Abstract
Transport and translation of mRNA are tightly coupled to ensure strict temporal and spatial expression of nascent proteins. Fragile X mental retardation protein (FMRP) has been shown to be involved in translational regulation and is found in ribonucleoprotein (RNP) granules that travel along dendrites of neurons. In this study, GFP-tagged Drosophila homologue of FMRP (dFMR) was used to visualize RNP granule movement in Drosophila S2 cells. GFP-dFMR form granules that contain both endogenous dFMR and mRNA. Live fluorescence microscopy revealed that dFMR-containing RNP granules move bidirectionally in thin processes formed by S2 cells in the presence of cytochalasin D. Knocking down the heavy chains of either kinesin-1 (kinesin heavy chain) or cytoplasmic dynein (dynein heavy chain) by RNA interference blocks the movement of the dFMR granules. In contrast, knockdown of kinesin light chain (KLC), which is typically necessary for movement of membrane organelles by kinesin-1, had no effect on the dFMR granule translocation. In immunoprecipitation assays, dFMR associates with both kinesin heavy chain and dynein heavy chain, but not KLC. Based on these findings, we conclude that dFMR-containing RNP granules are moved by both kinesin-1 and cytoplasmic dynein and that KLC is not essential and is likely missing from RNP-transporting kinesin-1.
Keywords: microtubules, molecular motors, cytoskeleton, RNA transport
RNA transport and localized protein synthesis indicate spatial and temporal regulation of protein expression. The biological functions of RNA localization range from establishing cell polarity to regulating synaptic plasticity (1–3). RNA localization may be achieved by various mechanisms such as diffusion, which relies on specific entrapment and anchoring, active transport that sorts and targets mRNAs to their destination, or a combination of both processes (4–6). Several different model systems have been used for dissecting RNA transport. In Drosophila oocytes, microtubules show gross polarity with the plus-end at the posterior and the minus-end at the anterior of the oocytes (7, 8). It has been shown that localization of oskar mRNA to the posterior pole is mediated by kinesin-1 (9, 10), whereas anterior-dorsal localization of gurken mRNA and anterior localization of bicoid mRNA is mediated by dynein (8, 11). However, how RNA attaches to the molecular motors remains elusive. More importantly, the identity of molecular motors involved in active transport of mRNA in somatic cells remains under scrutiny.
One example of localized mRNA in somatic cells is fragile X mental retardation 1 mRNA (12, 13), which encodes fragile X mental retardation protein (FMRP). The absence of FMRP causes the most common hereditary form of mental retardation, fragile X syndrome (14–16). In addition to FMRP, mammals have two other members of this family, FXR1 and FXR2. FMRP contains three RNA-binding domains, KH1, KH2 [heterogeneous nuclear ribonucleoprotein (RNP)-K homology], and an RGG box (arginine-glycine rich region), and shows binding specificity toward an estimated 4% of total brain RNA, including its own mRNA (17–19). By using numerous in vivo and in vitro assays, several putative protein and RNA-binding partners of FMRP have been described (20–28). Further examination of some target RNAs has indicated that FMRP may be involved in posttranscriptional regulation, including mRNA localization and translation (24, 29). Drosophila has only one member of the FMRP family, called Drosophila FMRP (dFMR; also called dFMR1 or dFXR) (30). dFMR shares the fundamental and characteristic molecular architecture with the mammalian homologues, implying functional conservation.
In mammalian cells, FMRP forms granules that contain its own RNA (13) and moves along the neurites of differentiated PC12 cells (31). In addition, ZBP/IMP-1, a protein that is involved in recognition of localized mRNA, and FMRP are able to recruit each other into RNP granules (32), suggesting that FMRP may be involved in RNA trafficking. Thus, FMRP is a good marker for RNP granules because it appears to be involved in both transport and translation of a specific subset of mRNAs. In this study, we show that in Drosophila S2 cells, GFP-tagged dFMR is able to form RNP granules that are moved by kinesin-1 and cytoplasmic dynein.
Materials and Methods
RNA Interference (RNAi) Treatment. RNAi treatment for S2 cells was as described (33). DNA templates of T7 promoter-containing dynein heavy chain (DHC), kinesin heavy chain (KHC), kinesin light chain (KLC), Klp64D, Klp68D, and Klp61F sequences (33) were amplified by PCR and subcloned into pCR2.1 vector (Invitrogen). dsRNA was synthesized by using a T7 RiboMax kit (Promega) following manufacturer's protocol. A total of 1 × 106 cells in 35-mm dishes were incubated with 30 μg of dsRNA for 3 days. Cells were split at a 1:3 ratio on the third day and incubated with a fresh aliquot of 30 μg of dsRNA. On day 5, cells were plated onto concanavalin A (ConA)-coated coverslips (34) with 5 μM cytochalasin-D added to induce process outgrowth. Images were taken on the sixth day after the initial RNAi treatment. The efficiency of RNAi was checked by immunoblotting.
Antibodies. The following antibodies were used: anti-DHC antibody (a gift of J. Scholey, University of California, Davis); SUK4, 5A11, 9E10.2 (anti-KHC, anti-dFMR, and anti-myc antibody, respectively; Developmental Studies Hybridoma Bank, Iowa City, IA); HD and KLC (anti-KHC and anti-pan-KLC antibody, respectively; gifts of A. Minin, Institute of Protein Research, Moscow); anti-Drosophila KLC antibody (a gift of J. Gindhart, University of Richmond, Richmond, VA); anti-Klp68D antibody (a gift of L. S. B. Goldstein, University of California at San Diego, La Jolla); anti-Klp61F antibody (a gift of J. Scholey, University of California, Davis; and G. Rogers, Albert Einstein College of Medicine, New York); actin (anti-actin polyclonal rabbit antibody, Sigma); polyclonal rabbit anti-GFP antibody (a gift of R. Vale, University of California, San Francisco); and monoclonal mouse anti-GFP antibody (generated at the Immunology Center, University of Illinois at Urbana–Champaign).
Image Acquisition, Particle Tracking, and Image Analysis. Images of live cells were acquired by using an inverted Nikon Eclipse U2000 microscope with a plan-Apo 60 × 1.4 numerical aperture objective. A 100-W halogen light source was used for epifluorescence illumination to minimize phototoxicity. Images were captured by using an Orca II-ER cooled charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan) run by metamorph software. Images were captured every 2 sec for 2 min. The coordinates of the cell body and each particle were recorded, and the behavior of the particles was tracked by using diatrack software (Semasopht, Chavannes, Switzerland). Particles that traveled away from the cell body were defined as moving in the anterograde direction, whereas particles that traveled toward the cell body were defined as moving in the retrograde direction. A threshold of 0.1 μm/sec was used; movements smaller than this minimum were excluded from the calculations. At least four independent experiments were recorded for each condition, and three to six cells from each experiment were randomly chosen for analysis. Raw tracking data were analyzed by using customized software (Ontash & Ermac, River Edge, NJ).
Details of methods for plasmid constructions, Drosophila cell culture and stable cell line selection, and immunoprecipitation (IP) and RT-PCR assays are described in Supporting Methods, which is published as supporting information on the PNAS web site.
Results
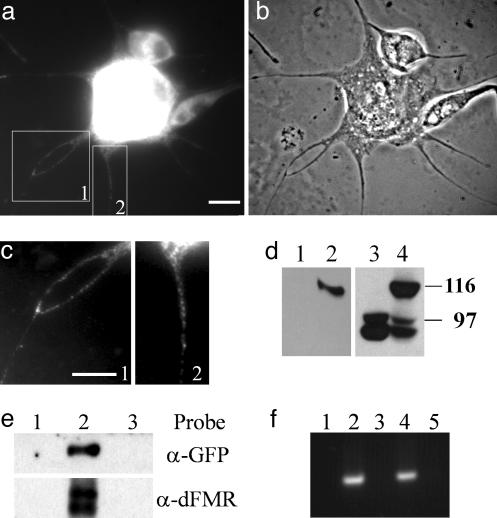
GFP-dFMR Recapitulates Endogenous dFMR Behavior. In Drosophila S2 cells, dFMR exhibits punctuate cytosolic distribution (Fig. 1 a and c), suggesting that dFMR forms RNP granules in Drosophila cells similar to its mammalian homologue. Similar to its untagged counterpart, GFP-dFMR forms granules in the cytosol, and the pattern is indistinguishable from endogenous dFMR staining (Fig. 2a). The expression level of GFP-dFMR in the stable cell line was confirmed by immunoblotting with both anti-GFP and anti-dFMR antibodies (Fig. 1c). Furthermore, when immunoprecipitated with the GFP antibody, GFP-dFMR pulls down endogenous dFMR, suggesting that GFP-tagged and untagged endogenous dFMR are incorporated into the same complex (Fig. 1d). In addition, the GFP-dFMR complex also contains dFMR mRNA as shown by RT-PCR (Fig. 1e). In IP and IP/RT-PCR experiments, two controls were used: (i) IP using lysate from untransfected cells and (ii) IP with antibodies that were blocked with an excess of recombinant GFP before addition to lysates from transfected cells. Neither controls showed signals for dFMR protein and mRNA, confirming the specificity of IP. Therefore, we concluded that GFP-dFMR forms RNP granules with native dFMR protein and its mRNA, and faithfully recapitulates the behavior of endogenous dFMR. This finding is consistent with previous reports (13, 31) that GFP-FMRP forms moving mRNA-containing granules when transfected into mammalian cells.
Fig. 1.
GFP-dFMR forms RNP granules with endogenous dFMR and contains RNA. (a and b) Immunofluorescent staining of untransfected S2 cells with an antibody to dFMR (a) and a corresponding phase-contrast image (b). Cells were treated with cytochalasin D and plated on a ConA-coated coverslip. Under these conditions, S2 cells form thin processes that contain dFMR granules. Areas 1 and 2 of a are shown at higher magnification and higher contrast (c) to better visualize dFMR granules in the processes. (Scale bars in a and c, 10 μm.) (d) Immunoblotting with anti-GFP and anti-dFMR antibody shows expression of GFP-dFMR. Lanes 1 and 3, untransfected cells; lanes 2 and 4, stable cell line expressing GFP-dFMR. Lanes 1 and 2 were probed with anti-GFP antibody and lanes 3 and 4 were probed with anti-dFMR antibody. (e) IP by using anti-GFP antibody. Lane 1, untransfected cells; lanes 2 and 3, GFP-dFMR cells. Monoclonal anti-GFP antibody was used for IP in Lanes 1 and 2, and a control monoclonal antibody was used for lane 3. The upper and lower portions of the blot were probed with anti-GFP antibody and anti-dFMR antibody, respectively. GFP-dFMR associates with endogenous dFMR. (f) RNA were extracted from S2 cells or from IP samples and subjected to RT-PCR. Lane 1, total RNA from S2 cells without reverse transcription; lane 2, total RNA from S2 cell with RT-PCR showed predicted size of dFMR fragment. Lanes 3–5 show the results of RT-PCR from GFP-IP assays. Lane 3, untransfected cells; lanes 4 and 5, GFP-dFMR stable line. In lane 5, GFP antibodies were blocked with excess recombinant GFP before incubation with cell lysate. GFP-dFMR associates with its own RNA.
Fig. 2.
Characterization of GFP-dFMR granule movement. (a) GFP-dFMR forms granules with a distribution similar to wild-type dFMR. These granules move bidirectionally in the processes (arrow). The start of the movie sequences is indicated by 0, with subsequent 2-sec intervals. (Scale bar, 5 μm.) Histograms of velocity (b) and run length (c) for GFP-dFMR particles indicate an unbiased bidirectional translocation of dFMR granules.
When S2 cells are attached onto coverslips coated with ConA, high-resolution microscopy can be used to observe flattened cells (34). Interestingly, S2 cells on ConA can be stimulated to form long, thin processes when treated with cytochalasin D (R. Vale, personal communication). When stained with anti-tubulin antibody, these processes contained long and unfragmented microtubules (Fig. 5, which is published as supporting information on the PNAS web site). Furthermore, because cytochalasin D destroys the network of actin filaments, the treatment enables assessment of microtubule-based transport without interference from the acto-myosin system (Fig. 5). The polarity of microtubules in the processes was examined by using EB1-GFP fusion protein that specifically localizes to the plus-ends of growing microtubules (34). Live microscopy revealed that the majority of EB1-GFP moved away from the cell body, indicating that microtubules in those processes are oriented primarily with their plus-ends away from cell body, with a small percentage of processes containing microtubules of mixed polarity (≈20% of processes examined). Both endogenous dFMR and GFP-dFMR granules were present in the processes (Figs. 1c and 2a). In addition, GFP-dFMR granules moved bidirectionally in the processes (Fig. 2a, and Movie 1, which is published as supporting information on the PNAS web site). At any moment, ≈20% of granules undergo saltatory movement. During the time of the recording some granules stopped, whereas others started moving, and a number of them changed direction. For quantitative analysis, the position of granules was automatically recorded by using diatrack 2.3 tracking software, and the velocity of movement and the lengths of runs in each direction were calculated. For the purpose of analysis, only movements with a rate of >0.1 μm/sec and a duration at least 2 sec were scored. The results of this analysis are presented in Fig. 2 b and c. GFP-dFMR/RNP particles displayed similar distributions of velocity and run length for both anterograde and retrograde movement. Furthermore, they exhibited an unbiased bidirectional motion, in that there was an equal probability of engaging in anterograde or retrograde movement (Fig. 2 b and c).
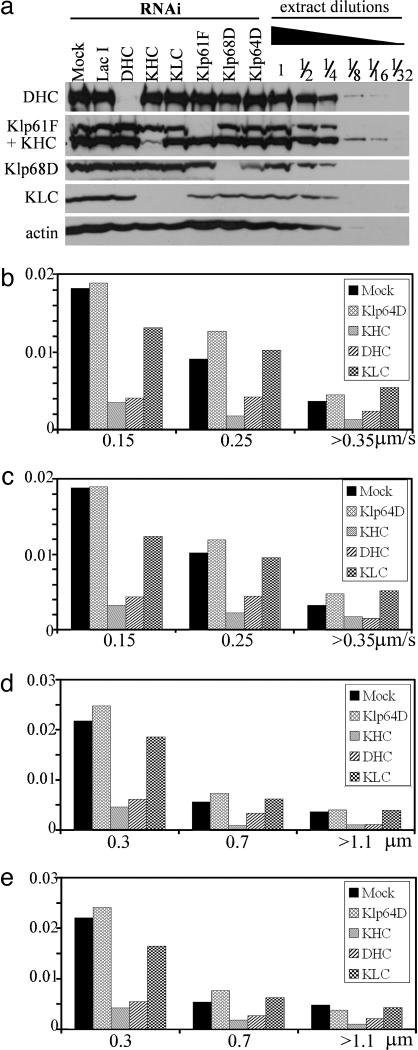
Kinesin-1 and Cytoplasmic Dynein Transport dFMR-Containing RNP Granules. FMRP has been shown to be colocalized with kinesin-1 in cultured PC12 cells (31), and biochemical analysis using brain homogenates also suggest an association between FMRP and kinesin-1 (35). However, direct functional evidence of kinesin involvement has been lacking. Because RNAi-mediated protein knockdown on S2 cells has been proven to be effective and specific (33, 36), we adopted this approach to identify components of the transport machinery for dFMR-containing RNP granules. GFP-dFMR cells were treated with dsRNA and the specificity and efficiency of RNAi knockdown was examined by immunoblotting (Fig. 3a). The target proteins were consistently reduced to <10% of the control level without affecting other proteins (with the exception of KLC reduction by KHC-RNAi, see below). When the heavy chain of kinesin-1 (KHC) was knocked down, the motility of GFP-dFMR-containing RNP granules was dramatically reduced (Movie 2, which is published as supporting information on the PNAS web site). Quantitative analysis of the movement demonstrates the KHC-RNAi treatment reduces the number of movements >5-fold compared with untreated cells. Analysis of the run length and velocities of movement demonstrates that KHC-RNAi decreases both parameters (Fig. 3 b–d). As an additional control for specificity, we used RNAi against two other members of the kinesin superfamily, Klp64D (a subunit of Drosophila kinesin-2) and Klp61F (a kinesin-5 family member). Neither of these two treatments had any effect on GFP-dFMR movement (Table 1).
Fig. 3.
Movements of GFP-dFMR granules after RNAi treatments. (a) Immunoblotting confirmed the specificity and efficiency of RNAi treatment. Control extracts were diluted to one-half, one-fourth, one-eighth, 1/16th, and 1/32nd to estimate the knockdown efficiency. The antibodies used were indicated at the left. Target proteins were consistently knockdown to <10% of the control level. Histograms of anterograde velocity (b), retrograde velocity (c), anterograde run length (d), and retrograde run length (e) for GFP-dFMR particles are expressed as relative frequency of total particles in various RNAi treatments. Total number of cells and particles analyzed are listed in Table 1. Mock-treated and Klp64D-RNAi showed comparable results, indicating that kinesin-2 is not involved in dFMR granule transport. Either KHC or DHC knockdown is sufficient to impede the movement bidirectionally. KLC-RNAi has no effect on RNP granule translocation.
Table 1. Effects of RNAi on GFP-dFMR granule movements.
| RNAi treatment | Relative no. of movements* | No. of cells analyzed | No. of particles tracked | Total no. of movements |
|---|---|---|---|---|
| Mock | 3.8 | 7 | 108 | 410 |
| KHC (kinesin-1) | 0.8 | 21 | 216 | 178 |
| Klp64D (kinesin-2) | 4.3 | 30 | 734 | 3,156 |
| Klp61F (kinesin-5) | 3.3 | 13 | 136 | 442 |
| KLC (kinesin-1) | 3.3 | 17 | 318 | 1,065 |
| DHC (dynein) | 1.3 | 12 | 135 | 169 |
Relative number of movements is defined as a ratio of total number of movements to the number of particles scored.
Interestingly, these measurements demonstrate that KHC RNAi inhibits not only anterograde (plus-end-directed) but also retrograde component of the transport (Fig. 3 b–d). To rule out the possibility that this inhibition of bidirectional transport is caused by general effects on microtubules, control cells and cells treated with RNAi were fixed and stained by an anti-tubulin antibody. The gross morphology of cells and distribution of microtubules, especially in the processes, was similar in all RNAi treatment (Fig. 6, which is published as supporting information on the PNAS web site). Therefore, inhibition of transport cannot be explained by microtubule depolymerization.
The bidirectional nature of GFP-dFMR RNP granule transport implies that a minus-end-directed microtubule motor might be involved in the observed retrograde movement. When DHC was knocked down, movement of particles was also strongly inhibited (Fig. 3 b–d and Table 1). As in the case of KHC-RNAi, DHC-RNAi also inhibited both retrograde and anterograde transport. This inhibition of bidirectional transport cannot be explained by simultaneous knockdown of DHC and KHC because the expression of either of these motors was not affected when the other was knocked down (Fig. 3a). Based on these results, we conclude that both kinesin-1 and cytoplasmic dynein are required for the transport of dFMR-containing RNP granules.
KLC Is Not Required for dFMR-RNP Transport. Kinesin-1 is composed of two heavy chains (KHC) and two light chains (KLC) (37, 38). The N terminus of KLC interacts with KHC, and the C-terminal tetratricopeptide repeat of KLC is thought to mediate the attachment of kinesin-1 to various membranous cargoes (reviewed in refs. 39 and 40). Therefore, we sought to determine whether KLC plays a similar role in attachment of kinesin-1 to dFMR granules. It is worth noting that KHC-RNAi results in depletion of not only KHC itself but also of KLC (Fig. 3a). This result can be explained by the fact that nascent KLC polypeptides associate with KHC cotranslationally, and KLC that is not associated with KHC may be rapidly degraded (41). Fortunately, this relationship is not reciprocal, and depletion of KLC by RNAi had no effect at the level of KHC (Fig. 3a), which made it possible to directly examine whether KLC is necessary for dFMRP transport. This analysis is further facilitated by the fact that KLC in Drosophila is encoded by a single gene and does not appear to be alternatively spliced (42). Two different anti-KLC antibodies were used to examine the knockdown efficiency of KLC (43, 44). Only a single band was recognized by these antibodies and showed dramatic reduction after RNAi treatment, indicating that only one KLC isoform exists in S2 cells (Fig. 3a and data not shown). KLC knockdown did not inhibit GFP-dFMR movement (Fig. 3 b–d and Table 1), suggesting that KLC is not required for dFMR-RNP granule transport. This idea is further strengthened by our biochemical data (see below).
To ensure that KLC-RNAi was able to affect transport in this preparation, we analyzed peroxisome transport, which uses kinesin-1 in Drosophila S2 cells (H. Kim, S.-C.L., and V.I.G., unpublished observations). GFP peroxisomes also exhibit bidirectional transport in the processes. The number of moving peroxisomes in the processes was dramatically decreased by KHC and KLC-RNAi (Fig. 7, which is published as supporting information on the PNAS web site). Therefore, peroxisome transport requires both KHC and KLC. By contrast, KLC is not required for RNP granule translocation.
dFMR Associates with KHC and DHC but Not with KLC. The foregoing functional data suggest that both KHC and DHC are directly involved in GFP-dFMR-containing RNP granule transport. Whether dFMR physically associates with motor complexes was examined by using IP. When the GFP-dFMR-containing complex was immunoprecipitated with GFP antibody, both KHC and DHC, but not KLC, were detected by immunoblotting (Fig. 4 a and b). Two controls were used for IP experiments; one used untransfected cells as the material for IP, and in the other experiment, the GFP antibody was blocked with excess recombinant GFP before incubating with extracts from GFP-dFMR-expressing cells. KHC and DHC could not be detected in either control, demonstrating that the interaction of RNP with KHC and DHC is specific. To ensure that kinesin–RNP interactions also occur also in untransfected cells, IPs by using two different antibodies against KHC [monoclonal antibody SUK4 (45) and polyclonal antibody HD (46)] were performed. When used on extracts of untransfected S2 cells, both antibodies were able to pull down dFMR (Fig. 4c), whereas no dFMR was present in control IPs (anti-myc antibody 9E10.2 was used as a control for SUK4 and rabbit IgG was used as a control for the polyclonal antibody HD). Therefore, the biochemical data support the functional evidence that dFMR indeed forms a complex with KHC and DHC in vivo, the attachment of RNP to KHC is independent of KLC, and KLC is likely absent from the dFMR-containing complex.
Fig. 4.
KHC and DHC, but not KLC, associates with dFMR-RNP. An IP assay was used to determine the physical association of dFMR-RNP with motor complexes. (a) Polyclonal GFP antibodies were used for IP. Lane 1, untransfected cells; lane 2, GFP-dFMR stable line; lane 3, GFP-dFMR stable cell lines, but the antibodies were blocked with recombinant GFP before incubation with cell extracts. GFP-dFMR pulls down endogenous dFMR, KHC, and DHC. (b) Lanes 1–3, IP from untransfected cells; lanes 4 and 5, IP from the GFP-dFMR stable line. Lane 1, IP with anti-myc antibody; lane 2, IP with SUK4 antibody (anti-KHC antibody); lanes 3 and 4, GFP-IP; lane 5, GFP-IP, but the antibodies were blocked with recombinant GFP. GFP-dFMR pulls down KHC, but not KLC. (c) Untransfected cells were used for IP. Lane 1, IP with anti-myc antibody; lane 2, IP with SUK4 antibody (anti-KHC antibody); lane 3, IP with normal rabbit IgG; lane 4, IP with HD antibody (anti-KHC antibody). Kinesin-1 can pull down endogenous dFMR. Anti-myc antibody and normal rabbit IgG were used as controls and they did not pull down dFMR.
Discussion
In this report, we have demonstrated that dFMR-containing RNP granules are transported bidirectionally in somatic Drosophila cells. Both kinesin-1 and cytoplasmic dynein are required for these translocations because knockdown of either of these motors is sufficient to inhibit the bidirectional movement. Thus, dFMR-RNP granules, like other RNA-RNP complexes in a number of systems from yeast to human that are moved and localized by motor proteins. In addition to kinesin-1 and dynein, other motors have been shown to participate in RNA transport in different systems (3–6). For example, it is well documented that Ash1 and a number of other mRNAs are moved in yeast cell by myo4p, a class V myosin (47). In higher eukaryotes, including Drosophila, Xenopus, and mammalian cells, mRNP complexes are transported along microtubules by members of kinesin-1, kinesin-2, and dynein families (8–11, 48–51).
Our results agree with a recent study by Kanai et al. (52). By using an elegant biochemical analysis, they showed that kinesin-1 (KIF5 and KHC), but not kinesin-2 (KIF3), interacts with several RNA-binding proteins, including Staufen and FMRP, and that KHC is involved in transporting RNP granules in the dendrites of neurons (52). Interestingly, kinesin-2 (KIF3A) is proposed to transport tau mRNA-RNP granules in the axons of differentiated p19 cells (53). Together, these results suggest that the transport of different populations of RNP granules may require different motor proteins to specify the site of localization.
One unusual aspect of dFMR-mRNP transport by kinesin-1 is that it does not depend on KLC. Our biochemical analysis shows that KLC is not part of the dFMR-RNP complex. This finding indicates that RNP granule is moved by KHC dimer alone and may interact with KHC directly. This result agrees with genetic data of Palacios and St Johnston (54), demonstrating that kinesin-1-dependent oskar mRNA localization in Drosophila oocytes is unaffected in KLC-null mutants. In a mammalian system, KLC was not found in RNP complexes pulled down by the tail fragment of KHC (52). Two other examples of KLC-independent kinesin-1-mediated transport include vesicles movement in Neurospora (55) and glutamate-receptor-interacting protein 1-containing vesicles in hippocampal neurons (56). Taking all of these results into consideration, dFMR-RNP appears to be transported by a special isoform of kinesin-1 that is devoid of light chains. Furthermore, it is likely that kinesin-1 transport of other RNAs might not employ light chains.
Our results demonstrate that knockdown of either KHC or DHC is sufficient to inhibit both the retrograde and anterograde components of dFMR-RNP transport. This interdependent relationship of plus- and minus-end-directed motors seems to be a general property of bidirectional transport (57). By using genetic approaches in Drosophila, bidirectional vesicle transport in axons was blocked in KHC, KLC, DHC, or p150/glued (dynactin complex) mutant backgrounds (58, 59). Similarly, the plus-end movement of lipid droplets in Drosophila embryos were severely impaired when either DHC or p150/glued was mutated (60). However, the molecular mechanism for coordination between opposite polarity motors is still unknown (57, 61). Further study of the regulation of this interdependent relationship between the plus- and minus-end-directed transport machinery remains a challenge. Robust functional assay combining live imaging with specific knockdown of target proteins should provide an excellent tool to delineate these issues.
Supplementary Material
Acknowledgments
We thank Gohta Goshima, Stephen Rogers, and Ronald Vale for sharing reagents and helpful suggestions, and we thank our colleagues who provided antibodies and other reagents (see Materials and Methods). The SUK4 (anti-KHC), 9E10 (anti-myc), and 5A11 (anti-dFMR) were obtained from the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences (Iowa City). This work was supported by grants from the National Institute of General Medical Sciences (to V.I.G.).
Author contributions: W.T.G. and V.I.G. designed research; S.-C.L. and P.S.F. performed research; S.-C.L., P.S.F., and V.I.G. analyzed data; and S.-C.L., W.T.G., and V.I.G. wrote the paper.
Abbreviations: FMRP, fragile X mental retardation protein; dFMR, Drosophila FMRP; RNP, ribonucleoprotein; KHC, kinesin heavy chain; KLC, kinesin light chain; DHC, dynein heavy chain; ConA, concanavalin A; IP, immunoprecipitation; RNAi, RNA interference.
References
- 1.Kiebler, M. A. & DesGroseillers, L. (2000) Neuron 25, 19–28. [DOI] [PubMed] [Google Scholar]
- 2.Kloc, M., Zearfoss, N. R. & Etkin, L. D. (2002) Cell 108, 533–544. [DOI] [PubMed] [Google Scholar]
- 3.Jansen, R. P. (2001) Nat. Rev. Mol. Cell Biol. 2, 247–256. [DOI] [PubMed] [Google Scholar]
- 4.Lopez de Heredia, M. & Jansen, R. P. (2004) Curr. Opin. Cell Biol. 16, 80–85. [DOI] [PubMed] [Google Scholar]
- 5.Van de Bor, V. & Davis, I. (2004) Curr. Opin. Cell Biol. 16, 300–307. [DOI] [PubMed] [Google Scholar]
- 6.Tekotte, H. & Davis, I. (2002) Trends Genet. 18, 636–642. [DOI] [PubMed] [Google Scholar]
- 7.Cha, B. J., Koppetsch, B. S. & Theurkauf, W. E. (2001) Cell 106, 35–46. [DOI] [PubMed] [Google Scholar]
- 8.MacDougall, N., Clark, A., MacDougall, E. & Davis, I. (2003) Dev. Cell 4, 307–319. [DOI] [PubMed] [Google Scholar]
- 9.Cha, B. J., Serbus, L. R., Koppetsch, B. S. & Theurkauf, W. E. (2002) Nat. Cell Biol. 4, 592–598. [DOI] [PubMed] [Google Scholar]
- 10.Brendza, R. P., Serbus, L. R., Duffy, J. B. & Saxton, W. M. (2000) Science 289, 2120–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnorrer, F., Bohmann, K. & Nusslein-Volhard, C. (2000) Nat. Cell Biol. 2, 185–190. [DOI] [PubMed] [Google Scholar]
- 12.Weiler, I. J., Irwin, S. A., Klintsova, A. Y., Spencer, C. M., Brazelton, A. D., Miyashiro, K., Comery, T. A., Patel, B., Eberwine, J. & Greenough, W. T. (1997) Proc. Natl. Acad. Sci. USA 94, 5395–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antar, L. N., Afroz, R., Dictenberg, J. B., Carroll, R. C. & Bassell, G. J. (2004) J. Neurosci. 24, 2648–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Donnell, W. T. & Warren, S. T. (2002) Annu. Rev. Neurosci. 25, 315–338. [DOI] [PubMed] [Google Scholar]
- 15.Oostra, B. A. & Willemsen, R. (2003) Hum. Mol. Genet. 12, R249–R257. [DOI] [PubMed] [Google Scholar]
- 16.Schaeffer, C., Beaulande, M., Ehresmann, C., Ehresmann, B. & Moine, H. (2003) Biol. Cell 95, 221–228. [DOI] [PubMed] [Google Scholar]
- 17.Ashley, C. T., Jr., Wilkinson, K. D., Reines, D. & Warren, S. T. (1993) Science 262, 563–566. [DOI] [PubMed] [Google Scholar]
- 18.Brown, V., Small, K., Lakkis, L., Feng, Y., Gunter, C., Wilkinson, K. D. & Warren, S. T. (1998) J. Biol. Chem. 273, 15521–15527. [DOI] [PubMed] [Google Scholar]
- 19.Siomi, H., Siomi, M. C., Nussbaum, R. L. & Dreyfuss, G. (1993) Cell 74, 291–298. [DOI] [PubMed] [Google Scholar]
- 20.Brown, V., Jin, P., Ceman, S., Darnell, J. C., O'Donnell, W. T., Tenenbaum, S. A., Jin, X., Feng, Y., Wilkinson, K. D., Keene, J. D., et al. (2001) Cell 107, 477–487. [DOI] [PubMed] [Google Scholar]
- 21.Bardoni, B., Schenck, A. & Mandel, J. L. (1999) Hum. Mol. Genet. 8, 2557–2566. [DOI] [PubMed] [Google Scholar]
- 22.Bardoni, B., Castets, M., Huot, M. E., Schenck, A., Adinolfi, S., Corbin, F., Pastore, A., Khandjian, E. W. & Mandel, J. L. (2003) Hum. Mol. Genet. 12, 1689–1698. [DOI] [PubMed] [Google Scholar]
- 23.Darnell, J. C., Jensen, K. B., Jin, P., Brown, V., Warren, S. T. & Darnell, R. B. (2001) Cell 107, 489–499. [DOI] [PubMed] [Google Scholar]
- 24.Miyashiro, K. Y., Beckel-Mitchener, A., Purk, T. P., Becker, K. G., Barret, T., Liu, L., Carbonetto, S., Weiler, I. J., Greenough, W. T. & Eberwine, J. (2003) Neuron 37, 417–431. [DOI] [PubMed] [Google Scholar]
- 25.Ceman, S., Brown, V. & Warren, S. T. (1999) Mol. Cell. Biol. 19, 7925–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceman, S., Nelson, R. & Warren, S. T. (2000) Biochem. Biophys. Res. Commun. 279, 904–908. [DOI] [PubMed] [Google Scholar]
- 27.Schenck, A., Bardoni, B., Moro, A., Bagni, C. & Mandel, J. L. (2001) Proc. Natl. Acad. Sci. USA 98, 8844–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaeffer, C., Bardoni, B., Mandel, J. L., Ehresmann, B., Ehresmann, C. & Moine, H. (2001) EMBO J. 20, 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zalfa, F., Giorgi, M., Primerano, B., Moro, A., Di Penta, A., Reis, S., Oostra, B. & Bagni, C. (2003) Cell 112, 317–327. [DOI] [PubMed] [Google Scholar]
- 30.Wan, L., Dockendorff, T. C., Jongens, T. A. & Dreyfuss, G. (2000) Mol. Cell. Biol. 20, 8536–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Diego Otero, Y., Severijnen, L. A., van Cappellen, G., Schrier, M., Oostra, B. & Willemsen, R. (2002) Mol. Cell. Biol. 22, 8332–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rackham, O. & Brown, C. M. (2004) EMBO J. 23, 3346–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goshima, G. & Vale, R. D. (2003) J. Cell Biol. 162, 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers, S. L., Rogers, G. C., Sharp, D. J. & Vale, R. D. (2002) J. Cell Biol. 158, 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohashi, S., Koike, K., Omori, A., Ichinose, S., Ohara, S., Kobayashi, S., Sato, T. A. & Anzai, K. (2002) J. Biol. Chem. 277, 37804–37810. [DOI] [PubMed] [Google Scholar]
- 36.Rogers, S. L., Wiedemann, U., Stuurman, N. & Vale, R. D. (2003) J. Cell Biol. 162, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bloom, G. S., Wagner, M. C., Pfister, K. K. & Brady, S. T. (1988) Biochemistry 27, 3409–3416. [DOI] [PubMed] [Google Scholar]
- 38.Kuznetsov, S. A., Vaisberg, E. A., Shanina, N. A., Magretova, N. N., Chernyak, V. Y. & Gelfand, V. I. (1988) EMBO J. 7, 353–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karcher, R. L., Deacon, S. W. & Gelfand, V. I. (2002) Trends Cell Biol. 12, 21–27. [DOI] [PubMed] [Google Scholar]
- 40.Kamal, A. & Goldstein, L. S. (2002) Curr. Opin. Cell Biol. 14, 63–68. [DOI] [PubMed] [Google Scholar]
- 41.Gyoeva, F. K., Sarkisov, D. V., Khodjakov, A. L. & Minin, A. A. (2004) Biochemistry 43, 13525–13531. [DOI] [PubMed] [Google Scholar]
- 42.Gauger, A. K. & Goldstein, L. S. (1993) J. Biol. Chem. 268, 13657–13666. [PubMed] [Google Scholar]
- 43.Khodjakov, A., Lizunova, E. M., Minin, A. A., Koonce, M. P. & Gyoeva, F. K. (1998) Mol. Biol. Cell 9, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gindhart, J. G., Jr., Desai, C. J., Beushausen, S., Zinn, K. & Goldstein, L. S. (1998) J. Cell Biol. 141, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingold, A. L., Cohn, S. A. & Scholey, J. M. (1988) J. Cell Biol. 107, 2657–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodionov, V. I., Gyoeva, F. K. & Gelfand, V. I. (1991) Proc. Natl. Acad. Sci. USA 88, 4956–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shepard, K. A., Gerber, A. P., Jambhekar, A., Takizawa, P. A., Brown, P. O., Herschlag, D., DeRisi, J. L. & Vale, R. D. (2003) Proc. Natl. Acad. Sci. USA 100, 11429–11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Betley, J. N., Heinrich, B., Vernos, I., Sardet, C., Prodon, F. & Deshler, J. O. (2004) Curr. Biol. 14, 219–224. [DOI] [PubMed] [Google Scholar]
- 49.Yoon, Y. J. & Mowry, K. L. (2004) Development (Cambridge, U.K.) 131, 3035–3045. [DOI] [PubMed] [Google Scholar]
- 50.Huang, Y. S., Carson, J. H., Barbarese, E. & Richter, J. D. (2003) Genes Dev. 17, 638–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chennathukuzhi, V., Morales, C. R., El-Alfy, M. & Hecht, N. B. (2003) Proc. Natl. Acad. Sci. USA 100, 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanai, Y., Dohmae, N. & Hirokawa, N. (2004) Neuron 43, 513–525. [DOI] [PubMed] [Google Scholar]
- 53.Aronov, S., Aranda, G., Behar, L. & Ginzburg, I. (2002) J. Cell Sci. 115, 3817–3827. [DOI] [PubMed] [Google Scholar]
- 54.Palacios, I. M. & St. Johnston, D. (2002) Development (Cambridge, U.K.) 129, 5473–5485. [DOI] [PubMed] [Google Scholar]
- 55.Seiler, S., Kirchner, J., Horn, C., Kallipolitou, A., Woehlke, G. & Schliwa, M. (2000) Nat. Cell Biol. 2, 333–338. [DOI] [PubMed] [Google Scholar]
- 56.Setou, M., Seog, D. H., Tanaka, Y., Kanai, Y., Takei, Y., Kawagishi, M. & Hirokawa, N. (2002) Nature 417, 83–87. [DOI] [PubMed] [Google Scholar]
- 57.Welte, M. A. (2004) Curr. Biol. 14, R525–R537. [DOI] [PubMed] [Google Scholar]
- 58.Hurd, D. D. & Saxton, W. M. (1996) Genetics 144, 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin, M., Iyadurai, S. J., Gassman, A., Gindhart, J. G., Jr., Hays, T. S. & Saxton, W. M. (1999) Mol. Biol. Cell 10, 3717–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gross, S. P., Welte, M. A., Block, S. M. & Wieschaus, E. F. (2002) J. Cell Biol. 156, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gross, S. P. (2003) Curr. Biol. 13, R320–R322. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.