Abstract
The nuclear-encoded plastid sigma factors are supposed to be a regulatory subunit of the multisubunit bacteria-type plastid RNA polymerase. We studied here whether or not three genes, PpSig1, PpSig2, and PpSig5 encoding plastid sigma factors, are controlled by the circadian clock and/or by blue light signaling in the moss Physcomitrella patens. Among the three PpSig genes, only PpSig5 was clearly controlled by the circadian clock. In contrast to the differential regulation on a daily timescale, a pulse of blue light induced the expression of all the three PpSig genes. This induction was significantly reduced in a knockout mutant that lacked the blue light photoreceptor cryptochromes PpCRY1a and PpCRY1b, indicating that PpCRY1a and/or PpCRY1b mediate the blue light signal that induces the expression of the PpSig genes. In a daily cycle of 12-h blue light/12-h dark, the timing of peak expression of PpSig5 and a chloroplast gene psbD, encoding the D2 subunit of photosystem II, advanced in the cryptochrome mutant relative to those in the wild type, suggesting the presence of regulatory interactions among the expression of PpSig5 and psbD, the circadian clock, and the blue light signaling mediated by the cryptochrome(s).
Plastids contain their own genome owing to their free-living bacterial ancestry. At least two distinct RNA polymerases transcribe plastid genes in higher plants: one is the nuclear-encoded plastid RNA polymerase and the other is the primarily plastid-encoded plastid RNA polymerase (PEP; Gray and Lang, 1998). Nuclear-encoded plastid RNA polymerase is a single-subunit bacteriophage-type enzyme and is thought to generally transcribe housekeeping genes including the PEP core enzyme genes (Stern et al., 1997; Weihe and Borner, 1999). On the other hand, PEP is a multisubunit bacteria-type enzyme and consists of four plastid-encoded subunits of the core enzyme, α, β, β′, β″, and a nuclear-encoded plastid sigma factor (Allison, 2000). The plastid sigma factor is supposed to define binding specificity to target promoter sequences, based on the functions of eubacterial counterpart sigma-70 family proteins (Allison, 2000). The bacterial genome encodes multiple sigma factors with different promoter specificities, enabling transcriptional activation of different sets of genes in response to various endogenous and environmental signals (Gruber and Gross, 2003). Recent studies have suggested this picture to be also applicable to the plastid sigma factors. The Arabidopsis (Arabidopsis thaliana) genome contains six distinct sigma genes from AtSig1 to AtSig6, for each of which, except for AtSig4, orthologous genes have been found from various higher plant species (Fujiwara et al., 2000; Tsunoyama et al., 2004). Therefore, the plant sigma factor family is subdivided into structurally, and presumably functionally, different groups beyond species. Recently, AtSIG2 and AtSIG5 have been shown to define respective sets of target chloroplast genes albeit with some overlaps; moreover, AtSIG5 mediates specific stress stimuli such as high light and low temperature to the target genes (Kanamaru et al., 2001; Hanaoka et al., 2003; Privat et al., 2003; Nagashima et al., 2004; Tsunoyama et al., 2004). On the other hand, the functions and target genes of sigma factors other than AtSIG2 and AtSIG5 remain unknown.
The circadian clock is an autonomous oscillator with an endogenous period of approximately 24 h, and it controls a wide variety of processes from gene expression to leaf movements for the adaptation to the environment that changes on a daily timescale (Young and Kay, 2001). In Arabidopsis, “clock genes” encoding the candidate components of the core clock machinery have been isolated (Hayama and Coupland, 2003). Currently, functions and interactions of clock genes and their encoded proteins (clock proteins) are being studied intensively to unravel the molecular mechanisms of the clock (Hayama and Coupland, 2003). The clock controls many genes (called clock-controlled genes [ccgs]) on the nuclear and chloroplast genomes, resulting in circadian rhythms of the transcript levels of the ccgs even under constant conditions (Nakahira et al., 1998; Harmer et al., 2000; Schaffer et al., 2001). Candidate clock proteins CCA1 (circadian clock-associated-1) and LHY (elongated hypocotyl), myb transcription factors, seem to control the rhythmic expression of certain sets of ccgs on the nuclear genome by directly binding to its promoters (Wang et al., 1997; Alabadí et al., 2001). By contrast, it is still almost totally unknown how temporal information is transmitted from the clock to chloroplast genes. In cyanobacteria, RpoD2, one of the sigma-70 family proteins, revealed to mediate between the clock and a set of downstream genes (Tsinoremas et al., 1996). Given that plastids arose through endosymbiosis from a photosynthetic bacterium closely related to extant cyanobacteria, the function of RpoD2 might be conserved in a plastid sigma factor(s). This idea is supported by the observations that some of the plastid sigma factor genes show circadian rhythms in transcript accumulations (Morikawa et al., 1999; Harmer et al., 2000), though its direct role on the chloroplast gene expression has not yet been examined. Nakahira et al. (1998) reported a robust circadian rhythm in the transcription from the blue light responsive promoter (BLRP) of the psbD gene, encoding the D2 protein of PSII. Since an unusual −35 sequence was critical for the rhythmic expression of psbD gene, they argued that a sigma factor(s), known to recognize the −35 and −10 sequences of a bacterial type promoter (Allison, 2000), might transmit temporal information from the clock to BLRP (Nakahira et al., 1998). BLRP is controlled not only by the clock, but also directly activated by blue light, as inferred from its name (Stern et al., 1997). The plastid sigma factors have been candidate proteins mediating light-inducible expression of plastid genes, because many sigma genes are light-inducible (Allison, 2000). Very recently, this was reported to be the case for BLRP; AtSIG5 protein mediates between a blue light signal from the blue light photoreceptor cryptochromes and transcription from BLRP (Nagashima et al., 2004; Tsunoyama et al., 2004). Collectively, the plastid sigma factor is expected to be key a regulator integrating different regulations such as the clock and light, and hence its study is critical for understanding the regulatory mechanisms underlying the daily expression profiles of plastid genes in daily light-dark cycles.
Physcomitrella patens is a newly established model plant to which gene targeting with exceptionally high efficiency for a plant can be easily applicable (Schaefer, 1994). In addition, P. patens is an interesting plant from an evolutionary point of view because it is classified into bryophyte, which divided from other plant lineages at a very early stage of the evolution of land plants (Heckman et al., 2001; Schaefer and Zrÿd, 2001). The regulatory system of plastid gene expression has drastically changed during the evolution of photosynthetic organisms (Sato, 2001). Based on these facts, we chose P. patens as the experimental plant for the functional study of plastid sigma factors (Hara et al., 2001a, 2001b). We isolated two distinct plastid sigma factor genes PpSig1 and PpSig2 encoding PpSIG1 and PpSIG2 proteins, respectively, from P. patens (Hara et al., 2001a, 2001b). PpSIG1 and PpSIG2 are closely related to the higher plant Sig1 and Sig2 groups, respectively; that is, the Sig1 and Sig2 groups were already present in the common ancestor of the bryophyte and higher plants (Hara et al., 2001a, 2001b). Here we report a newly isolated cDNA species PpSig5 encoding another P. patens plastid sigma factor, which is classified into the most diverged Sig5 group. We examined: (1) whether the expression of the PpSig genes is controlled by a circadian clock; and (2) how the blue light signaling mediated by cryptochromes affects the daily expression patterns of the PpSig genes using a mutant strain that lacked PpCry1a and PpCry1b, encoding the blue light photoreceptor cryptochromes. The results demonstrate that the expression of the newly isolated PpSig5 shows a unique feature among the three sigma factor genes in P. patens.
RESULTS
Identification of PpSig5 Gene in P. patens
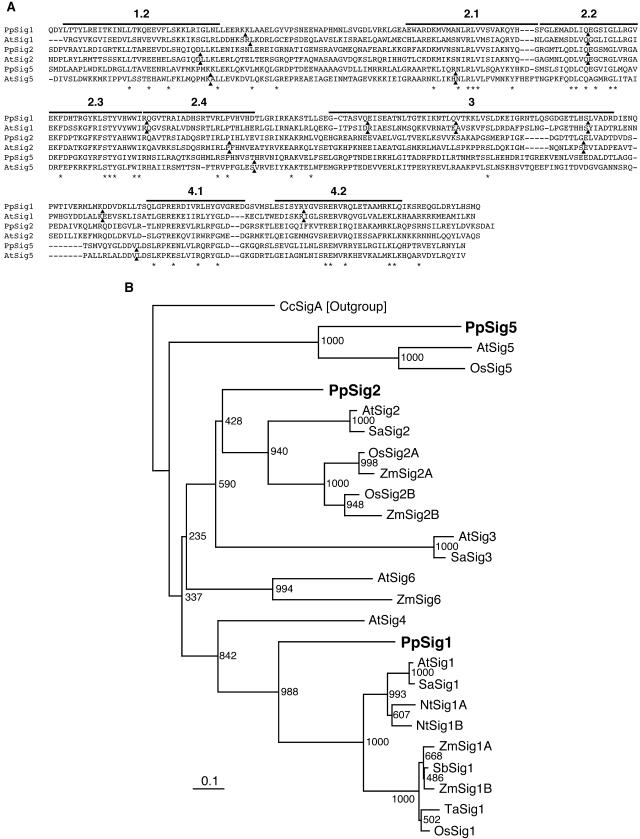
By a database (the Physcomitrella EST Programme) search using the BLAST program we found an expressed sequence tag (EST) sequence (accession no. BI894524) that was similar to plastid sigma factor sequences from various plants but was identical to neither PpSig1 nor PpSig2 sequences. We isolated the full-length cDNA by screening a cDNA library (a gift from the Physcomitrella EST Programme) using this EST sequence as a probe. The amino acid sequence deduced from the predicted coding region of the cDNA showed the highest similarity to Arabidopsis AtSIG5 sequence (identity and similarity were 37% and 58%, respectively). All the domains conserved among plant sigma factors (domains 1.2–4.2 [Fig. 1A]) were present in the novel P. patens sigma factor sequence. To examine the evolutionary relationships between this sigma factor and the plastid sigma factors reported to date, a phylogenetic tree was constructed based on the sequences of the novel sigma factor and 22 plastid sigma factors from various seed plants and P. patens. As described previously (Hara et al., 2001b), the plastid sigma factors were divided into some clusters in the tree (Fig. 1B). The novel sigma factor was clustered together with AtSIG5 and OsSIG5, indicating that it is classified into the Sig5 group of the plastid sigma factor family. This result was supported by a comparison of intron positions on the coding sequences of Arabidopsis and P. patens sigmas (Fig. 1A). The intron positions of the novel sigma factor and AtSIG5 were found completely at identical positions by the alignment of amino acid sequences. Based on these results, we speculated that the novel sigma cDNA encodes the third plastid sigma factor in P. patens and named the corresponding gene and its protein product PpSig5 and PpSIG5, respectively.
Figure 1.
A, Amino acid sequence alignment of the sigma factors from P. patens and Arabidopsis and comparison of intron sites. Sequences of conserved C-terminal regions through subdomains 1.2 to 4.2 of PpSIG1, PpSIG2, and PpSIG5 from P. patens and AtSIG1, AtSIG2, and AtSIG5 from Arabidopsis are aligned using the ClustalW program. Subdomains are indicated above the sequences. Intron positions are shown by black triangles. Identical amino acids are indicated by asterisks. B, A phylogenetic tree constructed using sigma factor sequences from various species. The tree was constructed by the neighbor-joining method using approximately 260 aligned residues corresponding to the conserved regions 1.2 and 2.1 to 4.2. Numbers at each node represent bootstrap values out of 1,000 bootstrap resamplings. Bar indicates the distance corresponding to 10 changes/100 amino acid positions. PpSIG1 (AB059354), PpSIG2 (B059356), and PpSIG5 (AB189171) from P. patens are indicated by bold letters. Other sigma factors are as follows: AtSig1 (AB019942), AtSig2 (AB019943), AtSig3 (AB019944), AtSig4 (AB021119), AtSig5 (AB021120), AtSig6 (AB029916) from Arabidopsis, SaSig1 (Y15899), SaSig2 (AJ276656), and SaSig3 (AJ276657) from Sinapis alba, NtSig1A (AB023571) and NtSig1B (AB023572) from Nicotiana tabacum, OsSig1 (AB005290), OsSig2A (AB095094), OsSig2B (AB095094), and OsSig5 (AB096071) from Oryza sativa, SbSig1 (Y14276) from Sorghum bicolor, TaSig1 (AJ132658) from Triticum aestivum, ZmSig1A (AF058708), ZmSig1B (AF058709), ZmSig2A (AF099110), ZmSig2B (AF099111), and ZmSig6 (AF099112) from Zea mays, and CcSigA (D83179) from Cyanidium caldarium. CcSigA was used as the out group.
Diurnal Changes in mRNA Accumulation of Three PpSig Genes and psbD gene in LD
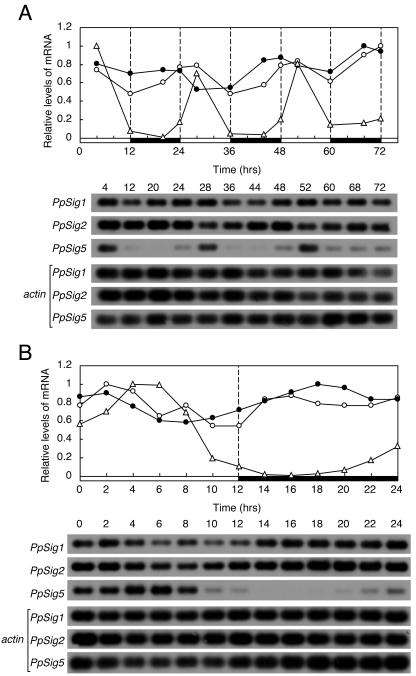
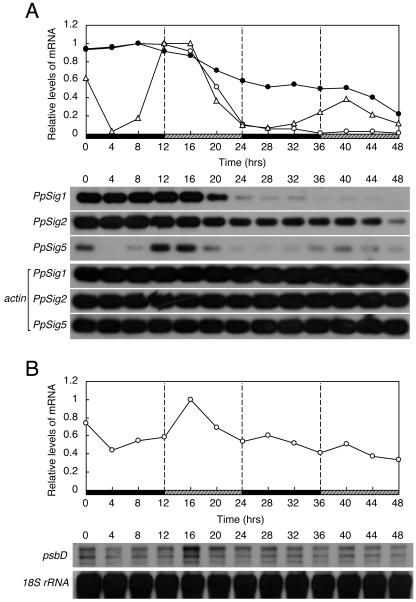
We examined the temporal patterns of the expression of three distinct P. patens sigma factor genes (PpSig1, PpSig2, and PpSig5) under three different light regimens to study if the sigma genes are under the control of the circadian clock. First, we measured the changes of the three PpSig mRNA accumulations for three 12-h-light/12-h-dark cycles (LD) by semiquantitative reverse transcription (RT)-PCR analyses (Fig. 2A). The PpSig1 and PpSig2 genes were expressed throughout a day, with fluctuations that suggest diurnal rhythms of mRNA levels with very low amplitude. By contrast, PpSig5 mRNA showed a very high-amplitude diurnal rhythm with peaks observed in light phases. In dark phases, the PpSig5 mRNA levels were close to the detection limit and started to increase before the onset of light. We measured the mRNA levels for 24 h at intervals of 2 h under LD (Fig. 2B). The PpSig1 mRNA levels showed slight increases at the onset of light and dark phases, whereas the PpSig2 mRNA levels showed a monophasic diurnal rhythm with a peak in the light phase and a trough in the dark phase. The PpSig5 peaked 4 to 6 h after the onset of light and showed a trough at 2 to 6 h in the dark. Then it started to increase around 4 h before the onset of light. The predawn “anticipatory” rise of PpSig5 expression, together with very high peak-to-trough ratio of the oscillation, suggests that PpSig5 is under the strong control of an endogenous circadian clock.
Figure 2.
Changes in abundance of mRNA for PpSig1, PpSig2, and PpSig5 under LD examined by semiquantitative RT-PCR. P. patens cells were maintained in LD for more than 3 weeks after which cells were harvested in LD at indicated times. Total RNA was extracted, reverse-transcribed, and amplified by PCR using a primer set specific to each PpSig gene or actin gene as internal control (Aoki et al., 2004). cDNA blots were generated and hybridized with the probe for each PpSig or actin gene. The top section shows the results of quantification of each PpSig mRNA after normalization to actin mRNA. The maximum level is expressed as 1.0. The bottom section shows the hybridized bands for each PpSig mRNA. The actin mRNA bands used as internal control are also shown below PpSig bands. A, Changes in mRNA abundance for 3 d. B, Changes in mRNA abundance with a finer time resolution for 24 h. White circles, PpSig1; black circles, PpSig2; white triangles, PpSig5. White and black bars indicate light (40 μmol m−2 s−1) and dark intervals, respectively. We obtained similar results in two independent experiments.
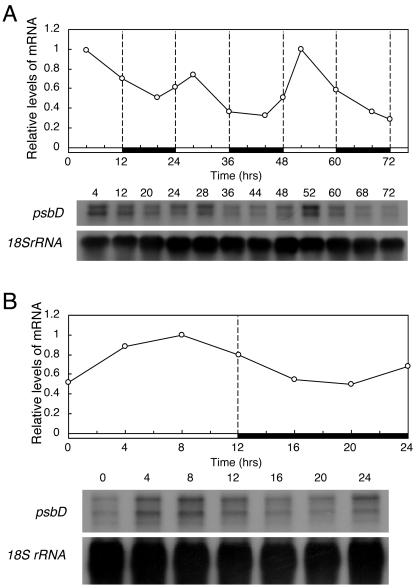
We also measured the changes in mRNA accumulation of a chloroplast gene psbD, which encodes the D2 protein of PSII. In wheat (Triticum aestivum), the expression of psbD is reported to be controlled by a circadian clock (Nakahira et al., 1998). The P. patens psbD showed a diurnal rhythm with peaks observed in light phases (Fig. 3, A and B). The predawn rise was also observed for psbD, suggesting that psbD is also controlled by the clock in P. patens.
Figure 3.
Changes in the abundances of psbD mRNA under LD examined by northern-blotting analyses. Culture and sampling of P. patens cells and RNA extraction were carried out as described in the legend for Figure 2. Northern blots were generated and hybridized with the DNA probe for psbD or 18S rRNA gene. The top section show the levels of psbD mRNA after normalization to those of 18S rRNA. The maximum level is expressed as 1.0. The bottom sections show the hybridized psbD and 18S rRNA bands. A, Changes in mRNA abundance for 3 d. B, Changes in mRNA with a finer time resolution abundance for 24 h. We obtained similar results in two independent experiments.
Changes in mRNA Accumulation of PpSig and psbD Genes in Constant Conditions
The most reliable diagnostic feature of circadian rhythms is that they persist under constant conditions. Therefore, we measured the expression of the PpSig and psbD genes in continuous light (LL; Fig. 4), or in continuous dark (DD; Fig. 5) after entraining the clock in LD.
Figure 4.
Changes in abundance of mRNA for three PpSigs and psbD under LL. P. patens cells were maintained in LD for more than 3 weeks after which cells were harvested in LL (40 μmol m−2 s−1) at indicated times for 2 d. The abundance of PpSigs mRNAs and psbD mRNAs were measured as described in legends for Figures 2 and 3, respectively. A, Changes in abundance of PpSig1, PpSig2, and PpSig5 mRNAs. White circles, PpSig1; black circles, PpSig2; white triangles, PpSig5. B, Changes in abundance of psbD mRNA. White and hatched bars indicate light intervals and subjective dark intervals, respectively. We obtained similar results in two independent experiments.
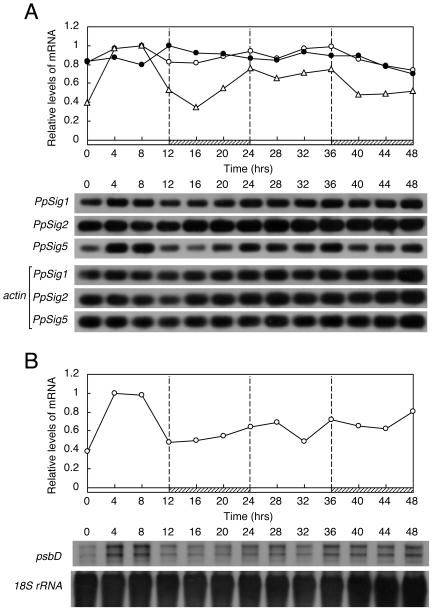
Figure 5.
Changes in abundance of mRNA for three PpSigs and psbD under DD. P. patens cells were maintained in LD for more than 2 weeks after which cells were harvested at indicated times during 48 h in DD. Abundance of mRNA was measured as described in legends for Figures 2 and 3. A, Changes in abundance of PpSig1, PpSig2, and PpSig5 mRNAs. White circles, PpSig1; black circles, PpSig2; white triangles, PpSig5. B, Changes in abundance of psbD mRNA. Black and gray hatched bars indicate dark intervals and subjective light intervals, respectively. We obtained similar results in two independent experiments.
In LL, we could not obtain any signs of circadian control on any of the genes tested (Fig. 4). The PpSig1 and PpSig2 mRNA levels did not show significant fluctuation throughout the monitoring period. The PpSig5 and psbD mRNA levels peaked immediately after the transfer from LD to LL, corresponding to the peaks observed in LD. However, the PpSig5 mRNA levels did not show severe reduction after this peak, unlike the patterns observed in LD.
In DD, the three PpSig genes showed characteristic patterns that were distinct from each other (Fig. 5A). The PpSig1 mRNA levels rapidly decreased in the latter half of the first day in DD, and only very low levels, if any, of expression were detected after 1 d in DD. The PpSig2 mRNA levels showed a gradual decrease in DD, while its expression was still observed even 2 d after the onset of DD. The PpSig5 mRNA levels showed a clear peak in the early half of the first subjective day phase (subjective day and night in continuous conditions correspond to light and dark phases, respectively, in the preceding light dark cycles) and a lower, but reproducible, peak in the second subjective day. These observations, together with those in LD and LL experiments, clearly indicate that three sigma genes are differentially regulated on the daily timescale. In particular, the rhythmic expression of PpSig5 in LD and DD indicates that the circadian clock controls this gene. In DD, the psbD gene also showed rapidly damping oscillation with phasing similar to that of PpSig5, though with a much lower amplitude, suggesting that psbD is also under the control of the clock (Fig. 5B).
Expression of PpRpoA Gene
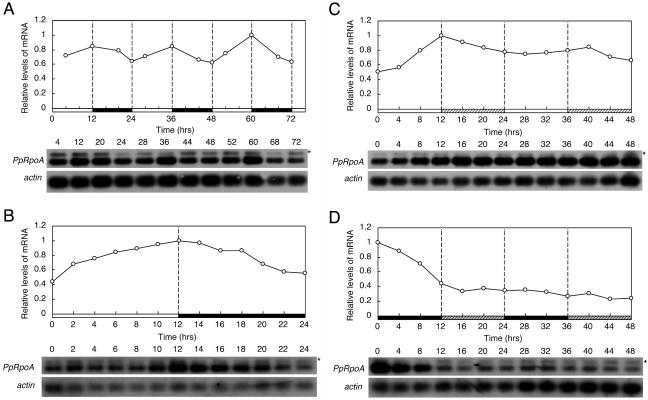
In P. patens, the nuclear gene PpRpoA encodes the α-subunit of PEP (Sugiura et al., 2003). We also measured the temporal changes in the PpRpoA mRNA levels (Fig. 6). In LD (Fig. 6, A and B), the PpRpoA mRNA levels gradually increased in light and decreased in dark, resulting in a diurnal rhythm with a very low amplitude with peaks and troughs observed at the offset and onset of light, respectively. In LL, after the gradual increase in the first 12 h, the mRNA levels did not fluctuate greatly (Fig. 6C). In DD, after the decrease in the first 12 h, the mRNA levels did not fluctuate and remained relatively low over the monitoring period (Fig. 6D). These expression patterns of PpRpoA suggested that the diurnal rhythm of PpRpoA in LD resulted from direct but weak up-regulation by light, not from the endogenous regulation by the clock. The results are consistent with those in wheat, where the expression of a sigma factor gene (TaSig1) showed a circadian rhythm while that of rpoA did not (Morikawa et al., 1999).
Figure 6.
Changes in abundance of PpRpoA mRNA under LD, LL, and DD conditions examined by semiquantitative RT-PCR. P. patens cells were maintained in LD for more than 2 weeks after which cells were harvested in LD, LL, or DD at indicated times. Experiments were carried out as in Figures 2, 4A, and 5A except that the primers and probe specific to PpRpoA (Sugiura et al., 2003) were used instead of those for PpSigs. A, Changes in abundance of PpRpoA mRNA in LD for 3 d. B, Changes in abundance of PpRpoA mRNA in LD for 24 h. C, Changes in abundance of PpRpoA mRNA in LL for 2 d. D, Changes in abundance of PpRpoA mRNA in DD for 2 d. Graphic representations are the same as in Figures 2, 4A, and 5A. Asterisks show genomic amplifications of PpRpoA gene. We obtained similar results in two independent experiments.
Effect of Blue Light Signaling Mediated by Cryptochromes on Induction of PpSig, PpRpoA, and psbD Genes
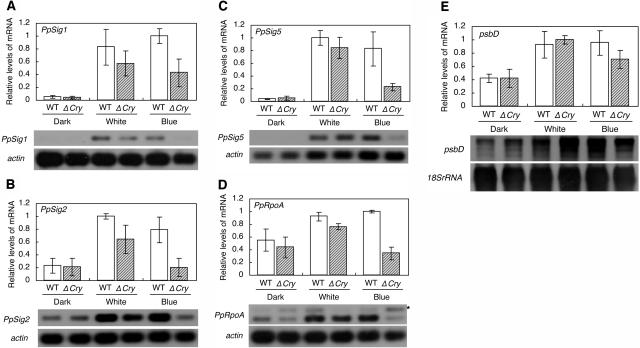
Recent studies indicated that the blue light signal mediated by cryptochromes induced the transcription from BLRP via AtSIG5 protein (Thum et al., 2001; Nagashima et al., 2004; Tsunoyama et al., 2004). We examined if the blue light signaling mediated by cryptochromes induces the PpSig, PpRpoA, and psbD genes in P. patens. First, we tested the light-inducibility of PpSig, PpRpoA, and psbD genes in wild-type cells. We previously demonstrated that white light induced the expression of PpSig1 and PpSig2 (Hara et al., 2001b). We observed here that white light induced PpSig5, PpRpoA, and psbD as well as PpSig1 and PpSig2 (Fig. 7). The induction rates of PpSig2, PpRpoA, and psbD were not as large as those of PpSig1 and PpSig5, reflecting that the former genes retained significant levels of expression even after exposure to darkness for 24 h (Fig. 5, A, B, and D). We also observed that a blue light pulse also induced the expression of all genes tested in wild-type cells (Fig. 7). Next, we tested the light-inducibility of the genes in a cryptochrome mutant. Imaizumi et al. (2002) identified from P. patens two cryptochrome genes, PpCry1a and PpCry1b, encoding PpCRY1a and PpCRY1b proteins, respectively. They demonstrated by the gene targeting experiments that PpCRY1a and PpCRY1b have redundant functions, being supported by the fact that there was only one amino acid difference between PpCRY1a and PpCRY1b (Imaizumi et al., 2002). Based on these observations, we used a double disruption mutant of PpCry1a and PpCry1b, where more severe phenotypes were expected than the single disruption mutant of either gene (Imaizumi et al., 2002). Imaizumi et al. (2002) developed this mutant strain by integrating the hpt (hygromycin B phosphotransferase) and nptII (neomycin phosphotransferase II) gene cassettes into the coding regions of PpCry1a and PpCry1b, respectively. In the mutant strain, the induction of the three PpSig genes and PpRpoA gene by blue light was significantly reduced relative to those in wild-type cells (Fig. 7, A–D), indicating that either or both of the cryptochromes mediated the blue light signal inducing these genes. The induction of PpSig1 and PpSig5 by blue light was only partially reduced in the mutant, indicating that a blue light photoreceptor(s) other than the two cryptochromes is involved in the induction. On the other hand, induction of the three PpSig genes and PpRpoA gene by white light was not, if at all, significantly reduced in the mutant relative to that observed in wild-type cells. This result indicated that photoreceptors sensing wavelengths other than blue light also contribute to the light induction of these genes. As for the psbD gene, we could not obtain data indicating the involvement of the cryptochrome(s) in the induction by blue light (Fig. 7E).
Figure 7.
Effects of cryptochrome mutations on light-induced expression of PpSigs, psbD, and PpRpoA genes. Protonema cells of wild-type (WT) and the cryptochrome double-knockout mutant cry1a cry1b (ΔCry) (Imaizumi et al., 2002) were grown under LL for 1 week, exposed to darkness for 24 h, and 2 h of white light (40 μmol m−2 s−1) or blue light (40 μmol m−2 s−1) or dark period was administered before the cells were sampled. Total RNAs were extracted, and the abundances of the PpSigs, psbD, and PpRpoA mRNAs were measured. Experiments were carried out as in previous Figures. The top sections show the relative levels of each mRNA. Values are means ± sd from three independent experiments. White and gray hatched bars indicate values for wild-type and cry1a cry1b cells, respectively. The bottom sections show the hybridized bands for each test gene and actin gene. A, PpSig1 mRNA; B, PpSig2 mRNA; C, PpSig5 mRNA; D, PpRpoA mRNA; E, psbD mRNA. Asterisks show genomic amplifications of PpRpoA.
Effect of Blue Light Signaling Mediated by Cryptochromes on the Daily Expression Profiles of PpSig, PpRpoA, and psbD Genes
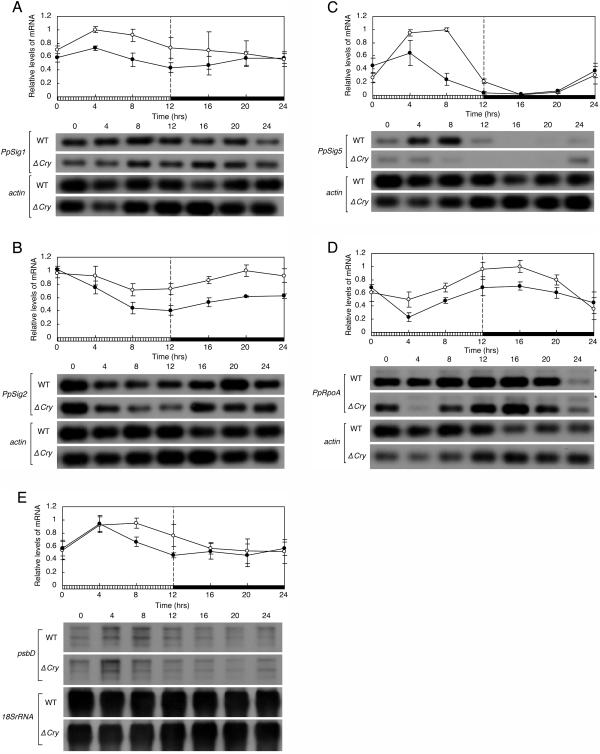
To study the effect of the blue light signal via cryptochromes on the diurnal expression profiles of PpSigs, PpRpoA, and psbD, we compared the changes in the mRNA levels of these genes in a 12-h-blue light/12-h-dark cycle (BLD) between wild-type cells and the cryptochrome mutant cells. The accumulation of all genes tested was reduced in a time-dependent manner in the disruption mutant (Fig. 8). Especially, the PpSig5 mRNA levels were more severely reduced in the latter half of the blue light period (hours 8 and 12) than at other time points, resulting in the apparent phase advance of the diurnal rhythm of PpSig5. The kinetics of psbD was also altered in a way similar to that of PpSig5 in the mutant (Fig. 8, C and E), although the reduction of the average level of expression of psbD was not as large as that of PpSig5. The results indicate that the blue light signaling via cryptochrome(s) influences the daily expression profiles of tested genes.
Figure 8.
Effects of cryptochrome mutations on diurnal expression profiles of PpSigs, psbD, and PpRpoA genes. Protonema cells of both wild-type (WT) and cry1a cry1b (ΔCry) were maintained in LD for more than 2 weeks and then transferred to a BLD. Abundances of the PpSigs, PpRpoA, and psbD mRNAs were measured in samples harvested at indicated times in BLD as described in previous figures. The top sections show the changes of the relative levels of each gene's mRNA. Values are means ± sd from three independent experiments. Striped and black bars indicate blue light intervals (40 μmol m−2 s−1) and dark intervals, respectively. White circles, wild-type; black circles, cry1a cry1b. The bottom sections show the hybridized bands for each test gene and actin or 18S rRNA gene. Asterisks show genomic amplifications of the PpRpoA gene. A, PpSig1; B, PpSig2; C, PpSig5; D, PpRpoA; E, psbD.
DISCUSSION
This is the first demonstration of the presence of a Sig5 ortholog gene in a plant other than angiosperms (Tsunoyama et al., 2004). The identification of PpSig1, PpSig2 (Hara et al., 2001b), and PpSig5 (this study) genes, together with the ancient phylogenetic origin of bryophytes, indicates that the members of the plant sigma family were diverged at very early stages in the evolution of land plants. It remains to be seen how much further the diversification of the sigma family can be traced back in the plant lineages. Interestingly, the Sig5 sequences are always found to be positioned outside all the other sigma groups in the phylogenetic trees (Fig. 1B; Allison, 2000; Fujiwara et al., 2000; Hara et al., 2001b; Tsunoyama et al., 2004), suggesting that the Sig5 group originated still earlier than any other sigma groups. This idea is supported by the fact that the distribution of intron positions of AtSig5 and PpSig5 is unique among the sigma factor genes from P. patens and Arabidopsis (Fig. 1A; Allison, 2000; Fujiwara et al., 2000; Tsunoyama et al., 2004).
This study demonstrated with a high-density time resolution that the expressions of the P. patens sigma genes are regulated differently from each other on a daily timescale, reflecting different dependency of the three genes on light and the circadian clock. In nature, environmental factors such as light and temperature, which strongly affect photosynthesis, change with a period of 1 d due to the earth's rotation. The dependency of respective photosynthesis genes on such environmental factors should differ from each other, depending on each gene's function. Therefore, to achieve efficient photosynthesis under alternating day and night cycles, it is supposed to be very important to activate a particular set(s) of chloroplast genes at a specific time of day. It is very likely that differential use of plastid sigma factors on a daily basis underlies such complex regulation. The most contrasting point in the expression of three sigma genes is that only PpSig5 is under the strong control of the circadian clock. The PpSig1 and PpSig2 genes were expressed in both day and night, though with low-amplitude fluctuations (Fig. 2), suggesting that PpSIG1 and PpSIG2 proteins might regulate the genes whose protein products are required throughout a day. The Arabidopsis AtSIG2 protein is known to activate the transcription of chloroplast tRNA genes (Kanamaru et al., 2001). This observation is consistent with the result obtained in P. patens, because tRNA genes should be expressed throughout a day for enabling translation of different mRNA species with various expression profiles. In sharp contrast to other two PpSig genes, PpSig5 showed very low levels of expression in the dark phases (Fig. 2), being controlled by the clock. The Arabidopsis AtSIG5 protein, when overexpressed, enhanced the expression of psbA, psaA, psbB, and psbD genes, indicating these photosystem genes to be possible targets of AtSIG5 (Tsunoyama et al., 2004). The P. patens psbD gene showed expression with temporal profiles similar to that of PpSig5 (Figs. 2–5), suggesting a possibility that psbD may be a target gene of Sig5 protein in P. patens as well as in Arabidopsis. It seems reasonable that a positive regulator of the photosystem genes shows a daily rhythm that ensures a sufficient level of photosystem proteins in the light. However, we do not yet have evidence that PpSIG5 regulates the expression of photosystem genes including psbD gene. To directly examine this issue, targeted disruption of PpSig5 followed by the expression analysis of chloroplast genes should be carried out.
An interesting possibility is that PpSig5 may be involved in adaptation to stress conditions. It is well known that the PSII reaction center is damaged by high light, especially when combined with other environmental stresses like low temperature (Giardi et al., 1997). Nagashima et al. (2004) postulated that AtSIG5 repairs such damaged PSII by inducing the expression of PSII genes in response to stresses such as high light and low temperature. The coincidence of strong light and low temperature, possibly damaging PSII, is expected to occur in the morning. If the function of AtSIG5 postulated by Nagashima et al. (2004) is conserved in PpSIG5, the rhythmic expression of PpSig5, peaking in the light phase with anticipatory predawn rise, is a plausible mechanism for protection against rhythmic changes of environmental stresses. We do not yet know the functions of PpSIG5. However, PpSig5 is induced by high light, whereas other PpSig genes are not (K. Ichikawa and S. Aoki, unpublished data), suggesting that Sig5 gene in P. patens may also be involved in the stress adaptation like its counterpart in angiosperm. In P. patens, not only gene disruption but also promoter replacement is applicable (Schaefer and Zrÿd, 2001). By using such a technical advantage of P. patens, it will be possible to generate strains with different phasing of the PpSig5 expression, with which the physiological significance of rhythmic expression of PpSig5 can be addressed.
Interestingly, neither PpSig5 nor psbD showed rhythmic expression in LL (Fig. 4), which is in contrast to higher plants, in which rhythmic expression of the sigma and psbD genes was clearly observed in LL (Nakahira et al., 1998; Morikawa et al., 1999). Likewise, we reported that the expression of other ccgs in P. patens (Lhcb and PpCOL1 genes) was arrhythmic in LL, whereas they showed damping oscillation in DD (Aoki et al., 2004; Shimizu et al., 2004). These observations are also in contrast to higher-plant counterpart genes, which were reported to show robust circadian oscillations in LL (Kay and Millar, 1993; Suarez-Lopez et al., 2001). To our knowledge, there has so far been no report of P. patens showing clear circadian rhythm in LL. It is an interesting question whether this arrhythmicity in LL is specific only to particular ccgs or is a fundamental property of the circadian clock itself in P. patens. This question is important especially from the evolutionary point of view, because if the latter is the case, it is indicated that regulatory mechanisms of the circadian clocks are significantly diverged between P. patens and higher plants. This issue will be clarified by cloning and expression analysis of clock genes in P. patens.
The three PpSig genes were all directly induced by light (Fig. 7). However, the profiles of the PpSig1 and PpSig2 expression in LD showed very little, if any, light responsiveness at the transition from dark to light (Fig. 2B). PpSig5 showed a clear peak in the light phase; however, judged from kinetics, this peak is unlikely to contain a component of the direct response to light, but is likely to be exclusively a circadian peak (Fig. 2B). Therefore, direct response to light cannot be clearly seen in any of the PpSig genes in LD. Because the responses to light shown in Figure 7 were induced after cells were kept in the dark for 24 h, the ability to respond to light may be only observed for cells in such dark-adapted cells. Alternatively, the response to light of the sigma genes may be circadian clock-dependent as reported for Lhcb genes in higher plants (Millar and Kay, 1996; Sugiyama et al., 2001), in which the amplitude and kinetics of the acute response changed depending on the circadian time at which a light pulse is administered.
The blue light signal, at least partly mediated by the cryptochrome(s) PpCRY1a and/or PpCRY1b, induced three PpSig genes and PpRpoA genes (Fig. 7). In addition, the results also indicated the involvement of a blue light photoreceptor(s) other than the two cryptochromes in light induction of PpSig1 and PpSig5 (Fig. 7). Very recently, Mochizuki et al. (2004) clearly demonstrated that both Cry1 and Cry2 mediate the blue light signal to induce the AtSig5 expression in Arabidopsis. The two P. patens cryptochromes are more similar to Arabidopsis Cry1 than to Cry2 (Lin and Shalitin, 2003). A yet-unidentified P. patens cryptochrome, possibly an ortholog of Arabidosis Cry2, or phototropin (Kasahara et al., 2004) might be involved in the blue light induction of PpSig1 and PpSig5. We also demonstrated that the blue light signal mediated by the cryptochrome(s) affected the daily expression profiles of all genes tested in a time-dependent manner (Fig. 8). In particular, PpSig5 and psbD genes, where a circadian clock control was revealed in this study, showed phase advance of the rhythmic expression (Fig. 8, C and E). These results are likely due to the effect of blue light signaling through a function of the circadian clock. In Arabidopsis, the expression of photoreceptor genes including two cryptochrome genes Cry1 and Cry2 are controlled by the circadian clock (Toth et al., 2001). Such rhythmic expression of photoreceptor genes may cause the time-dependent regulation by light on the expression of downstream genes.
In summary, three PpSig genes are differentially expressed on a daily timescale, largely due to different dependency of the three genes on light and the circadian clock. Moreover, blue light signaling mediated by a cryptochrome(s) fine-tunes the daily expression profiles of three PpSig genes. These regulations might be mechanisms that enable differential activation of plastid genes under natural day-night environmental cycles.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Light Sources
Physcomitrella patens subsp. patens (Ashton and Cove, 1977) was maintained in LD using white fluorescent lamps as the light source (light intensity approximately 40 μmol m−2 s−1) at 25°C. Protonema cells were grown on BCD medium supplemented with 1 mm CaCl2 and 5 mm ammonium tartrate (Nishiyama et al., 2000); the cells were collected every 7 d, and were ground with a homogenizer (Physcotron, Microtec Nition, Chiba, Japan) before they were applied to a new BCD medium supplemented with 1 mm CaCl2 and 5 mm ammonium tartrate agar plate. The light-emitting diodes (LEDs) used as blue light sources in the light induction and blue light-dark cycle experiments was STICK LED (λmax = 470 nm at 40 μmol m−2 s−1; Tokyo Rikakikai, Tokyo).
Molecular Cloning of PpSig5 cDNA and Gene from P. patens
We amplified a DNA fragment corresponding to an expressed sequence tag (accession no. BI894524) from P. patens cDNA by PCR using two primers: Sig5probeU2 (5′-GGGAAAATACAGCCCATGAATGA-3′) and Sig5probeL1 (5′-CCTCATCATTGCATGAACATGAC-3′). The PCR product with the expected size of 481 bp was used as a probe to screen a P. patens cDNA library (gift from the Physcomitrella EST Programme; 1 × 108 pfu/μL). The 5′-terminal portion of the Sig5 cDNA was obtained by the 5′-RACE method using the 5′-RACE system for amplification of cDNA ends, version 2.0 (Invitrogen, Carlsbad, CA) with primers Sig5-5′RACE-GSP1 (5′-TTGAGTTTGCGGAGCCGGATAGCTA-3′), Sig5-5′RACE-GSP2 (5′-AGCAACTTGCTGATCCTCGACCAC-3′), and Sig5-5′RACE-Nested (5′-CATTCTGCCTCCGACTGGAGCGTTT-3′). The full-length cDNA sequence was 2,625 bp in length and the largest open reading frame was predicted to encode a protein of 632 amino acids with an estimated molecular mass of 71.3 kD. The PpSig5 gene-containing region was amplified from P. patens genomic DNA using two primers, Sig5L2 (5′-TTGAGGTTGAGTCGGCTTGCTACTG-3′) and Sig5probeU2, based on the PpSig5 cDNA sequence. The PCR product with the length of 3,289 bp containing PpSig5 gene was cloned and sequenced. The exon-intron structure was predicted by comparison of the cDNA and gene sequences with the help of the program for splice site prediction (NetPlantGene, http://www.cbs.dtu.dk/services/NetPGene/).
Semiquantitative RT-PCR Analysis
Semiquantitative RT-PCR analysis was conducted as previously described (Aoki et al., 2004) with minor modifications. Protonema cells that had been cultured for more than 3 weeks in LD were harvested and frozen at intervals indicated in each figure. Total RNA was extracted from frozen cells with a kit (RNAeasy Plant Mini kit; Qiagen, Valencia, CA). The extracted RNA (1 μg) was reverse-transcribed by using M-MLV reverse transcriptase (Invitrogen) with oligo(dT)12-18 primer (total volume of the reaction mixture was 25 μL), and 1/173 of the reaction mixture was subjected to semiquantitative PCR analysis. The primers for PCR analysis were as follows: for PpSig1 hara5sita (5′-AAATCCGGCAGTCCGTCTGCTCGT-3′) and ESTprimer (5′-ACTGATGCTCTCTAGTGACA-3′); for PpSig2 PR991119/1 (5′-GTTGAATTGGATACAGAGGCT-3′) and Pr000905/1 (5′-GCTCCTGAACCAGCATTCGCTTTG-3′); for PpSig5 Sig5L5 (5′-CAAGTGGCTGAGGATCAGCAAGT-3′) and PW-U3forSig5 (5′-TTGGCGCGTTGGATATTCACTCT-3′); for PpRpoA RpoA1-f (5′-GTGAGAGGATTGAGACTGGTG-3′) and RpoA2-r (5′-AGGGAGTTGTAGGCTTTAGA-3′); for actin (accession no. AW698983) PpActin3U1 (5′-CGGAGAGGAAGTACAGTGTGTGGA-3′) and PpActin3D1 (5′-ACCAGCCGTTAGAATTGAGCCCAG-3′; Aoki et al., 2004). The optimal cycle number was 16 cycles for all PCR reactions. We designed all primer sets for PpSigs and PpRpoA genes based on sequences of both sides of an intron of each gene so that genomic amplification could be distinguished (Hara et al., 2001b). The resulting PCR products were fractionated on agarose gels, Southern blotted onto nylon membranes (nylon membrane positively charged; Roche Diagnostics, Mannheim), and probed with cDNA for each target sequence. The primer sets for the cDNA probes were as follows: for PpSig1 hara2sita (5′-TGGGTCGCATCAGGAAGGCAAAGT-3′) and ESTprimer; for PpSig2 PR991119/1 and Pr000905/1; for PpSig5 Sig5L5 and PW-U4forSig5 (5′-ACTTGTACGCTTGCGACAACACT-3′); for PpRpoA RpoA1-f and RpoA2-r; for actin PpActin3U1 and PpActin3D1. The cDNA probes amplified with these primer sets were labeled by using a digoxigenin (DIG) DNA labeling and detection kit (Roche Diagnostics). The digitized signals of the PpSigs and PpRpoA products were normalized by the signals of actin PCR products. In a control experiment where various known relative amounts of cDNA were used as PCR templates, the ratio between the PCR product of each reaction showed a linear relationship with the ratio of input amount of cDNAs.
Northern-Blotting Analysis
Each 0.3 μg of total RNA was electrophoresed with a formaldehyde-containing agarose gel, transferred onto the nylon membrane (nylon membrane positively charged; Roche Diagnostics), and hybridized with a DIG-labeled DNA probe for psbD or 18S rRNA. The DNA fragment corresponding to the coding region of psbD and 18S rRNA was amplified and then labeled with a DIG DNA labeling kit (Roche Diagnostics). The primer sets for the DNA probes were as follows: for psbD PsbdU1 (5′-TTGTAGGTTGGTCTGGTCTATTAC-3′) and PsbdD1 (5′-AATTCAGGGTCTTCAGCTGCACGA-3′); for 18S rRNA (accession no. X80986) 18SrRNA-U1 (5′-GTACTGTGAAACTGCGAATGGCT-3′) and 18SrRNA-L1 (5′-AGCTGATGACTCGCGCTTACTAGG-3′). We used a 100-ng aliquot of the DIG-labeled DNA probe in 4.0 mL of EasyHyb (Roche Diagnostics) for hybridization at 50°C and then detected psbD or 18S rRNA transcripts by chemiluminescence, by using a DIG nucleic acid detection kit (Roche Diagnostics). After detection of psbD signals, the blots were stripped with a solution containing 50% formamide (deionized), 5% SDS, and 50 mm Tris/HCl, pH 7.5, and then reprobed with 18S rRNA probe. The digitized signals of psbD transcripts were normalized by the signals of the 18S rRNA transcripts.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner under standard material transfer agreements for noncommercial research purposes.
Sequence data from this article has been deposited with the DDBJ/EMBL/GenBank data libraries under the accession number AB189171.
Acknowledgments
We thank Dr. M. Hasebe and Dr. Y. Hiwatashi (National Institute of Basic Biology, Aichi, Japan) for supplying materials and helpful advice, and Dr. T. Yamashino (Nagoya University, Nagoya, Japan) for the LED irradiation system. We also thank Dr. S. Bashiardes (University of Leeds, UK) for the P. patens cDNA library as part of the Physcomitrella EST Programme at the University of Leeds and Washington University (St. Louis) and Kyowa Hakko Kogyo for Driserase.
This work was supported by the Sasakawa Scientific Research Grant from The Japan Science Society (to K.I.) and by the Ministry of Education, Science, Sports, Culture and Technology (grant-in-aid no. 14340252 to M.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.053033.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Allison LA (2000) The role of sigma factors in plastid transcription. Biochimie 82: 537–548 [DOI] [PubMed] [Google Scholar]
- Aoki S, Kato S, Ichikawa K, Shimizu M (2004) Circadian expression of the PpLhcb2 gene encoding a major light-harvesting chlorophyll a/b-binding protein in the moss Physcomitrella patens. Plant Cell Physiol 45: 68–76 [DOI] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ (1977) The isolation and preliminary characterization of auxotrophic and analogue resistant mutants in the moss Physcomitrella patens. Mol Gen Genet 154: 87–95 [Google Scholar]
- Fujiwara M, Nagashima A, Kanamaru K, Tanaka K, Takahashi H (2000) Three new nuclear genes, sigD, sigE and sigF, encoding putative plastid RNA polymerase sigma factors in Arabidopsis thaliana. FEBS Lett 481: 47–52 [DOI] [PubMed] [Google Scholar]
- Giardi MT, Masojídek J, Godde D (1997) Effects of abiotic stresses on the turnover of the D1 reaction centre II protein. Physiol Plant 101: 635–642 [Google Scholar]
- Gray MW, Lang BF (1998) Transcription in chloroplasts and mitochondria: a tale of two polymerases. Trends Microbiol 6: 1–3 [DOI] [PubMed] [Google Scholar]
- Gruber TM, Gross CA (2003) Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57: 441–466 [DOI] [PubMed] [Google Scholar]
- Hanaoka M, Kanamaru K, Takahashi H, Tanaka K (2003) Molecular genetic analysis of chloroplast gene promoters dependent on SIG2, a nucleus-encoded sigma factor for the plastid-encoded RNA polymerase, in Arabidopsis thaliana. Nucleic Acids Res 31: 7090–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Morita M, Takahashi R, Sugita M, Kato S, Aoki S (2001. b) Characterization of two genes Sig1 and Sig2 encoding distinct plastid sigma factors in the moss Physcomitrella patens: phylogenetic relationships to plastid sigma factors in higher plants. FEBS Lett 499: 87–91 [DOI] [PubMed] [Google Scholar]
- Hara K, Sugita M, Aoki S (2001. a) Cloning and characterization of the cDNA for a plastid sigma factor from the moss Physcomitrella patens. Biochim Biophys Acta 1517: 302–306 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hayama R, Coupland G (2003) Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol 6: 13–19 [DOI] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB (2001) Molecular evidence for the early colonization of land by fungi and plants. Science 293: 1129–1133 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kadota A, Hasebe M, Wada M (2002) Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell 14: 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru K, Nagashima A, Fujiwara M, Shimada H, Shirano Y, Nakabayashi K, Shibata D, Tanaka K, Takahashi H (2001) An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant Cell Physiol 42: 1034–1043 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Kagawa T, Sato Y, Kiyosue T, Wada M (2004) Phototropins mediate blue and red light-induced chloroplast movements in Physcomitrella patens. Plant Physiol 135: 1388–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S, Millar AJ (1993) Circadian-regulated cab gene transcription in higher plants. In MW Young, ed, Molecular Genetics of Biological Rhythms. Marcel Dekker, New York, pp 73–90
- Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Kay SA (1996) Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA 93: 15491–15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki T, Onda Y, Fujiwara E, Wada M, Toyoshima Y (2004) Two independent light signals cooperate in the activation of the plastid psbD blue light-responsive promoter in Arabidopsis. FEBS Lett 571: 26–30 [DOI] [PubMed] [Google Scholar]
- Morikawa K, Ito S, Tsunoyama Y, Nakahira Y, Shiina T, Toyoshima Y (1999) Circadian-regulated expression of a nuclear-encoded plastid sigma factor gene (sigA) in wheat seedlings. FEBS Lett 451: 275–278 [DOI] [PubMed] [Google Scholar]
- Nagashima A, Hanaoka M, Shikanai T, Fujiwara M, Kanamaru K, Takahashi H, Tanaka K (2004) The multiple-stress responsive plastid sigma factor, SIG5, directs activation of the psbD blue light-responsive promoter (BLRP) in Arabidopsis thaliana. Plant Cell Physiol 45: 357–368 [DOI] [PubMed] [Google Scholar]
- Nakahira Y, Baba K, Yoneda A, Shiina T, Toyoshima Y (1998) Circadian-regulated transcription of the psbD light-responsive promoter in wheat chloroplasts. Plant Physiol 118: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Privat I, Hakimi MA, Buhot L, Favory JJ, Mache-Lerbs S (2003) Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3. Plant Mol Biol 51: 385–399 [DOI] [PubMed] [Google Scholar]
- Sato N (2001) Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci 6: 151–155 [DOI] [PubMed] [Google Scholar]
- Schaefer D (1994) Molecular genetic approaches to the biology of the moss Physcomitrella patens. PhD thesis. University of Lausanne, Lausanne, Switzerland
- Schaefer DG, Zrÿd JP (2001) The moss Physcomitrella patens, now and then. Plant Physiol 127: 1430–1438 [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, Ichikawa K, Aoki S (2004) Photoperiod-regulated expression of the PpCOL1 gene encoding a homolog of CO/COL proteins in the moss Physcomitrella patens. Biochem Biophys Res Commun 324: 1296–1301 [DOI] [PubMed] [Google Scholar]
- Stern DB, Higgs DC, Yang J (1997) Transcription and translation in chloroplasts. Trends Plant Sci 2: 308–315 [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sugiura C, Kobayashi Y, Aoki S, Sugita C, Sugita M (2003) Complete chloroplast DNA sequence of the moss Physcomitrella patens: evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res 31: 5324–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Izawa T, Oikawa T, Shimamoto K (2001) Light regulation of circadian clock-controlled gene expression in rice. Plant J 26: 607–615 [DOI] [PubMed] [Google Scholar]
- Thum KE, Kim M, Christopher DA, Mullet JE (2001) Cryptochrome 1, cryptochrome 2, and phytochrome a co-activate the chloroplast psbD blue light-responsive promoter. Plant Cell 13: 2747–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsinoremas NF, Ishiura M, Kondo T, Andersson CR, Tanaka K, Takahashi H, Johnson CH, Golden SS (1996) A sigma factor that modifies the circadian expression of a subset of genes in cyanobacteria. EMBO J 15: 2488–2495 [PMC free article] [PubMed] [Google Scholar]
- Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T (2004) Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSIG5. Proc Natl Acad Sci USA 101: 3304–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe A, Borner T (1999) Transcription and the architecture of promoters in chloroplasts. Trends Plant Sci 4: 169–170 [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA (2001) Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2: 702–715 [DOI] [PubMed] [Google Scholar]