Abstract
The plant plastidial thioredoxins (Trx) are involved in the light-dependent regulation of many enzymatic activities, owing to their thiol-disulfide interchange activity. Three different types of plastidial Trx have been identified and characterized so far: the m-, f-, and x-types. Recently, a new putative plastidial type, the y-type, was found. In this work the two isoforms of Trx y encoded by the nuclear genome of Arabidopsis (Arabidopsis thaliana) were characterized. The plastidial targeting of Trx y has been established by the expression of a Trx∷GFP fusion protein. Then both isoforms were produced as recombinant proteins in their putative mature forms and purified to characterize them by a biochemical approach. Their ability to activate two plastidial light-regulated enzymes, NADP-malate dehydrogenase (NADP-MDH) and fructose-1,6-bisphosphatase, was tested. Both Trx y were poor activators of fructose-1,6-bisphosphatase and NADP-MDH; however, a detailed study of the activation of NADP-MDH using site-directed mutants of its regulatory cysteines suggested that Trx y was able to reduce the less negative regulatory disulfide but not the more negative regulatory disulfide. This property probably results from the fact that Trx y has a less negative redox midpoint potential (−337 mV at pH 7.9) than thioredoxins f and m. The y-type Trxs were also the best substrate for the plastidial peroxiredoxin Q. Gene expression analysis showed that Trx y2 was mainly expressed in leaves and induced by light, whereas Trx y1 was mainly expressed in nonphotosynthetic organs, especially in seeds at a stage of major accumulation of storage lipids.
Thioredoxins (Trx) are small (approximately 12 kD) ubiquitous proteins involved in thiol-disulfide exchange reactions (Buchanan, 1980; Holmgren, 1989). In higher plant cells, they are found in cytosol, mitochondria, and plastids (Schürmann and Jacquot, 2000). All are nuclear encoded irrespective of their subcellular localization. The availability of the Arabidopsis (Arabidopsis thaliana) complete genome sequence revealed that higher plant Trxs belonged to a multigene family. Several different Trx types have been distinguished on the basis of sequence identity and intron positions, with several isoforms within each type (Meyer et al., 2002). The situation is particularly complex for plastidial Trx (cpTrx), where four different types of Trx have been found. Besides the long-time known thioredoxins m and f comprising 4 and 2 members respectively, a third type of cpTrx, named the x-type, has been recently studied (Collin et al., 2003) and found to be a very good reductant of plastidial 2-Cys peroxiredoxins (Prxs), whereas having almost no activity with the classical target enzymes of cpTrx m and f, i.e. NADP-malate dehydrogenase (NADP-MDH) and Fru-1,6-bisphosphatase (FBPase). The fourth type has been identified quite recently (Lemaire et al., 2003) during the analysis of the genomes of Chlamydomonas reinhardtii and of the cyanobacterium Synechocystis. While only one isoform is present in the green alga and the cyanobacterium, two isoforms were found in Arabidopsis. They were missed in previous genome-wide searches due to incorrect annotations in the Arabidopsis database. When aligned with the sequence of cyanobacterial Trx y, the Trxs y of photosynthetic eukaryotes display N-terminal extensions that are predicted to be signal peptides for plastidial targeting.
In this work we have characterized the two Trx y isoforms of Arabidopsis. Their subcellular localization was established by a transient expression of Trx y fused with green fluorescent protein (GFP). Both Trx y isoforms were produced as recombinant proteins and purified in their mature forms. Their ability to activate FBPase and NADP-MDH was measured. Their putative roles in oxidative stress responses have been tested by measuring their abilities to serve as electron donors to different plastidial Prxs. Finally, their expression profiles have been examined. Taken together, the results strongly suggest their involvement in oxidative stress responses and that they are not involved in the regulation of carbon metabolism.
RESULTS
Arabidopsis Trxs y Are Targeted to Plastids
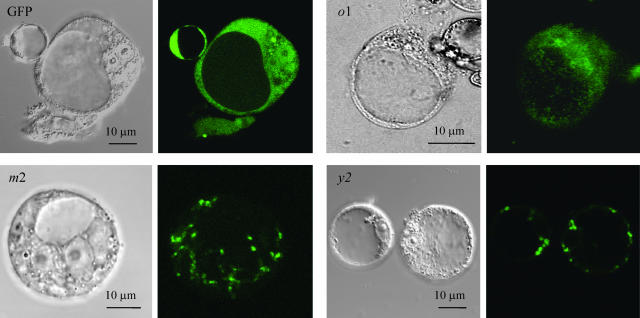
The cDNAs encoding thioredoxins y1 and y2 were predicted from the Arabidopsis genomic sequence, amplified by reverse transcription (RT)-PCR, and sequenced. The Trx y protein sequences deduced from the cDNA sequences confirmed the presence of an N-terminal extension in both isoforms when compared to the cyanobacterial Trx y. Analysis by different targeting prediction programs strongly suggested that Trxs y are plastidial thioredoxins. This point was verified by a transient expression system in Arabidopsis cell protoplasts. The cDNAs encoding pre-Trx y2 as well as the plastidial pre-Trx m2 and the mitochondrial pre-Trx o1 were fused with the cDNA encoding smGFP in a transient expression vector. The recombinant vectors were used to transform Arabidopsis cell protoplasts. The fluorescence of cells expressing Trx y2 fused with GFP was found in the same compartment as the fluorescence of cells expressing Trx m2 fused with GFP, i.e. the chloroplast (Fig. 1), whereas Trx o1∷GFP localized to punctuated compartments of smaller dimension and distinct of plastids. Considering the similarity of sequences of Trx y1 and Trx y2, and the presence of a transit peptide in both isoforms, it can be inferred that Trx y1 is also plastidial. This prompted us to check the activity of Trxs y exclusively with potential chloroplastic targets.
Figure 1.
Intracellular localization of putative cpTrxs. Subcellular localization of premature Trx-fused GFP proteins was monitored by confocal microscopy in Arabidopsis cell protoplasts. Trx types are indicated in the figures. For each Trx type transmission and green fluorescent images are shown side-by-side. GFP, nonaddressed GFP. o1, m2, y2, premature forms of the corresponding Trx isoforms fused to GFP.
The Plastidial FBPase Cannot Be Activated by Trx y
The putative mature Arabidopsis Trxs y1 and y2 were produced in a bacterial system. The recombinant proteins were then purified by anion-exchange chromatography and gel filtration and used to test the activation of plastidial FBPase. The effect of Trxs y on FBPase activation was compared to the effect of Trx f, which is well documented to be the most efficient activator of FBPase (Buchanan, 1980). No significant activation of this target enzyme was detected even at the highest test concentration (100 μm) of Trx y1 and y2 compared to Trx f1, which was efficient at 0.5 μm (data not shown).
NADP-MDH Is Partially Activated by Trx y
The activation assays of the plastidial MDH were performed on a recombinant sorghum (Sorghum vulgare) NADP-MDH. The activation efficiencies of Trxs y1 and y2 were compared to the activation obtained with the m-type Trx. A similar degree of activation of this target enzyme could be measured only at Trx y concentrations 3 to 6 times higher than the concentration used for Trx m (data not shown). The kinetic parameters of NADP-MDH activation reflected this poor efficiency. The half-saturation (S0.5) values were 250 μm and 400 μm for Trx y1 and Trx y2, respectively (data not shown), whereas the S0.5-value for Trx m4 was as low as 7 μm (Collin et al., 2003). The activation of the wild-type NADP-MDH by Trxs y was so slow that the t1/2 could not be determined, whereas the t1/2 for Trx m4, measured at its S0.5 Trx concentration, was 6 min. All these parameters confirmed that the y-type Trxs are inefficient activators of NADP-MDH.
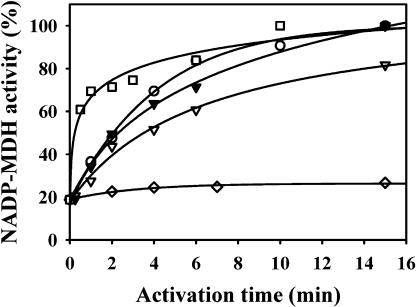
It is well known now that NADP-MDH activation requires the reduction of two different disulfides per monomer (the enzyme is a homo-dimer). One of the disulfides is located at the C-terminal extension that blocks the access to the active site (Ruelland and Miginiac-Maslow, 1999). It should be pointed out that activation of the enzyme also involves the Trx-dependent reduction of a third disulfide formed as an intermediate during the activation process. Redox potential determinations showed that the C-terminal disulfide is thermodynamically the most difficult to reduce, having a midpoint oxidation-reduction potential (Em) of −380 mV at pH 7.9 that is significantly more negative than the Em values of the other two regulatory disulfides (Hirasawa et al., 2000). In an attempt to identify the factors responsible for the inefficiency of y-type Trxs to activate NADP-MDH, we examined their ability to activate an NADP-MDH variant having no C-terminal disulfide, due to a site directed mutation of its two C-terminal Cys into Ser and no ability to form a C29-C207 transient disulfide, due to a C207A mutation (mutant MDH C207A/C365A/C377A; Ruelland et al., 1997). This mutated enzyme has a basal activity of about 20% in the oxidized form, increasing to 100% after reductive activation. The results clearly show that the y-type Trxs are able to activate this enzyme (Fig. 2). The activation is nearly as fast as the one obtained with Trx m1, a typical m-type Trx, but is not as efficient as the activation obtained with Trx f1. Apparently Trx y is able to reduce the N-terminal disulfide of the mutant NADP-MDH.
Figure 2.
Efficiencies of various plastidial thioredoxins in the activation of a mutant NADP-MDH where only the N-terminal disulfide is left (mutant C207A/C365A/C377A). Mutant NADP-MDH was activated in the presence of either 10 mm DTT (⋄), or DTT plus 20 μm Trxs f1 (□), or m1 (○), or y1 (▿), or y2 (▾). Aliquots (5 μg of enzyme) were withdrawn periodically for activity measurements.
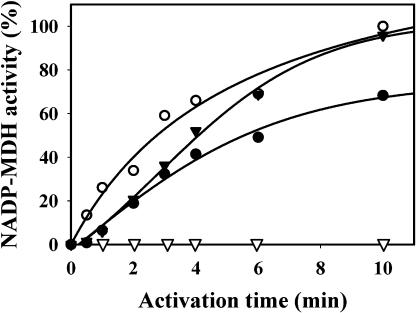
To verify the Trx y function in reduction of the N-terminal disulfide of the wild-type NADP-MDH as well, the mutant enzyme was preincubated with reduced Trx y1 (a treatment that does not confer any activity on the wild-type enzyme) before being activated by Trx f1 (Fig. 3). The concentration of Trx f1 used was suboptimal to slow down NADP-MDH activation. When the two Trx types were present together in the activation medium, without preincubation, the activation rate of NADP-MDH was higher than with Trx f1 alone. Thus, in the presence of Trx f1, Trx y participates in the activation process. When NADP-MDH was preincubated with Trx y1 for 10 min before the addition of Trx f1, the activation of NADP-MDH was faster than without preincubation and was devoid of the typical lag phase. This can be interpreted as a reduction of the N-terminal disulfide by Trx y1 during the preincubation, without any gain in activity, followed by the reduction of the C-terminal disulfide by Trx f, leading to activation. A similar effect was observed by Hatch and Agostino (1992) who could accelerate the Trx-dependent MDH activation by a pretreatment with high concentrations of mercaptoethanol that did not activate the enzyme; this led them to propose the existence of a “preregulatory” disulfide at a time where the existence of two different regulatory disulfides had not been yet demonstrated.
Figure 3.
Activation kinetics of wild-type sorghum NADP-MDH with 1 μm Trx f1, with or without 20 μm Trx y1. Trx y1 alone (▿); Trx f1 alone (•); Trx f1 and Trx y1 added at the beginning of activation (▾); preincubation with DTT-reduced 20 μm Trx y1 for 10 min, then addition of 1 μm Trx f1 (○). Aliquots (5 μg of enzyme) were withdrawn periodically for activity measurements.
Trxs y1 and y2 Have Somewhat Less Negative Redox Potentials
As mentioned above, the C-terminal disulfide of NADP-MDH is the most difficult to reduce. The result obtained for mutant NADP-MDH activation suggested that Trx y might have a less negative redox potential than the other Trxs. Thus, the redox potentials of y-type Trxs were measured (Table I). The Em values of Trx y1 and y2 at pH 7.9, both of which were measured to be −337 mV ± 10 mV, were significantly less negative than the Em values of typical Trxs m and f, which are −357 mV for Trx m1 and −351 mV for Trx f1 (Collin et al., 2003). This difference in redox potentials is significant and might well account for the inefficiency of Trxs y in NADP-MDH activation.
Table I.
Midpoint redox potentials of Trx y1 and Trx y2 determined at pH 7.9
| Thioredoxin | Em at pH 7.9 |
|---|---|
| mV | |
| AtTrx y1 | −335 |
| AtTrx y2 | −337 |
The values are the average of at least 2 independent titrations, with average deviations of ±5 mV. They were independent of the total concentration of DTT and of redox equilibration time.
y-Type Trxs Are Efficient Proton Donors for Plastidial Prx Q
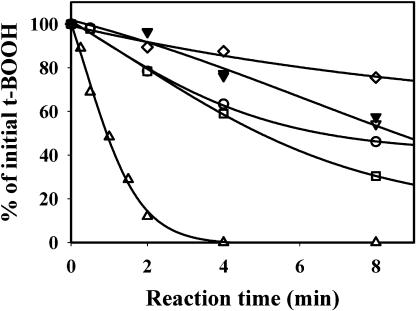
One of the physiological roles of Trx in antioxidant defense is electron donation to Prxs (Dietz, 2003). The ability of Trxs y to serve as electron donors to Prxs was tested on three different plastidial Prxs. The two first Prxs were classical 2-Cys Prxs: 2-Cys Prx A and 2-Cys Prx B (König et al., 2002). 2-Cys Prx B displayed a weak activity in the presence of the chemical reductant dithiothreitol (DTT; Fig. 4). This activity was slightly increased with Trx y1 or y2 to the same extent as with a typical Trx m (m1) or Trx f (f1). The maximal activity was obtained with Trx x, as previously described (Collin et al., 2003). The kinetics for 2-Cys Prx A obtained with the different Trxs were very similar to those obtained with 2-Cys Prx B (data not shown). Unlike Trx x, the y-type Trx were very poor substrates for the 2-Cys Prx.
Figure 4.
Activity of 2-Cys PRX B with various Trxs as proton donors. The enzyme (20 μm) 2-Cys Prx B was incubated with 400 μm DTT, 400 μm t-BOOH, and 10 μm Trx. The reduction of t-BOOH was measured by colorimetric titration and expressed as the remaining percentage of unreduced reagent. Control without Trx (⋄) or with Trx f1 (□), m1 (○), y1 (▿), y2 (▾), and x (▵). The actual initial rates of reduction, expressed in μmoles t-BOOH reduced min−1 μmol−1 Prx were, respectively, 0.37 (without Trx), 0.78 (Trx y1 or y2), 1.25 (Trx m1 or f1), and 10.00 (Trx x).
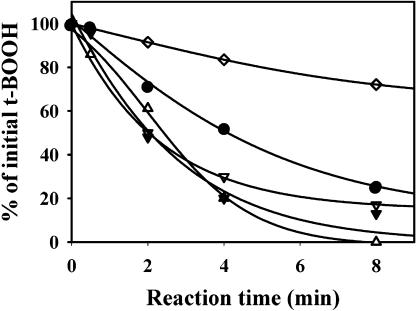
The third Prx tested was the plastidial Prx Q that is related to the bacterioferritin comigratory protein of bacteria (Horling et al., 2003). This Prx had also a weak activity in the presence of DTT (Fig. 5). The activity was increased with m-type Trxs, especially Trx m4, but the maximal activities were obtained in the presence of Trx x, Trx y1, and y2. As no significant differences could be observed between Trxs x and y on reaction kinetics, the different kinetic parameters of the reaction were measured (Table II). The Km value was close to a few micromolar for each Trx tested, but the lowest Km was obtained with y-type Trxs. The calculation of the catalytic efficiencies (kcat/Km) revealed that Prx Q activity was about 3 times higher with Trx y than with Trx x. Both y isoforms were equally efficient substrates for Prx Q with very close kcat/Km values.
Figure 5.
Activity of Prx Q with various Trxs as electron donors. Prx Q (5 μm) was incubated with 400 μm DTT, 400 μm t-BOOH, and 10 μm Trx. The reduction of t-BOOH was measured by colorimetric titration and expressed as the remaining percentage of unreduced reagent. DTT alone or Trx f1 (⋄), m4 (•), y1 (▿), y2 (▾), and x (▵). The actual initial rates of reduction, expressed in μmoles t-BOOH reduced min−1 μmol−1 Prx were, respectively, 2.0 (without Trx), 5.0 (Trx m4), 8.7 (Trx x), and 12.5 (Trx y1 or y2).
Table II.
Kinetic parameters of the chloroplastic peroxiredoxin Q with various chloroplastic thioredoxins
| Thioredoxin | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 (×103) | μm−1 s−1 (×103) | |
| AtTrx m4 | 8.4 ± 1.1 | 238 ± 83 | 29 ± 13 |
| AtTrx x | 5.7 ± 0.66 | 441 ± 79 | 77 ± 11 |
| AtTrx y1 | 1.0 ± 0.12 | 286 ± 78 | 283 ± 56 |
| AtTrx y2 | 2.6 ± 0.22 | 560 ± 19 | 215 ± 24 |
The determinations were done at a constant concentration of Prx (5 μm) and saturating t-BOOH (400 μm). The range of concentrations of Trx was varied from 1 to 30 μm for m4, 2 to 60 μm for x, and 0.5 to 12 μm for y1 and y2.
The Two Arabidopsis Trx y Isoforms Have Different Expression Profiles
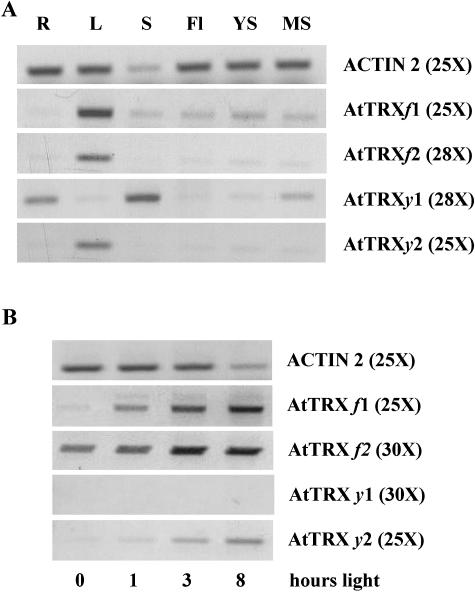
The presence of mRNAs encoding the two Trxs y was detected in different plant organs using RT-PCR (Fig. 6A). The mRNA encoding Trx y2 was detected in leaf extracts, whereas the mRNA encoding Trx y1 was detected in roots, mature siliques, and seed extracts. Light induction of gene expression was also tested in leaves (Fig. 6B). After 72 h of darkness the mRNAs for Trxs y were undetectable. After 1 h of illumination, the expression of Trx y2 was increased, whereas the mRNA of Trx y1 was still undetectable even after 8 h of illumination. Clearly, Trx y2 mRNA expression is very similar to the expression of typical chloroplastic thioredoxins, like Trx f1 and f2.
Figure 6.
Expression of Trx y genes in different plant organs and in dependence on light conditions. RNA levels were evaluated by semiquantitative RT-PCR using primers specific for Trx genes and reference gene (Actin 2). mRNA levels of Trx f isoforms are shown for comparison. A, RNA from R, roots; L, leaves; S, seeds; YS, young siliques; MS, mature siliques; and Fl, flowers were analyzed. B, After 72 h of dark adaptation, plants were transferred to white light and leaf RNA levels analyzed after 1, 3, and 8 h of exposure to light (180 μE m−2 s−1). Optimized number of PCR cycles is indicated in brackets.
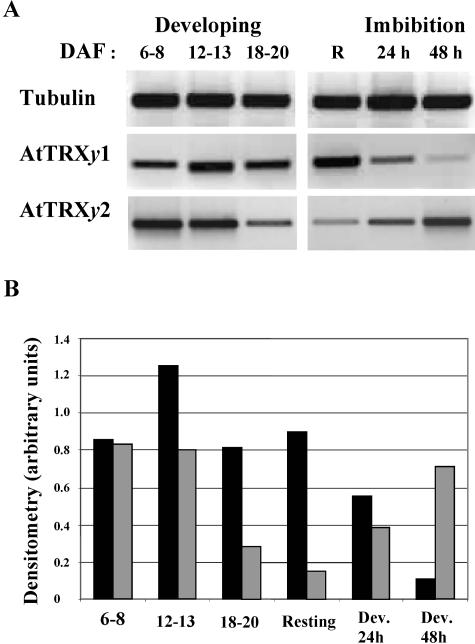
The presence of TRX y1 in mature siliques and seeds prompted us to examine more closely the expression of this Trx isoform in developing seeds. Typical stages of seed development have been chosen (Focks and Benning, 1998) corresponding to an early stage (6–8 d after flowering [DAF]), major accumulation of storage lipids (12–13 DAF), or final size, before desiccation (18–20 DAF). mRNA levels of TRX y1 increased at the storage lipids accumulation stage and decreased afterward, with a bell-shaped pattern. In contrast mRNA levels of y2 decreased steadily during seed development. A similar antagonistic variation was observed also during seed imbibition at the onset of germination where Trx y1 mRNA levels decreased and Trx y2 mRNA levels increased (Fig. 7). It should be noted that based on the Arabidopsis microarray database (www.genevestigator.ethz.ch) there exists a good correlation (r approximately 0.8) between mRNA levels of Prx Q and Trx y2 when tissue-specific expression or stress treatments are considered; high levels of Prx Q mRNA were usually correlated with high amounts of Trx y2, and low expression of both genes coincided as well, supporting the conclusion of a functional relationship between both redox proteins.
Figure 7.
Expression of y-type Trxs in Arabidopsis seeds. Semiquantitative RT-PCR with gene-specific primers for tubulin β 9 (constitutive), AtTRXy1, and AtTRXy2 were used to examine y-type-TRX gene expression in Arabidopsis developing seeds: 6 to 8, 12 to 13, or 18 to 20 DAF; in resting seeds (R); and in seeds after 24 and 48 h imbibition with water. A, Agarose gel. B, Densitometric analysis. Black bars, AtTRXy1; gray bars, AtTRXy2. A typical expression profile is presented.
DISCUSSION
The already large number of plant Trxs increased recently with the identification of Trx y in the genomes of Chlamydomonas and Arabidopsis. This study was devoted to the characterization of this new Trx type comprising two isoforms in Arabidopsis. The ability of recombinant Trxs to interact with known target proteins belonging to the same subcellular compartment gives a clue about possible physiological functions of these Trx. Thus, the first step of the study consisted of a localization of Trxs y.
Sequence comparison of Trxs y from Arabidopsis with the Trx y of the cyanobacterium Synechocystis PCC 6803, showing the presence of an N-terminal extension in the Arabidopsis proteins (Lemaire et al., 2003), suggested already a plastidial localization. This localization was firmly established in this study by a GFP-based approach. As three other types of Trx have also been localized in plastids, raising the total number of plastidial Trxs to nine, the question of the target specificity of Trx types in plastids becomes increasingly complex. As in our previous study (Collin et al., 2003), we produced and purified y-type recombinant proteins in their putative mature forms and tested their abilities to reduce selected plastidial target enzymes.
The first plastidial enzyme tested was FBPase, already known for its specific requirement for Trx f for activation. The preference was further accentuated in this study; both Trx y isoforms were very poor activators of FBPase, even at very high concentrations. Apparently, only the f-type Trx physiologically links activation of FBPase to photosynthetic electron transport.
The second plastidial enzyme tested was NADP-MDH, which is implicated in the transfer of excess reducing power from plastids to the cytosol (Scheibe, 2004). Trxs y proved to be very poor activators of this enzyme, with a S0.5 of several hundreds micromolar. These values are 25 times higher than the values obtained with classical m-type Trx and f-type Trx (Hodges et al., 1994; Collin et al., 2003), indicating that y-type Trxs cannot be physiological reductants of NADP-MDH. The activation mechanism of this target enzyme is rather complex (Miginiac-Maslow and Lancelin, 2002); two different disulfides have to be reduced to trigger a conformational change leading to the full activation of the enzyme. The ability of low concentrations of Trx y to activate a mutant version of NADP-MDH where only the N-terminal disulfide remains suggests that this Trx type might function in a prereduction step of the enzyme. This assumption is supported by the observation that the preincubation of wild-type NADP-MDH with Trx y increased the rate of NADP-MDH activation by Trx f and eliminated the characteristic activation lag phase. It can be surmised that Trxs y are able to reduce the N-terminal disulfide of NADP-MDH, keeping the enzyme in a preactivated, i.e. ready-for-activation, state in light conditions when the reducing capacity of the chloroplast stroma is not sufficient to activate the enzyme. Indeed, the C-terminal disulfide of the enzyme is known to be the most difficult of all the MDH regulatory disulfides to reduce, with an Em value of −380 mV at pH 7.9 (the pH of illuminated chloroplast stroma), whereas the N-terminal disulfide, with an Em value of −350 mV at pH 7.9, is equipotential with the disulfide bridge of most Trxs (Hirasawa et al., 2000). The redox potential of Trx y1 and y2 (−337 mV) is slightly, but significantly, less negative than the redox potential of m-type and f-type Trx (Hirasawa et al., 1999; Collin et al., 2003) and very similar to the redox potential of Trx x. This difference could account at least in part for the inability of Trx y to reduce the C-terminal disulfide of NADP-MDH, and hence, to achieve enzyme activation, but the charge and hydrophobicity of the amino acids of the contact area might be important as well.
Trxs are also known to be implicated in oxidative stress response (Issakidis-Bourguet et al., 2001), serving as proton donors to Prxs. Different plastidial Prxs of Arabidopsis have been identified. Two of them (2-Cys Prx A and 2-Cys Prx B) belong to the 2-Cys-Prx type, share a high identity with each other (König et al., 2002), and are homologs of the barley Prx BAS1 (Baier and Dietz, 1997). Our results show that the y-type Trxs are low efficiency reductants of these Prxs. Thus, the conclusion of our previous work on 2-Cys Prx A indicating that the x-type Trx is the best candidate for a physiological partnership with the 2-Cys Prxs still holds true (Collin et al., 2003). Another peroxiredoxin tested in this work was the Arabidopsis Prx Q (Horling et al., 2003). The activity of this enzyme was considerably increased in the presence of x-type and y-type Trx. The measurement of the catalytic efficiencies of Prx Q with different plastidial Trxs revealed that the y-type Trx are by far the best substrate of this enzyme. Moreover, both y-type isoforms were indistinguishable in peroxide detoxification tests with Prx Q. The ability of the Trx y from Chlamydomonas to donate electrons to poplar Prx Q has already been reported. However, in the heterologous system used by the authors, Trx y was not the most efficient electron donor among the Trxs tested (Rouhier et al., 2004). The predominant function of Prx Q in the chloroplast is not known at present. Interestingly, a major part of Prx Q protein was associated with the thylakoid membrane (Dietz et al., 2004), suggesting a role in antioxidant defense or redox signaling in context of primary reactions of photosynthesis.
In all the activities tested so far, the two isoforms of Trx y appeared to be equivalent. This result is not really surprising since an analysis of the putative mature proteins showed that both isoforms are highly conserved, sharing 81.7% identity. All these data raised the question of a possible functional redundancy of the two isoforms versus functional specificity in space and time. Very often, a functional redundancy is compensated by differential expression patterns. The hypothesis is supported by the distinct expression profiles of Trx y in different organs and light conditions. Similar to other plastidial Trxs, the Trx y2 mRNA was most abundant in leaves. Its level was increased when plants were transferred from dark to light. This result suggests that Trx y2 expression is light induced. In contrast, the mRNA encoding Trx y1 was more abundant in roots, mature siliques, and seeds, i.e. in nonphotosynthetic tissues. Moreover, no light induction of the gene expression could be detected, a situation rather unusual for Trxs. For example, in the green alga C. reinhardtii, even the cytosolic Trx h has been shown to be induced by light (Lemaire et al., 1999). The differences in gene expression show that the two isoforms of Trx y are not physiologically redundant in planta. This observation is strengthened by the observation that Trx y mRNA expression patterns in developing seeds showed opposite variations. The high levels of Trx y1 mRNA at the active oil synthesis period suggest the implication of this isoform in lipid synthesis. In plants, de novo fatty acid biosynthesis from acetyl-CoA occurs solely in plastids, and the first committed step of this synthesis is catalyzed by acetyl-CoA carboxylase (ACCase; Sasaki and Nagano, 2004). Redox dependence of plastidic heteromeric ACCase has been demonstrated, the enzyme being more efficiently activated in vitro by f-type than by m-type Trx (Sasaki et al., 1997). In Arabidopsis, Trx f subtype appears to be exclusively present in green tissues and AtTrx f1 and AtTrx f2 mRNA levels show very similar patterns as that of AtTRXy2 (Fig. 6), especially during seed development and germination (data not shown). In contrast, in developing seeds AtTrx y1 mRNA level increases exhibiting a bell-shaped pattern very similar to that previously found for subunits of heteromeric ACCase (Ke et al., 2000; Ruuska et al., 2002). Reductive activation of heteromeric ACCase, demonstrated in leaves, has not been established yet in nonphotosynthetic tissues. If such regulation occurs, data from gene expression profiling of target and plastidial Trxs strongly suggest that Trx y1 subtype would be a good candidate to fulfill this function. Numerous physiological functions of Trx in chloroplasts have been evidenced (Balmer et al., 2003), but no targets of cpTrx have been identified so far in nonphotosynthetic tissues. The situation of Trx y1 is shared to some extent by Trx m3, whose mRNA is mainly present in flower buds (Mestres-Ortega and Meyer, 1999). If indeed these Trxs function in nonphotosynthetic tissues, where no photosynthetic electron transfer exists, an important remaining problem will be the identification of the Trx reduction system in these nongreen tissues.
MATERIALS AND METHODS
Isolation and Cloning of cDNAs Encoding y-type Trxs
The total RNA from roots and leaves were prepared using SV Total RNA Isolation System from Promega (Madison, WI). cDNA synthesis was performed on 1 μg of RNA with oligo(dT) as primer and moloney murine leukemia virus reverse transcriptase by following the supplier's instructions (Promega). The Trx y1 and Trx y2 cDNAs were obtained by PCR using upstream and downstream oligonucleotides: Sensy1, 5′-CCAACGCCGAATCAAGAGATCG-3′ and Revy1, 5′-AAAACATCGCTATCACTAAGTCCTGAATAGG-3′ and oligonucleotides Sensy2, 5′-TCTTTAACTTAAAAACTCTCCCGTTTCGC-3′ and Revy2, 5′-TATACCGAGCAAACAATCAACAATGGC-3′, respectively, on root and leaf cDNA preparations. The specific full-length cDNAs were then cloned in a pGEM-T vector (Promega) and sequenced (big dye terminator kit v3; Hitachi 3100 Genetic Analyser from Applied Biosystems, Courtaboeuf, France).
Subcellular Localization of Trx-GFP Fusion Proteins
The protein sequences deduced from the cDNA sequences were analyzed by targeting programs Predotar, chloroP, Psort, and TargetP (http://www.inra.fr/Internet/Produits/Predotar; http://www.cbs.dtu.dk/services/ChloroP; http://psort.nibb.ac.jp; http://www.cbs.dtu.dk/services/TargetP). The cloned cDNA encoding Trx y2 in its unprocessed form was then amplified by PCR using upstream and downstream oligonucleotides 5′-AAAATCTAGAGCTTCTTCAATGGCGATTTCTCTCG-3′ and 5′-AAAAGGATCCCTGTTTCACTTGCAAAGAATTCTCAATACGC-3′. This cDNA was then cloned in XbaI-BamH1 restriction sites of the transient expression vector pBI/SmGFP (Jasinski et al., 2002). The control fusion constructs encoding plastidial and mitochondrial Trxs were described in our previous work (Collin et al., 2003). Arabidopsis (Arabidopsis thaliana) cell protoplasts were transformed following the polyethylene glycol (PEG) procedure described by Léon et al. (2002). Green fluorescence of chimeric cpTrxs fused to GFP, chlorophyll red autofluorescence, and transmission images were monitored using a confocal laser scanning microscope.
Recombinant Trx Production and Purification
The putative cleavage sites of the pre-Trxs were determined by using the prediction program SignalP (http://www.cbs.dtu.dk/services/SignalP) and comparing the sequences of Arabidopsis and cyanobacterial Trxs y (Lemaire et al., 2003). The sequence comparisons of putative mature Trxs y were obtained by the program align (http://xylian.igh.cnrs.fr/bin/align-guess.cgi). The cDNAs encoding mature Trxs y were obtained by a PCR amplification using upstream and downstream oligonucleotides 5′-AAAACCATGGAAGCCAAGAAGCAGACATTTGAC-3′ and 5′-AAAAGGATCCTATGGCTTCACTTTTAGAGAATCCTC-3′ for Trx y1, or 5′-AAAACCATGGCAGCAAAGAAGCAAACTTTCAACTC-3′ and 5′-AAAAGGATCCTACTGTTTCACTTGCAAAGAATTCTCAATAC-3′ for Trx y2. cDNAs were then cloned into Nco1-BamH1 restriction sites of the expression vector pET-3d. The recombinant proteins were obtained and purified as described in Collin et al. (2003).
FBPase Activation
The recombinant FBPase, produced and purified as described by Jacquot et al. (1995), was activated by Trx in presence of DTT. The activated FBPase was then quantified by measuring its activity in a coupling system as described previously (Collin et al., 2003). The reproducibility of the values was better than ±10%.
MDH Activation
The activations of Δ15-NADP-MDH (shortened by 15 amino acids at the N terminus) whose biochemical characteristics are identical to those of the full-length wild-type MDH, (Johansson et al., 1999), and of the mutated MDH C207A/C365A/C377A (Ruelland et al., 1997) were measured as described previously (Johansson et al., 1999). Each enzyme variant was activated by incubation with Trx in presence of 10 mm DTT, and the quantity and extent of enzyme activation was measured by its enzymatic activity. In the case of the coactivation of Δ15-NADP-MDH, activation assays were performed using a Trx f1 concentration of 1 μm and/or a concentration of Trx y1 of 20 μm. The average reproducibility of the values was better than ±10%.
Purification of Prx and Activity Tests
The Prx Q-cDNA encoding the mature protein was cloned in pCR-T7/NT-TOPO-vector (Invitrogen, Carlsbad, CA). The His-tagged protein was produced in BL21 (pLysS)DE3 (Invitrogen) and purified on Ni-NTA-agarose (Qiagen, Valencia, CA) column. The His-tagged 2-Cys Prx A and 2-Cys Prx B Prx were produced as described before (König et al., 2002). Prx activity measurements were performed in conditions previously described (Collin et al., 2003). The concentrations of 2-Cys Prx A and B used were, respectively, 20 and 10 μm, whereas the Prx Q was used at a concentration of 5 μm. The release of tert-butyl hydroperoxide (t-BOOH) was quantified at 560 nm using 0.5 mL of xylenol orange 125 μm in 100 mm sorbitol, 25 mm H2SO4, and 250 μm Fe(NH4)2(SO4)2. The average reproducibility of the values was better than ±10%.
Determination of the Redox Potentials of Recombinant Trxs
Oxidation-reduction titrations were carried out using monobromo bimane (mBBr) to monitor the reduction state of the thiol/disulfide of Trxs, the useful range of potentials being generated by different ratios of reduced to oxidized DTT. The experimental procedure was described by Hirasawa et al. (1999). The Em values were shown to be independent of the total DTT concentration and of the redox equilibration time. All determinations were performed in Tricine-NaOH buffer (100 mm, pH 7.9).
Semiquantitative RT-PCR
Total RNA was isolated from approximately 100 mg of plant tissue material using SV Total RNA Isolation system from Promega according to the instruction manual. A preliminary centrifugation step (15 min, 60,000 rpm, 4°C in a TL100 Beckman centrifuge [Fullterton, CA]) was added for seed extracts. Semiquantitative RT-PCR reactions were performed with oligonucleotides designed for priming from the 5′-untranslated region and/or the 3′-untranslated region to ensure gene-specific amplification that was verified by sequencing of PCR products. Oligonucleotides used for gene-specific RT-PCR amplifications were: ACT2up2, 5′-TTCCCTCAGCACATTCCAGCAG-3′ and ACT2do, 5′-TTAACATTGCAAAGAGTTTCAAGG-3′ for actin2; Tub.beta.9up, 5′-GAGGTTGATGAACAGATGATGAACGTCCAG-3′ and Tub.beta.9do, 5-GGAAATGAGTGCGACGAACTTACAGCC-3′ for tubulin-beta-9; for Trx y1 and y2, the primers were as for full-length cDNA isolation. PCR products were fractionated on agarose gels and visualized by ethidium bromide staining using the GeneGenius BioImaging System from SYNGENE (St Quentin-en-Yvelines, France). Densitometric analysis of the PCR products was performed using the GeneSnap software from the same company.
Acknowledgments
The authors are grateful to E. Keryer for technical assistance and to Dr. C. Bergounioux and her team for providing the T87 Arabidopsis cell cultures and for sharing their expertise in plant cell biology. Microscopy was done in the core facility of the Institut Fédératif de Recherche 87 (CNRS, Gif/Yvette, France) with the appreciated technical support of C. Talbot.
This work was supported in part by a Génoplante II Grant AF 2001 047 (to V.C., M.M-M., and E.I-B.), by the Robert A. Welch Foundation (grant no. D–0710 to D.B.K), and by the Deutsche Forschungsgemeindshaft (Di 346/6, FOR 387, TP 3, to P.L. and K.-J.D.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.052233.
References
- Baier M, Dietz K-J (1997) The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J 12: 179–190 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schürmann P, Buchanan BB (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA 100: 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB (1980) Role of light in the regulation of chloroplast enzymes. Annu Rev Plant Physiol 31: 341–374 [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278: 23747–23752 [DOI] [PubMed] [Google Scholar]
- Dietz KJ (2003) Plant peroxiredoxins. Annu Rev Plant Biol 54: 93–107 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Stork T, Finkemeier I, Lamkemeyer P, Li WX, El-Tayeb MA, Michel KP, Pistorius E, Baier M (2004) The role of peroxiredoxins in oxygenic photosynthesis of cyanobacteria and higher plants: peroxide detoxification or redox sensing. In B Demmig-Adams, WW Adams III, AK Mattoo, eds, Photoprotection, Photoinhibition, Gene Regulation, and Environment. Kluwer Academic Publishers, Dordrecht, The Netherlands (in press)
- Focks N, Benning C (1998) Wrinkled1: a novel low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD, Agostino A (1992) Bi-level disulfide group reduction in the activation of C4 leaf nicotinamide adenine dinucleotide phosphate malate dehydrogenase. Plant Physiol 100: 360–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa M, Ruelland E, Schepens I, Issakidis-Bourguet E, Miginiac-Maslow M, Knaff DB (2000) Oxidation-reduction properties of the regulatory disulfides of sorghum chloroplast nicotinamide adenine dinucleotide phosphate-malate dehydrogenase. Biochemistry 39: 3344–3350 [DOI] [PubMed] [Google Scholar]
- Hirasawa M, Schürmann P, Jacquot J-P, Manieri W, Jacquot P, Keryer E, Hartman FC, Knaff DB (1999) Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry 38: 5200–5205 [DOI] [PubMed] [Google Scholar]
- Hodges M, Miginiac-Maslow M, Decottignies P, Jacquot J-P, Stein M, Lepiniec L, Crétin C, Gadal P (1994) Purification and characterization of pea thioredoxin f expressed in Escherichia coli. Plant Mol Biol 26: 225–234 [DOI] [PubMed] [Google Scholar]
- Holmgren A (1989) Thioredoxin and glutaredoxin systems. J Biol Chem 264: 13963–13966 [PubMed] [Google Scholar]
- Horling F, Lamkemeyer P, König J, Finkemeier I, Kandlbinder A, Baier M, Dietz K-J (2003) Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol 131: 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issakidis-Bourguet E, Mouaheb N, Meyer Y, Miginiac-Maslow M (2001) Heterologous complementation of yeast reveals a new putative function for chloroplast m-type thioredoxin. Plant J 25: 127–135 [DOI] [PubMed] [Google Scholar]
- Jacquot J-P, Lopez-Jaramillo J, Chueca A, Cherfils J, Lemaire S, Chedozeau B, Miginiac-Maslow M, Decottignies P, Wolosiuk RA, Lopez-Gorge J (1995) High-level expression of recombinant pea chloroplast fructose-1,6-bisphosphatase and mutagenesis of its regulatory site. Eur J Biochem 229: 675–681 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Perennes C, Bergounioux C, Glab N (2002) Comparative molecular and functional analyses of the tobacco cyclin-dependent kinase inhibitor NtKIS1a and its spliced variant NtKIS1b. Plant Physiol 130: 1871–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson K, Ramaswamy S, Saarinen M, Lemaire-Chamley M, Issakidis-Bourguet E, Miginiac-Maslow M, Eklund H (1999) Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form. Biochemistry 38: 4319–4326 [DOI] [PubMed] [Google Scholar]
- Ke J, Wen TN, Nikolau BJ, Wurtele ES (2000) Coordinate regulation of the nuclear and plastidic genes fort he subunits of heteromeric acetyl-Coenzyme A carboxylase. Plant Physiol 122: 1057–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Baier M, Hörling F, Kahmann U, Harris G, Schürmann P, Dietz K-J (2002) The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc Natl Acad Sci USA 99: 5738–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire SD, Stein M, Issakidis-Bourguet E, Keryer E, Benoit V, Pineau B, Gérard-Hirne C, Miginiac-Maslow M, Jacquot J-P (1999) The complex regulation of the ferredoxin-thioredoxin related genes by light and the circadian clock. Planta 209: 221–229 [DOI] [PubMed] [Google Scholar]
- Lemaire SD, Collin V, Keryer E, Quesada A, Miginiac-Maslow M (2003) Characterization of thioredoxin y, a new type of thioredoxin identified in the genome of Chlamydomonas reinhardtii. FEBS Lett 543: 87–92 [DOI] [PubMed] [Google Scholar]
- Léon S, Touraine B, Briat J-F, Lobréaux S (2002) The AtNFS2 gene from Arabidopsis thaliana encodes a NifS-like plastidial cysteine desulphurase. Biochem J 336: 557–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestres-Ortega D, Meyer Y (1999) The Arabidopsis thaliana genome encodes at least four thioredoxins m and a new prokaryotic-like thioredoxin. Gene 240: 307–316 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Vignols F, Reichheld J-P (2002) Classification of plant thioredoxins by sequence similarity and intron position. Methods Enzymol 347: 394–402 [DOI] [PubMed] [Google Scholar]
- Miginiac-Maslow M, Lancelin J-M (2002) Intrasteric inhibition in light signaling: light activation of NADP-malate dehydrogenase. Photosynth Res 72: 1–12 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Gualberto JM, Jordy MN, De Fay E, Hirasawa M, Duplessis S, Lemaire SD, Frey P, Martin F, et al (2004) Poplar peroxiredoxin Q. A thioredoxin-linked chloroplast antioxidant functional in pathogen defense. Plant Physiol 134: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelland E, Miginiac-Maslow M (1999) Regulation of chloroplast enzyme activities by thioredoxins: activation or relief from inhibition? Trends Plant Sci 4: 136–141 [DOI] [PubMed] [Google Scholar]
- Ruelland E, Lemaire-Chamley M, Le Maréchal P, Issakidis-Bourguet E, Djukic N, Miginiac-Maslow M (1997) An internal cysteine is involved in the thioredoxin-dependent activation of sorghum leaf NADP-malate dehydrogenase. J Biol Chem 272: 19851–19857 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Hatano M (1997) Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc Natl Acad Sci USA 94: 11096–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Nagano Y (2004) Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotechnol Biochem 68: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Scheibe R (2004) Malate valves to balance cellular energy supply. Physiol Plant 120: 21–26 [DOI] [PubMed] [Google Scholar]
- Schürmann P, Jacquot J-P (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51: 371–400 [DOI] [PubMed] [Google Scholar]