Abstract
Porous graphitic carbon (PGC) is an important tool in a chromatographer’s armory that retains polar compounds with mass spectrometry (MS)-compatible solvents. However, its applicability is severely limited by an unpredictable loss of retention, which can be attributed to contamination. The solutions offered fail to restore the original retention and our observations of retention time shifts of gemcitabine/metabolites on PGC are not consistent with contamination. The mobile phase affects the ionization state of analytes and the polarizable PGC surface that influences the strength of dispersive forces governing retention on the stationary phase. We hypothesized that failure to maintain the same PGC surface before and after running a gradient is a cause of the observed retention loss/variability on PGC. Herein, we optimize the choice of mobile phase solvent in a gradient program with three parts: a preparatory phase, which allows binding of analytes to column; an elution phase, which gives the required separation/peak shape; and a maintenance phase, to preserve the required retention capacity. Via liquid chromatography/tandem mass spectrometry (LC-MS/MS) analysis of gemcitabine and its metabolites extracted from tumor tissue, we demonstrate reproducible chromatography on three PGC columns of different ages. This approach simplifies use of the PGC to the same level as that of a C-18 column, removes the need for column regeneration, and minimizes run times, thus allowing PGC columns to be used to their full potential.
There are well-known challenges posed by chromatographic separation of polar compounds such as nucleosides and nucleotides with subsequent detection by mass spectrometry (MS).1,2 Equally well documented are the drawbacks of possible solutions such as MS contamination (e.g., when using ion-pairing reagents) and reduced sensitivity.1,3 Chromatographic methods can be developed on porous graphitic carbon (PGC) using MS-compatible solvents that resolve nucleosides and nucleotides with excellent peak shapes.4,5 However, we and others have reported variability and a general loss of retention on PGC within a run and over a period of time that can limit its use.6−9 While hydrophilic interaction liquid chromatography (HILIC) can offer a useful alternative, its applications do not always overlap with those of PGC, so PGC remains an important tool for the chromatography of polar compounds.
The general loss of retention on PGC is thought to be mainly due to the column’s particularly high susceptibility to contamination,7 hence the suggestion to backflush regularly. Wash procedures involve the use of strong acids/bases and solvents such as tetrahydrofuran, acetone, trifluoroacetate, hydrochloric acid, and sodium hydroxide.7,10,11 In addition to contamination, the column’s capacity for redox reactions has been cited as an important contributor to variability of retention and poor peak shape. Preconditioning of the column with buffered hydrogen peroxide has been shown to prevent a loss of retention, because of reduction of the stationary phase.7,9 All these treatments add a significant amount of time to sample analysis and involve the use of significant volumes of hazardous chemicals but fail to restore the original retention. Thus, there is a need to better understand the mechanisms underlying analyte retention and to simplify robust method development on PGC.
A detailed description of PGC and the hitherto known mechanisms of retention have been well-reviewed elsewhere.12 Retention is thought to occur through a combination of factors with interactions between the mobile phase and analytes promoting or discouraging retention. Forces such as hydrogen bonds (that keep analytes in solution) discourage retention. On the other hand, hydrophobic interactions promote retention by pushing analytes out of solution toward the PGC surface. Nonpolar analytes interact with the PGC surface via dispersive forces while polar ones are retained via charge induction on the polarizable PGC surface. The mobile phase is a major determinant of retention because it influences the ionization state of both analytes and the PGC surface.13 We hypothesized that failure to maintain the same PGC surface before and after running a gradient is a cause of the observed retention loss/variability on PGC. We now report a method for robust maintenance of retention capacity of gemcitabine and its metabolites on PGC through careful selection of the mobile phase and sequence in a gradient elution program that obviates the need for washing.
Experimental Section
Reagents and Chemicals
Acetonitrile, methanol, water (HPLC grade), and ammonium hydroxide were purchased from Fisher Scientific (U.K.). Ammonium acetate (LC-MS grade) and CTP–15N2,13C were obtained from Sigma (U.K.). Gemcitabine (2′,2′-difluoro-2′-deoxycytidine, dFdC), was purchased from Tocris, while its metabolites—2′,2′-difluoro-2′-deoxyuridine (dFdU), gemcitabine 5′-triphosphate (dFdCTP), gemcitabine 5′-diphosphate choline (GdPC),14 stable labeled dFdU–13C,15N2, dFdC–13C,15N2, citicoline-d9—were purchased from Toronto Research Chemicals (Toronto, Canada).
Instrumentation
Samples were injected using a CTC PAL HTS-xt autosampler (CTC Analytics AG, Switzerland) with two wash stations: solvent 1 (50% methanol in water) and solvent 2 (water). Chromatography was performed with an Accela pump (Thermo Fisher Scientific, USA). Tandem mass spectrometry (MS/MS) analysis was performed using a TSQ Vantage triple-stage quadrupole mass spectrometer (Thermo Scientific, USA) fitted with a heated electrospray ionization (HESI-II) probe operated in positive and negative mode at a spray voltage of 2.5 kV, a capillary temperature of 150 °C, and a vaporizer temperature of 250 °C. Sheath and auxiliary gas pressures were set at 50 and 20 units, respectively. Quantitative data acquisition was performed using LC Quan2.5.6 (Thermo Fisher Scientific, USA). The MS scan parameters have been described previously.6,14
Chromatography
PGC Hypercarb (100 mm × 2.1 mm, ID 5 μm; Thermo Fisher Scientific) columns were kept at 40 °C and fitted with a guard column (Hypercarb, 10 mm × 2.1 mm, 5 μm; Thermo Fisher Scientific). Three different PGC columns were tested, each of which had previously been used for a period of 1–3 years.
Gemcitabine (dFdC) and its metabolites—dFdU, dFdCTP, and GdPC (200 ng/mL of each analyte)—were spiked into tumor tissue homogenate prepared from MIA PaCa-2 xenografts and extracted as previously described.6 The repeatability of retention and peak shape was assessed for 95 injections.
Repeatability of retention time was also assessed for gemcitabine and dFdU (spiked in water) on an Acquity UPLC T3 C-18 column (50 mm × 2.1 mm, ID 1.8 μm; Waters) maintained at 40 °C (for comparison with PGC). dFdCTP and GdPC are not retained on the C18 column and, therefore, were not analyzed. The gradient elution program comprised mobile phase A (0.1% formate in water) and mobile phase B (0.1% formate in 100% acetonitrile) at a flow rate of 200 μL/min. The gradient was initiated with 100% A for 1 min, followed by an increase over 1 min to 70% of B and back to 100% A over another minute. An additional minute of 100% A completed the gradient to give a total run time of 4 min.
Results and Discussion
Lack of Consistent Retention on PGC with Previous Methods
Problems with previous methods start when a new column is exposed to mobile phases other than methanol:water ((95:5), hereafter referred to as 95% methanol), which may modify the surface of the stationary phase resulting in altered retention properties. This causes failure to get reproducible retention from one injection to the next, or on two different columns with the same method (see Tables S1 and S2 in the Supporting Information). Two papers by Jansen et al.4,9 illustrate this: despite treating the columns with preconditioning buffer containing 0.05% hydrogen peroxide, the retention time of gemcitabine between the two studies differed by 2.73 min. Our previous gradient elution program (Figure S1 in the Supporting Information), which had at least 5% acetonitrile throughout,6 suffered from the same problems; gemcitabine retention capacity slowly reduced and limited the number of samples that could be analyzed in one batch. This meant that after approximately every 50 samples regeneration of the PGC column was required. This involved inversion and back-flushing with 50% tetrahydrofuran (THF), 0.1% trifluoroacetic acid (TFA), then 50% THF plus 0.1% sodium hydroxide, then water, then re-equilibration with 95% methanol prior to reuse.
Development of the Chromatography
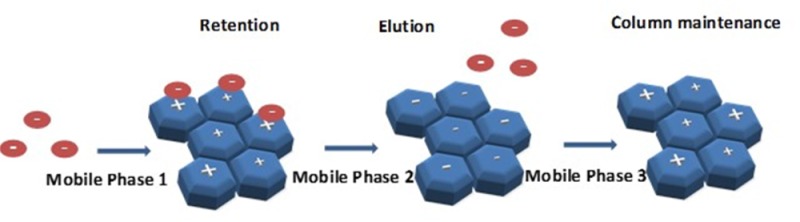
We hypothesized that understanding PGC surface-altering interactions of different mobile phases would enable development of a method that maintained the same PGC surface before and after each sample, standardizing the retention time. Polar analytes interact with the surface of the stationary phase via charge-induced interactions, while nonpolar analytes interact via dispersive forces whose strength may be affected by charge induction caused by the mobile phase.13 The PGC surface may be charged due to the effects of mobile phase pH on ionizable functional groups.13 In addition, the mobile phase may be involved in electron lone-pair interactions with the electron cloud of graphite15 that may expose analyte-retaining charge on the PGC surface. However, it is not clear if the mobile phase must be adsorbed to be involved in electron lone-pair interactions with the surface of the stationary phase. Therefore, a switch from one mobile phase to another may cause a fundamental change to the column’s surface resulting in a modification of retention. The extent of modification of the surface would be dependent on the mobile phases involved. A new PGC column comes stored in 95% methanol and using this as a mobile phase should ensure maintenance of retention without the need for any column regeneration. The hypothesis was tested by analysis of gemcitabine (dFdC) and three of its metabolites.
Under isocratic conditions (95% methanol) however, only dFdC and dFdU were retained and eluted from the column. dFdCTP and GdPC may have eluted together with the column void volume (Figure S2A in the Supporting Information). We reasoned that, consistent with previous studies,15,16 charge induction on the PGC by methanol could have occurred via its two electron lone pairs interacting with the electron cloud of graphite, resulting in the exposure of some positive charge on the column. Increasing pH would ensure ionization of the phosphate groups that could be important for retention of dFdCTP and, to a lesser extent, GdPC. Indeed, a switch to 10 mM ammonium acetate (pH 10) (pH adjusted using ammonium hydroxide) in 95% methanol resulted in retention of all four analytes (dFdC, dFdU, dFdCTP, and GdPC) on PGC (Figure S2B in the Supporting Information). In an attempt to increase retention further and to improve peak shape particularly for dFdCTP and GdPC, the percentage of methanol was reduced from 95% to 70% (Figure S3A in the Supporting Information) and finally to 50% (Figure S3B in the Supporting Information). Reducing the methanol concentration resulted in increased retention, but the peaks became broader. In order to increase retention and to make the peaks sharper, a gradient elution program was then tested.
Designing a Gradient Elution Program with a Maintenance Step
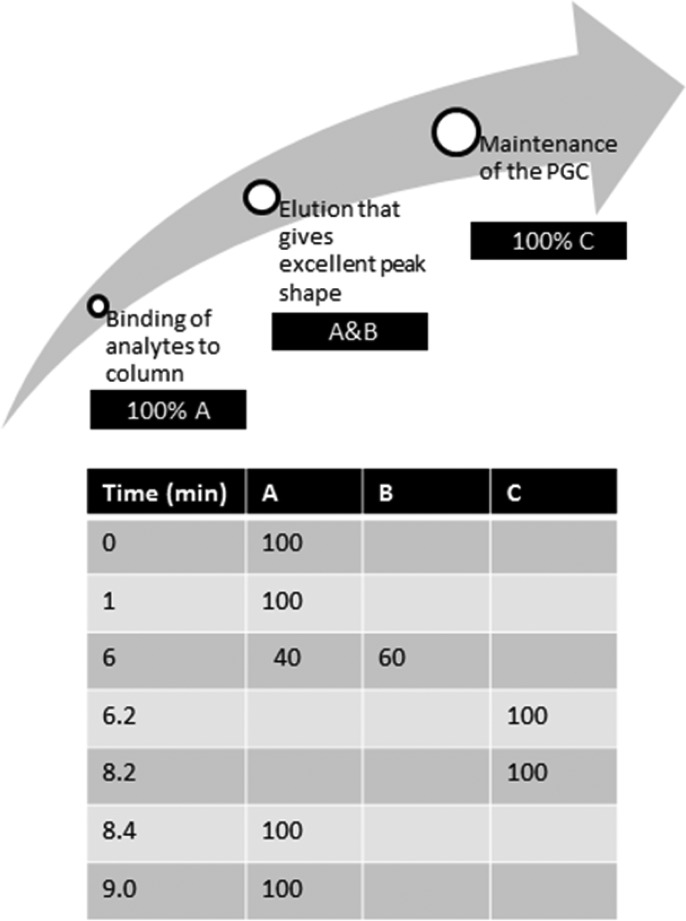
According to our hypothesis, water with its two electron lone pairs could interact with the graphite electrons in a manner similar to that of methanol. Adjusting the pH to 10 would also ensure maintenance of the negative charge on the phosphate groups of dFdCTP and GdPC. Therefore, water would be an excellent part of the preparatory phase, because it would allow binding of all analytes to the column. The second part of the gradient focused on elution of the analytes and, for this, we used acetonitrile to disrupt the charge induced by water and compete for dispersive interactions with the PGC surface. The acetonitrile gradient was optimized to give excellent peak shapes, and a final step of 95% methanol (in water) was added to ensure maintenance of the original retention capacity. The optimal elution gradient is shown in Figure 1.
Figure 1.
Gradient elution program on the PGC column (100 mm × 2.1 mm, ID 5 μm) thermostated at 40 °C. The mobile phase at a flow rate of 0.3 mL/min consisted of (A) 10 mM ammonium acetate, pH 10; (B) 100% acetonitrile; and (C) 95% methanol, 5% water.
Analytical Runs
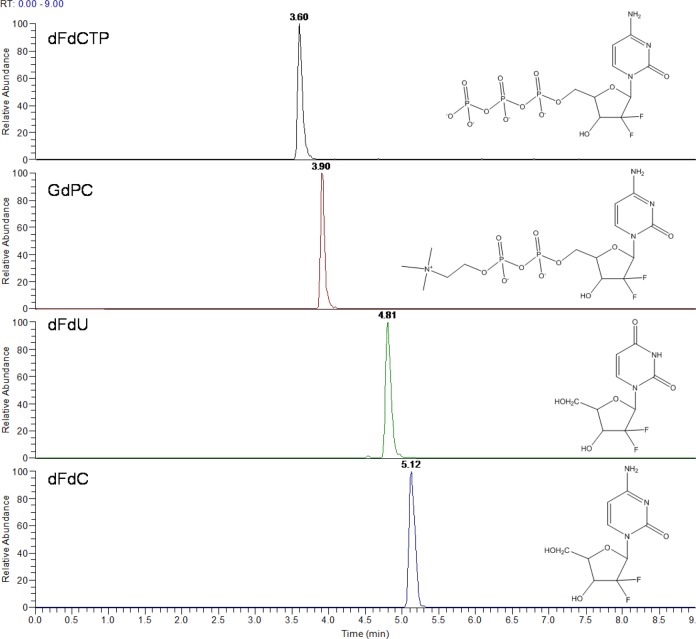
Analytical runs on PGC are notorious for very long equilibration times, with some groups reporting times up to 2 h,17 while others had to precondition the column using an acidic buffer before starting a run.9 In this study, an analytical run was started by ensuring the original methanol-specific retention capacity (running 95% methanol at 150 μL/min for just 5 min). This was followed by an equilibration of the column for only 2.5 min with mobile phase A (10 mM ammonium acetate pH 10 in water). The retention time stabilized after just one injection. Use of the mobile phases as shown in Figure 1 maintained retention capacity from one sample to the next, with typical peak shapes demonstrated in Figure 2. Thus, there was no need for additional steps such as injecting formic acid between each sample, which others have used to correct for loss of retention.9 The number of samples one can run on a PGC column before losing retention/peak shape has been a big issue; Jansen et al.9 were able to maintain retention for 60 injections, but this necessitated the use of hydrogen peroxide. Performance of our method was tested by extracting the analytes from tumor tissue homogenate and 95 injections were run on each of three different PGC columns of varying age and usage (over 3 years). Over the 95 injections, peak shape and retention were maintained: the maximum standard deviation for retention time was ±0.03 min for all four analytes on all three columns (coefficient of variation <1%) (see Table 1).
Figure 2.
Chemical structures and typical chromatograms, on PGC, of dFdC, dFdU, GdPC, and dFdCTP (200 ng/mL) spiked and extracted from tumor homogenate, separated using the gradient shown in Figure 1 and detected following heated electrospray ionization using a triple-stage quadrupole mass spectrometer.
Table 1. Average Retention Times (tR (min)) and Standard Deviation of 95 Injections of dFdC and Its Metabolites Extracted from Tumor Homogenate on Three Different PGC Columns, Using the Gradient Shown in Figure 1, Detected Following Heated Electrospray Ionization Using a Triple-Stage Quadrupole Mass Spectrometer.
| Average Retention Time, tR (min) | |||
|---|---|---|---|
| dFdC | dFdU | GdPC | dFdCTP |
| Column No. 10170107 | |||
| 5.09 ± 0.02 | 4.83 ± 0.02 | 3.93 ± 0.02 | 3.62 ± 0.03 |
| Column No. 0610524 V6 | |||
| 5.06 ± 0.03 | 4.86 ± 0.02 | 3.92 ± 0.03 | 3.65 ± 0.02 |
| Column No.: 10065922 | |||
| 5.14 ± 0.02 | 4.81 ± 0.02 | 3.91 ± 0.03 | 3.61 ± 0.03 |
In order to test how this gradient on PGC compared to the performance of a C-18 column using the same pump, a method was set up on an Acquity T3 C-18 column for gemcitabine and dFdU only (because the phosphorylated metabolites dFdCTP and GdPC are not retained on the C-18 column, eluting in the column void volume) (see Figure S-4 in the Supporting Information). Comparing Figure S-4 with Figure 2 and Table 1 indicates that the PGC is just as robust as a C-18 column with excellent peak shapes, with the added advantage of being able to retain dFdCTP and GdPC.
Use of Mobile Phases Other Than 95% Methanol for the Maintenance Step
The use of a 95% methanol maintenance step enables the performance of a column to be consistent over time, so that a column then only needs to be replaced due to a genuine deterioration in performance and not a reversible modification of its surface by a solvent other than 95% methanol. The column maintenance step could potentially be done using any suitable mobile phase and not just 95% methanol. We predicted that water would interact with the PGC surface in a similar way to methanol, so its impact on reproducibility of the chromatography was tested by replacing methanol with water in the maintenance step (solution C in Figure 1). Reproducible chromatography was obtained with water, but with slightly longer retention times than with 95% methanol (see Table S-3 in the Supporting Information). Using acetonitrile in the maintenance step did not work: dFdCTP and GdPC were no longer retained, illustrating the importance of choosing the correct mobile phase to use in a gradient elution program.
Conclusion
In conclusion, we have developed a liquid chromatography method on porous graphitic carbon (PGC) that overcomes a key challenge of variable retention and loss of retention. We applied the approach to analysis of gemcitabine and its metabolites 2′,2′-difluoro-2′-deoxyuridine (dFdU), gemcitabine 5′-triphosphate (dFdCTP), and gemcitabine 5′-diphosphate choline (GdPC). The class of nucleoside and nucleotide compounds has been of interest to bioanalysts for decades, because of their therapeutic uses as anticancer drugs (e.g., gemcitabine, cytarabine, capecitabine) and antiviral drugs (e.g., zidovudine, acyclovir). More recently, the importance of having robust methods for quantifying endogenous nucleotides and nucleosides has increased, because of a developing body of literature, demonstrating their roles in epigenetic control of gene expression18−20 and immune responses.21 The approach that we have described will allow such molecules to be quantified by liquid chromatography/tandem mass spectrometry (LC-MS/MS) in a robust and reproducible manner without the need for ion-pairing reagents. Identification of the correct mobile phase sequence in a gradient elution program that ensures analytes interact with the same stationary phase with every injection is key to maintaining reproducible retention on PGC. We believe methanol modifies the PGC surface (probably via electron lone-pair interaction with the graphite), allowing consistent analyte retention, indicating that methanol should form the preparatory and maintenance phases. Water is predicted to interact similarly with the PGC surface and may be used in place of methanol (although, in our example, retention times were longer with water). This approach simplifies use of the PGC to the same level as that of a C-18 column, removes the need for column regeneration, minimizes run times, and thus allows PGC columns to be used to their full potential.
Acknowledgments
We thank Drs. Luisa Pereira and Tony Edge (ThermoFisher Scientific) for useful discussions on the mechanism of retention. We are grateful to the PK/Bioanalytics Core Facility (CRUK Cambridge Institute) for access to the LC-MS system. This work was funded by the Cancer Research UK Cambridge Institute (Grant NO. C14303/A17197).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.6b01167.
Average retention times and number of runs on PGC as reported by others (Table S1); average retention times on different PGC columns (age/usage) and the same column after regeneration, as reported by others (Table S2); average retention times of gemcitabine and metabolites on PGC when water replaces 95% methanol in the column maintenance step (Table S3); gradient elution program from our previously published method6 (Figure S1); effect on retention and peak shape of changing from an isocratic mobile phase of 95% methanol to 10 mM ammonium acetate pH 10 in 95% methanol (Figure S2); effect on retention and peak shape of reducing methanol concentration (Figure S3); and typical chromatogram and average retention from a C18 column (Figure S4) (PDF)
Author Present Address
† Oncology iMED DMPK AstraZeneca UK, Ltd., HODGKIN C/o B310 Cambridge Science Park, Milton Road, Cambridge CB4 0WG, U.K.
The authors declare no competing financial interest.
Supplementary Material
References
- Jansen R. S.; Rosing H.; Schellens J. H.; Beijnen J. H. Mass Spectrom. Rev. 2011, 30, 321–343. 10.1002/mas.20280. [DOI] [PubMed] [Google Scholar]
- Wang J.; Lin T.; Lai J.; Cai Z.; Yang M. S. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2009, 877, 2019–2024. 10.1016/j.jchromb.2009.05.027. [DOI] [PubMed] [Google Scholar]
- Vela J. E.; Olson L. Y.; Huang A.; Fridland A.; Ray A. S. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2007, 848, 335–343. 10.1016/j.jchromb.2006.10.063. [DOI] [PubMed] [Google Scholar]
- Jansen R. S.; Rosing H.; Schellens J. H.; Beijnen J. H. Rapid Commun. Mass Spectrom. 2009, 23, 3040–3050. 10.1002/rcm.4212. [DOI] [PubMed] [Google Scholar]
- Agrofoglio L. A.; Bezy V.; Chaimbault P.; Delepee R.; Rhourri B.; Morin P. Nucleosides, Nucleotides Nucleic Acids 2007, 26, 1523–1527. 10.1080/15257770701544336. [DOI] [PubMed] [Google Scholar]
- Bapiro T. E.; Richards F. M.; Goldgraben M. A.; Olive K. P.; Madhu B.; Frese K. K.; Cook N.; Jacobetz M. A.; Smith D. M.; Tuveson D. A.; Griffiths J. R.; Jodrell D. I. Cancer Chemother. Pharmacol. 2011, 68, 1243–1253. 10.1007/s00280-011-1613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reepmeyer J. C.; Brower J. F.; Ye H. J. Chromatogr A 2005, 1083, 42–51. 10.1016/j.chroma.2005.05.092. [DOI] [PubMed] [Google Scholar]
- Huang L.; Lizak P.; Aweeka F.; Long-Boyle J. J. Pharm. Biomed. Anal. 2013, 86, 198–203. 10.1016/j.jpba.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. S.; Rosing H.; Schellens J. H.; Beijnen J. H. J. Chromatogr A 2009, 1216, 3168–3174. 10.1016/j.chroma.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Pabst M.; Altmann F. Anal. Chem. 2008, 80, 7534–7542. 10.1021/ac801024r. [DOI] [PubMed] [Google Scholar]
- Pabst M.; Grass J.; Fischl R.; Leonard R.; Jin C.; Hinterkorner G.; Borth N.; Altmann F. Anal. Chem. 2010, 82, 9782–9788. 10.1021/ac101975k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West C.; Elfakir C.; Lafosse M. J. Chromatogr A 2010, 1217, 3201–3216. 10.1016/j.chroma.2009.09.052. [DOI] [PubMed] [Google Scholar]
- De Matteis C. I.; Simpson D. A.; Euerby M. R.; Shaw P. N.; Barrett D. A. J. Chromatogr A 2012, 1229, 95–106. 10.1016/j.chroma.2011.12.090. [DOI] [PubMed] [Google Scholar]
- Bapiro T. E.; Frese K. K.; Courtin A.; Bramhall J. L.; Madhu B.; Cook N.; Neesse A.; Griffiths J. R.; Tuveson D. A.; Jodrell D. I.; Richards F. M. Br. J. Cancer 2014, 111, 318. 10.1038/bjc.2014.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepont C.; Gunatillaka A. D.; Poole C. F. Analyst 2001, 126, 1318–1325. 10.1039/b102719k. [DOI] [PubMed] [Google Scholar]
- Pereira L. J. Liq. Chromatogr. Relat. Technol. 2008, 31, 1687–1731. 10.1080/10826070802126429. [DOI] [Google Scholar]
- Antonopoulos A.; Favetta P.; Helbert W.; Lafosse M. J. Chromatogr A 2007, 1147, 37–41. 10.1016/j.chroma.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Zauri M.; Berridge G.; Thezenas M. L.; Pugh K. M.; Goldin R.; Kessler B. M.; Kriaucionis S. Nature 2015, 524, 114–118. 10.1038/nature14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. G.; Jiang Y.; Qiu R.; Rauch T. A.; Wang Y.; Schackert G.; Krex D.; Lu Q.; Pfeifer G. P. Cancer Res. 2011, 71, 7360–7365. 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt C. G.; Gnerlich F.; Smits A. H.; Pfaffeneder T.; Jansen P. W.; Bauer C.; Munzel M.; Wagner M.; Muller M.; Khan F.; Eberl H. C.; Mensinga A.; Brinkman A. B.; Lephikov K.; Muller U.; Walter J.; Boelens R.; van Ingen H.; Leonhardt H.; Carell T.; Vermeulen M. Cell 2013, 152, 1146–1159. 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Cekic C.; Linden J. Nat. Rev. Immunol. 2016, 16, 177–192. 10.1038/nri.2016.4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.