Abstract
Head and neck squamous cell carcinoma (HNSCC) is broadly classified into HNSCC associated with human papilloma virus (HPV) infection, and HPV negative HNSCC, which is typically smoking-related. A subset of HPV negative HNSCCs occur in patients without smoking history, however, and these etiologically ‘atypical’ HNSCCs disproportionately occur in the oral cavity, and in female patients, suggesting a distinct etiology.
To investigate the determinants of clinical and molecular heterogeneity, we performed unsupervised clustering to classify 528 HNSCC patients from The Cancer Genome Atlas (TCGA) into putative intrinsic subtypes based on their profiles of epigenetically (DNA methylation) deregulated genes.
HNSCCs clustered into five subtypes, including one HPV positive subtype, two smoking-related subtypes, and two atypical subtypes. One atypical subtype was particularly genomically stable, but featured widespread gene silencing associated with the ‘CpG island methylator phenotype’ (CIMP).
Further distinguishing features of this ‘CIMP-Atypical’ subtype include an antiviral gene expression profile associated with pro-inflammatory M1 macrophages and CD8+ T cell infiltration, CASP8 mutations, and a well-differentiated state corresponding to normal SOX2 copy number and SOX2OT hypermethylation. We developed a gene expression classifier for the CIMP-Atypical subtype that could classify atypical disease features in two independent patient cohorts, demonstrating the reproducibility of this subtype. Taken together, these findings provide unprecedented evidence that atypical HNSCC is molecularly distinct, and postulates the CIMP-Atypical subtype as a distinct clinical entity that may be caused by chronic inflammation.
Keywords: Head and neck squamous cell carcinoma, etiological subtypes, CpG island methylator phenotype, Multi-omics data analysis, antiviral gene signature
Highlights
-
•
We identified five etiologically distinct DNA methylation subtypes of head and neck squamous cell carcinoma.
-
•
One subtype appears not to be associated with smoking or HPV, and may represent a distinct etiological entity.
-
•
Distinctive molecularly features of this subtype include CIMP, CASP8 mutations, and antiviral immune response.
To identify factors that define clinical and biological variability in head and neck squamous cell carcinoma (HNSCC), we clustered patients into subtypes based on their epigenetic profiles, revealing five subtypes, including previously identified subtypes. We focus on discovery of a subtype that matches the clinical characteristics previously ascribed to ‘atypical’ HNSCC, i.e., HNSCC that is not caused by the classic HNSCC risk factors of smoking or HPV. This subtype is biologically distinct across multiple molecular data types, and was reproducible in independent patient populations. We postulate that this ‘CIMP-Atypical’ subtype represents a clinically distinct HNSCC subtype of unknown cause.
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer by incidence (Siegel et al., 2016), and a leading cause of cancer-related death (Belcher et al., 2014). HNSCC displays substantial variability in prognosis and response to standard therapies (Belcher et al., 2014), which may reflect underlying etiological and molecular heterogeneity. For example, it is now well-understood that HNSCCs caused by high risk strains of human papilloma virus (HPV) are molecularly distinct from HPV negative (HPV −) HNSCCs and that they have better prognosis and therapy response (Wise-Draper et al., 2012, Kimple et al., 2013). Smoking is the major risk factor for HPV − HNSCC, causing genetic mutations in tumor suppressor genes including TP53 (DeMarini, 2004, Vettore et al., 2015). Alcohol consumption is a modest risk factor for HNSCC and is thought to increase risk particularly in smokers (Dal Maso et al., 2016). However, there is an increasingly recognized incidence of HPV − HNSCCs in individuals with no smoking history, indicating that etiological factors other than smoking and HPV exist (Heaton et al., 2014, Harris et al., 2011, Chaturvedi et al., 2013, Toner and O'Regan, 2009, MacKenzie et al., 2000, Patel et al., 2011, Koch et al., 1999, Koo et al., 2013, Brown et al., 2012, Montero et al., 2012, Perry et al., 2015, Toporcov et al., 2015). HNSCCs lacking these classic risk factors, referred to as ‘atypical’ HNSCCs, are usually oral squamous cell carcinomas (OSCCs), occur with higher relative frequency in women (particularly women of low socioeconomic status (Conway et al., 2008, Conway et al., 2015)) than smoking-related HNSCCs, and may be increasing in incidence (Chaturvedi et al., 2013, Patel et al., 2011, Koch et al., 1999, Koo et al., 2013, Katzel et al., 2015). While molecular differences between HPV positive (HPV +) and HPV − HNSCC have been described in recent years (Lleras et al., 2013, Fertig et al., 2013, Seiwert et al., 2015), molecular attributes of smoking-related, atypical, or other etiological HNSCC subgroups have not been established.
Unsupervised clustering of molecular data, such as gene expression or DNA methylation, provides a method of classifying intrinsic subtypes within cancer populations (Heiser et al., 2012, Hoadley et al., 2014). Identification of cancer subtypes has provided insight into the etiological factors underlying molecular and clinical heterogeneity in other cancers and has provided clinical biomarkers to predict prognosis and subtype-specific therapeutic response (Heiser et al., 2012, Hoadley et al., 2014, Marisa et al., 2013). Four HNSCC subtypes have been identified by clustering of gene expression data (Chung et al., 2004, Keck et al., 2015, Lawrence et al., 2015, Walter et al., 2013). We have previously reported our identification of five HNSCC subtypes based on clustering of integrated DNA methylation and gene expression data from 310 HNSCCs from The Cancer Genome Atlas (TCGA) study (Gevaert et al., 2015).
DNA methylation, i.e., the covalent addition of methyl groups to CpG dinucleotides to form 5-methylcytosine (5mC), is the best-known epigenetic mechanism of transcriptional regulation, and is widely altered in virtually all cancers, as an early and potentially causative event (Jones and Baylin, 2002, Fernandez et al., 2012). Typical patterns of abnormal methylation in cancer include silencing of tumor suppressor genes by aberrant methylation (hypermethylation) of gene promoters, particularly at promoter CpG islands, as well as general loss of DNA methylation overall (hypomethylation), potentially resulting in genomic instability and reactivation of oncogenes (Jones and Baylin, 2002, Jones, 2012).
DNA methylation patterns are altered by smoking (Shenker et al., 2013, Massion et al., 2008), HPV (Lleras et al., 2013) and age (Xu and Taylor, 2014), and may therefore capture important information about etiological drivers of HNSCC. Moreover, cancer molecular subtypes tend to differ depending on the molecular analyte (such as DNA methylation and gene expression) used for clustering (Heiser et al., 2012). Therefore, we have investigated the clinical, etiological, and molecular attributes of DNA methylation HNSCC subtypes in the complete set of TCGA HNSCC patients (n = 528), in order to gain insight into the factors that drive intertumoral heterogeneity.
We have reproduced five DNA methylation subtypes, which differ from the reported gene expression subtypes, and which more clearly segregate with etiological subgroups defined by HPV status and smoking. As most research into molecular heterogeneity has focused on differences between HPV + and HPV − HNSCC, we have focused primarily on heterogeneity within HPV − HNSCC. Most importantly, we identified two atypical HNSCC subtypes, including a molecularly distinct subtype that is reproducible in additional data sets, providing molecular classification for atypical HNSCC.
2. Methods
2.1. Data processing
Preprocessed TCGA DNA methylation data (generated using the Illumina Infinium HumanMethylation450 BeadChip array), gene expression data (generated by RNA sequencing), somatic point mutation data (generated using genome sequencing), DNA copy number data (generated by microarray technology) and clinical data were downloaded using the Firehose pipeline (Samur, 2014). Preprocessing for these data sets was done according to the Firehose TCGA pipelines described elsewhere (Samur, 2014).
Mutation data was accessed as Mutation Analysis reports, generated using MutSig CV v2.0 (Lawrence et al., 2013). For analysis of individual significantly mutated genes (e.g., CASP8, NSD1), mutations predicted as silent by MutSig CV were removed. For analysis of smoking mutation signatures, G > T & C > A transversions in all genes, including silent mutations, were included. DNA copy number data was accessed as Copy Number reports, generated using GISTIC 2.0 (Mermel et al., 2011).
Additional data preprocessing of gene expression and DNA methylation data was done as follows: Genes and patients with > 10% missing values for gene expression, and > 20% missing values for DNA methylation, were removed. All remaining missing values were estimated using KNN impute (Troyanskaya et al., 2001). TCGA data were generated in batches, creating a batch effect for most data sets. Batch correction was done using Combat (Johnson et al., 2007).
TCGA assignments for gene expression subtypes for 279 HNSCCs were derived from Stransky et al. (2011). Assignments for HPV and other viral infection status for 304 HNSCCs was derived from Tang et al. (2013), a recent study in which TCGA cancers were screened for expression of viruses in RNA-Seq data. We compared these HPV status assignments with those derived using in situ hybridization and P16 staining for smaller numbers of samples in standard TCGA clinical data: While HPV status determined by RNA-Seq analysis was consistent with HPV status defined by in situ hybridization, as reported (Tang et al., 2013), three HNSCCs that were found HPV − based on RNA-Seq detection were found HPV + based on p16 staining. However, p16 staining is an indirect method of HPV detection, and is considered less accurate than measurement of HPV RNA expression (Mirghani et al., 2015), therefore RNA-Seq analysis was used as a primary measure of HPV status in our analysis.
Oral squamous cell carcinomas were defines as cancers of the alveolar ridge, buccal mucosa, floor of mouth, hard palate, oral cavity, oral tongue, and oropharynx.
2.2. Clustering of DNA methylation data
Methylation of neighboring CpG sites tends to be highly correlated. To reduce multiple testing of highly correlated CpG probes, and to reduce the dimensionality of the methylation array data, probes for each gene were clustered using hierarchical clustering with complete linkage, and average methylation was calculated for each CpG cluster.
2.3. Classification of abnormally methylated genes
MethylMix was applied to CpG cluster data to systematically identify CpG clusters (referred to as ‘MethylMix genes’) that are abnormally methylated in cancer versus normal tissue, where DNA methylation is inversely associated with RNA expression of the same gene, using beta-mixture models, as previously described (Gevaert, 2015). For each MethylMix gene, MethylMix ascribes either normal or abnormal (hypomethylated or hypermethylated) DNA methylation states to each patient, providing lists of hypermethylated and hypomethylated genes for each cancer.
2.4. Consensus clustering of MethylMix driver genes
Unsupervised Consensus clustering was applied to MethylMix gene DNA methylation state data for 528 HNSCCs, to identify robust patient clusters (Putative subtypes). Consensus clustering was performed using the ConsensusClusterPlus R package (Wilkerson and Hayes, 2010), using 1000 rounds of k-means clustering, with a maximum of k = 10 clusters. Selection of the best number of clusters was based on visual inspection of plots provided in the ConsensusClusterPlus output.
2.5. Survival analysis
To test for overall survival differences between the MethylMix subtypes, the chi-square statistic test for equality was used to compare survival curves for each subtype. Survival data was censored at five years, to exclude deaths that were not HNSCC-related.
2.6. Application of gene expression signatures
The xenobiotic metabolism gene signature included 95 human genes annotated to the term ‘xenobiotic metabolic process’ (GO:0006805), derived from the AmiGO web application (Carbon et al., 2009). Wilcoxon rank sum tests were used to test for differences in mean expression of these genes between MethylMix subtypes.
2.7. Development of a gene expression classifier to predict the CIMP-Atypical subtype
Prediction Analysis of Microarrays (PAM) was used to develop a gene expression classified to predict the CIMP-Atypical subtype, as previously described (Tibshirani et al., 2002). PAM analysis uses a nearest shrunken centroids machine learning method that predicts the class (MethylMix subtype) of an individual based on the squared distance of the gene expression profile for that individual to the centroids of known CIMP-Atypical and other subtype patient groups. Shrinkage is used to select the optimum number of genes for class prediction, such that the model selects only a subset of genes to develop the centroids. We first used PAM in combination with 10-fold cross validation to determine the ability of the gene expression data to predict the CIMP-Atypical subtype within TCGA data. For each fold of cross validation, the PAM model was trained on 90% of patients and assigned class probability for belonging to the CIMP-Atypical subtype to the each of the remaining 10% of patients based on the distance of the patient to its closest centroid. We used the area under the ROC curve (AUC) to evaluate the performance of the model in accurately predicting the class of samples.
We applied this gene expression classifier signature to two independent gene expression data sets to classify them into either a ‘Predicted CIMP-Atypical’ group or a ‘Not predicted CIMP-Atypical’ group, using gene expression data for the top 25% most varying genes (i.e., with the highest mean absolute deviation). These included GSE65858 (Wichmann et al., 2015) which included 253 primary HNSCCs, with gene expression measured using the Illumina HumanHT-12 V4.0 expression beadchip, and GSE39366 (Walter et al., 2013), which included 139 primary HNSCCs, with gene expression measured using an Agilent-UNC-custom-4X44K array. Normalized gene expression data for these data sets was accessed. To classify a new sample, its distance is calculated to each of the centroids by using the weights as an inner product, and the sample is classified to its closest centroid. We only used classification results when probabilities were > 60% or < 40%, excluding low confidence assignments for the remaining borderline individuals from analyses.
2.8. Inference of tumor associated leukocyte levels
CIBERSORT was applied to gene expression (RNA-Seq) data to infer the levels of specific TAL types, as previously described (Newman et al., 2015, Gentles et al., 2015b). Only patients for with estimation p-values < 0.05 (Indicating high confidence TAL estimation) were included in downstream analyses.
2.9. Identification of genes associated with the CIMP-Atypical subtype
SAM analysis (Tusher et al., 2001) was used to identify genes that were overexpressed and underexpressed in the CIMP-Atypical subtype relative to other subtypes (combined).
2.10. Functional gene set enrichment analysis
Functional gene set enrichment analysis (GSEA) was carried out using MSigDB (Subramanian et al., 2005), selecting all 18,026 gene-set libraries for comparison with the input prognostic gene-set. The top 100 most enriched gene sets were visualized as a network map generated using Enrichment Map as a plugin for Cytoscape (Isserlin et al., 2014).
3. Results
3.1. Unsupervised clustering of HNSCC patients
To investigate possible sources of clinical and biological heterogeneity in HNSCC, we subtyped 528 HNSCC cases according to their profiles of epigenetically deregulated genes. We applied MethylMix (Gevaert, 2015) to whole genome DNA methylation and gene expression data, to identify abnormally methylated genes in HNSCC, i.e., genes that are hypermethylated or hypomethylated in all or a subgroup of HNSCCs relative to normal adjacent tissue and where this methylation is associated with altered RNA expression. MethylMix identified 2227 differentially methylated ‘MethylMix genes’ (Supplementary Table 1, see methods for definition of MethylMix genes). MethylMix assigned each cancer to a categorical DNA methylation class (hypermethylated, hypomethylated or normal-like) for each gene, with differential methylation (DM) values indicating the direction and mean difference in methylation between cancer and normal tissue. We then performed Consensus clustering to DM value data to cluster HNSCCs into clusters or putative intrinsic subtypes.
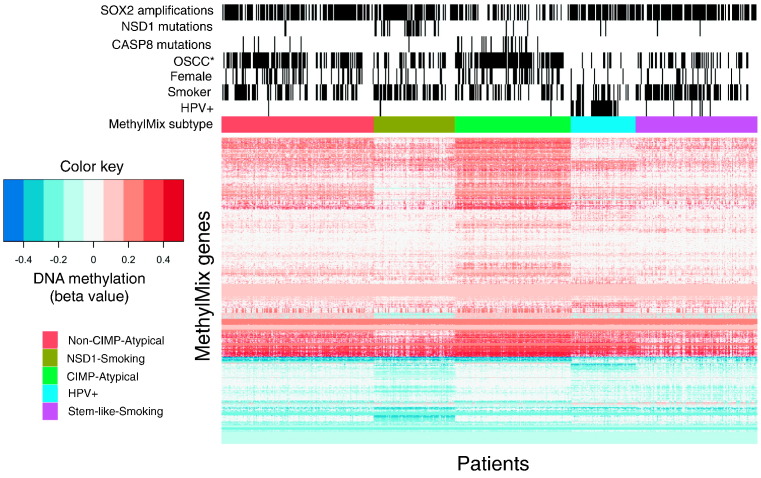
To identify the optimum number of HNSCC clusters, consensus clustering was performed iteratively with incremental numbers of clusters, and the cluster number at which consensus was strongest was chosen. Consistent with our previous report (Gevaert et al., 2015), greatest consensus was achieved for 5 clusters (Supplementary Fig. 1). These clusters were considered putative subtypes, including one HPV + subtype (subtype 4), and four HPV − subtypes (Fig. 1, Supplementary Table 2). We named these subtypes the ‘Non-CIMP-Atypical’, ‘NSD1-Smoking’, ‘CIMP-Atypical’, ‘HPV +’, and ‘Stem-like-Smoking’ subtypes, according to their most distinctive clinical and molecular attributes.
Fig. 1.
Heatmap indicating differential methylation and distribution of key etiological and molecular factors between MethylMix subtypes. MethylMix subtypes identified by consensus clustering of abnormally methylated genes, identified using MethylMix (Gevaert, 2015). ‘Smokers’ refers to current or reformed former smokers (< 15 years). OSCC: oral squamous cell carcinoma.
3.2. MethylMix subtypes differed from TCGA gene expression subtypes
Four gene expression subtypes have been identified for HNSCC, each associated with distinct etiological HNSCC features (Chung et al., 2004, Keck et al., 2015, Lawrence et al., 2015, Walter et al., 2013). We assessed the distribution of these expression subtypes within our MethylMix subtypes, based on assignments by the TCGA study for 279 cancers (Lawrence et al., 2015) (Supplementary Fig. 2). While our HPV + subtype was comprised almost entirely of the Lawrence et al. designated HPV-related ‘Atypical’ (AT) gene expression subtype, all other MethylMix subtypes were comprised of mixtures of multiple gene expression subtypes. Notably, the ‘Mesenchymal’ (ME) expression subtype distributed almost evenly between four MethylMix subtypes. This indicates that integrated DNA methylation and expression subtypes capture unique sources of molecular heterogeneity.
3.3. Identification of a single HPV positive subtype
A recent study inferred infection status for HPV and other human viruses by measuring expression of viral transcripts in tumor RNA-Seq data (Tang et al., 2013). We utilized this, the most complete data source in terms of patient numbers, as a measure of HPV status. 97% of patients within the HPV + MethylMix subtype were positive for either HPV16 (89% (32/36)) or HPV33 (8% (3/36)), compared with 0–7% of other subtypes (Fig. 1, Supplementary Fig. 3, Table 1). The HPV + subtype was enriched for classic features of HPV + HNSCC, including enrichment for base of tongue and tonsil cancer (Supplementary Fig. 4, Table 1), few smokers (Fig. 1, Fig. 2), and the lowest overall mutation burden (Table 1), with lack of TP53 and CDKN2A mutations (Fig. 1, Supplementary Fig. 5). The HPV + subtype was associated with significantly improved overall survival compared with other subtypes (Supplementary Fig. 6), consistent with previous reports (Sethi et al., 2012, Ang et al., 2010). Our MethylMix subtypes segregated more clearly with HPV status than the TCGA gene expression subtypes: While most HPV + HNSCCs occurred within the AT gene expression subtype, 57% HNSCCs within this subtype have had no detected virus (Supplementary Fig. 7). This suggests that integrated DNA methylation and gene expression is a more accurate biomarker of HPV status than gene expression alone and demonstrates that our MethylMix subtypes represent biologically meaningful groups. Moreover, separation of MethylMix subtypes according to HPV status allowed us to unambiguously investigate variability of other factors, such as smoking, between HPV − subtypes, independent of HPV.
Table 1.
Distribution of key clinical and etiological variables between MethylMix subtypes.
| MethylMix subtype |
||||||
|---|---|---|---|---|---|---|
| Non-CIMP-Atypical (N = 150) | NSD1-Smoking (N = 80) | CIMP-Atypical (n = 114) | HPV + (N = 64) | Stem-like-Smoking (N = 120) | ||
| Smoking status (n (%)) | Never | 34 (29%) | 6 (10%) | 28 (29%) | 18 (39%) | 10 (11%) |
| Former > 15 years | 17 (15%) | 6 (10%) | 24 (25%) | 5 (11%) | 11 (12%) | |
| Former < 15 years | 32 (27%) | 16 (27%) | 17 (18%) | 12 (26%) | 33 (35%) | |
| Current | 34 (29%) | 31 (53%) | 27 (28%) | 11 (24%) | 39 (42%) | |
| p-Valuesa | 0.162 | < 0.001 | – | 0.775 | < 0.001 | |
| HPV status (n (%)) | HPV − | 81 (99%) | 44 (100%) | 63 (98%) | 1 (3%) | 72 (92%) |
| HPV16 | 0 (0%) | 0 (0%) | 0 (0%) | 32 (89%) | 5 (6%) | |
| HPV33 | 1 (1%) | 0 (0%) | 1 (2%) | 3 (8%) | 1 (1%) | |
| p-Valuesb | 1 | 1 | – | < 0.001 | 0.197 | |
| Gender (n (%)) | F | 45 (37%) | 12 (20%) | 42 (43%) | 5 (11%) | 18 (19%) |
| M | 77 (63%) | 49 (80%) | 56 (57%) | 41 (89%) | 77 (81%) | |
| p-Valuesc | 0.446 | 0.005 | – | < 0.001 | < 0.001 | |
| Anatomic subsite (n (%)) | Base of tongue | 4 (3%) | 2 (3%) | 1 (1%) | 11 (24%) | 4 (4%) |
| Hypopharynx | 2 (2%) | 0 (0%) | 1 (1%) | 1 (2%) | 2 (2%) | |
| Larynx | 15 (12%) | 27 (44%) | 7 (7%) | 1 (2%) | 44 (46%) | |
| Lip | 1 (1%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (1%) | |
| Oral | 94 (77%) | 31 (51%) | 89 (91%) | 8 (17%) | 44 (46%) | |
| Tonsil | 6 (5%) | 1 (2%) | 0 (0%) | 25 (54%) | 0 (0%) | |
| p-Valuesd | 0.011 | < 0.001 | – | < 0.001 | < 0.001 | |
| Pathological grade (n (%)) | 1 | 15 (12%) | 5 (8%) | 20 (21%) | 1 (2%) | 5 (6%) |
| 2 | 82 (68%) | 39 (65%) | 59 (61%) | 15 (37%) | 60 (67%) | |
| 3 | 23 (19%) | 16 (27%) | 18 (19%) | 19 (46%) | 24 (27%) | |
| 4 | 0 (0%) | 0 (0%) | 0 (0%) | 6 (15%) | 1 (1%) | |
| p-Valuese | 0.152 | 0.069 | – | 0.014 | 0.005 | |
| Age (mean (IQR)) p-valuef | 61 (52–70) p = 0.406 | 58 (52–65) p = 0.016 | 63 (53–73) | 56 (50–62) p = 0.002 | 63 (56–68) p = 0.947 | |
| Overall mutation burden (mean (IQR)) p-valuef | 124 (62–115) p = 0.08 | 188 (97–252) p = 0.004 | 114 (695–145) | 99 (38–117) p = 0.004 | ||
| Smoking mutation rates (mean (IQR)) p-valuef | G > T transversions | 7 (3–9) p = 0.574 | 24 (6–34) p ≤ 0.001 | 6 (3–8) | 5 (2–5) p = 0.037 | 17 (5–20) p ≤ 0.001 |
| C > A transversions | 7 (3–8) p = 0.875 | 25 (6–36) p ≤ 0.001 | 7 (4–9) | 4 (1–7) p = 0.003 | 17 (5–21) p = 0.002 | |
| # CNAs (mean (IQR)) p-valuef | CNAScore | 10,420 (6145–14,060) p ≤ 0.001 | 11,850 (8055–15,560) p ≤ 0.001 | 6805 (2550–9738) | 6838 (3886–8006) p = 0.294 | 11,550 (8476–15,680) p ≤ 0.001 |
| Xenobiotic metabolism expression (mean (IQR)) p-valuef | − 0.3 (− 0.8–0.1) p ≤ 0.001 | 0.8 (0.2–1.3) p ≤ 0.001 | 0 | 0.2 (− 0.2–0.6) p ≤ 0.001 | 0.6 (0.1–1.2) p ≤ 0.001 | |
| Abnormally methylated genes (mean (IQR)) p-valuef | # hyper genes | 954 (878–1033) p ≤ 0.001 | 724 (621–804) p ≤ 0.001 | 1282 (1184–1362) | 866 (736–943) p ≤ 0.001 | 760 (674–850) p ≤ 0.001 |
| # hypo genes | 422 (384–461) p ≤ 0.001 | 587 (542–627) p ≤ 0.001 | 370 (344–394) | 462 (424–496) p ≤ 0.001 | 439 (380–494) p ≤ 0.001 | |
| IFN gene expression (mean (IQR)) p-valuef | 0.2 (− 0.4–0.9) p ≤ 0.001 | − 0.4 (− 1.1–0.4) p ≤ 0.001 | 0.8 (0.3–1.3) | − 0.2 (− 0.5–0.3) p ≤ 0.001 | − 0.6 (− 1.2–0.1) p ≤ 0.001 | |
Pearson chi-squared test for difference in distribution of non-smokers (never or former > 15 years) between CIMP-Atypical subtype and other subtypes.
Pearson chi-squared test for difference in distribution of HPV − patients between CIMP-Atypical subtype and other subtypes.
Pearson chi-squared test for difference in distribution of female patients between CIMP-Atypical subtype and other subtypes.
Pearson chi-squared test for difference in distribution of OSCC patients between CIMP-Atypical subtype and other subtypes.
Pearson chi-squared test for difference in distribution of grade 1 cancers between CIMP-Atypical subtype and other subtypes.
Wilcoxon rank sum test for mean difference in continuous variable between CIMP-Atypical subtype and other subtypes.
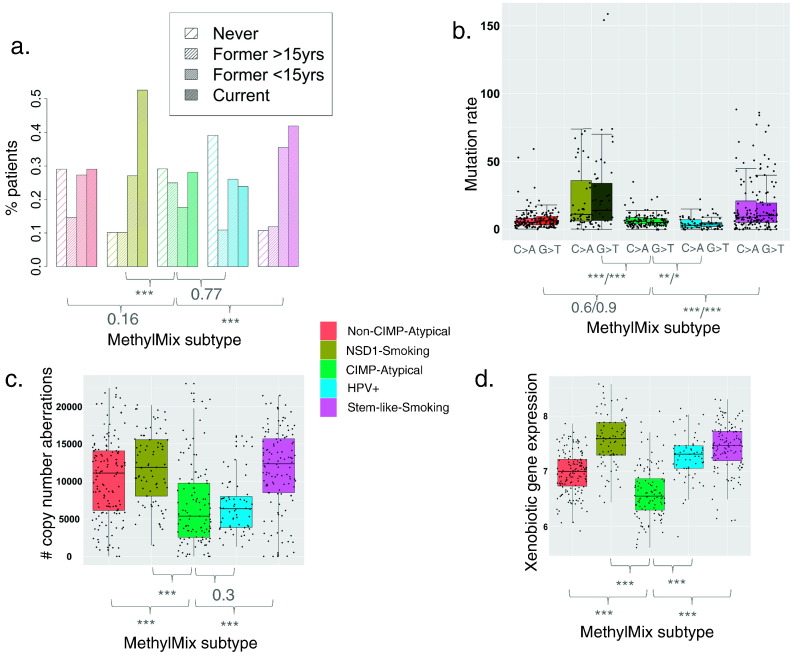
Fig. 2.
Differential distribution of smoking measures between MethylMix subtypes. Distribution of a) smoking status categories (Pearson's chi-squared test), b) smoking mutation signature rates (overall number of G > T and C > A transversions per individual) (Wilcoxon rank sum test, p-values are shown for C > A and G > T mutations signatures separately), c) copy number aberration rate (Wilcoxon rank sum test) and d) mean expression of xenobiotic metabolism genes (Wilcoxon rank sum test), between MethylMix subtypes. p-Values indicate significance of the differences in smoking variables between the CIMP-Atypical subtype (green) and each other subtype separately. *p < 0.05, **p < 0.01, ***p < 0.001.
3.4. Identification of smoking-related and atypical subtypes of HPV − HNSCC
Two HPV − subtypes, the NSD1-Smoking and Stem-like-Smoking subtypes, were overrepresented for current and recent former smokers (< 15 years), compared with the Non-CIMP-Atypical and CIMP-Atypical subtypes (Fig. 2a). Consistently, the NSD1-Smoking and Stem-like-Smoking subtypes had the highest overall mutation burden, primarily driven by significantly higher levels of C > A and G > T transversions, the primary mutation signatures associated with smoking (Helleday et al., 2014, Stransky et al., 2011, Hainaut and Pfeifer, 2001, Kandoth et al., 2013, Ding et al., 2008, Henderson et al., 2005) (Fig. 2b, Table 1). These subtype also displayed higher mean numbers of copy number aberrations (Fig. 2c, Table 1), also associated with smoking intensity in cancer (Huang et al., 2011). Moreover, the NSD1-Smoking and Stem-like-Smoking subtypes had higher levels of expression of xenobiotic metabolism genes (Fig. 2d, Table 1), a measure of cellular response to xenobiotic substances such as those found in tobacco smoke (Fang et al., 2013). Each of these smoking measures were correlated with smoking pack years (Supplementary Fig. 8), and with each other, but taken together, they provide platform-independent evidence that the NSD1-Smoking and Stem-like-Smoking subtypes are smoking related, and driven by smoking-related damage.
Conversely, the Non-CIMP-Atypical and CIMP-Atypical subtypes, but particularly the CIMP-Atypical subtype, had low levels of all smoking related measures that were more similar to the HPV + subtype, which is not primarily caused by smoking (Poling et al., 2014). Consistent with clinical descriptions of atypical HNSCC (Patel et al., 2011, Koo et al., 2013, Pickering et al., 2014), the CIMP-Atypical and Non-CIMP-Atypical subtypes were overrepresented for female patients (Supplementary Fig. 9, Table 1), and were enriched for OSCC (Primarily oral tongue and oral cavity), while the NSD1-Smoking and Stem-like-Smoking subtypes comprised of approximately equal numbers OSCC and laryngeal squamous cell carcinoma (LSCC) (Supplementary Fig. 4). Importantly, the differential distributions of these smoking-related features between subtypes were consistent in OSCC and LSCC analyzed separately (Supplementary Table 3), indicating that the degree of smoking-relatedness of subtypes is not simply driven by the proportions of different anatomic subsites.
Taken together, these findings indicate that our HPV − MethylMix subtypes differ in their degree of smoking-relatedness, and that the CIMP-Atypical subtype particularly matches the profile of atypical HNSCC.
3.5. DNA methylation profiles of MethylMix subtypes
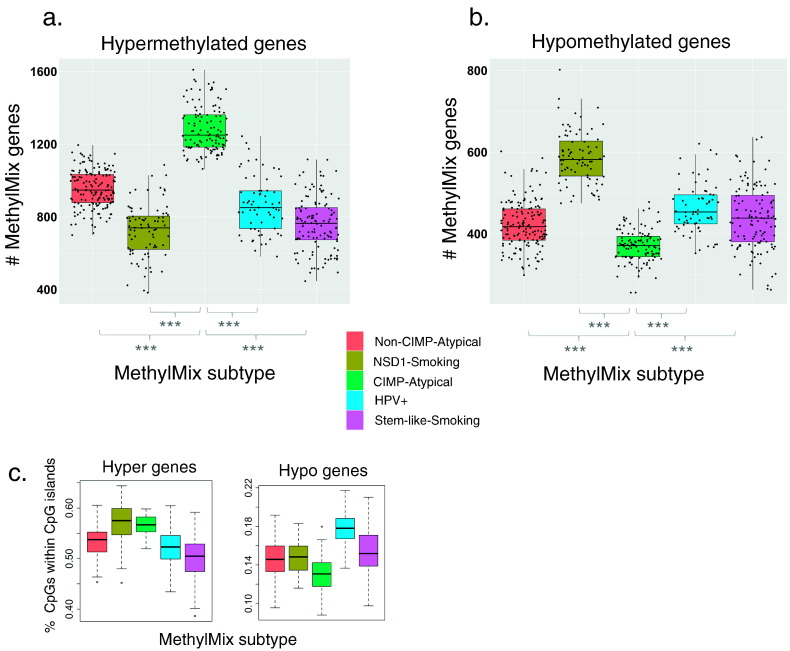
We next investigated the general patterns of DNA methylation perturbation that define each MethylMix subtype. These subtypes differed greatly in their average numbers of hypermethylated and hypomethylated genes (Fig. 3). The NSD1-Smoking subtype had a significantly higher number of hypomethylated genes than other subtypes, but the lowest number of hypermethylated genes. Conversely, the CIMP-Atypical subtype had a strikingly higher number of hypermethylated genes and lower number of hypomethylated genes compared with all other subtypes. The degree of hypermethylation in the CIMP-Atypical subtype was consistent when OSCC and LSCC were analyzed separately (Supplementary Table 3).
Fig. 3.
Different aberrant DNA methylation profiles associated with MethylMix subtypes. Variation in the mean number of a) hypermethylated and b) hypomethylated MethylMix genes per patient, between MethylMix subtypes, with a significantly higher number of hypermethylated genes, and lower number of hypomethylated genes in the CIMP-Atypical subtype (green) compared with each other subtype (Wilcoxon rank sum test). c) The proportion of CpG sites in hypermethylated genes that were within CpG islands was highest within the NSD1-Smoking (olive) and CIMP-Atypical (green) subtypes, while the number of hypomethylated CpG sites within CpG islands was highest within the HPV + subtype (blue). ***p < 0.001.
MethylMix ‘genes’ comprise clusters of concordantly methylated neighboring CpG sites. The proportion of hypermethylated CpG sites that were within CpG islands was highest in the NSD1-Smoking and CIMP-Atypical subtypes, while the proportion of hypomethylated CpG sites within CpG islands was highest in the HPV + subtype (Fig. 3c), suggesting different epigenetic mechanisms that define DNA methylation landscapes within different subtypes. The methylation profile of the CIMP-Atypical subtype, i.e., a strongly elevated number of hypermethylated CpG islands, matches the description of the ‘CpG island methylator phenotype’ (CIMP) (Hughes et al., 2013, Teodoridis et al., 2008). CIMP defines clinically distinct subtypes in other cancers (Hughes et al., 2013), and can be caused by various factors such as oncogenic viruses or IDH1/IDH2 mutations (Turcan et al., 2012, Figueroa et al., 2010), however CIMP is not well characterized in HNSCC. We found that the degree of CIMP, i.e., the overall number of hypermethylated CpG sites, was correlated with increasing age (Supplementary Fig. 10), suggesting that it is caused by age-related epigenetic instability, an etiological factor that increases with age, or increasing duration of exposure to a given cause.
3.6. Somatic mutation profiles differ between smoking-related and atypical HNSCC subtypes
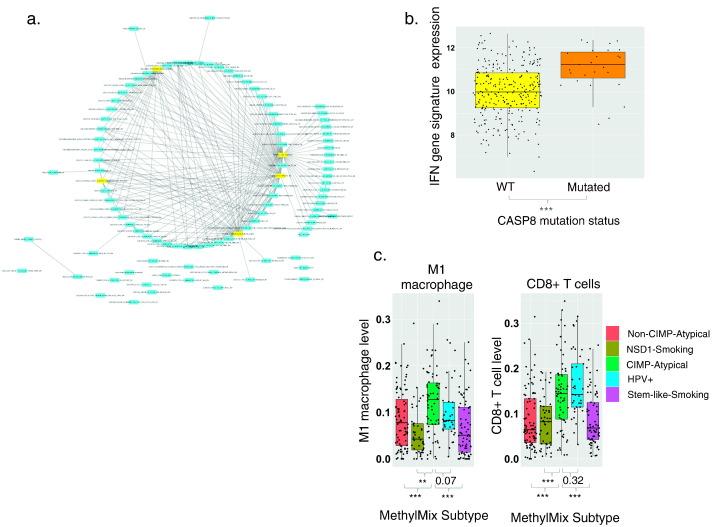
We next assessed the distribution of significantly mutated genes within our MethylMix subtypes. Of all genes, NSD1 mutations were most significantly differentially distributed between MethylMix subtypes (p = 2.2e − 16), occurring almost exclusively within the hypomethylated NSD1-Smoking subtype (Supplementary Fig. 5, Supplementary Table 4), consistent with our previous report (Gevaert et al., 2015). Our NSD1-Smoking subtype corresponding to the H3K36me-impared subtype of HNSCC reported by Papillon-Cavanagh et al. (2017). Thirty percent of patients within the CIMP-Atypical subtype featured CASP8 mutations, compared with 0–7% in other subtypes, a highly significant enrichment (Supplementary Fig. 5). Mutations in HRAS, a gene co-muted with CASP8, and NOTCH1, were also significantly enriched in the CIMP-Atypical subtype, though HRAS was only mutated in 8/66 (12%) of CIMP-Atypical subtype patients with mutation data. CASP8 and NOTCH1 mutations that were not in the CIMP-Atypical subtype were mostly found in the Non-CIMP-Atypical subtype, suggesting that they may be related to atypical, rather than smoking or HPV-related HNSCC etiology.
3.7. Driver copy number aberrations lacking in the CIMP-Atypical subtype
Given the depletion of CNAs in atypical MethylMix subtypes relative to smoking-related subtypes, we assessed the distribution between subtypes of frequent driver CNA events that were previously described in the TCGA patient cohort (Lawrence et al., 2015) (namely 3q26.33, 5p12, 8q11.21 and 8q24.21 amplifications, and 3p12.1, 3p24.1 and 8p23.2 deletions). The CIMP-Atypical subtype had fewer 5p12 amplifications, 3p12.1 deletions, and 3p24.1 deletions than other HPV − subtypes (Supplementary Fig. 11), indicating that this subtype lacks CNAs that are frequent in HNV − HNSCC, but lacking in more genomically stable HPV + HNSCC. Notably, the CIMP-Atypical subtype was depleted for 3q26.33 amplifications, among the most frequent and well studied focal CNA event in HNSCC (Maier et al., 2011), compared with all (both HPV + and HPV −) subtypes (Supplementary Fig. 11). 3q26.33 encompasses SOX2, a major oncogenic driver of stem-like gene expression in SCC (Maier et al., 2011, Boumahdi et al., 2014, Keysar et al., 2017).
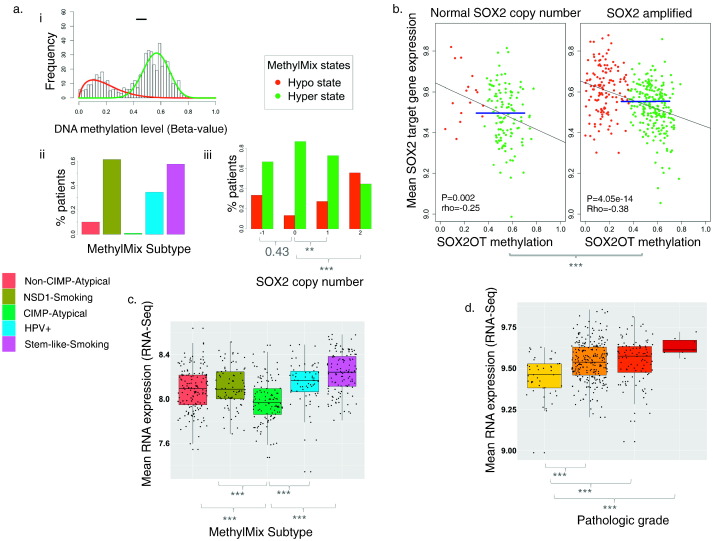
Intriguingly, SOX2 amplifications co-occurred with DNA hypomethylation of a region of the SOX2 overlapping transcript (SOX2OT) (Fig. 4a), lying adjacent to a SOX upstream enhancer (Catena et al., 2004). This SOX2OT region was hypomethylated in most patients within smoking-related and HPV + subtypes, but hypermethylated in atypical HNSCC subtypes (Supplementary Fig. 12).
Fig. 4.
SOX2OT hypomethylation and SOX2 amplifications drive SOX2 pathway expression, and are lacking in the CIMP-Atypical subtype. a) i) Mixture model plot indicating two abnormal SOX2OT DNA methylation states. Histogram illustrates the frequency of patients at levels of SOX2OT methylation in tumor. DNA methylation states (mixture model components) include a hypomethylated and hypermethylated state in tumor, indicated by red and green curves, respectively. The 95% confidence interval for the range of SOX2OT methylation in normal adjacent tissue is indicated by the black horizontal bar. ii) The SOX2OT hypomethylated state occurred in only one patient within the CIMP-Atypical subtype, but occurred in 10–61% of patients in other subtypes. iii) The SOX2OT hypomethylated state (red) was more frequent among patients with either monoallelic (Siegel et al., 2016) or biallelic (Belcher et al., 2014) SOX2 amplifications, but did not differ between patients with SOX2 deletions and normal SOX2 copy number (Pearson's chi-squared test). b) Mean expression of SOX2 target genes (Blue horizontal line) was higher in patients with SOX2 amplifications compared with patients without SOX2 amplifications (Wilcoxon rank sum test), and was negatively correlated with SOX2OT methylation in both groups, indicating that both mechanisms contribute independently to SOX2-related transcription in HNSCC. Linear regression lines and p values, as well as Spearman correlation coefficients (rho) are indicated. SOX2OT MethylMix methylation states are indicated by point colors. c) Mean expression of SOX2 target genes, i.e., genes with promoters bound by SOX2 in embryonic stem cells (ESCs) (Lee et al., 2006) was lower in the CIMP-Atypical subtype compared with each other subtypes (Wilcoxon rank sum test). d) Mean expression of SOX2 target genes displays a stepwise increase with increasing pathologic grade (Wilcoxon rank sum test). **p < 0.01, ***p < 0.001.
SOX2 amplification and SOX2OT methylation were each independently associated with expression of SOX2 (Supplementary Fig. 13a), SOX2 target genes, i.e., genes whose promoters were bound by SOX2 in human embryonic stem cells (ESCs) (Lee et al., 2006) (Fig. 4b), and ESC marker genes (Assou et al., 2007) (Supplementary Fig. 13b). Expression of SOX2 (Supplementary Fig. 13c), and both of these gene sets was lowest in the CIMP-Atypical subtype (Supplementary Figs. 4b and 13d, Fig. 5c). Taken together, these findings indicate that the CIMP-Atypical HNSCC subtype lacks both mechanisms that drive SOX2 overexpression in other HNSCC subtypes, that results in more stem-like transcriptional identity.
Fig. 5.
The CIMP-Atypical subtype features an inflammatory gene expression signature. a) Network map illustrating enrichment for immune response genes among genes overexpressed in the CIMP-Atypical subtype. Nodes represent enriched gene sets and edges represent mutual overlap between gene sets, indicating redundancy between enriched gene sets. Hub gene sets, i.e., the top five gene sets with the highest number of edges are highlighted yellow. The top 100 gene sets identified by gene set enrichment analysis were included in the Network Map. b) Higher mean expression of a reported IFN response gene expression signature (Moserle et al., 2008) in HNSCCs with CASP8 mutations, versus those without CASP8 mutations (Wilcoxon rank sum test). c) Levels of infiltrating M1 macrophages and CD8+ T cells, inferred using CIBERSORT (Newman et al., 2015) within MethylMix subtypes Wilcoxon rank sum test p values for difference in mean TAL levels between the CIMP-Atypical subtype and other subtypes are indicated. **p < 0.01, ***p < 0.001.
Expression of these SOX2 target and ESC marker gene sets was associated with a more undifferentiated state in breast and other cancer types (Ben-Porath et al., 2008). Consistently, we found that mean expression of SOX2 target genes is correlated with increasing pathological grade (Fig. 4d), and that the CIMP-Atypical included a higher frequency of well-differentiated HNSCCs than other subtypes (Supplementary Fig. 14).
3.8. The CIMP-Atypical subtype features an antiviral gene expression signature
In order to identify potential driver pathways and etiological factors associated with the CIMP-Atypical subtypes, we identified genes overexpressed in the CIMP-Atypical subtype (Supplementary Table 5), and performed gene expression enrichment analysis (GSEA) to identify gene sets that significantly overlapped with these genes. The most enriched gene sets represented genes that are activated by type I and II interferons (IFNs), i.e. genes activated during interferon-mediated innate immune response to viral or other pathogen infection (Supplementary Table 6).
A network map illustrating the top 100 most overexpressed gene sets revealed a dense cluster of partially redundant enriched gene sets (Fig. 5a). Three of the top five hub gene sets, i.e., gene sets with the highest number of edges/mutually overlapping genes between gene sets, represented IFNα responsive genes: These included sets of genes upregulated by treatment with IFNα in ovarian cancer side-population cells (Moserle et al., 2008), primary fibroblasts (Browne et al., 2001), and primary hepatocytes (Radaeva et al., 2002). Curiously, although the CIMP-Atypical subtype was not associated with any known oncogenic virus (see above, Supplementary Fig. 3), most of these enriched gene sets represent interferon-inducible antiviral response gene sets. The CIMP-Atypical gene expression signature is also remarkable similar to the set of genes upregulated by expression of double-stranded RNA (dsRNA) derived from reactivated endogenous retroviruses (ERVs), as a result of inhibition of DNA methylation in cancer (Chiappinelli et al., 2015, Roulois et al., 2015). Indeed, all of the IFN-inducible genes upregulated by DNA methylation inhibition in ovarian cancer (Chiappinelli et al., 2015) were upregulated in the CIMP-Atypical subtype (Supplementary Fig. 15).
The co-occurrence of IFN responsive gene overexpression and CASP8 mutations within the CIMP-Atypical subtype is intriguing, as CASP8 initiates apoptosis in response to type I and II interferon-mediated signaling (Mocarski et al., 2012, Kantari and Walczak, 2011, Parker et al., 2016, Tekautz et al., 2006). CASP8 was the only significantly mutated gene associated with an IFN response signature (the top hub enriched gene set (Moserle et al., 2008)) (Fig. 5b), and is therefore uniquely related to IFN response in HNSCC.
Immune gene expression signatures in solid tumors typically reflect the distribution of tumor infiltrating leukocytes (TALs) within the tumor (Gentles et al., 2015a). To gain insight into the immune transcriptional profile of the CIMP-Atypical subtype, we inferred the levels of specific immune cell types within each TCGA patient using CIBERSORT (Newman et al., 2015). Patients of the CIMP-Atypical subtype were enriched for pro-inflammatory M1 macrophages (Fig. 5c, Supplementary Table 7), consistent with upregulation of IFN responsive genes, as M1 macrophages are activated by IFNγ, and M1 activation or ‘polarization’ induces upregulation of many IFN responsive genes (Hu et al., 2008, Martinez et al., 2006).
The CIMP-Atypical subtype also featured elevated levels of CD8 positive (CD8 +) T cells (Fig. 5c, Supplementary Table 7), a marker of anti-cancer immune response, and a favorable prognostic marker in HNSCC (Balermpas et al., 2015), compared with other HPV − subtypes, and almost as high as the HPV + subtype.
As MethylMix filters differentially methylated genes to include only those at which methylation is associated with gene expression, the CIMP-Atypical subtype featured an elevated number of epigenetically silenced genes.
GSEA did not reveal any consistent themes among genes downregulated of hypermethylated in the CIMP-Atypical subtype (data not shown), suggesting that CIMP does not selectively silence any particular class of genes. However, genes that were downregulated and/or hypermethylated in the CIMP-Atypical subtype included tumor suppressor genes listed by TSgene (Zhao et al., 2016) and genes that are causally implicated in cancer, listed within COSMIC cancer Gene Census (Forbes et al., 2008) (Supplementary Table 8), indicating that CIMP alters cancer gene expression pathways.
3.9. Validation of the CIMP-Atypical subtype
Overall, our findings indicate that the CIMP-Atypical subtype is clinically atypical and molecularly distinct, and may therefore represent a distinct etiological and clinical entity. We therefore focused on validating this subtype in independent patient cohorts. We developed a gene expression classifier using prediction of microarrays (PAM) analysis (Tibshirani et al., 2002) to predict the CIMP-Atypical subtype based on gene expression data, and tested the ability of this model to classify the CIMP-Atypical subtype using 100-fold cross validation within the training (TCGA) data set. This classifier could classify the CIMP-Atypical subtype with an area under the curve (AUC) of 0.92 (95% confidence interval 0.89–0.94) (Supplementary Fig. 16). The genes used by the model to classify the CIMP-Atypical subtype were highly consistent across folds of cross-validation, and included 10 upregulated and 22 downregulated genes (in the CIMP-Atypical subtype relative to other subtypes) that were consistently used across all cross-validation folds (Supplementary Table 9). The upregulated genes generally comprised of IFN responsive genes, and included notable TAM expressed genes with important functions related to tumor-immune interactions, including VEGFC (Schoppmann et al., 2006), CD274 (PD-L1) (Schalper et al., 2015), and PDCD1LG2 (PD-L2) (Zhang et al., 2006).
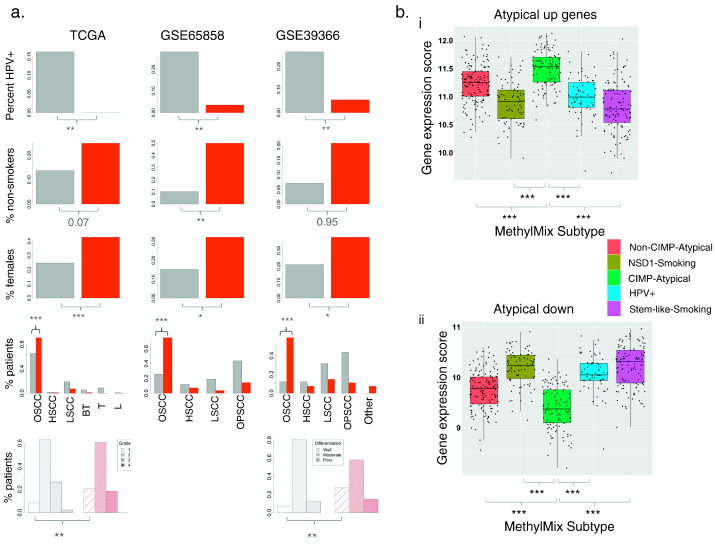
We then applied this classifier to the two largest independent patient cohorts with gene expression and relevant clinical data, GSE65858 (n = 253) (Wichmann et al., 2015) and GSE39366 (n = 138) (Walter et al., 2013). We found that 11% (28/253) patients within GSE65858, and 59% (81/138) patients within GSE39366, were predicted as the CIMP-Atypical subtype. Patients predicted as CIMP-Atypical were overrepresented for HPV − cancers, females, and OSCCs in both validations sets (Fig. 6, Supplementary Table 10). Within HPV − HNSCCs (to exclude confounding by HPV status), patients predicted as CIMP-Atypical were overrepresented for non-smokers in both data sets (Fig. 6a, Supplementary Table 10). In GSE39366 (for which differentiation status data were available), patients predicted as the CIMP-Atypical subtype were more likely to be ‘well-differentiated’, consistent with enrichment of grade 1 tumors in this subtype in the TCGA study (Supplementary Table 10, Fig. 6a, Supplementary Fig. 14). Thus, these findings demonstrate that the gene expression signature of the CIMP-Atypical subtype can robustly classify atypical HNSCC patients. Finally, we confirmed that the IFNα response gene expression signature (Moserle et al., 2008) (the top hub gene set enriched in the CIMP-Atypical subtype) was higher in atypical HNSCCs than smoking-related (HPV −) HNSCCs in GSE39366 and GSE65858 (Supplementary Fig. 17), though this only reached statistical significance in GSE39366.
Fig. 6.
Validation of the CIMP-Atypical subtype gene expression signature. a) Differences in the distribution of clinical features that define the CIMP-Atypical subtype between patients within (red) or not within (grey) the CIMP-Atypical subtype in the TCGA cohort (shown for reference), and in within patients predicted as belonging to the CIMP-Atypical subtype (red) or not (grey), by a gene expression classifier, in two additional patient cohorts (GSE65858 (Wichmann et al., 2015), GSE39366 (Walter et al., 2013)). There was a higher percentage of non-smokers* (never smokers or long-term reformed former smokers), female patients, OSCCs and well-differentiated/pathologic grade 1 tumors, among patients predicted as belonging to the CIMP-Atypical subtype. Pearson's chi-squared p values are indicated. b) Mean expression of genes reported as i) upregulated and ii) downregulated, in atypical HNSCC compared with typical HNSCC (smoking and alcohol-associated) (Farshadpour et al., 2012), was significantly higher and lower, respectively, within the CIMP-Atypical subtype (green) compared with within each other subtype (Wilcoxon rank sum test).
*Difference in the proportion of non-smokers was restricted to HPV − HNSCCs only, as HPV + HNSCC are frequently non-smokers.
Abbreviations for anatomic subsites: Oral squamous cell carcinoma (OSCC), hypopharyngeal squamous cell carcinoma (HSCC), laryngeal squamous cell carcinoma (LSCC), oropharyngeal squamous cell carcinoma (OPSCC), base of tongue (BT), tonsil (T) lip (L). *p < 0.05, **p < 0.01, ***p < 0.001.
3.10. Genes associated with atypical OSCC are consistent with a previous study
To further validate the CIMP-Atypical subtype, we examined a previously reported set of genes that were differentially expressed in non-smoking, non-alcohol related HNSCC relative to smoking and alcohol related HNSCC (Farshadpour et al., 2012) including 28 upregulated, and 21 downregulated genes. This study included only OSCC and oropharyngeal HNSCCs, sites primarily associated with HPV − HNSCC; therefore these gene signatures were considered putative atypical signatures: We confirmed this by validating these signatures as associated with atypical HNSCC within the TCGA, GSE65858, and GSE39366 studies (Supplementary Fig. 18).
Next, Mean expression of the upregulated and downregulated genes was found to be significantly higher and lower, respectively, in the CIMP-Atypical subtype, compared with all other subtypes (Fig. 6b), and this was consistent in OSCC and LSCC analyzed separately (Supplementary Table 11). This confirms the existence of gene expression patterns reproducibly associated with atypical HNSCC, and indicates that such expression patterns pertain particularly to the CIMP-Atypical HNSCC subtype.
4. Discussion
Herein, we confirmed our previous finding of five HNSCC MethylMix subtypes (Gevaert et al., 2015), now within the complete TCGA HNSCC data set. These MethylMix subtypes segregated with HPV status and smoking, the best-known risk factors for HNSCC, indicating that they represent biologically meaningful subtypes.
HPV + HNSCCs clustered into a single, almost ubiquitously HPV + MethylMix subtype, agreeing with previous studies reporting a clear HPV DNA methylation signature (Lleras et al., 2013, Anayannis et al., 2015). Moreover, our MethylMix subtypes segregated with HPV status more perfectly than gene expression subtypes, consistent with previous reports that HPV + HNSCCs occur in two gene expression subtypes (Keck et al., 2015), or make up a subset of the AT expression subtype (Chung et al., 2004, Lawrence et al., 2015). This provided proof of principle that our MethylMix subtypes capture key etiological heterogeneity in HNSCC, as HPV + HNSCC is known to be a clinically and biologically distinct subtype (Sethi et al., 2012, Poling et al., 2014). The original TCGA paper (Lawrence et al., 2015), and other reports (Seiwert et al., 2015); (Lawrence et al., 2015) have focused on molecular differences between HPV + and HPV − HNSCC, while Lleras et al. described DNA methylation features of HPV + HNSCC (Lleras et al., 2013). Therefore, we took advantage of the segregation of HPV + from HPV − HNSCC in our study to investigate less well-studied heterogeneity within the four HPV − subtypes.
Overall, our findings indicate that smoking is a major driver of molecular heterogeneity in HNSCC, as two HPV − subtypes were clearly more smoking-related, indicated by smoking behavior measures (smoking status, pack years), rates of smoking-related genetic damage (smoking mutation signatures, copy number aberration rate), and expression of xenobiotic response genes as a measure of cellular response to smoking.
Smoking mutation signatures provide a long-term historical record of damage caused by smoking, the primary mutagen in HNSCC, as these signatures are elevated in cancers of former smokers (Stransky et al., 2011, Kandoth et al., 2013). These smoking measures are correlated, but capture different facets of smoking as it relates to cancer, including exposure level and levels of biological response to smoking.
The major difference between the two smoking-related subtypes was a striking enrichment for NSD1 mutations in one subtype, associated with widespread DNA hypomethylation. This agrees with previous findings by ourselves and others in a subset (n = 258) of the cohort used here (Lawrence et al., 2015, Gevaert et al., 2015). Papillon-Cavanagh have recently reported on the NSD1 inactivated HNSCC subtype, which also features mutations in K36M-encoding mutations in histone 3 genes, and is defined by impairment of histone 3 lysine 36 (H3K36) methylation (Papillon-Cavanagh et al., 2017).
Two MethylMix subtypes lacked the classic risk factors, i.e., they were HPV −, yet had levels of smoking-related measures similar to the HPV + HNSCC. These subtypes comprised mostly OSCCs and had an overrepresentation of female patients, matching the clinical description of elusive atypical HNSCC (Chaturvedi et al., 2013, Toner and O'Regan, 2009, MacKenzie et al., 2000, Patel et al., 2011, Koch et al., 1999, Koo et al., 2013, Brown et al., 2012, Montero et al., 2012, Perry et al., 2015). Epidemiological evidence indicates that smoking is less of a risk factor for OSCC than it is for LSCC (Maasland et al., 2014, Maasland et al., 2015). Consistent with this, we have found that LSCCs are primarily (though not exclusively) found in the smoking-related MethylMix subsets, while OSCCs are more heterogeneous and can be smoking-related, HPV-related, or atypical.
We named the more atypical of these subtypes the CIMP-Atypical subtype, due to its hypermethylated CpG island phenotype, among other distinguishing molecular features. A gene expression signature for this subtype was predictive of atypical HNSCC features in independent patient population data sets from two previous studies (Walter et al., 2013, Wichmann et al., 2015). Moreover, the CIMP-Atypical subtype displayed differential expression of genes previously reported as markers of non-smoking, non-drinking OSCCs (Farshadpour et al., 2012), that we found to be reproducibly associated with atypical HNSCC. This confirms that atypical HNSCCs display distinct gene expression patterns independent of anatomic subsite. These atypical gene expression patterns pertain particularly to the CIMP-Atypical subtype, rather than non-smoking-related HNSCC in general, as they were altered in the CIMP-Atypical subtype relative to other (HPV + and HPV −) non-smoking related subtypes.
CIMP is a driver of cancer development, conferring epigenetic silencing of tumor suppressor genes (Teodoridis et al., 2008, Hill et al., 2014). CIMP has rarely been reported in HNSCC (Hughes et al., 2013) and only using methods based on methylation of a small panel of genes (Shaw et al., 2007). CIMP implies a distinct etiological basis for this subtype, as CIMP marks clinically relevant etiological subtypes of other cancers and is caused by key oncogenic drivers, including oncogenic viruses (Chang et al., 2006); (Goel et al., 2006) and driver mutations affecting DNA demethylation (Hughes et al., 2013).
Patients of the CIMP-Atypical subtype had few CNAs, in contrast with a previous report that oral tongue squamous cell carcinomas (OTSCC) of young non-smoking patients have similar CNA profiles to those of older smokers (Pickering et al., 2014). We found that the CIMP-Atypical subtype had lower frequency of CNAs that occur in other HPV − subtypes and are also infrequent in HPV + HNSCC. Particularly interesting, however, was a low frequency of SOX2 amplifications within the CIMP-Atypical subtype compared with all other subtypes. SOX2 is a major driver of pluripotency in ESCs and its overexpression maintains a cancer stem cell-like cellular population in HNSCC and other cancers (Maier et al., 2011, Boumahdi et al., 2014, Keysar et al., 2017). SOX2 protein expression is reproducibly associated with poor prognosis and development of lymph node metastases in HNSCC (Dong et al., 2014). Previous studies have shown that SOX2 amplification and protein overexpression more frequently occurs in a smoking-related gene expression HNSCC subtype in independent patient populations (Keck et al., 2015, Walter et al., 2013). We found that SOX2 amplifications co-occur with hypomethylation of SOX2OT, the long non-coding RNA that overlaps with SOX2 and positively regulates SOX2 expression (Shahryari et al., 2015), in both smoking and HPV + subtypes, driving overexpression of SOX2 and SOX2 target/ESC marker genes in these subtypes. SOX2OT hypomethylation has only been reported, to our knowledge, in systemic sclerosis (Altorok et al., 2014), but SOX2OT overexpression in lung cancers promotes proliferation (Hou et al., 2014). The CIMP-Atypical subtype had lower frequency of both mechanisms driving SOX2 overexpression, apparently resulting in overall lower expression of SOX2 target and ESC marker genes. This may be a mechanism underlying lower pathological grade of the CIMP-Atypical subtype, as SOX2 target gene expression was associated with higher grade, as previously reported in other cancers (Ben-Porath et al., 2008), and SOX2 protein expression of was associated with pathological grade in a previous report (He et al., 2014).
The CIMP-Atypical subtype features a gene expression signature characteristic of IFN immune response, overlapping with multiple gene sets related to viral infection, and this signature was reproducibly associated with atypical HNSCC in independent patient population. IFNs are best known as mediators of innate immune response to pathogens (Bekisz et al., 2013), but play diverse roles in cancer. This interferon signature coincide with infiltration of pro-inflammatory M1 macrophages, typifying an inflammatory signature, as M1 macrophages are activated by IFNγ, and M1 macrophage polarization stimulates expression of IFN responsive genes (Hu et al., 2008, Martinez et al., 2006).
Tumor associated macrophages (TAMs) are broadly classified into pro-inflammatory M1 TAMs and anti-inflammatory M2 TAMs. While M2 TAMs are generally considered to be pro-oncogenic, M1 TAMs are considered anti-oncogenic, as they can kill tumor cells and oncogenic pathogens (Costa et al., 2013, Ostuni et al., 2015). M1 TAMs co-occurred with CD8+ T lymphocytes in the CIMP-Atypical subtype, consistent with evidence that M1 TAMs stimulate activate CD8+ T cell immune response (Crouse et al., 2015, Duray et al., 2010). CD8+ T cells are the best known mediators of anti-cancer immune response, and CD8+ T cell levels are favorably prognostic in both HPV + and HPV − HNSCC (Balermpas et al., 2015, Balermpas et al., 2014).
We have found evidence that CD8+ T cell response may be inhibited in the CIMP-Atypical subtype, however, as CD274 (PD-L1) and PDCD1LG2 (PD-L2), ligands for immune checkpoint receptor PD-1, were overexpressed. Binding of PD-L1 and PDCD1LG2 to PD-1 on T CD8+ T cells is well known to inhibit CD8+ T cell response (Zhang et al., 2006; Schalper et al., 2015).
PD-L1 is expressed on both tumor cells and TAMs (Schalper et al., 2015), however we have recently show that M1 TAM levels are correlated with PD-L1 expression across multiple cancer types (Champion et al., Manuscript in preparation), suggesting that M1 TAMs represent an important source of PD-L1 expression in cancer. Given strong CD274 overexpression in CIMP-Atypical subtype, CIMP-Atypical HNSCCs may be uniquely sensitive to PD-1/PD-L1 checkpoint blockade.
Another key oncogene that is apparently expressed by M1 TAMs is VEGFC, which may contribute to metastasis by promoting lymphangiogenesis (Schoppmann et al., 2006). Investigation of potential pro-tumorigenic roles of M1 TAMs in CIMP-Atypical HNSCC is therefore warranted.
The CIMP-Atypical subtype featured enrichment of mutations in CASP8, a frequently mutated gene in OSCC (Vettore et al., 2015, Stransky et al., 2011, Cancer and Consortium, 2013). This is consistent with a previous report that CASP8 mutations marked a HNSCC molecular subtype featuring few CNAs (Pickering et al., 2013). CASP8 mutations reportedly driving growth, migration and invasion in HNSCC (Li et al., 2014). CIMP in glioblastoma (G-CIMP) can be caused by IDH1 mutations (Heiser et al., 2012). BRAF V600E mutations in colorectal cancer only found in cancers with CIMP, but approximately 50% of CIMP positive colorectal cancers have BRAF mutations, indicating that BRAF mutations are unlikely to cause CIMP (Hoadley et al., 2014). CASP8 mutations are less tightly associated with CIMP in HNSCC than either IDH1 or BRAF mutations in their respective cancers, occurring within 30% of the CIMP-Atypical subtype, while 26% of CASP8 mutations occurred in other subtypes, indicating that CASP8 mutations and CIMP are unlikely to be causally related. Nonetheless, the strong enrichment of CASP8 mutations in the CIMP-Atypical subtype suggests that they play a pathogenic role that is inherent to this subtype. We hypothesize that CASP8 mutations enable cell survival within the CIMP-Atypical subtype by blocking the normal apoptotic process induced by IFN signaling: IFNs induce apoptosis via the extrinsic pathway by activating the JAK-STAT pathway, in turn inducing expression and activation of CASP8/Caspase-8 Mocarski et al., 2012, Kantari and Walczak, 2011, Parker et al., 2016. Restoration of CASP8 function, or stimulation of its effectors may represent a therapeutic option for CASP8 inactivated HNSCCs.
The occurrence of an inflamed phenotype in CIMP-Atypical HNSCC raises the intriguing possibility that CIMP-Atypical HNSCC is caused by chronic inflammation: This corresponds to an emerging hypothesis that pathogen-related chronic inflammation causes some OSCCs, as periodontal disease is associated with increased OSCC incidence (Moergel et al., 2013, Tezal et al., 2009, Fitzpatrick and Katz, 2010, Feller et al., 2013). The inflammatory state may reflect innate immune response to an infectious agent that cause periodontal disease or oral inflammation (Whitmore and Lamont, 2014), given the role of M1 macrophages and IFNs response to infection (Liu et al., 2014, McNab et al., 2015). The notion that the CIMP-Atypical subtype may be caused by an infectious agent is further supported by the presence of CIMP, which is caused by viruses in some cancers (Lleras et al., 2013, Goel et al., 2006, Minarovits et al., 2016, Birdwell et al., 2014), and the low mutation burden, suggesting a non-carcinogenic origin. The IFN signature of the CIMP-Atypical subtype is strikingly similar to a reported ‘interferon-inducible antiviral signature’, caused by expression of double-stranded RNA (dsRNA) derived from reactivated ERVs as a result of inhibition of DNA methyltransferases in ovarian and colorectal cancer cell lines (Chiappinelli et al., 2015; Roulois et al., 2015). Moreover, this ERV-induced signature is associated with the CIMP subtype of colorectal cancer (Roulois et al., 2015). Given that many of the antiviral genes overexpressed in the CIMP-Atypical subtype, such as members of the 2′-5′-oligoadenylate synthase (OAS) family (Kristiansen et al., 2011) and DDX58 (Jang et al., 2015), sense viral dsRNA, and the ancient role played by DNA methylation in regulation of ERVs (Stoye, 2012), it seems plausible that reactivation of ERVs may explain the co-occurrence of CIMP and the anti-viral signature in the CIMP-Atypical subtype.
Whether the CIMP-Atypical antiviral signature reflects response to exogenous or endogenous retroviruses, other pathogens, or whether this signature reflects a more general IFN response to a non-pathogen stimulus such as age, obesity (Nishimura et al., 2009) or cancer itself, remain to be resolved by future molecular and epidemiological research.
Our findings provide conclusive evidence that atypical HNSCC is molecularly distinct from smoking and HPV-related HNSCC across multiple platforms/molecular levels, provide a molecular classification system for atypical HNSCC, and postulate that CIMP-Atypical HNSCC represents a distinct etiological and clinical entity. DNA methylation-based validation of the CIMP-Atypical subtype in independent patient cohorts is warranted, particularly as the DNA methylation signature may be used to develop molecular biomarkers to clinically diagnose and investigate CIMP-Atypical HNSCC.
The incidence of atypical OSCC in women appears to be rising, despite the decline of smoking-related HNSCC in males in western countries (Koo et al., 2013, Brown et al., 2012). Identification of the etiological and molecular drivers of atypical subtypes, and development of appropriate prevention and treatment strategies, remain a priority.
The following are the supplementary data related to this article.
Supplementary Fig. 1 Consensus plot indicating five DNA methylation clusters or subtypes: Visualization of consensus clustering (Wilkerson and Hayes, 2010) of 528 HNSCC patients into five clusters, with blue indicating high consensus and white indicating low consensus.
Supplementary Fig. 2 Distribution of TCGA gene expression subtypes between MethylMix subtypes: Bar plot indicates the distribution of gene expression subtype assignments for 279 HNSCCs, derived from Lawrence et al. (2015), between our five MethylMix subtypes.
Supplementary Fig. 3 Distribution of patients with tumor viruses between MethylMix subtypes: Differential distribution of HPV16, HPV33, and other viruses, between MethylMix subtypes, using data derived from Tang et al. (2013). Viral infection status for infection was inferred based on detection of viral RNA in tumor RNA-Seq data.
Supplementary Fig. 4 Distribution of HNSCC anatomic subsites between MethylMix subtypes: Bars indicate the percentages of HNSCCs within each anatomic subsite, for each MethylMix subtype separately.
Supplementary Fig. 5 Differential distribution of point mutations in significantly mutated genes between MethylMix subtypes: Mutations shown include all genes that were significantly mutated in HNSCC (based on a MutSig DB report), and that were significantly differentially distributed between MethylMix subtypes (FDR corrected Pearson's chi-squared test p-value < 0.05). Bars indicate the percentage patients with mutations in each subtype. *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Fig. 6 Kaplan Meier survival curve indicating variability in survival between MethylMix subtypes: Prolonged overall survival in the HPV + subtype compared with other MethylMix subtypes, based on a chi-square statistic test for equality, with survival data censored at five years. **p-value < 0.001.
Supplementary Fig. 7 Distribution of patients with human papilloma virus and other viral infections within TCGA gene expression subtypes: Differential distribution of HPV16, HPV33, and other viruses, between TCGA gene expression subtypes, based on expression subtype assignments derived from Lawrence et al. (2015) and viral infection data based on detection of viral RNA in tumor RNA-Seq data, derived from Tang et al. (2013).
Supplementary Fig. 8 Correlation of smoking mutation signatures with smoking exposure measures: Smoking pack years were positively correlated with a) G > T transversion rate, b) C > A transversion rate c) mean xenobiotic metabolism gene expression and d) overall copy number aberration (CNA) rate. Regression lines, linear regression p-values, and spearman correlation coefficients (Rho) are indicated.
Supplementary Fig. 9 Distribution of female patients between MethylMix subtypes.
Supplementary Fig. 10 Correlation of the degree of CIMP with increasing age: Scatter plot indicating the correlation of the number of hypermethylated MethylMix genes per patient with age at cancer diagnosis. Regression line, linear regression p-value, and spearman correlation coefficients (Rho) are indicated.
Supplementary Fig. 11 Frequency of common amplification and deletions in MethylMix subtypes: Frequency of 3q26.33 and 5qp12 amplifications, and 3p12.1 and 3p24.1 deletions within each subtype, represented by individual genes within each altered region (indicated in plot titles).
Supplementary Fig. 12 Hypermethylation of SOX2OT in atypical MethylMix subtypes, and hypomethylated in a subset of smoking-related MethylMix subtypes: SOX2OT methylation was higher in HNSC tumor compared with normal adjacent tissue, in patients of the Non-CIMP-Atypical and CIMP-Atypical subtypes (Wilcoxon rank sum test). Mean SOX2OT methylation did not differ significantly between tumor and normal tissue for patients within the NSD1-Smoking and Stem-like-Smoking subtypes, as SOX2OT hypomethylation occurred in some patients, but SOX2OT methylation increased or remained stable in others, within these subtypes. Point colors indicate SOX2OT MethylMix DNA methylation states. **p value < 0.001.
Supplementary Fig. 13 SOX2OT hypomethylation and SOX2 amplification associated with SOX2 expression in smoking and HPV-related HNSC subtypes: Expression of a) SOX2 and b) mean expression of embryonic stem cell (ESC) marker genes (indicated by blue horizontal lines) were higher in patients with SOX2 amplifications compared with patients with normal SOX2 copy number (Wilcoxon rank sum test). Both expression measures were negatively correlated with SOX2OT methylation in patients with SOX2 amplifications. Regression lines, linear regression p-values, and spearman correlation coefficients (Rho) are indicated. SOX2OT MethylMix methylation states are indicated by point colors. Expression of c) SOX2 and d) ESC marker genes was significantly lower in the CIMP-Atypical subtype (green) than smoking-related (olive & pink) and HPV + (blue) subtypes, but not atypical subtype 1 (red) (Wilcoxon rank sum test). ***p < 0.001.
Supplementary Fig. 14 Frequency of pathological grades in MethylMix subtypes. p-Values (Pearson's chi-squared test) for significance of differential distribution of grade 1 tumors between the CIMP-Atypical subtype and each other subtype are indicated. **p < 0.01.
Supplementary Fig. 15 Expression of interferon-inducible genes upregulated by azacytidine treatment (Chiappinelli et al., 2015), within MethylMix subtypes. Scaled mean expression of Interferon-inducible viral defense genes that were upregulated by azacytidine treatment in ovarian cancer cell lines, within each MethylMix subtype.
Supplementary Fig. 16 Performance of a gene-expression based supervised predictor in classifying the CIMP-Atypical subtype. ROC curve illustrating the performance of a gene-expression based supervised classifier in correctly classifying the CIMP-Atypical subtype, over 10 rounds of cross-validation within TCGA data. The classifier was determined using prediction of microarray (PAM) analysis (Tibshirani et al., 2002), which were differentially expressed between the CIMP-Atypical subtype and other subtypes combined.
Supplementary Fig. 17 Association of interferon response gene signature with atypical HNSCC in validation data sets. Mean expression (scaled) of an interferon response gene expression signature derived from Moserle et al. (2008) in atypical HNSCCs (HPV − non-smokers) and smoking-related HNSCCs (HPV − smokers) in two validation studies: GSE39366 (Walter et al., 2013) (n HPV − non-smoker = 18, n HPV − smoker = 96) and GSE65858 (Wichmann et al., 2015) (n HPV − non-smoker = 23, n HPV − smoker = 157).
Supplementary Fig. 18 Validation of previously reported gene expression signatures associated with atypical HNSCC, in three independent patient populations. Mean expression (scaled) of genes reported as being significantly overexpressed and underexpressed in atypical HNSCC (non-smoking, non-drinking patients) relative to smoking and drinking HNSCC patients (Farshadpour et al., 2012), was higher and lower, respectively in atypical HNSCC patients in the TCGA, GSE39366 (Walter et al., 2013) and GSE65858 (Wichmann et al., 2015) studies. The number of atypical (HPV negative (HPV −) non-smokers) and smoking related HNSCC patients (HPV − smokers) are: TCGA (n = 89 atypical, 161 smoking-related), GSE39366 (n = 18 atypical, 96 smoking-related), GSE65858 (n = 23 atypical, 157 smoking-related). Wilcoxon rank sum test p-values are indicated: *p < 0.05, ***p < 0.001.
MethylMix driver genes in the TCGA HNSCC study.
MethylMix subtypes and covariate data for TCGA HNSCC patients.
Distribution of key clinical and etiological variables between MethylMix subtypes (OSCC and LSCC separately).
Significantly mutated genes that are differentially distributed between MethylMix subtypes.
Genes upregulated and downregulated in the CIMP-Atypical subtype relative to other MethylMix subtypes.
Gene sets (GSEA) enriched in genes overexpressed in the CIMP-Atypical subtype.
Relative inferred levels of tumor infiltrating leukocyte (TAL) types in MethylMix subtypes.
Tumor suppressor and cancer mutated genes hypermethylate and/or downregulated within the CIMP-atypical subtype.
Genes stably used by the PAM classifier to classify the CIMP-Atypical subtype across all 100 folds of cross validation.
Validation of clinical attributes of the CIMP-Atypical subtype in two validation gene expression cohorts.
Levels of Farshadpour atypical HNSCC gene expression signatures (Farshadpour et al., 2012) in MethylMix subtypes (OSCC and LSCC separately).
Funding sources
Research reported in this publication was supported by the National Cancer Institute under Award Number U01 DE025188 and the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number R01 EB020527. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest
The authors declare that they do not have any conflicts of interest.
Author contributions
K.B. and O.G. conceived and designed the study. K.B., J.L.K., and O.G. accessed and analyzed the data. K.B., O.G., and J.B.S. provided biological interpretation of results. J.B.S. provided clinical consultation and interpretation of the results. A.J.G. performed CIBERSORT analysis. K.B., O.G., and J.B.S. wrote the manuscript. All authors revised the manuscript.
References
- Altorok N., Tsou P.-S., Coit P., Khanna D., Sawalha A.H. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann. Rheum. Dis. 2014:1–9. doi: 10.1136/annrheumdis-2014-205303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anayannis N.V.J., Schlecht N.F., Belbin T.J. Epigenetic mechanisms of human papillomavirus–associated head and neck cancer. Arch. Pathol. Lab. Med. 2015 doi: 10.5858/arpa.2014-0554-RA. [DOI] [PubMed] [Google Scholar]
- Ang K.K. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balermpas P., Rödel F., Weiss C., Rödel C., Fokas E. Tumor-infiltrating lymphocytes favor the response to chemoradiotherapy of head and neck cancer. Oncoimmunology. 2014;3:e27403. doi: 10.4161/onci.27403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balermpas P. CD8 + tumour-infiltrating lymphocytes in relation to HPV status and clinical outcome in patients with head and neck cancer after postoperative chemoradiotherapy: a multicentre study of the German cancer consortium radiation oncology group (DKTK-ROG) Int. J. Cancer. 2015 doi: 10.1002/ijc.29683. 10.1002/ijc.29683 (n/a-n/a) [DOI] [PubMed] [Google Scholar]
- Bekisz J. Immunomodulatory effects of interferons in malignancies. J. Interf. Cytokine Res. 2013;33:154–161. doi: 10.1089/jir.2012.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher R., Hayes K., Fedewa S., Chen A.Y. Current treatment of head and neck squamous cell cancer. J. Surg. Oncol. 2014;110:551–574. doi: 10.1002/jso.23724. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdwell C.E. Genome-wide DNA methylation as an epigenetic consequence of Epstein-Barr virus infection of immortalized keratinocytes. J. Virol. 2014;88:11442–11458. doi: 10.1128/JVI.00972-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumahdi S. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–253. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- Brown L.M., Check D.P., Devesa S.S. Oral cavity and pharynx cancer incidence trends by subsite in the United States: changing gender patterns. J. Oncol. 2012;2012 doi: 10.1155/2012/649498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne E.P., Wing B., Coleman D., Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 2001;75:12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer I., Consortium G. Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat. Commun. 2013;4:2873. doi: 10.1038/ncomms3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catena R. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J. Biol. Chem. 2004;279:41846–41857. doi: 10.1074/jbc.M405514200. [DOI] [PubMed] [Google Scholar]
- Chang M.-S. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin. Cancer Res. 2006;12:2995–3002. doi: 10.1158/1078-0432.CCR-05-1601. [DOI] [PubMed] [Google Scholar]
- Chaturvedi A.K. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J. Clin. Oncol. 2013;31:4550–4559. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli K.B. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C.H. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- Conway D.I. Socioeconomic inequalities and oral cancer risk: a systematic review and meta-analysis of case-control studies. Int. J. Cancer. 2008;122:2811–2819. doi: 10.1002/ijc.23430. [DOI] [PubMed] [Google Scholar]
- Conway D.I. Estimating and explaining the effect of education and income on head and neck cancer risk: INHANCE consortium pooled analysis of 31 case-control studies from 27 countries. Int. J. Cancer. 2015;136:1125–1139. doi: 10.1002/ijc.29063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa N.L. Tumor-associated macrophages and the profile of inflammatory cytokines in oral squamous cell carcinoma. Oral Oncol. 2013;49:216–223. doi: 10.1016/j.oraloncology.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Crouse J., Kalinke U., Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 2015;15:231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- Dal Maso L. Combined effect of tobacco smoking and alcohol drinking in the risk of head and neck cancers: a re-analysis of case–control studies using bi-dimensional spline models. Eur. J. Epidemiol. 2016;31:385–393. doi: 10.1007/s10654-015-0028-3. [DOI] [PubMed] [Google Scholar]
- DeMarini D.M. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat. Res. 2004;567:447–474. doi: 10.1016/j.mrrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Ding L. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Liu G., Huang B., Sun J., Wu D. SOX2 as prognostic factor in head and neck cancer: a systematic review and meta-analysis. Int. J. Clin. Exp. Med. 2014;134:1101–1108. [Google Scholar]
- Duray A., Demoulin S., Hubert P., Delvenne P., Saussez S. Immune suppression in head and neck cancers: a review. Clin. Dev. Immunol. 2010;2010(701657) doi: 10.1155/2010/701657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Netzer M., Baumgartner C., Bai C., Wang X. Genetic network and gene set enrichment analysis to identify biomarkers related to cigarette smoking and lung cancer. Cancer Treat. Rev. 2013;39:77–88. doi: 10.1016/j.ctrv.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Farshadpour F., Roepman P., Hordijk G.J., Koole R., Slootweg P.J. A gene expression profile for non-smoking and non-drinking patients with head and neck cancer. Oral Dis. 2012;18:178–183. doi: 10.1111/j.1601-0825.2011.01861.x. [DOI] [PubMed] [Google Scholar]
- Feller L., Altini M., Lemmer J. Inflammation in the context of oral cancer. Oral Oncol. 2013;49:887–892. doi: 10.1016/j.oraloncology.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Fernandez A.F. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012;22:407–419. doi: 10.1101/gr.119867.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertig E.J. Preferential activation of the hedgehog pathway by epigenetic modulations in HPV negative HNSCC identified with meta-pathway analysis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa M.E. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick S.G., Katz J. The association between periodontal disease and cancer: a review of the literature. J. Dent. 2010;38:83–95. doi: 10.1016/j.jdent.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Forbes S.A. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr. Protoc. Hum. Genet. 2008 doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21:1–12. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert O. MethylMix: an R package for identifying DNA methylation driven genes. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv020. 10.1093/bioinformatics/btv020 btv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert O., Tibshirani R., Plevritis S.K. Pancancer analysis of DNA methylation-driven genes using MethylMix. Genome Biol. 2015:1–13. doi: 10.1186/s13059-014-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A. Association of JC virus T-antigen expression with the methylator phenotype in sporadic colorectal cancers. Gastroenterology. 2006;130:1950–1961. doi: 10.1053/j.gastro.2006.02.061. [DOI] [PubMed] [Google Scholar]
- Hainaut P., Pfeifer G.P. Patterns of p53 G → T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001;22:367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- Harris S.L. Association of p16(INK4a) overexpression with improved outcomes in young patients with squamous cell cancers of the oral tongue. Head Neck. 2011;33:1622–1627. doi: 10.1002/hed.21650. [DOI] [PubMed] [Google Scholar]
- He K.-F. CD163 + tumor-associated macrophages correlated with poor prognosis and cancer stem cells in oral squamous cell carcinoma. Biomed. Res. Int. 2014;2014(838632) doi: 10.1155/2014/838632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton C.M., Durr M.L., Tetsu O., Van Zante A., Wang S.J. TP53 and CDKN2a mutations in never-smoker oral tongue squamous cell carcinoma. Laryngoscope. 2014;124 doi: 10.1002/lary.24595. [DOI] [PubMed] [Google Scholar]
- Heiser L.M. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T., Eshtad S., Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson P.T. Urea lesion formation in DNA as a consequence of 7,8-dihydro-8-oxoguanine oxidation and hydrolysis provides a potent source of point mutations. Chem. Res. Toxicol. 2005;18:12–18. doi: 10.1021/tx049757k. [DOI] [PubMed] [Google Scholar]
- Hill V.K. Stability of the CpG island methylator phenotype during glioma progression and identification of methylated loci in secondary glioblastomas. BMC Cancer. 2014;14:506. doi: 10.1186/1471-2407-14-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley K.A. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int. J. Biochem. Cell Biol. 2014;53:380–388. doi: 10.1016/j.biocel.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Hu X., Chakravarty S., Ivashkiv L. Regulation of IFN and TLR signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol. Rev. 2008;226:41–56. doi: 10.1111/j.1600-065X.2008.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-T. Cigarette smoking increases copy number alterations in nonsmall-cell lung cancer. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16345–16350. doi: 10.1073/pnas.1102769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L., A E. The CpG island methylator phenotype: what's in a name? Cancer Res. 2013;73:5858–5868. doi: 10.1158/0008-5472.CAN-12-4306. [DOI] [PubMed] [Google Scholar]
- Isserlin R., Merico D., Voisin V., Bader G.D. Enrichment Map - a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res. 2014;3:141. doi: 10.12688/f1000research.4536.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M.A. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten syndrome. Am. J. Hum. Genet. 2015;96:266–274. doi: 10.1016/j.ajhg.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Kandoth C. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantari C., Walczak H. Caspase-8 and Bid: caught in the act between death receptors and mitochondria. Biochim. Biophys. Acta. 2011;1813:558–563. doi: 10.1016/j.bbamcr.2011.01.026. [DOI] [PubMed] [Google Scholar]
- Katzel J.A., Merchant M., Chaturvedi A.K., Silverberg M.J. Contribution of demographic and behavioral factors on the changing incidence rates of oropharyngeal and oral cavity cancers in Northern California. Cancer Epidemiol. Biomark. Prev. 2015;24:978–984. doi: 10.1158/1055-9965.EPI-14-1416. [DOI] [PubMed] [Google Scholar]
- Keck M.K. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin. Cancer Res. 2015;21:870–881. doi: 10.1158/1078-0432.CCR-14-2481. [DOI] [PubMed] [Google Scholar]
- Keysar S.B. Regulation of head and neck squamous cancer stem cells by PI3K and SOX2. J. Natl. Cancer Inst. 2017;109 doi: 10.1093/jnci/djw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple R.J. Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 2013;73:4791–4800. doi: 10.1158/0008-5472.CAN-13-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W.M., Lango M., Sewell D., Zahurak M., Sidransky D. Head and neck cancer in nonsmokers: a distinct clinical and molecular entity. Laryngoscope. 1999;109:1544–1551. doi: 10.1097/00005537-199910000-00002. [DOI] [PubMed] [Google Scholar]
- Koo K., Barrowman R., McCullough M., Iseli T., Wiesenfeld D. Non-smoking non-drinking elderly females: a clinically distinct subgroup of oral squamous cell carcinoma patients. Int. J. Oral Maxillofac. Surg. 2013;42:929–933. doi: 10.1016/j.ijom.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Kristiansen H., Gad H.H., Eskildsen-Larsen S., Despres P., Hartmann R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J. Interf. Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- Lawrence M.S. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Egloff A.M., Sen M., Grandis J.R., Johnson D.E. Caspase-8 mutations in head and neck cancer confer resistance to death receptor-mediated apoptosis and enhance migration, invasion, and tumor growth. Mol. Oncol. 2014:1–11. doi: 10.1016/j.molonc.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-C., Zou X.-B., Chai Y.-F., Yao Y.-M. Macrophage polarization in inflammatory diseases. Int. J. Biol. Sci. 2014;10:520–529. doi: 10.7150/ijbs.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lleras R.A. Unique DNA methylation loci distinguish anatomic site and HPV status in head and neck squamous cell carcinoma. Clin. Cancer Res. 2013;19:5444–5455. doi: 10.1158/1078-0432.CCR-12-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maasland D.H.E., van den Brandt P.A., Kremer B., Goldbohm R.A.S., Schouten L.J. Alcohol consumption, cigarette smoking and the risk of subtypes of head-neck cancer: results from the Netherlands Cohort Study. BMC Cancer. 2014;14:187. doi: 10.1186/1471-2407-14-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maasland D.H.E., Van Den Brandt P.A., Kremer B., Goldbohm R.A., Schouten L.J. Consumption of vegetables and fruits and risk of subtypes of head-neck cancer in the Netherlands Cohort Study. Int. J. Cancer. 2015;136:E396–E409. doi: 10.1002/ijc.29219. [DOI] [PubMed] [Google Scholar]
- MacKenzie J. Increasing incidence of oral cancer amongst young persons: what is the aetiology? Oral Oncol. 2000;36:387–389. doi: 10.1016/s1368-8375(00)00009-9. [DOI] [PubMed] [Google Scholar]
- Maier S. SOX2 amplification is a common event in squamous cell carcinomas of different organ sites. Hum. Pathol. 2011;42:1078–1088. doi: 10.1016/j.humpath.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Marisa L. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F.O., Gordon S., Locati M., Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- Massion P.P. Smoking-related genomic signatures in non-small cell lung cancer. Am. J. Respir. Crit. Care Med. 2008;178:1164–1172. doi: 10.1164/rccm.200801-142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015:0–103. doi: 10.1038/nri3787. (TL -15, 15 VN-r) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel C.H. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minarovits J., Demcsák A., Banati F., Niller H.H. vol. 879. 2016. pp. 71–90. (Advances in Experimental Medicine and Biology). [DOI] [PubMed] [Google Scholar]
- Mirghani H. Diagnosis of HPV-driven head and neck cancer with a single test in routine clinical practice. Mod. Pathol. 2015;28:1518–1527. doi: 10.1038/modpathol.2015.113. [DOI] [PubMed] [Google Scholar]
- Mocarski E.S., Upton J.W., Kaiser W.J. Viral infection and the evolution of caspase 8-regulated apoptotic and necrotic death pathways. Nat. Rev. Immunol. 2012;12:79–88. doi: 10.1038/nri3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moergel M. Chronic periodontitis and its possible association with oral squamous cell carcinoma - a retrospective case control study. Head Face Med. 2013;9:39. doi: 10.1186/1746-160X-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero P.H. Changing trends in smoking and alcohol consumption in patients with oral cancer treated at Memorial Sloan-Kettering Cancer Center from 1985 to 2009. Arch. Otolaryngol. Head Neck Surg. 2012;138:817–822. doi: 10.1001/archoto.2012.1792. [DOI] [PubMed] [Google Scholar]
- Moserle L. The side population of ovarian cancer cells is a primary target of IFN-alpha antitumor effects. Cancer Res. 2008;68:5658–5668. doi: 10.1158/0008-5472.CAN-07-6341. [DOI] [PubMed] [Google Scholar]
- Newman A.M. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015:1–10. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S. CD8 + effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat. Med. 2009;15:914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- Ostuni R., Kratochvill F., Murray P.J., Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 2015;36:229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Papillon-Cavanagh S. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet. 2017;49:180–185. doi: 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker B.S., Rautela J., Hertzog P.J. Antitumour actions of interferons: implications for cancer therapy. Nat. Rev. Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- Patel S.C. Increasing incidence of oral tongue squamous cell carcinoma in young white women, age 18 to 44 years. J. Clin. Oncol. 2011;29:1488–1494. doi: 10.1200/JCO.2010.31.7883. [DOI] [PubMed] [Google Scholar]
- Perry B.J. Sites of origin of oral cavity cancer in nonsmokers vs smokers: possible evidence of dental trauma carcinogenesis and its importance compared with human papillomavirus. JAMA Otolaryngol. Head Neck Surg. 2015;141:5–11. doi: 10.1001/jamaoto.2014.2620. [DOI] [PubMed] [Google Scholar]
- Pickering C.R. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3:770–781. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering C.R. Squamous cell carcinoma of the oral tongue in young non-smokers is genomically similar to tumors in older smokers. Clin. Cancer Res. 2014;20:3842–3848. doi: 10.1158/1078-0432.CCR-14-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poling J.S. Human papillomavirus (HPV) status of non-tobacco related squamous cell carcinomas of the lateral tongue. Oral Oncol. 2014;50:306–310. doi: 10.1016/j.oraloncology.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radaeva S. Interferon-alpha activates multiple STAT signals and down-regulates c-Met in primary human hepatocytes. Gastroenterology. 2002;122:1020–1034. doi: 10.1053/gast.2002.32388. [DOI] [PubMed] [Google Scholar]
- Roulois D. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samur M.K. RTCGAToolbox: a new tool for exporting TCGA firehose data. PLoS One. 2014;9 doi: 10.1371/journal.pone.0106397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalper K. Clinical significance of PD-L1 protein expression on tumor-associated macrophages in lung cancer. J. Immunother. Cancer. 2015;3:P415. [Google Scholar]
- Schoppmann S.F. VEGF-C expressing tumor-associated macrophages in lymph node positive breast cancer: impact on lymphangiogenesis and survival. Surgery. 2006;139:839–846. doi: 10.1016/j.surg.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Seiwert T.Y. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin. Cancer Res. 2015;21:632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S. Characteristics and survival of head and neck cancer by HPV status: a cancer registry-based study. Int. J. Cancer. 2012;131:1179–1186. doi: 10.1002/ijc.26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahryari A., Jazi M.S., Samaei N.M., Mowla S.J. Long non-coding RNA SOX2OT: expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis. Front. Genet. 2015;6:1–9. doi: 10.3389/fgene.2015.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R.J. CpG island methylation phenotype (CIMP) in oral cancer: associated with a marked inflammatory response and less aggressive tumour biology. Oral Oncol. 2007;43:878–886. doi: 10.1016/j.oraloncology.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Shenker N.S. DNA methylation as a long-term biomarker of exposure to tobacco smoke. Epidemiology. 2013;24:712–716. doi: 10.1097/EDE.0b013e31829d5cb3. [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Stoye J.P. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 2012;10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- Stransky N. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K.-W., Alaei-Mahabadi B., Samuelsson T., Lindh M., Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat. Commun. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekautz T.M. Evaluation of IFN-γ effects on apoptosis and gene expression in neuroblastoma—preclinical studies. Biochim. Biophys. Acta. 2006;1763:1000–1010. doi: 10.1016/j.bbamcr.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Teodoridis J.M., Hardie C., Brown R. CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett. 2008;268:177–186. doi: 10.1016/j.canlet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- Tezal M. Chronic periodontitis and the incidence of head and neck squamous cell carcinoma. Cancer Epidemiol. Biomark. Prev. 2009;18:2406–2412. doi: 10.1158/1055-9965.EPI-09-0334. [DOI] [PubMed] [Google Scholar]
- Tibshirani R., Hastie T., Narasimhan B., Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toner M., O'Regan E.M. Head and neck squamous cell carcinoma in the young: a spectrum or a distinct group? Part 1. Head Neck Pathol. 2009;3:246–248. doi: 10.1007/s12105-009-0135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toporcov T.N. Risk factors for head and neck cancer in young adults: a pooled analysis in the INHANCE consortium. Int. J. Epidemiol. 2015;44:169–185. doi: 10.1093/ije/dyu255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya O. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Turcan S. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettore A.L. Mutational landscapes of tongue carcinoma reveal recurrent mutations in genes of therapeutic and prognostic relevance. Genome Med. 2015;7:98. doi: 10.1186/s13073-015-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter V. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013;8:e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore S.E., Lamont R.J. Oral bacteria and cancer. PLoS Pathog. 2014;10:1–3. doi: 10.1371/journal.ppat.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann Gunnar, Rosolowski Maciej, K K. The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. 2015;0 doi: 10.1002/ijc.29649. [DOI] [PubMed] [Google Scholar]
- Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise-Draper T.M. Future directions and treatment strategies for head and neck squamous cell carcinomas. Transl. Res. 2012;160:167–177. doi: 10.1016/j.trsl.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Taylor J.A. Genome-wide age-related DNA methylation changes in blood and other tissues relate to histone modification, expression and cancer. Carcinogenesis. 2014;35:356–364. doi: 10.1093/carcin/bgt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Regulation of T cell activation and tolerance by PDL2. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11695–11700. doi: 10.1073/pnas.0601347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Kim P., Mitra R., Zhao J., Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016;44:D1023–D1031. doi: 10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Consensus plot indicating five DNA methylation clusters or subtypes: Visualization of consensus clustering (Wilkerson and Hayes, 2010) of 528 HNSCC patients into five clusters, with blue indicating high consensus and white indicating low consensus.
Supplementary Fig. 2 Distribution of TCGA gene expression subtypes between MethylMix subtypes: Bar plot indicates the distribution of gene expression subtype assignments for 279 HNSCCs, derived from Lawrence et al. (2015), between our five MethylMix subtypes.
Supplementary Fig. 3 Distribution of patients with tumor viruses between MethylMix subtypes: Differential distribution of HPV16, HPV33, and other viruses, between MethylMix subtypes, using data derived from Tang et al. (2013). Viral infection status for infection was inferred based on detection of viral RNA in tumor RNA-Seq data.
Supplementary Fig. 4 Distribution of HNSCC anatomic subsites between MethylMix subtypes: Bars indicate the percentages of HNSCCs within each anatomic subsite, for each MethylMix subtype separately.
Supplementary Fig. 5 Differential distribution of point mutations in significantly mutated genes between MethylMix subtypes: Mutations shown include all genes that were significantly mutated in HNSCC (based on a MutSig DB report), and that were significantly differentially distributed between MethylMix subtypes (FDR corrected Pearson's chi-squared test p-value < 0.05). Bars indicate the percentage patients with mutations in each subtype. *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Fig. 6 Kaplan Meier survival curve indicating variability in survival between MethylMix subtypes: Prolonged overall survival in the HPV + subtype compared with other MethylMix subtypes, based on a chi-square statistic test for equality, with survival data censored at five years. **p-value < 0.001.
Supplementary Fig. 7 Distribution of patients with human papilloma virus and other viral infections within TCGA gene expression subtypes: Differential distribution of HPV16, HPV33, and other viruses, between TCGA gene expression subtypes, based on expression subtype assignments derived from Lawrence et al. (2015) and viral infection data based on detection of viral RNA in tumor RNA-Seq data, derived from Tang et al. (2013).
Supplementary Fig. 8 Correlation of smoking mutation signatures with smoking exposure measures: Smoking pack years were positively correlated with a) G > T transversion rate, b) C > A transversion rate c) mean xenobiotic metabolism gene expression and d) overall copy number aberration (CNA) rate. Regression lines, linear regression p-values, and spearman correlation coefficients (Rho) are indicated.
Supplementary Fig. 9 Distribution of female patients between MethylMix subtypes.
Supplementary Fig. 10 Correlation of the degree of CIMP with increasing age: Scatter plot indicating the correlation of the number of hypermethylated MethylMix genes per patient with age at cancer diagnosis. Regression line, linear regression p-value, and spearman correlation coefficients (Rho) are indicated.
Supplementary Fig. 11 Frequency of common amplification and deletions in MethylMix subtypes: Frequency of 3q26.33 and 5qp12 amplifications, and 3p12.1 and 3p24.1 deletions within each subtype, represented by individual genes within each altered region (indicated in plot titles).
Supplementary Fig. 12 Hypermethylation of SOX2OT in atypical MethylMix subtypes, and hypomethylated in a subset of smoking-related MethylMix subtypes: SOX2OT methylation was higher in HNSC tumor compared with normal adjacent tissue, in patients of the Non-CIMP-Atypical and CIMP-Atypical subtypes (Wilcoxon rank sum test). Mean SOX2OT methylation did not differ significantly between tumor and normal tissue for patients within the NSD1-Smoking and Stem-like-Smoking subtypes, as SOX2OT hypomethylation occurred in some patients, but SOX2OT methylation increased or remained stable in others, within these subtypes. Point colors indicate SOX2OT MethylMix DNA methylation states. **p value < 0.001.
Supplementary Fig. 13 SOX2OT hypomethylation and SOX2 amplification associated with SOX2 expression in smoking and HPV-related HNSC subtypes: Expression of a) SOX2 and b) mean expression of embryonic stem cell (ESC) marker genes (indicated by blue horizontal lines) were higher in patients with SOX2 amplifications compared with patients with normal SOX2 copy number (Wilcoxon rank sum test). Both expression measures were negatively correlated with SOX2OT methylation in patients with SOX2 amplifications. Regression lines, linear regression p-values, and spearman correlation coefficients (Rho) are indicated. SOX2OT MethylMix methylation states are indicated by point colors. Expression of c) SOX2 and d) ESC marker genes was significantly lower in the CIMP-Atypical subtype (green) than smoking-related (olive & pink) and HPV + (blue) subtypes, but not atypical subtype 1 (red) (Wilcoxon rank sum test). ***p < 0.001.
Supplementary Fig. 14 Frequency of pathological grades in MethylMix subtypes. p-Values (Pearson's chi-squared test) for significance of differential distribution of grade 1 tumors between the CIMP-Atypical subtype and each other subtype are indicated. **p < 0.01.
Supplementary Fig. 15 Expression of interferon-inducible genes upregulated by azacytidine treatment (Chiappinelli et al., 2015), within MethylMix subtypes. Scaled mean expression of Interferon-inducible viral defense genes that were upregulated by azacytidine treatment in ovarian cancer cell lines, within each MethylMix subtype.
Supplementary Fig. 16 Performance of a gene-expression based supervised predictor in classifying the CIMP-Atypical subtype. ROC curve illustrating the performance of a gene-expression based supervised classifier in correctly classifying the CIMP-Atypical subtype, over 10 rounds of cross-validation within TCGA data. The classifier was determined using prediction of microarray (PAM) analysis (Tibshirani et al., 2002), which were differentially expressed between the CIMP-Atypical subtype and other subtypes combined.
Supplementary Fig. 17 Association of interferon response gene signature with atypical HNSCC in validation data sets. Mean expression (scaled) of an interferon response gene expression signature derived from Moserle et al. (2008) in atypical HNSCCs (HPV − non-smokers) and smoking-related HNSCCs (HPV − smokers) in two validation studies: GSE39366 (Walter et al., 2013) (n HPV − non-smoker = 18, n HPV − smoker = 96) and GSE65858 (Wichmann et al., 2015) (n HPV − non-smoker = 23, n HPV − smoker = 157).
Supplementary Fig. 18 Validation of previously reported gene expression signatures associated with atypical HNSCC, in three independent patient populations. Mean expression (scaled) of genes reported as being significantly overexpressed and underexpressed in atypical HNSCC (non-smoking, non-drinking patients) relative to smoking and drinking HNSCC patients (Farshadpour et al., 2012), was higher and lower, respectively in atypical HNSCC patients in the TCGA, GSE39366 (Walter et al., 2013) and GSE65858 (Wichmann et al., 2015) studies. The number of atypical (HPV negative (HPV −) non-smokers) and smoking related HNSCC patients (HPV − smokers) are: TCGA (n = 89 atypical, 161 smoking-related), GSE39366 (n = 18 atypical, 96 smoking-related), GSE65858 (n = 23 atypical, 157 smoking-related). Wilcoxon rank sum test p-values are indicated: *p < 0.05, ***p < 0.001.
MethylMix driver genes in the TCGA HNSCC study.
MethylMix subtypes and covariate data for TCGA HNSCC patients.
Distribution of key clinical and etiological variables between MethylMix subtypes (OSCC and LSCC separately).
Significantly mutated genes that are differentially distributed between MethylMix subtypes.
Genes upregulated and downregulated in the CIMP-Atypical subtype relative to other MethylMix subtypes.
Gene sets (GSEA) enriched in genes overexpressed in the CIMP-Atypical subtype.
Relative inferred levels of tumor infiltrating leukocyte (TAL) types in MethylMix subtypes.
Tumor suppressor and cancer mutated genes hypermethylate and/or downregulated within the CIMP-atypical subtype.
Genes stably used by the PAM classifier to classify the CIMP-Atypical subtype across all 100 folds of cross validation.
Validation of clinical attributes of the CIMP-Atypical subtype in two validation gene expression cohorts.
Levels of Farshadpour atypical HNSCC gene expression signatures (Farshadpour et al., 2012) in MethylMix subtypes (OSCC and LSCC separately).