Abstract
Directed evolution of a cyanobacterial phytochrome was undertaken to elucidate the structural basis of its light sensory activity by remodeling the chemical environment of its linear tetrapyrrole prosthetic group. In addition to identifying a small region of the apoprotein critical for maintaining phytochrome's native spectroscopic properties, our studies revealed a tyrosine-to-histidine mutation that transformed phytochrome into an intensely red fluorescent biliprotein. This tyrosine is conserved in all members of the phytochrome superfamily, implicating direct participation in the primary photoprocess of phytochromes. Fluorescent phytochrome mutants also hold great promise to expand the present repertoire of genetically encoded fluorescent proteins into the near infrared.
Keywords: biliprotein, light signaling, tetrapyrrole, photoisomerization, fluorescence
The spectroscopic and photophysical properties of linear tetrapyrroles (bilins) are profoundly influenced by their chemical environment (1). In aqueous solution, bilins adopt cyclic porphyrin-like conformations, which strongly absorb light in the near-UV region and mainly dissipate absorbed light energy by radiationless processes. Upon association with proteins such as the phycobiliprotein antennae of blue-green, red, and cryptomonad algae (2) or the phytochrome photoreceptors of plants, cyanobacteria, and many nonphotosynthetic microorganisms (3–5), bilins assume more extended conformations that significantly increase their visible light absorption and alter the pathways used for light deexcitation. Adapted for efficient transfer of excitation energy to membrane-bound photosynthetic reaction centers, phycobiliproteins are intensely fluorescent, with fluorescence quantum yields (ΦF) near unity (6). Phytochromes, by contrast, are poorly fluorescent biliproteins with ΦF < 0.005 because of efficient Z-to-E double bond isomerization of their bilin prosthetic group (7). This reversible photoisomerization process, which triggers the interconversion between red- and far-red-light absorbing Pr and Pfr species of phytochrome (Fig. 1A), initiates transcriptional signaling cascades that regulate plant growth and development (8, 9).
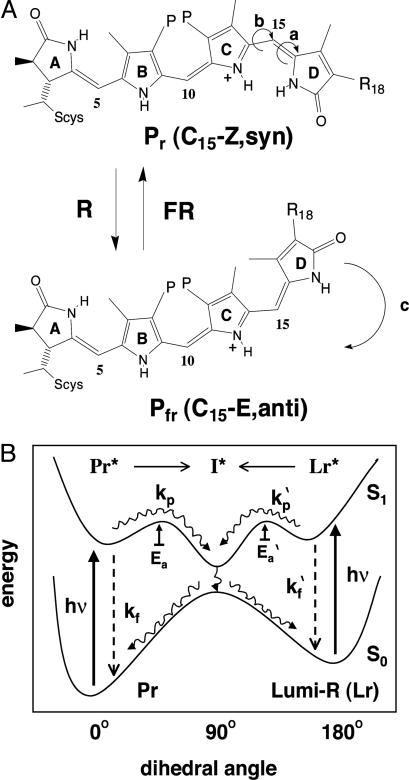
Fig. 1.
Structure and photointerconversions of the phytochrome chromophore. (A) The proposed structure of the Pr and Pfr chromophores of plant (R18, vinyl; P, propionate) and cyanobacterial (R18, ethyl; P, propionate) phytochromes are based on vibrational spectroscopy and semiempirical vibrational energy calculations (35, 36). The Pr to Pfr conversion is thought to involve large motions: a Z-to-E configurational isomerization of the C15 double bond (a) followed by rotation around the C14–C15 bond (b), whereas the reverse reaction appears to be a concerted configurational and conformational inversion (c). (B) Ground and excited state potential energies of the Pr to Lumi-R photointerconversion as a function of the C15 dihedral angle are based on the trans-to-cis photoisomerization processes of stilbene (37). In phytochromes, light excitation energy (hv) is mainly dissipated on the picosecond time scale by photoproduct formation (kp) involving the formation of a twisted C15 double bond, electronically excited intermediate (I*) that is gated by a thermal activation energy barrier (Ea) (7). This excited state intermediate decays radiationlessly into Pr (C15Z-isomer) and Lumi-R (C15E-isomer) in an ≈5:1 ratio. Lumi-R thermally converts to Pfr via a series of spectroscopically distinct intermediates in which the syn-to-anti conformational change occurs (data not shown). Lumi-R to Pr photoconversion is envisaged to proceed through the same intermediate (I*). It is proposed that the PR-1 Y176H mutation identified in this work leads to a larger thermal activation energy barrier (Ea), thereby greatly decreasing the rate of photoproduct formation (kp) and leading to enhanced fluorescence (kf).
The forward and reverse photoconversion pathways for plant phytochromes have been extensively analyzed spectroscopically, revealing that the two processes proceed by means of different intermediates (7, 10). 15Z/15E photoisomerization is the primary photoreaction that follows Pr excitation, a process that yields the Lumi-R intermediate within 5–30 ps (11, 12). The inverse temperature dependence of Pr fluorescence and Lumi-R formation indicates that a small activation barrier for excited state decay by photoproduct formation is responsible for the low fluorescence quantum yield of phytochromes (7). Together with evidence indicating that the photoconversion of Lumi-R to Pr proceeds through the same twisted C15 double bond excited state “intermediate,” a schematic representation of phytochrome's forward photoisomerization reaction coordinate has been formalized (Fig. 1B). This scheme fits the photochemical data for both plant phytochromes and the more recently discovered cyanobacterial phytochrome 1 (Cph1) (7, 13). The local protein environment of the bilin chromophore of phytochromes thus appears to have been preserved throughout billions of years of evolution.
The molecular basis of bilin photochemistry and spectral tuning by the phytochrome apoprotein has been the subject of extensive investigation. Approaches used to address this question include deletion/site-directed mutagenesis, comparative analysis of phytochromes from different organisms, and incorporation of unnatural bilin precursors into recombinant apophytochromes (reviewed in ref. 14). Such studies have established that the N-terminal “photosensory-input” domains of phytochromes are responsible for both photochemistry and spectral tuning of their bilin prosthetic groups, whereas the more variable C-terminal “regulatory-output” domains are completely dispensable. The highly conserved GAF and PHY photosensory domains of plant and cyanobacterial phytochromes appear essential to maintaining phytochrome's characteristic spectroscopic and photochemical properties (15, 16). The PAS-related photosensory domain and N-terminal serine-rich extension of plant phytochromes also play spectral tuning roles (4, 17). However, the absence of the PAS domain in some cyanobacterial phytochromes suggests that this domain does not contribute residues that directly participate in the primary photoisomerization process (4). Taken together, these studies indicate that the GAF-PHY domain pair plays a central role in bilin photochemistry and spectral tuning by all phytochromes.
The present study was undertaken to develop a global, high-throughput approach to identify specific residues within a phytochrome apoprotein responsible for spectral tuning and photochemical activity by using the technique of directed evolution. By exploiting a two-plasmid bacterial expression system that yields a photoactive deletion mutant of cyanobacterial phytochrome 1 (Cph1Δ) in living cells, the spectroscopic consequences of random mutations introduced into the cph1 gene were assessed in situ. Our studies identified a mutational “hot spot” within the PHY domain that is critical for maintaining native bilin–apoprotein interactions. In addition, we isolated an intensely red fluorescent Cph1Δ mutant whose primary photochemistry is severely impaired because of mutation of a conserved tyrosine residue found in the GAF domain. Owing to the conserved domain architecture of plant and cyanobacterial phytochromes (4), our results implicate participation of the region/residues identified by mutagenesis in the molecular mechanism of light signaling by all phytochromes.
Materials and Methods
Construction of the Initial PHY Subdomain Mutant Library. An errorprone PCR library of the PHY subdomain of Synechocystis sp PCC 6803 Cph1Δ (Fig. 2A) was generated by using the Diversify PCR Random Mutagenesis kit (Clontech) with the pBAD-Cph1Δ plasmid template (18), sense primer 5′-CTTCGATTACCGGGTGCAGCTG-3′, and antisense primer 5′-GCAAGCTTGTTCTTCTGCCTGGCG-3′. Six variations of MnSO4 and dGTP final concentrations were used: 0 and 40 μM, 320 and 40 μM, 480 and 40 μM, 640 and 80 μM, 640 and 120 μM, and 640 and 200 μM, respectively. PCR products from all six mutagenesis reactions were pooled and purified by using QiaQuick PCR columns (Qiagen, Valencia, CA). After restriction enzyme digestion with PvuII and HindIII, the purified DNA fragments were ligated with the PvuII- and HindIII-restricted pBAD-Cph1Δ plasmid from which the PHY domain had been removed. A library of ≈2,000 clones was generated.
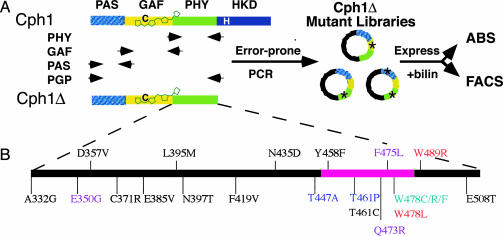
Fig. 2.
Directed evolution of the photosensory domains of Cph1Δ.(A) Phytochrome Cph1 consists of the three related PAS, GAF, and PHY photosensory domains terminated with the histidine kinase domain (HKD) module. The truncated Cph1Δ photosensory construct lacks the HKD domain. The conserved cysteine site of bilin attachment at position 259 is shown in black. Four mutagenesis libraries were generated from either single domains or a combination of the entire photosensory domain (PGP). These libraries were coexpressed in E. coli with a bilin biosynthetic plasmid for the production of holoCph1Δ mutants in vivo. Holoproteins were analyzed for alterations in absorbance (ABS) or fluorescence (FACS). (B) The position and residue change of the 21 mutants having single amino acid substitutions identified from the PHY library absorbance screen. The alleles are colored with respect to phenotypic class: black, WT; purple, mutant class I; red, mutant class II; blue, mutant class III; teal, mutant class IV. The pink bar designates the conserved HisG insert that is present in the PHY domain of all know phytochromes.
Construction of Additional Mutant Libraries and Site-Directed Mutants. The GeneMorph PCR Mutagenesis kit (Stratagene) was used to generate four additional Cph1Δ mutant libraries comprising individual PAS, GAF, or PHY subdomains or all three together, i.e., the PGP library (Fig. 2 A). The following variations in template DNA amount were used: 1 pg, 10 pg, 100 pg, and 2 ng. PCR cycling conditions were 95°C for 1 min; 95°C for 1 min, 55°C for 1 min, 72°C for 2.5 min. (30–40 cycles); and 72°C for 10 min. The PAS subdomain was amplified by using the sense primer 5′-GGGCTAACAGGAGGAATTAACCATG-3′ and the antisense primer 5′-CGGAAGTGTAGGCTGGCTCG-3′. The GAF domain was amplified by using the sense primer 5′-GACGGTTTATGGTATGTGAACTCG-3′ and the antisense primer 5′-CAATAAAACCGCTTCATGCTCCGCCAG-3′. The PHY domain was amplified with the primers described above for the PHY subdomain mutagenesis. The PGP library was constructed by amplifying the entire Cph1Δ gene with the PAS domain sense primer and the PHY domain antisense primer. The PCR products generated from the different mutagenesis reactions were gel-purified by using QIAQuick PCR columns (Qiagen), restricted, and ligated with the gel-purified pBAD-Cph1Δ fragment restricted in the same way. The estimated number of clones per library is as follows: PAS, 7,500; GAF, 1,000; PHY no. 2, 6,000; and PGP, 4,500. For some studies, the QuikChange Site-Directed Mutagenesis kit (Stratagene) was used to generate site-specific mutations in Cph1Δ. The mutagenesis reactions were carried out by using 10 ng of dsDNA template. The DNA sequences for all mutants studied were determined by Davis Sequencing (Davis, CA).
Recombinant Protein Expression and Spectroscopic Characterization. All mutants were expressed in phycocyanobilin (PCB)-producing, Cph1Δ-expressing cells and purified as described (18). Absorption and phytochrome difference spectra were obtained by using an HP8453 UV-visible spectrophotometer (18). Red (650 ± 5 nm) and far-red (720 ± 5 nm) light with fluence rates of 150 μmol·m–2·s–1 were used for measuring phytochrome photointerconversion. Corrected fluorescence excitation and emission spectra were obtained with an SLM Aminco Bowman AB2 fluorimeter with both monochromators adjusted to 4 nm bandpass. Quantum yield measurements were determined with a ratiometric method using purified cyanine dye (CY-5.18) as a standard (19, 20). Corrected fluorescence emission profiles (λex = 600 ± 4 nm) for various dilutions of phytofluor red-1 (PR-1) and CY-5 were integrated and plotted as a function of the absorbance at 600 nm below 0.05. The ratio of the slopes of PR-1 and CY-5 plots was used to determine the quantum yield of PR-1 based on the known quantum yield of CY-5.18, i.e., Φf = 0.27 in water (L. A. Ernst, personal communication).
Flow Cytometry. A Cytomation MoFlo high-speed cell sorting flow cytometer at the University of California, Davis, Optical Biology Laboratory was used to screen for fluorescent Cph1Δ mutants expressed in Escherichia coli cells. Fluorescence excitation was performed by using the 647-nm laser line of a tunable argon/krypton mixed gas laser. A HQ660lp long-pass filter (Chroma Technology, Rockingham, VT) was used to detect fluorescence at wavelengths longer than 660 nm. The mutant libraries were induced for holophytochrome expression before sorting (18). Cells were sorted with the pressure differential set to 1.0 at a rate of ≈5,000 events per second. For a direct comparison of the fluorescence properties of WT Cph1Δ and PR-1 in living cells, a Becton Dickinson FACSCalibur flow cytometer was used with the 635-nm red diode laser and the FL4 allophycocyanin emission window (650–670 nm).
Colony Fluorimaging. Small-scale holophytochrome expressions of WT Cph1Δ, the PR-1 mutant, or PR1-derived single or double mutants were carried out as described (18). Cell cultures were spotted onto a sterile 47-mm polyester 0.2-μm membrane (GE Osmonics, Trevose, PA) and placed on RM media containing 50 μg/ml ampicillin and 25 μg/ml kanamcyin. Membranes were incubated overnight at 30°C, transferred to induction media (RM containing 50 μg/ml ampicillin, 25 μg/ml kanamcyin, 1 mM IPTG, and 0.004% wt/vol l-arabinose) and incubated overnight at 30°C. After incubation at 37°C for 5 h, membranes were transferred to 1% agarose–PBS plates for imaging. Photographs were taken under UV light (365 nm) by using a Tiffen 67-mm yellow 12 UV-cutoff filter.
Results and Discussion
Cph1 Mutants with Altered Absorbance Properties Cluster to a Small Region of the PHY Subdomain. The truncated Cph1 monomer Cph1Δ, which lacks the regulatory histidine kinase “output” domain of full-length Cph1 (Fig. 2 A), was chosen for mutagenesis because it retains the ability to bind the native chromophore precursor PCB and yields a spectroscopically native holophytochrome (18). Because the PHY domain stabilizes both the extended configuration of the bilin prosthetic group and the FR absorbing form of phytochrome (15, 16), we initially analyzed a Cph1Δ library in which the PHY domain was mutagenized by error-prone PCR. Ninety-one clones were selected at random for characterization by DNA sequencing. The Cph1Δ clones containing amino acid substitutions (58 total) were expressed and purified as holoproteins for spectrophotometric analyses. Many of these mutants possessed Cph1Δ WT spectra (25 total) despite multiple mutations. Those with altered spectra (33 total) fell into four phenotypic classes, none of them showing enhanced fluorescence (Fig. 6, which is published as supporting information on the PNAS web site). All of the mutants displayed a decreased ratio of their red to near-UV absorption maxima, indicating a less extended chromophore conformation. Class I mutants, representing 51% of the mutants identified, exhibited 7- to 13-nm blue shifts in both the Pr and Pfr absorption maxima. Class II mutants (24%) displayed spectral shifts similar to class I, with the exception of a significant reduction in the amount of Pfr formed at photoequilibrium, a phenotype that is quite similar to phytochromes that lack the PHY domain entirely (15, 16). The final two classes displayed either blue-shifted Pr spectra, i.e., class III (12%), or blue-shifted Pfr spectra and a reduction in the amount of Pfr formed at photoequilibrium, i.e., class IV (12%), and both exhibited an increase in nonphotochemical dark reversion. Class IV mutants showed the most extreme decrease in the red to near-UV absorption maxima ratio (Fig. 6).
Because most of the spectrally shifted Cph1Δ mutants contained multiple amino acid changes (Fig. 7, which is published as supporting information on the PNAS web site), individual point mutations were introduced into WT to ascribe the spectral shifts to specific amino acid substitutions. This process was simplified by the observation that many of the same residues mutated in other Cph1Δ mutant clones had no effect on the spectrum (see Fig. 7, WT mutants). Single mutants responsible for the observed spectral changes were identified after their reintroduction into a WT template. The spectrally shifted mutations clustered in a small region of the PHY domain of Cph1Δ, between residues 440 and 490 (Fig. 2B). This mutation hot spot is of particular interest because it is related to a sequence found in the HisG family of ATP phosphoribosyltransferases, a sequence that appears to have inserted into the GAF-related PHY domains of all known phytochromes (4). A number of mutations that alter plant phytochrome function also fall within this region, i.e., phyA-112 (21), phyB-401 (22), and phyB-GFP-4 (23). Taken together, these results implicate a key role for the HisG insert of the PHY domain in phytochrome signaling (i.e., the stabilization of the active Pfr state of the photoreceptor). Saturation mutagenesis of this region should prove useful in understanding how the biochemical role of this region relates to the basic structure and function of phytochromes.
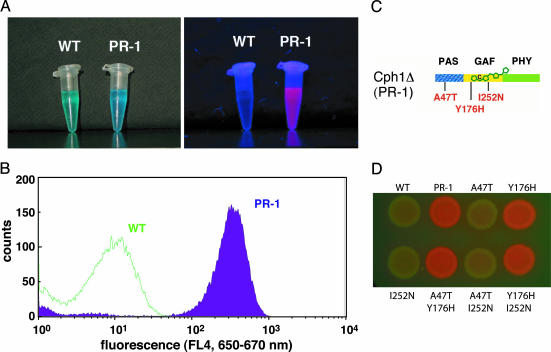
Identification of a Red Fluorescent Cph1 Mutant, PR-1. The lack of fluorescent mutants recovered from the PHY mutant library suggested that such gain-of-function mutations might be rare, possibly requiring multiple amino acid changes throughout the photosensory domain. Therefore, we screened new mutant libraries encompassing all three photosensory domains of Cph1Δ by using the higher throughput screening method of fluorescence-activated cell sorting (FACS). Four Cph1Δ libraries with mutations in the PAS, GAF, and/or PHY domains (Fig. 2 A) were introduced into a bacterial cell line engineered to express genes encoding two enzymes for the conversion of endogenous heme to PCB (18). After in vivo assembly of holoCph1Δ, fluorescent mutants were selected by FACS using red (647 nm) excitation and a 660-nm long-pass emission window. A total of 4.9 × 107 cells were sorted from the four Cph1Δ mutant libraries, from which we isolated 18 colonies for further analysis. Five of these cell lines produced holophytochrome, as determined by their deep blue color, only one of which was strongly fluorescent (Fig. 3A). This clone was named PR-1 by using the nomenclature proposed in an earlier study (20). Comparative flow cytometry showed that PR-1 expressing cells were resolved from the WT parental cell line by over two orders of fluorescence magnitude, thereby confirming the validity of the FACS screen (Fig. 3B). Plasmid DNA isolated from PR-1-expressing cells revealed three base pair mutations that resulted in three amino acid changes, i.e., A47T, Y176H, and I252N (Fig. 3C). To determine whether all three mutations were necessary for PR-1's red fluorescence, all single mutants and double mutant combinations were constructed by site-directed mutagenesis, and their corresponding holoproteins were expressed in PCB-producing bacterial cell lines. Fluorescence colony imaging revealed that the Y176H mutation was both necessary and sufficient for the enhanced red fluorescence of PR-1 (Fig. 3D). Optical measurements showed that all nonfluorescent mutant cell lines possessed photoactive holophytochrome, indicating that their lack of fluorescence was not caused by poor expression and/or bilin attachment (data not shown). Taken together, these results showed that the single Y176H mutation was sufficient to convert WT into an intensely red fluorescent biliprotein.
Fig. 3.
Properties of the red fluorescent phytochrome mutant PR-1. (A) Purified WT Cph1Δ and PR-1 PCB adducts imaged in white (Left) or UV(Right) light. (B) Comparative flow cytometry analysis of PR-1 and WT Cph1Δ in PCB-producing cell lines by using the 635-nm red diode laser as an excitation source. (C) The three amino acid substitutions identified in the original PR-1 mutant are shown in red. (D) Colony fluoroimaging assay of WT Cph1Δ, PR-1 and the six possible single and double amino acid substitution combinations as PCB adducts produced in E. coli. The membrane was imaged under UV light to observe fluorescence.
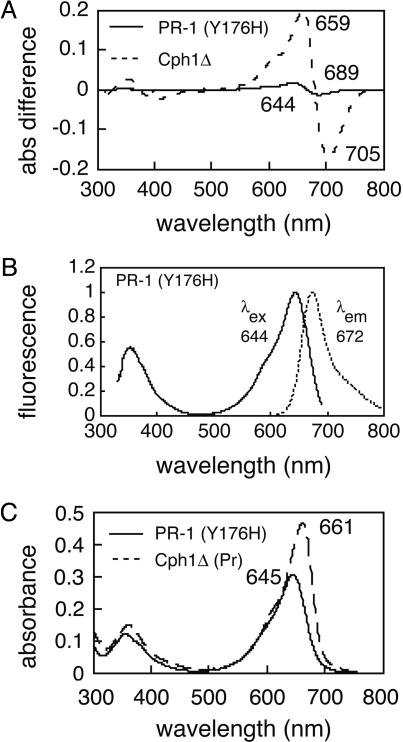
PR-1 Is Poorly Photoactive and Intensely Fluorescent. Both the parental PR-1 and the PR-1 (Y176H) single mutant were expressed in bacteria engineered to produce PCB, and the corresponding holoproteins were affinity-purified for biochemical and spectroscopic comparison with WT Cph1Δ. Because the properties of both proteins were qualitatively similar, only data for the PR-1 (Y176H) single mutant will be presented. SDS/PAGE and zinc-blot analyses established that the purified holoprotein was >95% pure and that a bilin chromophore was covalently attached (Fig. 8, which is published as supporting information on the PNAS web site). As expected, affinity-purified PR-1 was poorly photoactive, exhibiting greatly reduced phytochrome difference spectra <5% that of WT (Fig. 4A). This decreased photoconversion efficiency correlated with PR-1's enhanced fluorescence with an emission maximum at 672 nm (Fig. 4B). By using CY-5.18 dye as a quantum yield standard, we estimated a fluorescence quantum yield to be >0.1 for the PCB adduct of PR-1 (Y176H) at 25°C (data not shown). PR-1 (Y176H) also exhibited a single fluorescence lifetime of 1.8 ns (data not shown). Both values were similar to those reported previously for the nonphotoactive, orange-fluorescent phycoerythrobilin adduct of Cph1Δ (20). Fig. 4C shows that the absorption spectra of PR-1 is similar to that of the Pr form of WT Cph1Δ except that its long wavelength absorption maxima is slightly smaller, blue-shifted, and broadened. We estimate the molar absorption coefficient of the PCB adduct of PR-1 at 648 nm to be 73,000 M–1·cm–1 based on that of WT Cph1 (24). Altogether, the absorption spectra data indicates that the bilin chromophore of PR-1 and WT Cph1Δ have similar conformations.
Fig. 4.
Spectroscopic properties of the PCB adducts of WT Cph1Δ and PR-1 (Y176H). (A) Phytochrome difference absorption spectra for WT Cph1Δ (dashed line) and PR-1 Y176H single mutant (solid line). (B) Corrected fluorescence excitation (solid line) and emission (dotted line) spectra for the PR-1 Y176H single mutant. (C) Pr absorption spectra for WT Cph1Δ (dashed line) and PR-1 Y176H single mutant (solid line).
Structural Implications of the Spectral Tuning Mutations. The discovery that a single Y176H amino acid substitution can transform a phytochrome into a strongly red-emitting phytofluor is of great significance with regard to phytochrome structure and function. This result strongly implicates a direct role for this tyrosine residue in the primary Z-to-E photoisomerization reaction of phytochrome's bilin prosthetic group because it is found in GAF domains of all known phytochromes, and introduction of the Y176H mutation into a plant phytochrome also yields a red fluorescent holoprotein (data not shown). The enhanced fluorescence of the PR-1 mutant shows that the Y176H substitution strongly inhibits the rate of photoproduct formation. We hypothesize that this effect is caused by an increased activation energy barrier (Ea) for Pr* excited state decay via the twisted C15 double bond transition state “intermediate” (Fig. 1B). Although this could be accomplished by altering the C15 double bond planarity of either the Pr ground/excited state and/or the transition state “intermediate,” we prefer the hypothesis that Y176H substitution increases the energy barrier for rotation of the aromatic side-chain that must occur to enable the bilin D-ring to rotate into the “twisted” C15 double bond transition state. Therefore, we propose that Y176 acts as a molecular gate for this movement via phenol ring rotation, a process that would be significantly inhibited for a more highly H-bonded imidazole moiety of histidine (Fig. 5). Although it is possible that Y176 performs a structural role distant from the bilin chromophore itself, we favor a direct role because this conserved residue occurs within the β1-strand of the GAF domain that occupies a central position within its PAS-like fold (25, 26). Indeed, both GAF and PAS protein domains have been shown to comprise the location of planar aromatic ligand binding sites; residues within the β1 strands of both families have been shown to participate in ligand binding (25–27). By contrast with the direct role of Y176, an indirect role of the PHY domain in spectral tuning is more likely. Based on our absorption screen results, we envisage that the HisG insert of the PHY domain allosterically communicates with the bilin prothestic group. This communication could be accomplished in a manner similar to that proposed for phototropin signaling, in which light activation of its PAS domain bound flavin prosthetic group bound alters the association of a C-terminal “transducing” α-helix (28). Although phototropin photochemistry still occurs when this helix is removed, its spectral properties are altered (28, 29). Until a crystal structure for a phytochrome photosensory domain is solved, our proposed photochemical models must be considered speculative, albeit useful, starting points for further experimental studies. Introduction of the corresponding mutations identified in the present studies into other phytochromes should provide insight into the similarities as well as differences in light perception and molecular signaling by the extended phytochrome family of biliprotein photoreceptors.
Fig. 5.
Tyrosine gating hypothesis for Pr excited state decay. Tyrosine 176 is proposed to act as a molecule gate to photoisomerization of the C15 double bond. Upon photoexcitation to Pr*, simultaneous torsional rotation of the C15 double bond (a) and the tyrosine phenol ring (a′) must occur to generate the proposed excited state intermediate (I*). This intermediate vibrationally relaxes to ground state Lumi-R (Lr) or Pr via radiationless pathways labeled b and c, respectively. The activation barrier for Pr* to I* interconversion, i.e., Ea (WT), is proposed to be greater for the Y176H mutant because of H-bonding of the histidine imidazole side chain that would inhibit C15 photoisomerization (shown at Right). The increase in Ea leads to an increased lifetime and enhanced fluorescence from Pr*.
Applications of Red and Near-Infrared Fluorescent Phytochromes. Phytofluors, like members of the green fluorescent protein family (30–32), are genetically encoded fluorescent proteins that can be used as molecule probes within living cells (20). Unlike green fluorescent protein, two components are needed to produce a phytofluor: a phytochrome-derived apoprotein and a bilin that combine to produce a fluorescent biliprotein in situ. Therefore, phytofluors are more analogous to fluorescent antibody and RNA aptamer technologies that require a ligand whose fluorescence depends on macromolecule binding (33, 34). Through directed evolution, we have now extended the fluorescence emission range of phytofluors into the red region of the light spectrum. By using recently discovered phytochrome genes and additional rounds of mutagenesis, smaller phytofluors with emission extending into the nearinfrared are now within grasp. Moreover, exchange of the PCB chromophore of PR-1 with the more extended plant phytochrome chromophore precursor phytochromobilin leads to a red shift of the fluorescence emission maximum by >15 nm (unpublished data), indicating that alterations in the bilin component can be used to tune the fluorescence properties. Despite the emphasis on the enhanced fluorescence of the PR-1 mutant, we have observed that PR-1-derived phytofluors retain partial photochemical activity. This latent photochemistry offers the ability to study phytochrome function and dynamics in living cells by introducing the corresponding Y176H phytochrome mutant into a phy mutant background. Indeed, phytochrome-derived probes that are both photoregulatory and fluorescent clearly have a glowing future.
Supplementary Material
Acknowledgments
We thank Lauren A. Ernst (Carnegie Mellon University, Pittsburgh) for the gift of purified CY-5.18, for measurement of corrected fluorescence spectra, and for helpful discussions about quantum yield measurements; Abigail Yap for construction of site-directed PR-1 mutants; Nicole Baumgarth and Donna Lagarias for experimental design for the FACS mutant library and analytical flow cytometry; and Abigail Miller for fluorescence lifetime measurements. This work was supported in part by National Institutes of Health Grant GM068552-01, National Science Foundation Center for Biophotonics Science and Technology Grant PHY-0120999, and the University of California, Davis, Biotechnology Training Program.
Author contributions: A.J.F. and J.C.L. designed research; A.J.F. performed research; A.J.F. and J.C.L. analyzed data; and A.J.F. and J.C.L. wrote the paper.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2001.
Abbreviations: Cph1, cyanobacterial phytochrome 1; PCB, phycocyanobilin; PR-1, phytofluor red-1; FACS, fluorescence-activated cell sorting.
See accompanying Biography on page 17331.
References
- 1.Falk, H. (1989) The Chemistry of Linear Oligopyrroles and Bile Pigments (Springer, Vienna).
- 2.Glazer, A. N. (1988) Methods Enzymol. 167, 291–303. [DOI] [PubMed] [Google Scholar]
- 3.Smith, H. (2000) Nature 407, 585–591. [DOI] [PubMed] [Google Scholar]
- 4.Montgomery, B. L. & Lagarias, J. C. (2002) Trends Plant Sci. 7, 357–366. [DOI] [PubMed] [Google Scholar]
- 5.Vierstra, R. D. & Davis, S. J. (2000) Semin. Cell Dev. Biol. 11, 511–521. [DOI] [PubMed] [Google Scholar]
- 6.Glazer, A. N. (1999) in Chemicals from Microalgae, ed. Cohen, Z. (Taylor & Francis, London), pp. 261–280.
- 7.Sineshchekov, V. A. (1995) Biochim. Biophys. Acta Bioenerg. 1228, 125–164. [Google Scholar]
- 8.Quail, P. H. (2002) Nat. Rev. Mol. Cell Biol. 3, 85–93. [DOI] [PubMed] [Google Scholar]
- 9.Nagy, F. & Schafer, E. (2002) Annu. Rev. Plant Biol. 53, 329–355. [DOI] [PubMed] [Google Scholar]
- 10.Braslavsky, S. E. (2003) in Photochromism: Molecules and Systems, eds. H., D. & BouasLaurent, H. (Elsevier Science, Amsterdam), pp. 738–755.
- 11.Andel, F., Hasson, K. C., Gai, F., Anfinrud, P. A. & Mathies, R. A. (1997) Biospectroscopy 3, 421–433. [Google Scholar]
- 12.Bischoff, M., Hermann, G., Rentsch, S. & Strehlow, D. (2001) Biochemistry 40, 181–186. [DOI] [PubMed] [Google Scholar]
- 13.Sineshchekov, V., Koppel, L., Esteban, B., Hughes, J. & Lamparter, T. (2002) J. Photochem. Photobiol. B 67, 39–50. [DOI] [PubMed] [Google Scholar]
- 14.Gartner, W. & Braslavsky, S. E. (2004) in Photoreceptors and Light Signalling, ed. Batschauer, A. (Royal Soc. Chem., Cambridge, U.K.), Vol. 3, pp. 136–180. [Google Scholar]
- 15.Wu, S. H. & Lagarias, J. C. (2000) Biochemistry 39, 13487–13495. [DOI] [PubMed] [Google Scholar]
- 16.Oka, Y., Matsushita, T., Mochizuki, N., Suzuki, T., Tokutomi, S. & Nagatani, A. (2004) Plant Cell 16, 2104–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park, C. M., Bhoo, S. H. & Song, P. S. (2000) Semin. Cell Dev. Biol. 11, 449–456. [DOI] [PubMed] [Google Scholar]
- 18.Gambetta, G. A. & Lagarias, J. C. (2001) Proc. Natl. Acad. Sci. USA 98, 10566–10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Southwick, P. L., Ernst, L. A., Tauriello, E. W., Parker, S. R., Mujumdar, R. B., Mujumdar, S. R., Clever, H. A. & Waggoner, A. S. (1990) Cytometry 11, 418–430. [DOI] [PubMed] [Google Scholar]
- 20.Murphy, J. T. & Lagarias, J. C. (1997) Curr. Biol. 7, 870–876. [DOI] [PubMed] [Google Scholar]
- 21.Maloof, J. N., Borevitz, J. O., Dabi, T., Lutes, J., Nehring, R. B., Redfern, J. L., Trainer, G. T., Wilson, J. M., Asami, T., Berry, C. C., et al. (2001) Nat. Genet. 29, 441–446. [DOI] [PubMed] [Google Scholar]
- 22.Kretsch, T., Poppe, C. & Schafer, E. (2000) Plant J. 22, 177–186. [DOI] [PubMed] [Google Scholar]
- 23.Chen, M., Schwabb, R. & Chory, J. (2003) Proc. Natl. Acad. Sci. USA 100, 14493–14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamparter, T., Esteban, B. & Hughes, J. (2001) Eur. J. Biochem. 268, 4720–4730. [DOI] [PubMed] [Google Scholar]
- 25.Ho, Y. S. J., Burden, L. M. & Hurley, J. H. (2000) EMBO J. 19, 5288–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez, S. E., Wu, A. Y., Glavas, N. A., Tang, X. B., Turley, S., Hol, W. G. J. & Beavo, J. A. (2002) Proc. Natl. Acad. Sci. USA 99, 13260–13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor, B. L. & Zhulin, I. B. (1999) Microbiol. Mol. Biol. Rev. 63, 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harper, S. M., Neil, L. C. & Gardner, K. H. (2003) Science 301, 1541–1544. [DOI] [PubMed] [Google Scholar]
- 29.Crosson, S. & Moffat, K. (2002) Plant Cell 14, 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsien, R. Y. (1998) Annu. Rev. Biochem. 67, 509–544. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. (2002) Nat. Rev. Mol. Cell Biol. 3, 906–918. [DOI] [PubMed] [Google Scholar]
- 32.Matz, M. V., Lukyanov, K. A. & Lukyanov, S. A. (2002) BioEssays 24, 952–959. [DOI] [PubMed] [Google Scholar]
- 33.Simeonov, A., Matsushita, M., Juban, E. A., Thompson, E. H., Hoffman, T. Z., Beuscher, A. E. T., Taylor, M. J., Wirsching, P., Rettig, W., McCusker, J. K., et al. (2000) Science 290, 307–313. [DOI] [PubMed] [Google Scholar]
- 34.Stojanovic, M. N. & Kolpashchikov, D. M. (2004) J. Am. Chem. Soc. 126, 9266–9270. [DOI] [PubMed] [Google Scholar]
- 35.Remberg, A., Lindner, I., Lamparter, T., Hughes, J., Kneip, C., Hildebrandt, P., Braslavsky, S. E., Gartner, W. & Schaffner, K. (1997) Biochemistry 36, 13389–13395. [DOI] [PubMed] [Google Scholar]
- 36.Andel, F., Murphy, J. T., Haas, J. A., McDowell, M. T., van der Hoef, I., Lugtenburg, J., Lagarias, J. C. & Mathies, R. A. (2000) Biochemistry 39, 2667–2676. [DOI] [PubMed] [Google Scholar]
- 37.Waldeck, D. H. (1991) Chem. Rev. 91, 415–436. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.