Abstract
The evolution of pathways within plant secondary metabolism has been studied by using the pyrrolizidine alkaloids (PAs) as a model system. PAs are constitutively produced by plants as a defense against herbivores. The occurrence of PAs is restricted to certain unrelated families within the angiosperms. Homospermidine synthase (HSS), the first specific enzyme in the biosynthesis of the necine base moiety of PAs, was originally recruited from deoxyhypusine synthase, an enzyme involved in the posttranslational activation of the eukaryotic initiation factor 5A. Recently, this gene recruitment has been shown to have occurred several times independently within the angiosperms and even twice within the Asteraceae. Here, we demonstrate that, within these two PA-producing tribes of the Asteraceae, namely Senecioneae and Eupatorieae, HSS is expressed differently despite catalyzing the same step in PA biosynthesis. Within Eupatorium cannabinum, HSS is expressed uniformly in all cells of the root cortex parenchyma, but not within the endodermis and exodermis. Within Senecio vernalis, HSS expression has been previously identified in groups of specialized cells of the endodermis and the adjacent root cortex parenchyma. This expression pattern was confirmed for Senecio jacobaea as well. Furthermore, the expression of HSS in E. cannabinum is dependent on the development of the plant, suggesting a close linkage to plant growth.
Pyrrolizidine alkaloids (PAs) provide a suitable system for studying aspects of pathway evolution. They are a typical group of plant secondary compounds and are constitutively produced by various plant species as a defense against herbivores (Hartmann and Ober, 2000). PAs are part of a complex system of chemical ecological interactions between the plant and insect herbivores. Some adapted herbivores have even developed specific mechanisms to use these plant-derived compounds for their own defense against predators (Ober, 2003; Hartmann, 2004).
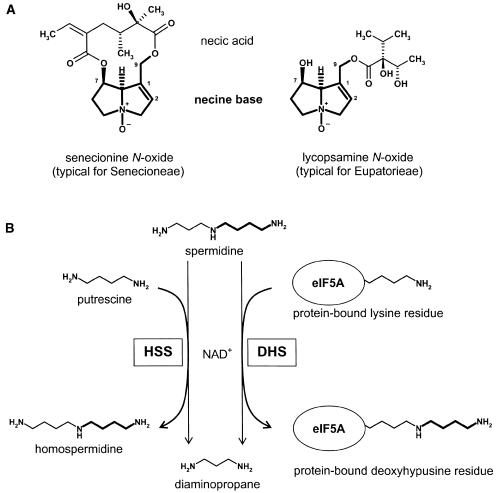
PAs are ester alkaloids composed of a necine base, the characteristic bicyclic structure that is common to all PAs, and one or more necic acids. In most plants, they occur in their N-oxide form (Fig. 1A). Homospermidine synthase (HSS) is the first pathway-specific enzyme in PA biosynthesis. HSS transfers the aminobutyl moiety of spermidine in an NAD+-dependent reaction to putrescine, resulting in homospermidine (Fig. 1B). Homospermidine formed by HSS is exclusively incorporated into the necine base of PAs (for review, see Hartmann and Ober, 2000; Ober and Hartmann, 2000). Recently, we have been able to trace back the evolutionary origin of HSS to deoxyhypusine synthase (DHS), an enzyme of primary metabolism involved in the posttranslational modification of the eukaryotic initiation factor 5A (eIF5A; Ober and Hartmann, 1999a). Although both enzymes are part of different metabolic processes (i.e. DHS modifies a regulatory protein and HSS catalyzes the first step in the formation of low-molecular weight defense compounds), both enzymes share the same reaction mechanism (Ober and Hartmann, 2000). Like HSS, DHS catalyzes the transfer of the aminobutyl moiety of spermidine but uses, as an acceptor substrate, a specific protein-bound Lys residue of the eIF5A precursor protein (Fig. 1B). Heterologous expression of HSS and DHS of Senecio vernalis has revealed that DHS is also able to accept putrescine as a second substrate to form homospermidine, whereas HSS is unable to bind the eIF5A substrate (Ober and Hartmann, 1999b). It is unknown whether the ability of DHS to form homospermidine is of any relevance in vivo, but this side activity is held responsible for the universal occurrence of traces of homospermidine in plants (Ober et al., 2003a). The HSS of S. vernalis can thus be regarded as a DHS that has lost its ability to bind the eIF5A precursor protein without any substantial modifications of enzyme kinetics (Ober et al., 2003b). Although activated eIF5A is essential for eukaryotes, its mode of action is not well understood. The hypusination of eIF5A is highly conserved within eukaryotes and archaea, indicating an essential role in cell life. Activated eIF5A appears to be essential for cell proliferation. Intervention at any of the four major steps in its biogenesis, i.e. synthesis of its substrate spermidine, expression of eIF5A precursor protein, deoxyhypusine synthesis, and deoxyhypusine hydroxylation, results in the growth arrest of eukaryotic cells (Kang and Hershey, 1994; Park et al., 1997; Caraglia et al., 2001, 2003). eIF5A appears not to be required for global protein biosynthesis, but may act as a nucleocytoplasmic shuttle protein, translocating specific mRNAs with relevance for the cell cycle progress (Rosorius et al., 1999; Xu and Chen, 2001). In plants, eIF5A obviously plays a central role in growth and development. The transcripts of plant DHS and eIF5A are up-regulated during senescence in response to stress (Wang et al., 2001, 2003; Chou et al., 2004), and tobacco (Nicotiana tabacum) plants transformed with the putative dhs promoter in fusion with the gus gene exhibit high levels of dhs expression during germination and seedling development (Moll et al., 2002). The different isoforms of eIF5A identified within many plant genomes are probably elements of a biological switch that mediates the pleiotropic effects of activated eIF5A (Wang et al., 2003; Thompson et al., 2004).
Figure 1.
Structural types of PAs within the Asteraceae and the reactions catalyzed by HSS and DHS. A, Senecionine N-oxide and lycopsamine N-oxide as typical structures of PAs found within the Senecioneae and Eupatorieae, respectively. The necine base moiety is a bicyclic structure that is common to all PAs. B, HSS and DHS catalyze the transfer of an aminobutyl moiety of spermidine to putrescine (HSS) or to a specific protein-bound Lys residue (DHS).
The occurrence of PAs is restricted to the angiosperms. More than 95% of the more than 400 known structures are found within the Asteraceae (the tribes Senecioneae and Eupatorieae), some genera of the Boraginaceae and Apocynaceae, within the Fabaceae (mainly in the genus Crotalaria), and in some genera of the Orchidaceae. Isolated occurrences within a few species have also been described within the Celastraceae, Convolvulaceae, Ranunculaceae, Rhizophoraceae, Santalaceae, and Sapotaceae (Hartmann and Witte, 1995). A scattered occurrence within the different angiosperm families seems to be a common feature of the major alkaloid classes (Facchini et al., 2004). For virtually all of them, it remains an open question whether the ability to produce a special class of alkaloids indicates a common ancestor early in angiosperm evolution followed by several independent losses in many lineages, or whether this ability arose several times independently. Considering that each alkaloid biosynthesis implies the recruitment not only of the structural genes but also of the regulatory elements that orchestrate the expression of all relevant genes makes it easier to imagine the loss of an ancestral pathway as a result of gene mutations rather than their independent origins (Facchini et al., 2004). However, the HSS-coding gene of PA biosynthesis has been shown to be recruited independently from the DHS-coding gene in at least four lineages within the angiosperms. Two of them concern the Eupatorieae and the Senecioneae, the two tribes within the Asteraceae family that are able to produce PAs (Reimann et al., 2004). This independent origin of HSS is supported by phylogenetic analyses of the Asteraceae family based on morphological and chemical data (Karis, 1993), and on chloroplast and ribosomal data (Kim and Jansen, 1995; Bayer and Starr, 1998; Goertzen et al., 2003) showing that the two PA-producing tribes are not sister groups. However, the origin of the regulatory elements remains enigmatic in all these cases.
In this study, we have compared the tissue- and cell-specific expression of HSS in Eupatorium cannabinum belonging to the Eupatorieae with the HSS of the Senecioneae, namely of Senecio jacobaea and S. vernalis. In all three species of the Asteraceae family, HSS catalyzes the first pathway-specific step in PA biosynthesis in the roots. Despite this functional identity in both tribes, different cell types are involved in HSS expression. These data further support the independent recruitment of the HSS-coding gene within these two tribes and also imply the recruitment of individual regulatory elements.
RESULTS
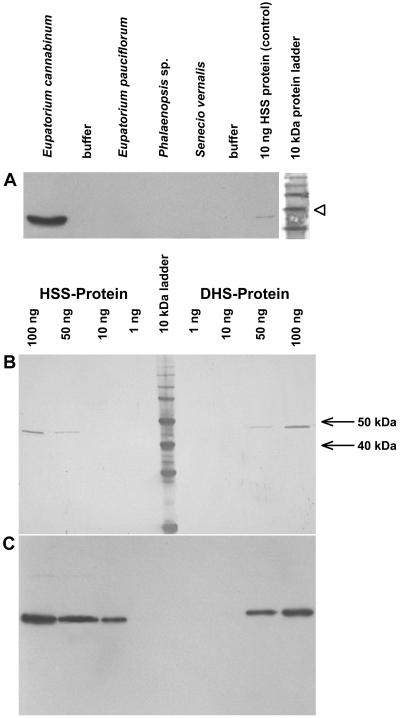
Antibody Preparation and Specificity
To purify the polyclonal antibody serum, HSS of E. cannabinum was expressed heterologously in Escherichia coli, purified by using a His tag, and immobilized for affinity chromatography purification. To test the specificity of the resulting HSS-specific antibody, extracts of PA-producing root cultures of related Asteraceae species and an extract of the aerial root tips of the Orchidaceae Phalaenopsis sp. that were previously characterized as a site of PA biosynthesis (Böttcher et al., 1993; Hartmann, 1994; N. Nurhayati, S. Anke, C. Frölich, and D. Ober, unpublished data) were separated and detected by immunoblot analysis (Fig. 2A). The affinity-purified antibody only detects the HSS of E. cannabinum root cultures. It is inefficient with HSS from other sources, although the various HSS proteins share significant amino acid sequence identities (80% between HSS of E. cannabinum and HSS of S. vernalis; 72% between HSS of E. cannabinum and Phalaenopsis sp.; calculated according to the data of Reimann et al. [2004] ). Because HSS and DHS of E. cannabinum share a high degree of amino acid sequence identity (83%), we tested the cross-reactivity of the affinity-purified HSS antibody against DHS in another immunoblot. We prepared two SDS-PAGE gels, each with identical amounts of purified recombinant HSS and DHS of E. cannabinum. Both gels were blotted before one of them was stained with Indian ink and the other developed as an immunoblot. Figure 2, B and C, shows that the HSS-specific antibody also binds the DHS protein, but with an approximately 10-fold lower intensity.
Figure 2.
Specificity of the affinity-purified antibody against the HSS of E. cannabinum. A, Immunoblot of proteins extracted from PA-biosynthesizing tissues of various plant species with the antibody specific for HSS of E. cannabinum. Twenty-microgram aliquots of each extract were applied to the gel, and the protein size marker was stained separately with Indian ink. The 50-kD band is indicated by an arrowhead. B, Membranes with identical amounts of pure HSS and DHS after protein staining with Indian ink. C, Membrane identical to B after immunoblot detection with the affinity-purified antibody against the HSS of E. cannabinum.
hss Gene Expression in E. cannabinum Is Restricted to Young Roots
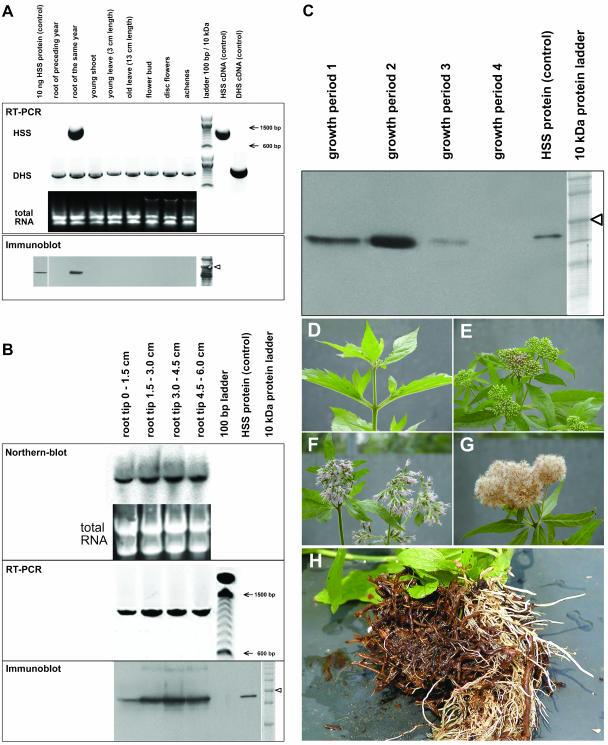
Various tissues were collected from an E. cannabinum plant just before flowering. The tissues included previous year roots (brown roots), newly grown roots of the same vegetation period (white roots), leaves, stems, and flower buds. Open-disc flowers and fully developed achenes were collected from the same plant several weeks later. Total RNA was extracted from all tissues and applied to reverse transcription (RT)-PCR. To ensure comparability of the resulting band intensities, spectrophotometric quantitation of RNA was confirmed by applying equal amounts of total RNA to an agarose gel. For amplification, primers specific for HSS and DHS, respectively, were used (Fig. 3A). mRNA coding HSS was only detected in young white roots that had emerged within the same vegetation period; roots of the preceding vegetation period (characterized by their brown color) and all other analyzed tissues of the plant did not express HSS. DHS-coding mRNA was detectable in all analyzed tissues with a comparable intensity. In a comparison of the band intensities of the HSS- and DHS-positive controls (each with 50 pg purified plasmid DNA as template) that were coamplified with the tissue samples, HSS was obviously expressed at a much higher rate within the young roots than DHS in the same or any other tissue. An immunoblot of protein extracts of exactly the same tissues (Fig. 3A) and an RNA gel-blot analysis (data not shown) confirmed that the HSS protein and the respective mRNA were found exclusively in the young roots of the same year. To test whether HSS expression was restricted to certain developmental phases of the young root, root sections defined by their distance from the root tip were analyzed separately. Figure 3B shows the results of an RNA gel-blot analysis and semiquantitative RT-PCR and immunoblot analysis. All three methods confirmed the expression of HSS at the mRNA and protein levels in all analyzed root segments. Only within the first segment (0–1.5 cm from the root tip) was expression of HSS slightly weaker than within the basipetal following segments, most probably because of incomplete tissue differentiation in the root tip.
Figure 3.
Expression analyses of HSS and DHS in E. cannabinum in various tissues (A), in segments of the root tip (B), and at different seasonal stages (C–G). A, For RT-PCR, cDNAs resulting from reverse transcription of identical amounts of total RNA from various tissues were used as templates for PCR with primers specific for HSS and DHS, respectively. As a control, 50 pg of plasmid DNA carrying the respective cDNAs were coamplified to ensure specificity of the primers used. A 100-bp DNA size marker is shown. For the immunoblot, 20 μg of soluble protein extracted from the same tissues as used for RT-PCR were applied. The 50-kD band of the 10-kD protein size marker is indicated by an arrowhead. B, For northern-blot analysis, total RNA was isolated from 15-mm consecutive segments of the root tip. Ten micrograms of each segment were used as loading control and for hybridization with a probe specific for HSS-coding cDNA. RT-PCR was performed with cDNA from each segment with primers specific for HSS. For immunoblot, proteins were extracted from each segment and detected with the affinity-purified antibody against the HSS of E. cannabinum. The 50-kD band of the 10-kD protein size marker is indicated by an arrowhead. C, Immunoblot of proteins extracted from E. cannabinum roots at different seasonal states of the plant. The 50-kD band of the protein size marker is indicated by an arrowhead. D to G illustrate the growth periods 1 to 4, respectively, as given in C. H, Roots of field-grown E. cannabinum plants with older brown roots and newly grown white roots.
HSS Expression Is Shut Down Depending on the Flowering Stage of the Plant
To test whether HSS expression is dependent on the seasonal state of the E. cannabinum plant, young roots from individual plants from the following stages (termed growth periods 1–4, respectively) were harvested and stored at −80°C (1) before inflorescences emerged, (2) before the flower buds within the inflorescences opened, (3) with all flowers blooming, and (4) with achenes. Protein extracts were used for immunoblot analysis. HSS expression increased over the growth period of E. cannabinum plants, peaked in growth period 2, was switched off when the flower buds opened (growth period 3), and was no longer detectable in growth period 4 when fruits were produced (Fig. 3C).
Immunolocalization Identifies the Root Cortex Parenchyma as the Exclusive Site of HSS Expression in E. cannabinum
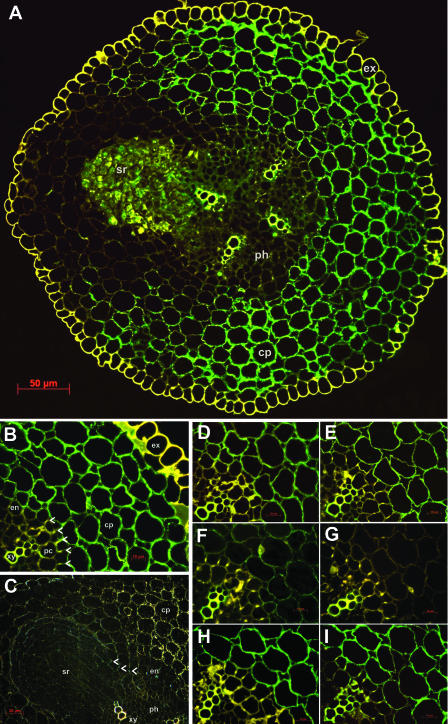
Using the protocol of Moll et al. (2002), we observed that the white roots became brown during fixation, probably because of the oxidation of phenolic compounds within the sample. To avoid any interference of these phenolics with the antigenicity of the HSS protein, we modified the fixative. After addition of Triton X-100 and soluble polyvinylpyrrolidone to facilitate tissue infiltration and to bind the phenolics, respectively, the roots remained white during fixation. Cross-sections of the young roots of E. cannabinum were labeled with the affinity-purified polyclonal antibody raised against the recombinant HSS. Figure 4A shows such a cross-section after detection with a fluoresceinisothiocyanate (FITC)-labeled secondary antibody by fluorescence microscopy. In the tetrarch vascular bundle, the four xylem rays exhibit yellow autofluorescence, as do the exodermis as the outermost cell layer of the root and the casparian strip in the radial cell walls of the endodermis (Fig. 4B, marked with arrowheads). From the pericycle above one of the xylem rays, a lateral root is just emerging, penetrating the cortex parenchyma. The green immunofluorescence of the FITC label shows the localization of HSS within the whole cortex parenchyma, with the exception of those cell layers that surround the emerging lateral root. The exodermis as the outermost and the endodermis as the innermost cell layers of the root cortex do not show any label (Fig. 4B). The same labeling pattern has been observed by using silver enhancement with a gold-labeled secondary antibody. Figure 4C shows the emerging lateral root in detail. In the dark field, the gold-labeled antibody appears yellow on a black background, whereas the casparian strip of the pierced endodermis appears in blue. Again, those layers of the cortex parenchyma that surround the emerging root and the lateral root itself are devoid of any label.
Figure 4.
Immunolabeling of HSS in root sections of E. cannabinum. A, FITC labeling of HSS within the cortex parenchyma (cp; green). The four xylem rays in the central cylinder and the exodermis (ex) show yellow autofluorescence. A lateral root (sr) is emerging from the pericycle above the xylem ray in the upper left part of the section. B, FITC labeling of HSS in a cutout of A. HSS expression is restricted to cells of the cortex parenchyma (cp); cells of the endodermis (en) and of the exodermis (ex) do not show any label. The casparian strip (arrowheads), the xylem (xy), and the exodermis (ex) show yellow autofluorescence. pc, Pericycle. C, Immunogold detection of the cross-section with the emerging lateral root in detail. Labeled HSS appears as golden grains in the cortex parenchyma (cp); the incrustation of the casparian strip in the radial cell walls of the endodermis (en) shows a faint blue color (indicated by arrowheads). D to I, Cutouts of B that were labeled with HSS-specific antibody according to the standard protocol (D), and after preincubation of the antibody with BSA (E), HSS (F and G), and DHS (H and I) in a molar ratio of antibody to added protein of 10:1 (F and H) and 1:3 (E, G, and I).
To test for the specificity of the label, we replaced the primary antibody by phosphate-buffered saline (PBS). Incubation with the FITC-labeled secondary antibody resulted in no labeling, as did the incubation with preimmune serum (data not shown). Because the HSS-specific antibody cross-reacts with DHS (Fig. 2C), we excluded the possibility that the label may have been partially attributable to the detection of DHS within the tissue. We repeated the labeling of serial cross-sections with antibody that had been preincubated with two different concentrations of purified HSS, DHS, and bovine serum albumin (BSA), respectively. If the labeling had arisen, at least to some extent, to cross-reactivity of the antibody with DHS, we would have expected the label to be attenuated after preincubation with DHS. In the other case, only preincubation with HSS should have resulted in the attenuation of the label, whereas preincubations with DHS and BSA should have had no effects. Figure 4, D to I, confirms that the labeling intensity is not affected by preincubation with BSA (Fig. 4E) and with DHS (Fig. 4, H and I) in comparison to the control (Fig. 4D). By contrast, preincubation with HSS in a molar ratio of antibody to added HSS of 10:1 reduces (Fig. 4F) and even completely blocks (1:3 ratio) the label (Fig. 4G).
HSS of E. cannabinum Is a Cytosolic Protein That Is Not Expressed in the Endodermis
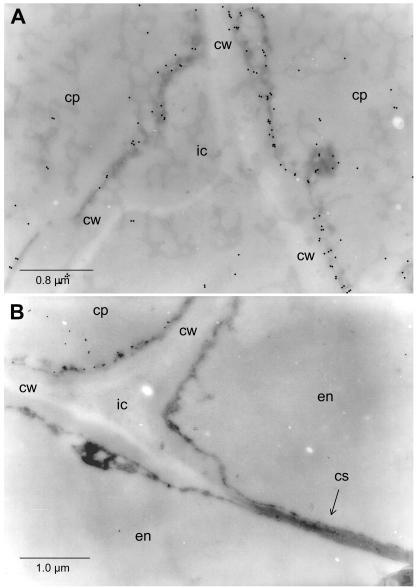
To confirm that HSS is expressed only in cells of the root cortex parenchyma but not within the endodermis, we analyzed, by transmission electron microscopy, cross-sections labeled with 18-nm gold particles after incubation with the affinity-purified antibody against HSS. Figure 5A shows cells of the cortex parenchyma adjoining an intercellular space. The label is found only in the cytosolic fringe; no label is associated with intracellular structures. In Figure 5B, two types of cells are shown: two endodermis cells with the characteristic incrustation of the casparian strip in their joint radial cell wall, and one cell belonging to the cortex parenchyma. Gold label is detectable only in the latter.
Figure 5.
Electron micrographs of in situ immunogold-labeled HSS in roots of E. cannabinum. A, Cells of the cortex parenchyma (cp) adjoining an intercellular space (ic). cw, Cell wall. B, Two cells of the endodermis (en) separated by a cell wall (cw) carrying the casparian strip (cs), an intercellular (ic), and an adjoining cell of the cortex parenchyma (cp) exclusively being labeled by the HSS-specific antibody.
Localization of HSS in S. jacobaea
The expression of HSS within S. vernalis was previously localized in groups of specialized cells of the root in the endodermis and the adjacent cortex parenchyma with a close spatial correlation to the phloem tissue (Moll et al., 2002). To test whether this expression in specialized cells is also found in other species of the tribe Senecioneae, we localized HSS in root cross-sections of S. jacobaea using an affinity-purified antibody against HSS of S. jacobaea. Also, in this perennial plant, HSS expression is restricted to the same type of cells that form four distinct groups due to the tetrarch organization of the central cylinder in S. jacobaea (data not shown).
DISCUSSION
HSS catalyzes the first specific step in PA biosynthesis. Despite its identical position within the pathway yielding the necine base of this class of alkaloids, the expression of HSS is individually regulated within the two different PA-containing tribes of the Asteraceae, namely the Senecioneae exemplified with S. vernalis (Moll et al., 2002) and S. jacobaea and the Eupatorieae exemplified with E. cannabinum, as described here. This reflects the independent evolutionary origin of HSS and the individual recruitment and adjustment of the regulatory elements in these two PA-producing lineages of the Asteraceae (Reimann et al., 2004).
Antibody Specificity
The antibody against HSS of E. cannabinum that we used was affinity purified and showed high specificity in a western-blot analysis of HSS-expressing tissues of selected PA-producing plants. Even the HSS of a close relative of E. cannabinum, namely Eupatorium pauciflorum, was not detected by the antibody (Fig. 2A). But the antibody showed some cross-reactivity with DHS, the protein with which HSS shares a common ancestor. Phylogenetic analyses suggest that the HSS-coding gene was recruited from the DHS-coding gene independently within the two PA-producing tribes of the Asteraceae (Reimann et al., 2004). This close relationship between both proteins of E. cannabinum is apparent in their high amino acid sequence identity (83%), which is the reason for the observed cross-reactivity. Our RT-PCR results show that the dhs gene is transcribed in all tested tissues, whereas the hss gene is exclusively transcribed in the roots. Several control experiments were performed to ensure that the observed label was not influenced by at least a partial interaction of the HSS-specific antibody with DHS expressed in the roots. We were able to confirm the root-specific expression of HSS in a western-blot analysis of several tissues of E. cannabinum (Fig. 3A). No label was detectable in any other tissue, although DHS is transcribed in all tested tissues as shown by RT-PCR (Fig. 3A). Obviously, the much lower expression level of DHS in comparison with HSS (Fig. 3A) and the 10-fold weaker labeling intensity of the DHS by the HSS-specific antibody (Fig. 2, B and C) were sufficient to eliminate cross-reactivity of the HSS-antibody with DHS. Nevertheless, we established the specificity of the immunohistological detection by control experiments with antibody that had been preincubated with soluble HSS, DHS, and BSA, respectively.
Tissue-Specific Expression of HSS
Tracer experiments and biochemical analyses have identified the root as the site of PA biosynthesis and as the tissue expressing active HSS in both PA-producing tribes of the Asteraceae (Toppel et al., 1987; Hartmann et al., 1989; Böttcher et al., 1993, 1994). Root cultures of species belonging to these tribes have thus proven to be excellent systems in which to study PA biosynthesis (Toppel et al., 1987; Sander and Hartmann, 1989; Weber et al., 1999); however, they have been found less suitable for histological studies within E. cannabinum because of incomplete tissue differentiation. Immunohistological analyses of tissues of whole plants show HSS expression in young roots in all cells of the cortex parenchyma. The exodermis and the endodermis, respectively, are devoid of any label. This cell-specific expression of HSS is different from that previously described for S. vernalis (Moll et al., 2002). Within S. vernalis, two different tissues are involved in HSS expression, but both only partially. HSS is restricted to groups of specialized cells of the endodermis and the adjoining cortex parenchyma that are localized directly opposite of the phloem tissues of the triarch central cylinder. Since, in Senecio, PAs are transported through the phloem, from the roots as the site of synthesis to the aerial parts as the site of accumulation (Hartmann et al., 1989), HSS expression seems to be adapted to the functional needs of alkaloid transport. This conclusion is supported by the observation that, also in S. jacobaea, the groups of HSS-expressing cells are opposite of the phloem, although S. jacobaea has a tetrarch organization of the central cylinder. Such a spatial correlation with one of the vascular tissues is not detectable in Eupatorium roots, although PAs within E. cannabinum are also preferentially accumulated in the aerial parts of the plant with the highest concentrations within the flower heads (P. Hülsmeyer and T. Hartmann, personal communication). HSS expression appears to be strictly linked to the development of the plant. In E. cannabinum, HSS expression is switched off when the flowers open (see below). S. vernalis as an annual plant fading away shortly after fruit dispersal may not have the need to shut down HSS expression during its vegetation period. Moreover, the down-regulation of HSS expression in those cortex parenchyma cells of E. cannabinum surrounding the lateral root primordia has not previously been observed. Most interestingly, a similar observation has been made for the expression of tropinone reductase and hyoscyamine 6β-hydroxylase, two enzymes involved in the biosynthesis of tropane alkaloids (Nakajima and Hashimoto, 1999).
The tissue- and cell-specific expression of alkaloid-specific enzymes has also been observed for monoterpene indole alkaloids (St-Pierre et al., 1999; Burlat et al., 2004), benzylisoquinoline alkaloids (Facchini and De Luca, 1995; Bird et al., 2003), tropane alkaloids (Hashimoto et al., 1991; Kanegae et al., 1994; Nakajima and Hashimoto, 1999; Suzuki et al., 1999), and nicotine (Shoji et al., 2000, 2002). As in PA-producing Asteraceae, plants producing tropane alkaloids and nicotine synthesize these alkaloids exclusively in the roots. Putrescine N-methyltransferase (PMT) and hyoscyamine 6β-hydroxylase as the first and the last enzyme in the biosynthesis of the tropane alkaloid scopolamine, respectively, are expressed exclusively in the pericycle. Surprisingly, tropinone reductase I, another enzyme of tropane alkaloid biosynthesis, is localized in the endodermis and the outer cortical layer, but not in the pericycle (Nakajima and Hashimoto, 1999). This differential compartmentation suggests that pathway intermediates have to be transported between different cell types, as has been postulated for intermediates of the monoterpene indole alkaloid biosynthesis in leaves of Catharanthus roseus (St-Pierre et al., 1999; Burlat et al., 2004). In E. cannabinum, we have to postulate the transport of intermediates, or at least of the biosynthetic end products, from the cortex parenchyma to the central vascular tissue in the central cylinder. Although little is known about the cell-to-cell transport of secondary metabolites, apoplastic transport appears unlikely. Root cultures of Eupatorium that are able to synthesize PAs never lose traces of PA N-oxides into the medium (Sander and Hartmann, 1989). The situation is different in Nicotiana root cultures, which are known to secrete a significant portion of the nicotine produced to the culture medium (Hibi et al., 1994). Because the weakly basic nicotine is able to permeate membranes passively, it is likely that it is transported within the root merely by diffusion (Shoji et al., 2000). The PMT, the first pathway-specific enzyme in nicotine biosynthesis in Nicotiana sylvestris, has recently been localized by promoter-GUS fusions within the outer cortex, the endodermis, and xylem tissue (Shoji et al., 2000). These results have been confirmed by the immunohistochemical analysis of PMT, which has shown the same cell-specific expression as for an isoflavone reductase with a putative role in downstream nicotine biosynthesis (Shoji et al., 2002).
HSS Expression in E. cannabinum Is Correlated to Plant Growth
During angiosperm evolution, the PA-specific HSS has been recruited several times independently from DHS, an enzyme involved in the posttranslational activation of eIF5A in primary metabolism (Reimann et al., 2004). The hypusination mechanism of eIF5A is highly conserved within eukaryotes and archaea and is essential for cell proliferation (Park et al., 1997). Moreover, a correlation with protein biosynthesis and growth in the root apices has been described for the PA biosynthesis (Sander and Hartmann, 1989). One may speculate that, during HSS recruitment, not only the structural dhs gene has been copied but also parts of the regulatory elements of the dhs gene. Although HSS expression is not restricted to the apices of young roots (Fig. 3B), it is strictly limited to newly grown, white roots with close correlation to the growth of the plant (Fig. 3C). HSS is only detectable in these young roots of E. cannabinum in the first half of the year, until the produced biomass peaks at the time when the flowers open. With the end of the vegetation period, these parts of the plant die off, requiring de novo alkaloid production at the beginning of the next growth period. This coupling of plant growth and PA biosynthesis warrants constant alkaloid concentrations in the tissues of the plant (Sander and Hartmann, 1989) as a prerequisite for the successful constitutive chemical defense of the plant. Work is in progress to identify those regulatory elements that regulate the hss promoter in E. cannabinum and that are dependent on plant and root growth.
MATERIALS AND METHODS
Plant Material
Eupatorium cannabinum plants were grown in the Institute garden. Achenes of the perennial herb were collected, and the pappus was removed and sown in February in the greenhouse to produce seedlings. If the plant material was not used directly, samples were frozen in liquid nitrogen and stored at −80°C.
Heterologous Expression and Purification of Proteins
Coding regions of HSS and DHS of E. cannabinum, each with a C-terminal His tag, were inserted into the expression vector pET3a-mod (Reimann et al., 2004) by using XhoI and BamHI restriction sites introduced by the PCR with Pfx DNA polymerase (Invitrogen, Carlsbad, CA). cDNAs coding for HSS and DHS (Reimann et al., 2004) were used as templates with primers P1/P2 and P3/P4, respectively (P1, 5′-dATCTAGACTCGAGATGGCGGCAGCAATTAAAGAAG-3′; P2, 5′-dTAGGATCCTAATGATGATGATGATGATGTGCGGAACATAGTTTCTTCT CAA-3′; P3, 5′-dATCTAGACTCGAGATGGGGGAACCCACTAAAGAAG-3′; P4, 5′-dTAGGATCCTAATGATGATGATGATGATGTCCAGAACTTGGTTTTGAATC-3′). Both constructs were introduced into Escherichia coli BL21(DE3) for heterologous expression as described previously (Ober and Hartmann, 1999a). E. coli cells were harvested by centrifugation, suspended in lysis buffer (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole), and broken open by sonication. From the supernatant, the His-tagged proteins were purified with nickel-nitrilotriacetic acid-agarose (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Preparation and Purification of Polyclonal Antibodies
Polyclonal serum was raised in rabbits against heterologously expressed and purified HSS of E. cannabinum by Bioscience (Göttingen, Germany). Purified recombinant HSS coupled to CNBr-activated Sepharose 4B (Amersham Biosciences, Buckinghamshire, UK), according to the manufacturer's instructions, was used for affinity purification of the serum. For this purpose, the matrix was incubated overnight at room temperature with the serum, washed with 0.1 m sodium acetate, pH 4.5, containing 0.5 m NaCl, and eluted with 0.2 m sodium acetate, pH 2.7, containing 0.5 m NaCl. The eluting purified antibodies were rebuffered in PBS, concentrated, and stored at −20°C until further use. An antibody against the HSS of Senecio jacobaea was raised using the heterologously expressed cDNA (Reimann et al., 2004) and purified as described for the antibody against HSS of E. cannabinum.
Protein Gel-Blot Analysis
Plant tissue samples were frozen in liquid nitrogen, ground in a mortar to fine powder, and extracted for 30 min on ice in PBS supplemented with 2.5% (w/v) polyvinylpyrrolidone and 2.5% (w/v) sodium ascorbate. Cell debris and the insoluble polyvinylpyrrolidone were removed by centrifugation. Soluble proteins were quantified (Bradford, 1976) and separated on 12% (w/v) SDS-PAGE gels by using a discontinuous buffer system (Laemmli, 1970) at 200 V constant voltage. Protein gels were electroblotted onto polyvinylidene fluoride membrane (Immobilon P; Millipore, Bedford, MA) with a current density of 2.5 mA cm−2. The lane with the blotted protein weight standard was cut off and stained with Indian ink (Hancock and Tsang, 1983). The remaining blots were blocked for 1 h at room temperature with Tris-buffered saline (TBS) supplemented with 0.1% (v/v) Tween 20 (TBS-T) containing 5% (m/v) milk powder, washed with PBS (5 × 1 min), and incubated with the affinity-purified polyclonal antibody (diluted 1:10,000 [v/v] in blocking solution) for 1 h at room temperature. After being washed with TBS-T (3 × 7 min), the membrane was incubated with a goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (diluted 1:3,300 [v/v]; Dianova, Hamburg, Germany) for 1 h. The washing steps were repeated before chemiluminescence detection was performed with the ECL Western Blotting System (Amersham Biosciences) and documented on XAR5 x-ray film (Eastman-Kodak, Rochester, NY).
RNA Isolation and Gel-Blot Analysis
Collected tissue samples were frozen in liquid nitrogen and stored at −80°C until use. Total RNA was extracted with the RNeasy Plant mini kit (Qiagen). Total RNA (10 μg per sample) was separated on a formaldehyde-agarose gel and transferred onto positively charged nylon membranes (Roche Diagnostics, Mannheim, Germany) by capillary blotting under alkaline conditions in 5× SSC (75 mm sodium citrate, 0.75 m NaCl, 10 mm NaOH). After prehybridization in high SDS buffer (5× SSC supplemented with 7% SDS, 50% deionized formamide, 50 mm sodium phosphate, pH 7.0, 0.1% N-lauroylsarcosine, 2% blocking reagent [Roche Diagnostics]), RNA gel blots were hybridized overnight at 39°C in high SDS buffer with HSS and DHS probes. Probes were digoxigenin labeled by using the PCR DIG Probe Synthesis kit (Roche Diagnostics) with the primers P1/P2 and P3/P4 for HSS and DHS, respectively. Chemiluminescent detection was performed with CSPD (Roche Diagnostics) according to the manufacturer's instructions. Exposure time was 10 min.
Semiquantitative RT-PCR
For each sample, 1 μg of total RNA was used as a template for cDNA synthesis with an oligo(dT)17 primer (Ober and Hartmann, 1999a) by using SuperScriptII Reverse Transcriptase (Invitrogen) in a total volume of 25 μL. Primer pairs P1/P2 and P3/P4 that had previously been used to amplify the full-length cDNA of HSS and DHS, respectively, were used to perform PCR with Taq DNA polymerase (Invitrogen). Aliquots of the reactions were analyzed after 34 cycles with an annealing temperature of 58°C by agarose gel electrophoresis.
Tissue Fixation and Embedding for Immunohistochemistry
For fixation of tissues, a previously published procedure (Moll et al., 2002) was used but slightly modified for tissues of E. cannabinum. Briefly, small segments of various plant organs (0.5–1.0 cm) were cut and fixed immediately for 2 h under reduced pressure in ice-cold buffered fixative (2% [w/v] formaldehyde freshly prepared from paraformaldehyde, 0.1% [v/v] glutaraldehyde, 0.1% Triton X-100 [w/v], and 2% soluble polyvinylpyrrolidone [Kollidon 30; BASF, Ludwigshafen, Germany] in 0.05 m potassium phosphate buffer, pH 7.2). The tissues were then washed twice for 10 min with 0.1 m potassium phosphate, pH 7.2, and dehydrated in a graded ethanol series. Embedding in Technovit 7100 resin (Heraeus-Kulzer, Hanau, Germany) for light microscopic analyses and embedding in Unicryl resin (Plano, Wetzlar, Germany) for analyses by transmission electron microscopy were performed according to the manufacturer's instructions. Polymerization of Unicryl was achieved under UV light at 4°C within 3 d. Sections of 3 to 4 μm were cut on a microtome and mounted on glass slides coated with Teflon (Roth, Karlsruhe, Germany).
Immunocytochemical Localization by UV and Light Microscopy
To remove excessive fixative, sections were washed successively with 50 mm ammonia chloride and 50 mm Gly (20 min each, at 37°C). After being blocked with 10% (w/v) BSA and 0.1% fish gelatin, sections were washed with PBS (3 × 7 min) before incubation with affinity-purified primary antibody (1:100 dilution in PBS) at 37°C for 3 h in a humid chamber. After being washed with PBS (3 × 10 min), the sections were incubated for 1 h in the dark at 37°C with a secondary goat anti-rabbit antibody labeled with FITC for fluorescence detection (1:100 [v/v] dilution in PBS; Sigma, St. Louis) and with 18-nm gold particles for immunogold labeling (1:75 [v/v] dilution in PBS; Dianova). FITC-labeled sections were protected with mounting medium (Citiflour; Agar Scientific, Essex, UK) against bleaching by UV light. Gold-particle-labeled sections were exposed to silver enhancement reagent according to the manufacturer's instructions (Amersham Biosciences). To view FITC labeling, the sections were excitated by UV light of 450 to 490 nm on an Axioskop 2 epifluorescence microscope (Zeiss, Göttingen, Germany) and recorded with an AxioCam HRc digital camera (Zeiss). The same equipment was used to take images of the silver-enhanced sections. To exclude cross-reactivity, serial cross-sections were labeled with antibody that had been preincubated for 30 min at 4°C with soluble purified recombinant HSS, DHS, and BSA, respectively. Molar ratios of antibody to added protein were 10:1 and 1:3.
Immunocytochemical Localization by Transmission Electron Microscopy
Sections (80 nm thick) of tissue embedded in Unicryl, as described above, were cut on an ultramicrotome and mounted onto nickel grids (300 mesh; Plano; Moll et al., 2002). Blocking and antibody incubation (goat anti-rabbit with 18-nm gold particles as secondary antibody) were performed as described for light microscopy, but at room temperature. After being poststained with 2% (w/v) aqueous uranyl acetate for 2 × 10 min, the sections were analyzed on a transmission electron microscope (300 EM; Philips, Eindhoven, The Netherlands).
Acknowledgments
We are grateful to Thomas Hartmann for helpful discussions during the preparation of this manuscript and to Bettina Hause for the establishment of the immunolocalization techniques. We thank Doris Glindemann for her excellent technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.052357.
References
- Bayer RJ, Starr JR (1998) Tribal phylogeny of the Asteraceae based on two noncoding chloroplast sequences, the trnL intron and trnL/trnF intergenic spacer. Ann Mo Bot Gard 85: 242–256 [Google Scholar]
- Bird DA, Franceschi VR, Facchini PJ (2003) A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15: 2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher F, Adolph RD, Hartmann T (1993) Homospermidine synthase, the first pathway-specific enzyme in pyrrolizidine alkaloid biosynthesis. Phytochemistry 32: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher F, Ober D, Hartmann T (1994) Biosynthesis of pyrrolizidine alkaloids: Putrescine and spermidine are essential substrates of enzymatic homospermidine formation. Can J Chem 72: 80–85 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Burlat V, Oudin A, Courtois M, Rideau M, St-Pierre B (2004) Co-expression of three MEP pathway genes and geraniol 10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J 38: 131–141 [DOI] [PubMed] [Google Scholar]
- Caraglia M, Marra M, Giuberti G, D'Alessandro AM, Baldi A, Tassone P, Venuta S, Tagliaferri P, Abbruzzese A (2003) The eukaryotic initiation factor 5A is involved in the regulation of proliferation and apoptosis induced by interferon-α and EGF in human cancer cells. J Biochem (Tokyo) 133: 757–765 [DOI] [PubMed] [Google Scholar]
- Caraglia M, Marra M, Giuberti G, D'Alessandro AM, Budillon A, del Prete S, Lentini A, Beninati S, Abbruzzese A (2001) The role of eukaryotic initiation factor 5A in the control of cell proliferation and apoptosis. Amino Acids 20: 91–104 [DOI] [PubMed] [Google Scholar]
- Chou W-C, Huang Y-W, Tsay W-S, Chiang T, Huang D-D, Huang H-J (2004) Expression of genes encoding the rice translation initiation factor, eIF5A, is involved in developmental and environmental responses. Physiol Plant 121: 50–57 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Bird DA, St-Pierre B (2004) Can Arabidopsis make complex alkaloids? Trends Plant Sci 9: 116–122 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, De Luca V (1995) Phloem-specific expression of tyrosine/dopa decarboxylase genes and the biosynthesis of isoquinoline alkaloids in opium poppy. Plant Cell 7: 1811–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goertzen LR, Cannone JJ, Gutell RR, Jansen RK (2003) ITS secondary structure derived from comparative analysis: implications for sequence alignment and phylogeny of the Asteraceae. Mol Phylogenet Evol 29: 216–234 [DOI] [PubMed] [Google Scholar]
- Hancock K, Tsang VC (1983) India ink staining of proteins on nitrocellulose paper. Anal Biochem 133: 157–162 [DOI] [PubMed] [Google Scholar]
- Hartmann T (1994) Senecio spp.: biochemistry of the formation of pyrrolizidine alkaloids in root cultures. In YPS Bajaj, ed, Biotechnology in Agriculture and Forestry, Vol 26, Medicinal and Aromatic Plants VI. Springer, Berlin, pp 339–355
- Hartmann T (2004) Plant-derived secondary metabolites as defensive chemicals in herbivorous insects: a case study in chemical ecology. Planta 219: 1–4 [DOI] [PubMed] [Google Scholar]
- Hartmann T, Ehmke A, Eilert U, von Borstel K, Theuring C (1989) Sites of synthesis, translocation and accumulation of pyrrolizidine alkaloid N-oxides in Senecio vulgaris L. Planta 177: 98–107 [DOI] [PubMed] [Google Scholar]
- Hartmann T, Ober D (2000) Biosynthesis and metabolism of pyrrolizidine alkaloids in plants and specialized insect herbivores. In FJ Leeper, JC Vederas, eds, Topics in Current Chemistry, Vol 209. Springer, Berlin [u.a.], pp 207–244
- Hartmann T, Witte L (1995) Pyrrolizidine alkaloids: chemical, biological and chemoecological aspects. In SW Pelletier, ed, Alkaloids: Chemical and Biological Perspectives, Vol 9. Pergamon Press, Oxford, pp 155–233
- Hashimoto T, Hayashi A, Amano Y, Kohno J, Iwanari H, Usuda S, Yamada Y (1991) Hyoscyamine 6β-hydroxylase, an enzyme involved in tropane alkaloid biosynthesis, is localized at the pericycle of the root. J Biol Chem 266: 4648–4653 [PubMed] [Google Scholar]
- Hibi N, Higashiguchi S, Hashimoto T, Yamada Y (1994) Gene expression in tobacco low-nicotine mutants. Plant Cell 6: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanegae T, Kajiya H, Amano Y, Hashimoto T, Yamada Y (1994) Species-dependent expression of the hyoscyamine 6-β-hydroxylase gene in the pericycle. Plant Physiol 105: 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HA, Hershey JWB (1994) Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem 269: 3934–3940 [PubMed] [Google Scholar]
- Karis PO (1993) Morphological phylogenetics of the Asteraceae-Asteroideae, with notes on character evolution. Plant Syst Evol 186: 69–93 [Google Scholar]
- Kim KJ, Jansen RK (1995) ndhF sequence evolution and the major clades in the sunflower family. Proc Natl Acad Sci USA 92: 10379–10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Moll S, Anke S, Kahmann U, Hänsch R, Hartmann T, Ober D (2002) Cell-specific expression of homospermidine synthase, the entry enzyme of the pyrrolizidine alkaloid pathway in Senecio vernalis, in comparison with its ancestor, deoxyhypusine synthase. Plant Physiol 130: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Hashimoto T (1999) Two tropinone reductases that catalyze opposite stereospecific reductions in tropane alkaloid biosynthesis are localized in plant root with different cell-specific patterns. Plant Cell Physiol 40: 1099–1107 [DOI] [PubMed] [Google Scholar]
- Ober D (2003) Chemical ecology of alkaloids exemplified with the pyrrolizidines. In JT Romeo, ed, Integrative Phytochemistry: From Ethnobotany to Molecular Ecology, Vol 37. Pergamon, Amsterdam, pp 203–230
- Ober D, Gibas L, Witte L, Hartmann T (2003. a) Evidence for general occurrence of homospermidine in plants and its supposed origin as by-product of deoxyhypusine synthase. Phytochemistry 62: 339–344 [DOI] [PubMed] [Google Scholar]
- Ober D, Harms R, Witte L, Hartmann T (2003. b) Molecular evolution by change of function: alkaloid-specific homospermidine synthase retained all properties of deoxyhypusine synthase except binding the eIF5A precursor protein. J Biol Chem 278: 12805–12812 [DOI] [PubMed] [Google Scholar]
- Ober D, Hartmann T (1999. a) Deoxyhypusine synthase from tobacco: cDNA isolation, characterization, and bacterial expression of an enzyme with extended substrate specificity. J Biol Chem 274: 32040–32047 [DOI] [PubMed] [Google Scholar]
- Ober D, Hartmann T (1999. b) Homospermidine synthase, the first pathway-specific enzyme of pyrrolizidine alkaloid biosynthesis, evolved from deoxyhypusine synthase. Proc Natl Acad Sci USA 96: 14777–14782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober D, Hartmann T (2000) Phylogenetic origin of a secondary pathway: the case of pyrrolizidine alkaloids. Plant Mol Biol 44: 445–450 [DOI] [PubMed] [Google Scholar]
- Park MH, Lee YB, Joe YA (1997) Hypusine is essential for eukaryotic cell proliferation. Biol Signals 6: 115–123 [DOI] [PubMed] [Google Scholar]
- Reimann A, Nurhayati N, Backenköhler A, Ober D (2004) Repeated evolution of the pyrrolizidine alkaloid-mediated defense system in separate angiosperm lineages. Plant Cell 16: 2772–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosorius O, Reichart B, Kraetzer F, Heger P, Dabauvalle MC, Hauber J (1999) Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J Cell Sci 112: 2369–2380 [DOI] [PubMed] [Google Scholar]
- Sander H, Hartmann T (1989) Site of synthesis, metabolism and translocation of senecionine N-oxide in cultured roots of Senecio erucifolius. Plant Cell Tissue Organ Cult 18: 19–32 [Google Scholar]
- Shoji T, Winz R, Iwase T, Nakajima K, Yamada Y, Hashimoto T (2002) Expression patterns of two tobacco isoflavone reductase-like genes and their possible roles in secondary metabolism in tobacco. Plant Mol Biol 50: 427–440 [DOI] [PubMed] [Google Scholar]
- Shoji T, Yamada Y, Hashimoto T (2000) Jasmonate induction of putrescine N-methyltransferase genes in the root of Nicotiana sylvestris. Plant Cell Physiol 41: 831–839 [DOI] [PubMed] [Google Scholar]
- St-Pierre B, Vazquez-Flota FA, De Luca V (1999) Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell 11: 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Yamada Y, Hashimoto T (1999) Expression of Atropa belladonna putrescine N-methyltransferase gene in root pericycle. Plant Cell Physiol 40: 289–297 [DOI] [PubMed] [Google Scholar]
- Thompson JE, Hopkins MT, Taylor C, Wang T-W (2004) Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development. Trends Plant Sci 9: 174–179 [DOI] [PubMed] [Google Scholar]
- Toppel G, Witte L, Riebesehl B, von Borstel K, Hartmann T (1987) Alkaloid patterns and biosynthetic capacity of root cultures from some pyrrolizidine alkaloid producing Senecio spp. Plant Cell Rep 6: 466–469 [DOI] [PubMed] [Google Scholar]
- Wang TW, Lu L, Wang D, Thompson JE (2001) Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eukaryotic translation initiation factor 5A from tomato. J Biol Chem 276: 17541–17549 [DOI] [PubMed] [Google Scholar]
- Wang T-W, Lu L, Zhang C-G, Taylor C, Thompson JE (2003) Pleiotropic effects of suppressing deoxyhypusine synthase expression in Arabidopsis thaliana. Plant Mol Biol 52: 1223–1235 [DOI] [PubMed] [Google Scholar]
- Weber S, Eisenreich W, Bacher A, Hartmann T (1999) Pyrrolizidine alkaloids of the lycopsamine type: biosynthesis of trachelanthic acid. Phytochemistry 50: 1005–1014 [Google Scholar]
- Xu A, Chen KY (2001) Hypusine is required for a sequence-specific interaction of eukaryotic initiation factor 5A with postsystematic evolution of ligands by exponential enrichment RNA. J Biol Chem 276: 2555–2561 [DOI] [PubMed] [Google Scholar]