Abstract
Disease outbreaks of verotoxin-producing Escherichia coli (VTEC) O157:H7 and non-O157 serotypes associated with leafy green vegetables are becoming a growing concern. A better understanding of the behavior of VTEC, particularly non-O157 serotypes, on lettuce under stress conditions is necessary for designing more effective control strategies. Hydrogen peroxide (H2O2) can be used as a sanitizer to reduce the microbial load in leafy green vegetables, particularly in fresh produce destined for the organic market. In this study, we tested the hypothesis that H2O2 treatment of contaminated lettuce affects in the same manner transcription of stress-associated and virulence genes in VTEC strains representing O157 and non-O157 serotypes. Six VTEC isolates representing serotypes O26:H11, O103:H2, O104:H4, O111:NM, O145:NM, and O157:H7 were included in this study. The results indicate that 50 mM H2O2 caused a population reduction of 2.4–2.8 log10 (compared to non-treated control samples) in all six VTEC strains present on romaine lettuce. Following the treatment, the transcription of genes related to oxidative stress (oxyR and sodA), general stress (uspA and rpoS), starvation (phoA), acid stress (gadA, gadB, and gadW), and virulence (stx1A, stx2A, and fliC) were dramatically downregulated in all six VTEC serotypes (P ≤ 0.05) compared to not treated control samples. Therefore, VTEC O157:H7 and non-O157 serotypes on lettuce showed similar survival rates and gene transcription profiles in response to 50 mM H2O2 treatment. Thus, the results derived from this study provide a basic understanding of the influence of H2O2 treatment on the survival and virulence of VTEC O157:H7 and non-O157 serotypes on lettuce.
Keywords: verotoxin-producing Escherichia coli, non-O157 serotypes, lettuce decontamination, hydrogen peroxide treatment, gene transcription
Introduction
Verotoxin-producing Escherichia coli (VTEC), also referred to as Shiga toxin (Stx)-producing E. coli (STEC), often cause life-threatening diseases, such as hemorrhagic colitis (HC) and hemolytic uremic syndrome (HUS) (Croxen et al., 2013; Karpman and Stahl, 2014; Karmali, 2017). Although VTEC serotype O157:H7 is commonly identified in human diseases, non-O157 serogroups have been increasingly associated with serious outbreaks and were recently responsible for more than 50% of STEC illness in U.S. (Luna-Gierke et al., 2014; Parsons et al., 2016; CDC, 2017). In 2015, the incidence of confirmed VTEC non-O157 infections was 40% higher than in 2012–2014 (Huang et al., 2016). More than 70% of infections linked to non-O157 VTEC were caused by serotypes O26, O45, O103, O111, O121, and O145 (termed the Top 6; Saito et al., 1998; Brooks et al., 2005; Folster et al., 2011; Bradley et al., 2012; Brown et al., 2012). In 2011, enteroaggregative E. coli O104:H4 caused the biggest outbreak in Germany. This strain produces Stx2 and is one of the most virulent strains of non-O157 VTEC (Buchholz et al., 2011; Zangari et al., 2013; Karmali, 2017).
Fresh leafy green vegetables are an important part of a healthy diet due to their richness in minerals, vitamins, and phytochemicals. However, leafy green vegetables can be contaminated by pathogenic bacteria such as VTEC during growth, harvesting, and transportation leading to subsequent illnesses and outbreaks (Wachtel et al., 2002; Solomon et al., 2003; Islam et al., 2004; Delaquis et al., 2007). A number of surveys have shown an increase in foodborne outbreaks linked to contaminated leafy green vegetables (Harris et al., 2003; Sivapalasingam et al., 2004; Rangel et al., 2005; Lynch et al., 2009; Painter et al., 2013; Herman et al., 2015). Although the contamination can be minimized by preventing produce exposure to sources of pathogenic bacteria, including contaminated water, soil, and animals, the occasional contamination of leafy green vegetables on farms by VTEC can still occur, which leads to contaminated produce entering the processing lines (Olaimat and Holley, 2012). Therefore, effectively reducing the contamination during processing is crucial to ensure the safety of fresh leafy green vegetables.
Chlorine has been widely used as a sanitizer to reduce the microbial load in fresh-cut vegetables (Beuchat et al., 1998). However, chlorine may react to form potentially carcinogenic or mutagenic products (Hurst, 1995). In addition, the by-products formed when sodium hypochlorite (NaOCl—another sanitizer commonly used in the fresh produce industry) reacts with organic compounds, have been shown to increase the risk of bladder cancer (Villanueva et al., 2004). Hydrogen peroxide (H2O2) is a potential alternative to chlorine treatment and breaks down to ecologically friendly water and oxygen (Linley et al., 2012). In addition, H2O2 has been approved for use in organic postharvest processing systems. As such, this sanitizer could be used for decontamination of fresh produce destined for organic market.
To date, several studies have been conducted on the gene expression in pure culture of E. coli O157:H7 under oxidative stress (Wang et al., 2009; Allen and Griffiths, 2012; Mei et al., 2015). In addition, using H2O2 as a sanitizer on fresh produce to reduce E. coli O157:H7 was investigated (Sapers et al., 2000; Huang et al., 2012). However, the survival and gene transcription of non-O157 VTEC serotypes under stress conditions on lettuce remain largely unknown. In this study, the behaviors of O157:H7 and non-O157 VTEC serotypes in response to H2O2 treatment on lettuce were evaluated.
Materials and Methods
Bacterial Strains and Growth Conditions
Six VTEC strains were tested in this study: E. coli O157:H7 (EDL933) (ATCC 700972), O26:H11 (EC20070549), O103:H2 (EC19970811), O104:H4 (NML#11-3088), O111:NM (EC20070546), and O145:NM (EC19970355). E. coli EDL933 was included in the present study because it is prototype O157:H7 strain. Serotypes O26, O103, O111, and O145 are predominant non-O157 serotypes worldwide, therefore strains representing these serotypes have been investigated in this study. In addition VTEC O104:H4 isolated from German outbreak was selected, because this strain is highly virulent and is linked to fresh produce outbreak. All the strains except EDL933 were of human origin. E. coli O157:H7 (EDL933) was isolated from raw hamburger meat linked to an outbreak of HC. Further details regarding these strains have been published previously (Mei et al., 2015). E. coli O157:H7 (EDL933) possesses genes encoding Stx1 and Stx2, intimin gene (eae) and flagellin gene (fliC). VTEC O26:H11 (EC20070549) and O103:H2 (EC19970811) do not have stx2 genes. VTEC strains O111:NM (EC20070546) and O145:NM (EC19970355) do not have genes encoding Stx2 and flagellin. Enteroaggregative E. coli strain O104:H4 (NML#113088) does not possess genes encoding Stx1 and intimin. For simplicity, VTEC strains will be referred to by their serotype as each strain has its distinct serotype. All the strains were grown in Tryptic soy broth (TSB) at 37°C with shaking (180 rpm) for 18 h. Subsequently 0.5 mL of 18 h culture of each VTEC strain was inoculated into 50 mL pre-warmed TSB and incubated for 3 h at 37°C with shaking (180 rpm). The bacterial cells in logarithmic phase were collected, washed twice with sterile distilled water and used to inoculate lettuce samples.
Hydrogen Peroxide Treatment of Contaminated Lettuce
Romaine lettuce was purchased from a local grocery store in Guelph. The outer leaves were removed and the remaining leaves were cut into 4 × 10 cm slices. Twenty-five grams of the leaves were weighed, placed into sterile polyethylene stomacher bags, and inoculated with 5 mL of bacterial suspension (108 CFU/g). The leaves were massaged for 2 min to distribute the bacterial suspension evenly on the leaves. Subsequently, 195 mL of H2O2 was added to the bag to achieve a final concentration of 50 mM. The negative control sample contained lettuce leaves with 5 mL of autoclaved distilled water and 195 mL H2O2. The non-treated control sample contained 5 mL of bacterial suspension and 195 mL of autoclaved distilled water. The bags were incubated at room temperature for 40 min. After incubation, 1 mL of culture was taken from each stomacher bag and serially diluted with 0.1% (w/v) buffered peptone water. Subsequently, 100 μL of each dilution was spread on cefixime/tellurite—Sorbitol MacConkey agar (CT-SMAC) plates in duplicate. The plates were incubated for 18 h at 37°C, and the CFU/mL was calculated. Additionally, 4 mL samples from controls and each stomacher bag treated with 50 mM H2O2 were collected for RNA extraction.
RNA Extraction and cDNA Synthesis
Bacterial cells were concentrated by centrifugation before RNA extraction. After taking samples from controls and each stomacher bag treated with 50 mM H2O2, each 4 mL sample was added to 8 mL (2 volume) of RNAprotect Bacteria Reagent (Qiagen). The samples were centrifuged for 10 min at 5000×g and supernatants were decanted prior to RNA extraction. RNA was isolated using the RNeasy Mini Kit (Qiagen, Mississauga, ON, Canada), following the manufacturer’s instructions for Gram-negative bacteria. Contaminating genomic DNA was removed from each RNA preparation using the Turbo DNA-freeTM kit (Ambion, Cambridge, UK), according to the manufacturer’s instructions for DNase treatments. Total RNA concentration was determined using a Thermo Scientific Nanodrop 2000 (ON, Canada). Subsequently, 0.3 μg of RNA was reverse-transcribed using SuperScript III (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. A no reverse transcriptase control (No RT) was included for each DNase-treated RNA sample.
Real-Time PCR
All stress-associated and virulence genes tested in this study are described in Table 1. Transcription of key stress and virulence genes (oxyR, sodA, soxR, uspA, rpoS, phoA, dps, cspA, cspC, cspE, gadA, gadB, gadW, mutS as well as eae, stx1A, stx2A, and fliC) was evaluated using a comparative real-time PCR. Each 25-μL reaction contained 1 μL of reverse-transcribed cDNA, 12.5 μL of Power SYBR® Green PCR Master Mix, 0.25 U AmpErase® Uracil N-Glycosylase (UNG; Applied Biosystems), 2.0 μM of each primer and 6.25 μL nuclease-free water. All primers used in the study were previously described (Mei et al., 2015). Amplification and detection were carried out on a MX3500® Multiplex Quantitative PCR System (Stratagene, La Jolla, CA, USA) with an initial temperature of 50°C for 2 min. Following denaturation at 95°C for 10 min, reactions were cycled 40 times as follows: amplification at 95°C for 15 s, annealing at 60°C for 30 s, extension at 72°C for 30 s. Subsequent melt curve analysis involved heating the products to 95°C for 1 min, followed by cooling to 55°C for 30 s, and heating to 95°C while monitoring fluorescence. No template control and No RT were included for each assay and no Ct values were obtained for all negative controls after 40 cycles of PCR (data not shown). Several well-known candidate reference genes including 16S rRNA, tufA/B, mdh, pyrC, gatB, recA, serC, frr, rpsU, udp, mdoG, rpoA, and arcA were tested for expression stability as previously described (Mei et al., 2015). Only 16S rRNA gene expression was stable in all VTEC strains on lettuce under experimental conditions. Therefore, 16S rRNA was used as reference gene in this study to normalize the data. Serial dilutions of the cDNA template were examined with 16S rRNA primers. Using 50-fold dilutions the Ct value was around 12, and with other primers for different target genes all the Ct values were between 20 and 35. Therefore, 50-fold dilutions of cDNA samples were used in the experiment.
Table 1.
List of stress-associated and virulence genes tested for differential gene transcription upon treatment of VTEC strains on lettuce with hydrogen peroxide.
| Stress-associated and virulence genes | Protein encoded/function |
|---|---|
| Oxidative stress | |
| oxyR | DNA-binding transcriptional regulator OxyR |
| sodA | Manganese-containing superoxide dismutase |
| soxR | Redox-sensitive transcriptional activator of oxidative stress regulon |
| General stress | |
| uspA | Universal stress global response regulator |
| rpoS | Regulator of the general stress response (σS) |
| Starvation | |
| phoA | Alkaline phosphatase |
| dps | DNA-binding protein from starved cells |
| Cold shock | |
| cspA | Cold shock protein A |
| cspC | Cold shock protein C |
| cspE | Cold shock protein E |
| UV | |
| mutS | DNA mismatch repair protein—Mutator S |
| Acid resistance | |
| gadA | Glutamate decarboxylase A |
| gadB | Glutamate decarboxylase B |
| gadW | DNA-binding transcriptional activator GadW |
| Intimin | |
| eae | Intimin (adherence protein) |
| Toxin | |
| stx1A | Shiga-like toxin 1 A subunit |
| stx2A | Shiga-like toxin 2 A subunit |
| Motility | |
| fliC | Flagellin |
Statistical Analysis
The effect of H2O2 treatment on survival and gene transcription of VTEC strains present on lettuce was investigated by at least three independent experiments. Each biological sample was run in duplicate on real-time RT PCR. Relative mRNA levels were determined according to the method described by Hellemans et al. (2007). Gene transcription data were analyzed using Student’s t-test. Data on reduction of VTEC populations on lettuce following exposure to H2O2 were analyzed using one-way ANOVA.
Results
Treatment of contaminated lettuce with 50 mM H2O2 for 40 min reduced the populations of all VTEC strains tested in this study by 2.4–2.8 log10 (Table 2). The differences in sensitivity to H2O2 treatment on lettuce between VTEC O157:H7 and non-O157 serotypes were not statistically significant (P > 0.05).
Table 2.
Effect of H2O2 treatment on populations of six VTEC strains on lettuce.
| E. coli strain | Mean log reduction ± SE |
|---|---|
| O157:H7 (EDL933) | 2.42 ± 0.15 |
| O26:H11 (EC20070549) | 2.73 ± 0.25 |
| O103:H2 ((EC19970811) | 2.81 ± 0.15 |
| O104:H4 (NML#11-3088) | 2.53 ± 0.18 |
| O111:NM (EC20070546) | 2.82 ± 0.25 |
| O145:NM (EC19970355) | 2.52 ± 0.26 |
Data are expressed as mean log reduction ± SE.
In addition, the influence of H2O2 treatment on stress and virulence gene transcription of six VTEC strains on romaine lettuce was investigated. This study focused on well-known virulence genes including genes encoding intimin (eae), Stx genes stx1A and stx2A and flagellin genes (fliC) (Figures 1–6). Transcription of key stress-associated genes such as genes involved in response to oxidative damage (oxyR, sodA, and soxR), general stress (uspA and rpoS), and starvation (phoA and dps) was investigated. The study also focused on the effects of H2O2 treatment of contaminated lettuce on transcription of acid stress genes (gadA, gadB, and gadW), cold shock (cspA, cspC, and cspE), and gene related to UV radiation stress (mutS) (Figures 1–6). A fold change cutoff of 1.5 was applied in this study.
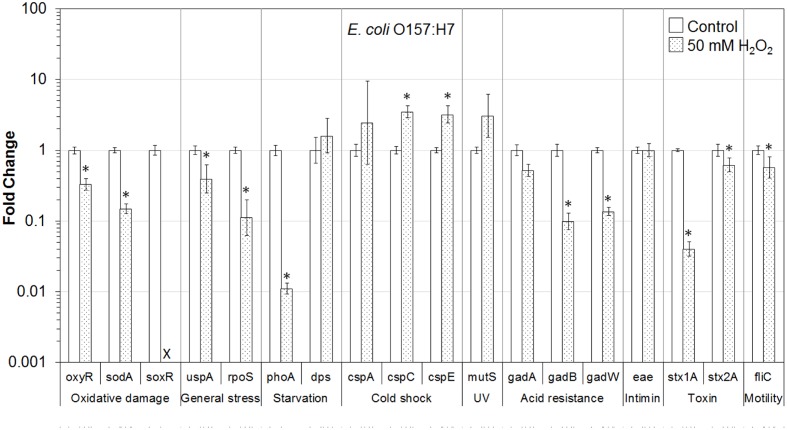
FIGURE 1.
Effect of H2O2 on gene transcription of E. coli O157:H7. Relative gene transcription represents the change in transcription compared to the bacteria without H2O2 treatment (control, value of 1.0). The transcription of each gene was normalized to the 16S rRNA transcription in each sample. Data are expressed as the means ± SE for RNA extracted in three biological replicates. × denotes no transcription of soxR by E. coli O157:H7 after exposure to H2O2 on lettuce, however, soxR is present in this strain. ∗denotes the significant change of gene transcription between treatment and control.
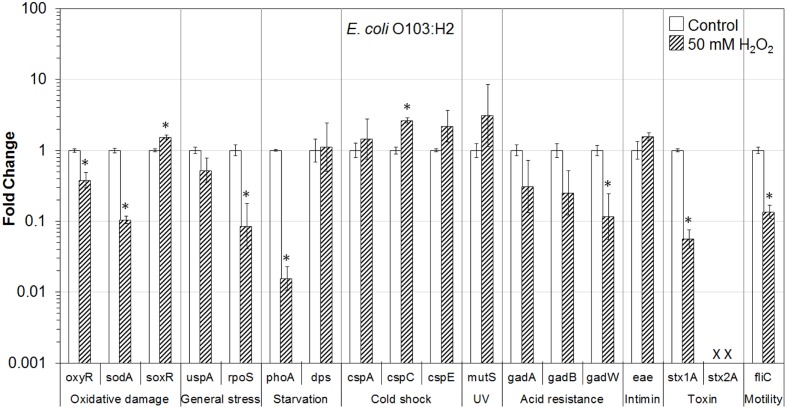
FIGURE 6.
Effect of H2O2 on gene transcription of E. coli O103:H2. Relative gene transcription represents the change in transcription compared to the bacteria without H2O2 treatment (control, value of 1.0). The transcription of each gene was normalized to the 16S rRNA transcription in each sample. Data are expressed as the means ± SE for RNA extracted in three biological replicates. × denotes absence of stx2A gene in E. coli O103:H2. ∗indicates the significant change of gene transcription between treatment and control.
In E. coli O157:H7 present on lettuce, the genes associated with oxidative stress (oxyR and sodA), universal stress (uspA and rpoS), starvation (phoA), acid stress (gadB and gadW), and virulence (stx1A, stx2A, and fliC) were significantly (P ≤ 0.05) downregulated after exposure to 50 mM H2O2. The gadA gene was downregulated 1.9-fold and the expression of soxR was below the detectable level. Interestingly, two genes related to cold shock (cspC and cspE) were significantly (P ≤ 0.05) upregulated. Genes related to starvation (dps), cold shock (cspA), and mismatch repair (MMR; mutS) were also upregulated (Figure 1).
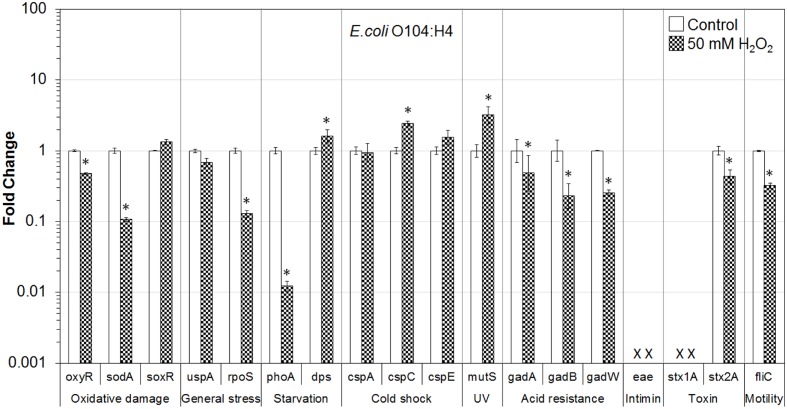
In the case of E. coli O104:H4 on lettuce, H2O2 treatment caused significant (P ≤ 0.05) downregulation of genes related to oxidative stress (oxyR and sodA), general stress (rpoS), acid resistance (gadA, gadB, and gadW), and virulence (stx2A and fliC). The uspA gene was also downregulated. Transcription of dps, cspC, and mutS was significantly (P ≤ 0.05) upregulated. In addition, gene encoding cold shock protein (cspE) was upregulated (Figure 2).
FIGURE 2.
Effect of H2O2 on gene transcription of E. coli O104:H4. Relative gene transcription represents the change in transcription compared to the bacteria without H2O2 treatment (control, value of 1.0). The transcription of each gene was normalized to the 16S rRNA transcription in each sample. Data are expressed as the means ± SE for RNA extracted in three biological replicates. × denotes absence of eae and stx1A genes in E. coli O104:H4. ∗indicates the significant change of gene transcription between treatment and control.
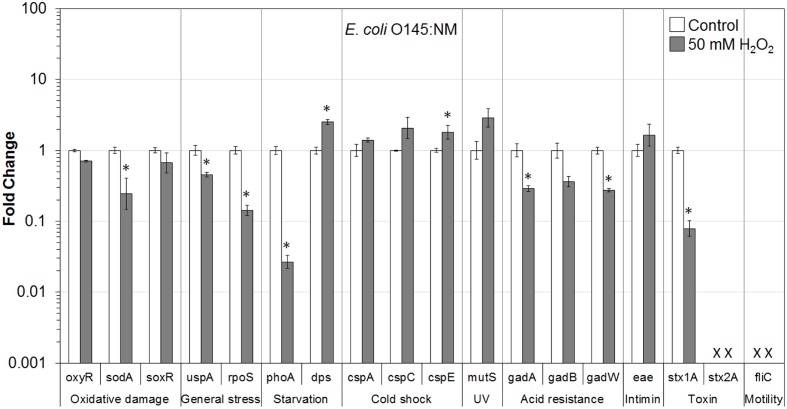
In E. coli O145:NM present on lettuce, genes related to oxidative damage (sodA), general stress (uspA and rpoS), starvation (phoA), acid resistance (gadA and gadW), and virulence (stx1A) were significantly (P ≤ 0.05) downregulated. The soxR and gadB genes were also downregulated. Only two genes (dps and cspE) were significantly (P ≤ 0.05) upregulated. Furthermore, genes encoding cold shock protein (cspC), MMR sensor (mutS), and intimin (eae) were upregulated (Figure 3).
FIGURE 3.
Effect of H2O2 on gene transcription of E. coli O145:NM. Relative gene transcription represents the change in transcription compared to the bacteria without H2O2 treatment (control, value of 1.0). The transcription of each gene was normalized to the 16S rRNA transcription in each sample. Data are expressed as the means ± SE for RNA extracted in three biological replicates. × denotes absence of stx2A and fliC genes in E. coli O145:NM. ∗indicates the significant change of gene transcription between treatment and control.
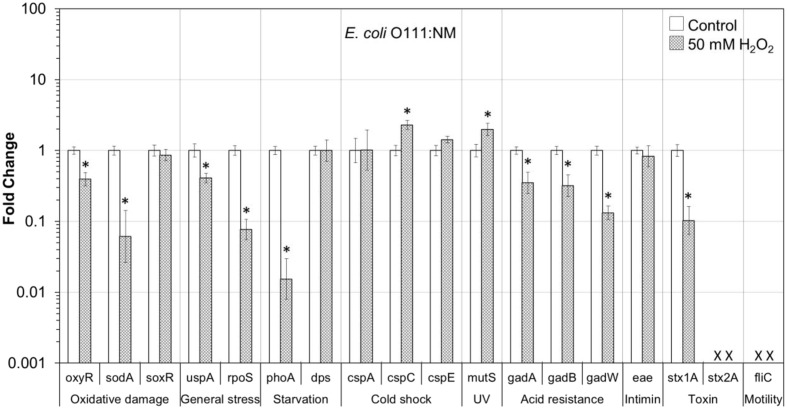
In E. coli O111:NM on lettuce, nine genes, responsible for oxidative damage (oxyR and sodA), general stress (uspA and rpoS), starvation (phoA), acid resistance (gadA, gadB, and gadW) and virulence (stx1A), were significantly (P ≤ 0.05) downregulated. Whereas, genes for cold shock (cspC) and MMR (mutS) were significantly (P ≤ 0.05) upregulated (Figure 4).
FIGURE 4.
Effect of H2O2 on gene transcription of E. coli O111:NM. Relative gene transcription represents the change in transcription compared to the bacteria without H2O2 treatment (control, value of 1.0). The transcription of each gene was normalized to the 16S rRNA transcription in each sample. Data are expressed as the means ± SE for RNA extracted in three biological replicates. × denotes absence of stx2A and fliC genes in E. coli O111:NM. ∗indicates the significant change of gene transcription between treatment and control.
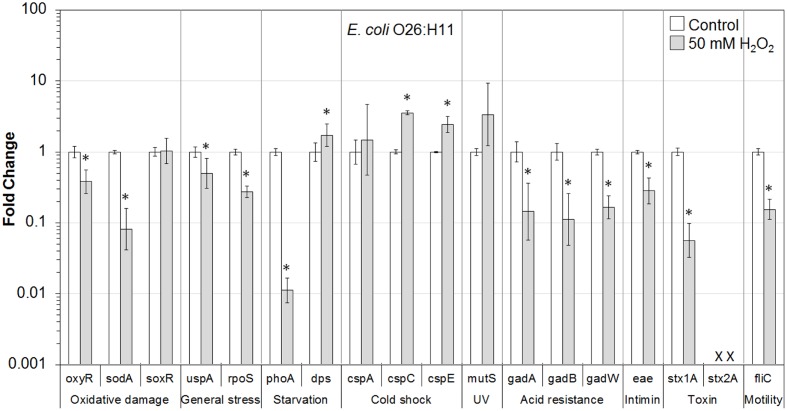
In E. coli O26:H11 present on lettuce, 9 out of 17 genes, including genes associated with oxidative damage (oxyR and sodA), general stress (uspA and rpoS), starvation (phoA), acid resistance (gadA, gadB, and gadW), and virulence (eae, stx1A, and fliC), were significantly (P ≤ 0.05) downregulated. Three genes, dps (starvation-related), cspC and cspE (cold shock-related), were significantly (P ≤ 0.05) upregulated. In addition, genes for cold shock (cspA) and DNA damage repair (mutS) were upregulated (Figure 5).
FIGURE 5.
Effect of H2O2 on gene transcription of E. coli O26:H11. Relative gene transcription represents the change in transcription compared to the bacteria without H2O2 treatment (control, value of 1.0). The transcription of each gene was normalized to the 16S rRNA transcription in each sample. Data are expressed as the means ± SE for RNA extracted in three biological replicates. × denotes absence of stx2A gene in E. coli O26:H11. ∗indicates the significant change of gene transcription between treatment and control.
In E. coli O103:H2 on lettuce, genes related to oxidative stress (oxyR and sodA), general stress (rpoS), starvation (phoA), acid resistance (gadW), and virulence (stx1A and fliC) were significantly (P ≤ 0.05) downregulated. In addition, two genes associated with acid resistance—gadA and gadB, as well as uspA gene were downregulated. Interestingly, soxR and cspC, were significantly (P ≤ 0.05) upregulated. Genes encoding cold shock protein (cspA and cspE), and intimin (eae) were also upregulated (Figure 6).
Overall, most of the genes associated with stress and virulence were downregulated in O157:H7 and non-O157 serotypes on lettuce treated with 50 mM H2O2. Only three genes, associated with cold shock (cspC and cspE) and MMR (mutS), were upregulated in all VTEC strains tested (Figures 1–6).
Discussion
The effect of H2O2 on the survival on lettuce of six VTEC strains, representing O26:H11, O103:H2, O104:H4, O111:NM, O145:NM, and O157:H7 serotypes, was investigated in this study. A population reduction of 2.4–2.8 log10 was observed for all VTEC strains on lettuce following treatment with 50 mM H2O2 (Table 1). A previous study in our laboratory, using the same VTEC strains suspended in TSB, reported greater sensitivity to H2O2. For instance treatment, of pure broth culture of VTEC O104:H4 with 1 mM H2O2 resulted in a 2.7 log reduction and 2.5 mM H2O2 caused a 3.7 log reduction in population of E. coli O104:H4 (Tang and Kostrzynska, 2012). Therefore, the results derived from these studies demonstrate that H2O2 treatment is less effective in lettuce decontamination compared to its effect on reducing VTEC populations in pure cultures. Previous investigations showed that several factors, such as organic loads of fresh-cut produce (Gonzalez et al., 2004), whole- or cut-leaf wash (Nou and Luo, 2010), and leaf age (Brandl and Amundson, 2008), can influence the efficacy of sanitizer to inactivate E. coli O157:H7 on fresh produce. These studies indicate that the lettuce leaves may help to protect bacteria, making it harder to eliminate pathogens from fresh-cut lettuce. Indeed, bacterial cells on lettuce may be physically sequestered from H2O2 exposure. In addition, organic molecules released by fresh-cut lettuce will react with H2O2 and therefore reduce the effective exposure to H2O2. Therefore, in order to choose an effective treatment to reduce the pathogens on leafy greens, it is important to understand the behavior of bacteria on lettuce under different stress conditions.
To get a better understanding of the response to H2O2 treatment in different VTEC serotypes, in the present study, the transcription of genes related to stress and virulence in O157:H7 and non-O157 serotypes on lettuce was evaluated. Different gene transcription profiles following exposure to H2O2 were observed in VTEC strains on lettuce compared to pure broth cultures (Mei et al., 2015). H2O2 caused dramatic downregulation of stress-associated and virulence genes in VTEC strains present on lettuce, including O157 and non-157 serotypes. Ten genes were downregulated in all six VTEC strains on lettuce (Figures 1–6). However, only three genes were upregulated. These genes belong to different regulation systems in E. coli and respond to various environmental conditions by adjusting behavior accordingly to protect cells from damaging.
Anti-Oxidant System Response (oxyR, sodA, and soxR)
Superoxide dismutases (SODs) and catalases are employed by E. coli to respond to superoxide and peroxide stress. There are three SODs, including MnSOD (sodA), FeSOD (sodB), and CuZnSOD (sodC), and two catalases (HPI and HPII, encoded by katG and katE, respectively) in E. coli. These SODs and catalase genes are regulated by two major oxidative stress regulons, OxyR and SoxRS (Chiang and Schellhorn, 2012). In the presence of oxidative stress, OxyR can sense H2O2 and be converted to the oxidized form, subsequently activating transcription of the OxyR regulon genes, such as katG, dps, and oxyR, which protect the cell from H2O2 toxicity (Storz et al., 1990a; Farr and Kogoma, 1991; Chiang and Schellhorn, 2012). In this study, the transcription of oxyR was downregulated in all VTEC strains (Figures 1–6), this may cause a reduction in catalase expression, increasing peroxide sensitivity.
SoxRS is another key regulon triggered under oxidative stress in E. coli. It is positively regulated by superoxide-generating agents such as paraquat and constitutes a two-stage regulatory system, in which SoxR, activated as a transcriptional activator, induces the expression of soxS and the resulting increased levels of SoxS protein regulate the transcription of the various genes important for responding to oxidative stress (Demple and Amabile-Cuevas, 1991; Nunoshiba et al., 1992; Chiang and Schellhorn, 2012). The genes controlled by soxRS include sodA (Mn-containing SOD), nfo (DNA repair endonuclease IV), micF (antisense regulator of ompF), and zwf (glucose-6-phosphate dehydrogenase) (Christman et al., 1985; Greenberg et al., 1990; Tsaneva and Weiss, 1990). In the present study, following exposure of VTEC strains on lettuce to 50 mM H2O2, the transcription of soxR in E. coli O103:H2 slightly increased (1.5-fold), whereas, the transcription of soxR decreased to below the detectable level in O157:H7. Therefore, SodA reduction may be attributed to the reduction or only slight induction of soxR. Manchado et al. (2000) reported that the expression of OxyR-regulated genes (katG and dps) were induced when the concentration of H2O2 was from 1 to 100 μM, while the higher concentration (≥500 μM H2O2) resulted in the upregulation of soxS and sodA. That study suggests that the expression of oxyR or soxRS is dose-dependent. In addition, downregulation of soxR was also observed in VTEC broth cultures exposed to oxidative stress (Mei et al., 2015), however, the transcription of sodA was significantly (P ≤ 0.05) increased in pure cultures of all VTEC strains. The inconsistent results of expression of soxR and sodA may indicate that (1) sodA can react to superoxide stress independently; (2) SoxRS is not the sole regulator of sodA gene.
General Stress Response (uspA and rpoS) and Cold Shock Response (cspA, cspC, and cspE)
Escherichia coli contains a large CspA family, consisting of nine homologous cold inducible proteins, CspA to CspI, among which, CspA is the major one produced at 10–24°C, and is negatively downregulated by cspC. Whereas, CspC and CspE are constitutively produced at 37°C and are not temperature regulated (Yamanaka et al., 1994; Etchegaray et al., 1996; Phadtare and Inouye, 2001). It has been reported that CspC and CspE are important regulators of the expression of RpoS, a global stress response regulator, and UspA, universal stress protein A responding to various general stresses (Nystrom and Neidhardt, 1992, 1994). In addition, RpoS-controlled genes such as dps and katG are upregulated or downregulated by the overexpression or deletion of cspC and cspE (Phadtare and Inouye, 2001; Phadtare et al., 2006). Therefore, CspC and CspE play important roles in the stress response of E. coli. Upregulation of cspC and cspE in all six VTEC strains on lettuce was observed in the present study (Figures 1–6). However, uspA and rpoS were downregulated, and a minor change in the transcription of dps was observed. Interestingly, the transcription of cspC was downregulated in broth cultures of all VTEC strains exposed to 2.5 mM H2O2 at 37°C (Mei et al., 2015). On the other hand, in the present study cspC was upregulated in VTEC strains on lettuce treated with 50 mM H2O2 at room temperature. Previous investigation showed that CspC and CspE cannot be induced under some stress conditions, like 0.5 M NaCl, 0.5 M KCl, 5% ethanol, pH 10, pH 4, temperature of 15 or 50°C (Phadtare and Inouye, 2001). It is possible that a different concentration of H2O2 used for lettuce decontamination as well as environmental factors such as H2O2 released from fresh-cut lettuce leaves as well as nutrients present in lettuce samples and natural microbiota contributed to the induction of cspC and cspE. More interestingly, contrary results of transcription of cspC and cspE observed in response to H2O2 treatment of VTEC pure cultures and in the same strains present on lettuce, suggest that VTEC may react to the same stress differently in different environments. However, it is not possible to rule out that induction of cold stress genes in present study was more of the effect of temperature change combined with peroxide treatment. Centrifugation of bacterial cells using refrigerated centrifuge, following VTEC exposure on lettuce to H2O2 could influence transcription of cold shock genes. Further studies are required to gain a better understanding of the stress conditions that affect the cspC and cspE gene transcription in VTEC present on lettuce.
Starvation Stress Response (phoA and dps)
The gene phoA, encoding alkaline phosphatase, was induced under a phosphate-limited condition, but was not synthesized in normal growth medium (Han and Lee, 2006). The phoA is a member of pho regulon, regulated by a two-protein system PhoR–PhoP (Sola-Landa et al., 2003). The phoA was significantly downregulated in all the VTEC strains on lettuce tested in this study (Figures 1–6). Furthermore, downregulation of phoA was observed in most VTEC strains when pure cultures were treated with H2O2 (Mei et al., 2015). Therefore, further studies are needed to test, if H2O2 treatment changes the sensitivity of VTEC strains to phosphate-limited condition.
The H2O2 can cause lethality of the bacterial cells through several mechanisms. It has been proposed that the primary cause of cell inactivation by H2O2 or other oxidative agents is DNA damage (Storz et al., 1990b). Thus, it is important for the cells to cope with stresses by inducing the production of a variety of DNA repair enzymes as well as catalases. Glutathione also acts to protect cells from oxidative stress. In addition, Dps—the DNA binding protein from starved cells, protects cells during environmental stresses, including oxidative stress and nutritional deprivation (Almiron et al., 1992; Calhoun and Kwon, 2011). Dps protects cells from harmful oxidative radicals by DNA binding, iron storage, and by binding and oxidizing Fe ions at ferroxidase centers. Furthermore, Dps may regulate the expression of DNA repair enzymes and catalases necessary for stress resistance. Zheng et al. (2001) reported that the expression of dps considerably increased when the cells were exposed to 1 mM H2O2 for 10 min. However, only minor upregulation of dps was observed in this study for most of VTEC strains on lettuce (Figures 1–6).
Mismatch Repair Response (mutS)
MutS is a member of methylation-dependent MMR system which helps to maintain chromosome stability in E. coli. MutS binds to the mismatches and initiates the long-patch MMR on daughter DNA strands (Modrich and Lahue, 1996; Schofield and Hsieh, 2003). MMR has also been shown to be involved in the repair of oxidative DNA damage which causes spontaneous lesion 7,8-dihydro-8-oxo-guanine (8-oxoG or GO; Michaels and Miller, 1992; Tchou and Grollman, 1993; Lu et al., 2001). Studies have shown that the amount of MutS in E. coli remarkably decreased when cells were in the stationary phase and under starvation stress and the expression of MutS repair protein was negatively regulated by the RpoS and Hfq global regulators (Feng et al., 1996; Tsui et al., 1997; Li et al., 2003). In the present study, mutS was upregulated following exposure of VTEC on lettuce to H2O2 (Figures 1–6). This upregulation may be caused by significantly downregulated expression of RpoS, which is a negative regulator of mutS.
Acid Response (gadA, gadB, and gadW)
Food-borne pathogenic E. coli must be able to survive in the extremely acidic environment of the stomach and resist very low pH (1.5–3.0) for several hours (Peterson et al., 1989). Previous studies have shown that a total of 12 genes comprise an acid fitness island, including a glutamate decarboxylase (GAD) system and three transcriptional regulators (GadE, GadX, and GadW) of the GAD enzymes, which renders E. coli the ability to survive strong acidic stress (Hommais et al., 2001, 2004; Ma et al., 2002; Bergholz et al., 2007; Mates et al., 2007). E. coli produce two isozymes of GAD encoded by the gadA and gadB genes, which are induced by GadX at any pH, while GadW represses expression of gadX. GadW activates the expression of gadA and gadB only in the absence of GadX (Ma et al., 2002; Tucker et al., 2003). In addition, it has been reported that the expression of gadA and gadBC were induced when bacteria were cultured in acidified medium, treated with acetate, and during entry into stationary phase (Selinger et al., 2000; Arnold et al., 2001; Tucker et al., 2002). However, the expression of regulators GadX and GadW, under the same conditions, were unknown. In the present study, gadA, gadB, and gadW were significantly downregulated in all six VTEC strains on lettuce (Figures 1–6). We suspect that the expression of gadX was also downregulated in response to H2O2, which subsequently resulted in the downregulation of gadA and gadB. Furthermore, the results indicate that following H2O2 treatment, VTEC on lettuce may become more sensitive to acid stress, which decreases the viability of pathogens under low pH.
Virulence Factors (Encoded by eae, stx1A, stx2A, and fliC Genes)
Production of Stx is the definitive virulence factor of VTEC O157:H7 and non-O157 serotypes (Croxen et al., 2013; Scheutz, 2014; Smith et al., 2014; Franz et al., 2015). Stx produced by VTEC can be classified into two types Stx1 and Stx2. There are three subtypes of Stx1 (a, c, d) and seven subtypes of Stx2 (a, b, c, d, e, f, g). Although both toxins could cause bloody diarrhea and HUS, a specific subset of stx2 subtypes (stx2a, stx2c, and stx2d) have a higher association with HC and HUS than stx1 subtypes or other stx2 subtypes (Scheutz, 2014). Stx are encoded on bacteriophage genomes that are integrated into bacterial chromosome. As such, the biology of the Stx-encoding phages influences the expression of stx1 and stx2. Production and release of toxins depend on induction of these bacteriophages (Croxen et al., 2013; Scheutz, 2014). Amongst the VTEC strains tested in this study, E. coli O157:H7 produced both Stx1 and Stx2, E. coli O104:H4 produced only Stx2, and the remaining four strains produced only Stx1. In this study, both stx1A and stx2A genes were dramatically downregulated in all VTEC strains on lettuce following exposure to H2O2 (Figures 1–6).
Many VTEC strains produce attaching and effacing (AE) lesions, which are controlled by the locus of enterocyte effacement (LEE; Croxen et al., 2013; Scheutz, 2014). Although AE lesions are not essential for bloody diarrhea and HUS, the majority of strains implicated in these syndromes are LEE-positive. Intimin, encoded by eae (E. coli attaching and effacing) gene, is responsible for intimate adhesion of VTEC to the intestinal epithelium and formation of AE lesions. Intimin binds to the cell receptor Tir, which is translocated by the pathogen to the enterocyte via a type III secretion system (Croxen et al., 2013). The presence of eae gene is strongly correlated to the presence of genes encoding many other virulence factors (Franz et al., 2015). In addition, VTEC strains with eae and stx2 genes have been associated with HUS and bloody diarrhea (Scheutz, 2014). In the present study, the transcription of eae (except for E. coli O104:H4 which is LEE-negative) changed only slightly following H2O2 treatment (Figures 1–6). Thus, further studies about eae regulation and the intimin-related virulence of VTEC strains under different stress conditions are needed.
Production of flagella is another contributor to the pathogenicity of VTEC (Gyles, 2007). Flagella are mainly responsible for motility, chemotaxis, and secretion of virulence factors. The key structural component of the flagellum filament is encoded by the fliC gene. In our studies, the transcription of fliC was dramatically downregulated in all four motile VTEC strains on lettuce treated with H2O2 (Figures 1–6). In addition, fliC was downregulated in pure broth cultures exposed to H2O2 (Mei et al., 2015). These results suggest that H2O2 treatment affects motility of VTEC strains.
Conclusion
H2O2 treatment caused less dramatic reduction in VTEC populations on lettuce compared to pure broth cultures, which indicates that VTEC are protected from H2O2 on leafy greens. Consequently higher concentration of H2O2 or other sanitizers are required to reduce and eliminate VTEC on lettuce. As such, the results derived from this study showed the importance of food matrix in studying the effect of sanitizers on survival rate of VTEC. In addition, VTEC strains on lettuce showed similar transcription patterns (regardless of serotype) in response to treatment with 50 mM H2O2. The transcription of genes related to oxidative stress, general stress, acid stress and some virulence genes were significantly downregulated in six VTEC serotypes. Consequently, VTEC strains on lettuce may become more sensitive to acid stress following H2O2 treatment. Interestingly, different gene transcription patterns in response to H2O2 were observed in VTEC strains on lettuce compared to pure broth cultures (Mei et al., 2015). Particularly, sodA gene encoding manganese SOD was significantly upregulated in pure cultures of all VTEC serotypes, however, this gene was downregulated in the same strains present on lettuce. In addition, Stx genes were upregulated in broth culture of E. coli O157:H7 and downregulated in most VTEC serotypes (including O157:H7) on lettuce following H2O2 treatment. It is possible that factors released from fresh-cut lettuce leaves and natural microbiota present in lettuce samples contributed to different gene transcription patterns in VTEC strains on lettuce compared to broth cultures.
Author Contributions
All authors have made substantial direct and intellectual contributions to this work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors would like to thank Dr. Roger Johnson of Public Health Agency of Canada and Dr. Pascal Delaquis (AAFC) for non-O157 VTEC strains.
Footnotes
Funding. This research was supported by the Agriculture and Agri-Food Canada.
References
- Allen K. J., Griffiths M. W. (2012). Impact of hydroxyl- and superoxide anion-based oxidative stress on logarithmic and stationary phase Escherichia coli O157:H7 stress and virulence gene expression. Food Microbiol. 29 141–147. 10.1016/j.fm.2011.09.014 [DOI] [PubMed] [Google Scholar]
- Almiron M., Link A. J., Furlong D., Kolter R. (1992). A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6 2646–2654. 10.1101/gad.6.12b.2646 [DOI] [PubMed] [Google Scholar]
- Arnold C. N., McElhanon J., Lee A., Leonhart R., Siegele D. A. (2001). Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183 2178–2186. 10.1128/JB.183.7.2178-2186.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergholz T. M., Tarr C. L., Christensen L. M., Betting D. J., Whittam T. S. (2007). Recent gene conversions between duplicated glutamate decarboxylase genes (gadA and gadB) in pathogenic Escherichia coli. Mol. Biol. Evol. 24 2323–2333. 10.1093/molbev/msm163 [DOI] [PubMed] [Google Scholar]
- Beuchat L. R., Nail B. V., Adler B. B., Clavero M. R. (1998). Efficacy of spray application of chlorinated water in killing pathogenic bacteria on raw apples, tomatoes, and lettuce. J. Food Prot. 61 1305–1311. 10.4315/0362-028X-61.10.1305 [DOI] [PubMed] [Google Scholar]
- Bradley K. K., Williams J. M., Burnsed L. J., Lytle M. B., McDermott M. D., Mody R. K.et al. (2012). Epidemiology of a large restaurant-associated outbreak of Shiga toxin-producing Escherichia coli O111:NM. Epidemiol. Infect. 140 1644–1654. 10.1017/S0950268811002329 [DOI] [PubMed] [Google Scholar]
- Brandl M. T., Amundson R. (2008). Leaf age as a risk factor in contamination of lettuce with Escherichia coli O157:H7 and Salmonella enterica. Appl. Environ. Microbiol. 74 2298–2306. 10.1128/AEM.02459-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. T., Sowers E. G., Wells J. G., Greene K. D., Griffin P. M., Hoekstra R. M., et al. (2005). Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J. Infect. Dis. 192 1422–1429. 10.1086/466536 [DOI] [PubMed] [Google Scholar]
- Brown J. A., Hite D., Gillim-Ross L. A., Maguire H. F., Bennett J. K., Patterson J. J., et al. (2012). Outbreak of Shiga toxin-producing Escherichia coli serotype O26: H11 infection at a child care center in Colorado. Pediatr. Infect. Dis. J. 31 379–383. 10.1097/INF.0b013e3182457122 [DOI] [PubMed] [Google Scholar]
- Buchholz U., Bernard H., Werber D., Bohmer M. M., Remschmidt C., Wilking H., et al. (2011). German outbreak of Escherichia coli O104:H4 associated with sprouts. N. Engl. J. Med. 365 1763–1770. 10.1056/NEJMoa1106482 [DOI] [PubMed] [Google Scholar]
- Calhoun L. N., Kwon Y. M. (2011). Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J. Appl. Microbiol. 110 375–386. 10.1111/j.1365-2672.2010.04890.x [DOI] [PubMed] [Google Scholar]
- CDC (2017). Reports of Selected E. coli Investigations. Available at: http://www.cdc.gov/ecoli/outbreaks.html [accessed Feb 1 2017]. [Google Scholar]
- Chiang S. M., Schellhorn H. E. (2012). Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525 161–169. 10.1016/j.abb.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Christman M. F., Morgan R. W., Jacobson F. S., Ames B. N. (1985). Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41 753–762. 10.1016/S0092-8674(85)80056-8 [DOI] [PubMed] [Google Scholar]
- Croxen M. A., Law R. J., Scholz R., Keeney K. M., Wlodarska M., Finlay B. B. (2013). Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26 822–880. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaquis P., Bach S., Dinu L. D. (2007). Behaviour of Escherichia coli O157:H7 in leafy vegetables. J. Food Prot. 70 1966–1974. 10.4315/0362-028X-70.8.1966 [DOI] [PubMed] [Google Scholar]
- Demple B., Amabile-Cuevas C. F. (1991). Redox redux: the control of oxidative stress responses. Cell 67 837–839. 10.1016/0092-8674(91)90355-3 [DOI] [PubMed] [Google Scholar]
- Etchegaray J. P., Jones P. G., Inouye M. (1996). Differential thermoregulation of two highly homologous cold-shock genes, cspA and cspB, of Escherichia coli. Genes Cells 1 171–178. 10.1046/j.1365-2443.1996.d01-231.x [DOI] [PubMed] [Google Scholar]
- Farr S. B., Kogoma T. (1991). Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55 561–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G., Tsui H. C., Winkler M. E. (1996). Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J. Bacteriol. 178 2388–2396. 10.1128/jb.178.8.2388-2396.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folster J. P., Pecic G., Taylor E., Whichard J. (2011). Characterization of isolates from an outbreak of multidrug-resistant, Shiga toxin-producing Escherichia coli O145 in the United States. Antimicrob. Agents Chemother. 55 5955–5956. 10.1128/AAC.05545-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz E., van Hoek A. H., Wuite M., van der Wal F. J., de Boer A. G., Bouw E. I., et al. (2015). Molecular hazard identification of non-O157 Shiga toxin-producing Escherichia coli (STEC). PLoS ONE 10:e0120353 10.1371/journal.pone.0120353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R. J., Luo Y., Ruiz-Cruz S., McEvoy J. L. (2004). Efficacy of sanitizers to inactivate Escherichia coli O157:H7 on fresh-cut carrot shreds under simulated process water conditions. J. Food Prot. 67 2375–2380. 10.4315/0362-028X-67.11.2375 [DOI] [PubMed] [Google Scholar]
- Greenberg J. T., Monach P., Chou J. H., Josephy P. D., Demple B. (1990). Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87 6181–6185. 10.1073/pnas.87.16.6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyles C. L. (2007). Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85 E45–E62. 10.2527/jas.2006-508 [DOI] [PubMed] [Google Scholar]
- Han M. J., Lee S. Y. (2006). The Escherichia coli proteome: past, present, and future prospects. Microbiol. Mol. Biol. Rev. 70 362–439. 10.1128/MMBR.00036-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L. J., Farber J. N., Beuchat L. R., Parish M. E., Suslow T. V., Garrett E. H., et al. (2003). Outbreaks associated with fresh produce: incidence, growth, and survival of pathogens in fresh and fresh-cut produce. Compreh. Rev. Food Sci. Food Saf. 2 78–141. 10.1111/j.1541-4337.2003.tb00031.x [DOI] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management of automated analysis of real-time quantitative PCR data. Genome Biol. 8 R19. 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman K. M., Hall A. J., Gould L. H. (2015). Outbreaks attributed to fresh leafy vegetables, United States, 1973-2012. Epidemiol. Infect. 143 3011–3021. 10.1017/S0950268815000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommais F., Krin E., Coppee J. Y., Lacroix C., Yeramian E., Danchin A., et al. (2004). GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150(Pt 1), 61–72. 10.1186/gb-2007-8-2-r19 [DOI] [PubMed] [Google Scholar]
- Hommais F., Krin E., Laurent-Winter C., Soutourina O., Malpertuy A., Le Caer J. P., et al. (2001). Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40 20–36. 10.1046/j.1365-2958.2001.02358 [DOI] [PubMed] [Google Scholar]
- Huang J. Y., Henao O. L., Griffin P. M., Vugia D. J., Cronquist A. B., Hurd S., et al. (2016). Infection with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance — Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2012–2015. MMWR 65 368–371. 10.15585/mmwr.mm6514a2 [DOI] [PubMed] [Google Scholar]
- Huang Y., Ye M., Chen H. (2012). Efficacy of washing with hydrogen peroxide followed by aerosolized antimicrobials as a novel sanitizing process to inactivate Escherichia coli O157:H7 on baby spinach. Int. J. Food Microbiol. 153 306–313. 10.1016/j.ijfoodmicro.2011.11.018 [DOI] [PubMed] [Google Scholar]
- Hurst W. C. (1995). “Disinfection methods: a comparison of chlorine dioxide, ozone and ultraviolet light alternatives. Cutting edge, fall issue,” in Proceedings of the International Fresh-cut Produce Association, Alexandria, VA, 4–5. [Google Scholar]
- Islam M., Doyle M. P., Phatak S. C., Millner P., Jiang X. (2004). Persistence of enterohemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. J. Food Prot. 67 1365–1370. 10.4315/0362-028X-67.7.1365 [DOI] [PubMed] [Google Scholar]
- Karmali M. A. (2017). Emerging public health challenges of Shiga toxin-producing Escherichia coli related to changes in the pathogen, the population, and the environment. Clin. Infect. Dis. 64 371–376. 10.1093/cid/ciw708 [DOI] [PubMed] [Google Scholar]
- Karpman D., Stahl A.-L. (2014). Enterohemorrhagic Escherichia coli pathogenesis and the host response. Microbiol. Spectr. 2. 10.1128/microbiolspec.EHEC-0009-2013 [DOI] [PubMed] [Google Scholar]
- Li B., Tsui H. C., LeClerc J. E., Dey M., Winkler M. E., Cebula T. A. (2003). Molecular analysis of mutS expression and mutation in natural isolates of pathogenic Escherichia coli. Microbiology 149 1323–1331. 10.1099/mic.0.26213-0 [DOI] [PubMed] [Google Scholar]
- Linley E., Denyer S. P., McDonnell G., Simons C., Maillard J. Y. (2012). Use of hydrogen peroxide as a biocide: new consideration of its mechanisms of biocidal action. J. Antimicrob. Chemother. 67 1589–1596. 10.1093/jac/dks129 [DOI] [PubMed] [Google Scholar]
- Lu A. L., Li X., Gu Y., Wright P. M., Chang D. Y. (2001). Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem. Biophys. 35 141–170. 10.1385/CBB:35:2:141 [DOI] [PubMed] [Google Scholar]
- Luna-Gierke R. E., Griffin P. M., Gould L. H., Herman K., Bopp C. A., Strockbine N., et al. (2014). Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol. Infect. 142 2270–2280. 10.1017/S0950268813003233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. F., Tauxe R. V., Hedberg C. W. (2009). The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol. Infect. 137 307–315. 10.1017/S0950268808001969 [DOI] [PubMed] [Google Scholar]
- Ma Z., Richard H., Tucker D. L., Conway T., Foster J. W. (2002). Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184 7001–7012. 10.1128/JB.184.24.7001-7012.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado M., Michan C., Pueyo C. (2000). Hydrogen peroxide activates the SoxRS regulon in vivo. J. Bacteriol. 182 6842–6844. 10.1128/JB.182.23.6842-6844.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mates A. K., Sayed A. K., Foster J. W. (2007). Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J. Bacteriol. 189 2759–2768. 10.1128/JB.01490-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei G.-Y., Tang J., Carey C., Bach S., Kostrzynska M. (2015). The effect of oxidative stress on gene expression of Shiga toxin-producing Escherichia coli (STEC) O157:H7 and non-O157 serotypes. Int. J. Food Microbiol. 215 7–15. 10.1016/j.ijfoodmicro.2015.07.029 [DOI] [PubMed] [Google Scholar]
- Michaels M., Miller J. H. (1992). The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174 6321–6325. 10.1128/jb.174.20.6321-6325.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., Lahue R. (1996). Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65 101–133. 10.1146/annurev.bi.65.070196.000533 [DOI] [PubMed] [Google Scholar]
- Nou X., Luo Y. (2010). Whole-leaf wash improves chlorine efficacy for microbial reduction and prevents pathogen cross-contamination during fresh-cut lettuce processing. J. Food Sci. 75 M283–M290. 10.1111/j.1750-3841.2010.01630.x [DOI] [PubMed] [Google Scholar]
- Nunoshiba T., Hidalgo E., Amabile Cuevas C. F., Demple B. (1992). Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 174 6054–6060. 10.1128/jb.174.19.6054-6060.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T., Neidhardt F. C. (1992). Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 6 3187–3198. 10.1111/j.1365-2958.1992.tb01774.x [DOI] [PubMed] [Google Scholar]
- Nystrom T., Neidhardt F. C. (1994). Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11 537–544. 10.1111/j.1365-2958.1994.tb00334 [DOI] [PubMed] [Google Scholar]
- Olaimat A. N., Holley R. A. (2012). Factors influencing the microbial safety of fresh produce: a review. Food Microbiol. 32 1–19. 10.1016/j.fm.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Painter J. A., Hoekstra R. M., Ayers T., Tauxe R. V., Braden C. R., Angulo F. J., et al. (2013). Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998-2008. Emerg. Infect. Dis. 19 407–415. 10.3201/eid1903.111866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons B. D., Zelyas N., Berenger B. M., Chui L. (2016). Detection, characterization, and typing of Shiga toxin-producing Escherichia coli. Front. Microbiol. 7:478 10.3389/fmicb.2016.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson W. L., Mackowiak P. A., Barnett C. C., Marling-Cason M., Haley M. L. (1989). The human gastric bactericidal barrier: mechanisms of action, relative antibacterial activity, and dietary influences. J. Infect. Dis. 159 979–983. 10.1093/infdis/159.5.979 [DOI] [PubMed] [Google Scholar]
- Phadtare S., Inouye M. (2001). Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183 1205–1214. 10.1128/JB.183.4.1205-1214.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadtare S., Tadigotla V., Shin W. H., Sengupta A., Severinov K. (2006). Analysis of Escherichia coli global gene expression profiles in response to overexpression and deletion of CspC and CspE. J. Bacteriol. 188 2521–2527. 10.1128/JB.188.7.2521-2527.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel J. M., Sparling P. H., Crowe C., Griffin P. M., Swerdlow D. L. (2005). Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11 603–609. 10.3201/eid1104.040739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Yatsuyanagi J., Kinouchi Y., Sato H., Miyajima Y., Morita M. (1998). A familial outbreak of verotoxin-producing Escherichia coli O103:H2 infection in which a calf was the suspected infectious source. Kansenshogaku Zasshi 72 707–713. 10.11150/kansenshogakuzasshi1970.72.707 [DOI] [PubMed] [Google Scholar]
- Sapers G. M., Miller R. L., Jantschke M., Mattrazzo A. M. (2000). Factors limiting the efficacy of hydrogen peroxide washes for decontamination of apples containing Escherichia coli. J. Food Sci. 65 529–532. 10.1111/j.1365-2621.2000.tb16041 [DOI] [Google Scholar]
- Scheutz F. (2014). Taxonomy meets public health: the case of Shiga toxin-producing Escherichia coli. Microbiol. Spectrum 2. 10.1128/microbiolspec.EHEC-0019-2013 [DOI] [PubMed] [Google Scholar]
- Schofield M. J., Hsieh P. (2003). DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57 579–608. 10.1146/annurev.micro.57.030502.090847 [DOI] [PubMed] [Google Scholar]
- Selinger D. W., Cheung K. J., Mei R., Johansson E. M., Richmond C. S., Blattner F. R., et al. (2000). RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18 1262–1268. 10.1038/82367 [DOI] [PubMed] [Google Scholar]
- Sivapalasingam S., Friedman C. R., Cohen L., Tauxe R. V. (2004). Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 67 2342–2353. 10.4315/0362-028X-67.10.2342 [DOI] [PubMed] [Google Scholar]
- Smith J. L., Fratamico P. M., Gunther N. W. IV. (2014). Shiga toxin-producing Escherichia coli. Adv. Appl. Microbiol. 86 145–197. 10.1016/B978-0-12-800262-9.00003-2 [DOI] [PubMed] [Google Scholar]
- Sola-Landa A., Moura R. S., Martin J. F. (2003). The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. U.S.A. 100 6133–6138. 10.1073/pnas.0931429100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon E. B., Pang H. J., Matthews K. R. (2003). Persistence of Escherichia coli O157:H7 on lettuce plants following spray irrigation with contaminated water. J. Food Prot. 66 2198–2202. 10.4315/0362-028X-66.12.2198 [DOI] [PubMed] [Google Scholar]
- Storz G., Tartaglia L. A., Ames B. N. (1990a). Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248 189–194. 10.1126/science.2183352 [DOI] [PubMed] [Google Scholar]
- Storz G., Tartaglia L. A., Farr S. B., Ames B. N. (1990b). Bacterial defenses against oxidative stress. Trends Genet. 6 363–368. 10.1016/0168-9525(90)90278-E [DOI] [PubMed] [Google Scholar]
- Tang J., Kostrzynska M. (2012). “Sensitivity of various VTEC serotypes to oxidative stress,” in Proceedings of the 10th Annual OMAFRA Food Safety Research Forum, Guelph, ON. [Google Scholar]
- Tchou J., Grollman A. P. (1993). Repair of DNA containing the oxidatively-damaged base, 8-oxoguanine. Mutat. Res. 299 277–287. 10.1016/0165-1218(93)90104-L [DOI] [PubMed] [Google Scholar]
- Tsaneva I. R., Weiss B. (1990). soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J. Bacteriol. 172 4197–4205. 10.1128/jb.172.8.4197-4205.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H. C., Feng G., Winkler M. E. (1997). Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179 7476–7487. 10.1128/jb.179.23.7476-7487.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker D. L., Tucker N., Conway T. (2002). Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 184 6551–6558. 10.1128/JB.184.23.6551-6558.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker D. L., Tucker N., Ma Z., Foster J. W., Miranda R. L., Cohen P. S., et al. (2003). Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 185 3190–3201. 10.1128/JB.185.10.3190-3201.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva C. M., Cantor K. P., Cordier S., Jaakkola J. J., King W. D., Lynch C. F., et al. (2004). Disinfection byproducts and bladder cancer: a pooled analysis. Epidemiology 15 357–367. 10.1097/01.ede.0000121380.02594.fc [DOI] [PubMed] [Google Scholar]
- Wachtel M. R., Whitehand L. C., Mandrell R. E. (2002). Association of Escherichia coli O157:H7 with preharvest leaf lettuce upon exposure to contaminated irrigation water. J. Food Prot. 65 18–25. 10.4315/0362-028X-65.5.741 [DOI] [PubMed] [Google Scholar]
- Wang S., Deng K., Zaremba S., Deng X., Wang Q., Lin C., et al. (2009). Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl. Environ. Microbiol. 75 6110–6123. 10.1128/AEM.00914-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Mitani T., Ogura T., Niki H., Hiraga S. (1994). Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Mol. Microbiol. 13 301–312. 10.1111/j.1365-2958.1994.tb00424.x [DOI] [PubMed] [Google Scholar]
- Zangari T., Melton-Celsa A. R., Panda A., Boisen N., Smith M. A., Tatarov I., et al. (2013). Virulence of the Shiga toxin type 2-expressing Escherichia coli O104:H4 German outbreak isolate in two animal models. Infect. Immun. 81 1562–1574. 10.1128/IAI.01310-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., Storz G. (2001). DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183 4562–4570. 10.1128/JB.183.15.4562-4570.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]