Abstract
Although the majority of acute myeloid leukemia (AML) patients initially respond to chemotherapy, many patients subsequently relapse; the mechanistic basis for AML persistence following chemotherapy has not been delineated. Recurrent somatic mutations in DNA methyltransferase 3A (DNMT3A), most frequently at arginine 882 (DNMT3Amut), are observed in AML1–3 and in individuals with clonal hematopoiesis in the absence of leukemic transformation4,5. DNMT3Amut AML patients have an inferior outcome when treated with standard-dose daunorubicin-based induction chemotherapy6,7, suggesting that DNMT3Amut cells persist and drive relapse8. Here we show that Dnmt3amut induces hematopoietic stem cell (HSC) expansion, cooperates with Flt3ITD and Npm1c to induce AML in vivo, and promotes resistance to anthracycline chemotherapy. In AML patients, DNMT3AR882 mutations predict for minimal residual disease (MRD), underscoring their role in AML chemoresistance. DNMT3Amut cells show impaired nucleosome eviction and chromatin remodeling in response to anthracyclines, resulting from attenuated recruitment of histone chaperone SPT-16 following anthracycline exposure. This defect leads to an inability to sense and repair DNA torsional stress, which results in increased mutagenesis. Our studies identify a critical role for DNMT3AR882 mutations in driving AML chemoresistance, and highlight the importance of chromatin remodeling in response to cytotoxic chemotherapy.
Mutations in genes which regulate DNA and histone modifications are commonly observed in human cancers9, including AML10. Genetic studies of elderly subjects with clonal hematopoiesis and of functionally defined pre-leukemic clones identified recurrent mutations in epigenetic regulators4,5,8,11,12, suggesting, together with studies in murine models13–16, that they increase hematopoietic stem/progenitor cell (HSPC) fitness and predispose to subsequent malignant transformation. DNA methyltransferase 3A (DNMT3A) is the most commonly mutated epigenetic regulatory gene in AML2,3,6. Approximately half of all AML-associated DNMT3A mutations are monoallelic nonsense or frameshift alterations. Notably, almost half of all DNMT3A mutations occur at a single hotspot, arginine 882, which is mutated to histidine (R882H) or cysteine (R882C)1,17. DNMT3AR882 mutations are the most prevalent somatic mutations observed in individuals with clonal hematopoiesis4,5. Biochemical studies suggest DNMT3AR882 can function as dominant negative with respect to methyltransferase activity18–20, however their role in leukemia pathogenesis and in the response to anti-leukemic therapies has not been elucidated.
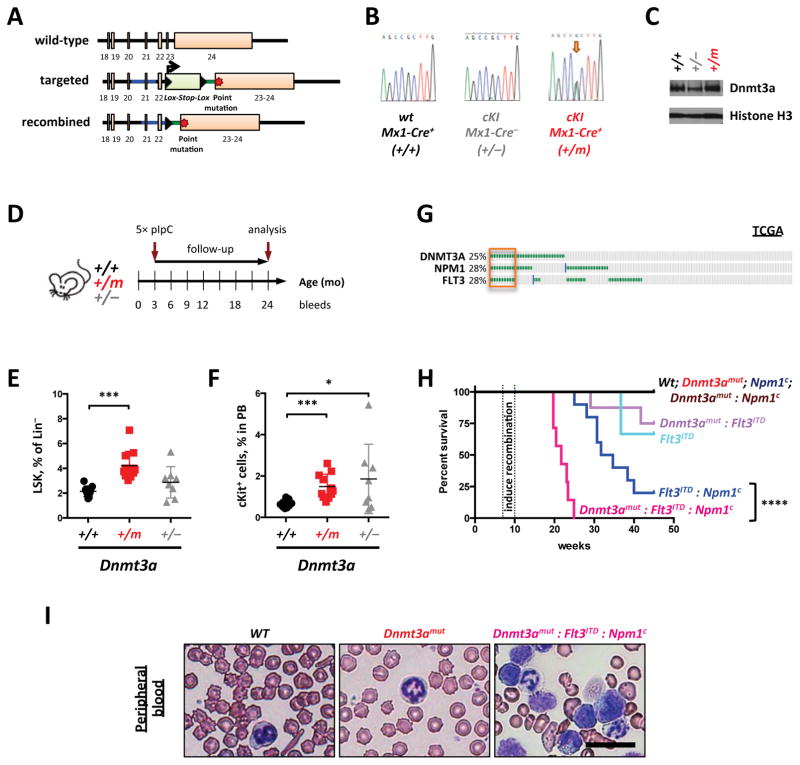
To address these questions we generated a mouse model that conditionally expresses Dnmt3aR878H (mouse homolog to DNMT3AR882H) from the endogenous locus (Figure 1A–B). PolyI-polyC-treated Mx1-Cre:Dnmt3aR878H mice (referred to as Dnmt3amut) expressed equal levels of Dnmt3aR878H and wild-type Dnmt3a, with physiologic protein expression (Figure 1C). Mice expressing Dnmt3amut in the absence of other disease alleles did not develop leukemia (Figure 1D, H) but were characterized by the accumulation of Lineage−Sca1+cKit+ (LSK) cells (Figure 1E and Supplementary Fig. 1A), and by increased percentage of circulating c-Kit-positive progenitor cells (Figure 1F) consistent with HSPC expansion (Supplementary Figure 1B). Dnmt3amut bone marrow cells exhibited enhanced serial colony-forming potential in vitro (Supplementary Fig. 1C). We observed impaired erythroid differentiation in the bone marrow (Supplementary Fig. 1D) and a modest increase in myeloid bias (Supplementary Fig. 1E–F) of Dnmt3amut mice. These data demonstrate that expression of Dnmt3amut in hematopoietic cells expands HSPC and alters differentiation in vivo.
Figure 1. Dnmt3aR878H mutation augments HSC stem cell function and cooperates with co-occurring AML disease alleles in vivo.
(A–C) Generation of the conditional Dnmt3aR878H knock-in (cKI) allele (A) and validation of its expression on mRNA level by Sanger-sequencing of cDNA generated from peripheral blood nucleated cells (B) and protein expression levels in spleens (C). Wild-type Dntm3a allele is denoted as “+”, non-recombined cKI allele functions as a null allele and is denoted as “−”, Cre-mediated recombination results in the mutant cKI allele denoted as “m”. Histone H3, loading control; data representative of at least two independent litters of mice. D) Timeline of an experiment to evaluate biological effect of Dnmt3aR878H expression. Five injections of poly(I:C) (5× pIpC) were used to induce recombination of the cKI allele. (E, F) Bone marrow (E) and peripheral blood (F) from aged (18–24 months) mice treated as in (D) were analyzed by flow cytometry. Lineage−Sca1+cKit+ (LSK) cells (E) and c-Kit+ cells (F) were quantified (n=8–12; * p<0.05, *** p<0.01). (G) Co-occurrence of DNMT3A, NPM1 and FLT3 mutations in 166 AML cases according to TCGA (p<1.9×10−6). Survival (H) and peripheral blood cytopathology (I) of recipient mice transplanted with 1×106 bone marrow cells of indicated genotypes (H, **** p<0.0001). Bar – 50 μm; images are representative of at least 5 random fields.
We hypothesized that expression of Dnmt3amut would cooperate with other disease alleles to promote leukemic transformation. Analysis of AML TCGA and other data1,21 revealed a significant co-association of DNMT3A mutations with FLT3 internal tandem duplications (FLT3ITD) and NPM1c mutations; notably all 3 mutations were often concurrent (Figure 1G; p<1.9×10−6). Therefore, we generated mice expressing Flt3ITD, Dnmt3amut and/or Npm1c and assessed the ability of different combinatorial permutations to induce an AML phenotype (Figure 1H). Concurrent expression of Flt3ITD, Dnmt3amut and Npm1c resulted in a fully penetrant leukemic phenotype, whereas any single or pair of disease alleles either led to longer latency, incompletely penetrant disease (Flt3ITD/Npm1c, Flt3ITD/Dnmt3amut or Flt3ITD alone) or no leukemic phenotype (Dnmt3amut/Npm1c or Dnmt3a and Npm1 single mutants, Figure 1H). Dnmt3amut:Flt3ITD:Npm1c AML was characterized by circulating large myeloblasts without myeloid dysplasia (Figure 1I and Supplementary Fig. 2A), a hypercellular bone marrow with proliferating leukemic blasts, obliteration of splenic architecture and hepatic portal infiltration (Supplementary Fig. 2A). Dnmt3amut contributed to leukemic transformation due to, at least in part, augmented stem cell function as seen by enhanced competitive transplantability (Supplementary Fig. 2B–C) and enhanced myeloid-to-lymphoid engraftment ratio in non-competitive transplantation studies (Supplementary Fig. 2D).
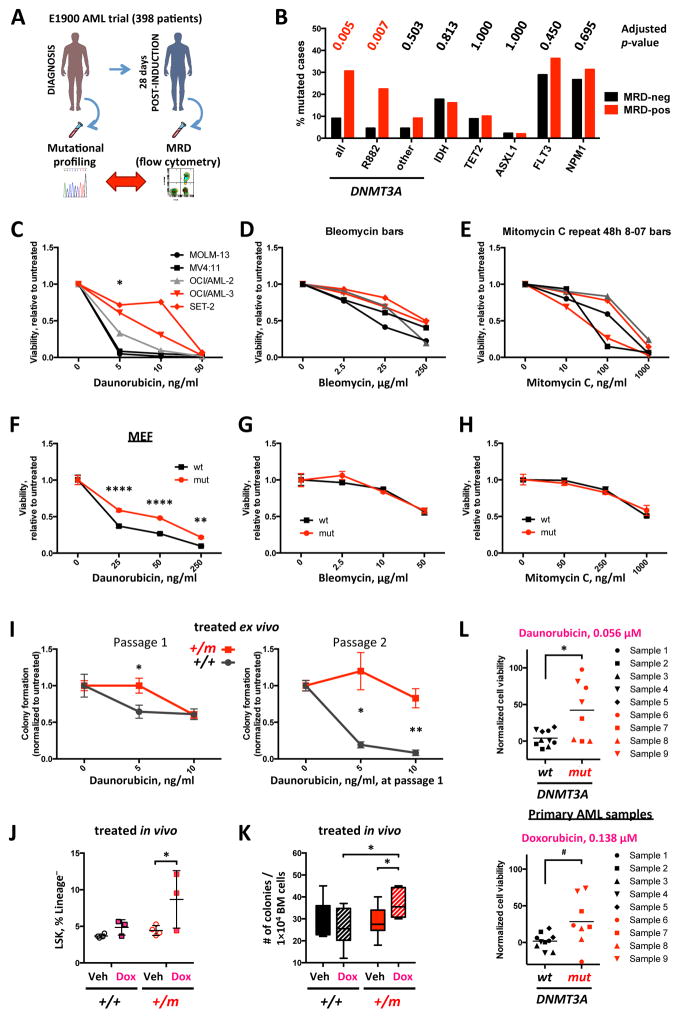
We next investigated the relevance of DNMT3A mutations to the response to anti-leukemic therapy. We previously showed that DNMT3A-mutant patients in the ECOG 1900 clinical trial had a poor outcome with standard-dose daunorubicin-based induction consistent with other clinical studies22–25; however the adverse prognostic impact of DNMT3A mutations was mitigated by daunorubicin dose-intensification6,7. These data suggested that DNMT3A mutations could promote resistance to anthracycline-based chemotherapy. We investigated whether mutations in DNMT3A or in other AML disease alleles were associated with the presence or absence of flow-cytometrically defined minimal residual disease (MRD) after induction chemotherapy in the ECOG 1900 clinical trial cohort (Figure 2A), as the MRD 28 days after induction chemotherapy has prognostic value in AML26–30. In a multivariate analysis DNMT3AR882 mutations, but not non-R882 DNMT3A mutations or mutations in other AML genes, robustly predicted for the presence of MRD following induction chemotherapy (p=0.007, Figure 2B and Supplementary Table 1).
Figure 2. Expression of mutant DNMT3A leads to anthracycline resistance.
(A) Design of the multivariate analysis study correlating mutational frequency with incidence of flow cytometric minimal residual disease in ECOG E1900 patient cohort. (B) Frequency of mutations in indicated genes in a cohort of 398 AML patients stratified according to presence of MRD 28 days after induction chemotherapy. (C–E) Sensitivity to torsional stress and topoisomerase II inhibitor daunorubicin (C), double-strand break-inducer bleomycin (D), and DNA cross-linking agent mitomycin C (E) after 48 hours of drug exposure in a panel of cell lines harboring DNMT3A R882 (red, OCI/AML-3 and SET-2), non-R882 (gray, OCI/AML-2) or no (black, MOLM-13 and MV4:11) mutations, relative to untreated control (* p<0.05, all pairwise comparisons between DNMT3A wild-type and mutant cell lines). Data are representative of at least 3 independent experiments. (F–H) Sensitivity to daunorubicin (F), bleomycin (G), and mitomycin C (H) in immortalized mouse embryonic fibroblasts (MEF) derived from Dnmt3aR878H and wild-type animals, relative to untreated control, representative of two replicate experiments (** p<0.01, **** p<0.0001). (I) Clonogenic survival in MethoCult M3434 of BM cells derived from 3 Dnmt3aR878H and 3 wild-type mice in duplicate after ex vivo exposure to daunorubicin at first plating. Number of colonies relative to untreated control (* p<0.05, ** p<0.01). (J, K) Fraction of LSK cells within Lineage− population (J) and clonogenic potential (number of colonies per 104 unfractionated cells plated in MethoCult M3434 in triplicate) of bone marrow cells (K) derived from Dnmt3amut and wild-type mice after one-time in vivo exposure to 3mg/kg doxorubicin (n=3–4, * p<0.05). Recipient mice were engrafted with whole bone marrow derived from young newly-recombined Dnmt3amut or wild-type control congenic donors and allowed 3 months to reconstitute steady-state hematopoiesis, when they were treated with a single dose of doxorubicin (3 mg/kg) or vehicle and analyzed 24 hours later. Data representative of 2 independent transplant experiments. (L) Ex vivo sensitivity to anthracyclines daunorubicin (top panel) and doxorubicin (bottom panel) at indicated concentrations in 9 primary FLT3ITD-NPM1c AML samples depending on their DNMT3A status (black – DNMT3A wild-type, n=5, and red – DNMT3A mutant, n=4; * p<0.05, # p=0.06, t-test with Welch’s correction).
We next performed mutational analysis on 9 paired samples (from diagnosis and 28 days post-induction) from our E1900 AML cohort with DNMT3AR882 mutations and MRD analysis by flow-cytometry. In 8 of 9 cases, mutational studies showed that the R882 mutant disease allele was present at a detectable variant allele frequency (VAF) at the time of clinical remission (average sequencing depth 2246×). In cases with flow-positive MRD, we see the small leukemic clone (DNMT3A and other mutations) overlaid on a larger (majority of the cells) DNMT3A-mutant pre-leukemic population, or a persisting bulk DNMT3A-mutant pre-leukemic population, and in both scenarios we observed subsequent relapse. A persisting DNMT3A-mutant pre-leukemic subpopulation was detected in MRD-negative cases (Supplementary Fig. 3A, and Supplementary Table 2). These data indicate that DNMT3AR882 mutations are associated with AML chemoresistance and with persistence of leukemic/pre-leukemic DNMT3AR882 mutant cells following chemotherapy.
AML cell lines with DNMT3AR882 mutations (OCI/AML-3 and SET-2) were less sensitive to daunorubicin than non-R882 DNMT3A-mutant (OCI/AML-2) and DNMT3A-wild-type (MOLM-13 and MV4:11) AML cells (Figure 2C, Supplementary Fig. 3B). By contrast, DNMT3A mutational status did not influence the sensitivity of AML cells to DNA-damaging agents with other mechanisms of action, including bleomycin and mitomycin C (Figure 2D, E). Expression of DNMT3AR882H reduced sensitivity to daunorubicin in MOLM-13 cells (Supplementary Fig. 3C), and Dnmt3amut mouse embryonic fibroblasts (MEF) showed reduced sensitivity to daunorubicin but not to other DNA damaging agents in vitro (Figure 2F–H). We found decreased sensitivity to anthracyclines (daunorubicin/doxorubicin) in Dnmt3amut bone marrow plated in methylcellulose (Figure 2I), which was enhanced with serial passage. In vivo studies with doxorubicin demonstrated an enrichment of immature LSK cells in the bone marrow of Dnmt3amut-engrafted mice that translated into enhanced colony-forming potential ex vivo (Figure 2J, K). Anthracycline treatment of Dnmt3aWT mice reduced quiescence and increased short-term (ST)-HSCs numbers at the expense of long-term (LT)-HSCs in vivo, which was not observed in Dnmt3amut animals (Supplementary Fig. 3D–E). We next performed studies in primary AML samples with FLT3ITD and NPM1c mutations showing that samples harboring DNMT3A mutations had reduced sensitivity to anthracyclines ex vivo compared to DNMT3A wild-type cases (Figure 2L), which was lost at higher concentrations (data not shown).
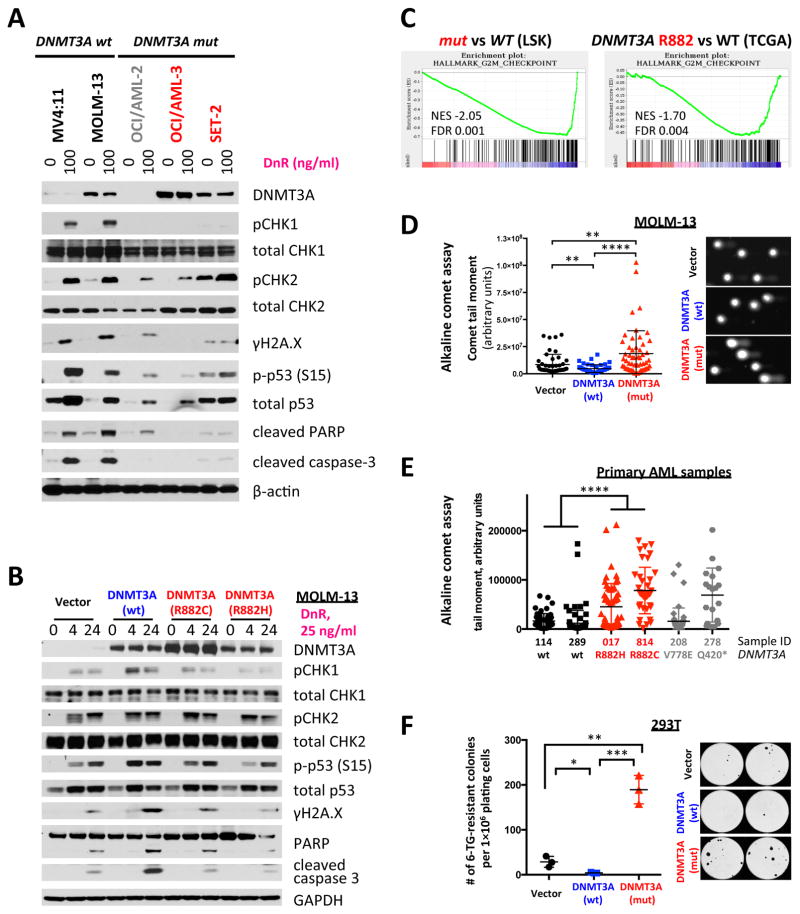
The modest DNA hypomethylation relative to other AML alleles with altered DNA methylation31 as assessed by enhanced reduced representation bisulfite sequencing (ERRBS)32,33, the paucity of genes with expression changes correlating with changes in DNA methylation, and the lack of enrichment of pathways relevant to cell survival or drug metabolism in mouse Dnmt3amut LSK cells compared to wild-type control, suggest that alterations in DNA methylation at specific loci are unlikely to explain anthracycline resistance in DNMT3AR882 mutant cells (Supplementary Fig. 3F–H). Anthracycline resistance was not due to differential drug efflux, metabolism, or intracellular compartmentalization of anthracyclines (Supplementary Fig. 4A–F). These data suggest a novel mechanism for anthracycline chemoresistance in AML cells with DNMT3AR882 mutations. We next investigated whether DNMT3AR882 mutant cells had an intact DNA damage response (DDR) to anthracyclines. Daunorubicin induces DNA torsional stress at lower doses, and can inhibit topoisomerase II leading to double-strand breaks (DSBs) at higher concentrations34. DNMT3A-mutant AML cells showed attenuated CHK1 phosphorylation and downstream signaling, including reduced phosphorylated histone H2A.X (γH2A.X), p53 phosphorylation/stabilization and apoptotic signaling (cleaved PARP and caspase 3) in response to daunorubicin, compared to DNMT3A wild-type cells (Figure 3A–B). CHK2 activation remained intact, suggesting a specific defect in the ATR/CHK1 pathway, which was specific to daunorubicin and not etoposide, a DNA-topoisomerase II inhibitor (Supplementary Fig. 5A). Expression of DNMT3Amut, but not wild-type DNMT3A in AML cells modestly attenuated the p53 response (Figure 3A–B, and Supplementary Fig. 5B) and γH2A.X accumulation in AML cells and in Dnmt3amut mice in vivo after anthracycline exposure (Figure 3A, B and Supplementary Figure 5 C–D). Gene expression analysis of Dnmt3amut LSK cells revealed negative enrichment of the Chk1-regulated G2/M checkpoint signature35; the same signature was also significantly attenuated in DNMT3AR882-mutant primary AML samples from the TCGA dataset (Figure 3C). The defect in checkpoint activation was not seen in Dnmt3a haploinsufficient LSKs or in DNMT3A non-R882 mutant AML patients (Supplementary Fig. 5E).
Figure 3. Cells with mutant DNMT3A have a DNA damage signaling defect in response to anthracyclines.
(A) Western blot analysis of DNA damage response pathways 16 hours after daunorubicin exposure in a panel of cell lines with different DNMT3A status (red – R882 mutation, gray – non-R882 mutation, black – no mutation). Data are representative of at least 3 replicate experiments. (B) Analysis of DNA damage response activation in MOLM-13 cells retrovirally transduced with DNMT3Amut, wild-type or empty vector control at different timepoints after daunorubicin treatment as indicated, representative of at least three independent experiments. (C) Gene set enrichment analysis (GSEA) of the G2/M checkpoint signature from the Hallmark collection in the bone marrow LSK cells from Dnmt3amut mice compared to Dnmt3aWT animals and in peripheral blood mononuclear cells of DNMT3Amut AML patients compared to DNMT3AWT cases from TCGA; gene expression by RNA-sequencing. (D) Detection of all types of DNA breaks and changes in DNA looping by alkaline comet assay in MOLM-13 cells retrovirally transduced with DNMT3Amut, wild-type or empty vector control after 16 hours of daunorubicin (25 ng/ml) exposure in vitro (** p<0.01, **** p<0.0001, Mann-Whitney rank sum test). (E) Alkaline comet assay in a panel of primary AML samples with different DNMT3A mutational status (black – no mutations, red – R882 mutations, gray – other non-R882 mutations) after 16 hour exposure to 25 ng/ml daunorubicin in vitro (**** p<0.0001, Mann-Whitney rank sum test). Sample #208 with DNMT3A V778E substitution appears more similar to DNMT3AWT samples (p=0.2310), while AML sample #278 with a truncating Q420* mutation is more similar to DNMT3A R882-mutated cases (p=0.4003). (F) Random mutagenesis measured by number of 6-TG-resistant colonies in 293T cells retrovirally transduced with DNMT3Amut or wild-type, or empty vector control (* p<0.05, ** p<0.01, *** p<0.001). Data are representative of at least 3 independent biological replicates.
We next investigated possible changes in the DNA state leading to the DDR signaling defect after daunorubicin in DNMT3AR882 mutant cells by comet assay, which reads out DNA single- and double-stranded breaks and other conditions leading to relaxation of DNA supercoiling, such as presence of labile apurinic sites or changes in nucleosomal density36. DNMT3AR882-transduced cells had increased alkaline comet signal in response to daunorubicin (Figure 3D), which was also observed in TP53-mutant AML cells (Supplementary Fig. 5F). We found a similar increase in comet signal in primary FACS-sorted multipotent progenitors from Dnmt3amut bone marrow compared to wild-type controls (Supplementary Fig. 5G) and in primary AML patient samples harboring DNMT3AR882 mutations compared to DNMT3A-wild-type AML cells (Figure 3E). DNMT3A non-R882 mutant cells had variable response to daunorubicin in the comet assay suggesting other, less common DNMT3A mutations might have a differential response to anthracyclines. Expression of DNMT3AR882, but not wild-type DNMT3A increased the rate of secondary mutations as measured by the number of 6-thioguanine-resistant HPRT-mutant colonies in hematopoietic and non-hematopoietic cells (Figure 3F and Supplementary Fig. 5H), which may be explained by inadequate repair of increased DNA damage due to impaired signaling. Taken together, these data suggest DNMT3AR882 mutant cells fail to initiate CHK1-mediated DDR after anthracycline treatment.
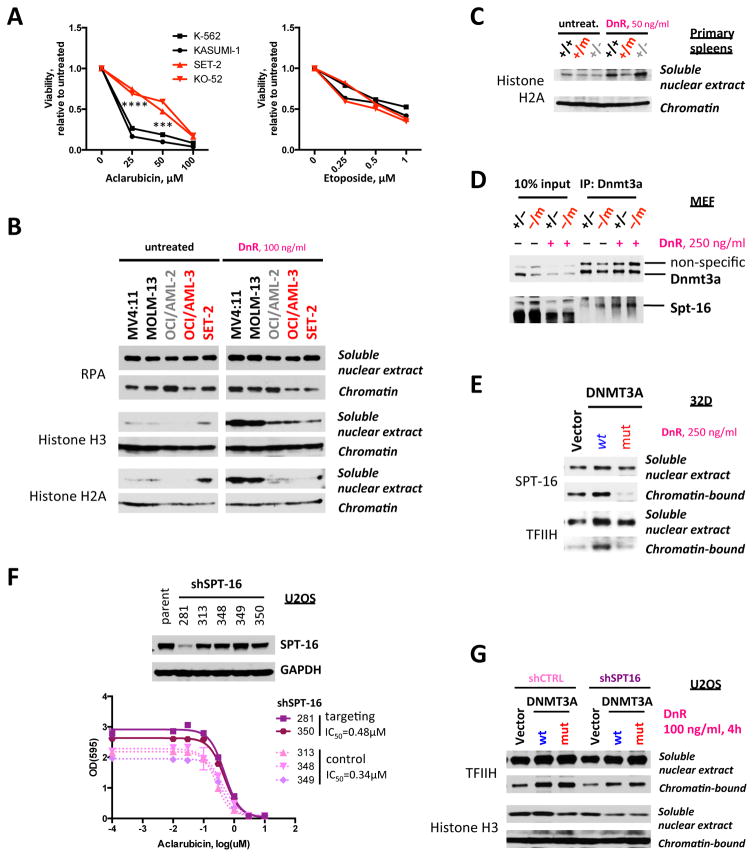
We noted that DNMT3AR882 cells showed reduced sensitivity to aclarubicin, but not to etoposide (Figure 4A, and Supplementary Fig. 6A–B). Given aclarubicin induces torsional stress and etoposide inhibits topoisomerase II, while daunorubicin acts through both mechanisms, these data suggest a specific mechanism of resistance to DNA damage. Previous studies demonstrated torsional stress imposed by anthracyclines destabilizes nucleosomes, which facilitates remodeling and histone eviction as the first step immediately after topological DNA strain, contributing to cytotoxicity34,37. We evaluated whether DNMT3AR882 mutant cells exhibited impaired chromatin remodeling and reduced histone eviction in response to anthracyclines. Daunorubicin induced significant histone eviction and an increase in free nuclear histones in DNMT3A-wild-type AML cells, which was markedly attenuated in DNMT3Amut cells (Figure 4B). A similar defect in histone eviction was observed in Dnmt3amut MEFs (Supplementary Fig. 6C) and in hematopoietic cells from Dnmt3amut mice (Figure 4C). This mechanism was specific to anthracyclines as we observed no difference in histone release after etoposide treatment in cells expressing wild-type or mutant forms of DNMT3A (Supplementary Fig. 6D).
Figure 4. Expression of mutant DNMT3A impairs chromatin remodeling in response to DNA damage.
(A) Sensitivity to aclarubicin (torsional stress) and etoposide (topoisomerase II inhibition) in leukemia cell lines with different DNMT3A mutational status (red – R882 mutations, black – no mutation; *** p<0.001, **** p<0.0001, pair-wise comparisons between DNMT3A wild-type and mutant cell lines). (B) Distribution of histones H2A and H3 and of DNA single-strand binding protein RPA between chromatin-bound fraction and soluble nuclear extract after in vitro daunorubicin treatment in a panel of cell lines with different DNMT3A status synchronized in early S-phase by double thymidine block (red – R882 mutation, gray – non-R882 mutation, black –f no mutation). Protein abundance in each of the cellular compartments was analyzed by Western blotting. (C) Distribution of histone H2A between chromatin-bound fraction and soluble nuclear extract after 4 hours of daunorubicin treatment in splenocytes derived from Dnmt3amut, wild-type, and haploinsufficient animals. (D) Co-immunoprecipitation (co-IP) with Dnmt3a-specific antibodies using nuclease-treated extracts from MEF cells expressing either one wild-type copy of Dnmt3a (+/−) or one mutant copy of Dnmt3a (−/m) analyzed for the presence of endogenous Dnmt3a and Spt-16 by Western blotting. Cells were exposed to 250 ng/ml daunorubicin for 4 hours before harvest where indicated. (E) Distribution between chromatin-bound and free nuclear states of SPT-16 and of DNA helicase TFIIH in 32D cells retrovirally transduced with DNMT3AR882 or wild-type, or empty vector control, after 4 hours of treatment with 250 ng/ml daunorubicin. (F) Sensitivity to aclarubicin after shSUPT16H-mediated knock-down of SPT-16 in U2OS cells (IC50 0.48μM in cells with SPT-16 knock-down plotted as solid lines, compared to 0.34μM in cells with non-targeting shRNAs plotted as dotted lines). Data from a single experiment. Top panel – Western blotting analysis of knock-down efficiency of 5 different SPT-16-specific shRNAs compared to parental cells. (G) Distribution between chromatin-bound and free nuclear states of TFIIH and of histone H3 in U2OS cells ectopically expressing wild-type or mutant DNMT3A, or empty vector control, with and without SPT-16 knock-down, after exposure to 100 ng/ml daunorubicin for 4 hours. Data are representative of two independent experiments.
We next sought to identify the specific proteins involved in the impaired nucleosome turnover and histone eviction in DNMT3AR882 cells. Protein pull-down assays in cell nuclear extracts using a synthetic peptide corresponding to DNMT3A N-terminal domain followed by mass-spectroscopy identified the facilitates chromatin transcription (FACT) complex subunit SPT-16, which is a histone H2A/H2B dimer chaperone also involved in DNA replication and repair38–40, encoded by the SUPT16H gene. This interaction was validated by co-immunoprecipitation studies in cells expressing endogenous and ectopic levels of DNMT3A (Figure 4D and Supplementary Fig. 7A–B). Reciprocal co-immunoprecipitation experiments confirmed direct protein-protein binding between DNMT3A and SPT-16 (Supplementary Figure 7C–D). We observed daunorubicin-induced SPT-16 recruitment to chromatin in cells expressing wild-type DNMT3A, which was abrogated by expression of DNMT3AR882 (Figure 4E). Consistent with these data, shRNA-mediated knock-down of SUPT16H decreased the sensitivity of cells to aclarubicin, similar to effects seen in DNMT3A-mutant cells (Figure 4F), and abrogated the effect of wild-type DNMT3A on histone eviction after daunorubicin (Figure 4G). Of note, both wild-type and mutant DNMT3A could bind SPT-16, suggesting a defect in the ability of DNMT3AR882 cells to bind DNA and recruit SPT-16 to nucleosomes. Chromatin immunoprecipitation sequencing (ChIP-seq) analysis in MEFs expressing either wild-type or mutant Dnmt3a, but not both, demonstrated a marked reduction in DNA binding by the Dnmt3a-mutant compared to Dnmt3a wild-type or haploinsufficient cells (Supplementary Fig. 7E–F).
Our study shows that leukemia-associated DNMT3AR882 mutations promote HSC function and cooperate with FLT3ITD and NPM1c to induce AML. DNMT3Amut AML cells show increased resistance to anthracycline-based chemotherapy, which results from a proximal defect in nucleosome remodeling that attenuates DDR (Supplementary Fig. 7G). Our data describe a novel mechanism of chemoresistance in AML driven by a specific mutation in an epigenetic regulator, and provide insights that can be used to interrogate DNA damage pathways as a therapeutic vulnerability in this common, poor-risk AML subtype.
Online Methods
Generation of the conditional Dnmt3aR878H allele in mice
To achieve inducible expression of the Dnmt3aR878H allele from its endogenous locus, a minigene combining exons 23 and 24 carrying a point mutation was inserted in place of endogenous Dnmt3a exon 23, preceded by a Lox-Stop-Lox NeoR (LSL) cassette (Figure 1A). After selection on G418, targeted mouse ES cell clones were screened by PCR and verified by Southern blotting; positive clones were expanded and injected into blastocysts. Removal of the stop cassette by Cre-mediated excision allows for the expression of the mutant Dntm3aR878H mRNA and protein, confirmed by Sanger sequencing of the cDNA derived from peripheral blood mononuclear cell RNA.
Animals, bone marrow transplantation, and flow-cytometric analysis
Laboratory mice were housed at the Memorial Sloan Kettering Cancer Center animal facility; all animal procedures were approved by the Institutional Animal Care and Use Committee. To achieve inducible hematopoietic-specific excision, Dnmt3a+/LSL-R878H animals crossed to Mx1-Cre-deletor received 4 intraperitoneal injections of poly(I:C) (Amersham). For phenotyping of primary animals, 9 Dnmt3a+/+:Mx1-Cre+, 12 Dnmt3a+/LSL-R878H:Mx1-Cre+, and 8 Dnmt3a+/LSL-R878H:Mx1-Cre− were used. The number of animals was chosen to ensure 90% power with 5% error based on observed standard deviation. For re-transplantation/engraftment studies 1×106 mononuclear cells from freshly harvested bone marrow were injected through tail veins into lethally irradiated (9.5 Gy) CD45.1 recipients; Mx1-Cre-driven recombination was induced by poly(I:C) injections in the recipients and was confirmed by Sanger sequencing of cDNA generated from peripheral blood mononuclear cells. For competitive bone marrow transplantation studies, 1×106 CD45.2 test bone marrow cells were competed against an equal number of CD45.1 wild-type cells and monitored by CD45.1/CD45.2 peripheral blood chimerism every 2 months; Mx1-Cre-driven recombination was induced by poly(I:C) injections in the recipients and was confirmed by Sanger sequencing of cDNA generated from peripheral blood mononuclear cells. Animals that failed to engraft (<1% CD45.2 chimerism in peripheral blood) or were lost due to poly(I:C) toxicity were excluded from analysis. For in vivo Dox treatment studies lethally-irradiated CD45.1 recipients were engrafted with 1×106 CD45.2 test bone marrow cells recombined in the donor; randomization was done by conducting CBC analysis prior to the start of drug administration and confirming that WBC count averages were equivalent in treatment and vehicle groups. Blinding was not done in these experiments. Peripheral blood was collected by submandibular puncture. Complete blood counts were obtained using IDEXX ProCyte DX automated hemocytometer. Flow cytometric analysis of mature blood lineages was performed as described41. Analysis of the hematopoietic stem and myeloid progenitor populations was performed on single-cell suspensions prepared from bone marrow, spleens and livers by flow cytometry after red blood cell lysis. Populations were defined as follows: long-term (LT)-HSC – Lineage−Sca1+c-Kit+ (LSK) CD150+CD48−, short-term (ST)-HSC – LSK CD150+CD48+, multipotent progenitors (MPP) – LSK CD150−CD48+, common myeloid progenitors (CMP) – Lineage−Sca1−c-Kit+ (LK) CD16/32−CD34+, granulocyte/macrophage progenitors (GMP) – LK CD16/32+CD34+, megakaryocyte/erythroid progenitors (MEP) – LK CD16/32−CD34−42. For immunophenotypic analysis of erythroid maturation the red blood cell lysis step was omitted; erythroblastic progenitor populations were defined as described43,44. All antibodies were from eBioscience or BioLegend: NK1.1 (PK136), CD11b (M1/70), CD45R (RA3-6B2), CD3 (17A2), Gr-1 (RB6-8C5), Ter119 (TER119), CD19 (6D5), CD4 (GK1.5), CD8 (53-6.7), cKit (2B8), Sca-1 (D7), CD150 (TC15-12F12.2), CD48 (HM48-1), CD16/32 (93), CD34 (RAM34), CD71 (R17217), Ki67 (SolA15), CD45.1 (A20), CD45.2 (104). Cell viability was monitored by propidium-iodide (PI) exclusion, DNA content was measured in formaldehyde-fixed and Triton X-100 permeabilized cells by DAPI staining. Peripheral blood smears were stained by Wright-Giemsa method. For histological analysis spleens, livers and sterna were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned and stained with hematoxylin and eosin (H/E). Slides were scanned using PerkinElmer Panoramic FLASH scanner and a CIS VCC color CCD camera.
Clonogenic potential in semisolid media
Freshly isolated 1×104 whole bone marrow cells were plated in MethoCult M3434 medium (StemCell) containing daunorubicin where indicated, in duplicate or triplicate. Colonies were counted after 10–14 days, when cells were harvested and replated, 1×104 cells per well in replicate, for an indicated number of passages or until colony-forming potential was exhausted.
Minimal residual disease in AML patients
Presence of minimal residual disease was assessed centrally in the ECOG-ACRIN Leukemia Translational Research Laboratory (LTRL) on day 28 after induction chemotherapy by multi-parameter flow cytometry using a FACSCanto II cytometer operated with FACSDiva software for both acquisition and analysis. Given the significantly lower MRD levels in blood than bone marrow in AML45, submission of aspirates was requested at all MRD time-points. Residual leukemic cells were identified based on leukemia-associated immunophenotypic features found at diagnosis. Heparinized bone marrow aspirates were shipped to the LTRL by overnight delivery on cool-packs and processed within 24 hours of collection. MRD was determined in whole, unseparated samples and expressed as percent of nucleated white blood cells based on SYTO® 16 green fluorescent nucleic acid staining (Invitrogen). Antibody panels for MRD determination were based on the findings at diagnosis. If more than one immunophenotypic clone were detected at diagnosis, antibody panels for MRD assessment were adjusted accordingly to cover all the antigen combinations of interest. Whenever possible, a minimum of 200,000 events were acquired. To reach this goal, aspirates with very low white blood cell count were subjected to red cell lysis with a solution of ammonium chloride, potassium bicarbonate and EDTA at room temperature for 10 minutes. In agreement with the literature, the threshold for MRD positivity was set at ≥0.1% of cells which stained for leukemia associated features46.
Molecular persistence of DNMT3A-mutant clones and other AML-associated mutations 28 days post-induction was assessed by hybridization capture-based next-generation sequencing of patient DNA purified from PBMCs for alterations included in the MSKCC-IMPACT panel as previously described47. The average sequencing depth was 2246× to ensure that even the lowest covered regions in the panel are covered at >500×. See also Supplementary Table 2.
Mouse embryonic fibroblasts (MEF)
MEFs were generated from E13.5 embryos by trypsin dissociation after removal of neural and fetal liver tissues. Fibroblasts were grown in DME supplemented with 4.5 g/L glucose, non-essential aminoacids (Gibco), 10% fetal calf serum (FCS), and penicillin/streptomycin and immortalized by SV40 large T-antigen by direct transfection. Expression of the Dnmt3aR878H allele was achieved by lentivirally-mediated expression of the Cre recombinase. For generation of MEFs that only expressed Dnmt3aWT or Dnmt3aR878H allele, but not both, Dnmt3a+/LSL-R878H animals were crossed with Dnmt3af/f conditional knock-out line48. Complete excision and expression of the Dnmt3amut allele was verified by cDNA Sanger sequencing.
Cell lines
Cell line authentication by STR (short tandem repeat) profiling was performed at Genetica DNA Laboratories – LabCorp. Cell line identity was confirmed at 100% except OCI/AML-3 and SET-2, which showed minor genetic drift within acceptable range (94.1% and 96.3% match, respectively). MOLM-13 cells were authenticated by a collaborator. KO-52 line was recently procured from a cell repository, therefore testing was not performed. Prior to experimental work all cell lines were maintained on 50 μg/ml plasmocin (InvivoGen) over the course of 3 weeks for mycoplasma prevention. Mycoplasma testing was performed using a PCR-based kit according to manufacturer’s instructions (Applied Biological Materials Inc.). DNMT3A R882 mutant status was validated by Sanger sequencing of DNMT3A exon 23 on genomic DNA. Human leukemia cell lines were grown in RPMI supplemented with 10–20% FCS and penicillin/streptomycin. 293T cells were maintained in DME with 10% FCS and penicillin/streptomycin.
Ectopic expression of wild-type and mutant DNMT3A and of shRNAs targeting SUPT16H
cDNA coding human wild-type DNMT3A was a kind gift from Dr. F. Fuks (Free University of Brussels). DNMT3A open reading frame was PCR amplified and subcloned into pMIGR1 (MSCV-IRES-GFP) retroviral vector. R882H substitution was introduced by site-directed mutagenesis using QuickChange Lightning kit (Agilent Technologies) according to manufacturer’s instructions. Viral particles were produced in 293T cells using pPAX2 packaging system and pseudotyped with VSV-G. Virus-containing supernatants were concentrated by PEG-8000 precipitation. Target cells transduced with DNMT3Awt or DNMT3AR882H viral particles were FACS-sorted based on GFP expression. SUPT16H (a gene coding SPT-16) targeting constructs were cloned in the pLKO.1 backbone from the TRC 2.0 collection: TRCN0000293281, TRCN0000293313, TRCN0000293348, TRCN0000293349, TRCN0000293350. Viral particles were generated as described above and used to transduce U2OS cells. Transduced cell populations were selected on puromycin for 7 days. Knock-down efficiency was assessed by immunoblotting.
Cell viability and proliferation assays
Cells were plated in 96-well plates at 1×104/well density and exposed to drugs in duplicate or triplicate as indicated. Relative cell numbers (compared to vehicle-treated control) were estimated after 48 hours by measuring ATP levels by CellTiter Glo (Promega) for suspension cultures, or by nucleic acids content by methylene blue retention for adherent cultures. As ATP levels as well as DNA and especially RNA content differ between cell lines, normalization to untreated was necessary for adequate comparison. CellTiter Glo assay was performed according to manufacturer’s instructions. For methylene blue staining cells were first thoroughly washed in PBS, then simultaneously fixed and stained by 0.5% methylene blue (Sigma) in 50% methanol, and plates were extensively washed under running water and let to air-dry. Retained methylene blue was solubilized by 0.1% SDS, and OD595 was measured by SpectraMax plate reader. Data are representative of at least two independent experiments in each cellular system.
For the ex vivo anthracycline dose-response studies on primary AML samples, 50 nl of each compound were dispensed via acoustic-based transfer into clear 384-well plates dissolved in DMSO at 1000-fold their respective screening concentration. 5000 cells were seeded in each well in 50 μl media on top of these compounds and incubated for 72 hours. ATP levels were measured as surrogate for cell viability (CellTiter Glo, Promega) according to manufacturer’s protocol. Data for each AML sample were normalized to the 32 negative control (DMSO) wells on each plate (100% viability) and the 32 positive control wells (1 μM bortezomib, set to 0% viability) after outlier removal. Clinical characteristics of patient samples are summarized in Supplementary Table 3. Banking and ex vivo drug testing of leukemic patient samples was approved by the ethical committee of the Medical University of Vienna # EK Nr: 2008/2015.
Comet assay
Alkaline and neutral comet assays were performed using CometSlides and Comet Assay kit (Trevigen) according to manufacturer’s instructions. Slides were stained with SYBR Gold and imaged using Zeiss Axio2Imaging upright microscope equipped with a 10× or a 20× Plan-NEOFLUAR 0.3NA objective and an AxioCam MRm camera through a FITC-compatible filter. At least 5 random fields containing a minimum of 40 non-overlapping comets in each group total were photographed. Images were analyzed using ImageJ software (NIH) and a CometAssay plugin calculating comet tail moment (arbitrary units) based on comet head and tail sizes (measured in pixels) and their integral intensity. The magnitude of these parameters depends on time of electrophoresis, staining brightness and image magnification, which were constant within each assay but varied between experiments. Blinding was not performed. Comet assays were repeated once in each of the following cell lines with similar results: MOLM-13, U-937, 32D, and independently confirmed in primary FACS-sorted MPPs from one animal of each genotype and from previously banked primary AML samples (please see Supplementary Table 4 for mutational information and clinical correlates). Statistical significance was determined by non-parametric Mann-Whitney rank sum test using GraphPad Prism software version 6.
Mutation frequency
Mutagenesis rate within HPRT locus was measured by 6-thioguanine (6-TG)-resistant colony formation as described49. HPRT (hypoxantine-guanine phosphoribosyltransferase, encoded by the HPRT1 gene) is an enzyme in the purine salvage pathway, which is critical for the generation of purine nucleotides. It is also involved in converting 6-thioguanine into a toxic antimetabolite that is subsequently incorporated into nascent DNA in replicating cells. Therefore cells lacking functional HPRT are resistant to 6-TG, forming the basis for the HPRT mutagenesis assay. However cells grown in conventional culture media do not depend on the purine salvage pathway and therefore are not hindered from accumulating HPRT-negative subclones. These pre-existing HPRT− cells can confound the analysis and need to be selected out prior to 6-TG assay. This can be done by selection on HAT (hypoxantine-aminopterin-thymidine) supplement. Aminopterin is a powerful inhibitor of folate metabolism and blocks de novo DNA synthesis. In this case DNA replication can be rescued by purine salvage pathway, which relies on hypoxantine and thymidine as substrates. Only cells with wild-type HPRT can use this metabolic pathway for survival.
HPRT1wt 293T and MOLM-13 cells were preselected on media containing HAT supplement (Sigma-Aldrich), then lentivirally transduced to express DNMT3A wild-type or R882H mutant, or with empty vector control. All constructs expressed GFP as a selectable marker. Transduced cells were FACS-sorted based on GFP expression, and grown exponentially for 1 month to allow accumulation of mutations. 1×106 293T cells were seeded on gelatin-coated p100 plates in triplicate, allowed to attach overnight, and selected on media containing 10 μg/ml 6-TG for 2 weeks. 6-TG-resistant colonies were fixed, stained with methylene blue and automatically counted by GelCount plate scanner and colony counter (Oxford Optronix) using embedded software. Data are representative of 3 independent experiments. 0.5×106 MOLM-13 cells were plated in 0.5ml/well of a 12-well plate of ClonaCell TSC methylcellulose-based medium (StemCell Technology) supplemented with 5 μg/ml 6-TG in triplicate. Colony counts were corrected for plating efficiency calculated based on a number of colonies formed by plating cell at clonal densities (100–200 cells per well or dish) in the absence of selective pressure.
Analysis of soluble nuclear and chromatin-bound proteins
Cells were synchronized in early-mid S-phase by double thymidine block (4 hours after second release) according to standard methods57, exposed to daunorubicin at time of second release where indicated, and equal numbers of cells were subjected to Subcellular Protein Fractionation procedure with the corresponding kit (Pierce). Soluble nuclear extracts and chromatin-bound fractions were resolved by SDS-PAGE and analyzed by Western blotting. Similar results showing differences in histone eviction in Dnmt3amut/DNMT3Amut-expressing or control cells were observed in each of 3 different cellular systems (leukemia cell lines, MEFs, or primary mouse splenocytes).
Co-immunoprecipitation (co-IP) and immunoblotting and analyses
After extensive washing in ice-cold PBS cells were lysed in lysis and digestion buffer (150 mM NaCl, 50 mM Tris HCl pH7.4, 0.2% NP-40) supplemented with protease and phosphatase inhibitors and 2.5 mM MgCl2, and 50 U/ml benzonase nuclease (Santa Cruz Biotechnology) were used to digest genomic DNA at 4°C with constant rotation. Reaction was stopped by adding EDTA to 5mM final concentration, insoluble material pelleted by centrifugation, and 0.5 mg whole cell lysate was incubated with either 5 μg DNMT3A-specific antibody ab13888 (Abcam) or 5 μg anti-SPT-16 H-300 antibody (Santa Cruz) overnight at 4°C with constant rotation. Immunocomplexes were captured by Protein A/G Plus Agarose (Santa Cruz Biotechnology) according to manufacturer’s instructions, washed, and eluted by boiling in Laemmli buffer. For target detection by Western blotting proteins were resolved on 4–12% Bis-Tris gels (Invitrogen) by SDS-PAGE, blotted onto PVDF membranes, and probed by standard methods using the following antibodies: DNMT3A (#3598), pCHK1 (#2341), total CHK1 (#2345), pCHK2 (#2661), total CHK2 (#2662), p-p53 (#9284), pH2A.X (#9718), PARP (#9542), Caspase-3 (#9662), β-actin (#4970), GAPDH (#2118) from Cell Signaling Technology; total p53 (DO-1), SPT-16 (H-300), TFIIH p89 (S-19) from Santa Cruz Biotechnology; and histone H2A (ab18255), histone H3 (ab1791), H3K36me3 (ab9050), P-glycoprotein (ab3366), MRP1 (ab3369), and RPA32 (ab16850) from Abcam. Co-immunoprecipitation experiment with DNMT3A-specific antibodies was performed in 5 different cellular systems (MEFs, stably transduced 293T cells, transiently transfected 293T cells, stably transduced U2OS cells, a panel of 5 DNMT3A wild-type or mutant cells lines) with similar results. Reciprocal co-IP was performed in MEFs and stably transduced MOLM-13 cells.
Peptide pull-down coupled with mass-spectroscopy
DNMT3A AC-MEGSRGRLRGGLGWEC peptide mapping to the N-terminal domain was synthesized, quantified, and conjugated to SulfoLink agarose (Pierce) according to the manufacturer’s instructions. Ten micrograms of peptide bound to the agarose beads were used in a pull-down reaction with 10 mg of MOLM-13 cell nuclear extract as previously reported58. The bound proteins were then eluted with 1×SDS-sample buffer and resolved on a 4–12% Bis-Tris gel (Invitrogen). After silver staining, recovered proteins from 2 independent experiments were identified by LC-MS/MS at the Proteomics and Microchemistry Core Facility at MSKCC. Data analysis was performed using Scaffold software. A total of 1840 unique peptides corresponding to 216 proteins were identified; 17 keratins and various cytoskeletal components among them were excluded. Among top-scoring proteins SPT-16 was represented by 11 unique peptides with 13.7% sequence coverage. See also Supplementary Table 5.
Statistical Analysis
Statistical significance was determined by unpaired Student’s t-test after testing for normal distribution. For samples with significantly different variances Welch’s correction was applied. Samples with non-normal distribution (continuous variables) were compared using non-parametric Mann-Whitney rank sum test. For categorical data Fisher’s exact t-test was used. Data were plotted using GraphPad Prism 7 software as mean values, error bars represent standard deviation; for measurements done in duplicate error bars were not plotted. For select figure panels graphs were generated using R. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
The relationship between MRD and individual mutations was examined using a four-fold contingency table and Fisher’s exact test. To this end, each mutation was coded as a binary (wildtype or mutant) variable as previously described6. Minimal residual disease was defined as detection of ≥0.1% of cells that stained for AML-associated surface markers by flow cytometry as described above. There were 144 patients with full mutational and MRD data, summarized in Supplementary Table 1. To ensure that family-wise error rate is controlled at the 5% level we used a resampling-based method to adjust the p-values. This method is more powerful than traditional step-down Holm method, taking into account the discreteness and the multivariate distribution of the test statistics60. The resulting p-values not only control the marginal (comparison-wise) false positive rate at the nominal level of 5% but also the overall (family-wise) error rate at 5%. In other words, if none of the mutations were associated with MRD, the probability of finding at least one association is less than 5%. See also Supplementary Table 1.
Supplementary Material
Acknowledgments
This work was supported by NCI K99 grant CA178191 and Lauri Strauss Leukemia Foundation award to OAG, by NCI K08 grant CA169055 and an American Society of Hematology (ASHAMFDP-20121) under the ASH-AMFDP partnership with The Robert Wood Johnson Foundation to FEG-B, by a Hyundai Hope On Wheels award to BS, by a Gabrielle’s Angel Fund grant to RLL and AMM, by NCI grant CA172636 to RLL and AMM, and by the Samuel Waxman Cancer Research Center. ASM is supported by NIH grants T32GM007739 and F30CA18349. AMM is a Burroughs Wellcome Clinical Translational Scholar, and is supported by the Sackler Center for Biomedical and Physical Sciences. RLL is a LLS Scholar. PBS is supported by the Austrian Research Foundation (#P27132) and the Oesterreichische Nationalbank (OeNB) Anniversary Fund (#15936). MSKCC cores used in these studies are supported by the P30 Core Grant CA008748. ECOG-ACRIN Integrated Leukemia Translational Research Center is funded by NCI U10 grant CA180827 to EMP et al. The authors are grateful to C. Sheridan, J. Gandara and Y. Neelamraju for technical support, to Weill Cornell Medicine Epigenomics Core for sequencing services, to H. Erdjument-Bromage (MSKCC Proteomics Core), to S. Zha (Columbia University Medical Center) and to A. Ciccia (Columbia University Medical Center) for critical reading of the manuscript and helpful suggestions.
Footnotes
Accession codes
RNA-seq, ERRBS, and ChIP-seq raw and processed data were deposited into GEO database under bundle ID GSE72883.
Author contributions
O.A.G. and K.S. performed most experiments. L.B., M.D.K., D.T., E.L., A.R.W., A.A.R.G., E.P., F.P., and A.S.M. assisted with many experiments. C.Z., J.R., M.S.T., O.A.-W., M.G., E.M.P., A.M.M., and R.L.L. designed the clinical study, collected and analyzed patient data. A.G.C., A.V.K., R.P.K. and S.A.A. designed, conducted, and analyzed ChIP-sequencing experiments. B.S., F.E.G.-B., and A.M.M. assisted with RNA-sequencing, ERRBS, and data analysis. B.S. mined TCGA data. L.L. and S.D.N. designed and performed peptide pull-down assay. B.H.D. assisted with animal hematopathology. A.M. and M.E.A. assisted with patient DNA resequencing. G.H., W.R.S., S.K., and P.B.S. designed and conducted ex vivo drug studies on primary AML samples. J.C. designed and oversaw small-molecule quantification by mass-spectroscopy. S.A.R. and S.M. provided Dnmt3a cKO mice, S.M. contributed to study conception. O.A.-W. assisted with study design. O.A.G. and R.L.L. conceived the study, designed experiments, analyzed data, and wrote the manuscript. All authors read, edited, and approved the manuscript.
Statement of competing financial interests
Authors declare no competing financial interests.
References
- 1.Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nature genetics. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 4.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. The New England journal of medicine. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. The New England journal of medicine. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sehgal AR, et al. DNMT3A Mutational Status Affects the Results of Dose-Escalated Induction Therapy in Acute Myelogenous Leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:1614–1620. doi: 10.1158/1078-0432.CCR-14-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leiserson MD, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nature genetics. 2015;47:106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busque L, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nature genetics. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jan M, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Science translational medicine. 2012;4:149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature genetics. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nature reviews. Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holz-Schietinger C, Matje DM, Reich NO. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. The Journal of biological chemistry. 2012;287:30941–30951. doi: 10.1074/jbc.M112.366625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russler-Germain DA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SJ, et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013;122:4086–4089. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papaemmanuil E, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaidzik VI, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG) Blood. 2013;121:4769–4777. doi: 10.1182/blood-2012-10-461624. [DOI] [PubMed] [Google Scholar]
- 23.Ostronoff F, et al. Mutations in the DNMT3A exon 23 independently predict poor outcome in older patients with acute myeloid leukemia: a SWOG report. Leukemia. 2013;27:238–241. doi: 10.1038/leu.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribeiro AF, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119:5824–5831. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
- 25.Tie R, et al. Association between DNMT3A mutations and prognosis of adults with de novo acute myeloid leukemia: a systematic review and meta-analysis. PLoS One. 2014;9:e93353. doi: 10.1371/journal.pone.0093353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, et al. Relation of clinical response and minimal residual disease and their prognostic impact on outcome in acute myeloid leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:1258–1264. doi: 10.1200/JCO.2014.58.3518. [DOI] [PubMed] [Google Scholar]
- 27.Ganzel C, et al. Minimal Residual Disease Assessment By Flow Cytometry in AML Is an Independant Prognostic Factor Even after Adjusting for Cytogenetic/Molecular Abnormalities. Blood. 2014;124 [Google Scholar]
- 28.Ivey A, et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N Engl J Med. 2016;374:422–433. doi: 10.1056/NEJMoa1507471. [DOI] [PubMed] [Google Scholar]
- 29.Othus M, et al. Effect of measurable (‘minimal’) residual disease (MRD) information on prediction of relapse and survival in adult acute myeloid leukemia. Leukemia. 2016 doi: 10.1038/leu.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walter RB, et al. Comparison of minimal residual disease as outcome predictor for AML patients in first complete remission undergoing myeloablative or nonmyeloablative allogeneic hematopoietic cell transplantation. Leukemia. 2015;29:137–144. doi: 10.1038/leu.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shih AH, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer cell. 2015;27:502–515. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akalin A, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS genetics. 2012;8:e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garrett-Bakelman FE, et al. Enhanced reduced representation bisulfite sequencing for assessment of DNA methylation at base pair resolution. Journal of visualized experiments : JoVE. 2015 doi: 10.3791/52246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang F, Teves SS, Kemp CJ, Henikoff S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim Biophys Acta. 2014;1845:84–89. doi: 10.1016/j.bbcan.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stracker TH, Usui T, Petrini JH. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 2009;8:1047–1054. doi: 10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins AR, et al. The comet assay: topical issues. Mutagenesis. 2008;23:143–151. doi: 10.1093/mutage/gem051. [DOI] [PubMed] [Google Scholar]
- 37.Pang B, et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun. 2013;4:1908. doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinant C, et al. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell. 2013;51:469–479. doi: 10.1016/j.molcel.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belotserkovskaya R, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 41.Mayle A, Luo M, Jeong M, Goodell MA. Flow cytometry analysis of murine hematopoietic stem cells. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2013;83:27–37. doi: 10.1002/cyto.a.22093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 43.Chen K, et al. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17413–17418. doi: 10.1073/pnas.0909296106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Socolovsky M, et al. Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 45.Paietta E. Should minimal residual disease guide therapy in AML? Best Pract Res Clin Haematol. 2015;28:98–105. doi: 10.1016/j.beha.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 46.Paietta E. Minimal residual disease in acute myeloid leukemia: coming of age. Hematology Am Soc Hematol Educ Program. 2012;2012:35–42. doi: 10.1182/asheducation-2012.1.35. [DOI] [PubMed] [Google Scholar]
- 47.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236:1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- 49.Havre PA, Yuan J, Hedrick L, Cho KR, Glazer PM. p53 inactivation by HPV16 E6 results in increased mutagenesis in human cells. Cancer research. 1995;55:4420–4424. [PubMed] [Google Scholar]
- 50.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 55.Akalin A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome biology. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li S, et al. An optimized algorithm for detecting and annotating regional differential methylation. BMC bioinformatics. 2013;14(Suppl 5):S10. doi: 10.1186/1471-2105-14-S5-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Molecular biology of the cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wysocka J. Identifying novel proteins recognizing histone modifications using peptide pull-down assay. Methods. 2006;40:339–343. doi: 10.1016/j.ymeth.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deshpande AJ, et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer cell. 2014;26:896–908. doi: 10.1016/j.ccell.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westfall PH, Young SS. P-Value Adjustments for Multiple Tests in Multivariate Binomial Models. J Am Stat Assoc. 1989;84:780–786. doi: 10.2307/2289666. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.