Abstract
Fragile X mental retardation is caused by absence of the RNA-binding protein fragile X mental retardation protein (FMRP), encoded by the FMR1 gene. There is increasing evidence that FMRP regulates transport and modulates translation of some mRNAs. We studied neurotransmitter-activated synaptic protein synthesis in fmr1-knockout mice. Synaptoneurosomes from knockout mice did not manifest accelerated polyribosome assembly or protein synthesis as it occurs in wild-type mice upon stimulation of group I metabotropic glutamate receptors. Direct activation of protein kinase C did not compensate in the knockout mouse, indicating that the FMRP-dependent step is further along the signaling pathway. Visual cortices of young knockout mice exhibited a lower proportion of dendritic spine synapses containing polyribosomes than did the cortices of wild-type mice, corroborating this finding in vivo. This deficit in rapid neurotransmitter-controlled local translation of specific proteins may contribute to morphological and functional abnormalities observed in patients with fragile X syndrome.
Keywords: dendrites, metabotropic glutamate receptor, mRNA, plasticity, ultrastructure
Fragile X mental retardation syndrome is an inherited, X-linked disorder. In most patients, methylation of an extreme expansion (200–1,000 repeats) of a (CGG)n trinucleotide repeat in the 5′ UTR of the FMR1 gene blocks transcription of fmr1 mRNA (1). The resulting absence of fragile X mental retardation protein (FMRP) causes the syndrome, which is characterized by mental retardation, macroorchidism, and behavioral abnormalities (2). The brains of these patients exhibit an unusual, spindly appearance of the dendritic spines as well as an overabundance of spines (3, 4), a morphology that resembles early postnatal tissue.
The function of FMRP is unknown; in neurons much of the protein is found in dendrites (5). FMRP contains RNA-binding elements (6) and is associated with actively translating polyribosomes in the brain (7–9). Several laboratories have described sets of mRNAs bound by FMRP (10–12), and specific motifs involved in FMRP binding of some mRNAs have been identified (13, 14). Recently, we demonstrated (10) that several members of a subset of mRNAs bound by FMRP in intact cells are differentially distributed and/or translated in dendritic, as compared to somatic, subcellular domains. This finding suggests direct involvement of FMRP in transport and/or translation of mRNA in dendrites. Antar et al. (15) have demonstrated rapid transport of FMRP into dendrites upon KCl depolarization. We report here that a dynamic aspect of translation, neurotransmitter-induced rapid initiation, is directly impacted by the absence of FMRP.
Protein translation in dendrites was suggested by early descriptions of postsynaptic polyribosomal aggregates (PRAs) during synaptogenesis and in the visual cortex of rats reared in complex environments, indicating the importance of local translation for synaptic plasticity (16, 17). Components necessary for translation are present postsynaptically, and protein synthesis has been described in synaptosomes as well as in dendrites in culture (18–24). As suggested by postsynaptic polyribosome up-regulation in association with synaptic plasticity (25), dendritic protein synthesis appears to be activity-regulated. We have shown that rapid association of mRNAs with ribosomes, accompanied by accelerated protein translation, can be elicited by K+ depolarization and by specific agonists of group I metabotropic glutamate receptors (mGluRs 1 and 5; ref. 26). Moreover, this response is mimicked by 1-oleoyl-2-acetyl-sn-glycerol (OAG), the cell-permeable diacyl glycerol analog and synthetic stimulant of PKC, indicating that the signaling pathway involves PKC. Among the proteins synthesized near synapses in response to activity and activation of mGluR1/5 is FMRP (15, 27–29).
To examine directly the role of FMRP in neurotransmitter-activated dendritic translation, we used an fmr1-knockout (KO) mouse model in which no full-length FMRP is produced (30). These mice show immature dendritic spine morphology similar to that observed in human fragile X patients (4, 31). Here, we investigate rapid neurotransmitter-induced translation in synaptoneurosomes of fmr1-KO and WT mice. We report that, unlike those from WT mice, synaptoneurosomes from fmr1-KO mice do not exhibit neurotransmitter-induced rapid formation of polyribosomes or accelerated methionine incorporation into proteins. The deficit in translation initiation appears to be downstream of PKC, because OAG does not induce protein synthesis in KO preparations but does in WT preparations. PKC levels do not differ between WT and KO synaptoneurosomes. Moreover, we report that developing KO mice display a lower proportion of cortical dendritic spines with postsynaptic polyribosomes; this finding is an in vivo corroboration of the measurement of translation in synaptoneurosomes in vitro. Thus, one role of FMRP in normal brains might involve selective facilitation of a rapid, localized translational response to synaptic activation.
Materials and Methods
WT and fmr1-KO mice of the FVB.129P2-Fmr1tm1Cgr strain were used in this study. The majority of these experiments used “sighted” mice in which the Pde6b gene [a mutation in this gene codes for retinal degeneration in FVB mice (32)] had been selectively replaced by crossing with strains carrying the nondefective allele (V. Errijgers and R. F. Kooy, unpublished work). However, some earlier synaptoneurosome activation experiments used a “blind” variant of this strain that still possessed the Pde6b mutation. For synaptoneurosome preparations, animals are killed before retinal degeneration is complete, suggesting that this trait would have little effect on experimental results, but for some tests, offspring of an F1 hybrid cross with C57BL/6Hsd, ICR mice (Harlan–Sprague–Dawley) were used to verify that retinal degeneration had not biased the results.
Synaptoneurosome Preparations. Occipital and parietal cortices were removed from groups of six to eight mice (postnatal day 12 to postnatal day 15) and homogenized in a glass homogenizer in chilled homogenization buffer (50 mM Hepes/125 mM NaCl/20 mM potassium acetate/5 mM MgCl2/75 mM sucrose, pH 7.1); the resulting population of subcellular particles was size-selected through a series of filters, with the smallest pore size being 10 μm (33). The suspension was centrifuged briefly (1 min at 4,000 × g at 4°C) to remove heavy particles. Synaptoneurosomes were continuously stirred for aeration and treated with 10–6 M tetrodotoxin (for 5 min at 4°C and for 5 min at room temperature) to decrease spontaneous synapse firing. Immediately before activation, duplicate baseline (t = 0 min) samples of ≈0.5 ml were removed and lysed in 1.2% Triton X-100.
Synaptic Activation. Aliquots were stimulated with either 40 mM K+ or 5 × 10–6 M (S)3,5-dihydroxyphenylglycine (DHPG, a group 1 mGluR agonist, Tocris Cookson, Ellisville, MO). In some experiments, parallel pools of WT and KO synaptoneurosomes were stimulated with 7.5 μg/ml freshly prepared OAG. From both stimulated and unstimulated pools, samples of 1 ml were removed at 1, 2, and 5 min and lysed in 1.2% Triton X-100. Lysates were spun for 30 sec at 13,600 × g, and supernatants were layered over 1 M sucrose in Ross–Kobs polysome buffer (1 mM potassium acetate, pH 6/10 mM Tris·HCl, pH 7.6/1.5 mM MgCl2) and then centrifuged for 11 min at 400,000 × g at 4°C. The polyribosomal pellet was resuspended in 0.15 ml of 0.5 M KCl. The RNA content of supernatant and pellet samples was measured by OD260. At each time point, polyribosomal RNA content was expressed as a proportion of total RNA (pellet plus supernatant) and normalized to the average proportion of polyribosomal RNA in the sample taken immediately before stimulation (t = 0 min).
Repeated-measures ANOVA was used to evaluate the difference in proportion of polyribosomal RNA between experimental groups. For each biochemical agonist and within each genotype, the response of stimulated synaptoneurosomes was compared with the response of unstimulated synaptoneurosomes. We tested for a between-subject effect of stimulation, a within-subject effect of time, and a stimulation–time interaction. The polyribosomal RNA content of each time point's stimulated sample was then divided by that of the corresponding unstimulated sample, and the response of KO synaptoneurosomes was compared with the response of WT synaptoneurosomes. Here we tested for a between-subject effect of genotype, a within-subject effect of time, and a genotype–time interaction. When a significant effect was observed, post hoc analysis of stimulation or genotype differences at individual time points was done with the Student–Newman–Keuls test (P < 0.05). Because of missing data at longer and shorter intervals, only the 2- and 5-min intervals were included in the statistical analyses for K+, and only 1-, 2-, and 5-min intervals were included for the DHPG and OAG experiments.
For determination of amino acid incorporation, synaptoneurosomal pellets from WT or KO mice were resuspended in Eagle's medium, treated with 10–6 M tetrodotoxin (for 5 min at 4°C) then 5 × 10–5 M 2-amino-5-phosphonovaleric acid (for 15 min at room temperature) with continuous stirring. [35S]Methionine [35 μCi/ml, Amersham Pharmacia (1 Ci = 37 GBq)] was added, and, at t = 0 min, triplicate 0.05-ml samples were pipetted into 1 ml of cold 10% trichloroacetic acid. The remaining suspension was divided into two separate, stirred aliquots: one stimulated with 5 × 10–6 M DHPG and the other unstimulated. At 1, 2, and 5 min after stimulation, triplicate 0.05-ml samples were taken. All samples were incubated for 1 h on ice, filtered, washed with 5% trichloroacetic acid, dried, and counted for methionine incorporation. Paired t tests were used to test for the effect of DHPG stimulation on WT or KO samples at each time point.
Western Blots. For comparison of PKC levels in KO and WT mice, synaptoneurosomes were prepared from cortices of individual mice (n = 4 per group; postnatal day 12 to postnatal day 15). Preparations were lysed in a buffer containing 1.6% SDS in 50 mM Tris (pH 8), 2 mM EDTA, 2 mM EGTA, 5 mM NaF, 50 mM NaCl, 1 mM PMSF, 0.1 mM sodium orthovanadate, and protease inhibitor mixture (Sigma). For each sample, 40 μg of total protein was separated on 8% PAGE gels, blotted, and stained with monoclonal antibody to PKC isoforms α, β, and γ (Santa Cruz Biotechnology). A goat antimouse secondary antibody (Sigma) was applied, followed by chemiluminescence (Sigma). Subsequent staining for β-actin (Sigma) provided a loading standard. The relative optical densities of the 80-kDa PKC band and the 42-kDa actin band were measured with image 4.0.2 (Scion, Frederick, MD). The ratio of PKC:actin was calculated, and a t test was used to test for group differences in PKC levels.
Electron Microscopy. To examine the abundance of postsynaptic polyribosomes in KO vs. WT mice in vivo, eight WT and five fmr1-KO sighted mice (on postnatal day 15) and six WT and seven KO sighted mice (on postnatal day 25) were deeply anesthetized and intracardially perfused. Perfusion and tissue-preparation procedures for electron microscopy have been described (34). Layer IV of the visual cortex was examined by raters blind to experimental group, and synapses were identified in aligned, overlapping areas of serial 60-nm-thick sections (final magnification ×26,400) with as criteria the presence of at least three vesicles in the presynaptic process and the presence of a postsynaptic density. Synapses on dendritic spines (axospinous synapses) were counted with the unbiased stereological “physical disector” method (35) in double disector mode (all synapses that terminated within the series of sections counting in either direction through the series were analyzed). Each axospinous synapse was evaluated to determine whether it contained at least one polyribosome (containing at least three ribosomes) in the spine, head, or neck or within 0.23 μm of the base of the spine (6 mm on printed micrographs) as it was visually reconstructed from the micrograph series. For each animal, the proportion of axospinous synapses with PRAs was calculated of all axospinous synapses evaluated. ANOVA with genotype and age as main effects was used to detect differences between groups.
Synaptoneurosome Electron Microscopy. To assess possible differences in postsynaptic size in KO vs. WT synaptoneurosomes, synaptoneurosomes were prepared as described above and centrifuged, and pellets enriched in synaptoneurosomes were fixed for 24 h in 2% glutaraldehyde/2% paraformaldehyde in 0.14 M sodium cacodylate buffer (pH 7.3) and prepared for electron microscopy, as was the brain tissue above. Sections of 60 nm were taken from the embedded pellets and examined with electron microscopy. Random pictures of 15 fields of view were taken from each pellet at a final magnification of ×26,400. At least 50 synaptoneurosomes per pellet were analyzed by raters blind to group. The volume of the postsynaptic component was measured with the “nucleator,” an unbiased stereological tool (36). χ2 analysis was used to assess the effect of genotype on frequency of synaptoneurosomes in each of nine size bins.
Results
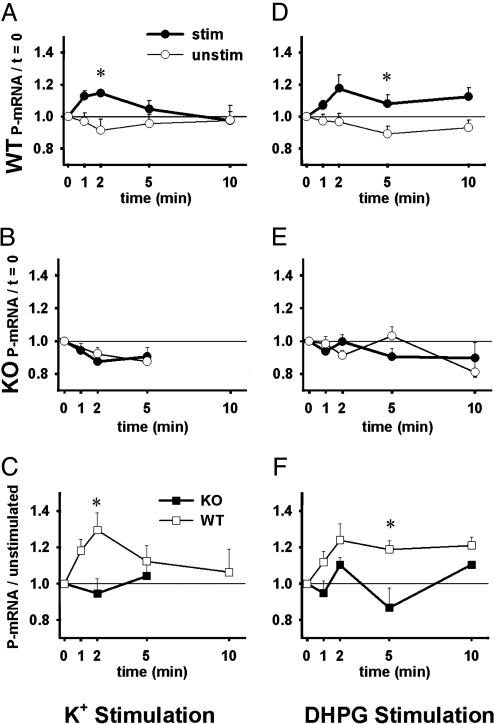
Stimulation of Synaptoneurosomes Induces Translation in WT but Not fmr1-KO Mice. In WT samples, (Fig. 1A), K+ stimulation elicited a rapid peak of polyribosomal aggregation above the basal level displayed by unstimulated synaptoneurosomes [ANOVA: main effect stimulation (P < 0.05); stimulation–time interaction (P < 0.05)]. Post hoc analysis revealed that levels of mRNA incorporation into polyribosomes [polyribosomal mRNA (P-mRNA)] were significantly higher in WT stimulated samples at t = 2 min than were the levels in unstimulated samples at t = 2 min. In KO samples (Fig. 1B), K+ stimulation did not elicit increased P-mRNA above unstimulated levels (P > 0.05). When each time point's stimulated sample is normalized to its corresponding unstimulated sample, we can compare KOs with WTs and test for genotype differences in the synaptoneurosome response to the agonist (Fig. 1C). Here, the overall difference between WT and KO response to K+ stimulation approaches statistical significance (P = 0.06), and P-mRNA levels diverge between WT and KO samples over time (genotype–time interaction; P < 0.05). At t = 2 min, WT levels of P-mRNA are significantly higher than KO levels. These data indicate that protein synthesis was increased after K+ stimulation in WT but not in fmr1-KO mice.
Fig. 1.
K+ or DHPG stimulation initiates translation in WT but not in fmr1-KO synaptoneurosomes. Graphs depict mRNA incorporation into polyribosomes (P-mRNA) after stimulation. Each point represents polyribosomal RNA as a fraction of total RNA divided by the baseline proportion (at t = 0). (A) WT levels of P-mRNA after K+ stimulation (•, n = 7 experiments) were significantly increased overall and at t = 2 min compared with WT unstimulated samples (○, n = 7) (all samples normalized to t = 0). (B) Response of KO samples stimulated with K+ (•, n = 6) did not differ from KO unstimulated samples (○, n = 6) (all samples normalized to t = 0). Because limited sample was available, the 10-min time point was omitted here. (C) When K+-stimulated samples are normalized to unstimulated samples, WT synaptoneurosomes (□, n = 7) exhibit increased P-mRNA at t = 2 min compared with KO samples (▪, n = 6). (D) WT P-mRNA levels after DHPG stimulation (•, n = 13 experiments) were significantly increased overall and at t = 5 min compared with WT unstimulated samples (○, n = 9) (all samples normalized to t = 0). (E) Response of KO samples stimulated with DHPG (•, n = 10) did not differ from KO unstimulated samples (○, n = 6) (all samples normalized to t = 0). (F) When DHPG-stimulated samples are normalized to corresponding unstimulated samples, WT synaptoneurosomes (□, n = 8) exhibit increased P-mRNA overall and at t = 5 min compared with KO samples (▪, n = 6). Error bars indicate SEMs; *, P < 0.05.
To test whether the progressive retinal rod degeneration to which FVB mice are subject beginning around postnatal day 12 (32) might have played a role in the outcome of these early experiments, synaptoneurosomes from WT mice of sighted F1 hybrid (FVB × C57BL/6) litters were also tested. Results for F1 hybrid mice were analogous to those observed with FVB.129 mice (data not shown), and experiments with F1 hybrid animals were therefore included in results for DHPG and OAG stimulation.
We then stimulated WT and KO synaptoneurosomes with the group I mGluR-specific glutamate analog DHPG. In WT samples, DHPG stimulation resulted in increased P-mRNA levels compared with levels in unstimulated samples overall (P < 0.05) and at t = 5 min (Fig. 1D). In KO samples (Fig. 1E), no effect was observed from DHPG stimulation (P > 0.05). Testing for genotype differences in the DHPG response reveals that synaptoneurosomes from WT mice exhibit an overall increase in incorporation over that of KO synaptoneurosomes (P < 0.05; Fig. 1F), including a significantly higher response at t = 5 min. The overall pattern indicates that WT protein synthesis was increased by DHPG treatment whereas KO protein synthesis was not. Methionine incorporation data described below further support this conclusion.
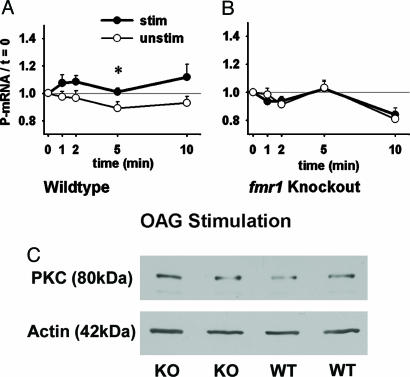
Because some proteins, including glutamate receptors, might be down-regulated in fmr1-KO mice, we next used OAG to activate PKC more directly (26). Fig. 2A shows that P-mRNA levels in WT synaptoneurosomes stimulated with OAG are elevated overall (P < 0.05) and at t = 5 min compared with WT unstimulated samples. Synaptoneurosomes from KO mice (Fig. 2B) do not exhibit this response to OAG stimulation (P > 0.05). Because PKC stimulation was being tested here, we examined whether KO and WT mice differ in synaptic PKC levels. Western blot analysis (Fig. 2C) demonstrated that the relative optical density of PKC (normalized to actin) was 0.64 ± 0.04 (mean ± SEM.) for KO synaptoneurosome preparations (n = 4) and 0.67 ± 0.10 in WTs (n = 4). Levels of PKC did not differ between WT and KO synaptoneurosomes (P > 0.05). Therefore, differential expression of PKC does not account for the absent OAG response in KO synaptoneurosomes. Together, these data indicate that the absence of DHPG response in KO mice appears to involve some deficit downstream of PKC.
Fig. 2.
Stimulation of PKC with OAG (diacylglycerol analog) initiates translation in WT but not in fmr1-KO synaptoneurosomes. (A) WT levels of P-mRNA after OAG stimulation (•, n = 12 experiments) were significantly increased overall and at t = 5 min compared with WT unstimulated samples (○, n = 9) (all samples normalized to t = 0). (B) Response of KO samples stimulated with OAG (•, n = 10) did not differ from KO unstimulated samples (○, n = 6) (all samples normalized to t = 0). (C) Western blot analysis reveals that levels of PKC (normalized to actin) do not differ between WT and KO synaptoneurosomes (P > 0.05; n = 4 per group; two per group shown). Error bars indicate SEMs; *, P < 0.05.
Methionine Incorporation Increases Rapidly After Stimulation in WT but Not fmr1-KO Synaptoneurosomes. Incorporation of [35S]methionine was next used to measure protein synthesis. As depicted in Table 1, synaptoneurosomes from WT mice exhibited a burst of translational activity such that at t = 5 min after DHPG stimulation, incorporation had increased 2.06-fold from t = 0, compared with a 1.61-fold increase from t = 0 to t = 5 min in WT unstimulated samples (seven experiments, P < 0.05). In KO mice, incorporation increases (from t = 0 levels) were less pronounced after 5 min in both the stimulated (1.12-fold) and unstimulated (1.19-fold) conditions and did not differ between these conditions (seven experiments, P > 0.05). These differences were not due to differential protein concentration in the synaptoneurosome preparations (data not shown).
Table 1. Incorporation of 35S methionine.
| Ratio of cpm (t = n/t = 0)
|
||
|---|---|---|
| Condition | WT | KO |
| Unstimulated | ||
| 1 min | 1.21 | 1.05 |
| 2 min | 1.17 | 1.21 |
| 5 min | 1.61 | 1.19 |
| Stimulated | ||
| 1 min | 1.35 | 1.06 |
| 2 min | 1.62 | 1.14 |
| 5 min | 2.06 | 1.12 |
Synaptoneurosomes from WT and KO mice were incubated with 35S methionine, and incorporation into polypeptides (cpm) was measured in samples removed at short intervals from parallel unstimulated and DHPG-stimulated aliquots. WT synaptoneurosomes exhibit a significant (P < 0.05) increase in 35S methionine incorporation (cpm) 5 min after DHPG stimulation, compared with WT unstimulated samples. KO synaptoneurosomes do not exhibit this increase after stimulation (all samples normalized to cpm at t = 0).
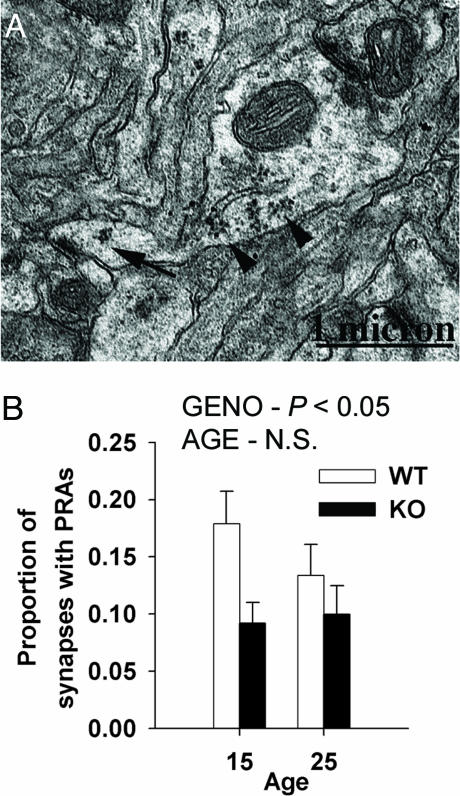
More Cortical Spine Synapses Have Polyribosomes in WT than in fmr1-KO Mice. We examined visual cortices with electron microscopy to compare the abundance of postsynaptic PRAs in fmr1-KO and WT mice (Fig. 3). For this anatomical study, WT and KO mice were tested for the Pde6b mutation (32) and were confirmed sighted. Axospinous synapses (see Fig. 3A) in the neuropil of layer IV visual cortices were examined on postnatal days 15 and 25, ages at which PRAs in spines are elevated, compared to PRAs in adulthood (H. M. Hwang and W.T.G., unpublished work). The proportion of axospinous synapses with PRAs was significantly higher in WT mice compared with KOs (Fig. 3B).
Fig. 3.
In vivo translational deficit in fmr1-KO mice. (A) Electron microscopic image of a dendrite in the neuropil of layer IV of visual cortex (FVB.129 sighted WT mouse). A spine on this dendrite forms a synaptic contact and contains a PRA (arrow). Numerous PRAs (arrowheads) are in the dendrite itself. (B) Unbiased stereological estimate of the proportion of axospinous synapses associated with PRAs from the visual cortex neuropil of sighted WT and fmr1-KO mice on postnatal days 15 and 25. KO mice exhibit a significantly lower proportion of synapses with PRAs than do WT mice (ANOVA: main effect of genotype, P < 0.05), indicating a deficit in synaptic protein synthesis. There was no significant effect of age. Error bars indicate SEMs. Preliminary data from this project were published in ref. 37.
Because of the abnormal dendritic spine morphology observed in human fragile X patients and fmr1-KO mice (3, 4, 31, 38), we next considered whether postsynaptic compartments in KO mice might reseal into smaller synaptic structures than those of WT mice, altering the probability that they contain translation components. Synaptoneurosome preparations were examined by electron microscopy for possible differences in the size or shape of the postsynaptic components. χ2 analysis revealed no statistical differences in the size of postsynaptic components between synaptoneurosomes of KO and WT mice (P > 0.2). Postsynaptic volumes thus provide no physical basis for a difference in the postsynaptic presence of translational machinery.
Discussion
We have previously demonstrated that stimulation of group 1 mGluRs triggers the assembly of polyribosomes and results in a rapid, transient burst of protein translation (26). The principal implication of the results presented here is that FMRP, an mRNA-binding protein, is essential to this rapid initiation of synaptically driven protein synthesis. Using an fmr1-KO mouse, we tested directly for a possible role of FMRP in protein synthesis, using cerebral cortical synaptoneurosomes. These KO mice are unable to respond to neurotransmitter stimulation with a typical burst in translation initiation as indicated by the lack of rapid polyribosome formation and the lack of change in [35S]methionine incorporation, in contrast with WT mice. In addition, we describe an in vivo corroboration of the synaptoneurosome results: In the absence of FMRP, there is a lower proportion of spine synapses containing postsynaptic PRAs in the visual cortices of KO mice.
Two groups (8, 9) have recently demonstrated that FMRP is associated with translating polyribosomes in neurons. It is not yet clear, however, just how FMRP might function in the context of what is known about the mechanisms that orchestrate control of postsynaptic translation. Some proteins whose synthesis is locally enabled or blocked by FMRP may be among those whose mRNAs are proposed to bind specifically to FMRP (10, 13, 14). Largely nonoverlapping lists of these FMRP-associated mRNAs have been identified in lymphoblastoid cells and neuronal tissues (10–12, 39). FMRP might affect transport and translation of largely separate sets of proteins in different cell types. Our in vivo studies (10) of mRNAs bound by FMRP in neurons show that some proteins are up-regulated, and others are down-regulated in dendritic regions of FMRP–KO brains.
Using an in vitro translation system derived from reticulocytes, Laggerbauer et al. (40) and Li et al. (41) found that the addition of substantial amounts of FMRP represses translation of some proteins. This action is to be expected of an mRNA-binding protein. We did not see an inhibitory effect. Rather, the phenomenon we describe here involves activity-dependent protein synthesis in WT synaptoneurosomes that did not occur in KO preparations, suggesting a pivotal role for FMRP in the control of this phase of translation. The electron microscopy evidence for a lower proportion of PRA-associated dendritic spines in KO mice supports the conclusion that, in the absence of FMRP, synaptically localized translation of some proteins is down-regulated.
It is likely that FMRP is required for the translational response of only a subset of dendritic mRNAs, such as those shown to be associated with FMRP in neurons (10). Both targeting and translational roles would be compatible with FMRP's RNA-binding capacity (14) and its association with polyribosomal complexes (7, 27, 42, 45). Indeed, the glucocorticoid receptor whose mRNA binds to FMRP is differentially localized in subcellular regions of hippocampal CA1 pyramidal neurons in fmr1-KO mice compared with WT mice (10). The strong phenotypic effect of a patient's spontaneous Ile304Asn mutation in one of FMRP's RNA-binding domains has supported the view that RNA binding is crucial to the function of FMRP (46). We suggest that the roles of FMRP are (i) to sequester and prevent the translation of certain mRNAs and (ii) upon receiving the appropriate synaptic signal, to release these mRNAs from sequestration, thus allowing translation (illustrated in detail in Fig. 4). A regulatory role for FMRP may involve conformational changes, as suggested by observation of phosphorylation effects by Ceman et al. (47). We predict that there are, then, specific mRNAs whose translation in certain subcellular regions, such as the synapse, is increased upon demand (e.g., refs. 44 and 48), possibly at the expense of some housekeeping proteins. The mRNAs whose translation is impaired in KO mice may include components of the ribosomal complex (i.e., 60S ribosomal protein L13a, an mRNA-target of FMRP; ref. 10) whose expression, if dysregulated, could contribute to an overall change in neurotransmitter-activated protein synthesis (8).
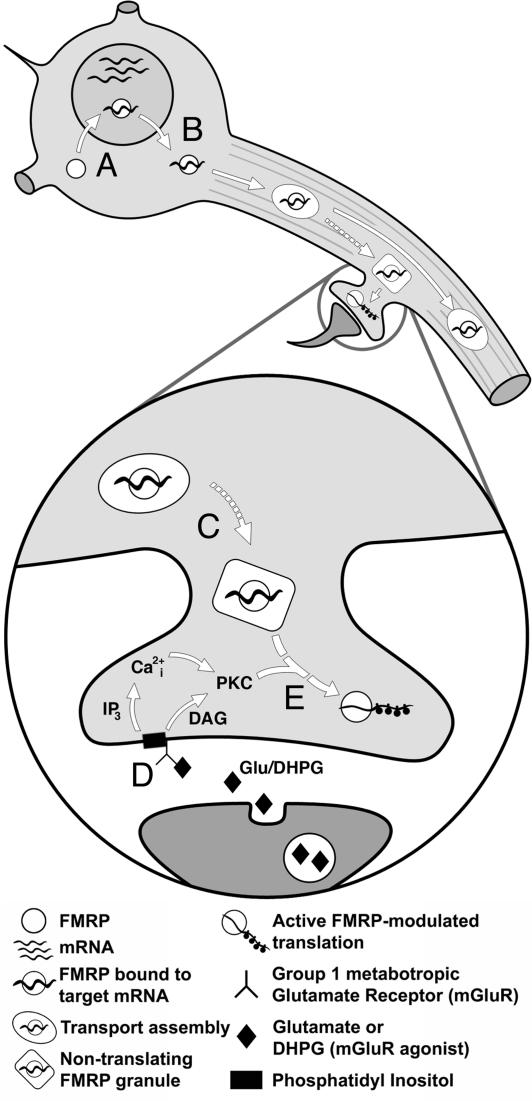
Fig. 4.
A putative role for FMRP in synaptic protein synthesis. (A) FMRP binds to its target mRNAs in the nucleus and helps export them to somatic cytoplasm (7, 42). (B) FMRP and its target mRNA are packaged into transport assemblies and travel, likely by way of microtubules, down dendrites toward synapses (15). (C) FMRP–mRNA transport assemblies may take the form of nontranslating granules near synapses where they await some synaptic signal (43, 44). (D) In response to activation of group 1 mGluRs (stimulated here with DHPG), phosphatidylinositol is cleaved into diacyl glycerol (DAG) and inositol triphosphate (IP3), initiating the release of intracellular calcium (Ca2+i) and activation of PKC (26). (E) PKC activation triggers an enzyme cascade that, by means of many intermediates (broken arrow), signals nontranslating granules to release FMRP-bound target mRNA for translation.
Because FMRP is an mRNA-binding protein, its absence might affect the expression of neurotransmitter receptors at KO mouse synapses. Any deficit in glutamate receptor level would be expected to have marked effects on the ability of the synapse to respond to neurotransmitter stimulation. Group I mGluR activation results in PKC activation along the biochemical pathway to protein synthesis initiation (49). We found no significant differences in PKC levels between KO and WT mice. Direct activation of this signaling pathway by the addition of a PKC stimulator, OAG, should, theoretically, bypass the need for mGluRs to initiate translation. OAG elicited protein synthesis in synaptoneurosomes from WT mice but not in synaptoneurosomes from KO mice. Hence, the FMRP-related defect appears to lie downstream of PKC.
The use of a synaptoneurosome preparation enriched in synapses enables us to obtain a rapid series of samplings from identical, parallel-treated samples. Some investigators (e.g., Steward and Schuman, ref. 50) have expressed concerns regarding the validity of results obtained with synaptoneurosomes. For example, some glial elements are included in the synaptoneurosome preparation because glia are intimately associated with synapses. Although it is not impossible that some of the methionine-incorporating proteins in this preparation are of glial origin, there is so far no evidence suggesting a rapid neurotransmitter-triggered translational increase in astrocytes. Our parallel in vivo finding of reduced postsynaptic polyribosomes also serves to dispel these concerns.
The absence of rapid neurotransmitter-stimulated protein synthesis in the synapse-enriched preparation from fmr1-KO mice and the reduction in synapse-associated polyribosomes in intact KO mice point to a common underlying process: the requirement for FMRP to regulate or enable neurotransmitter-activated dendritic protein synthesis. This finding is particularly relevant in light of recent descriptions of FMRP association with translating polyribosomes in neurons. A deficit in some aspect of localized rapid translation of certain proteins in dendritic processes of neurons might well underlie the deficiency in synaptic maturation observed both in KO mice and in humans affected by the fragile X syndrome.
Acknowledgments
We gratefully acknowledge Karen P. Solis, Roberto Galvez, Deborah Miller, Ann Benefiel, Amy Drew, Louis Lin, Cyrus Press, Claire Henson, and Georgina Aldridge for their assistance in various aspects of this research. We thank Prof. Ben A. Oostra (Erasmus University, Rotterdam) and Prof. Frank Kooy (University of Antwerp, Antwerp, Belgium) for the fmr1-KO mouse strains and Mark Flider (Imaging Technology Group, Beckman Institute) for creating Fig. 4. This work was supported by the Fragile X (FRAXA) Research Foundation, the Illinois–Eastern Iowa District of Kiwanis International Spastic Paralysis Research Foundation, and National Institutes of Health Grants HD37175, MH35321, and RO3HD35565-01.
Author contributions: I.J.W., C.C.S., A.Y.K., A.W.G., S.H.K., and W.T.G. designed research; I.J.W., C.C.S., A.Y.K., A.W.G., S.H.K., V.B.-A., H.K., F.E.d.V., F.A.E.L., F.H., and C.K.B. performed research; A.W.G. analyzed data; and I.J.W., A.W.G., and W.T.G. wrote the paper.
Abbreviations: KO, knockout; mGluR, metabotropic glutamate receptor; PRA, polyribosomal aggregate; OAG, 1-oleoyl-2-acetyl-sn-glycerol; DHPG, (S)3,5-dihydroxyphenylglycine; P-mRNA, polyribosomal mRNA.
See Commentary on page 17329.
References
- 1.Warren, S. T. & Nelson, D. L. (1994) J. Am. Med. Assoc. 271, 536–542. [Google Scholar]
- 2.Hagerman, R. J. & Hagerman, P. J. (2002) Fragile X Syndrome: Diagnosis, Treatment, and Research (Johns Hopkins Univ. Press, Baltimore).
- 3.Hinton, V. J., Brown, W. T., Wisniewski, K. & Rudelli, R. D. (1991) Am. J. Med. Genet. 41, 289–294. [DOI] [PubMed] [Google Scholar]
- 4.Irwin, S. A., Patel, B., Idupulapati, M., Harris, J. B., Crisostomo, R. A., Larsen, B. P., Kooy, F., Willems, P. J., Cras, P., Kozlowski, P. B., et al. (2001) Am. J. Med. Genet. 98, 161–167. [DOI] [PubMed] [Google Scholar]
- 5.Devys, D., Lutz, Y., Rouyer, N., Bellocq, J. P. & Mandel, J. L. (1993) Nat. Genet. 4, 335–340. [DOI] [PubMed] [Google Scholar]
- 6.Siomi, H., Siomi, M. C., Nussbaum, R. L. & Dreyfuss, G. (1993) Cell 74, 291–298. [DOI] [PubMed] [Google Scholar]
- 7.Feng, Y., Gutekunst, C. A., Eberhart, D. E., Yi, H., Warren, S. T. & Hersch, S. M. (1997) J. Neurosci. 17, 1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khandjian, E. W., Huot, M. E., Tremblay, S., Davidovic, L., Mazroui, R. & Bardoni, B. (2004) Proc. Natl. Acad. Sci. USA 101, 13357–13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefani, G., Fraser, C. E., Darnell, J. C. & Darnell, R. B. (2004) J. Neurosci. 24, 9272–9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyashiro, K. Y., Beckel-Mitchener, A., Purk, T. P., Becker, K. G., Barret, T., Liu, L., Carbonetto, S., Weiler, I. J., Greenough, W. T. & Eberwine, J. (2003) Neuron 37, 417–431. [DOI] [PubMed] [Google Scholar]
- 11.Sung, Y. J., Conti, J., Currie, J. R., Brown, W. T. & Denman, R. B. (2000) Biochem. Biophys. Res. Commun. 275, 973–980. [DOI] [PubMed] [Google Scholar]
- 12.Brown, V., Jin, P., Ceman, S., Darnell, J. C., O'Donnell, W. T., Tenenbaum, S. A., Jin, X., Feng, Y., Wilkinson, K. D., Keene, J. D., et al. (2001) Cell 107, 477–487. [DOI] [PubMed] [Google Scholar]
- 13.Schaeffer, C., Bardoni, B., Mandel, J. L., Ehresmann, B., Ehresmann, C. & Moine, H. (2001) EMBO J. 20, 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell, J. C., Jensen, K. B., Jin, P., Brown, V., Warren, S. T. & Darnell, R. B. (2001) Cell 107, 489–499. [DOI] [PubMed] [Google Scholar]
- 15.Antar, L. N., Afroz, R., Dictenberg, J. B., Carroll, R. C. & Bassell, G. J. (2004) J. Neurosci. 24, 2648–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steward, O. & Falk, P. M. (1986) J. Neurosci. 6, 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenough, W. T., Hwang, H. M. & Gorman, C. (1985) Proc. Natl. Acad. Sci. USA 82, 4549–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asaki, C., Usuda, N., Nakazawa, A., Kametani, K. & Suzuki, T. (2003) Brain Res. 972, 168–176. [DOI] [PubMed] [Google Scholar]
- 19.Tiruchinapalli, D. M., Oleynikov, Y., Kelic, S., Shenoy, S. M., Hartley, A., Stanton, P. K., Singer, R. H. & Bassell, G. J. (2003) J. Neurosci. 23, 3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiedge, H. & Brosius, J. (1996) J. Neurosci. 16, 7171–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torre, E. R. & Steward, O. (1992) J. Neurosci. 12, 762–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin, Y., Edelman, G. M. & Vanderklish, P. W. (2002) Proc. Natl. Acad. Sci. USA 99, 2368–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crino, P. B. & Eberwine, J. (1996) Neuron 17, 1173–1187. [DOI] [PubMed] [Google Scholar]
- 24.Job, C. & Eberwine, J. (2001) Proc. Natl. Acad. Sci. USA 98, 13037–13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostroff, L. E., Fiala, J. C., Allwardt, B. & Harris, K. M. (2002) Neuron 35, 535–545. [DOI] [PubMed] [Google Scholar]
- 26.Weiler, I. J. & Greenough, W. T. (1993) Proc. Natl. Acad. Sci. USA 90, 7168–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiler, I. J., Irwin, S. A., Klintsova, A. Y., Spencer, C. M., Brazelton, A. D., Miyashiro, K., Comery, T. A., Patel, B., Eberwine, J. & Greenough, W. T. (1997) Proc. Natl. Acad. Sci. USA 94, 5395–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todd, P. K. & Mack, K. J. (2000) Brain Res. Mol. Brain Res. 80, 17–25. [DOI] [PubMed] [Google Scholar]
- 29.Irwin, S. A., Swain, R. A., Christmon, C. A., Chakravarti, A., Weiler, I. J. & Greenough, W. T. (2000) Neurobiol. Learn. Mem. 73, 87–93. [DOI] [PubMed] [Google Scholar]
- 30.Bakker, C. E., Verheij, C., Willemsen, R., van der Helm, R., Oerlemans, F., Vermey, M., Bygrave, A., Hoogeveen, A. T., Oostra, B. A., Reyniers, E., et al. (1994) Cell 78, 23–33.8033209 [Google Scholar]
- 31.Irwin, S. A., Idupulapati, M., Gilbert, M. E., Harris, J. B., Chakravarti, A. B., Rogers, E. J., Crisostomo, R. A., Larsen, B. P., Mehta, A., Alcantara, C. J., et al. (2002) Am. J. Med. Genet. 111, 140–146. [DOI] [PubMed] [Google Scholar]
- 32.Bowes, C., Li, T., Frankel, W. N., Danciger, M., Coffin, J. M., Applebury, M. L. & Farber, D. B. (1993) Proc. Natl. Acad. Sci. USA 90, 2955–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollingsworth, E. B., McNeal, E. T., Burton, J. L., Williams, R. J., Daly, J. W. & Creveling, C. R. (1985) J. Neurosci. 5, 2240–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klintsova, A. Y., Matthews, J. T., Goodlett, C. R., Napper, R. M. & Greenough, W. T. (1997) Alcohol. Clin. Exp. Res. 21, 1257–1263. [PubMed] [Google Scholar]
- 35.Sterio, D. C. (1984) J. Microsc. 134, 127–136. [DOI] [PubMed] [Google Scholar]
- 36.Gundersen, H. J., Bagger, P., Bendtsen, T. F., Evans, S. M., Korbo, L., Marcussen, N., Moller, A., Nielsen, K., Nyengaard, J. R., Pakkenberg, B., et al. (1988) APMIS 96, 857–881. [DOI] [PubMed] [Google Scholar]
- 37.Greenough, W. T., Klintsova, A. Y., Irwin, S. A., Galvez, R., Bates, K. E. & Weiler, I. J. (2001) Proc. Natl. Acad. Sci. USA 98, 7101–7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Comery, T. A., Harris, J. B., Willems, P. J., Oostra, B. A., Irwin, S. A., Weiler, I. J. & Greenough, W. T. (1997) Proc. Natl. Acad. Sci. USA 94, 5401–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ceman, S., Brown, V. & Warren, S. T. (1999) Mol. Cell. Biol. 19, 7925–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laggerbauer, B., Ostareck, D., Keidel, E. M., Ostareck-Lederer, A. & Fischer, U. (2001) Hum. Mol. Genet. 10, 329–338. [DOI] [PubMed] [Google Scholar]
- 41.Li, Z., Zhang, Y., Ku, L., Wilkinson, K. D., Warren, S. T. & Feng, Y. (2001) Nucleic Acids Res. 29, 2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corbin, F., Bouillon, M., Fortin, A., Morin, S., Rousseau, F. & Khandjian, E. W. (1997) Hum. Mol. Genet. 6, 1465–1472. [DOI] [PubMed] [Google Scholar]
- 43.Mazroui, R., Huot, M. E., Tremblay, S., Filion, C., Labelle, Y. & Khandjian, E. W. (2002) Hum. Mol. Genet. 11, 3007–3017. [DOI] [PubMed] [Google Scholar]
- 44.Krichevsky, A. M. & Kosik, K. S. (2001) Neuron 32, 683–696. [DOI] [PubMed] [Google Scholar]
- 45.Tamanini, F., Meijer, N., Verheij, C., Willems, P. J., Galjaard, H., Oostra, B. A. & Hoogeveen, A. T. (1996) Hum. Mol. Genet. 5, 809–813. [DOI] [PubMed] [Google Scholar]
- 46.De Boulle, K., Verkerk, A. J., Reyniers, E., Vits, L., Hendrickx, J., Van Roy, B., Van den Bos, F., de Graaff, E., Oostra, B. A. & Willems, P. J. (1993) Nat. Genet. 3, 31–35. [DOI] [PubMed] [Google Scholar]
- 47.Ceman, S., O'Donnell, W. T., Reed, M., Patton, S., Pohl, J. & Warren, S. T. (2003) Hum. Mol. Genet. 12, 3295–3305. [DOI] [PubMed] [Google Scholar]
- 48.Huber, K. M., Gallagher, S. M., Warren, S. T. & Bear, M. F. (2002) Proc. Natl. Acad. Sci. USA 99, 7746–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoepp, D., Bockaert, J. & Sladeczek, F. (1990) Trends Pharmacol. Sci. 11, 508–515. [DOI] [PubMed] [Google Scholar]
- 50.Steward, O. & Schuman, E. M. (2001) Annu. Rev. Neurosci. 24, 299–325. [DOI] [PubMed] [Google Scholar]