Abstract
Many common diseases show wide phenotypic variation. We present a statistical method for determining whether phenotypically defined subgroups of disease cases represent different genetic architectures, in which disease-associated variants have different effect sizes in the two subgroups. Our method models the genome-wide distributions of genetic association statistics with mixture Gaussians. We apply a global test without requiring explicit identification of disease-associated variants, thus maximising power in comparison to a standard variant by variant subgroup analysis. Where evidence for genetic subgrouping is found, we present methods for post-hoc identification of the contributing genetic variants.
We demonstrate the method on a range of simulated and test datasets where expected results are already known. We investigate subgroups of type 1 diabetes (T1D) cases defined by autoantibody positivity, establishing evidence for differential genetic architecture with thyroid peroxidase antibody positivity, driven generally by variants in known T1D associated regions.
Introduction
Analysis of genetic data in human disease typically uses a binary disease model of cases and controls. However, many common human diseases show extensive clinical and phenotypic diversity which may represent multiple causative pathophysiological processes. Because therapeutic approaches often target disease-causative pathways, understanding this phenotypic complexity is valuable for further development of treatment, and the progression towards personalised medicine. Indeed, identification of patient subgroups characterised by different clinical features can aid directed therapy [1] and accounting for phenotypic substructures can improve ability to detect causative variants by refining phenotypes into subgroups in which causative variants have larger effect sizes [2].
Such subgroups may arise from environmental effects, reflect population variation in non-disease related anatomy or physiology, correspond to partitions of the population in which disease heritability differs, or represent different causative pathological processes. Our method tests whether there exist a subset of disease-associated SNPs which have different effect sizes in case subgroups, determining whether heterogeneity corresponds to differential genetic pathology.
Our test is for a stronger assertion than the question of whether subgroups of a disease group exhibit any genetic differences at all, as these may be entirely disease-independent: for example, although there will be systematic genetic differences between Asian and European patient cohorts with type 1 diabetes (T1D), these differences will not generally relate to the pathogenesis of disease.
Rather than attempting to analyse SNPs individually for differences between subgroups, a task for which GWAS are typically underpowered, we model allelic differences across all SNPs using mixture multivariate normal models. This can give insight into the structure of the genetic basis for disease. Given evidence that there exists some subset of SNPs that both differentiate controls and cases and differentiate subgroups, we can then reassess test statistics to search for single-SNP effects.
Results
Summary of proposed method
We jointly consider allelic differences between the combined case group and controls, and allelic differences between case subgroups independent of controls. Specifically, we establish whether the data support a hypothesis (H1) that a subset of SNPs associated with case-control status have different underlying effect sizes (and hence underlying allele frequencies) in case subgroups. This assumption has been used previously for genetic discovery [3].
H1 encompasses several potential underlying mechanisms of heterogeneity. A set of SNPs may be associated with one case subgroup but not the other; the same set of SNPs may have different relative effect sizes in subgroups, or heritability may differ between subgroups. These scenarios are discussed in the supplementary note.
Our overall protocol is to fit two bivariate Gaussian mixture models, corresponding to null and alternative hypotheses, to summary statistics (Z scores) derived from SNP data. We assume a group of controls and two non-intersecting case subgroups, and jointly consider allelic differences between the combined case group and controls, and allelic differences between case subgroups independent of controls (figure 1). Heterogeneity in cases can also be characterised by a quantitative trait, rather than explicit subgroups.
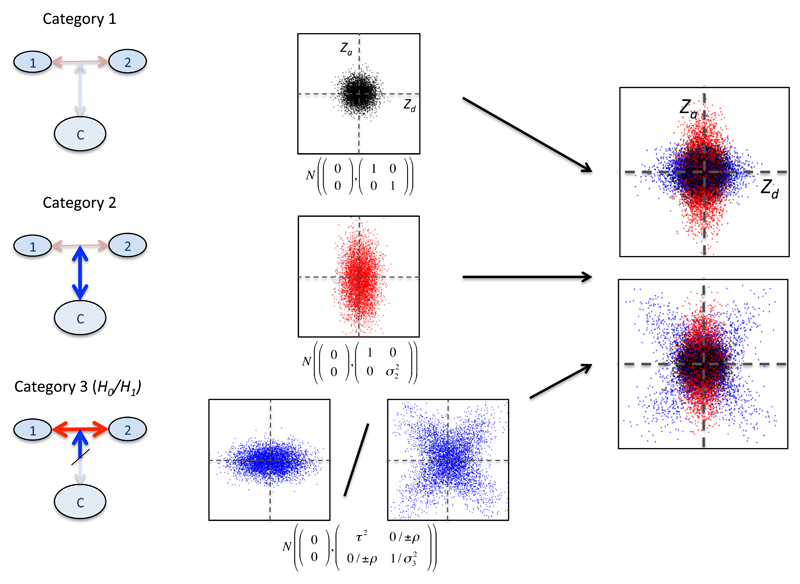
Figure 1.
Overview of three-categories model. Zd and Za are Z scores derived from GWAS p-values for allelic differences between case subgroups (1 vs 2), and between cases and controls (1+2 vs C) respectively (left). Within each category of SNPs, the joint distribution of (Zd,Za) has a different characteristic form. In category 1, Z scores have a unit normal distribution; in category 2, the marginal variance of Za can vary. The distribution of SNPs in category 3 depends on the main hypothesis. Under H0 (that all disease-associated SNPs have the same effect size in both subgroups), only the marginal variance of Zd may vary; under H1 (that subgroups correspond to differential effect sizes for disease-associated SNPs), any covariance matrix is allowed. The overall SNP distribution is then a mixture of Gaussians resembling one of the rightmost panels, but with SNP category membership unobserved. Visually, our test determines whether the observed overall Zd, Za distribution more closely resembles the bottom rightmost panel than the top.
For a given SNP we denote by μ1, μ2, μ12 and μc the population minor allele frequencies for each of the two case subgroups, the whole case group and the control group respectively, and Pd, Pa GWAS p-values for comparisons of allelic frequency between case subgroups and between cases and controls, under the null hypotheses μ1 = μ2 and μ12 = μc respectively (or similarly for quantitative heterogeneity). We then derive absolute Z scores |Zd| and |Za| from these p-values (see figure 1). We consider the values |Zd|,|Za| as absolute values of observations of random variables (Zd,Za) which are samples from a mixture of three bivariate Gaussians. Further details are given in the supplementary note.
We consider each SNP to fall into one of three categories, with each category corresponding to a different joint distribution of Zd, Za:
SNPs which do not differentiate subgroups and are not associated with the phenotype as a whole (μc = μ1 = μ2)
SNPs which are associated with the phenotype as a whole but which are not differentially associated with the subgroups (μc≠μ12; μ1 = μ2 = μ12)
SNPs which have different population allele frequencies in subgroups, and may or may not be associated with the phenotype as a whole (μ1≠μ2)
If the SNPs in category 3 are not associated with the disease as a whole (null hypothesis, H0), we expect Zd, Za to be independent and the variance of Za to be 1. If SNPs in category 3 are also associated with the disease as a whole (alternative hypothesis, H1), the joint distribution of (Zd,Za) will have both marginal variances greater than 1, and Za, Zd may co-vary. Our test is therefore focussed on the form of the joint distribution of (Zd,Za) in category 3. Importantly, we allow that the correlation between Zd and Za may be simultaneously positive at some SNPs and negative at others. This allows for a subset of SNPs to specifically alter risk of one subgroup, and another subset to alter risk for the other subgroup. To accommodate this, we only consider absolute Z scores and model the distribution of SNPs in category 3 with two mirror-image bivariate Gaussians.
Amongst SNPs with the same frequency in disease subgroups (categories 1 and 2), Za and Zd are independent and the expected standard deviation of Zd is 1. We therefore model the overall joint distribution of (Zd,Za) as a Gaussian mixture in which the pdf of each observation (Zd,Za) is given by
| (1) |
where NΣ(d,a) denotes the density of the bivariate normal pdf centered at 0 with covariance matrix Σ at (d,a). Θ is the vector of values (π1,π2,τ,σ2,σ3,ρ). Under H0, we have ρ = 0 and σ3 = 1. The values (π1,π2,π3) represent the proportion of SNPs in each category, with Σ πi = 1 (see table 1). Patterns of (Zd,Za) for different parameter values are shown in supplementary table 1.
Table 1.
Interpretation of parameter values in the fitted model. Parameters τ, σ2 and σ3 are dependent on sample sizes, but can be converted to sample-size independent forms (see supplementary note)
| Model | Interpretation | |
|---|---|---|
| π1 | H0 / H1 | Proportion of SNPs not associated with case/control (category 1) |
| π2 | H0 / H1 | Proportion of SNPs associated with case/control status but not subgroup status (category 2) |
| π3 | H0 / H1 | Proportion of SNPs associated with subgroup status (category 3) |
| τ | H0 / H1 | Standard deviation of observed Zd scores (effect sizes for subgroup status) in category 3 |
| σ2 | H0 / H1 | Standard deviation of observed Za scores (effect sizes for case/control status) in category 2 |
| σ3 | H1 only | Standard deviation of observed Za scores (effect sizes for case/control status) in category 3 |
| ρ | H1 only | ‘Absolute covariance’ between Zd scores (effect sizes for subgroup status) and Za scores (effect sizes for case/control status) in category 3 |
We use the product of values of the above pdf for a set of observed Zd , Za as an objective function (‘pseudo-likelihood’, PL) to estimate the values of parameters. This is not a true likelihood as observations are dependent due to linkage disequilibrium (LD), although because we minimise the degree of LD between SNPs using the LDAK method [4], the PL is similar to a true likelihood.
Model fitting and significance testing
We fit parameters π1, π2, π3 (= 1 - π1 - π2), σ2, σ3, τ and ρ under H1 and H0. Under H0, (ρ,σ3) = (0,1).
We then compare the fit of the two models using the log-ratio of PLs, giving an unadjusted pseudo-likelihood ratio (uPLR). We subtract a term depending only on Za to minimise the influence of the Za score distribution, and add a term log(π1π2π3) to ensure the model is identifiable [5]. We term the resultant test statistic the pseudo-likelihood ratio (PLR). The distribution of the PLR is minorised by a distribution of the form:
| (2) |
The value γ arises from the weighting derived from the LDAK procedure causing a scale change in the observed PLR. The mixing parameter κ corresponds to the probability that ρ = 0, (approximately ½).
We estimate γ and κ by sampling random subgroups of the case group. Such subgroups only cover the subspace of H0 with τ = 1 (no systematic allelic differences between subgroups), causing the asymptotic approximation of PLR by equation 2 to be poor. We thus estimate γ and κ from the distribution of a similar alternative test statistic, the cPLR (see methods section and supplementary note), which is well-behaved even when τ ≈ 1 and which majorises the distribution of PLR.
A natural next step is to search for the specific variants contributing to the PLR. An effective test statistic for testing subgroup differentiation for single SNPs is the Bayesian conditional false discovery rate (cFDR) [6, 7] applied to Zd scores ‘conditioned’ on Za scores. However, this statistic alone cannot capture all the means by which the joint distribution of (Za,Zd) can deviate from H0, and we also propose three other test statistics, each with different advantages, and compare their performance (supplementary note).
Power calculations, simulations, and validation of method
We tested our method by application to a range of datasets, using simulated and resampled GWAS data. First, to confirm appropriate control of type 1 error rates across H0, we simulated genotypes of case and control groups under H0 for a set of 5 × 105 autosomal SNPs in linkage equilibrium (supplementary note). Quantiles of the empirical PLR distribution were smaller than those for the empirical cPLR distribution and the asymptotic mixture-χ2, indicating that the test is conservative when τ > 1 (estimated type 1 error rate 0.048, 95% CI 0.039-0.059) and when τ ≈ 1 (estimated type 1 error rate 0.033, 95% CI 0.022-0.045) as expected; see figure 2. The distribution of cPLR closely approximated the asymptotic mixture-χ2 distribution across all values of τ (supplementary note).
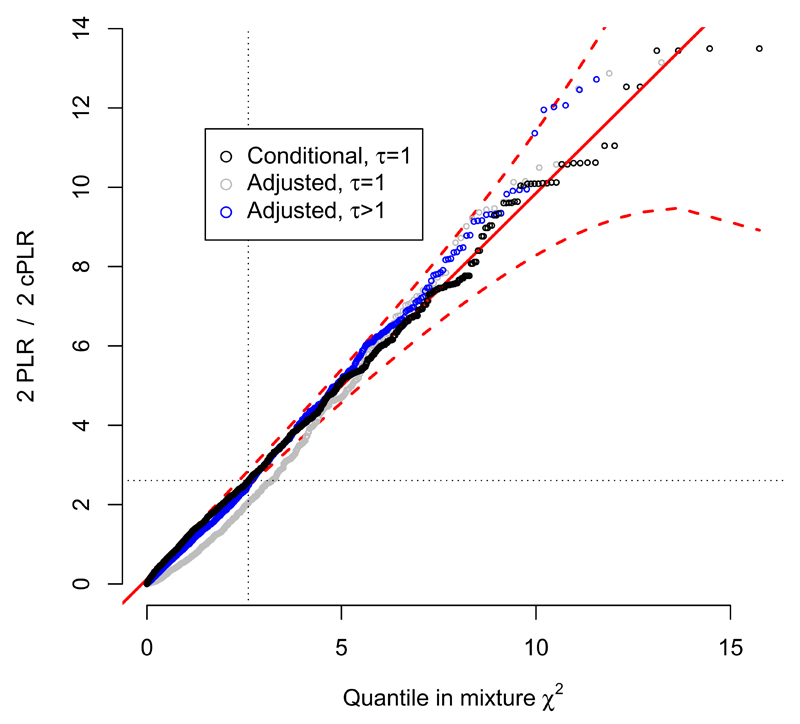
Figure 2.
QQ plot from simulations demonstrating type 1 error rate control of PLR test. PLR values for test subgroups under H0 with either τ = 1 (random subgroups; grey) or τ > 1 (genetic difference between subgroups, but independent of main phenotype; blue) with cPLR values for random subgroups (black) and against proposed asymptotic distribution under simulation solid red line; 99% confidence limits dashed red line). The distribution of cPLR for random subgroups majorises the distribution of PLR, meaning the PLR-based test is conservative. Further details are shown in the supplementary note.
We then established the suitability of the test when SNPs are in LD and when there exist genetic differences between subgroups that are independent of disease status overall. First, we used a dataset of controls and autoimmune thyroid disease (ATD) cases and repeatedly choose subgroups such that several SNPs had large allelic differences between subgroups. We found good FDR control at all cutoffs (supplementary note) and the overall type 1 error rate at α = 0.05 was 0.041 (95% CI 0.034-0.050). Second, we analysed a dataset of T1D cases with subgroups defined by geographical origin. Within the UK, there is clear genetic diversity associated with region [9]. As expected, Zd scores for geographic subgroups showed inflation compared to for random subgroups (supplementary figure 1). None of the derived test statistics reached significance at a Bonferroni-corrected p < 0.05 threshold (min. corrected p value > 0.8, supplementary figure 2).
To examine the power of our method, we used published GWAS data from the Wellcome Trust Case Control Consortium [10] comprising 1994 cases of Type 1 diabetes (T1D), 1903 cases of rheumatoid arthritis (RA), 1922 cases of type 2 diabetes (T2D) and 2953 common controls. We established that our test could differentiate between any pair of diseases, considered as subgroups of a general disease case group (all < 1 × 10-8, table 2).
Table 2.
Fitted parameter values for models of T1D/RA, T1D/T2D, T2D/RA, and GD/HT. H1 is the null hypothesis (under which σ3 = 1, ρ = 0) that SNPs differentiating the subgroups are not associated with the overall phenotype; H1 is the alternative (full model). p values for pseudo-likelihood ratio tests are also shown.
| π1 | π2 | π3 | σ2 | σ3 | τ | ρ | p-val | ||
|---|---|---|---|---|---|---|---|---|---|
| T1D/RA | H1 | 0.997 | 5.69 × 10-4 | 2.06 × 10-3 | 2.76 | 1.39 | 1.74 | 1.82 | 3.2 × 10-12 |
| H0 | 0.997 | 6.26 × 10-4 | 2.48 × 10-3 | 2.71 | - | 1.67 | - | ||
| T1D/T2D | H1 | 0.573 | 0.426 | 9.63 × 10-4 | 1.00 | 2.03 | 2.25 | 1.68 | 1.6 × 10-9 |
| H0 | 0.578 | 0.421 | 8.91 × 10-4 | 1.00 | - | 2.21 | - | ||
| T2D/RA | H1 | 0.573 | 0.426 | 8.71 × 10-4 | 1.00 | 2.23 | 1.75 | 1.69 | 5.1 × 10-9 |
| H0 | 0.910 | 8.05 × 10-4 | 0.0892 | 2.25 | - | 0.97 | - | ||
| GD/HT | H1 | 0.506 | 0.487 | 0.007 | 1.12 | 2.90 | 1.65 | 2.61 | 2.2 × 10-15 |
| H0 | 0.493 | 0.079 | 0.428 | 1.68 | - | 1.03 | - | ||
T1D and RA have overlap in genetic basis [10, 11, 7], as well as non-overlapping associated regions. T1D and T2D have less overlap [11] and T2D and RA less still. This was reflected in the fitted values (table 2, figure 3). The fitted values parametrizing category 2 in the full model for T1D/RA (π2, σ2) were consistent with a subset of SNPs associated with case/control status (T1D+RA vs control) but not differentiating T1D/RA. By contrast, the parametrization of category 2 for T1D/T2D and T2D/RA had marginal variance σ2 approximately 1, suggesting that a subset of SNPs associated with case/control status but not with ‘subgroup’ status did not exist in these cases. The rejection of H0 for the comparisons entails the existence of a set of SNPs associated both with case/control and subgroup status. The H0 model does not allow such a set of SNPs, forcing the parametrisation of Zd, Za scores for such SNPs to be ‘squashed’ into a category shape permitted under H0, with one marginal variance being 1: either category 2 (as happens in T2D/RA since π2|H0 ≈ π3|H1, σ2|H0 ≈ σ3|H1 in T2D/RA) or category 3 (as in T1D/T2D, where π3|H0 ≈ π3|H1, τ|H0 ≈ τ|H1).
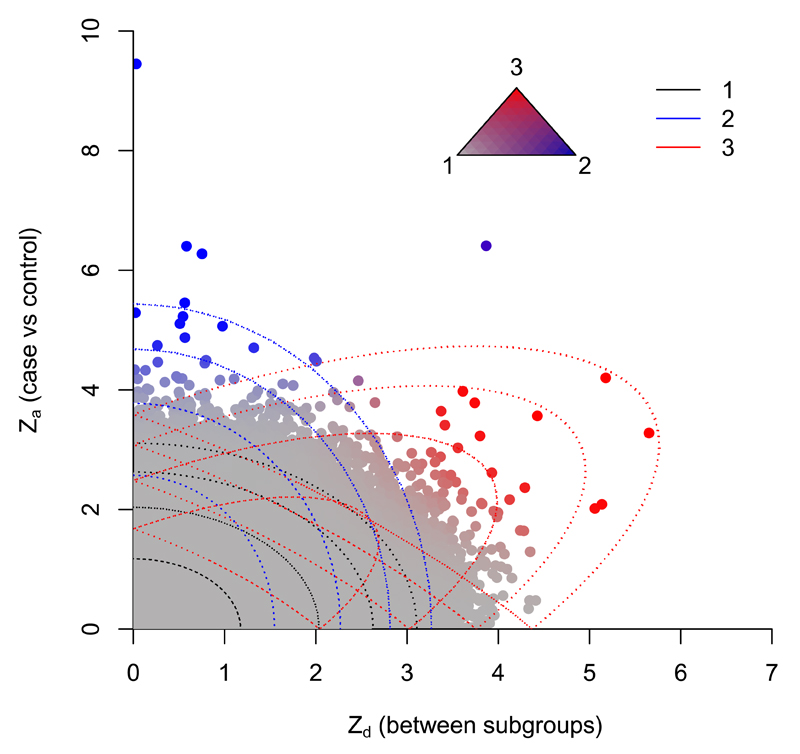
Figure 3.
Observed absolute Za and Zd for T1D/RA. Colourings correspond to posterior probability of category membership under full model (see triangle): grey - category 1, blue - category 2, red -category 3. Contours of the component Gaussians in the fitted full model are shown by dotted lines.
To determine the power of our test more generally, we showed that power depends on the number of SNPs in category 3 and on the underlying parameters of the true model, depending on the number of samples through the fitted model parameters (supplementary note). We therefore estimated the power of the test for varying numbers of SNPs in category 3 and for varying values of the parameters σ3, τ, and ρ. (figure 4; supplementary figure 3). As expected, power increases with an increasing number of SNPs in category 3, reflecting the proportion of SNPs which differentiate case subgroups and are associated with the phenotype as a whole. Power also increases with increasing τ, σ3, and absolute correlation (ρ/(σ3τ)) as high values enable better distinction of SNPs in the second and third categories.
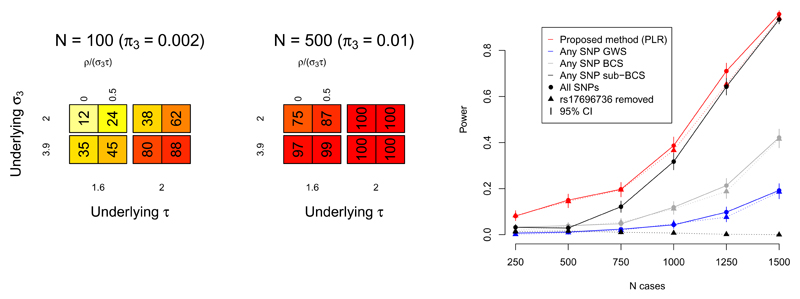
Figure 4.
Power of PLR to reject H0 (genetic homogeneity between subgroups) depends on the number of SNPs in category 3 and the underlying values of model parameters σ2, σ3, τ, ρ. Dependence on number of case/control samples arises through the magnitudes of σ3 and τ (supplementary note). Leftmost figure shows power estimates for various values of π3, σ3, τ, ρ. Value N is the approximate number of SNPs in category 3, (∝π3). Each simulation was on 5 × 104 simulated autosomal SNPs in linkage equilibrium. Value ρ/(σ3τ) is the absolute correlation between Za and Zd in category 3. Also see supplementary figure 3.
Rightmost figure shows power of PLR to detect differences in genetic basis of T1D and RA subgroups of a combined autoimmune dataset, downsampling to varying numbers of cases (X axis). PLR is compared with: power to find ≥ 1 SNP with Zd score reaching genome-wide significance (GWS, blue; p ≤ 5 × 10-8) or Bonferroni-corrected significance (BCS, green; p ≤ 0.05/(total # of SNPs)); and power to detect any SNP with Za score reaching genome-wide significance and Zd score reaching Bonferroni-corrected significance (sub-BCS, grey; p ≤ 0.05/(total # of SNPs with Za reaching GWS)). Error bars show 95% CIs. Circles/solid lines for each colour show power for all SNPs, triangles/dashed lines for all SNPs except rs17696736. Power for sub-BCS drops dramatically but power for PLR is not markedly affected, indicating relative robustness of PLR to single-SNP effects.
We explored the dependence of power on sample size by sub-sampling the WTCCC data for RA and T1D (figure 4) and compared the power of the PLR with the power to find any single SNP which differentiated the two diseases in several ways (see figure legend). Although the power of the PLR-based test was limited at reduced sample sizes, it remained consistently higher than the power to detect any single SNP which differentiated the two diseases. We then repeated the analysis removing the known T1D- and RA- associated SNP rs17696736. The power to detect a SNP with significant Zd score (Bonferroni-corrected) amongst SNPs with GW-significant Za score dropped dramatically, though the power of PLR was only slightly reduced. This illustrated the robustness of the PLR test to inclusion or removal of single SNPs with large effect sizes, a property not shared by single-SNP approaches.
Estimating power requires an estimate of the underlying values of several parameters: the expected total number of SNPs in the pruned dataset with different population MAF in case subgroups, and the distribution of odds-ratios such SNPs between subgroups and between cases/controls. With sparse genome-wide cover, such as that in the WTCCC study, > 1250 cases per subgroup are necessary for 90% power (discounting MHC region). If SNPs with greater coverage for the disease of interest are used (such as the ImmunoChip for autoimmune diseases) values of π3, σ3 and τ are correspondingly higher, and around 500-700 cases per subgroup may be sufficient.
Application to autoimmune thyroid disease and type 1 diabetes
Autoimmune thyroid disease (ATD) takes two major forms: Graves’ disease (GD; hyperthyroidism) and Hashimoto’s Thyroiditis (HT; hypothyroidism). Differential genetics of these conditions have been investigated. Detection of individual variants with different effect sizes in GD and HT is limited by sample size (particularly HT); however, the TSHR region shows evidence of differential effect [12]. T1D is relatively clinically homogenous with no major recognised subtypes, although heterogeneity arises between patients in levels of disease-associated autoantibodies, and disease course differs with age at diagnosis [3]. We analysed both of these diseases.
For ATD, we were able to confidently detect evidence for differential genetic bases for GD and HT (p = 2.2 × 10-15). Fitted values are shown in table 2. The distribution of cPLR statistics from random subgroups agreed well with the proposed mixture χ2 (supplementary figure 4b).
For T1D, we considered four subgroupings defined by plasma levels of the T1D-associated autoantibodies thyroid peroxidase antibody (TPO-Ab, n=5780), insulinoma-associated antigen 2 antibody (IA2-Ab, n=3197), glutamate decarboxylase antibody (GAD-Ab, n=3208) and gastric parietal cell antibodies (PCA-Ab, n=2240). A previous GWAS study on autoantibody positivity in T1D identified only two non-MHC loci at genome-wide significance: 1q23/FCRL3 with IA2-Ab and 9q34/ABO with PCA-Ab [3].
We tested each of the subgroupings retaining and excluding the MHC region. Fitted values for models with and without MHC are shown in supplementary table 2, and plots of Za and Zd scores are shown in supplementary figure 5. Retaining the MHC region, we were able to confidently reject H0 for subgroupings based on TPO-Ab, IA-2Ab and GAD-Ab (all p-values < 1.0 × 10-20). Although there was evidence that SNPs in the dataset were associated with PCA-Ab level (τ ≈ 2.5, null model), the improvement in fit in the full model was not significant, and we conclude that such SNPs determining PCA-Ab status are not in general T1D-associated. This can be seen by in the plot of Za against Zd (supplementary figure 5) where SNPs with high Zd values do not have higher than expected Za values.
With MHC removed, the subgrouping on TPO-Ab was significantly better-fit by the full model (p = 1.5 × 10-4). There was weaker evidence to reject H0 for GAD-Ab (p = 0.002) and IA2-Ab (p = 0.008) (Bonferroni-corrected threshold at α < 0.05: 0.006). Fitted values of τ in both the full and null models for GAD-Ab were ≈ 1, indicating absence of evidence for a category of non-MHC T1D-associated SNPs additionally associated with GAD-Ab positivity. Collectively, this indicates that differential genetic basis for T1D with GAD-Ab and IA2-Ab positivity is driven principally by the MHC region, and although PCA-Ab status is partially genetically determined, the set of causative variants is independent of T1D causative pathways.
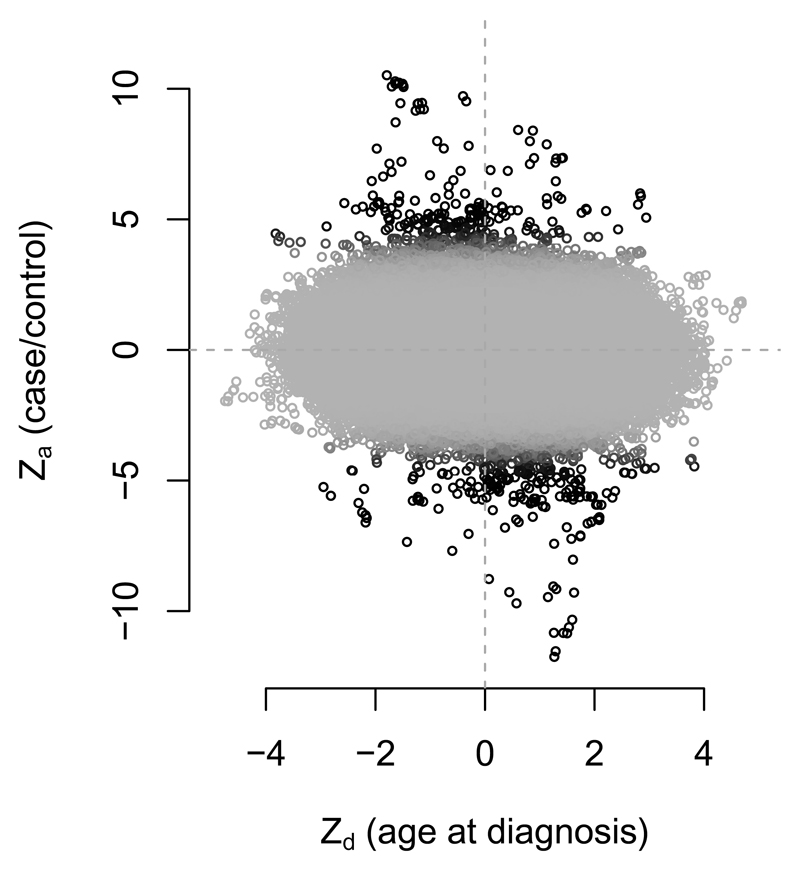
The variation in genetic architecture of T1D with age is not fully understood, but previous studies have suggested larger observed effects at known loci in patients diagnosed at a younger age [13, 14, 15, 16]. We investigated whether these differences were indicative of widespread differences in variant effect sizes with age-at-diagnosis, possibly due to differential heritability (see supplementary note). We applied the method to T1D dataset with Zd defined by age at diagnosis (quantitative trait). Fitted values are shown in supplementary table 3 and Za and Zd scores in supplementary figure 6. The hypothesis H0 could be rejected confidently when retaining or removing the MHC region (p values < 1.0 × 10-20 and 0.007 respectively). Signed Zd and Za scores for age at diagnosis showed a visible negative correlation (rg method 2; p = 0.002) amongst Zd and Za scores for disease-associated SNPs (figure 5). This is consistent with a higher genetic liability with lower age at diagnosis; the basis for differential genetic basis with age may be differing heritability with the same relative variant effect sizes.
Figure 5.
Za and Zd scores for age at diagnosis in T1D, excluding MHC region. Colour corresponds to posterior probability of category 2 membership in null model (since categories in full model are assigned on the basis of correlation), with black representing a high probability. Zd and Za are negatively correlated (p = 8.7 × 10-5 with MHC included, p = 0.002 with MHC removed) after accounting for LD using LDAK weights, and weighting by posterior probability of category 2 membership in the null model, to prioritise SNPs further from the origin
Assessment of individual SNPs
Many SNPs which discriminated subgroups were in known disease-associated regions (supplementary tables 4, 5, and 6). In several cases, our method identified disease-associated SNPs which have reached genome-wide significance in subsequent larger studies but for which the Za score in the WTCCC study was not near significance. For example, the SNP rs3811019, in the PTPN22 region, was identified as likely to discriminate T1D and T2D (p = 3.046 × 10-6; supplementary table 5), despite a p value of 3 × 10-4 for joint T1D/T2D association.
For GD and HT, SNPs near the known ATD-associated loci PTPN22 (rs7554023), CTLA4 (rs58716662), and CEP128 (rs55957493) were identified as likely to be contributing to the difference (see supplementary table 7). The SNPs rs34244025 and rs34775390 are not known to be ATD-associated, but are in known loci for inflammatory bowel disease and ankylosing spondylitis, and our data suggest they may differentiate GD and HT (FDR 0.003).
We searched for non-MHC SNPs with differential effect sizes with TPOA positivity in T1D, the subgrouping of T1D for which we could most confidently reject H0. Previous work [3] identified several loci potentially associated with TPO-Ab positivity by restricting attention to known T1D loci, enabling use of a larger dataset than was available to us. We list the top ten SNPs for each summary statistic for TPO-Ab positivity in supplementary table 8. Subgroup-differentiating SNPs included several near known T1D loci: CTLA4 (rs7596727), BACH2 (rs11755527), RASGRP1 (rs16967120) and UBASH3A (rs2839511) [17]. These loci agreed with those found by Plagnol et al [3], but our analysis used only available genotype data, without external information on confirmed T1D loci. We were not able to replicate the same p-values due to reduced sample numbers.
Finally, we analysed non-MHC SNPs with varying effect sizes with age at diagnosis in T1D (supplementary table 9). This implicated SNPs in or near CTLA4 (rs2352551), IL2RA (rs706781), and IKZF3 (rs11078927).
Discussion
The problem we address is part of a wider aim of adapting GWAS to complex disease phenotypes. As the body of GWAS data grows the analysis of between-disease similarity and within-disease heterogeneity has led to substantial insight into shared and distinct disease pathology [6, 7, 2, 20, 21]. We seek in this paper to use genomic data to infer whether such disease subtypes exist. Our problem is related to the question of whether two different diseases share any genetic basis [18] but differs in that the implicit null hypothesis relates to genetic homogeneity between subgroups rather than genetic independence of separate diseases.
Our test strictly assesses whether a set of SNPs have different effect sizes in case subgroups. We interpret this as ‘differential causative pathology’, which encompasses several disease mechanisms, discussed in the supplementary note. In some cases, if subgroups are defined on the basis of the presence or absence of a known disease risk factor, the heritability of the disease will differ between subgroups, with corresponding changes in variant effect sizes.
We use ‘absolute covariance’ ρ preferentially (see supplementary table 1) because we expect that Za and Zd will frequently co-vary positively and negatively at different SNPs in the same analysis; for instance, if some variants are deleterious only for subgroup 1 and others only for subgroup 2. A potential advantage of our symmetric model is the potential to generate Zd scores from ANOVA-style tests for genetic homogeneity between three or more subgroups, in which case reconstructed Z scores would be directionless.
Aetiologically and genetically heterogeneous subgroups within a case group correspond to substructures in the genotype matrix. Information about such substructures is lost in a standard GWAS, which only uses the column-sums (MAFs) of the matrix (linear-order information). Data-driven selection of appropriate case subgroups and corresponding analyses of these subgroups can use more of the remaining quadratic-order information the matrix contains. Indeed a ‘two-dimensional’ GWAS approach (using Za and Zd) instead of a standard GWAS (using only Za) may improve SNP discovery, as we found for PTPN22 in RA/T2D. However, this can only be the case if the subgroups correspond to different variant effect sizes; for other subgroupings, a two-dimensional GWAS will only add noise.
While it seems appealing to use this method to search for some ‘optimal’ partition of patients, we prefer to focus on testing subgroupings derived from independent clinical or phenotypic data. Firstly, it is difficult to characterise subgroupings as ‘better’ or ‘worse’, and no one parameter can parametrise the degree to which two subgroups differ; parameters π3, τ, and ρ all contribute, and attempts to test the hypothesis using a single measure such as genetic correlation have serious shortcomings (supplementary note). Secondly, even if subgroups could meaningfully be ranked, the search space of potential subgroupings of a case group is prohibitively large (2N for N cases), making exhaustive searches difficult.
We demonstrated that effect sizes of T1D-causative SNPs differ with age at disease diagnosis. The strong negative correlation observed (figure 5) was consistent with an increased total genetic liability in samples with earlier age of diagnosis, a finding supported by candidate gene studies [14, 15, 16] and epidemiological data [13]. Such a pattern arises naturally from a liability threshold model where total liability depends additively on both genetic effects and environmental influences which accumulate with age (supplementary note).
Our method necessarily dichotomises the multitude of mechanisms of heterogeneity, although there are many diverse forms (supplementary table 1, supplementary note). There is potential to further dissect the mechanisms of disease heterogeneity by incorporating estimations of genetic correlation [18] or assessing evidence for liability threshold models [22]. Similar mixture-Gaussian approaches may also be adaptable to this purpose, by assessing other families of effect size distributions.
Our method adds to the current body of knowledge by extracting additional information from a disease dataset over a standard GWAS analysis, and determines if further analysis of disease pathogenesis in subgroups is justified. Our approach is analogous to the intuitive method of searching for between-subgroup differences in SNPs with known disease associations [3] but does not restrict attention to strong disease associations, enabling use of information from disease-associated SNPs which do not reach significance. Our parametrisation of effect size distributions allows insight into the structure of the genetic basis of the disease and potential subtypes, improving understanding of genotype-phenotype relationships.
Methods
Ethics Statement
This paper re-analyses previously published datasets. All patient data were handled in accordance with the policies and procedures of the participating organisations.
Joint distribution of variables Za, Zd
We assume that SNPs may be divided into three categories, as described in the results section (figure 1). Under these assumptions, Za and Zd scores have the joint pdf given by equation 1. We define Θ is the vector of values (π1,π2,π3,τ,σ2,σ3,ρ). Z scores Za and Zd are reconstructed from GWAS p-values for SNP associations. In practice, since our model is symmetric, we only require absolute Z scores, without considering effect direction.
For sample sizes n1, n2 and 97.5% odds-ratio quantile α, the expected observed standard deviation of Z scores (that is, σ2, σ3, and τ) is given by
| (3) |
Definition and distribution of PLR statistics
For a set of observed Z scores Z = (Za, Zd) we define the joint unadjusted pseudo-likelihood PLda(Z|Θ) as
| (4) |
where the term C log(π1π2π3) is included to ensure identifiability of the model [5], and weights wi are included to adjust for LD (see below).
We now set
| (5) |
recalling that H0 is the subspace of the parameter space H1 satisfying σ3 = 1 and ρ = 0.
If data observations are independent, uPLR reduces to a likelihood ratio. Under H0, the asymptotic distribution of uPLR is then
| (6) |
according to Wilk’s theorem extended to the case where the null value of a parameter lies on the boundary of H1 (since ρ = 0 under H0) [23].
The empirical distribution of uPLR may substantially majorise the asymptotic distribution when τ ≈ 1. In the full model, the marginal distribution of Za has more degrees of freedom (four; π1, π2, σ2, σ3) than it does under the null model (two; π2, σ2, as σ3 ≡ 1). This can mean that certain distributions of Za can drive high values of uPLR independent of the values of Zd (supplementary note), which is unwanted as the values Za reflect only case/control association and carry no information about case subgroups. If observed uPLRs from random subgroups (for which τ = 1 by definition) are used to approximate the null uPLR distribution, this effect would lead to serious loss of power when τ >> 1.
This effect can be managed by subtracting a correcting factor based on the pseudo-likelihood of Za alone, which reflects the contribution of Za values to the uPLR. We define
| (7) |
that is, the marginal likelihood of Za. Given as defined above, we define
| (8) |
We now define the PLR as
| (9) |
The action of f(Za) leads to the asymptotic distribution of PLR slightly minorising the asymptotic mixture-χ2 distribution of uPLR, to differential degrees dependent on the value of τ (see supplementary note).
We define the similar test statistic cPLR:
| (10) |
noting that the expression
can be considered as a likelihood conditioned on the observed values of Za. Now
| (11) |
The empirical distribution of cPLR for random subgroups majorises the empirical distribution of PLR (supplementary note). Furthermore, the approximation of the empirical distribution of cPLR by its asymptotic distribution is good, across all values of τ; that is, across the whole null hypothesis space.
Our approach is to compare the PLR of a test subgroup to the cPLR of random subgroups, which constitutes a slightly conservative test under the null hypothesis (see supplementary note).
Allowance for linkage disequilibrium
The asymptotic approximation of the pseudo likelihood-ratio distribution breaks down when values of Za, Zd are correlated due to LD. One way to overcome this is to ‘prune’ SNPs by hiererarchical clustering until only those with negligible correlation remain. A disadvantage with this approach is that it is difficult to control which SNPs are retained in an unbiased way without risking removal of SNPs which contribute greatly to the difference between subgroups.
We opted to use the LDAK algorithm [4], which assigns weights to SNPs approximately corresponding to their ‘unique’ contribution. Denoting by ρij the correlation between SNPs i, j, and d(i,j) their chromosomal distance, the weights wi are computed so that
| (12) |
is close to constant for all i, and wi > 0 for all i. The motivation for this approach is that Σ i≠j ρij2 represents the replication of the signal of SNP i from all other SNPs.
This approach has the advantage that if n SNPs are in perfect LD, and not in LD with any other SNPs, each will be weighted 1/n, reducing the overall contribution to the likelihood to that of one SNP. In practice, the linear programming approach results in many SNP weights being 0. Using the LDAK algorithm therefore allows more SNPs to be retained and contribute to the model than would be retained in a pruning approach.
A second advantage of LDAK is that it homogenises the contribution of each genome region to the overall pseudo-likelihood. Many modern microarrays fine-map areas of the genome known or suspected to be associated with traits of interest [24] which could theoretically lead to peaks in the distribution of SNP effect sizes, disrupting the assumption of normality. LD pruning and LDAK both reduce this effect by homogenising the number of tags in each genomic region.
We adapted the pseudo-likelihood function to the weights by multiplying the contribution of each SNP to the log-likelihood by its weight (equation 4), essentially counting the ith SNP wi times over. Adjusting using LDAK was effective in enabling the distributions of PLR to be well-approximated by mixture-χ2 distributions of the form 2 (supplementary plots 4a, 4b, 4c).
E-M algorithm to estimate model parameters
We use an expectation-maximisation algorithm [25, 26] to fit maximum-PL parameters. Given an initial estimate of parameters Θ0 = (π10,π20,τ0,σ20,σ20,ρ0) we iterate three main steps:
-
Define for SNP s with Z scores Zd(s), Za(s)
(13) - For g ∈ (1,2,3), and LDAK weight ws for SNP s set
(14) - Set
(15)
Step 3 is complicated by the lack of closed form expression for the maximum likelihood estimator of ρ (because of the symmetric two-Gaussian distribution of category 3), requiring a bisection method for computation. The algorithm is continued until |PLR(Zd,Za|Θi) - PLR(Zd,Za|Θi-1)| < ϵ; we use ϵ = 1 × 10-5.
The algorithm can converge to local rather than global minima of the likelihood. We overcome this by initially computing the pseudo-likelihood of the data at 1000 points throughout the parameter space, retaining the top 100, and dividing these into 5 maximally-separated clusters. The full algorithm is then run on the best (highest-PL) point in each cluster.
An appropriate choice of Θ0 can speed up the algorithm considerably; for simulations, we begin the model at previous maximum-PL estimates of parameters for earlier simulations.
Maximum-cPL estimations of parameters were made using generic numerical optimisation with the optim function in R. Prior to applying the algorithm, parameters π2 and σ2 are estimated as maximum-PL estimators of the objective function
| (16) |
where wi is the weight for SNP i (see supplementary note for rationale). The conditional pseudo-likelihood was maximised over the remaining parameters.
The algorithm and other processing functions are implemented in an R package available at https://github.com/jamesliley/subtest
Properties and assumptions of the PLR test
Our assumption that (Za,Zd) follows a mixture Gaussian is generally reasonable for complex phenotypes with a large number of associated variants [8] and our adjustment for the distribution of Za (essentially conditioning on observed Za) reduces reliance on this assumption. If subgroup prevalence is unequal between the study group and population, our method can still be used with adaptation (supplementary note).
Our test is robust to confounders arising from differential sampling to the same extent as conventional GWAS. For example, if subgroups were defined based on population structure, and population structure also varied between the case and control group, SNPs which differed by ancestry would also appear associated with the disease, leading to a loss of control of type-1 error rate. However, the same study design would also lead to identification of spurious association of ancestry-associated SNPs with the phenotype in a conventional GWAS analysis. As for GWAS, this effect can be alleviated by including the confounding trait as a covariate when computing p-values (supplementary note).
Prioritisation of single SNPs
An important secondary problem to testing H0 is the determination of which SNPs are likely to be associated with disease heterogeneity. Ideally, we seek a way to test the association of a SNP with subgroup status (ie, Zd), which gives greater priority to SNPs potentially associated with case/control status (ie, high Za).
An effective test statistic meeting these requirements is the Bayesian conditional false discovery rate (cFDR) [6]. It tests against the null hypothesis H'0 that the population minor allele frequencies of the SNP in both case subgroups are equal (ie, that the SNP does not differentiate subgroups), but responds to association with case/control status in a natural way by relaxing the effective significance threshold on |Zd|. This relaxation of threshold only occurs if there is systematic evidence that high |Zd| scores and high |Za| scores typically co-occur. The test statistic is direction-independent.
Given a set of observed Za and Zd values Za(i), Zd(i), with corresponding two-sided p values pai, pdi, the cFDR for SNP j is defined as
| (17) |
The value gives the false-discovery rate for SNPs whose p-values fall in the region [0,pdj] × [0,paj]; this can be converted into a false-discovery rate amongst all SNPs for whom X4 passes some threshold [7].
We discuss three other single-SNP test statistics in the supplementary note, which test against different null hypotheses. If the hypothesis H0' is to be tested, then we consider the cFDR the best of these.
Contour plots of the test statistics for several datasets are shown in supplementary figures 7, 8, and 9.
Genetic correlation testing
Given the correlation between Zd and Za in the age-at-diagnosis analysis, methods to estimate narrow-sense genetic correlation (rg) [18, 19] may be adaptable to the subgrouping question by estimating rg across a set of SNPs between case/control traits of interest, with the potential advantage of characterising heterogeneity using a single widely-interpretable metric. This may be between Z scores derived from comparing the control group to each case subgroup, testing under the null hypothesis rg = 1 (method 1); or between the familiar Za and Zd, under the null hypothesis rg = 0 (method 2).
We explored these methods in supplementary note. We show that method 1 leads to systematically high false positive rates, as rg is also reduced from 1 in subgroupings that are independent of the overall disease process (e.g. hair colour in T2D). We show that method 2 is considerably less powerful than our method because it tests a narrower definition of H1 which does not take account of the marginal variances of the distribution of Zd, Za in category 3, and requires that correlation between Zd and Za be always positive or always negative, in contrast to our symmetric model (figure 1). Indeed, parameter ρ estimates an analogue of rg accounting for simultaneous correlation and anticorrelation.
Methods to compute rg were not explicitly proposed as a method for subgroup testing, and our analysis does not indicate any general shortcomings. However, comparison with rg based approaches places our method in the context of established methodology, demonstrating the necessity of considering both variance parameters (τ, σ3) and covariance parameters (ρ) in testing a subgrouping of interest.
Description of GWAS datasets
ATD samples were genotyped on the ImmunoChip [24] a custom array targeting putative autoimmune-associated regions. Data were collected for GWAS-like analyses of dense SNP data [12]. The dataset comprised 2282 cases of Graves’ disease, 451 cases of Hashimoto’s thyroiditis, and 9365 controls.
T1D samples were genotyped on either the Illumina 550K or Affymetrix 500K platforms, gathered for a GWAS on T1D [17]. We imputed between platforms in the same way as the original GWAS. The dataset comprised genotypes from 5908 T1D cases and 8825 controls, of which all had measured values of TPO-Ab, 3197 had measured IA2-Ab, 3208 had measured GAD-Ab, and 2240 had measured PCA-Ab. Comparisons for each autoantibody were made between cases positive for that autoantibody, and cases not positive for it. We did not attempt to perform comparisons of individuals positive for different autoantibodies (for instance, TPO-Ab positive vs IA2-Ab positive) because many individuals were positive for both.
To generate summary statistics corresponding to geographic subgroups, we considered the subgroup of cases from each of twelve regions and each pair of regions against all other cases (78 subgroupings in total). To maximise sample sizes, we considered T1D cases as ‘controls’ and split the control group into subgroups.
Quality control
Particular care had to be taken with quality control, as Z-scores had to be relatively reliable for all SNPs assessed, rather than just those putatively reaching genome-wide significance. For the T1D/T2D/RA comparison, which we re-used from the WTCCC, a critical part of the original quality control procedure was visual analysis of cluster plots for SNPs reaching significance, and systematic quality control measures based on differential call rates and deviance from Hardy-Weinberg equilibrium (HWE) were correspondingly loose [10]. Given that we were not searching for individual SNPs, this was clearly not appropriate for our method.
We retained the original call rate (CR) and MAF thresholds (MAF ≥ 1%, CR ≥ 95% if MAF ≥ 5%, CR ≥ 99% if MAF <5%) but employed a stricter control on Hardy-Weinberg equilibrium, requiring p ≥ 1 × 10-5 for deviation from HWE in controls. We also required that deviance from HWE in cases satisfied p ≥ 1.91 × 10-7, corresponding to |z|≤ 5. The looser threshold for HWE in cases was chosen because deviance from HWE can arise due to true SNP effects [27]. We also required that call rate difference not be significant (p ≥ 1 × 10-5) between any two groups, included case-case and case-control differences. Geographic data was collected by the WTCCC and consisted of assignment of samples to one of twelve geographic regions (Scotland, Northern, Northwestern, East and West Ridings, North Midlands, Midlands, Wales, Eastern, Southern, Southeastern, and London [10]). In analysing differences between autoimmune diseases, we stratified by geographic location; when assessing subgroups based on geographic location, we did not.
For the ATD and T1D data, we used identical quality control procedures to those employed in the original paper [12, 17]. We applied genomic control [28] to computation of Za and Zd scores except for our analysis of ATD (following the original authors [12]) and our geographic analyses (as discussed above). In all analyses except where otherwise indicated we removed the MHC region with a wide margin (≈ 5Mb either side).
Supplementary Material
Acknowledgments
We acknowledge the help of the Diabetes and Inflammation Laboratory Data Service for access and quality control procedures on the datasets used in this study. The JDRF/Wellcome Trust Diabetes and Inflammation Laboratory is in receipt of a Wellcome Trust Strategic Award (107212, JAT) and receives funding from the JDRF (5-SRA-2015-130-A-N, JAT) and the NIHR Cambridge Biomedical Research Centre. The research leading to these results has received funding from the European Union’s 7th Framework Programme (FP7/2007-2013, JAT) under grant agreement no. 241447 (NAIMIT). JL is funded by the NIHR Cambridge Biomedical Research Centre and is on the Wellcome Trust PhD programme in Mathematical Genomics and Medicine at the University of Cambridge. CW is funded by the Wellcome Trust (089989,107881, CW) and the MRC (MC_UP_1302/5, CW). The Cambridge Institute for Medical Research (CIMR) is in receipt of a Wellcome Trust Strategic Award (100140). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflicts of Interest
The JDRF/Wellcome Trust Diabetes and Inflammation Laboratory receives funding from Hoffmann La Roche and Eli-Lilly and Company.
Author contributions
AJL: conceived the statistical methods, wrote the software, performed the analyses, analysed the data, and wrote the manuscript.
JAT: analysed the data and edited the manuscript.
CW: conceived the study, analysed the data, and wrote the manuscript
Code availability
Code available from https://github.com/jamesliley/subtest (R package)
Data availability
This paper re-analyses previously published datsets. WTCCC data access for T1D/T2D/RA and controls [10] is described at https://www.wtccc.org.uk/info/access_to_data_samples.html. ATD data are available on request to the original study authors [12]. T1D genetic data from [17] is available at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000180.v3.p2 which we combined with autoantibody data available from study authors [3].
References
- [1].Li L, Cheng WY, Glicksberg BS, Gottesman O, Tamler R, et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Science translational medicine. 2015;7:311ra174–311ra174. doi: 10.1126/scitranslmed.aaa9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Morris AP, Lindgren CM, Zeggini E, Timpson NJ, Frayling TM, et al. A powerful approach to sub-phenotype analysis in population-based genetic association studies. Genetic Epidemiology. 2009;34:335–343. doi: 10.1002/gepi.20486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Plagnol V, Howson JMM, Smyth DJ, Walker N, Hafler JP, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLOS Genetics. 2011;7 doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. American Journal of Human Genetics. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen H, Chen J, Kalbfleisch JD. A modified likelihood ratio test for homogeneity in finite mixture models. Journal of the Royal Statistical Society, series B (methodological) 2001;63:19–29. [Google Scholar]
- [6].Andreassen OA, Thompson WK, Schork AJ, Ripke S, Mattingsdal M, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLOS Genetics. 2013;9(4) doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liley J, Wallace C. A pleiotropy-informed Bayesian false discovery rate adapted to a shared control design finds new disease associations from gwas summary statistics. PLOS Genetics. 2015 doi: 10.1371/journal.pgen.1004926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lo PR, Tucker G, Bulik-Sullivan BK, Vilhjalmsson BJ, Finucane HK, et al. Efficient Bayesian mixed model analysis increases association power in large cohorts. Nature Genetics. 2015;47:284–90. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Leslie S, Winney B, Hellenthal G, Davison D, Boumertit A, et al. The fine-scale genetic structure of the British population. Nature. 2015;519:309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].The Wellcome trust case control consortium. Genome-wide association study of 14000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fortune MD, Guo H, Burren O, Schofield E, Walker NM, et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nature Genetics. 2015;47:839–846. doi: 10.1038/ng.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cooper JD, Simmonds MJ, Walker NM, Burren O, Brand OJ, et al. Seven newly identified loci for autoimmune thyroid disease. Human Molecular Genetics. 2012;21:5202–5208. doi: 10.1093/hmg/dds357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hyttinen V, Kaprio J, Kinnunen L, Koskenvuo M, Tuomilehto J. Genetic liability of type 1 diabetes and the onset age among 22, 650 young Finnish twin pairs in a nationwide follow up study. Diabetes. 2003;52:1052–1055. doi: 10.2337/diabetes.52.4.1052. [DOI] [PubMed] [Google Scholar]
- [14].Howson JMM, Walker NM, Smyth DJ, Todd JA. Analysis of 19 genes for association with type 1 diabetes in the type 1 diabetes genetics consortium families. Genes and Immunity. 2009;10:S74–S84. doi: 10.1038/gene.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Howson JM, Rosinger S, Smyth DJ, Boehm BO, Todd JA, et al. Genetic analysis of adult-onset autoimmune diabetes. Diabetes. 2011;60:2645–2653. doi: 10.2337/db11-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Howson JM, Cooper JD, Smyth DJ, Walker NM, Stevens H, et al. Evidence of gene-gene interaction and age-at-diagnosis effects in type 1 diabetes. Diabetes. 2012;61:3012–3017. doi: 10.2337/db11-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nature genetics. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, et al. An atlas of genetic correlations across human diseases and traits. bioRxiv. 2015 doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Traylor M, Bevan S, Rothwell PM, Sudlow C, 2 WTCCC et al. Using phenotypic heterogeneity to increase the power of genome-wide association studies: Application to age at onset of ischaemic stroke subphenotypes. Genetic Epidemiology. 2013;37:495–503. doi: 10.1002/gepi.21729. [DOI] [PubMed] [Google Scholar]
- [21].Wen Y, Lu Q. A multiclass likelihood ratio approach for genetic risk prediction allowing for phenotypic heterogeneity. Genetic epidemiology. 2013;37:715–725. doi: 10.1002/gepi.21751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chatterjee N, Carroll RJ. Semiparametric maximum likelihood estimation exploiting gene-environment independence in case-control studies. Biometrika. 2005;92:399–418. [Google Scholar]
- [23].Self SG, Liang KY. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. Journal of the American Statistical Association. 1987;82:605–610. [Google Scholar]
- [24].Cortes A, Brown MA. Promise and pitfalls of the ImmunoChip. Arthritis Research and Therapy. 2011;13 doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the em algorithm. Journal of the Royal Statistical Society, series B (methodological) 1977;39:1–38. [Google Scholar]
- [26].Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. Springer Series in Statistics. Springer; 2001. [Google Scholar]
- [27].Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, et al. Data quality control in genetic case-control association studies. Nature protocols. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theoretical Population Biology. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.