Significance
Despite concerted efforts to identify causal genes that drive breast cancer (BC) initiation and progression, we have yet to establish robust signatures to stratify patient risk. Here we used in vivo transposon-based forward genetic screening to identify potentially relevant BC driver genes. Integrating this approach with survival prediction analysis, we identified six gene pairs that could prognose human BC subtypes into high-, intermediate-, and low-risk groups with high confidence and reproducibility. Furthermore, we identified susceptibility gene sets for basal and claudin-low subtypes (21 and 16 genes, respectively) that stratify patients into three relative risk subgroups. These signatures offer valuable prognostic insight into the genetic basis of BC and allow further exploration of the interconnectedness of BC driver genes during disease progression.
Keywords: breast cancer, Sleeping Beauty, cancer susceptibility, prognostic gene signature, survival prediction analysis
Abstract
Robust prognostic gene signatures and therapeutic targets are difficult to derive from expression profiling because of the significant heterogeneity within breast cancer (BC) subtypes. Here, we performed forward genetic screening in mice using Sleeping Beauty transposon mutagenesis to identify candidate BC driver genes in an unbiased manner, using a stabilized N-terminal truncated β-catenin gene as a sensitizer. We identified 134 mouse susceptibility genes from 129 common insertion sites within 34 mammary tumors. Of these, 126 genes were orthologous to protein-coding genes in the human genome (hereafter, human BC susceptibility genes, hBCSGs), 70% of which are previously reported cancer-associated genes, and ∼16% are known BC suppressor genes. Network analysis revealed a gene hub consisting of E1A binding protein P300 (EP300), CD44 molecule (CD44), neurofibromin (NF1) and phosphatase and tensin homolog (PTEN), which are linked to a significant number of mutated hBCSGs. From our survival prediction analysis of the expression of human BC genes in 2,333 BC cases, we isolated a six-gene-pair classifier that stratifies BC patients with high confidence into prognostically distinct low-, moderate-, and high-risk subgroups. Furthermore, we proposed prognostic classifiers identifying three basal and three claudin-low tumor subgroups. Intriguingly, our hBCSGs are mostly unrelated to cell cycle/mitosis genes and are distinct from the prognostic signatures currently used for stratifying BC patients. Our findings illustrate the strength and validity of integrating functional mutagenesis screens in mice with human cancer transcriptomic data to identify highly prognostic BC subtyping biomarkers.
Breast cancer (BC) is the most prevalent cancer in women in North America, representing nearly one in three cancers diagnosed (1). BC is classified clinically into three basic groups, based primarily on receptor expression, that are valuable from a therapeutic perspective: (i) patients with estrogen receptor-positive (ER+) cancer receive endocrine therapy, such as tamoxifen, which targets the ER; (ii) patients with amplified human epidermal growth factor receptor 2 (HER2, also called “ERBB2”) are treated with therapeutic agents against HER2, such as trastuzumab; and (iii) patients with triple-negative cancer [lacking the expression of ER, progesterone receptor (PGR), and HER2] are treated with chemotherapy.
Gene-expression patterns classify human BC into six major molecular subgroups: luminal A, luminal B, normal breast tissue-like, basal-like, HER2 (2), and claudin-low; the last was most recently discovered and is linked with poor prognosis (3). However, recent molecular subtyping analyses of the integrated copy number and transcriptomic datasets of 2,000 BC patients have revealed even further complexity, with 10 distinct subgroups that partially overlap with the previous subtypes (4). These classifications underscore the complexity of BC tumorigenesis, particularly the clinical heterogeneity within the intermediate and high histological grades and triple-negative tumors, which are generally associated with poor disease outcomes. The biological behaviors of these molecular subtypes are driven by aberrant (pro-oncogenic and tumor suppressor) signaling of regulatory pathways, but how this dysregulation relates to prognosis and treatment outcomes is still unclear (5). The systematic assessment of prognostic gene signatures for BC shows the distinct influence of time and ER status (6). Although the genetic aspects of BC have been studied for decades, BRCA1 (7) and BRCA2 (8) were identified only in the early 1990s as BC susceptibility genes (BCSGs) derived from mutations. Since then, several other BC driver genes have been identified, including TP53 (9), CHEK2 (10), PIK3CA (11–14), PTEN (15), CASP8, FGFR2, and MAP3K1 (16). Extensive mutational profiling by exome sequencing of 100 tumors recently highlighted more than 40 BCSGs, including nine that were previously unrecognized (17). Each BC can carry, on average, one mutation per megabase (11), and a normal human cell can acquire 7–15 somatic mutations before malignant transformation (18–21). Thus, most mutations in BC are likely to be passenger mutations that do not contribute to tumorigenesis or tumor progression. Functional screens that can identify the driver mutations in BC thus are distinctly warranted. Sleeping Beauty (SB) transposon-based insertional mutagenesis screening in mice has emerged as a powerful, functional approach for the identification of BCSGs. SB overcomes the limitations of previous tools (such as retroviral insertional mutagenesis) and has been applied successfully to a number of solid tumor types, including colorectal cancers (22), intestinal cancers (19, 23), hepatocellular cancers (24, 25), pancreatic adenocarcinoma (26), and peripheral nerve sheet tumors (27). The method harnesses the use of DNA cut-and-paste transposons that are engineered to elicit either loss- or gain-of-function mutations in somatic tissues to accelerate the formation of specific tumors in mice. Such transposon insertions can cause multiple dysfunctions in tumor-suppressor genes and proto-oncogenes: Tumor suppressors may be inactivated by loss-of-function mutations, or, in some cases, the mutation could change the function or interaction network of the genes and cause pro-oncogenic functions. Gain-of-function mutations in proto-oncogenes could lead to the activation of oncogenic pathways. As such, mapping the SB insertion sites will unveil the relevant BCSG(s).
In this study, we performed SB transposon-based forward genetic screening in mice to identify functionally relevant BC driver genes. We used a K5-Cre transgene that was expressed in both luminal and basal cells to induce transposition and drive the formation of different mammary tumor subtypes. We also used the K5-N57β-cat transgenic mouse line to introduce a stabilized N-terminally truncated β-catenin gene as a sensitizing mutation; the expression of activated β-catenin from the K5 promoter promotes basal-like mammary tumor formation in vivo (28). Through this approach, we identified 134 mouse BC susceptibility genes (mBCSGs) from 129 common integration loci. Of these, 126 human orthologs were identified as human BC susceptibility genes (hBCSGs). Through integrated data analyses we found that most of these hBCSGs are mutated in human BC and more commonly are tumor-suppressor genes. We identified a six-gene-pair signature that could be used to prognose disease-free and overall survival (OS) and to stratify all BC subtypes into three different risk groups. Within the basal-like and claudin-low tumor subtypes, we further defined two prognostic gene signatures (21-hBCSGs and 16-hBCSGs, respectively) that could be used to stratify patients with each tumor subtype reliably into groups at relatively low, medium, and high risk of disease development.
Results
SB Transposon Mutagenesis Promotes Mammary Tumor Formation and Induces a Broad Spectrum of Mammary Tumor Types.
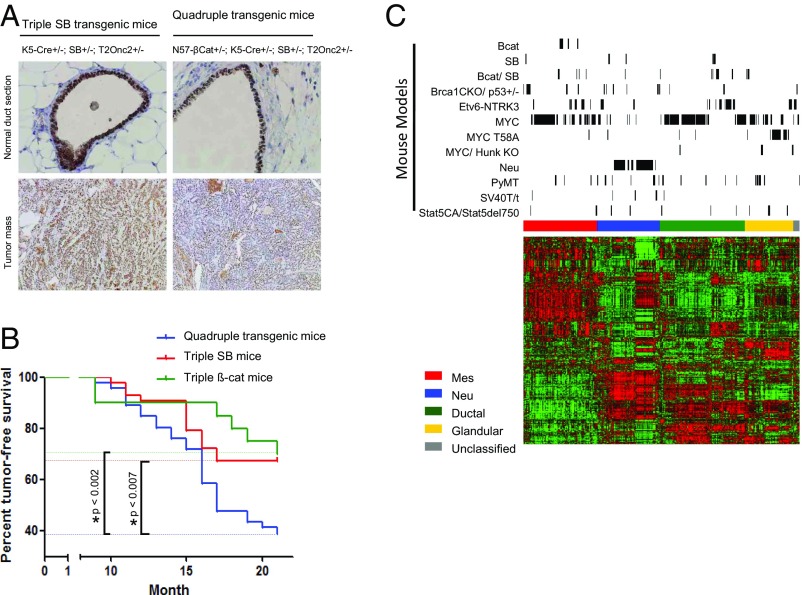
To identify BCSGs, we performed SB transposon-mediated mutagenesis screening in a K5-∆N57-β-catenin (N57β-cat) mouse model together with a K5-Cre line to activate Cre-inducible transposon mutagenesis in the mammary epithelium. The K5 promoter in the N57β-cat model drives the expression of stabilized N-terminally truncated β-catenin in the basal cell layer of the mammary epithelium (28), resulting in basal-like mammary tumors. For activation of the SB transposase, we used a K5-Cre transgene (29) that expresses Cre in both the luminal and basal epithelia from 18 d of gestation to the early postnatal period (Fig. S1). To generate the experimental cohorts, we first generated a K5-Cre+/−; N57β-cat+/− sensitizer line that was bred to compound homozygous SB mice carrying the Rosa26 Cre-inducible SB transposase (LSL-SB) and a high copy mutagenic transposon array (T2Onc2) (Fig. S2): (i) N57β-cat+/−, K5-Cre+/−, SB+/−, T2Onc2+/− mice expressing the N-terminally truncated β-catenin and activated SB transposase; (ii) K5-Cre+/−; SB+/−; T2Onc2+/−mice expressing the activated SB transposase; and (iii) N57β-cat+/−; SB+/−; T2Onc2+/− mice expressing only the N-terminally truncated β-catenin. From the experimental crosses, we aged 46 quadruple N57β-cat/SB transgenic mice (N57β-cat+/−; K5-Cre+/−; SB+/−; T2Onc2+/−), 43 triple SB transgenic mice (K5-Cre+/-; SB+/−; T2Onc2+/−), and 20 triple transgenic N57β-cat mice (N57β-cat+/−; SB+/−; T2Onc2+/−) and monitored them for mammary tumor formation over 24 mo. SB induced by the K5-Cre transgene was activated in the quadruple transgenic mice and triple SB mice (Fig. 1A). We observed that, on average, the time for tumor development did not differ significantly (log-rank P > 0.05) among the three cohorts (quadruple N57β-cat/SB mice: 14.2 mo; triple SB mice: 15.5 mo; triple N57β-cat mice: 13 mo). However, the incidence of mammary tumors was significantly higher in the quadruple N57β-cat/SB transgenic mice (61%, n = 28/46) than in the triple SB mice (33%, n = 14/43) or triple N57β-cat mice (30%, n = 6/20) (Fig. 1B). Thus, SB transposon mutagenesis is sufficient to initiate mammary tumor formation and can cooperate with N-terminally truncated N57β-catenin to promote a higher incidence of mammary tumorigenesis.
Fig. 1.
SB mutagenesis drives mammary tumorigenesis in the mouse. (A) SB was activated in both the luminal and basal cell layers of the mouse mammary gland. (B) Tumor incidence rates among three different groups of mice across time. (C) Heatmap of mouse mammary tumor subtypes and mRNA expression for our genes of interest across mouse (green indicates low expression; red indicates high expression). Genes and samples are represented along the rows and columns, respectively. Mouse tumors and genes are aligned based on consensus clustering results. The positions of mouse mammary tumors from different mouse models are indicated by the black bars on top. The color bar on top of the heatmap indicates the subtypes of mouse mammary tumors: red, mesenchymal (Mes); blue, HER2/Neu; green, ductal; yellow, glandular; gray, unclassified). Bcat, β-catenin; mmT, mouse mammary tumor.
Next, we examined the histological subtypes of the mammary tumors induced in the different transgenic mice groups (n = 34; 25 from quadruple N57β-cat/SB mice; nine from triple SB mice) (Dataset S1). Consistent with previous studies, mammary tumors from triple N57β-cat mice were exclusively squamous carcinoma. In contrast, mammary tumors from quadruple N57β-cat/SB mice and triple SB mice were a mixture of squamous carcinoma (50 and 38%, respectively), adenocarcinoma (23 and 38%, respectively), and adenosquamous carcinoma (32 and 25%, respectively) (Fig. S3 and Dataset S1), indicating that SB transposon mutagenesis may disrupt/overexpress driver genes that are involved in the differentiation of a broad spectrum of mammary tumor subtypes. To verify this hypothesis, we performed unsupervised clustering of expression profiles from the β-catenin– and SB-induced mammary tumors and compared them with published expression datasets of different mouse mammary tumors (Materials and Methods and Fig. 1C). Tumors induced by β-catenin expression from the K5 promoter were exclusively of the mesenchymal subtype. In contrast, SB-induced tumors showed a wider spectrum of subtypes [mesenchymal, human epidermal growth factor receptor 2 (Neu), ductal, and glandular] (Fig. 1C). These findings indicate that transposon mutagenesis promoted the development of different mammary tumor subtypes.
Identification of mBCSGs by Deep Sequencing of Tumors and Analysis of Transposon Integration Sites.
To identify candidate driver genes, we performed linker-mediated PCR and deep sequencing of tumors to map the transposon insertion sites. Among 41 sequenced samples only 34 mammary tumors with histology were used for analysis of common insertion site (CIS) genes. After filtering for mapped sequences that contain the transposon sequence–genome junctions at TA-motif sites in the mouse genome, we identified 11,664 transposon insertions, of which 11,457 (98.2%) were unique. Next, we used the Gaussian kernel convolution (GKC) algorithm (30) to identify CIS that were statistically overrepresented in the samples, signifying likely driver genes. To maximize statistical power to detect CIS genes, the insertions from both quadruple and triple SB transgenic mice were pooled for analysis using a range of 15- to 250-kB scales in the GKC algorithm to identify insertion peaks for both small and large genes. As an additional step to reduce false-positive calls of CIS peaks, we excluded insertions that mapped to chromosome 1 where the high-copy transposon array donor site is located and where higher insertion frequencies are observed because of local hopping induced by SB-mediated transposition (22, 30, 31).
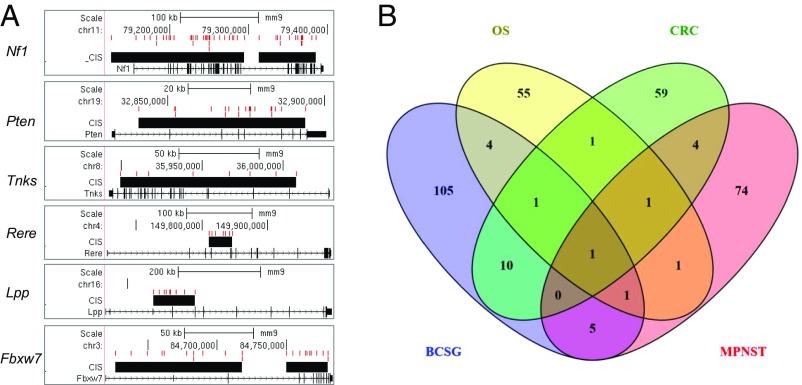
From the GKC analysis, we identified 129 CIS peaks; 128 were located in or near 133 coding genes, and one was located near a microRNA cluster (Dataset S2). To identify the putative effect of these insertions, we mapped integration sites and CIS peaks onto gene loci to visualize the pattern of integrations (Fig. 2A). For example, we identified Nf1 as the CIS-associated gene with the highest number of transposon insertion sites in mammary tumors induced by SB transposon mutagenesis (Fig. 2A and Dataset S2). Inspection of the integration sites showed that the insertions occurred in both directions, suggesting a loss-of-function disruption in the Nf1 gene. This finding is consistent with previous studies showing Nf1 loss-of-function mutations in mouse mammary tumors and NF1 loss-of-function mutations in human BCs (32). In further support of this association, the NF1 gene is deleted or mutated in 27.7% of all breast carcinomas (33), and women with neurofibromatosis type I disease (caused by NF1 loss-of-function mutations) have an increased risk of BC (34). Examination of the other CIS genes with frequent integrations (Pten, Tnks, Rere, Lpp, and Fbxw7) (Fig. 2A) showed random integrations along each of the gene loci similar to Nf1, indicating possible loss-of-function disruptions in these genes. Consistent with these observations, FBXW7 and PTEN are frequently mutated or deleted in BC (35). There is limited information for RERE, but NF1 (36), PTEN (37), TNKS (38), LPP (39) and FBXW7 (40) are each associated with tumor-suppression functions in human BCs. Taken together, the identification of genes with known tumor-suppressor function within or near CIS genes suggests the efficiency of our transposon-based screens.
Fig. 2.
Analysis of CIS of SB mutagenesis-induced BC. (A) Representation of insertions of the mutagenic transposon and CIS on six genes, including Nf1, Pten, Tnks, Rere, Lpp, and Fbxw7. The transposon insertion sites and CIS gene are represented in red and black, respectively. (B) Comparison of CIS-associated gene sets for different cancer types: BCSG, CRC, OST, and MPNST.
Comparison of SB Mutagenesis mBCSGs with Other SB Transposon Cancer Screens.
To understand the similarities and differences in CIS genes identified in the SB transposon screens, we compared the mBCSGs with previous SB transposon screens for other cancer types (22, 25, 27, 41). We examined the 127 mBCSGs (Dataset S2) together with CIS-associated susceptibility genes previously identified in SB studies of colorectal cancer (CRC) (77 genes) (22), osteosarcoma (OST) (65 genes) (41), and malignant peripheral nerve sheath tumors (MPNST) (87 genes) (27).
We observed that 22 of the 127 CIS-associated mBCSGs have been reported previously as CIS-associated genes in CRC, OST, MPNST, or hepatocellular carcinoma. The remaining 105 mBCSGs could be considered BC CIS-associated genes identified via SB mutagenesis screening (Fig. 2B). We found that Pten is the most common in four CIS-associated datasets (BC, OST, CRC, and MPNST), and Nf1 and Was were common in three datasets (BC, OST, and MPNST and BrCa, OST, and CRC), respectively). These observations suggest that Pten, Nf1, and Was have important common roles as the most frequent targets of the SB mutagenesis across different tissues.
Interestingly, among the 22 CIS-associated mBCSGs, 54% (12/22) were common to those of CRC, and 10 these 12 CIS genes (Fbxw7, Matr3, Tnks, Sfi1, Myst3, Pum1, Bmpr1a, Tcf12, Pik3r1, and Ppp1r12a) are observed only in BC and CRC. These observations suggest that common driver genes and their associated oncogenic pathways may play similar roles in malignancy occurrence and progression in humans, especially in BC and CRC, and that these cancers may be targetable with similar therapeutics strategies.
Correlation of mBCSG-Orthologous hBCSGs with Somatic Mutations in Human BC.
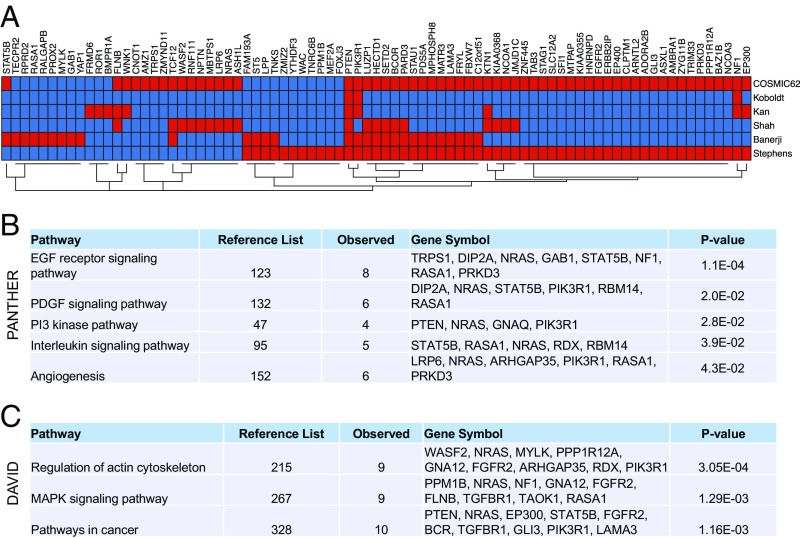
To examine the correlation of candidate driver genes from the transposon screen to somatic mutations in human BC, we first identified human orthologs of mBCSGs. Of 134 mBCSGs, we found 126 human protein-coding genes, defined by National Center for Biotechnology Information (NCBI) Entrez annotation (hereafter referred to as hBCSGs) (Dataset S3). We next evaluated the overlap between hBCSGs and candidate BCSGs identified in previous studies of human BC mutations (17, 33, 35, 42, 43). We observed that ∼64% (81/126) of hBCSGs are mutated in human BC (Fig. 3A and Dataset S4). Of note, PTEN, a CIS gene with frequent transposon integrations, was mutated in all datasets; other hBCSGs were mutated at varying frequencies (in one to four of five datasets). The high concordance of hBCSGs (∼64%) with existing somatic mutations suggests that we could identify relevant candidate driver mutations in human BC from our transposon screen.
Fig. 3.
Mutations and pathways of hBCSGs. (A) Somatic mutations in 81 of the 126 BCSGs have been previously reported for human BC tissues and cell lines (red color indicates the presence of a gene in the publications shown in Dataset S4). (B) PANTHER pathway enrichment analysis of 123 hBCSGs (multivariate-corrected P value cutoff <0.05 by Bonferroni). (C) Pathway enrichment analysis of 126 hBCSGs by DAVID Bioinformatics tools (multivariate-corrected P value cutoff <0.05 by Benjamini). (See also Dataset S5.)
Signaling Pathways and Gene Networks Regulated by hBCSGs.
To gain insight into the possible biological pathways regulated by hBCSGs, we performed gene ontology (GO) analysis of the 126 hBCSGs using the PANTHER (Protein ANalysis THrough Evolutionary Relationships) classification (44) and DAVID (Database for Annotation, Visualization and Integrated Discovery) (45) GO tools. Using the PANTHER tool, we identified five potential signaling pathways: the epidermal growth factor receptor (EGFR) signaling pathway, the PDGF signaling pathway, the PI3K pathway, the IL signaling pathway, and angiogenesis ( Fig. 3B and Dataset S5). Using the DAVID tool, we identified potential regulation by hBCSGs through the actin cytoskeletal and MAPK signaling pathways (Fig. 3C and Dataset S5). The concordance of EGFR and MAPK signaling pathways identified using both approaches is consistent with the known roles of EGFR/MAPK signaling in human BC progression, indicating that functionally relevant genes were uncovered in the screen.
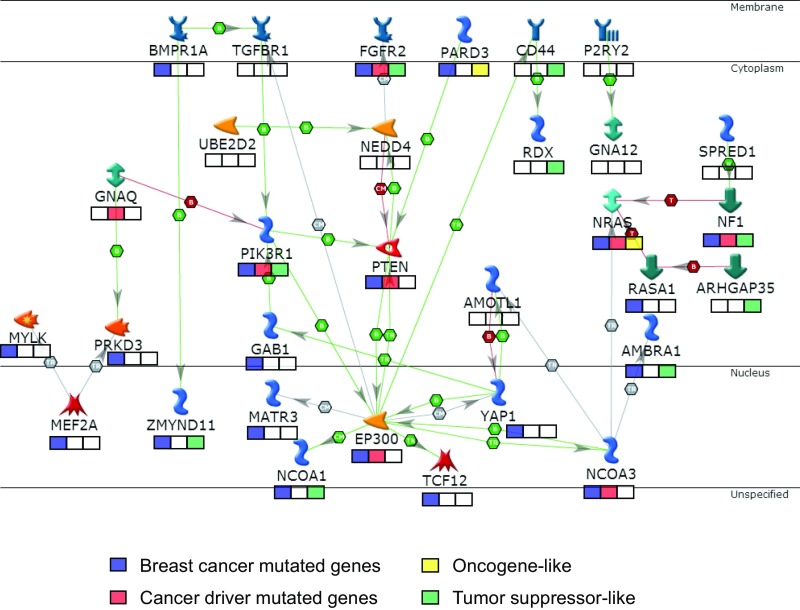
Next, we examined the potential gene networks within the hBCSG gene set using the MetaCore analysis package (https://portal.genego.com/). We found an interaction network involving 31 of the 126 hBCSGs (∼25%) with EP300 as a hub (Fig. 4). Most notably, 20 genes in the network are mutated in human BC (Fig. 3A and Dataset S4), and eight (FGFR2, GNAQ, NRAS, NCOA3, NF1, PIK3R1, PTEN, and EP300) among these 20 have been considered as cancer-driver genes (46). The identification of EP300 as a gene-network hub suggests a potential point of intersection in the signaling networks involved in the human BC oncogenic pathway driving cancer progression.
Fig. 4.
Thirty-one of 126 hBCSGs code for proteins involved in a tumor-suppression network with EP300 as the hub. The boxes below the gene names indicate the annotation of those genes: blue boxes denote BC-mutated genes (see Dataset S4), red boxes denote cancer-driver mutated genes (47), green boxes denote tumor-suppressor–like genes, and yellow boxes denote oncogene-like genes.
hBCSGs Can Classify Human BC Subtypes.
To determine whether hBCSGs could be used to distinguish the different molecular subtypes in BC, we performed unsupervised hierarchical clustering of both the mouse (n = 394) and human (n = 1345) expression datasets using BCSGs (Fig. S4 A and B and Dataset S6 B and C). To facilitate comparison, we clustered the human BC using hBCSGs into four subtypes corresponding to the mouse subtypes (Fig. S4B). Subsequently, we assessed the association of the four mouse subtypes with the human subtypes using a two-tailed Fisher’s exact test (Fig. S4C). The basal-like and claudin-low subtypes in human tumors appeared most similar to the mesenchymal subtype in mouse tumors (P = 5.65E-44). To a lesser extent, we observed common transcriptional patterns between the mouse ductal subtype and the human luminal-A subtype (P = 1.02E-18) and between the mouse glandular subtype and the human luminal-B subtype (P = 1.16E-17). The association of the ERBB2+ and normal-like subtypes with the corresponding mouse subtypes is unclear. These results indicate that mBCSGs and hBCSGs are differentially expressed in the different molecular subtypes of mouse and human BCs, respectively. These results support the notion that the hBCSGs identified in our mouse forward genetic screen are highly relevant in human BC.
Identification of High-Confidence Gene Signatures for Prediction of Risk Groups in BC Disease-Free Survival.
To determine if the 126 hBCSGs could be used to stratify risk groups of BC patients, we used a data-driven selection method (47, 48) to analyze the expression datasets of 2,333 BC samples together with their survival outcomes (Dataset S6 D and E). Table S1 shows common clinical characteristics available for the studied datasets. Fig. S5 shows a workflow of our pipeline, which selects the most significant prognostic variables, converts these variables into discrete variables according to their prognostic pattern, and combines these variables as prognostic vectors to construct our combine prognostic model. In our workflow (Fig. S5), to study whether the hBCSGs could be used to define gene signatures that determine the prognosis of human BC patients in terms of disease-free survival (DFS), the 126 hBCSGs were individually analyzed using our 1D data-driven grouping (1D-DDg) method (47, 48). This method allows the identification of the prognostic variables (e.g., gene-expression values or microarray hybridization signal intensity values) that stratify patients into different risk groups (SI Materials and Methods and Fig. S5). We found 70 significant survival genes (log-rank test, P < 0.01) in hBCSGs whose expression levels were characterized by their transcripts (detected by Affymetrix probe sets) (Dataset S7A). About two-thirds of these genes showed tumor-suppressive–like behavior, and higher expression levels were associated with better prognosis (Dataset S7B). Functionally, these genes may be considered as tumor suppressors. When we increased the stringency to log-rank P < 1.54 × 10−5, we observed 21 high-confidence survival-prognostic hBCSGs. Among these 21 genes, 14 (CD44, FGFR2, FLNB, KAT6A, MPHOSPH8, NCOA1, NF1, PCBP2, PIK3R1, RERE, STAT5B, TNKS, WASF2, and ZMYND11) were proposed to act in BC as tumor suppressors, and seven (ADORA2B, BCR, CNOT1, MAPRE1, NRAS, PARD3, and PDS5A) were proposed to act as pro-oncogenes (Dataset S7 A and B).
Next, we evaluated the prognostic value of these 1D-DDg–selected BCSGs as part of a multigene prognostic signature using the 2D-DDg method (48). The 2D-DDg method is a way to identify the most survival significant and synergistic pairs of the 1D-DDg predictors that are able to predict patients’ DFS as low- or high-risk subgroups. The most significant 2D-DDg predictors were subjected to a statistically weighted voting grouping (SWVg) method, which combines the results of the survival stratification prediction of the patients based on the 2D-DDg variables to build a more integrative, robust, and discriminative survival prediction model (SI Materials and Methods and Fig. S5) (48, 49).
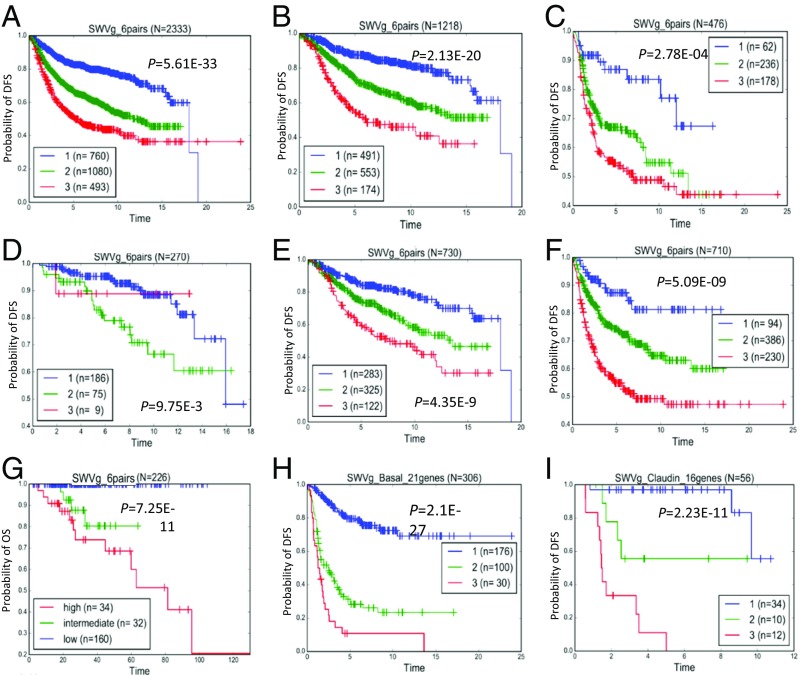
Thus, from the 70 survival-significant genes at P < 0.01, we identified a prognostic classifier comprising six gene pairs (TNKS–WASF2, FLNB–NRAS, NCOA1–RERE, MAPRE1–STAT5B, PARD3–ZMYND11, and ADORA2B–FGFR2) (Fig. 5A, Fig. S6, and Dataset S7C) that stratified our combined metadata cohort of 2,333 BC patients (Dataset S6 D and E) into three prognostic groups with high confidence (P = 5.6 × 10−33) (Fig. 5A).
Fig. 5.
Survival stratification based on SWVg analysis. (A) The six-gene-pair BC prognostic classifier found three BC subclasses in the 2,333 patients of the metadataset. The classifier was specified for the prediction of disease-free survival (DFS) time-to-event prediction. (B and C) Risk subgroups within ER+ (n = 1218) (B) and ER− (n = 476) (C) BC patients. (D–F) Risk subgroups within histological grade 1 (HG1; n = 270), histological grade 2 (HG2; n = 730), and histologic grade 3 (HG3; n = 710) patients, respectively. (G) Validation of the six-gene-pair BC prognostic classifier. SWVg found three BC (mostly invasive ductal carcinoma) subclasses in 226 TCGA BC patients who received systemic therapy (hormone therapy, chemotherapy, and combine therapy). OS data was available and used in this analysis. (H) The 21-gene prognostic signature found three distinct basal-like BC subtypes in the 306 patients of the metadataset. (I) The 16-gene prognostic signature found three distinct claudin-low BC subtypes in 56 patients of the metadataset.
We then performed SWVg analysis for the 12 individual genes that had formed the six pairs in our prognostic risk classifier. Our results revealed that, discretely, these 12 genes were unable to stratify the high- and intermediate-risk group patients, and the discrimination ability of 12 individual genes [defined by –log (P value)] was essentially smaller than that of the original six-gene-pair prognostic classifier [P = 8.22E−22 (Fig. S7) vs., P = 5.6 × 10−33 (Fig. 5A)]. Thus, the six-gene-pair prognostic classifier provides a significant synergistic/interaction effect from the specific combinations of the gene pairs selected by SWVg, leading to high-confidence partitioning of BC patients into three distinct risk subgroups. The details of the genes from the six-gene-pair prognostic classifier are presented in Dataset S7 C, 3.
To assess the robustness of the six-gene-pair BC prognostic classifier and its parameters, we applied our methods to five randomly derived and mutually independent patient subgroups with the same sample size. For each set, the six-gene-pair prognostic classifier stratified patients into three distinct risk subsets (Fig. S8) with statistical significance in all analyses (P ≤ 9E-08). The parameters of 1D-DDg and 2D-DDg (e.g., the expression cutoff values discriminating patients into the different risk subgroups) were relatively robust across the randomly sampled subsets.
Robustness of the Six-Gene-Pair Classifier in Clinicopathological BC Subgroups.
We applied the six-gene-pair prognostic model signature separately to the patients with ER+ and ER− tumor status and to patients with tumors of different histologic grades. Using the original parameters (cutoff gene expression value, survival patterns), of the six-gene-pair BC prognostic classifier derived by SWVg using the BC patient metadata, we found low-, moderate-, and high-risk subgroups within ER+ patients (Fig. 5B) and even in ER− patients (Fig. 5C) in the BC metadataset. Similarly, using histologic grading classification, we found low- and moderate-risk subgroups among patients with histological grade 1 (HG1) (Fig. 5D) and low-, moderate-, and high-risk subgroups among patients with histological grade 2 (HG2) (Fig. 5E) and histological grade 3 (HG3) (Fig. 5F) BC.
Thus, our prognostic model discriminates the patients in the risk groups within each of these clinical categories without the need for retraining or additional optimization. These findings support our basic strategy of identifying the causal genes and their expression patterns that drive cancer initiation and progression.
Reproducibility of the Six-Gene-Pair Prognostic Classifier.
To assess the reproducibility of the six-gene-pair BC prognostic classifier, we analyzed gene expression and clinical data of BC patients from the Cancer Gene Atlas (TCGA) database. Using the Agilent microarray data and OS data of these patients, we applied the prognostic workflow model used for the 2,333 patients of metadata cohort (Fig. S5). The 1D-DDg, 2D-DDg, and SWVg analyses all resulted in high-confidence prognostication of the BC patients using this six-gene-pair classifier (see Fig. S15 and see Dataset S9 C and E). SWVg specified three high-confidence prognostic groups (Fig. 5G).
Identification of Gene Signatures to Stratify Prognosis in Basal-Like and Claudin-Low BC Tumor Subtypes.
To determine whether these hBCSGs could further stratify patients within a specific BC subtype, we analyzed the expression profiles of 306 basal-like and 56 claudin-low tumor samples (Dataset S7D) with 1D-DDg analysis to define the prognostically significant hBCSGs in each cohort (Fig. S5 and Dataset S7 E and F). Then we used SWVg (49), as described in Fig. S5, to define the optimal number of 1D-DDg–defined hBCSGs (Dataset S7 E and F) in new prognostic signatures (SWVg predictors) that would categorize patients with basal-like (Fig. S9 and Dataset S7G) and claudin-low BC subtypes (Fig. S10 and Dataset S7H) into three risk subgroups. Both the 21-gene and 16-gene BC prognostic signatures identified three distinct subtypes in the 306 patients with basal-like BC (Fig. 5H) and in the 56 patients with claudin-low BC (Fig. 5I) in the metadataset, allowing us to define prognostic signatures for both subtypes (Dataset S7 G and H). Figs. S11 and S12 show the basic statistical characteristics of the individual genes of the prognostic signatures, across three risk groups of patients with the basal-like and claudin-low BC subtypes, respectively. The trends of the mean value across prognostic subgroups specify pro-oncogenic or tumor-suppressor–like expression patterns, defined by the 1D-DDg method for the prognostically significant genes.
We noted that there were six common genes (RERE, CLPTM1, WNK1, CD44, TCF12, and PTEN) between the basal-like and claudin-low BC prognostic signatures (Dataset S7I). However, only two (WNK1 and TCF12) exhibited similar 1D-DDg–defined (pro-oncogenic) functional patterns, possibly indicating common driver genes. Comparative analysis of the expression data of these genes among the three subgroups predicted by SWVg agreed with the results of the 1D-DDg analysis (Figs. S11 and S12). In contrast, CLPTM1, RERE, PTEN, and CD44 had different functional prognostic patterns (Figs. S11 and S12). Fifteen genes (NEDD4, GRLF1, RASA1, ST5, STAG1, PDS5A, GAB1, NCOA3, CGGBP1, MYLK, MAU2, RNF111, LUZP1, FLNB, and WAC) were present only in the basal-like tumor subtype prognostic signature, and 10 genes (PARD3, TAOK1, STAT5B, FGFR2, FNDC3A, NCOA1, STAU1, MBTPS1, TRIM33, and PUM1) were present only in the claudin-low tumor subtype prognostic signature (Fig. S13).
Using the SurvExpress tool (50), we confirmed the reproducibility and robustness of both gene signatures with high-confidence stratification of the TCGA BC cohort (502 patients) into three risk groups (Fig. S14 A and B). Note that TCGA data were derived using an Agilent microarray and data for OS time as an endpoint of disease outcome. These findings support the technical reproducibility, biological importance, and clinical significance of these hBCSG-defined predictors.
Univariate and Multivariate Analyses.
To confirm the validity of our six-gene-pair prognostic classifier, we carried out univariate and multivariate analysis of the SWVg-derived prognostic classifiers using clinical prognostic variables (ER, PGR, and lymph node status; tumor mass; and stage) (Dataset S8). Using clinical and microarray data of the 2333 BC patient and also results of our six-gene-pair patient's stratification (Dataset S8A), the univariate and multivariate analyses shown strong statistical significance and highest confidence values for our signature compared with other prognostic factors (Dataset S8 B and C). The multivariate regression of our 21-gene prognostic signature for basal-like BC was similarly significant, even after adjusting for commonly used clinical indicators (Dataset S8D). However, the results for our 16-gene prognostic classifier for claudin-low BC were not informative because of the small number of samples with data for calculating the SWVg categories.
Finally, we validated our six-gene-pair prognostic classifiers using an independent dataset of 226 BC patients from the TCGA database, which includes microarray expression, several clinical prediction factors, and systemic therapy information (SI Materials and Methods and Dataset S9A). Results of the 1D-DDg, 2D-DDg, and SWVg based classifiers (Dataset S8 C–E) were used as the input data sets for univariate and multivariate analyses. The significance was confirmed with the univariate and multivariate analyses, which showed the high significance and prevalence of the SWVg-derived six-gene-pair prognostic classifier, independent of most of the clinical factors and systemic treatment methods (Fig. S15 and Dataset S9 B, F, and G).
The hBCSG Subset as a Source of BC Prognostic Genes.
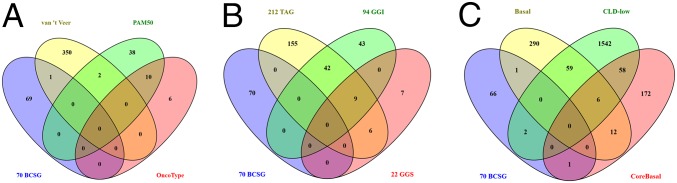
Overall, we identified 70 prognostic genes from our hBCSG list; with the exception of GLI3, none of these genes on our list hBCSGs appears on the commercial prognostic signatures (Fig. 6A and Dataset S7J) (51–53). Furthermore, these 70 hBCSGs are mostly unrelated to cell cycle/mitosis or genes in the oncogenic pathway (Fig. 6B and Dataset S7J) (12, 14) and are not common among other highly prognostic signatures for the stratification of basal-like and claudin-low BC subtypes (Fig. 6C and Dataset S7J) (3, 54, 55). We propose that our hBCSG-defined predictors contain highly prognostic and significant BC subtyping biomarkers.
Fig. 6.
The 70 prognostic hBCSGs are (A) mostly unique (69/70) in hBCSGs compared with known commercial prognostic signatures; (B) mostly unrelated to cell cycle/mitosis and genetic grading oncogenic pathway; and (C) not common, with high-prognostic signatures for stratification of basal-like and claudin-low BC subtypes.
Discussion
SB transposon mutagenesis is an unbiased approach for identifying candidate BC driver genes. We successfully induced mammary tumors in mice using the K5 promoter driving SB alone or together with stabilized N-terminally truncated β-catenin targeted to the basal layer of the mammary gland (28). Because the K5-Cre promoter is activated in both the luminal and basal cell layers of the mammary gland, transposition also occurs in both layers and not solely in the basal cell layer, as initially observed with the transgenic line expressing the truncated β-catenin. Not surprisingly, mammary tumors induced by our SB system represented all BC histological subtypes, consistent with the premise that the cell of origin for BC derives from either the luminal or basal layers of mammary glands (56, 57).
The SB mouse model provides a unique experimental basis for the identification of BC-associated susceptible genes relevant to the tumor subtypes. To understand the molecular subtypes of tumors induced in our transposon screen, we performed nonsupervised clustering of the expression profiles of tumors together and in combination with a collection of 394 murine expression profiles of other transgenic models of mammary tumors. Mouse mammary tumors could be regrouped into four clusters corresponding to human molecular subtypes: (i) a ductal cluster similar to the human luminal A subtype; (ii) a glandular cluster similar to the human luminal B subtype; (iii) a mesenchymal cluster analogous to the human basal-like and claudin-low subtypes; and (iv) a Neu cluster bearing closest similarity to the human HER2 subtype. Our transposon-driven tumor samples were distributed into all four clusters, indicating that mutagenesis drove the initiation and progression of mammary tumors from the different lineages. In contrast, β-catenin–driven tumor samples were restricted to the mesenchymal cluster, as expected from a previous study (28). Other tumor models, such as Myc- and Etv6-NTRK3–driven tumors, also induce all tumor molecular subtypes, whereas other tumor models produce a more limited set. For instance, Neu-driven tumors are uniquely restricted to the Neu cluster, and Brca1 conditional knockout and p53+/− transgenic mice do not form tumors of the glandular molecular subtype. From these and similar comparisons, it is evident that our transposon screen is sufficient to drive the initiation and progression of tumors within the four major molecular subtypes.
We identified 129 CIS genes in our screen and found 126 orthologous human genes that represented possible hBCSGs. Functional annotation of their characteristics was undertaken by translating the observations from the mouse to human BCs. The highest number of insertions was found in NF1, a well-known tumor suppressor reported by the TCGA consortium to be deleted or mutated in ∼30% of human BCs. Similarly, Pten, of which the human ortholog is a known BC gene, was another CIS gene with frequent insertions. The identification of these known tumor suppressors supports the validity of our screen to uncover driver genes in human BC progression. Further pathway analyses (PANTHER and DAVID analyses) highlighted several enriched pathways known to play key roles in the oncogenesis of BCs: angiogenesis, cytoskeleton, and signaling, such as the PI3K (58), EGFR (59, 60), IL (61), and MAPK (62) signaling pathways.
We constructed a protein interaction network of the 126 hBSCGs and identified a network of genes, including FGFR2, GNAQ, NRAS, NCOA3, NF1, PIK3R1, and PTEN, that is organized around EP300. We propose that the complex crosstalk among the hBCSGs may be critical for BC initiation and progression. Interestingly, we found several cell membrane-associated genes (BMPR1A, TGFBR1, FGFR2, PARD3, CD44, and P2RY2) that may be organized into a pro-oncogenic network around EP300. For example, BMPR1A, a member of the TGFB receptor superfamily, has been implicated as a tumor suppressor in ovarian tumor development (63), and an inactivating mutation in BMPR1A has been reported to cause juvenile polyposis (64, 65). Another membrane-associated gene, CD44, interacts with EP300 and contributes to the chemoresistance of BC cells (66); furthermore, CD44 regulates the ERK, AKT, and Hippo pathways in cell-cycle progression and in the maintenance of tumor-initiating cells (67). These different membrane-associated genes could be organized in a pro-oncogenic network around EP300, and the crosstalk among them may provide clues for the development of combinatorial targeted therapies (Dataset S10). The interconnectedness of the hBCSGs whose mutational profiles and expression patterns are significant for survival suggests an attractive paradigm to guide the design of stratified cancer development and outcome prognosis and of precise therapeutic strategies.
Gene-expression signatures are used to select patients likely to respond to adjuvant systemic therapy. For instance, MammaPrint and OncotypeDX are two commercially available prognostic platforms for BC, based on the 70-gene Amsterdam signature (68) and a 21-gene signature (52), respectively. Other signatures, such as the Rotterdam 60- and 76-gene signatures, also have been developed for prognostication (69). However, the predictive abilities of these signatures are limited to specific BC patient groups, which are predominantly ER+/PR+ and lymph node-negative, with a prognostic time extrapolation of less than 5 y after diagnosis. In contrast, our prognostic classification model is based on the initiation of explicit malignancy genes, includes hBCSGs, and can be tested, refuted, or confirmed.
According to our paradigm, multiple BC susceptibilities, related to the development of different tumor subtypes, can be involved in the initiation of pro-oncogenic or possible tumor-suppression pathways in normal breast epithelium cells. To specify such multigene subsets, our feature selection and prognostic pipeline, including the 1D-DDg, 2D-DDg, and SWVg methods, selects the most significant prognostic variables (expressed genes) and their critical cutoff values, optimizes the number of genes or gene pairs, and differentiates their expression patterns with significance, robustness, and synergistic disease-outcome prognostic ability. Intriguingly, of the 12 genes, three—MAPRE1 (3, 70, 71), NRAS (3, 72, 73), and STAT5B (14)—are found in other multigene signatures, characterizing certain BC subclasses and the gene expression patterns associated with BC treatment response.
The six-gene-pair classifier is also significant in univariate and multivariate analyses in very heterogeneous datasets and can prognose independent TCGA cohorts that use a different RNA expression platform (Agilent Microarray Technology) and a more stringent disease-outcome event (OS time). Indeed, these univariate and multivariate analyses—including clinical, prognostic, and predictive factors—revealed the independent and reproducible prognostic value of our molecular classifier in stratifying TCGA BC patients receiving systemic postsurgical therapy. Our six-gene-pair classifier also provided highly confident and reproducible stratification of patients into three risk groups within histological grades 1–3 of clinically defined cancer aggressiveness and into subgroups by treatment-predictive status (ER+ or ER−). Thus, the predictive ability of our signature is scalable within current clinical classifications and treatment groups. These findings can provide actionable insights for prognosis and treatment of BCs.
The identification of prognostic gene pairs suggests a functional and structural interconnectedness among BCSGs that perhaps modifies the biological behavior of the tumor or creates an “interaction effect” that influences prognosis. This hypothesis was supported by our network and statistical analyses. In the FLNB–NRAS gene pair, high FLNB expression and low NRAS expression is linked with a good prognosis, indicating a tumor-suppressive function of FLNB and an oncogenic function of NRAS in BC. The role of FLNB in breast tumorigenesis is largely unknown. FLNB, which functions as an F-actin cross-linking protein, undergoes a high frequency of skipping exon events in luminal cell lines compared with basal-like cells (74). Others have shown that FLNB suppresses tumor growth and metastasis by regulating the activity of matrix metalloproteinase 9 (MMP-9) and the secretion of VEGF-A, which is mediated by the RAS/ERK pathway. Specifically, Flnb deficiency in mouse embryonic fibroblasts results in increased proteolytic activity of MMP-9 and cell invasion through RAS/ERK signaling. Similarly, silencing FLNB in multiple human cancer cells increases the proteolytic activity of MMP-9 and tumor cell invasion (75).
In the NCOA1–RERE pair, high expression of both genes is associated with a favorable prognosis, indicating the tumor-suppressive functions of both genes in BC development. Consistently, patients with breast tumors with high NCOA1 expression have significantly longer OS and disease-free intervals (76). Although the function of RERE in BC is unclear, its overexpression enhances apoptosis and activates cell death (77), indicating a putative tumor-suppressor function.
Regarding MAPRE1–STAT5B, a good prognosis is linked with high STAT5B expression and low MAPRE1 expression, suggesting that STAT5B has a tumor-suppressive function and that MAPRE1 has an oncogenic function. MAPRE1 can act as a potential oncogene via activation of the β-catenin/TCF pathway to promote cellular growth and inhibit apoptosis (78). The role of STAT5 is less clear, because the loss of Nuc-pYStat5 is associated with poor prognosis in node-negative BC (79), but the inhibition of STAT5B also can block tumor growth (80). In other cancer types, such as gastric cancer, both STAT5B and MAPRE1 are characterized as oncogenic proteins (81, 82). Thus, the effect of STAT5B in cancers is likely to be contextual and dependent on the tumor (sub)type and perhaps on other gene alterations within the network.
In the PARD3–ZMYND11 pair, high ZMYND11 expression along with low PARD3 expression favors a good prognosis, supporting a tumor-suppressive function of ZMYND11 in such expression pattern combinations. PARD3 and ZMYND11 are also found in our designed tumorigenic gene network. ZMYND11 acts as a repressor of a transcriptional program that is essential for tumor cell growth (83) and is mutated in human cancers; its low expression in BC is correlated with worse prognosis. ZMYND11 overexpression inhibits the growth of different cancer cell types in vitro and breast tumorigenesis in mice (83). PARD3, on the other hand, controls cell polarity and contributes to cell migration and proliferation. Inhibiting PARD3 causes a loss of cell polarity and induces breast tumorigenesis and metastasis (84, 85). Others suggest that PARD3 activates YAP/TAZ to promote cell growth (86) and may function as an oncogene (87). Thus, PARD3 likely has dual cancer type-specific functions. It encodes multiple protein isoforms with varying regulatory functions; however, their roles in BC prognosis are poorly studied. According to our single-gene prognostic analysis, ZMYND11 and PARD3 provided strongly significant tumor-suppressive and pro-oncogenic prognostic patterns (at P < 5 × 10−6), respectively. As a prognostic gene pair, PARD3–ZMYND11 could categorize BC patients into two groups [Kaplan–Meier (KM) functions at P = 8.3 × 10−12].
Finally, for ADORA2B–FGFR2, a good prognosis is found with high FGFR2 expression and low ADORA2B expression. These findings indicate the tumor-suppressive function of FGFR2 and the pro-oncogenic prognostic function of ADORA2B. FGFR2 is a known BCSG (86). Pharmacological blockade of ADORA2B has been shown to inhibit the invasion of BC cells and reduce tumor outgrowth in the lungs (88), suggesting the pro-oncogenic prognostic potential of ADORA2B.
Overall, our six-gene-pair signature can stratify BC patients into three different risk subgroups with high confidence and reproducibility, and the gene pairs provide highly reliable predictive factors and clues to the interconnectedness between genes driving BC progression.
Although most of the clinically used signatures are strong risk predictors in the early follow-up intervals for low-grade, ER+/PR+, or HER2+ tumors, there is an urgent need to improve risk stratifications for long-term prognosis and for high-grade and triple-negative BCs. Through an analysis of basal-like and claudin-low tumor subtypes, we identified the most representative prognostically significant genes from among the 126 hBCSGs that could stratify patients into three distinct risk groups with high confidence. Our 21-gene and 16-gene signatures could stratify patients into three distinct risk subgroups for basal-like and claudin-low BC subtypes, respectively. Interestingly, in the different patient groups, the prediction signatures of alternative isoforms of a same gene could be included and play alternative roles in the context of disease outcome prognosis. For instance, according to 1D-DDg and SWVg, the RERE isoform defined by the 200939_s_at probe sets demonstrated a tumor-suppressor–like prognostic pattern in a metacohort (2,333 patients) with a basal-like BC subtype; however, in patients with the claudin-low BC subtype, another isoform of the RERE (defined by the 221643_s_at probe sets) was computationally selected and showed a significant oncogenic-like expression pattern. Comparing our 21- and 16-gene signature lists, we observed five additional common genes (CLPTM1, WNK1, CD44, TCF12, and PTEN). However, of these, only WNK1 and TCF12 showed common prognostic patterns. This commonality suggests that these subtypes may share similar genetic pro-oncogenic BCSG drivers, consistent with their association with triple-negative BCs. This hypothesis needs further study and validation.
In sum, unbiased forward genetic screens in mice can reveal functionally important gene networks that do not critically depend on highly variable and rapidly evolving genomic alteration profiles and transiently actionable point mutations. This method should complement strategies that rely on deep genomic sequencing and lead to therapeutic strategies that are more generic than those that rely on a limited number of mutations. These survival-significant BCSGs, their expression patterns identified in the current study, and the gene-based networks with signatures for stratifying BC risk groups could provide valuable information for understanding the genetic basis of breast tumorigenesis and tumor progression as well as for developing promising targeted therapeutics for BC treatment.
Materials and Methods
We used the following alleles to generate a mouse mammary tumor model: C57BL/6-K5-ΔN57-βcat (28), K5-Cre (29), T2Onc2 (6113) (89), and Rosa26-LSL-SB11 (90). The mouse-breeding scheme is shown in Fig. S1. All animals were genotyped and monitored monthly. The palpable mammary tumors were collected, and the samples were snap-frozen or fixed in formaldehyde and sent to the histopathology core facility of the Institute of Molecular and Cell Biology for paraffin embedding. One veterinarian pathologist (J.M.W.) and the senior principal investigator (J.P.T.) reviewed the H&E-stained sections for histotype annotation (Fig. S3). All procedures were carried out according to Institutional Animal Care and Use Committee guidelines of Biological Resource Centre at Agency for Science, Technology, and Research, Singapore. Approve no. 070238.
Methods for identifying transposon insertion sites, transcriptomic analysis of SB mammary tumors and data processing, subtyping of mouse mammary tumors and human BCs, metadata for identifying and validating the survival-significant genes, univariate and multivariate survival prediction, statistical tests, and software are presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported in part by a core grant from the Biomedical Research Council, Agency for Science, Technology, and Research, Singapore and The Cancer Prevention Institute of Texas (CPRIT) (to J.P.T.). N.G.C. and N.A.J. are CPRIT Scholars in Cancer Research. Bioinformatics Institute teamwork was supported by the core budget of the Bioinformatics Institute, Agency for Science, Technology, and Research, Singapore.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701512114/-/DCSupplemental and at web.bii.a-star.edu.sg/archive/vk/pnas_codes.zip.
References
- 1.DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin. 2011;61(6):409–418. doi: 10.3322/caac.20134. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtis C, et al. METABRIC Group The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fadoukhair Z, et al. Evaluation of targeted therapies in advanced breast cancer: The need for large-scale molecular screening and transformative clinical trial designs. Oncogene. 2016;35(14):1743–1749. doi: 10.1038/onc.2015.249. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X, et al. Systematic assessment of prognostic gene signatures for breast cancer shows distinct influence of time and ER status. BMC Cancer. 2014;14:211. doi: 10.1186/1471-2407-14-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall JM, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250(4988):1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 8.Wooster R, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265(5181):2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 9.Osborne RJ, et al. Mutations in the p53 gene in primary human breast cancers. Cancer Res. 1991;51(22):6194–6198. [PubMed] [Google Scholar]
- 10.Nevanlinna H, Bartek J. The CHEK2 gene and inherited breast cancer susceptibility. Oncogene. 2006;25(43):5912–5919. doi: 10.1038/sj.onc.1209877. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aswad L, et al. Genome and transcriptome delineation of two major oncogenic pathways governing invasive ductal breast cancer development. Oncotarget. 2015;6(34):36652–36674. doi: 10.18632/oncotarget.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivshina AV, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66(21):10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 14.Sotiriou C, et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 15.Petrocelli T, Slingerland JM. PTEN deficiency: A role in mammary carcinogenesis. Breast Cancer Res. 2001;3(6):356–360. doi: 10.1186/bcr322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Closas M, Chanock S. Genetic susceptibility loci for breast cancer by estrogen receptor status. Clin Cancer Res. 2008;14(24):8000–8009. doi: 10.1158/1078-0432.CCR-08-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens PJ, et al. Oslo Breast Cancer Consortium (OSBREAC) The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beerenwinkel N, et al. Genetic progression and the waiting time to cancer. PLOS Comput Biol. 2007;3(11):e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.March HN, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet. 2011;43(12):1202–1209. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller DG. On the nature of susceptibility to cancer. The presidential address. Cancer. 1980;46(6):1307–1318. doi: 10.1002/1097-0142(19800915)46:6<1307::aid-cncr2820460602>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 21.Schinzel AC, Hahn WC. Oncogenic transformation and experimental models of human cancer. Front Biosci. 2008;13:71–84. doi: 10.2741/2661. [DOI] [PubMed] [Google Scholar]
- 22.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323(5922):1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starr TK, et al. A Sleeping Beauty transposon-mediated screen identifies murine susceptibility genes for adenomatous polyposis coli (Apc)-dependent intestinal tumorigenesis. Proc Natl Acad Sci USA. 2011;108(14):5765–5770. doi: 10.1073/pnas.1018012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bard-Chapeau EA, et al. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat Genet. 2014;46(1):24–32. doi: 10.1038/ng.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27(3):264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann KM, et al. Australian Pancreatic Cancer Genome Initiative Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 2012;109(16):5934–5941. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahrmann EP, et al. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet. 2013;45(7):756–766. doi: 10.1038/ng.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teulière J, et al. Targeted activation of beta-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development. 2005;132(2):267–277. doi: 10.1242/dev.01583. [DOI] [PubMed] [Google Scholar]
- 29.Ramirez A, et al. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 2004;39(1):52–57. doi: 10.1002/gene.20025. [DOI] [PubMed] [Google Scholar]
- 30.de Ridder J, Uren A, Kool J, Reinders M, Wessels L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLOS Comput Biol. 2006;2(12):e166. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uren AG, et al. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat Protoc. 2009;4(5):789–798. doi: 10.1038/nprot.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace MD, et al. Comparative oncogenomics implicates the neurofibromin 1 gene (NF1) as a breast cancer driver. Genetics. 2012;192(2):385–396. doi: 10.1534/genetics.112.142802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madanikia SA, Bergner A, Ye X, Blakeley JO. Increased risk of breast cancer in women with NF1. Am J Med Genet A. 2012;158A(12):3056–3060. doi: 10.1002/ajmg.a.35550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 36.Brems H, Beert E, de Ravel T, Legius E. Mechanisms in the pathogenesis of malignant tumours in neurofibromatosis type 1. Lancet Oncol. 2009;10(5):508–515. doi: 10.1016/S1470-2045(09)70033-6. [DOI] [PubMed] [Google Scholar]
- 37.Saal LH, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40(1):102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCabe N, et al. Targeting Tankyrase 1 as a therapeutic strategy for BRCA-associated cancer. Oncogene. 2009;28(11):1465–1470. doi: 10.1038/onc.2008.483. [DOI] [PubMed] [Google Scholar]
- 39.Ngan E, Northey JJ, Brown CM, Ursini-Siegel J, Siegel PM. A complex containing LPP and α-actinin mediates TGFβ-induced migration and invasion of ErbB2-expressing breast cancer cells. J Cell Sci. 2013;126(Pt 9):1981–1991. doi: 10.1242/jcs.118315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, et al. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett. 2012;586(10):1409–1418. doi: 10.1016/j.febslet.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriarity BS, et al. A Sleeping Beauty forward genetic screen identifies new genes and pathways driving osteosarcoma development and metastasis. Nat Genet. 2015;47(6):615–624. doi: 10.1038/ng.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: Expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44(D1):D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motakis E, Ivshina AV, Kuznetsov VA. Data-driven approach to predict survival of cancer patients: Estimation of microarray genes’ prediction significance by Cox proportional hazard regression model. IEEE Eng Med Biol Mag. 2009;28(4):58–66. doi: 10.1109/MEMB.2009.932937. [DOI] [PubMed] [Google Scholar]
- 48.Motakis E, Kuznetsov VA. 2009. Genome-scale identification of survival significant genes and gene pairs. Wcecs 2009: World Congress on Engineering and Computer Science, Vols I and Ii, pp 41–46.
- 49.Tang Z, Ow GS, Thiery JP, Ivshina AV, Kuznetsov VA. Meta-analysis of transcriptome reveals let-7b as an unfavorable prognostic biomarker and predicts molecular and clinical subclasses in high-grade serous ovarian carcinoma. Int J Cancer. 2014;134(2):306–318. doi: 10.1002/ijc.28371. [DOI] [PubMed] [Google Scholar]
- 50.Aguirre-Gamboa R, et al. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8(9):e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korde LA, et al. Gene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancer. Breast Cancer Res Treat. 2010;119(3):685–699. doi: 10.1007/s10549-009-0651-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 53.van ’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 54.Sabatier R, et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res Treat. 2011;126(2):407–420. doi: 10.1007/s10549-010-0897-9. [DOI] [PubMed] [Google Scholar]
- 55.Taube JH, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107(35):15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Asselin-Labat ML, et al. Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor caspase-14. Mol Cell Biol. 2011;31(22):4609–4622. doi: 10.1128/MCB.05766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 58.Baselga J. Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. Oncologist. 2011;16(Suppl 1):12–19. doi: 10.1634/theoncologist.2011-S1-12. [DOI] [PubMed] [Google Scholar]
- 59.Lo HW, Hsu SC, Hung MC. EGFR signaling pathway in breast cancers: From traditional signal transduction to direct nuclear translocalization. Breast Cancer Res Treat. 2006;95(3):211–218. doi: 10.1007/s10549-005-9011-0. [DOI] [PubMed] [Google Scholar]
- 60.Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: A model for targeted therapy. Clin Cancer Res. 2006;12(18):5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 61.Guo Y, Xu F, Lu T, Duan Z, Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev. 2012;38(7):904–910. doi: 10.1016/j.ctrv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 62.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26(22):3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 63.Edson MA, et al. Granulosa cell-expressed BMPR1A and BMPR1B have unique functions in regulating fertility but act redundantly to suppress ovarian tumor development. Mol Endocrinol. 2010;24(6):1251–1266. doi: 10.1210/me.2009-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Calva-Cerqueira D, et al. Discovery of the BMPR1A promoter and germline mutations that cause juvenile polyposis. Hum Mol Genet. 2010;19(23):4654–4662. doi: 10.1093/hmg/ddq396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howe JR, et al. The prevalence of MADH4 and BMPR1A mutations in juvenile polyposis and absence of BMPR2, BMPR1B, and ACVR1 mutations. J Med Genet. 2004;41(7):484–491. doi: 10.1136/jmg.2004.018598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bourguignon LY, Xia W, Wong G. Hyaluronan-mediated CD44 interaction with p300 and SIRT1 regulates beta-catenin signaling and NFkappaB-specific transcription activity leading to MDR1 and Bcl-xL gene expression and chemoresistance in breast tumor cells. J Biol Chem. 2009;284(5):2657–2671. doi: 10.1074/jbc.M806708200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu S, et al. Adhesion glycoprotein CD44 functions as an upstream regulator of a network connecting ERK, AKT and Hippo-YAP pathways in cancer progression. Oncotarget. 2015;6(5):2951–2965. doi: 10.18632/oncotarget.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buyse M, et al. TRANSBIG Consortium Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98(17):1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 70.Chang HY, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS Biol. 2004;2(2):E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takahashi S, et al. Prediction of breast cancer prognosis by gene expression profile of TP53 status. Cancer Sci. 2008;99(2):324–332. doi: 10.1111/j.1349-7006.2007.00691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hannemann J, et al. Changes in gene expression associated with response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2005;23(15):3331–3342. doi: 10.1200/JCO.2005.09.077. [DOI] [PubMed] [Google Scholar]
- 73.Yu JX, et al. Pathway analysis of gene signatures predicting metastasis of node-negative primary breast cancer. BMC Cancer. 2007;7:182. doi: 10.1186/1471-2407-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shapiro IM, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7(8):e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bandaru S, et al. Targeting filamin B induces tumor growth and metastasis via enhanced activity of matrix metalloproteinase-9 and secretion of VEGF-A. Oncogenesis. 2014;3:e119. doi: 10.1038/oncsis.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Green AR, et al. The prognostic significance of steroid receptor co-regulators in breast cancer: Co-repressor NCOR2/SMRT is an independent indicator of poor outcome. Breast Cancer Res Treat. 2008;110(3):427–437. doi: 10.1007/s10549-007-9737-y. [DOI] [PubMed] [Google Scholar]
- 77.Waerner T, Gardellin P, Pfizenmaier K, Weith A, Kraut N. Human RERE is localized to nuclear promyelocytic leukemia oncogenic domains and enhances apoptosis. Cell Growth Differ. 2001;12(4):201–210. [PubMed] [Google Scholar]
- 78.Liu M, et al. EB1 acts as an oncogene via activating beta-catenin/TCF pathway to promote cellular growth and inhibit apoptosis. Mol Carcinog. 2009;48(3):212–219. doi: 10.1002/mc.20471. [DOI] [PubMed] [Google Scholar]
- 79.Peck AR, et al. Loss of nuclear localized and tyrosine phosphorylated Stat5 in breast cancer predicts poor clinical outcome and increased risk of antiestrogen therapy failure. J Clin Oncol. 2011;29(18):2448–2458. doi: 10.1200/JCO.2010.30.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Joung YH, et al. Enhancement of hypoxia-induced apoptosis of human breast cancer cells via STAT5b by momilactone B. Int J Oncol. 2008;33(3):477–484. [PubMed] [Google Scholar]
- 81.Kim K, et al. Epigenetic regulation of microRNA-10b and targeting of oncogenic MAPRE1 in gastric cancer. Epigenetics. 2011;6(6):740–751. doi: 10.4161/epi.6.6.15874. [DOI] [PubMed] [Google Scholar]
- 82.Nishigaki R, et al. Proteomic identification of differentially-expressed genes in human gastric carcinomas. Proteomics. 2005;5(12):3205–3213. doi: 10.1002/pmic.200401307. [DOI] [PubMed] [Google Scholar]
- 83.Wen H, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508(7495):263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCaffrey LM, Montalbano J, Mihai C, Macara IG. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell. 2012;22(5):601–614. doi: 10.1016/j.ccr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xue B, Krishnamurthy K, Allred DC, Muthuswamy SK. Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat Cell Biol. 2013;15(2):189–200. doi: 10.1038/ncb2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fletcher MN, et al. Master regulators of FGFR2 signalling and breast cancer risk. Nat Commun. 2013;4:2464. doi: 10.1038/ncomms3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iden S, et al. Tumor type-dependent function of the par3 polarity protein in skin tumorigenesis. Cancer Cell. 2012;22(3):389–403. doi: 10.1016/j.ccr.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 88.Desmet CJ, et al. Identification of a pharmacologically tractable Fra-1/ADORA2B axis promoting breast cancer metastasis. Proc Natl Acad Sci USA. 2013;110(13):5139–5144. doi: 10.1073/pnas.1222085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436(7048):221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 90.Dupuy AJ, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69(20):8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.