Abstract
Genetic diversity is greater in Africa than in other continental populations. Genetic variability in genes encoding drug metabolizing enzymes may contribute to the high numbers of adverse drug reactions reported in Africa. We reviewed publications (1995–April 2016) reporting frequencies of known cytochrome P450 (CYP) variants in African populations. Using principal components analysis (PCA) we identified CYP alleles of potential clinical relevance with a marked difference in distribution in Africa, compared with Asian and Caucasian populations. These were CYP2B6*6, CYP2C8*2, CYP2D6*3, CYP2D6*17, CYP2D6*29, CYP3A5*6, and CYP3A5*7. We show clearly that there is greater diversity in CYP distribution in Africa than in other continental populations and identify a need for optimization of drug therapy and drug development there. Further pharmacogenetic studies are required to confirm the CYP distributions we identified using PCA, to discover uniquely African alleles and to identify populations at a potentially increased risk of drug-induced adverse events or drug inefficacy.
Keywords: Pharmacogenetics, CYP450, Genetic diversity, Drug metabolism, Africa, Drug development
Highlights
-
•
Greater diversity in CYP450 distribution in Africa vs Europe or Asia was demonstrated.
-
•
CYP alleles with a marked difference in distribution in Africa were identified.
-
•
CYP alleles that may be uniquely African were highlighted.
-
•
These data may help to identify regions at a greater risk of adverse drug reactions.
Genetic diversity is greater in Africa than in other populations, yet in clinical therapeutics and drug development Africa is often treated as a homogenous, single entity. Genetic diversity in cytochrome P450 (CYP) genes may contribute to high numbers of adverse drug reactions reported in Africa. Reports on polymorphisms in drug metabolizing enzymes in African populations are limited. Our study demonstrates greater genetic diversity in CYPs in Africa and identifies CYP alleles with markedly different distribution in Africa compared with other populations. Our data can help identify African regions at potentially greater risk of adverse drug reactions or drug inefficacy.
1. Introduction
Modern humans have inhabited Africa for longer than any other geographic region (> 200,000 years; Tishkoff et al., 2009); the processes of random mutation, meiotic recombination, and genetic drift have therefore led to the accumulation of a relatively large pool of genetic variation in Africa compared with elsewhere. Approximately 100,000 years ago, a limited number of African subgroups migrated from Africa to other continents; many genotypes are therefore found only in Africa (Tishkoff et al., 2009, Fujikara et al., 2015, Kozyra et al., 2016, Wright et al., 2016). Based on a study calculating worldwide disease burden for 21 regions, the African continent has 15.5% of the global population but carries approximately 25% of the global disease burden (Murray et al., 2012). Furthermore, the African population carries a high burden of adverse drug reactions owing partly to the use of old, poorly-optimized drugs for the treatment of parasitic infections (Prentis et al., 1988, Ampadu et al., 2016), and also possibly owing to high levels of genetic diversity resulting in a greater proportion of patients having adverse reactions to therapeutic drugs.
Drug metabolism involves a complex interplay between multiple metabolic enzymes, often with overlapping substrate specificity. One significant class of enzymes involved in therapeutic drug metabolism is the cytochrome P450 (CYP) family. CYP1, CYP2, and CYP3 are the key sub-families involved in phase I drug metabolism. Single nucleotide polymorphisms (SNPs) in CYP genes have been extensively reported (Fujikara et al., 2015) with over 400 allelic variants recorded across the CYP1, CYP2, and CYP3 sub-families (The human cytochrome P450 allele nomenclature database http://cypalleles.ki.se/ accessed 28 October 2016). Variations in gene sequence and protein structure give rise to alleles conferring no enzyme function, decreased enzyme function, normal enzyme function or increased enzyme function. Individuals who carry allelic variants may demonstrate one of the following metabolic phenotypes: poor metabolizers (PM; individuals with a combination of no function or decreased function alleles, and little to no enzyme activity); intermediate metabolizers (IM; individuals with a combination of normal and decreased function alleles conferring decreased enzyme activity); normal metabolizers (NM; individuals with fully functional enzyme activity) or rapid and ultra-rapid metabolizers (UM; individuals with two increased function alleles or more than two normal function alleles) (Caudle et al., 2016, Gaedigk et al., 2016). Genotype information can be used to guide appropriate therapeutic drug doses to reduce the risk of drug-induced adverse reactions in PMs or drug resistance in UMs for drugs with inactive metabolites, or of drug-induced adverse reactions in UMs and drug resistance in PMs for pro-drugs that require metabolic activation (Ingeman-Sundberg et al., 2007).
The majority of pharmacogenetic studies are performed in Asian and Caucasian populations; data generated in African-American populations are sometimes extrapolated to represent the population of the African continent. However, the ancestry of African-Americans is predominantly from Niger-Kordofanian (~ 71%), European (~ 13%), and other African (~ 8%) populations (Tishkoff et al., 2009). It is therefore unlikely that the African-American population will be representative of the many different populations present in Africa. Raising awareness of the greater genetic variability in Africa and its relevance to drug metabolism and efficacy, and the requirement for further pharmacogenetic and clinical studies in African populations, has the potential to result in modifications to drug regimens that could reduce the risk of adverse drug reactions and the overall disease burden.
Here, we provide a comprehensive and up-to-date review of the frequencies of known CYP variants in African populations. In addition, we use principal components analysis to compare these with data from Asian and Caucasian populations. Such analyses can identify CYP variants demonstrating a marked difference in distribution in Africa, and African regions or populations that may be at a higher risk of drug toxicity or inefficacy. The findings of our study may help to support the future pharmacogenetic profiling of Africa which may be of relevance to both clinical therapeutics and drug research and development.
2. Methods
2.1. Search Strategy and Selection Criteria
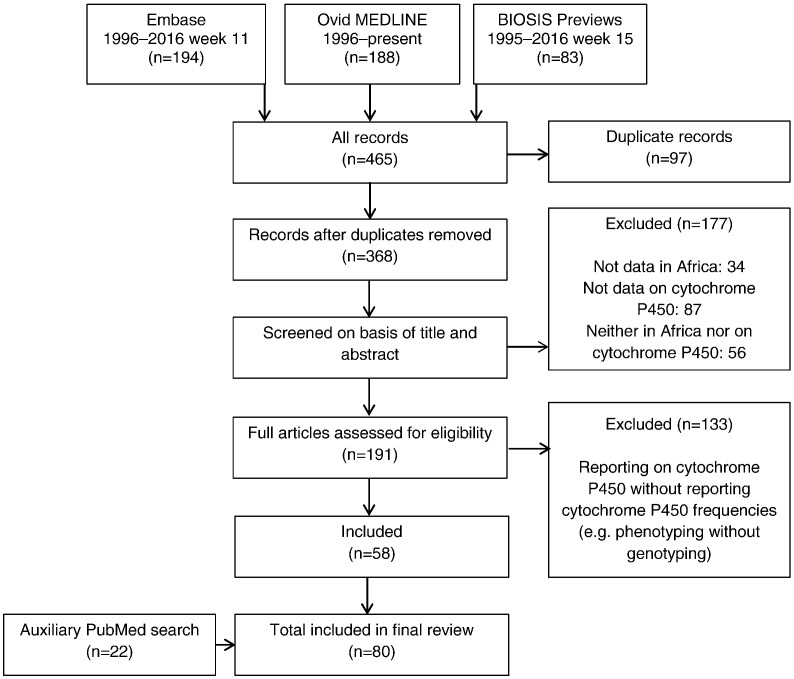
Embase, Ovid MEDLINE and BIOSIS Previews were searched for publications (1995–April 2016) reporting CYP allele frequencies in African populations using the search terms ‘genetic polymorphism’ or ‘DNA polymorphism’ or ‘genetic variability’ and ‘drug metabolizing enzyme’ and ‘Africa’. Following the selection of relevant publications from the initial search, PubMed was used to perform additional targeted searches to provide data on specific CYP alleles and/or populations for which data were minimal or absent in the initial search. Our review focused on reports describing the frequencies of the key subfamilies involved in phase I drug metabolism (CYP1, CYP2 and CYP3). Criteria used to include or exclude studies are summarized in Fig. 1.
Fig. 1.
Flow diagram of study selection.
2.2. Principal Components Analysis
Principal components analysis (PCA) is a statistical technique that systematically identifies underlying variables, or principal components, that best differentiate a set of data. In total, 17 CYP variants were selected for PCA (Supplemental Table 1, Table 1). These variants have been previously studied in a number of global populations and have a known association with significant functional changes in metabolic activity. Additional targeted searches of PubMed were performed to obtain CYP frequency data for Asian and Caucasian populations for comparison with African data. Search terms included a specific CYP variant (e.g. CYP2B6*6) and a specific population (e.g. Asians or Caucasians). Studies reporting CYP frequencies were selected only if they included > 50 participants, in order to exclude misleading frequency data. A minimum of ten studies were selected for Asian populations and Caucasian populations; once sufficient data had been obtained, searches were terminated. Search results were reviewed in order of publication date (as displayed in PubMed). To ensure that data for Asian and Caucasian populations were obtained for a minimum of 50% of the 17 CYP variants, frequencies from multiple studies of a specific population were pooled if required. When more than one study investigated the same variant in the same population, a weighted mean of the frequencies was calculated, accounting for study participant number. For an Asian or Caucasian population to be included in the PCA, data for 50% (i.e. at least 9 out of 17) of the selected variants must have been available. As a limited number of variants have been studied in African populations, it was not always possible to obtain frequency data for 50% of the selected CYP variants for individual populations; data for 40% (i.e. at least 7 out of 17) of the selected variants were therefore required for the inclusion of an African population in the PCA. The selection of a lower cut-off value for African populations allowed the inclusion of more populations in order to gain the best accuracy. PCA was performed using SIMCA version 14.1 (MKS Data Analytics Solutions, San José, CA, USA).
Table 1.
Highest and lowest frequencies (%) reported for 17 CYP allelic variants.
| CYP variant | Highest reported frequency (%) (population) | Lowest reported frequency (%) (population) |
|---|---|---|
| CYP2B6*6 | 69 (Uganda) | 9 (South Africa MA) |
| CYP2C8*2 | 22 (Senegal) | 11 (Uganda) |
| CYP2C8*3 | 5 (Mozambique) | 0 (Tanzania) |
| CYP2C8*4 | 0.6 (Tanzania) | 0 (Ghana) |
| CYP2C9*2 | 19 (Morocco) | 0 (multiple populations)a |
| CYP2C9*3 | 35 (Libya) | 0 (multiple populations)b |
| CYP2C19*2 | 33 (Igbo) | 6 (Ghana) |
| CYP2C19*3 | 7 (South Africa MA) | 0 (multiple populations)c |
| CYP2D6*3 | 9 (San) | 0 (multiple populations)d |
| CYP2D6*4 | 9 (San) | 0 (DRC, Kenya, Ethiopia, Kikuyu, South Africa Bantu, Tanzania, Yoruba) |
| CYP2D6*9 | 0 (All populations) | 0 (All populations) |
| CYP2D6*10 | 19 (South Africa Venda) | 0 (DRC, Kenya, South Africa Bantu, Zimbabwe Shona) |
| CYP2D6*17 | 34 (Shona) | 0 (DRC) |
| CYP2D6*29 | 29 (Igbo) | 2 (San) |
| CYP3A5*3 | 92 (Morocco) | 4 (Uganda) |
| CYP3A5*6 | 33 (North Sudan, Malawi Ngoni) | 4 (Morocco) |
| CYP3A5*7 | 23 (Gambia) | 0 (North Sudan, Ethiopia) |
Data are from Supplementary Table 1, Table 2.
CYP, cytochrome P450; DRC, Democratic Republic of Congo; MA, mixed ancestry.
Benin, Central African Republic, Democratic Republic of Congo, Ghana, Mozambique, Namibia, Senegal, South Africa.
Benin, Central African Republic, Democratic Republic of Congo, Ghana, Kenya (Luo), Namibia, Nigeria, Senegal, South Africa, Sudan, Tanzania.
African-American, Benin, Ghana, Hausa, Igbo, Kikuyu, Luo, Maasai, San, Shona, South Africa, (Venda, Xhosa), Yoruba.
Central African Republic, Democratic Republic of Congo, Ethiopia, Ghana, Kenya, Namibia, Yoruba, Senegal, South Africa (Bantu, Black, Venda).
3. Results
The initial literature search returned a total of 368 publications, 58 of which reported CYP frequencies in African populations. An additional 22 publications were obtained from the PubMed search (Fig. 1). CYP frequencies obtained for African populations are summarized in Supplemental Tables 1 and 2. Clinically relevant CYP substrates are detailed in Supplemental Tables 1 and 3.
Of the 74 papers cited as the basis of population-specific CYP allele frequencies in Supplemental Table 1 (Table 1), only 12 utilized genotyping methodologies capable of detecting novel sequence variations (Bains et al., 2013, Xiong et al., 2011, Chen et al., 2008, Klein et al., 2005, Park et al., 2008, Lee et al., 2006, Leathart et al., 1998, Gaedigk and Coetsee, 2008, Dickmann et al., 2001, Drögemöller et al., 2010, Wright et al., 2010, Dodgen et al., 2013). Of these, four were in Asian populations (two Chinese [Xiong et al., 2011, Chen et al., 2008]; two Korean [Park et al., 2008, Lee et al., 2006]) and eight involved Africans or African-Americans (Bains et al., 2013, Klein et al., 2005, Gaedigk and Coetsee, 2008, Leathart et al., 1998, Dickmann et al., 2001, Drögemöller et al., 2010, Wright et al., 2010, Dodgen et al., 2013).
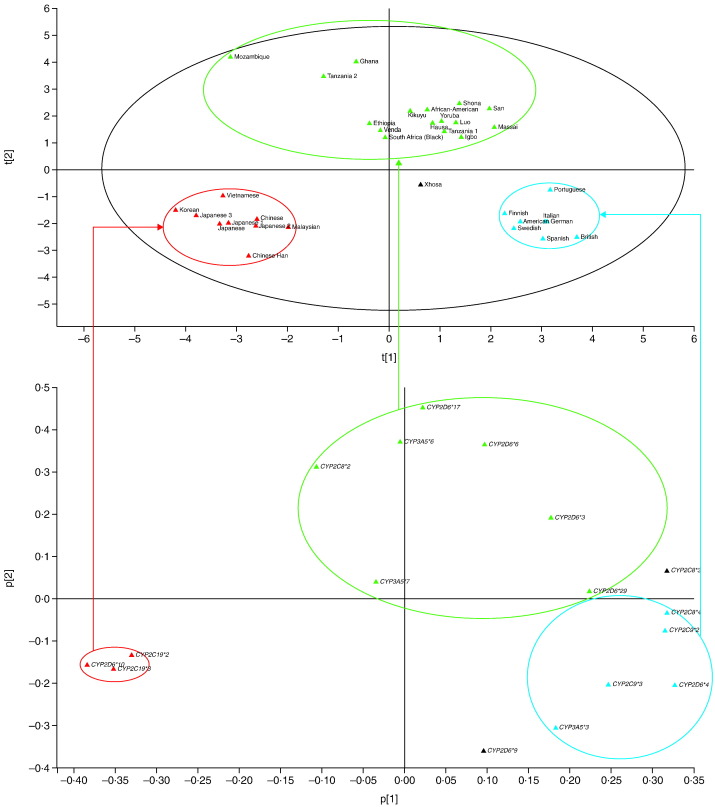
The analysis in SIMCA used three principal components; PC1 and PC2 described most of the variance in the data set and were therefore further analyzed. PCA demonstrated distinct clustering of African, Asian and Caucasian populations (Fig. 2), with the exception of Xhosa (South Africa; positioned outside of the three clusters). The lack of available CYP frequency data for admixed populations in South Africa prevented further analysis of this result. While Caucasian and Asian populations demonstrated tight clustering, a considerably greater variability in CYP frequencies was observed across Africa as indicated by the widespread distribution of the African populations on the PCA plot (Fig. 2). The following CYP variants showed a marked difference in distribution in African populations compared with other global populations: CYP2B6*6, CYP2C8*2, CYP2D6*3, CYP2D6*17, CYP2D6*29, CYP3A5*6 and CYP3A5*7 (Fig. 2).
Fig. 2.
Pharmacogenetics PCA cluster of global populations according to different CYP allele frequencies.
PCA analysis identifies X-variables (CYP variants) that most differentiate Y-objects (African/Asian/Caucasian populations). In this analysis, the software projects the populations into clusters based on shared characteristics with respect to the frequency and/or absence of the CYP variants analyzed. Green ovals indicate African populations. Blue ovals indicate Caucasian populations. Red ovals indicate Asian populations. Kikuyu, Luo and Maasai are regional populations of Kenya. Hausa, Igbo and Yoruba are regional populations of Nigeria. San and Shona are regional populations of Zimbabwe. Xhosa is a regional population of South Africa.
CYP, cytochrome P450; PCA, principal components analysis.
Our literature review also identified reports of polymorphisms in CYP2A6 (*17) and CYP2B6 (*33–*37) that have not been reported in populations outside of Africa (Supplemental Table 1, Table 2). In addition, African populations with the highest and lowest frequencies for certain CYP alleles are presented in Table 1. Of the 17 CYP variants selected for PCA, the greatest differences in reported frequencies were observed for CYP2B6*6 (highest reported frequency: Uganda, 69%; lowest reported frequency: South Africa mixed ancestry, 9%), CYP2C9*3 (Libya, 35%; Ghana, 0%), CYP2D6*17 (Shona, 34%; Democratic Republic of Congo, 0%) and CYP3A5*3 (Morocco, 92%; Uganda, 4%). Frequencies were very similar among African populations for CYP2D6*9 (0% in all populations investigated) and CYP2C8*4 (Tanzania, 0.6%; Ghana, 0%).
Table 2.
The 25 most common drugs (international non-proprietary names) reported in all drug adverse events in Africa.
| Drug | Total count of adverse events |
|---|---|
| Nevirapinea | 1195 |
| Efavirenza | 1099 |
| Sulfamethoxazole and trimethoprimb, c | 1068 |
| Lamivudine | 859 |
| Stavudine | 713 |
| Zidovudinea | 690 |
| Ribavirin | 682 |
| Diclofenaca | 679 |
| Lamivudine and zidovudineb | 634 |
| Ciprofloxacinc | 631 |
| Peginterferon alfa-2a | 623 |
| Tenofovir | 595 |
| Lamivudine, nevirapine, and zidovudineb | 565 |
| Ethambutol, isoniazid, pyrazinamide, and rifampicina, b, d |
548 |
| Carbamazepinea | 546 |
| Isoniazida | 523 |
| Amoxicillina | 512 |
| Insulin glargine | 499 |
| Paracetamola | 492 |
| Amodiaquine and artesunateb | 460 |
| Ceftriaxonea | 457 |
| Acetylsalicylic acida | 441 |
| Valproic acida | 439 |
| Docetaxela, d | 426 |
| Rifampicina, d | 423 |
Data are from VigiBase® (June 2016). VigiBase® is the World Health Organization Global Individual Case Safety Reports database, containing reports of adverse reactions received from member countries since 1968. Data in this table are from 33 countries in Africa (Angola, Benin, Botswana, Burkina Faso, Cameroon, Cape Verde, The Democratic Republic of the Congo, Côte d'Ivoire, Egypt, Eritrea, Ethiopia, Ghana, Guinea, Kenya, Liberia, Madagascar, Mali, Mauritius, Morocco, Mozambique, Namibia, Niger, Nigeria, Rwanda, Senegal, Sierra Leone, South Africa, United Republic of Tanzania, Togo, Tunisia, Uganda, Zambia and Zimbabwe). CYP, cytochrome P450.
Metabolized by CYP enzymes.
Combination therapies.
CYP inhibitor.
CYP inducer.
4. Discussion
Our study provides a comprehensive review and statistical analysis of CYP frequency data in global populations including Africa. We have demonstrated that genetic variation is greater in African populations than in Asian and Caucasian populations, and therefore that Africa cannot be treated as a single entity in drug research and development. PCA identified allelic variants showing a marked difference in distribution in African populations which may be of clinical significance and are discussed below. Moreover, our literature review identified reports of polymorphisms in CYP genes that have not been described in populations outside of Africa.
4.1. CYP2B6*6
CYP2B6 is involved in the metabolism of numerous therapeutic drugs including antiretrovirals (efavirenz, nevirapine), chemotherapeutics (cyclophosphamide), antidepressants (bupropion), and analgesics (ketamine, methadone; Flockhart Table™ http://medicine.iupui.edu/CLINPHARM/ddis/main-table accessed 28 October 2016). CYP2B6*6 is a decreased function allele and has been reported at very high frequencies of 66–68% in Uganda and Zimbabwe (Supplemental Table 1, Table 2), which is approximately tenfold higher than the reported frequency in South Africa (mixed ancestry; 9%; Supplemental Table 1, Table 2). Frequencies reported in Caucasian populations are considerably lower (21–29%; Supplemental Table 1, Table 1). Sub-Saharan Africa accounts for two-thirds of the global total of new HIV infections, with 25.6 million individuals living with HIV in 2015 (World Health Organization HIV/AIDS Fact Sheet http://www.who.int/mediacentre/factsheets/fs360/en/ accessed 28 October 2016). Many African patients who are prescribed antiretrovirals may be carriers of CYP2B6*6 and are therefore potentially at a greater risk of drug-induced adverse events than patients from other continents. Furthermore, nevirapine and efavirenz are the two most common drugs reported in all reports of drug-induced adverse events in Africa (Table 2; Ampadu et al., 2016). To reduce the frequency of adverse events reported for these (and potentially other) drugs, prospective genotyping of patients should be performed so that appropriate dose adjustments can be made, particularly given the very high frequencies of CYP2B6*6 (sometimes > 50%) observed in some African populations. In resource-limited regions where prospective genotyping may not be feasible, more frequent therapeutic monitoring should be performed in patients who are prescribed CYP2B6 substrates such as efavirenz or nevirapine, and a dose adjustment could be considered according to a patient's response.
4.2. CYP2C8*2
This variant has been reported at frequencies of 10–22% in populations from Uganda, Burkina Faso, Tanzania, Zanzibar, Madagascar, Mozambique, Ghana, and Senegal (Supplemental Table 1, Table 2), whereas lower frequencies (0–1.6%) have been reported in Asian and Caucasian populations (Supplemental Table 1, Table 1). CYP2C8 is involved in the metabolism of antimalarial drugs (e.g. amodiaquine; Li et al., 2002), the anti-cancer drug paclitaxel, and the anti-tuberculosis drug rifampicin (Flockhart Table™ http://medicine.iupui.edu/CLINPHARM/ddis/main-table accessed 28 October 2016). The *2 variant is a decreased function allele. In 275 individuals from Burkina Faso with malaria, a sixfold reduction in the intrinsic clearance of amodiaquine was observed in carriers of the mutant allele compared with individuals who were homozygous for the wild-type allele (Parikh et al., 2007). Additionally, our literature review identified a number of CYP2C8 variants for which frequency data were not available in many African populations (Supplemental Table 1, Table 1). Malaria is the disease with the highest health burden in central and western Africa (Alessandrini and Pepper, 2014); therefore, it is concerning that there are few reports of CYP2C8 profiling in these regions. This highlights the requirement for further data collection in African populations to identify those at a potentially increased risk of drug-induced adverse events or drug inefficacy.
4.3. CYP2D6*3, CYP2D6*17 and CYP2D6*29
CYP2D6 is involved in the metabolism of > 25% of registered drugs; the major classes are antidepressants, β-blockers, antipsychotics, and antiarrhythmics (Flockhart Table™ http://medicine.iupui.edu/CLINPHARM/ddis/main-table accessed 28 October 2016). *17 and *29 are decreased function alleles; *17 has been reported at a frequency of 34% in a Zimbabwean (Shona) population (Supplemental Table 1, Table 2), compared with very low frequencies (0–0.9%) in Asian and Caucasian populations (Supplemental Table 1, Table 1). *29 has been reported at a frequency of 20% in a Tanzanian population compared with a low frequency of 0% in Asian and Caucasian populations (Supplemental Table 1, Table 1). The CYP2D6*3 variant confers no enzyme activity and has been reported at frequencies of 0–9% in African populations (9% in a Zimbabwe San population) compared with frequencies of 0–3.5% in Asian and Caucasian populations (Supplemental Table 1, Table 1). As CYP2D6 is the main enzyme involved in converting the anti-cancer drug tamoxifen into its most potent anti-estrogenic metabolites, endoxifen and 4-hydroxytamoxifen, polymorphisms in the CYP2D6 gene may influence tamoxifen metabolism (Dean, 2016). High plasma levels of endoxifen require the presence of fully functional CYP2D6 alleles; in PMs endoxifen levels are decreased (Dean, 2016). The *3 non-functional allele, and the *17 (Masimirembwa et al., 1996) and *29 decreased function alleles of CYP2D6 may therefore potentially be associated with poor treatment outcomes for patients with breast cancer who are treated with tamoxifen in some African populations, but further clinical studies are required to establish if treatment outcomes are affected.
To date, 109 allelic variants of CYP2D6 have been identified (The human cytochrome P450 allele nomenclature database http://cypalleles.ki.se/ accessed 28 October 2016) and the phenotypic effects of many of these are well-characterized. Dosing guidelines have been developed by the Clinical Pharmacogenetics Implementation Consortium (CPIC) for many drug substrates of CYP2D6; for example codeine (Crews et al., 2012), fluvoxamine (Hicks et al., 2015), nortriptyline (Hicks et al., 2013, Hicks et al., 2016), and more recently, the antiemetic drugs ondansetron and tropisetron (Bell et al., 2016). Patients with an IM phenotype are recommended to receive a 25% reduction in dose, whereas those with PM or UM phenotypes are recommended to consider an alternative drug. CPIC guidelines and dosage guidelines published by the Royal Dutch Association for the Advancement of Pharmacy – Pharmacogenetics Working Group are freely available and can be accessed via The Pharmacogenomic Knowledgebase (PharmGKB; https://www.pharmgkb.org/view/dosing-guidelines.do?source=CPIC accessed 15 December 2016). These guidelines need to be publicized and practiced to a greater extent, especially in regions of Africa demonstrating high frequencies of the *3, *17 and *29 genotypes, or other clinically relevant CYP2D6 polymorphisms. The use of prospective genotyping to identify African patients with such genotypes would help to increase treatment efficacy and to reduce the risk of adverse events in response to drugs for which there are dosage guidelines available. In the absence of genotype data, close therapeutic drug monitoring should be carried out by clinicians prescribing drugs known to be affected by well-characterized CYP polymorphisms.
Genotype–phenotype discordance has been observed in some studies investigating CYP2D6 in African populations. In two South African populations (n = 96 and n = 98, both San Bushmen), 19% of individuals were identified as having a CYP2D6 PM phenotype when debrisoquine was used as the probe drug (Sommers et al., 1988), whereas only 4.1% were identified as having a PM phenotype when metoprolol was used (Sommers et al., 1989). A study in a Nigerian population reported a 9% prevalence of PM phenotypes of debrisoquine, whereas a later study reported no PMs of the same drug in the same study group (Mbanefo et al., 1980, Iyun et al., 1986). Such inconsistencies have not been observed in studies of Caucasian and Asian populations, in which the prevalence of PMs is reported to be 5–10% and 0–1%, respectively, regardless of the probe drug used (Mizutani, 2003). Factors contributing to genotype–phenotype discordance in African populations may include insufficiently large study populations, poor participant compliance, lack of comprehensive laboratory methods, effects of other metabolic enzymes, environmental factors (e.g. diet), and undiscovered allelic variants.
4.4. CYP3A5*6 and CYP3A5*7
CYP3A5 is involved in the metabolism of antimalarials, antiretrovirals, and immunosuppressants (Flockhart Table™ http://medicine.iupui.edu/CLINPHARM/ddis/main-table accessed 28 October 2016). The functional consequences of the *6 and *7 alleles on metabolic phenotype are not fully understood; hence the CPIC guideline for the immunosuppressant drug tacrolimus recommends therapeutic drug monitoring in patients with an IM phenotype (*1/*6 or *1/*7) or a PM phenotype (*6/*6 or *7/*7) (Birdwell et al., 2015). Considering the higher frequencies of the *6 and *7 alleles reported in Africa compared with other global populations (33% in Malawi and Sudan for *6; 22.9% in Gambia for *7; 0% in Caucasians for both; Supplemental Table 1, Table 2), further investigation into the effect of these polymorphisms on metabolic activity is required.
4.5. Proposed Actions to be Taken in Africa
Myrand et al. (2008) demonstrated that metabolic activity of polymorphic CYP enzymes is driven by genotype rather than by ethnicity, and that drug or metabolite concentration data from one region may therefore be extrapolated to other regions provided that a range of genotypes was investigated in the initial region; this would obviate the need for traditional ethnic sensitivity studies. However, the possible contribution to drug metabolism of genotypes that are unique to other regions must be considered before accepting this. 1000 Genomes Project data have shown that African-Americans have many rare pharmacogenetic variants that are not found in Europeans (Kozyra et al., 2016, Wright et al., 2016). Genome sequencing studies have indicated the difficulty of predicting allele frequencies in highly admixed populations (Wright et al., 2016). Furthermore, as the clinical utility of pharmacogenetic information continues to increase, it is likely that pharmacogenetic testing will need to replace colloquial categories of race and ethnicity in support of prescribing decision (Suarez-Kurtz et al., 2014).
Considering the great genetic variability across Africa, it is possible that there may be some as yet unidentified variations in genes coding for metabolizing enzymes that are unique to particular African subpopulations. Such undescribed, clinically relevant variants may also contribute to the high levels of drug-induced adverse events reported in Africa. Our literature review identified polymorphisms in CYP genes that to our knowledge have not been reported in populations outside of Africa: CYP2A6*17 (di Iulio et al., 2009), and CYP2B6*33–*37 (Radloff et al., 2013); Supplemental Table 1, Table 2). CYP2A6*17 has been investigated but not found in other global populations; however, the *33–*37 variants of CYP2B6 have been reported only in a Rwandese population and it is yet to be established if they are present in other global populations. Where novel, Africa-specific alleles have been discovered, there is a need for follow-up studies to identify their potential phenotypic effects. We also identified a number of countries or regions in Africa for which there were no available data on any CYP frequencies (Burundi, Cape Verde, Chad, Comoros, Djibouti, Equatorial Guinea, Eritrea, Lesotho, Mauritania, Mauritius, Niger, Somalia, Swaziland, Togo, Western Sahara and Zambia).
The majority of pharmacogenomic studies to date have either analyzed individual candidate genes or a subset of genetic variants using genotyping assays. Current sequencing technologies allow an assessment of the full spectrum of variation present in a given population, and the determination of the effect of rare genetic variation on genes of pharmacogenomic significance. Populations for which data are limited can now be studied on a scale not previously possible. Wright et al. (2016) highlight that sequencing technologies provide the best means for capturing rare variants, and that African populations have the highest number of polymorphic sites in their pharmacogenes. The intricate technological challenges associated with high-throughput sequencing that include highly repetitive DNA sequences, pseudo-pharmacogenes and detection of copy number variants, may be solved by massively-parallel sequencing methods (Ng et al., 2016). Massively parallel sequencing is a powerful engine for discovery of rare variants in pharmacogenes. In a study interrogating 127 variants in 13 pharmacogenes designated as clinically important by CPIC owing to their potential to alter prescribing decisions, genotypic concordance of exome and genome sequencing was 99.99% when using a clinically-validated pharmacogene microarray performed in a Clinical Laboratory Improvement Amendments–certified laboratory (Yang et al., 2016). It is therefore a reasonable first choice technology with which global populations including Africans may be screened for novel pharmacogenetic variants. Sole reliance on massively parallel, short-read sequencing may, however, prove insufficient despite the optimization of scientific protocols and the technology itself. It will be important to utilize long-read sequencing methods such as Pacific Biosystems (Mardis, 2017) to cross-validate and assess the sensitivity of short-read sequence systems. However, massively parallel sequencing technology's pharmacogenetic performance appears suitable as the first-line equipment for discovery of novel variants, and potentially, with further validation, clinical use within limits that will need to be clearly specified. Such technology has only become available in recent years, is costly and is not widely available in Africa. Therefore, to further facilitate clinical uptake of sequencing, it will be important to address the data analysis burden and to develop a variant interpretation system that includes drug response prediction.
4.5.1. Recommendations for Pharmacogenetic Profiling in Africa
Pharmacogenetic studies often focus on established clinically relevant polymorphisms, and individuals who do not carry at least one of the defined SNPs are identified as carriers of the wild-type genotype (Čolić et al., 2014). Consequently, new SNPs that may affect therapeutic drug metabolism are not routinely identified. Researchers and clinicians should be encouraged to interrogate existing databases containing reports of unexpected responses to treatment with a given drug in African patients, e.g. VigiBase® (Shankar, 2016), with a view to linking genotype data to the reported phenotypes. A registry of unexplained adverse events in African patients would be very valuable because it could allow further pharmacogenetic investigations to be undertaken, greatly facilitating the identification of clinically relevant events that could have pharmacogenetic underpinnings. Recent research demonstrates burgeoning interest and improving infrastructure for serious adverse drug event reporting in Africa, but pharmacogenetics has not yet been integrated into case investigations (Ampadu et al., 2016). Region-specific pharmacogenetic profiling of a list of prioritized CYP alleles in Africa, influenced by disease burden and the extent of available data for that region, could be carried out (Alessandrini and Pepper, 2014). The integration of existing data from resources such as the 1000 Genomes project and the exome sequencing project (Kozyra et al., 2016) into future studies may help to ensure adequate profiling of clinically relevant CYP alleles in all regions of Africa.
Few investigations of CYP variants have been interrogated for ethnic-specific novel variants by pharmacogene resequencing methods. The research focus to date has been mainly on surveys of the allele frequency distributions of known polymorphisms in various national and ethnic subpopulations. Similar to the findings of Gaedigk et al. (2016), only 12 of the 74 papers cited in Supplemental Table 1 (Table 1) utilized genotyping methodologies capable of detecting novel sequence variations. Small clinical trials are poorly powered to permit causal interference of the effect of rare variants on pharmacokinetic variation; large-scale pharmacogenetic epidemiological approaches will therefore be essential for creating a catalog of functionally important rare variants. Advances in methodology, combined with coordinated multi-national collaboration, promises to fill the knowledge gaps surrounding the significance of rare variants in CYP450.
Clinical studies performed in Africa are often limited by small study populations (which reduces the chance of identifying pharmacokinetic outliers), use of different probe drugs for a particular CYP across different studies, a lack of comprehensive genotyping, and a lack of reproducibility in data obtained from the same populations. The standardization of methods in terms of study population size and probe drug panels used will help to improve the quality and reproducibility of the data generated. Furthermore, the development of local policies will help to address issues such as participant recruitment and sample collection.
4.5.2. Funding and Training for Pharmacogenetic Research in Africa
Africa is a resource-limited setting. It is important that researchers in Africa are provided with the relevant training and facilities to conduct pharmacogenetic studies. Furthermore, the cultural diversity in Africa necessitates additional considerations relating to sample collection, participant understanding of informed consent, and community engagement in order to build trust and help participants to understand the research taking place (Wright et al., 2013). The Africa Genome Variation Project (AGVP; Gurdasani et al., 2015) and The Human Heredity and Health in Africa (H3 Africa) initiative (http://h3africa.org/ accessed 28 October 2016; Mulder et al., 2016) which has received funding from the Wellcome Trust and the US National Institutes of Health, have been established with the aims of developing local resources, improving training and research capacity, and increasing pan-African and global research collaborations. The AGVP has assessed genetic diversity among 1481 individuals from 18 ethnolinguistic groups in Sub-Saharan Africa to facilitate further genomic research and to provide a global resource for researchers (Gurdasani et al., 2015).
The pharmaceutical industry could become more involved in raising awareness of the genetic diversity in Africa and the need for more pharmacogenetic data, and could support local clinicians and researchers. We would further propose the establishment of a consortium of pharmaceutical companies that will collaborate with local (African) and global regulatory authorities, other relevant government groups, and expert academic and clinical centers to develop recommendations for the practical use of pharmacogenetics data to optimize the treatment of patients across Africa. The difference in frequency and distribution of some alleles may also influence the outcome of clinical trials and consequently the clinical development of drugs in Africa.
5. Conclusions
In summary, our review of the published literature on CYP polymorphisms has clearly demonstrated and confirmed that genetic variation is greater in African populations than in Asian and Caucasian populations. The African continent cannot, therefore, be treated as a single entity in drug research and development, nor can African-American populations be considered an adequate proxy for pharmacogenetic differences across Africa. By use of PCA, we identified allelic variants showing a marked difference in distribution in African populations; however, more research is required to confirm these distributions and to identify populations at a potentially greater risk of drug-induced adverse events or drug inefficacy. We also identified CYP alleles that may be unique to Africa or for which data in African populations are limited, and regions where further data need to be collected. Here, we focused on CYP alleles, but it is likely that polymorphisms in other enzymes and transporters contribute to the diversity in drug response observed in Africa. Furthermore, the potential effect of genetic variability on drug pharmacodynamics, and the subsequent effect on drug responses in Africa, were not addressed in this study. There are also challenges in translating research findings from the laboratory to the clinical environment. The involvement of clinicians in genomic research will facilitate this translation process, helping to ensure that patients are treated with efficacious doses of therapeutic drugs. The involvement of groups such as H3Africa, AGVP and the proposed consortium of pharmaceutical companies could help to implement these changes, with an aim of reducing the burden of disease and adverse drug reactions in Africa.
Funding
Funding for this review was provided by Novartis Pharma AG for the study design, literature searches, data collection, data analysis, data interpretation and editorial assistance for the development of the manuscript.
Conflicts of Interest
Iris Rajman is an employee of Novartis Institutes for Biomedical Research, Novartis, Basel, Switzerland. Laura Knapp is an employee of PharmaGenesis London, London, UK which has received funding from Novartis. Thomas Morgan is an employee of Novartis Institutes for Biomedical Research, Cambridge, MA, USA. Collen Masimirembwa is an employee of the African Institute of Biomedical Science and Technology, Harare, Zimbabwe.
Author Contributions
Iris Rajman: study design, data analysis, data interpretation and manuscript preparation.
Laura Knapp: literature search, data collection, data analysis, data interpretation, figures and manuscript preparation.
Thomas Morgan: literature search, data analysis, data interpretation, figures and manuscript preparation.
Collen Masimirembwa: study design, data analysis, data interpretation, figures and manuscript preparation.
Acknowledgments
The authors would like to thank Christiane Michele, Michael Petrin and Gopal Krishna Sharma (Novartis) for their assistance with pharmacovigilance data collection, and Jens Praestgaard (Novartis) for statistical and PCA assistance.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.02.017.
Appendix A. Supplementary Data
Supplemental Tables Tab 1: Frequency data for principal components analysis Tab 2: Frequencies of CYP450 allelic variants in different African populations Tab 3: Clinically relevant substrates of CYP450 enzymes Supplemental References.
References
- Alessandrini M., Pepper M.S. Priority pharmacogenetics for the African continent: focus on CYP450. Pharmacogenomics. 2014;15:385–400. doi: 10.2217/pgs.13.252. [DOI] [PubMed] [Google Scholar]
- Ampadu H.H., Hoekman J., De Bruin M.L., Pal S.N., Olsson S., Sartori D., Leufkens H.G., Dodoo A.N. Adverse drug reaction reporting in Africa and a comparison of individual case safety report characteristics between Africa and the rest of the world: analyses of spontaneous reports in VigiBase®. Drug Saf. 2016;39:335–345. doi: 10.1007/s40264-015-0387-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains R.K., Kovacevic M., Plaster C.A., Tarekegn A., Bekele E., Brandman N.N., Thomas M.G. Molecular diversity and population structure at the cytochrome p450 3A5 gene in Africa. BMC Genet. 2013;14:1–18. doi: 10.1186/1471-2156-14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G.C., Caudle K.E., Whirl-Carillo M., Gordon R.J., Hikino K., Prows C.A., Gaedigk A., Agundez J.A., Sadhasivam S., Klein T.E., Schwab M. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for the CYP2D6 genotype and use of ondansetron and tropisetron. Clin. Pharmacol. Ther. 2016 doi: 10.1002/cpt.598. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdwell K.A., Decker B., Barbarino J.M., Peterson J.F., Stein C.M., Sadee W., Wang D., Vinks A.A., He Y., Swen J.J., Leeder J.S., Van Schaik R., Thummel K.E., Klein T.E., Caudle K.E., Macphee I.A. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 genotype and tacrolimus dosing. Clin. Pharmacol. Ther. 2015;98 doi: 10.1002/cpt.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudle K.E., Dunnenberger H.M., Freimuth R.R., Peterson J.F., Burlison J.D., Whirl-Carillo M., Scott S.A., Rehm H.L., Williams M.S., Klein T.E., Relling M.V., Hoffman J.M. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC) Genet. Med. 2016 doi: 10.1038/gim.2016.87. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Qin S., Xie J., Tang J., Yang L., Shen W., Zhao X., He G., Feng G., He L., Xing Q. Genetic polymorphism analysis of CYP2C19 in Chinese Han populations from different geographical areas of mainland China. Pharmacogenomics. 2008;9:691–702. doi: 10.2217/14622416.9.6.691. [DOI] [PubMed] [Google Scholar]
- Čolić A., Alessandrini M., Pepper M.S. Pharmacogenetics of CYP2B6, CYP2A6 and UGT2B7 in HIV treatment in African populations: focus on efavirenz and nevirapine. Drug Metab. Rev. 2014;47:111–123. doi: 10.3109/03602532.2014.982864. [DOI] [PubMed] [Google Scholar]
- Crews K.R., Gaedigk A., Dunnenberger H.M., Klein T.E., Shen D.D., Callaghan J.T., Kharasch E.D., Skaar T.C. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin. Pharmacol. Ther. 2012;91:321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean L. Medical Genetics Summaries. National Center for Biotechnology Information (US); 2012; Bethesda (MD): 2016. Tamoxifen therapy and CYP2D6 genotype.https://www.ncbi.nlm.nih.gov/books/NBK61999/ Internet. Available from: [Google Scholar]
- Dickmann L.J., Rettie A.E., Kneller B.M., Kim R.B., Wood A.J.J., Stein C.M., Wilkinson G.R., Schwarz U.I. Identification and functional charactertization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol. Pharmacol. 2001;60:382–387. doi: 10.1124/mol.60.2.382. [DOI] [PubMed] [Google Scholar]
- di Iulio J., Fayet A., Arab-Alameddine M., Rotget M., Lubomirov R., Cavassini M., Furrer H., Günthard H.F., Colombo S., Csajka C., Eap C.B., Decosterd L.A., Telenti A., the Swiss HIV Cohort Study In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet. Genomics. 2009;19:300–309. doi: 10.1097/FPC.0b013e328328d577. [DOI] [PubMed] [Google Scholar]
- Dodgen T.M., Hochfeld W.E., Fickl H., Asfaha S.M., Durandt C., Rheeder P., Drögemöller B.I., Wright G.B., Warnich L., Labuschagne C.D.J., van Schalkwyk A., Gaedigk A., Pepper M.S. Introduction of the AmpliChip CYP450 test to a South African cohort: a platform comparitive prospective cohort study. BMC Med. Genet. 2013;14:20–35. doi: 10.1186/1471-2350-14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drögemöller B.I., Wright G.E., Niehaus D.J., Koen L., Malan S., Da Silva D.M., Hillerman-Rebello R., La Grange A.M., Venter M., Warnich L. Characterization of the genetic profile of CYP2C19 in two South African populations. Pharmacogenomics. 2010;11:1095–1103. doi: 10.2217/pgs.10.90. [DOI] [PubMed] [Google Scholar]
- Fujikara K., Ingelman-Sundberg M., Lauschke V.M. Genetic variation in the human cytochrome P450 supergene family. Pharmacogenet. Genomics. 2015;25:584–594. doi: 10.1097/FPC.0000000000000172. [DOI] [PubMed] [Google Scholar]
- Gaedigk A., Coetsee C. The CYP2D6 gene locus in South African Coloureds: unique allele distributions, novel alleles and gene arrangements. Eur. J. Clin. Pharmacol. 2008;64:465–475. doi: 10.1007/s00228-007-0445-7. [DOI] [PubMed] [Google Scholar]
- Gaedigk A., Sangkuhl K., Whirl-Carillo M., Klein T., Leeder J.S. Prediction of CYP2D6 phenotype from genotype across world populations. Genetics in Medicine. 2016;19:69–76. doi: 10.1038/gim.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdasani D., Carstensen T., Tekola-Ayele F., Pagani L., Tachmazidou I., Hatzikotoulas K., Karthikeyan S., Iles L., Pollard M.O., Choudury A., Ritchie G.R., Xue Y., Asimit J., Nsubuga R.N., Young E.H., Pomilla C., Kivenen K., Rockett K., Kamali A., Doumatey A.P., Asiki G., Seeley J., Sisay-Joof F., Jallow M., Tollman S., Meknonnen E., Ekong R., Oljira T., Bradman N., Bojang K., Ramsay M., Adeyemo A., Bekele E., Motala A., Norris S.A., Pirie F., Kaleebu P., Kwiatkowski D., Tyler-Smith C., Rotimi C., Zeggini E., Sandhu M.S. The African Genome Variation Project shapes medical genetics in Africa. Nature. 2015;517:327–332. doi: 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J.K., Bishop J.R., Sangkuhl K., Ji Y., Leckband S.G., Leeder J.S., Graham R.L., Chiulli D.L., Llerena A., Skaar T.C., Scott S.A., Stingl J.C., Klein T.E., Caudle K.E., Gaedigk A. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 2015;98:127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J.K., Sangkuhl K., Swen J.J., Ellingrod V.L., Müller D.J., Shimoda K., Bishop J.R., Kharasch E.D., Skaar T.C., Gaedigk A., Dunnenberger H.M., Klein T.E., Caudle K.E., Stingl J.C. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC®) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin. Pharmacol. Ther. 2016 doi: 10.1002/cpt.597. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J.K., Swen J.J., Thorn C.F., Sangkuhl K., Kharasch E.D., Ellingrod V.L., Skaar T.C., Müller D.J., Gaedigk A., Stingl J.C. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin. Pharmacol. Ther. 2013;93:402–408. doi: 10.1038/clpt.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingeman-Sundberg M., Sim S.C., Gomez A., Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Iyun A.O., Lennard M.S., Tucker G.T., Woods H.F. Metoprolol and debrisoquin metabolism in Nigerians: lack of evidence for polymorphic oxidation. Clin. Pharmacol. Ther. 1986;40:389–394. doi: 10.1038/clpt.1986.195. [DOI] [PubMed] [Google Scholar]
- Klein K., Lang T., Saussele T., Barbosa-Sicard E., Schunck W.H., Eichelbaum M., Schwab M., Zanger U.M. Genetic variability of CYP2B6 in populations of African and Asian origin: allele frequencies, novel functional variants, and possible implications for anti-HIV therapy with efavirenz. Pharmacogenet. Genomics. 2005;15:861–873. doi: 10.1097/01213011-200512000-00004. [DOI] [PubMed] [Google Scholar]
- Kozyra M., Ingeman-Sundberg M., Lauschke V.M. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet. Med. 2016 doi: 10.1038/gim.2016.33. [DOI] [PubMed] [Google Scholar]
- Leathart J.B., London S.J., Steward A., Adams J.D., Idle J.R., Daly A.K. CYP2D6 phenotype-genotype relationships in African-Americans and Caucasians in Los Angeles. Pharmacogenetics. 1998;8:529–541. doi: 10.1097/00008571-199812000-00010. [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Sohn K.M., Ryu J.Y., Yoon Y.R., Shin J.G., Kim J.W. Sequence-based CYP2D6 genotyping in the Korean population. Ther. Drug Monit. 2006;28:382–387. doi: 10.1097/01.ftd.0000211823.80854.db. [DOI] [PubMed] [Google Scholar]
- Li X.Q., Bjorkman A., Andersson T.B., Ridderstrom M., Masimirembwa C.M. Amodiaquine clearance and its metabolism to N-desethylamodiaquine is mediated by CYP2C8: a new high affinity and turnover enzyme-specific probe substrate. J. Pharmacol. Exp. Ther. 2002;300:399–407. doi: 10.1124/jpet.300.2.399. [DOI] [PubMed] [Google Scholar]
- Mardis E.R. DNA sequencing technologies: 2006–2016. Nat. Protoc. 2017;12:213–218. doi: 10.1038/nprot.2016.182. [DOI] [PubMed] [Google Scholar]
- Masimirembwa C., Persson I., Bertilsson L., Hasler J., Ingeman-Sundberg M. A novel mutant variant of the CYP2D6 gene (CYP2D6*17) common in a black African population: association with diminished debrisoquine hydroxylase activity. Br. J. Clin. Pharmacol. 1996;42:713–719. doi: 10.1046/j.1365-2125.1996.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbanefo C., Bababunmi E.A., Mahgoub A., Sloan T.P., Idle J.R., Smith R.L. A study of the debrisoquine hydroxylation polymorphism in a Nigerian population. Xenobiotica. 1980;10:811–818. doi: 10.3109/00498258009033811. [DOI] [PubMed] [Google Scholar]
- Mizutani T. PM frequencies of major CYPs in Asians and Caucasians. Drug Metab. Rev. 2003;35:99–106. doi: 10.1081/dmr-120023681. [DOI] [PubMed] [Google Scholar]
- Mulder N.J., Adebiyi E., Alami R., Benkahla A., Brandful J., Doumbia S., Everett D., Fadlelmola F.M., Gaboun F., Gaseitsiwe S., Ghazal H., Hazelhurst S., Hide W., Ibrahimi A., Fakim Y.J., Jongeneel V.C., Joubert F., Kassim S., Kayondo J., Kumuthini J., Lyantagaye S., Makani J., Alzohairy A.M., Masiga D., Moussa A., Nash O., Oukem-Boyer O.O.M., Owusu-Dabo E., Panji S., Patterton H., Radouani F., Sadki K., Seghrouchni F., Bishop O.T., Tiffin N., Ulenga N., The H3ABioNet Consortium H3ABioNet, a sustainable, pan-African bioinformatics network for human heredity and health in Africa. Genome Res. 2016;26:271–277. doi: 10.1101/gr.196295.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C.J., Vos T., Lozano R., Naghavi M., Flaxman A.D., Michaud C., Ezzati M., Shibuya K., Salomon J.A., Abdalla S., Aboyans V., Abraham J., Ackerman I., Aggarwal R., Ahn S.Y., Ali M.K., Alvarado M., Anderson H.R., Anderson L.M., Andrews K.G., Atkinson C., Baddour L.M., Bahalim A.N., Barker-Collo S., Barrero L.H., Bartels D.H., Basanez M.G., Baxter A., Bell M.L., Menjamin E.J., Bennett D., Brnabe E., Bhalla K., Bhandari B., Bikbov B., Bin Abdulhak A., Birbeck G., Black J.A., Blencowe H., Blore J.D., Blyth F., Bolliger I., Bonaventure A., Boufous S., Bourne R., Boussinesq M., Braithwaite T., Brayne C., Bridgett L., Brooker S., Brooks P., Brugha T.S., Bryan-Hancock C., Bucello C., Buchbinder R., Buckle G., Budke C.M., Burch M., Burney P., Burstein R., Calabria B., Campbell B., Canter C.E., Carabin H., Carapetis J., Carmona L., Cella C., Charlson F., Chen H., Cheng A.T., Chou D., Chugh S.S., Coffeng L.E., Colan S.D., Colquhoun S., Colson K.E., Condon J., Connor M.D., Cooper L.I., Corriere M., Cortinovis M., de Vaccaro K.C., Couser W., Cowie B.C., Criqui M.M., Cross M., Dabhadkar K.C., Dahiya M., Dahodwala N., Damsere-Derry J., Danaei G., Davis A., de Leo D., Degenhardt L., Dellavalle R., Delossantos A., Denenberg J., Derrett S., Des Jarlais D.C., Dharmaratne S.D. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- Myrand S.P., Sekiguchi K., Man M.Z., Lin X., Tzeng R.Y., Teng C.H., Hee B., Garrett M., Kikkawa H., Lin C.Y., Eddy S.M., Dostalik J., Mount J., Azuma J., Fujio Y., Jang I.J., Shin S.G., Bleavins M.R., Williams J.A., Paulauskis J.D., Wilner K.D. Pharmacokinetics/genotype associations for major cytochrome P450 enzymes in native and first- and third-generation Japanese populations: comparison with Korean, Chinese, and Caucasian populations. Clin. Pharmacol. Ther. 2008;84:347–361. doi: 10.1038/sj.clpt.6100482. [DOI] [PubMed] [Google Scholar]
- Ng D., Hong C.S., Singh L.N., Johnston J.J., Mullikin J.C., Biesecker L.G. Assessing the capability of massively parallel sequencing for opportunistic pharmacogenetic screening. Genet. Med. 2016 doi: 10.1038/gim.2016.105. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S., Ouedaogo J.B., Goldstein J.A., Rosenthal P.J., Kroetz D.L. Amodiaquine metabolism is impaired by common polymorphisms in CYP2C8: implications for malaria treatment in Africa. Clin. Pharmacol. Ther. 2007;82:197–203. doi: 10.1038/sj.clpt.6100122. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Kang Y.S., Jeong M.S., Yoon H.K., Han K.O. Frequencies of CYP3A5 genotypes and haplotypes in a Korean population. J. Clin. Pharm. Ther. 2008;33:61–65. doi: 10.1111/j.1365-2710.2008.00879.x. [DOI] [PubMed] [Google Scholar]
- Prentis R.A., Lis Y., Walker S.R. Pharmaceutical innovation by the seven UK-owned pharmaceutical companies (1964–1985) Br. J. Clin. Pharmacol. 1988;25:387–396. doi: 10.1111/j.1365-2125.1988.tb03318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Gras A., Zanger U.M., Masquelier C., Arumugam K., Karasi J.C., Arendt V., Seguin-Devaux C., Klein K. Novel CYP2B6 enzyme variants in a Rwandese population: functional characterization and assessment of in silico prediction tools. Hum. Mutat. 2013;34:725–734. doi: 10.1002/humu.22295. [DOI] [PubMed] [Google Scholar]
- Shankar P.R. VigiAccess: promoting public access to VigiBase. Indian J. Pharmacol. 2016;48:606–607. doi: 10.4103/0253-7613.190766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommers D.K., Moncrieff J., Avenant J. Polymorphism of the 4-hydroxylation of debrisoquine in the San Bushmen of southern Africa. Hum. Toxicol. 1988;7:273–276. doi: 10.1177/096032718800700308. [DOI] [PubMed] [Google Scholar]
- Sommers D.K., Moncrieff J., Avenant J. Metoprolol alpha-hydroxylation polymorphism in the San Bushmen of southern Africa. Hum. Toxicol. 1989;8:39–43. doi: 10.1177/096032718900800107. [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G., Paula D.P., Struchiner C.J. Pharmacogenomic implications of population admixture: Brazil as a model case. Pharmacogenomics. 2014;15:209–219. doi: 10.2217/pgs.13.238. [DOI] [PubMed] [Google Scholar]
- Tishkoff S.A., Reed F.A., Friedlaender F.R., Ehret C., Ranciaro A., Froment A., Hirbo J.B., Awomoyi A.A., Bodo J.M., Doumbo O., Ibrahim M., Juma A.T., Kotze M.J., Lema G., Moore J.H., Mortenesen H., Nyambo T.B., Omar S.A., Powell K., Pretorius G.S., Smith M.W., Thera M.A., Wambebe C., Weber J.L., Williams S.M. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G.E.B., Niehaus D.J.H., Drögemöller B.I., Koen L., Gaedigk A., Warnich L. Elucidation of CYP2D6 genetic diversity in a unique African populations: implications for the future application of pharmacogenetics in the Xhosa population. Ann. Hum. Genet. 2010;74:340–350. doi: 10.1111/j.1469-1809.2010.00585.x. [DOI] [PubMed] [Google Scholar]
- Wright G.E., Koornhof P.G., Adeyemo A.A., Tiffin N. Ethical and legal implications of whole genome and whole exome sequencing in African populations. BMC Med. Ethics. 2013;14:21–36. doi: 10.1186/1472-6939-14-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G.E.B., Carleton B., Hayden M.R., Ross C.J.D. The global spectrum of protein-coding pharmacogenomic diversity. Pharmacogenomics J. 2016 doi: 10.1038/tpj.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Wang M., Fang K., Xing Q., Feng G., Shen L., He L., Qin S. A systematic genetic polymorphism analysis of the CYP2C9 gene in four different geographical Han populations in mainland China. Genomics. 2011;97:227–281. doi: 10.1016/j.ygeno.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Yang W., Wu G., Broeckel U., Smith C.A., Turner V., Haidar C.E., Wang S., Carter R., Karol S.E., Neale G., Crews K.R., Yang J.J., Mullighan C.G., Downing J.R., Evans W.E., Relling M.V. Comparison of genome sequencing and clinical genotyping for pharmacogenes. Clin. Pharmacol. Ther. 2016;100:360–368. doi: 10.1002/cpt.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Tables Tab 1: Frequency data for principal components analysis Tab 2: Frequencies of CYP450 allelic variants in different African populations Tab 3: Clinically relevant substrates of CYP450 enzymes Supplemental References.