Abstract
Huntington disease (HD) is a fatal dominantly inherited neurodegenerative disorder caused by CAG repeat expansion (>36 repeats) within the first exon of the huntingtin gene. Although mutant huntingtin (mHTT) is ubiquitously expressed, the brain shows robust and early degeneration. Current RNA interference-based approaches for lowering mHTT expression have been efficacious in mouse models, but basal mutant protein levels are still detected. To fully mitigate expression from the mutant allele, we hypothesize that allele-specific genome editing can occur via prevalent promoter-resident SNPs in heterozygosity with the mutant allele. Here, we identified SNPs that either cause or destroy PAM motifs critical for CRISPR-selective editing of one allele versus the other in cells from HD patients and in a transgenic HD model harboring the human allele.
Keywords: CRISPR/Cas9, genome editing, Huntington’s disease, gene therapy, AAV
Monteys and colleagues show that CrispR/Cas9 editing can reduce expression of the mutant huntingtin allele in human cell lines from Huntington’s disease patients and in the brains of a mouse model of Huntington’s disease, advancing the therapeutic utility of genome editing for this and other dominantly inherited CNS disorders.
Introduction
Huntington’s disease (HD) is a fatal neurodegenerative disorder caused by CAG repeat expansion in the huntingtin (HTT) gene. Although huntingtin is ubiquitously expressed, the neuropathology of HD is characterized by early striatal atrophy followed by volume loss in other brain areas.1, 2 There is no cure for HD and treatments are focused on symptom management.3 Earlier studies using genetically modified mouse models showed that HD-like phenotypes can be resolved if mutant huntingtin expression is eliminated, even at advanced disease stages,4, 5 suggesting that therapeutic strategies focused on eliminating mutant huntingtin expression will be highly beneficial. As examples, knockdown strategies using RNAi or antisense oligonucleotides, which reduce mutant huntingtin expression either alone or together with the normal huntingtin, are beneficial in various mouse models.6, 7, 8, 9 Other strategies, such as genome editing with zinc finger nucleases targeted to the CAG-repeat expansion region, have also shown promise.10
Genome editing with the recently discovered CRISPR/Cas9 system represents an exciting alternative for tackling dominantly inherited genetic disorders such as HD.11, 12, 13 The most recent system advancements involve expressing Cas9 along with a single guide RNA molecule (sgRNA). When co-expressed, sgRNAs bind and recruit Cas9 to a specific genomic target sequence where it mediates a double-strand DNA (dsDNA) break, activating the dsDNA break repair machinery. Targeted gene deletions by non-homologous end joining (NHEJ) can be made when a pair of sgRNA/Cas9 complexes bind in proximity and produce dsDNA breaks.13, 14, 15
Given the potency and sequence specificity of the CRISPR/Cas9 targeting, and the fact that huntingtin is an important protein for several cellular functions,16 the use of CRISPR/Cas9 to direct allele-specific genome editing is an attractive alternative to the partial reduction approach using ASOs or RNAi methods. Targeting specificity of the CRISPR/Cas9 complex is regulated by two different elements, first, the binding complementarity between the targeted genomic DNA sequence (genDNA) and the 20 nt-guiding sequence of the sgRNA, and, second, the presence of a protospacer-adjacent motif (PAM) juxtaposed to the genDNA/sgRNA complementary region.11, 13, 17 While previous studies have shown that nucleotide mismatches at positions 1–10 on the sgRNA-target site interface are not well tolerated for cleavage, sequence context at this region is crucial to determine which nucleotide positions are more effective to influence cleavage.11, 14, 17, 18, 19 However, the preservation of an intact PAM motif appears to be critical and genome wide studies searching for Cas9 off-target cleavage events demonstrate that mutations on the PAM motif result in an important reduction of cleavage efficacy.20, 21, 22, 23, 24 Therefore, allele-specific gene editing could be achieved by taking advantage of prevalent SNPs that either eliminate or create a PAM sequence. In HD, polyglutamine repeat expansion occurs within exon-1 of HTT.1 Because the main regulatory elements for HTT expression reside within the first two kilobase 5′ of the transcription start site,25 SNP-dependent PAMs in heterozygosity with the mutation are natural CRISPR/Cas9 targets for allele-specific editing. We therefore screened genomic regions adjacent to HTT exon-1 to identify SNPs that were prevalent, and were within the critical position for CRISPR/Cas9- or CRISPR/Cpf1-directed editing, and tested their utility for allele-specific editing in HD patient cell lines and a mouse model expressing full length mutant human HTT.
Results
Screening SNP-Derived PAM Motifs in the HTT Locus
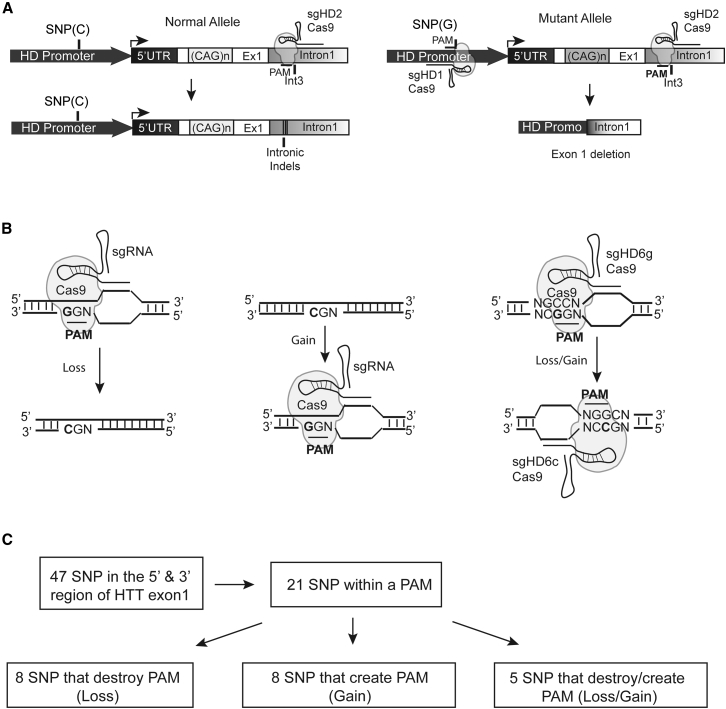
Our goal was to delete the mutant HTT allele using SNP-dependent PAMs flanking HTT exon-1 that, when present in heterozygosity, would tether the Cas9 protein to the mutant, but not the normal allele (Figure 1A). The CRISPR/SpCas9 system from Streptococcus pyogenes is the most widely used, and its PAM sequence (NRG, where N represents any nucleotide, R a purine, and G the conserved guanine) has been fully characterized.11, 13 SNPs present at the third PAM nucleotide position could generate, remove, or simultaneously do both in a strand specific way (Figure 1B). Using the NCBI website and the 1000 Genomes database, we identified 47 SNPs with a prevalence of more than 5%, located upstream (within ∼5 kilobase, Promoter/5′ UTR) and downstream (6.5 kilobase, Intron1) of HTT exon-1. Of these, 21 were present at the conserved third nucleotide of the NRG PAM sequence of SpCas9 (Figures 1C and S1; Table S1). NAG PAMs were included in our screen, although SpCas9 recognition for NAG PAM is less efficient than NGG PAM.26, 27 Overall, the nucleotide variation caused the loss (eight SNPs), gain (eight SNPs), or simultaneously the loss in one DNA strand and a gain on the opposite strand (five SNPs, Loss/Gain) (Figure 1C; Table S1).
Figure 1.
SNP-Dependent Editing for Huntington Disease Therapy
(A) Cartoon depicting the allele-specific editing strategy to abrogate mutant HTT expression. SNPs within PAM sequences upstream of HTT exon-1 permit specific targeted deletions of the mutant allele when present in heterozygosity. After DNA repair, mutant HTT exon-1 is deleted by a pair of sgRNA/Cas9 complexes binding upstream and downstream of exon-1 (right), whereas intronic indels could be generated by a single dsDNA break in the normal allele (left). (B) The nucleotide variation of a SNP within a PAM alters Cas9 recognition resulting in the loss (left), the gain (middle), or the simultaneous loss of a PAM in one DNA strand and the gain of a PAM on the opposite strand (right). (C) There are 21 out of 47 prevalent SNPs flanking HTT exon-1 that are located within predicted critical positions of a PAM sequence for the CRISPR/SpCas9 system analyzed. The minor frequency allele either mediates the loss (eight SNPs), gain (eight SNPs), or a loss/gain (five SNPs) of a PAM motif.
Experimental Validation of HTT Promoter SNP-Dependent PAM Motifs
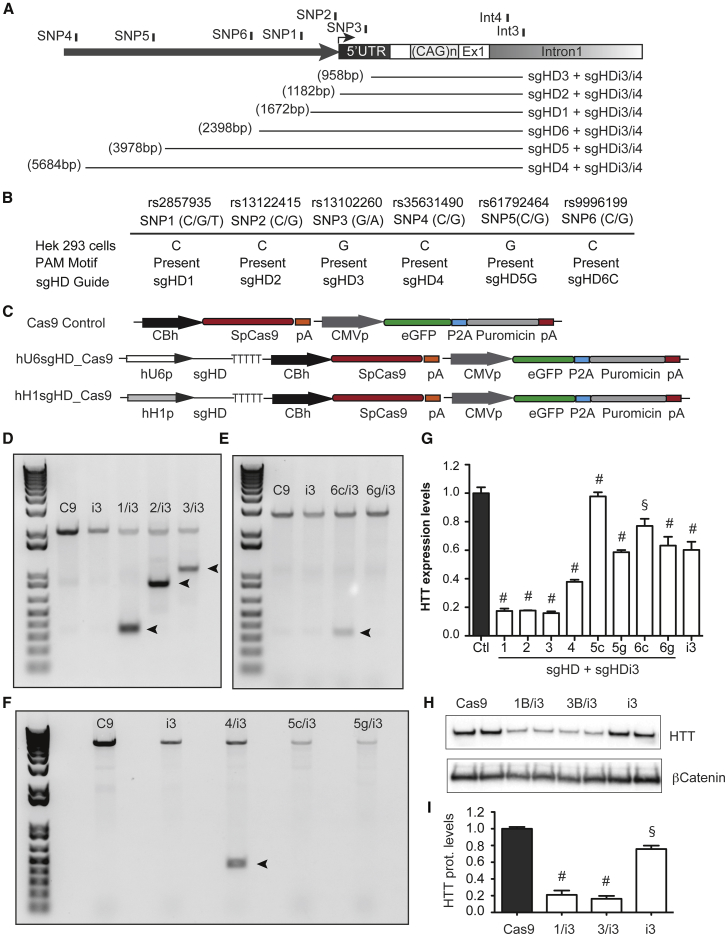
We next developed single-guide RNAs (sgRNAs, all ≤20 nt) to six of the identified SNP-dependent PAMs upstream of HTT exon-1 to test as candidates for CRISPR/Cas9 cleavage in HEK293 cells. There were five SNPs that were located within the ∼5 kilobase of the HTT promoter region (SNPs 1, 2, 4, 5, and 6) and one at the 5′ UTR near the HTT transcription start site (SNP3) (Figures 2A, 2B, and S2). These SNPs have a minor allele frequency of >10% in the general population, and the nucleotide variations cause the Loss or a Loss/Gain of the PAM motif (Table S2). Common sgRNAs were also designed to target sequences within HTT intron-1 (sgHDi3 and sgHDi4; Figures 2A and S2). The sgRNAs were cloned downstream of the hU6 or hH1 promoter, along with other elements as depicted (Figure 2C). HEK293 cells, which are homozygous for the targeting SNPs (Figure 2B), were transfected with SpCas9 and sgRNA expression plasmids and genomic deletion assessed. DNA products of the anticipated size were amplified in most of the sgRNA/SpCas9 pair complexes tested (Figures 2D–2F). Sanger sequencing of the small-amplified PCR products confirmed HTT exon-1 deletion and dsDNA repair (Figure S4). As expected, HTT remained intact in cells expressing SpCas9 or a single sgRNA sequence (sgHDi3; Figure 2, sgHD1, sgHD2, and sgHD3; Figure S5A) or co-expressing sgHDi3 with a sgRNA sequence for which a PAM sequence is absent in the HTT promoter (sgHD5c/i3 and sgHD6g/i3; Figure 2). We did not detect HTT exon-1 cleavage in cells transfected with sgHD5g/i3, in spite of the presence of the PAM. Both sgHD1 and sgHD5g have a 17 nt complementary sequence, yet sgHD1/i3 eliminated HTT exon-1, while sgHD5/i3 did not. Interestingly, sgHD1 has eight guanines, six cytosines, and one adenosine, whereas sgHD5g has four guanines, three cytosines, and three adenosines. This is consistent with earlier work showing a direct correlation between the sequence composition of the sgRNA complementary region to sgRNA activity, with the most active sequences enriched for guanine and cytosine and depleted of adenosine.28
Figure 2.
Cleavage of SNP-Dependent sgHD/SpCas9 Complexes in HEK293 Cells
(A) Cartoon depicting the relative position of the six prevalent SNP-dependent PAMs upstream of HTT exon-1 and two common PAMs within HTT intron-1. The estimated size of the targeted deleted sequence is indicated. (B) The genotype of the prevalent SNPs within the HTT promoter in HEK293 cells is shown. All SNPs were homozygous for the nucleotide variation and the PAM motif was present for the sgRNA indicated. (C) A diagram of the CRISPR expression systems transfected into HEK293 cells is shown. (D–F) A genomic PCR showing HTT exon-1-targeted deletion by sgRNA/SpCas9 pair complexes binding upstream and downstream of the target sequence is shown in the images. (G) RT-qPCR analysis of HTT mRNA levels in HEK293 cells transfected with sgHD/SpCas9 expression cassettes targeting upstream promoter SNPs and the common intronic sgHDi3 sequence is shown. All of the samples are normalized to human GAPDH, and the results are the mean ± SEM relative to cells transfected with plasmids containing the SpCas9 only control (n = 6 independent experiments; §p < 0.001, #p < 0.0001, and one-way ANOVA followed by a Bonferroni’s post hoc). (H) sgHD1/i3/SpCas9, sgHD3/i3/SpCas9, and sgHDi3/SpCas9 expression cassettes were transfected into HEK293 cells, and endogenous HTT protein levels were determined after puromycin selection and expansion. Cells transfected with Cas9 only were used as a control and beta catenin served as a loading control. (I) The quantification of HTT protein levels after treatment with sgHD/SpCas9 complexes is shown. The data are the mean ± SEM relative to cells transfected with plasmids containing SpCas9 only control (n = 6 independent experiments; #p < 0.0001, §p < 0.001, and one-way ANOVA followed by Bonferroni’s post hoc).
HTT mRNA and protein levels were reduced in cells following editing, as determined by qPCR and western blot, respectively (Figures 2G–2I and S3). Reduction of HTT mRNA levels was greater in cells expressing sgRNA/SpCas9 complex pairs that generated small targeted deletions, suggesting that HTT exon-1 removal efficacy may be influenced by the distance between the two dsDNA breaks (compare sgHD1, 2, and 3 versus sgHD4 and 6) (Figure 2G). Also, our results corroborate previous studies showing preference of SpCas9 for NGG over NAG PAM sequences (compare sgHD1, 2, 3, and 4 [NGG] versus sgHD6c [NAG]) (Figure 2G).26, 27 Interestingly, cells expressing a single sgRNA sequence alone, or sgHDi3 in combination with sgHD6g or sgHD5g, also showed reduced HTT mRNA and protein levels, albeit not to as great an extent as those where HTT exon-1 was removed (Figures 2G–2I, S5B, and S5C). Because these sgRNA/Cas9 complexes did not remove HTT exon-1, it suggests that elements within the first intron (sgHDi3) and the promoter region (sgHD1, sgHD2, and sgHD3) might be disrupted as result of indels generated after DNA repair (Figures 2G–2I and S5D).
The generation of short N-terminal fragments as a result of mutant HTT protein cleavage is one of the pathogenic hallmarks of HD. Whereas toxicity of N-terminal fragments has been widely demonstrated, several studies suggest that truncated C-terminal fragments resulting from mutant HTT proteolysis may also contribute to HD pathogenesis.29, 30 Importantly, our data indicate that truncated C-terminal fragments are also eliminated in HEK293 cells edited with our most effective sgRNA sequences, as determined by qRT-PCR or western blot (Figures S6B–S6D).
Assessment of Editing Specificity in HD Human Fibroblasts
Next, we aimed to determine whether allele-specific editing could be achieved for a single allele using the SNP-dependent PAMs in the HTT promoter region. There were 23 lines of fibroblast cell lines from HD patients that were screened for SNP heterozygosity using direct Sanger sequencing of PCR amplified products. There were 11 lines that were heterozygous for SNP1; one line was heterozygous for SNP2, SNP4, and SNP6; and two lines were heterozygous for SNP3 and SNP5 (Table S3).
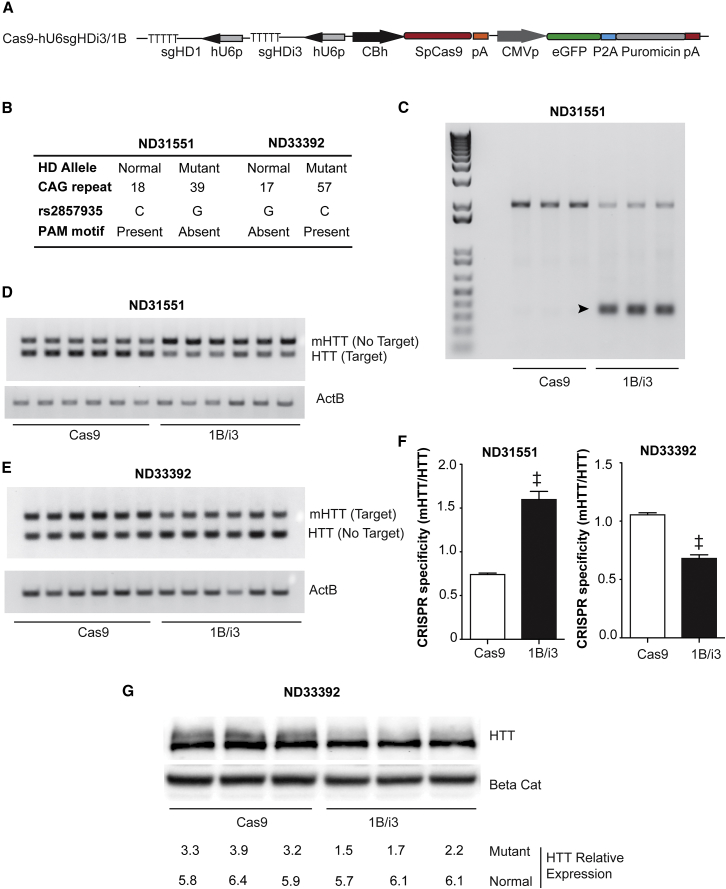
We focused on the sgHD1/i3 Cas9 complex pair, since it was one of the most active sgRNA/Cas9 pairs, generated a larger HTT promoter deletion than sgHD2/i3 and sgHD3/i3, and the SNP within the PAM was the most prevalent among the HD fibroblast lines tested and is present in heterozygosity for more than 20% in the population. Expression vectors for sgHD1/i3 and SpCas9, or SpCas9 only, were generated (Figure 3A) for testing in HD fibroblast cell lines. Two lines, ND31551 and ND33392, which are heterozygous for the SNP1 on opposite alleles, were chosen for specificity testing (Figure 3B; Table S4). PCR of genomic DNA showed target cleavage in cells transfected with plasmids expressing sgHD1/i3 and SpCas9 relative to those lacking sgRNAs (Figure 3C). Semiquantitative PCR for the normal and mutant HTT mRNAs showed target mRNA knockdown (Figures 3D–3F), which for ND31551 is the normal allele, and for ND33392 is the mutant allele. Western blot for protein confirmed allele-specific reduction of the target allele (Figures 3G and 3H).
Figure 3.
Assessment of Allele-Specific Cleavage in Human HD Fibroblasts
(A) Cartoon depicting the CRISPR expression plasmid used to co-express sgHD1 and sgHDi3 expression cassettes. SpCas9 and the selective reporter eGFP/puromycin expression cassettes present in the same plasmid are also shown. (B) ND31551 and ND33392 HD fibroblasts lines were selected to determine allele-specific deletion of HTT. CAG repeat length, nucleotide variation, and the allele location of the PAM motif are indicated in the image. (C) A representative genomic PCR showing HTT exon-1 deletion of DNA harvested from the electroporated ND31551 HD fibroblast cell line is shown in the image. The arrow indicates the expected PCR amplification product resulting from allele-specific deletion. (D and E) A semiquantitative PCR reaction showing the reduction of the targeted allele containing the conserved PAM sequence is shown in the image. For ND31551 fibroblasts, the PAM sequence is conserved in the normal allele, while for ND33392 fibroblasts, the PAM sequence is in the mutant allele. The expression levels are reduced only on the PAM-containing allele. (F) The quantification of mRNA reduction in treated HD fibroblasts is shown. The data show the ratio between mRNA levels of the mutant with respect to the normal allele, relative to cells electroporated with vectors expressing only the Cas9 control. The results are mean ± SEM relative to cells transfected with plasmids containing SpCas9 only control (n = 6; individual electroporations per group, ‡p < 0.01, Mann-Whitney t test). (G) A representative western blot showing allele-specific depletion (upper band of HTT doublet consisting of normal [lower] and expanded polyQ-containing proteins) after electroporation of HD fibroblasts with sgHD/SpCas9 expression vectors is shown in the image. The numbers below the image refer to relative HTT levels.
Assessment of Off-Target Cleavage Sites in Edited HD Fibroblasts
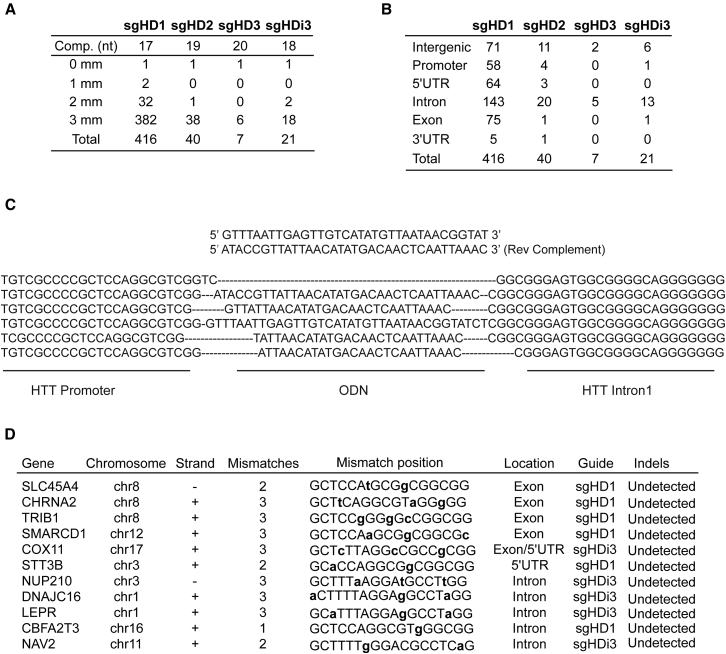
Although truncated sgRNA sequences (<20 nt) are reported to have higher selectivity for the on-target site, any sgRNA/Cas9 complex can also generate unwanted dsDNA breaks at off-target sites that resemble the on-target sequence.18, 22, 26, 31 We used the Cas9-Off finder algorithm to predict the number of potential off-target sites for the most effective sgRNAs (sgHD1, sgHD2, sgHD3, and sgHDi3) and the UCSC genome browser for mapping their location in the human genome.32 Our screen identified 416 sites for sgHD1, whereas 40, 21, and 7 off-targets are predicted for sgHD2, sgHDi3, and sgHD3, respectively (Figure 4A). Of note, sgHD1 has the shortest complementary sequence (17 nt), which could explain its higher frequency for genomic off targets. Importantly, all guides showed full complementarity only to HTT and more than 90% of the off-targets have three mismatches. As expected, they occur in the promoter, 5′ UTR, exons, introns, 3′ UTR, and intergenic regions. The highest number was predicted within introns (Figure 4B). HD fibroblasts were electroporated with vectors expressing SpCas9 and sgHD1/i3, along with a short ODN sequence for mapping off-target dsDNA breaks.22 As expected, the ODN sequence was incorporated within the HTT gene locus (Figure 4C), but it was not detected in any of the 11 top off-target sites tested (Figure 4D).
Figure 4.
Assessing Off-Target Activity of sgHD1 and sgHDi3/Cas9
(A) Table depicting the number of off-target sites for the most active sequences predicted to bind with 1, 2, or 3 mismatches. The nucleotide length of the complementary guide sequence is also indicated in the table. (B) Table highlighting the number of off-target sites binding at different genomic regions using the UCSC genome browser is shown. (C) The HD fibroblasts were electroporated with plasmids expressing sgHD1/i3 and SpCas9 along with an ODN sequence. The Sanger sequencing results showed the incorporation of the ODN sequence at the DNA cleavage site. A HTT promoter sequence and a HTT intron sequence outside the ODN sequence are also depicted. (D) Sanger sequencing results from 11 predicted off-target sites are shown. The gene name, chromosome position, DNA strand, number of mismatches and position within the guide, gene location, sgRNA sequence, and indel presence or absence are indicated.
Editing HTT Exon-1 In Vivo
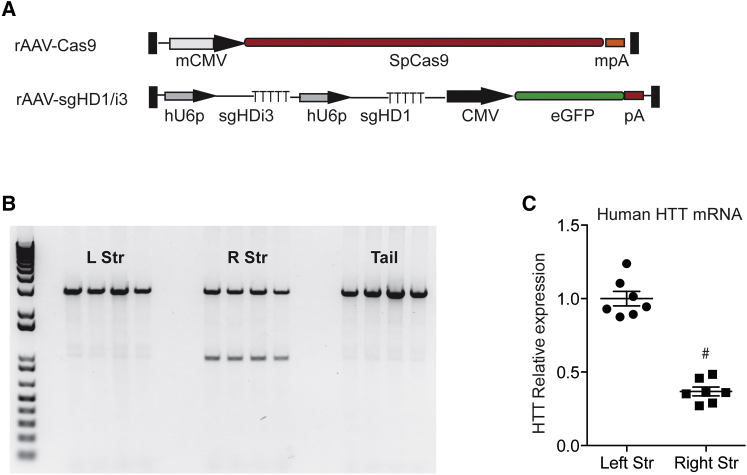
BacHD mice are transgenic for a modified human HD allele,33 which fortuitously contains SNPs 1, 2, and 3. These mice were used to first evaluate the efficacy of mutant HTT editing in vivo at the genomic level. For this, recombinant AAVs (rAAVs) expressing either SpCas9 (rAAV.SpCas9) or the sgRNAs (rAAV.sgHD1/i3) were generated, which effectively delete human HTT exon-1 in vitro (Figures 5A and S7). Mice were injected on the right hemisphere with rAAV.SpCas9 plus rAAV.sgHD1B/i3. The left hemisphere was used as a control and left uninjected. Brains were harvested 3 weeks later and genomic DNA isolated. PCR amplification of genomic DNA demonstrates cleavage in the setting of Cas9 and sgRNA expressing AAVs only (Figure 5B). Accordingly, HTT mRNA levels were reduced on the right, but not the left, hemisphere in concordance with DNA cleavage (Figure 5C). Interestingly, mouse Htt mRNA levels were also reduced on the injected hemisphere, although to a lesser degree than the human HTT allele. Several binding sites for sgHD1 and sgHDi3 were identified within the mouse Htt locus. Three binding sites were identified for sgHD1, all within exon-1 and containing five mismatches. In contrast, a single binding site with two mismatches within the first intron was predicted for sgHDi3 (Figure S8). We hypothesize that indels within the intron caused by sgHDi3 may disrupt transcription factor binding sites and, similar to HEK293 cells expressing sgHDi3 alone (Figure 2G), modestly reduce mouse Htt expression.
Figure 5.
In Vivo Gene Editing of the Mutant HTT Allele
(A) Cartoon depicting rAAV shuttle vectors containing the SpCas9 and sgHD1/i3 expression cassettes (mCMV, minimal CMV promoter; mpA, minimal polyA and hU6p, human U6 promoter; pA, SV40 polyA). (B) PCR of isolated genomic DNA showing human HTT exon-1 targeted deletion after injection of vectors expressing SpCas9 and sgRNA sequences is shown. left striatum, LStr; right striatum, RStr. (C) qRT-PCR analysis of HTT mRNA levels in striatum samples harvested 3 weeks after SpCas9 and sgHD1/i3 delivery is shown. All of the samples were normalized to beta actin and the results are mean ± SEM relative to uninjected striatal samples (n = 7 animals per group, #p < 0.0001, and unpaired t test).
Other Cas9 Systems for Silencing the HTT Allele
Finally, we aimed to broaden the approach we describe here for allele-specific editing of HTT to the other CRISPR/Cas9 and CRISPR/Cpf1 systems recently described. Thus, we screened which of the 47 SNPs flanking HTT exon-1 were contained within the conserved PAM nucleotide positions for the other systems. Engineered SpCas9 variants from Streptococcus pyogenes with altered PAM specificities have been generated (SpCas9_VQR, SpCas9_EQR, and SpCas9_VRER).21 The SpCas9 VQR variant strongly recognizes sequences bearing the NGAN PAM and, with lower efficiency, those sites with a NGNG motif. SpCas9_EQR is more specific for an NGAG PAM. In contrast, SpCas9_VRER has a strong selectivity for a NGCG PAM sequence with no cleavage activity when this is varied. For SpCas9_VQR, the SNP could be positioned at the second or the third nucleotide of the NGAN PAM, as well as at the second and fourth nucleotide of the NGNG PAM sequence. In contrast, because of the selectivity of the SpCas9_EQR for NGAG and SpCas9_VRER for NGCG sequences, the SNP could be permitted at any position of their PAM (Table S1; Figure S1). The discovery of SaCas9 from Staphylococcus aureus has extended the number of CRISPR/Cas9 systems, with the advantage that a SaCas9-encoding transgene can be easily packaged into AAV viral vectors.24 SaCas9 primarily recognizes a NNGRRT PAM, although dsDNA breaks are also observed at DNA targets adjacent to NNGRR motifs. For SaCas9, only those SNPs positioned at the third nucleotide of the PAM would allow for allele specificity (Table S1; Figure S1). A new Class 2 CRISPR system was recently identified that contains Cpf1 as effector protein to mediate dsDNA breaks.23 Unlike Cas9 that recognizes a G-rich PAM motif, the Cpf1 PAM motif is T-rich. Currently, 16 Cpf1-family proteins have been characterized, but only the Cpf1 proteins from Acidaminococcus (AsCpf1) and Lachnospiraceae (LbCpf1) have shown robust DNA interference activity when expressed in mammalian cells. AsCpf1 has strong selectivity for a TTTN PAM and does not recognize any sequence variants. Therefore, SNPs present at any position of the TTTN PAM could disrupt AsCpf1 recognition. In contrast, LbCpf1 recognizes multiple T-rich PAMs, albeit with different cleavage activity. Thus, for LbCpf1, only those SNPs where the variant nucleotide did not generate any other PAM that could be recognized above the LbCpf1 cleavage threshold activity could be considered for allele discrimination (Table S1; Figure S1).23
Overall, we identified 36 SNPs located within the specific PAM positions described above. Again, we found instances where the nucleotide variation caused the loss (12 SNPs), gain (11 SNPs), or a simultaneous loss in one DNA strand and a gain on the opposite strand (13 SNPs) (Table S1; Figure S1). Of special interests are the SNPs that generate a loss/gain, since CRISPR complexes could be designed for any of the two possible nucleotides linked to the mutant allele. Of note, we found instances where the same Cas9 protein would be predicted to target each nucleotide variation using a different sgRNA sequence or, alternatively, a different CRISPR effector protein could be used to target each nucleotide variant. Two interesting observations also arose from our screen. First, in the rs113331544 SNP, for which the minor allele contains a six-nucleotide insertion, the same PAM sequence is present on both alleles, but a different sgRNA sequence could be designed to tether SpCas9 to the mutant allele depending on the nucleotide variation. Second, for the rs28393280 and the rs28583447 SNPs, the nucleotide variation causes the gain of two PAM motifs on the same allele, one on the positive and the other on the negative DNA strand. Those SNPs could be appropriate for targeting with a nickase effector protein, which would efficiently generate on-target dsDNA breaks without detectable damage at potential off-target sites.34
Discussion
Currently, reduction of HTT mRNA levels with RNAi and ASOs are the leading therapeutic options for HD.9, 35 However, it is unknown whether these treatments when administered to human patients will be as beneficial as observed in animal models. Here, we investigated the possibility of using the CRISPR/Cas9 technology to target and inactivate the mutant HTT allele. Targeted gene deletions can be generated when two sgRNA/Cas9 complexes cause dsDNA breaks followed by DNA repair.13, 15 Given the potency of CRISPR/Cas9 and the high likelihood of cleaving both HTT alleles, the role of HTT protein on important cellular functions,16 and the fact that is unknown if complete loss of the huntingtin gene in human brain cells would also be tolerated as reported in adult mice,36 allele-specific gene editing provides an important strategy to investigate. Indeed, recent studies demonstrated allele-specific editing by taking advantage of SNP positioned at these conserved PAM nucleotides.37, 38, 39
We designed guide RNAs that bind and tether SpCas9 to six prevalent SNPs located 5′ of HTT exon-1, which in combination with a guide binding within the first HTT intron, effectively eliminate expression of the HTT protein. We found that the distance between upstream and downstream guides influenced editing efficacy, as well as confirmed the SpCas9 preference in HD cell lines. Our studies also suggest that intronic transcription binding sites may effect HTT gene expression, since indels generated by SpCas9 within the first HTT intron modestly reduced gene expression. This is important when designing intronic guide sequences that are not allele specific, since expression of the normal allele could also be affected. Moreover, we demonstrate that by eliminating the HTT proximal promoter in addition to HTT exon-1, we not only terminate the production of toxic HTT-exon-1 proteins, but also truncated C-terminal HTT proteins.30
In HD fibroblast cell lines for which these SNPs are present in heterozygosity, we observed HTT exon-1 excision only on the allele where the nucleotide variation did not disrupt the PAM motif. Notably, SNP1 (rs2857935) has a prevalence of 22% among the human population. In the HD fibroblast lines tested here, we observed that 9 out of 11 were heterozygous for the SNP and the PAM was linked to the mutant allele. This raises the exciting possibility that this SNP is in linkage disequilibrium with the mutant allele in the general HD population and warrants further study.
While this manuscript was in preparation, Shin and colleagues published a similar study using CrispR/Cas9 to inactivate the mutant HTT allele.39 While both studies demonstrate the efficacy of CRISPR/Cas9 to eliminate HTT exon-1, there are notable differences. First, Shin and colleagues generate a 44 kilobase deletion to inactivate HTT expression, whereas we create small-targeted deletions that are sufficient to terminate HTT expression. Second, we show that allele-specific HTT exon-1 deletion could be achieved using a single SNP-dependent PAM in the HTT promoter in combination with a common guide in intron 1, achieving elimination of N-terminal and C-terminal protein fragments. And third, we demonstrate for the first time, that sgRNA/Cas9 complexes are also effective in vivo in an HD mouse model. We found rAAV delivery of the sgRNA/SpCas9 complexes reduced human mutant HTT expression to 40% in treated hemispheres, a level of reduction known to provide benefit by RNAi or ASOs.6, 7, 9
The translation of this approach to humans will require delivery systems that allow for transient expression so as to reduce the risk of off-target consequences from lasting expression of the editing machinery, such as occurs in the setting of viral-mediated gene transfer systems. Additionally, the Cas9 protein is of bacterial origin and, if an immune response were elicited in human brain, edited cells could be effectively eliminated, mitigating the positive consequences of removing the mutant allele. Therefore, efforts should be extended to ensure that Cas9 and probably also the sgRNAs are transiently expressed.40, 41
The importance of on-target selectivity is crucial when using gene-editing approaches. In our strategy, we used truncated sgRNA guides, which have been shown to minimize unintended dsDNA breaks.42 We screened for potential off-targets from our guides using a combination of an in silico and PCR-based approach and did not detect disruption of the top predicted off-target genes. Importantly, additional tools with significant on-target selectivity such as the high fidelity Cas9 proteins and the Cas9 nickases, developed during the course of our study, could be tested for efficacy using our strategy.34, 43, 44
In summary, we developed and confirmed a strategy for allele-specific genome-editing of mutant HTT based on CRISPR/Cas9 technology that takes advantage of highly prevalent SNPs in the HTT locus for guiding mutant allele-specific cleavage and show its effectiveness in reducing the expression from mutant HTT alleles in human HD fibroblasts and mice brain.
Materials and Methods
Prediction of SNP-Dependent PAM Motifs
SNPs with a prevalence of ≥5% located upstream (6.5 kilobase) and downstream (Intron1) of HTT exon-1 were obtained from the 1000 Genomes database using the NCBI variation viewer website (http://www.ncbi.nlm.nih.gov/variation/view/?q=HTT&filters=source:dbsnp&assm=GCF_000001405.25). To predict SNP-dependent PAM motifs, SNPs were screened against the consensus PAM sequences of Streptococcus pyogenes (SpCas9, NGG or NAG) and Staphylococcus aureus (SaCas9 NNGRRT) or the CRISPR/Cpf1 systems of Acidaminococcus, NTTT) and Lachnospiraceae (LbCpf1, heterogeneous PAMs). Only those SNPs positioned in a conserved nucleotide PAM position in which the nucleotide variation disrupted the consensus PAM were predicted as SNP-dependent PAM motifs.
Cell Culture and Transfection
HEK293 cells (obtained from CHOP Research Vector Core stock) were maintained in DMEM media containing 10% fetal bovine serum (FBS), 1% L-Glutamine, and 1% penicillin/streptomycin at 37°C with 5% CO2. Cells were cultured in 24 well plates and transfected at 80%–90% confluence using Lipofectamine 2000 transfection reagent, according to the manufacturer’s protocol. Human HD patient fibroblasts (obtained from Coriell Institute for Medical Research cell repository) were maintained on MEM media supplemented with 15% FBS, 1% MEM non-essential amino acids, 1% penicillin/streptomycin, and 1% L-Glutamine at 37°C with 5% CO2. DNA transfection was done by electroporation using Invitrogen Neon transfection reagent using the electroporation conditions (ND31551: 1650V, 10 ms, and three pulses and ND33392: 1450V, 20 ms, and two pulses) and following the guidelines provided by manufacturer. Cells were not authenticated or tested for Mycoplasma by the investigators since they previously passed the quality controls of CHOP Research Vector Core and the Coriell Institute for Medical Research cell repository. None of the cells used in the study were listed in ICLAC database of commonly misidentified cell lines.
sgRNA and Cas9 Plasmid Construction
The plasmid pX330 containing the SpCas9 and sgRNA expression cassettes was kindly provided by Dr. Feng Zhang and used as a template for further modifications. To determine transfection efficacy and for selecting positive transfected cells, a CMV reporter cassette expressing eGFP/P2A/puromycin fusion protein was cloned downstream of the SpCas9 expression cassette. For all sgRNAs, the guide complementary sequences were cloned using a single cloning step with a pair of partially complementary oligonucleotides. The oligo pairs encoding the genomic complementary guide sequences were annealed and ligated into the BbsI cloning site upstream and in frame with the invariant scaffold of the sgRNA sequence. A hU6 or the hH1 Pol3 promoter was use to drive expression of the sgRNA sequences depending on the presence (hU6) or absence (hH1) of a guanine nucleotide at the transcription start site of the sgRNA sequence.
Genomic DNA Extraction, SNP Genotyping, and Genome Editing Analysis
Genomic DNA from HD fibroblast and HEK293 cell lines was extracted using a DNeasy Blood & Tissue Kit (QIAGEN) according to manufacturer’s instructions. SNPs were genotyped by direct Sanger sequencing of PCR amplified products containing the SNPs and using the primers listed in Table S5. To determine which nucleotide variation of SNP1 (rs2857935) was linked to the normal or the mutant allele, the genomic sequence containing SNP1 and the CAG repeat was amplified by PCR and cloned into TOPO plasmids using the TOPO TA Cloning Kit and subsequently transformed into DH5alpha competent cells. Individual colonies were analyzed using Sanger sequencing to determine which nucleotide variant is associated with the normal or mutant allele. Deletions of HTT exon-1 were confirmed on genomic DNA samples by PCR, using primers binding outside the intervening segment cleaved by the sgRNA/SpCas9 complex pair (Table S5).
RNA Extraction, qRT-PCR, and SQ-PCR of HTT Expression Levels
Total RNA was extracted using TRIzol (Life Technologies) according to the manufacturer’s protocol, with the exception of 1 μL Glycoblue (Life Technologies) in addition to the aqueous phase on the isopropanol precipitation step and a single wash with cold 70% ethanol. RNA samples were quantified by spectrophotometry and subsequently cDNAs were generated from 1 μg of total RNA with random hexamers (TaqMan RT reagents, Applied Biosystems). To determine human HTT expression levels in HD fibroblasts and HEK293 cells, we used TaqMan probes for human HTT and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNAs obtained from Applied Biosystems. For determining human and mouse HTT expression levels in BACHD mice samples, we used TaqMan Probes for human HTT mRNA, mouse Htt mRNA, and mouse beta actin mRNA obtained from Applied Biosystems. Relative HTT gene expression was determined using the ddCt method. Allele-specific editing was determined by a Semiquantitative PCR amplification of the CAG repeat within HTT exon-1. RT-PCR experiments were carried out with cDNAs generated from 1 μg of total RNA and using 80 ng for PCR amplification. The RT-exponential phase was determined on 25–30 cycles to allow semiquantitative (SQ) comparison of cDNAs developed from identical reactions with BIOLASE Taq Polymerase (Bioline). A SQ-PCR reaction for ActinB (20 cycles) was used as a reference gene to determine loading differences between samples. The primers and conditions are shown in Table S5.
Huntingtin Western Blots
HEK293 cells were transfected with sgRNA/SpCas9 expression cassettes, selected for 2 days with puromycin (3 μM) and expanded until cells reached confluence. Then, cells were rinsed once with PBS and lysed with Passive Lysis Buffer (PBL, Promega). Protein concentrations were determined using the DC protein assay (Bio-Rad) and 15 μg of protein loaded on a 3%–8% NuPAGE Tris-Acetate Gel (Novex Life Technologies). HD fibroblast cells were electroporated with sgRNA/SpCas9 expression cassettes, selected for 2 days with puromycin (2 μM), and expanded until cells reached confluence. Cells were then rinsed with iced-cold PBS, deattached, pelleted, snap froze, and lysed with SDP lysis buffer (50 mM Tris pH8.0, 150 mM NaCl, 1% NP40, 1× complete protease inhibitors, 1× phosphatase inhibitors, and 100 mM PMSF), followed by incubation on ice for 20 min with occasional vortexing. Debris was removed by centrifugation (15 min, 20,000 g 4°C) and the supernatant retained. Protein concentrations were determined using the DC protein assay (Bio-Rad). Samples (25 μg) were prepared for immunoblotting by denaturing the lysates in LDS sample buffer (Invitrogen) with 2× reducing agent (100 mM DTT, Invitrogen) and heating to 70°C for 10 min. Samples were resolved on a 10% low-Bis acrylamide gels (200:1 acrylamide:Bis) with Tris-glycine running buffer (25 mM Tris, 190 mM Glycine, and 0.1% SDS) containing 10.7 mM Beta mercaptoethanol. Gels were ran on ice for 40 min at 90 V through the stack, then at 190 V. Proteins were transferred o/n at 30 V and 4°C onto polyvinylidene fluoride (PVDF) membranes with NuPAGE Transfer Buffer (Invitrogen: 25 mM Bicine, 25 mM Bis-Tris, 1.025 mM EDTA, 5% MeOH, and pH7.2). Membranes were blocked with 5% milk in PBS-T and then blotted with a mouse anti-HTT (MAB2166. dilution: 1:5,000; Millipore) or rabbit anti beta-catenin (Ab2365. dilution: 1:5,000; Abcam) antibodies followed by horseradish peroxidase-coupled antibodies (Goat anti-mouse: 115-035-146. dilution: 1:10,000 or Goat anti-Rabbit: 111-035-144. dilution: 1:50,000; Jackson ImmunoResearch). Blots were developed with ECL Plus reagents (Amersham Pharmacia). HTT reduction was determined by densitometry (n = 3 independent experiments) of protein levels relative to beta-catenin with the ChemiDoc Imaging System (Bio-rad) and Image Lab analysis software.
rAAV Vector Design and Production
For in vivo studies, two different rAAV vectors were generated. One expressed SpCas9 and one the sgRNAs expression cassettes. Viruses were generated through the Research Vector Core at the Raymond G Perelman Center for Cellular and Molecular Therapeutics at The Children’s Hospital of Philadelphia. SpCas9 was expressed under the control of a minimal cytomegalovirus immediate-early gene enhancer/promoter region (CMV promoter) and cloned upstream of a minimal poly A sequence (FBAAV-Cas9). The sgRNA expression cassettes were moved into an AAV shuttle plasmid containing an eGFP gene under the control of the CMV promoter and upstream of an SV40pA signal. All rAAV plasmid shuttles have AAV2 inverted terminal repeat sequences. RAAV vectors were produced by the standard calcium phosphate transfection method in HEK293 cells by using the AdHelper, AAV1 trans-packaging, and AAV shuttle plasmids. Vector titers were determined by RT-PCR and were 1 × 1013 vg/mL. Vector purity was also tested by silver stain.
Off-Target Analysis
Potential off-target loci for sgHD guide sequences in the human genome were determined using the Cas9-Off finder algorithm previously described by Bae et al.32 To determine potential off-target sites, HD human fibroblasts were electroporated with an sgHD1/i3_Cas9 expression cassette and a 35 nt ODN sequence as described by Tsai et al.22 At 1 week after transfection, genomic DNA was obtained and amplicons generated with Phusion polymerase using PCR primers flanking the potential site. Amplicons were subjected to Sanger sequencing to determine mutations in the cleavage site using specific primers, as well as cloned into TOPO-cloning system for sequence confirmation using 3–4 colonies/site.
Mouse Studies
Animal protocols were approved by The Children’s Hospital of Philadelphia Institutional Animal Care and Use Committee. The 6-week-old male BacHD mice (FVB/N-Tg(HTT*97Q)IXwy/J transgenic mice) were obtained from Jackson Laboratories. Mice were housed in a temperature-controlled environment on a 12 hr light/dark cycle. Food and water were provided ad libitum. Mice were injected at 8 weeks of age with a combination 1:1 of rAAV2/1-SpCas9 virus and rAAV-hU6sgRNA/eGFP virus. For rAAV injections, mice were anesthetized with isoflurane and 5 μL of rAAV mixture injected unilaterally into the right striata at 0.2 μL/min (coordinates: +0.86 mm rostral to Bregma, −1.8 mm lateral to medial, and −2.5 mm ventral from brain surface). After 3 weeks, mice were anesthetized with a ketamine and xylazine mix and perfused with 18 mL of 0.9% cold saline mixed with 2 mL RNAlater (Ambion) solution. Brains were removed, blocked, and cut into 1-mm-thick coronal slices. Tissue punches from striata were taken using a tissue corer (1.4-mm in diameter; Zivic Instruments). All tissue punches were flash frozen in liquid nitrogen and stored at −80°C until use.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism v5.0 software. For all in vitro and in vivo studies, the appropriate experimental sample size was confirmed by performing a power analysis with α = 0.05 resulting on β > 0.8. Outlier samples were detected using the Grubb’s test (α = 0.05). Normal distribution of the samples was determined by using the Kolmogorov-Smirnov normality test. All data with normal distribution were analyzed using one-way ANOVA followed by a Bonferroni’s post hoc or an unpaired t test. Otherwise, data without normal distribution were analyzed using a Mann-Whitney test as indicated. Statistical significance was considered *p < 0.05, ‡p < 0.01, §p < 0.001, and #p < 0.0001. All results are shown as the mean ± SEM.
Author Contributions
A.M.M. and B.L.D. developed the study, designed the experiments, and analyzed the data. A.M.M. and S.A.E. carried out CRISPR-Cas9 related experiments and analyzed data. M.S.K. performed the rAAV injections and assisted with necropsies. A.M.M. and B.L.D. wrote the manuscript with input from all authors.
Acknowledgments
Funding support was provided by Hoppy’s Hope Foundation (A.M.M.), Philly Cure HD (B.L.D.), The Leslie Gehry Brenner Prize (B.L.D.), The Foerderer Grant for Excellence (A.M.M.), the NIH (NS084475 and NS076631; B.L.D.), and The Children’s Hospital of Philadelphia Research Institute. The authors would like to thank Luis Tecedor for statistical consultation.
Footnotes
Supplemental Information includes eight figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2016.11.010.
Contributor Information
Alex Mas Monteys, Email: monteysam@email.chop.edu.
Beverly L. Davidson, Email: davidsonbl@email.chop.edu.
Supplemental Information
References
- 1.Walker F.O. Huntington’s disease. Semin. Neurol. 2007;27:143–150. doi: 10.1055/s-2007-971176. [DOI] [PubMed] [Google Scholar]
- 2.Hicks R.R., Smith D.H., Lowenstein D.H., Saint Marie R., McIntosh T.K. Mild experimental brain injury in the rat induces cognitive deficits associated with regional neuronal loss in the hippocampus. J. Neurotrauma. 1993;10:405–414. doi: 10.1089/neu.1993.10.405. [DOI] [PubMed] [Google Scholar]
- 3.Johnson C.D., Davidson B.L. Huntington’s disease: progress toward effective disease-modifying treatments and a cure. Hum. Mol. Genet. 2010;19(R1):R98–R102. doi: 10.1093/hmg/ddq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto A., Lucas J.J., Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 5.Díaz-Hernández M., Torres-Peraza J., Salvatori-Abarca A., Morán M.A., Gómez-Ramos P., Alberch J., Lucas J.J. Full motor recovery despite striatal neuron loss and formation of irreversible amyloid-like inclusions in a conditional mouse model of Huntington’s disease. J. Neurosci. 2005;25:9773–9781. doi: 10.1523/JNEUROSCI.3183-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drouet V., Perrin V., Hassig R., Dufour N., Auregan G., Alves S., Bonvento G., Brouillet E., Luthi-Carter R., Hantraye P., Déglon N. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 9.Kordasiewicz H.B., Stanek L.M., Wancewicz E.V., Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garriga-Canut M., Agustín-Pavón C., Herrmann F., Sánchez A., Dierssen M., Fillat C., Isalan M. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc. Natl. Acad. Sci. USA. 2012;109:E3136–E3145. doi: 10.1073/pnas.1206506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuccato C., Valenza M., Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 17.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuscu C., Arslan S., Singh R., Thorpe J., Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 20.Anders C., Niewoehner O., Duerst A., Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coles R., Caswell R., Rubinsztein D.C. Functional analysis of the Huntington’s disease (HD) gene promoter. Hum. Mol. Genet. 1998;7:791–800. doi: 10.1093/hmg/7.5.791. [DOI] [PubMed] [Google Scholar]
- 26.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Ge X., Yang F., Zhang L., Zheng J., Tan X., Jin Z.B., Qu J., Gu F. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci. Rep. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochaba J., Lukacsovich T., Csikos G., Zheng S., Margulis J., Salazar L., Mao K., Lau A.L., Yeung S.Y., Humbert S. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. USA. 2014;111:16889–16894. doi: 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Daher M.T., Hangen E., Bruyère J., Poizat G., Al-Ramahi I., Pardo R., Bourg N., Souquere S., Mayet C., Pierron G. Huntingtin proteolysis releases non-polyQ fragments that cause toxicity through dynamin 1 dysregulation. EMBO J. 2015;34:2255–2271. doi: 10.15252/embj.201490808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 32.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray M., Shirasaki D.I., Cepeda C., André V.M., Wilburn B., Lu X.H., Tao J., Yamazaki I., Li S.H., Sun Y.E. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride J.L., Pitzer M.R., Boudreau R.L., Dufour B., Hobbs T., Ojeda S.R., Davidson B.L. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington’s disease. Mol. Ther. 2011;19:2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G., Liu X., Gaertig M.A., Li S., Li X.J. Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. Proc. Natl. Acad. Sci. USA. 2016;113:3359–3364. doi: 10.1073/pnas.1524575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Mendiratta S., Ehrhardt K., Kashyap N., White M.A., Bleris L. Exploiting the CRISPR/Cas9 PAM constraint for single-nucleotide resolution interventions. PLoS ONE. 2016;11:e0144970. doi: 10.1371/journal.pone.0144970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Courtney D.G., Moore J.E., Atkinson S.D., Maurizi E., Allen E.H., Pedrioli D.M., McLean W.H., Nesbit M.A., Moore C.B. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2016;23:108–112. doi: 10.1038/gt.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin J.W., Kim K.H., Chao M.J., Atwal R.S., Gillis T., MacDonald M.E., Gusella J.F., Lee J.M. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum. Mol. Genet. 2016 doi: 10.1093/hmg/ddw286. Published online September 15, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahdar M., McMahon M.A., Prakash T.P., Swayze E.E., Bennett C.F., Cleveland D.W. Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc. Natl. Acad. Sci. USA. 2015;112:E7110–E7117. doi: 10.1073/pnas.1520883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.