 | (38.5) |
| BRAF-mutated | 5 | (2.1) | 2 | (20.0) | 2 | (15.4) |
| TP53-mutated | 118 | (45.0) | 5 | (35.7) | 7 | (53.9) |
| KRAS-mutated | 113 | (44.0) | 3 | (21.4) | 4 | (26.7) |
The fold change (FC) between miRNA expression in tumor tissue by polyp type is shown in Table 2 for those miRNAs that were statistically significant after adjustment for multiple comparisons and for whom the FC was <0.75 or >1.34 for any of the comparisons. There were 70 miRNAs that met these criteria. The major differences in miRNA expression were observed between AD and SSP and AD and HP, with few differences in expression noted between SSP and HP. Additionally, we evaluated significant differences in differential expression between polyp and normal mucosa by polyp subtype and identified an additional 19 miRNAs beyond those associated with miRNA expression in polyps that were differentially expressed between AD and HP (let-7i-5p, miR-1229-5p, miR-1234-5p, miR-1249, miR-1268B, miR-1275, miR-194-5p, miR-215, miR-2392, miR-30b-5p, miR-331-3p, miR-3653, miR-3960, miR-4281, miR-4689, mR-4739, miR-518a-5p, miR-6510-5p, and miR-939-5p) when considering a FC of >1.34 or <0.75; there were no differences in differential expression between SSP and HP (data not shown in table).
Table 2.
Comparison of miRNA expression by adenoma type
| miRNA |
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean Level of Expression | Adenomatous vs SSP | Adenomatous vs Hyperplastic | Hyperplastic vs SSP | |||||||
|
| ||||||||||
| Normal | Adenomatous | SSP | Hyperplastic | FC | Adjusted p value | FC | Adjusted p value | FC | Adjusted p value | |
| hsa-let-7a-5p | 166.36 | 178.99 | 98.54 | 87.35 | 1.82 | 0.03 | 2.05 | 0.005 | 1.13 | 0.74 |
| hsa-let-7b-5p | 223.1 | 224.82 | 164.32 | 134.83 | 1.37 | 0.18 | 1.67 | 0.01 | 1.22 | 0.40 |
| hsa-let-7c | 54.52 | 51.77 | 40.45 | 32.01 | 1.28 | 0.39 | 1.62 | 0.02 | 1.26 | 0.41 |
| hsa-let-7d-5p | 24.64 | 28.05 | 14.61 | 13.73 | 1.92 | 0.04 | 2.04 | 0.006 | 1.06 | NA |
| hsa-let-7f-5p | 84.3 | 95.38 | 47.46 | 42.8 | 2.01 | 0.02 | 2.23 | 0.003 | 1.11 | 0.87 |
| hsa-let-7g-5p | 34.46 | 37.86 | 18.5 | 14.26 | 2.05 | 0.04 | 2.66 | 0.005 | 1.30 | NA |
| hsa-miR-103a-3p | 35.15 | 44.69 | 22.15 | 23.35 | 2.02 | 0.03 | 1.91 | 0.005 | 0.95 | NA |
| hsa-miR-107 | 22.23 | 26.82 | 12.98 | 13.21 | 2.07 | 0.02 | 2.03 | 0.005 | 0.98 | NA |
| hsa-miR-10a-5p | 17.83 | 23.97 | 13.51 | 8.78 | 1.77 | 0.20 | 2.73 | 0.008 | 1.54 | NA |
| hsa-miR-1225-5p | 3269.87 | 3320.42 | 3587.1 | 4478.36 | 0.93 | 0.18 | 0.74 | 0.04 | 0.80 | 0.35 |
| hsa-miR-1234-3p | 43.95 | 40.34 | 40.03 | 57.69 | 1.01 | 0.88 | 0.70 | 0.03 | 0.69 | 0.07 |
| hsa-miR-1246 | 393.84 | 519.46 | 379.29 | 403.11 | 1.37 | 0.0006 | 1.29 | 0.005 | 0.94 | 0.47 |
| hsa-miR-125b-5p | 32.85 | 8.68 | 18.7 | 11.95 | 0.46 | 0.009 | 0.73 | 0.08 | 1.57 | NA |
| hsa-miR-1323 | 6.47 | 7.01 | 9.51 | 7.93 | 0.74 | 0.009 | 0.88 | 0.43 | 1.20 | NA |
| hsa-miR-141-3p | 21.1 | 31.79 | 12.46 | 11.45 | 2.55 | 0.008 | 2.78 | <0.0001 | 1.09 | NA |
| hsa-miR-145-5p | 185.82 | 55.7 | 102.07 | 105.15 | 0.55 | 0.0008 | 0.53 | 0.0004 | 0.97 | 0.69 |
| hsa-miR-1470 | 4.26 | 3.28 | 4.73 | 5.14 | 0.69 | 0.02 | 0.64 | 0.009 | 0.92 | NA |
| hsa-miR-15b-5p | 17.39 | 22.27 | 8.85 | 7.86 | 2.51 | 0.02 | 2.83 | 0.006 | 1.13 | NA |
| hsa-miR-16-5p | 46.88 | 50.58 | 27.93 | 26.56 | 1.81 | 0.02 | 1.90 | 0.004 | 1.05 | 0.95 |
| hsa-miR-17-5p | 12.09 | 23.68 | 8.29 | 9.42 | 2.86 | 0.004 | 2.51 | 0.003 | 0.88 | NA |
| hsa-miR-192-5p | 99.98 | 96.27 | 49.44 | 43.32 | 1.95 | 0.01 | 2.22 | 0.002 | 1.14 | 0.99 |
| hsa-miR-1973 | 789.22 | 929.03 | 525.84 | 496.47 | 1.77 | 0.0009 | 1.87 | <0.0001 | 1.06 | 0.97 |
| hsa-miR-200a-3p | 15.63 | 23.97 | 7.3 | 5.62 | 3.28 | 0.003 | 4.26 | 0.0006 | 1.30 | NA |
| hsa-miR-200b-3p | 103.56 | 168.92 | 53.91 | 50.18 | 3.13 | 0.0004 | 3.37 | <0.0001 | 1.07 | 0.98 |
| hsa-miR-200c-3p | 95.42 | 153.38 | 61.12 | 61.38 | 2.51 | 0.0002 | 2.50 | <0.0001 | 1.00 | 0.72 |
| hsa-miR-21-5p | 97.57 | 154.87 | 73.59 | 73.37 | 2.10 | 0.009 | 2.11 | 0.005 | 1.00 | 0.86 |
| hsa-miR-222-3p | 7.69 | 9.04 | 12.84 | 12.88 | 0.70 | 0.01 | 0.70 | 0.002 | 1.00 | 0.63 |
| hsa-miR-22-3p | 10.79 | 6.13 | 8.94 | 9.51 | 0.69 | NA | 0.64 | 0.02 | 0.94 | NA |
| hsa-miR-23a-3p | 60.92 | 94.23 | 41.69 | 51.37 | 2.26 | 0.004 | 1.83 | 0.01 | 0.81 | 0.28 |
| hsa-miR-23b-3p | 52.29 | 55.89 | 29.78 | 32.11 | 1.88 | 0.05 | 1.74 | 0.02 | 0.93 | NA |
| hsa-miR-24-3p | 45.68 | 62.08 | 34.39 | 38.8 | 1.81 | 0.002 | 1.60 | 0.006 | 0.89 | 0.30 |
| hsa-miR-25-3p | 7.87 | 12.94 | 5.37 | 5.51 | 2.41 | NA | 2.35 | 0.02 | 0.97 | NA |
| hsa-miR-26a-5p | 70.89 | 74.86 | 41.99 | 38.33 | 1.78 | 0.04 | 1.95 | 0.01 | 1.10 | 0.89 |
| hsa-miR-27a-3p | 14.6 | 27.42 | 9.13 | 10.52 | 3.00 | NA | 2.61 | 0.008 | 0.87 | NA |
| hsa-miR-27b-3p | 18.27 | 23.79 | 9.82 | 8.59 | 2.42 | 0.01 | 2.77 | 0.007 | 1.14 | NA |
| hsa-miR-29a-3p | 31.9 | 51.07 | 23.27 | 23.43 | 2.19 | 0.01 | 2.18 | 0.007 | 0.99 | 0.83 |
| hsa-miR-29b-3p | 5.42 | 8.54 | 3.91 | 3.82 | 2.18 | NA | 2.24 | 0.02 | 1.02 | NA |
| hsa-miR-30d-5p | 24.06 | 23.44 | 15.59 | 15.45 | 1.50 | 0.001 | 1.52 | 0.0008 | 1.01 | 0.94 |
| hsa-miR-3174 | 11.76 | 12.52 | 9.58 | 10.61 | 1.31 | 0.01 | 1.18 | 0.04 | 0.90 | 0.11 |
| hsa-miR-3177-5p | 4.67 | 4.18 | 5.85 | 5.6 | 0.71 | 0.05 | 0.75 | 0.04 | 1.05 | NA |
| hsa-miR-3196 | 1524.08 | 1612.26 | 1831.32 | 2337.48 | 0.88 | 0.10 | 0.69 | 0.01 | 0.78 | 0.20 |
| hsa-miR-320d | 40.15 | 41.42 | 32.7 | 30.94 | 1.27 | 0.03 | 1.34 | 0.01 | 1.06 | 0.62 |
| hsa-miR-330-3p | 4.76 | 4.1 | 6.75 | 7.67 | 0.61 | 0.008 | 0.54 | 0.01 | 0.88 | NA |
| hsa-miR-342-3p | 12.79 | 6.34 | 13.01 | 9.39 | 0.49 | 0.007 | 0.67 | 0.02 | 1.38 | NA |
| hsa-miR-3651 | 21.94 | 42.02 | 16.74 | 15.88 | 2.51 | 0.0003 | 2.65 | 0.0002 | 1.05 | 0.88 |
| hsa-miR-375 | 42.33 | 41.12 | 21.06 | 25.46 | 1.95 | 0.01 | 1.62 | 0.09 | 0.83 | 0.22 |
| hsa-miR-424-3p | 23.54 | 32.36 | 22.69 | 20.18 | 1.43 | 0.0008 | 1.60 | <0.0001 | 1.12 | 0.30 |
| hsa-miR-4284 | 1161.35 | 1465.29 | 753.46 | 836.4 | 1.94 | 0.002 | 1.75 | 0.002 | 0.90 | 0.28 |
| hsa-miR-431-5p | 46.86 | 45.5 | 53.7 | 63.98 | 0.85 | 0.10 | 0.71 | 0.007 | 0.84 | 0.29 |
| hsa-miR-4323 | 11.19 | 8.77 | 9.57 | 14.29 | 0.92 | 0.26 | 0.61 | <0.0001 | 0.67 | 0.00 |
| hsa-miR-4449 | 13.41 | 22.01 | 12.3 | 11.63 | 1.79 | <0.0001 | 1.89 | <0.0001 | 1.06 | 0.27 |
| hsa-miR-4472 | 12.15 | 12.69 | 17.51 | 13.64 | 0.72 | 0.04 | 0.93 | 0.42 | 1.28 | 0.21 |
| hsa-miR-4485 | 1185.77 | 1294.33 | 804.72 | 823.47 | 1.61 | 0.0007 | 1.57 | 0.0008 | 0.98 | 0.83 |
| hsa-miR-4502 | 18.19 | 16.27 | 22.16 | 18.54 | 0.73 | 0.03 | 0.88 | 0.05 | 1.20 | 0.39 |
| hsa-miR-4526 | 4.06 | 3.73 | 5.4 | 4.74 | 0.69 | 0.0002 | 0.79 | 0.07 | 1.14 | NA |
| hsa-miR-4707-3p | 10.78 | 10.06 | 13.23 | 14.44 | 0.76 | 0.02 | 0.70 | 0.02 | 0.92 | 0.97 |
| hsa-miR-4749-3p | 13.45 | 12.67 | 17.38 | 20.56 | 0.73 | 0.003 | 0.62 | 0.002 | 0.85 | 0.47 |
| hsa-miR-4787-3p | 104.13 | 95.09 | 98.92 | 131.05 | 0.96 | 0.82 | 0.73 | 0.03 | 0.75 | 0.17 |
| hsa-miR-492 | 12.4 | 9.51 | 23.08 | 37.84 | 0.41 | <0.0001 | 0.25 | <0.0001 | 0.61 | 0.00 |
| hsa-miR-501-3p | 2.63 | 5.32 | 2.49 | 2.66 | 2.14 | 0.003 | 2.00 | 0.005 | 0.93 | NA |
| hsa-miR-5585-5p | 4.68 | 4.42 | 6.57 | 7.18 | 0.67 | 0.08 | 0.62 | 0.03 | 0.91 | NA |
| hsa-miR-650 | 15.94 | 9.15 | 17.64 | 10.03 | 0.52 | 0.002 | 0.91 | 0.19 | 1.76 | 0.02 |
| hsa-miR-654-5p | 28.25 | 27 | 31.91 | 37.32 | 0.85 | 0.10 | 0.72 | 0.02 | 0.86 | 0.47 |
| hsa-miR-663b | 32.65 | 49.56 | 35.33 | 38.08 | 1.40 | 0.0008 | 1.30 | 0.01 | 0.93 | 0.55 |
| hsa-miR-664a-5p | 9.97 | 11.9 | 5.58 | 2.92 | 2.13 | 0.006 | 4.08 | NA | 1.91 | NA |
| hsa-miR-664b-3p | 18.8 | 21.3 | 15.25 | 11.52 | 1.40 | 0.03 | 1.85 | 0.0002 | 1.32 | 0.10 |
| hsa-miR-664b-5p | 39.56 | 46.98 | 28.48 | 22.07 | 1.65 | 0.001 | 2.13 | <0.0001 | 1.29 | 0.09 |
| hsa-miR-758-5p | 31.78 | 36.59 | 25.83 | 26.61 | 1.42 | 0.0009 | 1.37 | 0.0002 | 0.97 | 0.46 |
| hsa-miR-92a-3p | 32.05 | 53.77 | 20.79 | 19.35 | 2.59 | 0.0004 | 2.78 | <0.0001 | 1.07 | 0.98 |
| hsa-miR-93-5p | 10.54 | 21.32 | 7.62 | 8.25 | 2.80 | 0.006 | 2.59 | 0.006 | 0.92 | NA |
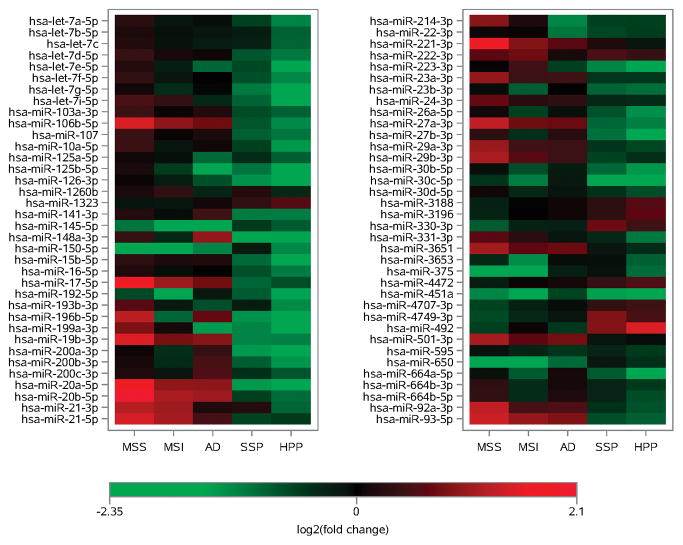
Evaluation of the top dysregulated miRNAs by tissue type showed that miRNAs in AD polyp tissue were more likely to be up-regulated from normal colonic mucosa, while miRNAs in SSP and HP tissue were more likely to be down-regulated from normal colonic mucosa (Figure 1). While the AD is often not different from the normal mucosa, the SSP and HP are downregulated and the MSI tumor contain a mixture of up-regulated, down-regulated and not differentially expressed miRNAs. Looking at a similar comparison that includes MSS tumors miRNA expression in the SSP and HP polyps almost uniformly go in opposite directions from the MSS tumor miRNA expression.
Figure 1.
Comparison of miRNA expression levels by polyp subtype and by MSI and MSS tumors
We assessed molecular pathways that were uniquely associated with AD and with SSP/HP differential expression. In these analyses we included miRNAs that were significantly dysregulated with a fold change of 1.35 or greater or 0.74 or smaller and were significantly different between AD and SSP/HP adenoma subtype. There were many more pathways identified for SSP/HP than for AD. After adjustment for multiple comparisons there were nine canonical pathways with an adjusted −log2 p value of 2.00E00 for AD (See Supplement 1) while there were 143 canonical pathways associated with genes regulated by differentially expressed miRNAs for SSP/HP. There was overlap in these pathways. For instance, both polyp types were associated with molecular mechanisms of cancer and TP53. However for AD the TP53 pathway appeared to be upregulated while for SSP/HP the pathway appeared to be downregulated based on the up and down regulation of the miRNAs in the adenoma tissue compared to normal colonic mucosa.
Discussion
We observed differences in miRNA expression by polyp type, with adenomatous polyps being upregulated from normal colonic mucosa while miRNA expression in SSP and HP was usually downregulated. There were few differences in miRNA expression between SSP and HP. However, examining polyps in people who had both AD and HP, we observed fewer differences in miRNA expression by polyp type.
As possible precursors to colorectal carcinomas, polyps are potentially a major component of the carcinogenic process. While adenomatous polyps represent the majority of polyps that evolve into cancers, it is thought that some sessile serrated polyps contribute to unique disease pathways that include MSI tumors. Serrated polyps can be divided into hyperplastic polyps, considered to have no malignant potential, and the possibly pre-malignant sessile serrated polyp. Sessile serrated polyps are defined by their histologic appearance [15] and were described in the 1990s based on their serrated glandular configuration and other clinical-pathological features [16]
Our findings suggest the miRNA profiles of HP and SSP are very similar and support the conclusions of Sandmeier and colleagues that HP and SSP have similar genomic characteristics [17]. These polyps differ in miRNA expression profiles from AD and are more likely to have down-regulated miRNA expression whereas AD are more likely to have up-regulated miRNA expression. These miRNAs also were more likely to be downregulated relative to MSI tumors and inconsistently up- and down-regulated with MSS tumors. Also of interest is the larger number of dysregulated miRNAs in SPP/HP than observed for AD and therefore the larger possible number of targeted dysregulated genes and their associated pathways.
At the population level, when comparing all AD to all SSP/HP we saw a large number of dysregulated miRNAs between these polyp types. However, for those few individuals that had both an AD and a SSP/HP we saw fewer miRNAs that were differentially expressed between the polyp types. It is possible that some of the differences could be from confounding variables that were associated with miRNA expression profile. Within an individual these factors, such as cigarette smoking, would have been controlled and fewer differences in dysregulation might be observed. Fewer statistical findings for this subset of individuals could also be the result of a very small sample resulting in limited statistical power.
We have a large set of polyps with paired carcinoma and normal colorectal mucosa tissue. All paired samples came from the same tumor location. We believe that these are strengths of the study. Additionally, by using the Agilent miRNA platform we were able to assess expression of over 2000 miRNAs in colorectal tissue. In addition to the miRNA data, we have data on tumor molecular phenotype and other lifestyle factors. However, the study has limitations. First the polyps are synchronous to the carcinoma and are not the precursor lesion. Thus we are able to look at characteristics of polyps with a certain miRNA profile and compare it to the miRNA profiles of other synchronous polyps and carcinoma miRNA profiles. Additionally given the number of targeted genes by these dysregulated miRNA, it is difficult to identify specific pathways that are unique to various polyp subtypes. This limitation is a limitation of the state of the field of miRNA bioinformatics at this time. Additionally we report results based on an arbitrary fold change cutoff of <0.75 or greater than 1.34 as well as statistical significance. It should be acknowledged that the biologically important fold change for each miRNA is not known and that miRNAs with lower levels of expression can have an absolute change in expression that is minimal, yet have a large fold change.
Supplementary Material
Supplemental Table 1. Canonical pathways associated with AD polyps
Supplemental Table 2. Canonical pathways associated with SSP
Acknowledgments
Supported by: This study was supported by NCI grants CA163683 and CA48998 from the National Cancer Institute at the National Institutes of Health.
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute. We acknowledge Sandra Edwards for data oversight and Dr. Bette Caan and the staff at the KPMCP for their contribution to data collection, Erica Wolff and Michael Hoffman for miRNA analysis, and Brett Milash and the Bioinformatics Shared Resource of the Huntsman Cancer Institute and University of Utah for miRNA and mRNA bioinformatics data processing, and Daniel Pellatt for statistical consultation.
References
- 1.Jass JR. Molecular heterogeneity of colorectal cancer: Implications for cancer control. Surg Oncol. 2007;16(Suppl 1):S7–9. doi: 10.1016/j.suronc.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 2.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50(1):113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol. 2008;18(2):103–110. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Slattery ML, Herrick JS, Pellatt DF, Stevens JR, Mullany LE, Wolff E, Hoffman MD, Samowitz WS, Wolff RK. MicroRNA profiles in colorectal carcinomas, adenomas and normal colonic mucosa: variations in miRNA expression and disease progression. Carcinogenesis. 2016 doi: 10.1093/carcin/bgv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slattery ML, Herrick JS, Mullany LE, Wolff E, Hoffman MD, Pellatt DF, Stevens JR, Wolff RK. Colorectal tumor molecular phenotype and miRNA: expression profiles and prognosis. Mod Pathol. 2016 doi: 10.1038/modpathol.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slattery ML, Potter J, Caan B, Edwards S, Coates A, Ma KN, Berry TD. Energy balance and colon cancer--beyond physical activity. Cancer research. 1997;57(1):75–80. [PubMed] [Google Scholar]
- 8.Slattery ML, Caan BJ, Benson J, Murtaugh M. Energy balance and rectal cancer: an evaluation of energy intake, energy expenditure, and body mass index. Nutr Cancer. 2003;46(2):166–171. doi: 10.1207/S15327914NC4602_09. [DOI] [PubMed] [Google Scholar]
- 9.Slattery ML, Edwards SL, Palmer L, Curtin K, Morse J, Anderson K, Samowitz W. Use of archival tissue in epidemiologic studies: collection procedures and assessment of potential sources of bias. Mutat Res. 2000;432(1–2):7–14. doi: 10.1016/s1383-5726(99)00010-2. [DOI] [PubMed] [Google Scholar]
- 10.Pellatt DF, Stevens JR, Wolff RK, Mullany LE, Herrick JS, Samowitz W, Slattery ML. Expression Profiles of miRNA Subsets Distinguish Human Colorectal Carcinoma and Normal Colonic Mucosa. Clin Transl Gastroenterol. 2016;7:e152. doi: 10.1038/ctg.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slattery ML, Curtin K, Anderson K, Ma KN, Ballard L, Edwards S, Schaffer D, Potter J, Leppert M, Samowitz WS. Associations between cigarette smoking, lifestyle factors, and microsatellite instability in colon tumors. J Natl Cancer Inst. 2000;92(22):1831–1836. doi: 10.1093/jnci/92.22.1831. [DOI] [PubMed] [Google Scholar]
- 12.Agilent GeneSpring User Manual
- 13.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 15.Rex DK, Ahnen DJ, Baron JA, Batts KP, Burke CA, Burt RW, Goldblum JR, Guillem JG, Kahi CJ, Kalady MF, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107(9):1315–1329. doi: 10.1038/ajg.2012.161. quiz 1314, 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longacre TA, Fenoglio-Preiser CM. Mixed hyperplastic adenomatous polyps/serrated adenomas. A distinct form of colorectal neoplasia. Am J Surg Pathol. 1990;14(6):524–537. doi: 10.1097/00000478-199006000-00003. [DOI] [PubMed] [Google Scholar]
- 17.Sandmeier D, Benhattar J, Martin P, Bouzourene H. Serrated polyps of the large intestine: a molecular study comparing sessile serrated adenomas and hyperplastic polyps. Histopathology. 2009;55(2):206–213. doi: 10.1111/j.1365-2559.2009.03356.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Canonical pathways associated with AD polyps
Supplemental Table 2. Canonical pathways associated with SSP