Abstract
Laminin-111 protein complex links the extracellular matrix to integrin α7β1 in sarcolemma, thus replacing in dystrophic muscles links normally insured by the dystrophin complex. Laminin-111 injection in mdx mouse stabilized sarcolemma, restored serum creatine kinase to wild-type levels, and protected muscles from exercised-induced damages. These results suggested that increased laminin-111 is a potential therapy for DMD. Laminin subunit beta 1 and laminin subunit gamma 1 are expressed in adult human muscle, but laminin subunit alpha 1 (LAMA1) gene is expressed only during embryogenesis. We thus developed an alternative method to laminin-111 protein repeated administration by inducing expression of the endogenous mouse Lama1 gene. This was done with the CRSPR/Cas9 system, i.e., by targeting the Lama1 promoter with one or several gRNAs and a dCas9 coupled with the VP160 transcription activation domain. Lama1 mRNA (qRT-PCR) and proteins (immunohistochemistry and western blot) were not detected in the control C2C12 myoblasts and in control muscles. However, significant expression was observed in cells transfected and in mouse muscles electroporated with plasmids coding for dCas9-VP160 and a gRNA. Larger synergic increases were observed by using two or three gRNAs. The increased Lama1 expression did not modify the expression of the α7 and β1 integrins. Increased expression of Lama1 by the CRISPR/Cas9 system will have to be further investigated by systemic delivery of the CRISPR/Cas9 components to verify whether this could be a treatment for several myopathies.
Keywords: CRISPR/Cas9, laminin-111, dCas9-VP160, Duchenne muscular dystrophy, DMD, transcription factor, transcription, induction of transcription
Introduction
Duchenne muscular dystrophy (DMD) is a severe recessive X-linked muscular dystrophy characterized by rapid muscle degeneration.1 DMD is due to a mutation of the dystrophin gene, leading to the absence of this protein.2 Dystrophin is thought to strengthen muscle cells by anchoring elements of the internal cytoskeleton (actin) to dystrophin complex present in the membrane.3 β-dystroglycan, one of the dystrophin associated proteins, provides a mechanical link with the extracellular matrix (ECM).4 This attachment stabilizes the membrane and protects the sarcolemma from the stresses during muscle contraction. In DMD patients, due to the absence of dystrophin, β-dystroglycan is absent from the muscle fiber membrane, and the linkage between the contracting apparatus and the extracellular matrix is lost. This causes muscle fiber damage during contraction. The damaged muscle fibers are repaired by the proliferation of muscle specific stem cells, the satellite cells, located in a niche containing laminin and in close contact with the muscle fibers. Although several therapeutic avenues are under investigation, there is currently no effective DMD treatment.5, 6, 7, 8, 9, 10 An increase of Lama1 also improved α7 integrin congenital myopathy and the viability of a mouse model of merosin-deficient congenital muscular dystrophy.11, 12
The laminin family of glycoproteins is a major constituent of the ECM.13, 14 The known laminin isoforms are formed by combinations of α, β, and γ chains. The trimeric protein complex binds to other molecules in the ECM and in the cell membrane. Laminins bind to integrin receptors, which are heterodimers composed of α and β subunits. Eighteen α subunits and eight β subunits can assemble in 24 different combinations that have overlapping binding specificity and cell-type-specific expression patterns.15, 16, 17, 18 Integrins are involved in mechanical linkage with the various cytoskeletal networks.19 The interactions of laminins with their integrin receptors induce modifications in the organization of the cytoskeleton and the phosphorylation of signaling proteins and activate signaling pathways.20, 21, 22, 23, 24, 25, 26 The laminin/integrin interactions influence cell survival, proliferation, differentiation, adhesion, and migration. Laminin α1β1γ1 (laminin-111) is the most widely studied isoform; it is expressed during embryonic development but absent in adult skeletal muscles, which, however, still express the β1 and the γ1 laminin chains. Laminin-111 binds to integrins α6β1 and α7β1.27 The Burkin’s group28, 29, 30 showed that intramuscular (i.m.) and intraperitoneal (i.p.) injection of Lam111 protein complex into the mdx mouse model of DMD2 increased expression of integrin α7β1, stabilized the sarcolemma, restored serum creatine kinase to wild-type levels, and protected muscle from exercised-induced damages. Our research group has shown that i.m. injections of mouse laminin-111 protected the mdx muscles during eccentric contractions and significantly increased the strength,31 independently confirming the results published by Burkin’s group.29 Laminin-111 makes links between the extracellular matrix and the integrin α7β1 in the muscle fibers thus replacing in embryonic dystrophic muscles the links between the matrix and the membrane normally insured by the dystrophin complex. It is not clear whether the beneficial effects of laminin-111 are due to the upregulation of the integrin α7β1 receptors and the consequent changes of intracellular signaling or to the new physical links between the laminin-111 in the extracellular matrix and the integrin α7β1 receptors in the muscle fiber membrane. Indeed, it has previously been shown that the absence of integrin α7 in dystrophin-deficient mice causes a myopathy similar to DMD.28, 30, 32 These results suggested that laminin-111 is a potential therapeutic agent for DMD.
The normal embryonic muscle fiber membrane is thus attached to the ECM by two important protein complexes: the dystrophin complex (via β-dystroglycan) and the laminin-111 complex (via integrin α7β1 receptor). Since Lama1 is expressed only during embryonic development, the links are insured only by the dystrophin complex in the normal adult muscles but are completely absent in the dystrophinopathies.
To be applicable to DMD patients, the positive results obtained in mdx require the GMP production of laminin-111 protein and repeated administrations. Although the Lama1 gene is expressed only during embryogenesis, the β1 and the γ1 chains are still expressed by adult human muscle fibers. Thus, the production and injection of only Lama1 would be sufficient, but the GMP production of this large protein is difficult and repeated administration would be expensive. Moreover, it is not sure whether the laminin-111 complex would be formed by the injection of only Lama1. Thus in the present project, we have developed an alternative to the repeated Lama1 protein administration by inducing the expression of the endogenous Lama1 gene. This was done by targeting the Lama1 promoter with the new CRISPR technology, i.e., one or several guide RNAs (gRNAs) and a non-functional Cas9 (i.e., dCas9) coupled with the VP160 transcription activation domain. Large induction of Lama1 expression was obtained in vitro in cultured mouse myoblasts and in vivo in mdx mouse muscles.
Results
Testing of gRNAs Targeting Lama1 Promoter
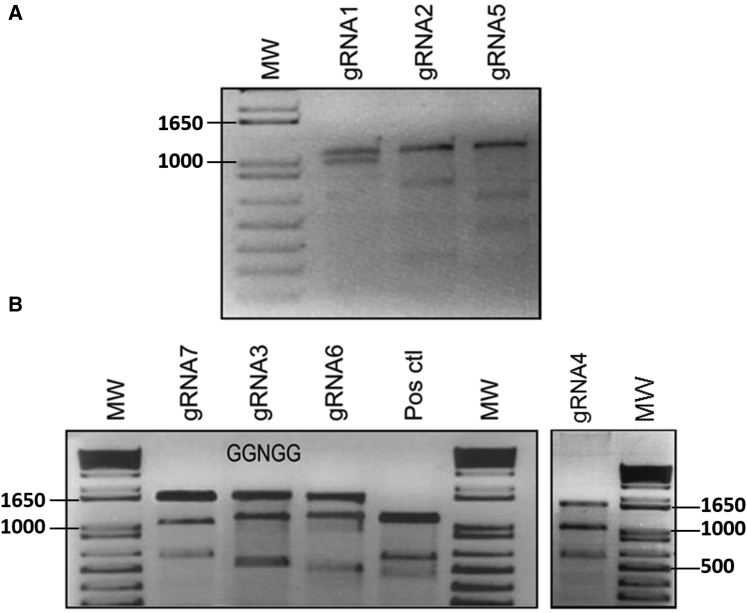
We designed seven gRNAs to target the mouse Lama1 promoter (Figure 1). Plasmids pX330-U6-Chimeric_BB-CBh-hSpCas9 coding for one of these gRNAs and the SpCas9 gene were transfected in C2C12 mouse myoblasts. The gRNAs correctly accessing the promoter induced a DNA double-strand break (DSB), which was spontaneously repaired by nonhomologous end joining (NHEJ). This is an imperfect repair system that results in micro-insertions or micro-deletions (i.e., INDELs). As shown by the Surveyor enzyme test in Figure 2, the seven gRNAs induced cuts in the amplicons indicating that they were able to effectively target the Lama1 promoter. It is gRNA3 that produced the most DSBs as indicted by the Surveyor enzyme test.
Figure 1.
Target Sites of gRNAs in the Promoter of the Mouse Lama1 Gene
The figure illustrates part of the sequence of the Lama1 promoter. The start signal (ATG) is highlighted in yellow. The positions of two pairs of primers are highlighted in gray and in brown. The first pair produces a 1,261-bp amplicon and the second pair a 1,602-bp amplicon. The positions of six different gRNA target sequences are highlighted in various colors. The PAM sequence is underlined. The positions of two transcription factors (KLF4 and SP1) are also highlighted, respectively, in green and in light blue. These two sites are partially overlapped, respectively, the gRNA4 and gRNA5 target sites.
Figure 2.
Results of Surveyor Enzyme Assay
The plasmids coding for SpCas9 and each of the seven gRNAs were transfected in C2C12 cells. 48 hr later, the DNA was extracted from the cells and amplified with either primer set #1 (in A producing a 1,261-bp amplicon) or primer set #2 (in B producing a 1,602-bp amplicon). Two additional bands were detected by gel electrophoresis following digestion of the amplicons with the Surveyor enzyme (CelI) indicating that all gRNAs were able to access the Lama1 promoter and had induced INDELs. The positive control is a mixture of amplicons containing a mismatch so that they are cut by the Surveyor enzyme.
gRNAs in Combination with dSpCas9-VP160-2A-Puro Increased the Expression of the Lama1 mRNA
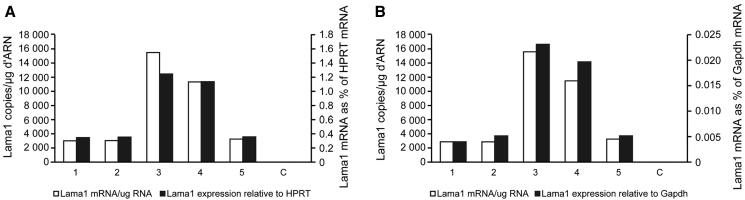
Five gRNAs were then individually inserted in the plasmid pAC154-dual-dCas9VP160-sg Expression (pAC154) coding for dCas9-VP160, a 2A peptide, a puromycin resistance gene and a gRNA. One microgram of each of these five plasmids was transfected in C2C12 myoblasts. Following puromycin selection, Lama1, HPRT, and GAPDH mRNAs were quantified 3 days later by qRT-PCR. Lama1 mRNA was not detected in the negative control cells transfected with pAC154 not containing a gRNA. However, between 3,000 and 16,000 copies of Lama1 mRNA per microgram of total RNA were detected in the cells transfected with pAC154 coding for one gRNA (Figure 3). The results of Lama1 mRNAs were also normalized with the HPRT mRNAs (Figure 3A) and the GAPDH mRNA (Figure 3B). The Lama1 mRNAs were up to 1.2% of the HPRT mRNAs and up to 0.023% of the GAPDH mRNAs. With all methods of presenting the results, gRNAs 3 and 4 produced the highest increases of the Lama1 mRNA.
Figure 3.
Increased Expression of Lama1 by dCas9-VP160 and Individual gRNA1–5
C2C12 cells were transfected with five different pAC154 each coding for dCas9-VP160 and for a gRNA (i.e., gRNAs 1–5). The control cells (C) were transfected with a plasmid coding for eGFP. The Lama1 mRNA was quantified by qRT-PCR and expressed either as the number of copies per microgram of total RNA (left axis and white bars) or normalized with HPRT mRNA (A, right axis, black bars) or GAPDH mRNA (B, right axis, black bar).
Combinations of Two or Three gRNAs and dCas9-VP160 Further Increased the Expression of the Lama1 mRNA
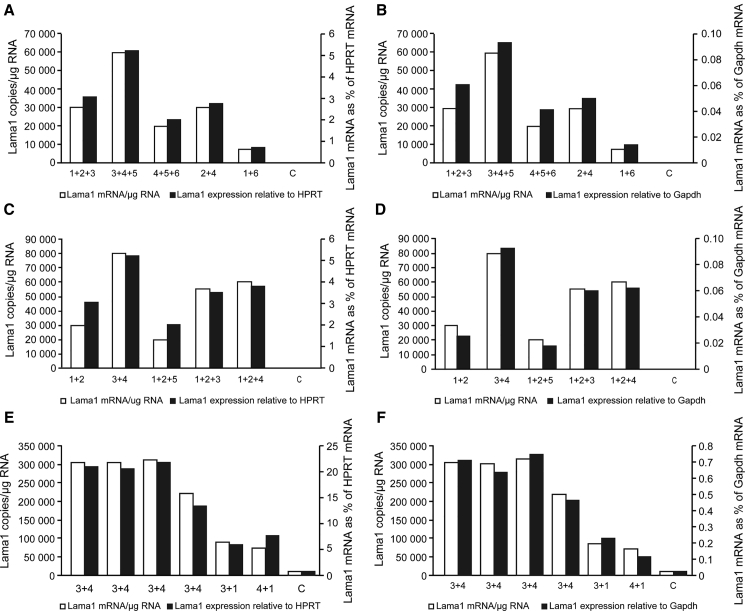
C2C12 cells were transfected with two or three pAC154 plasmids (equal amount of each plasmid for a total of 4 μg) coding for different gRNAs and dCas9-VP160. As in the previous experiment, the Lama1 mRNA was quantified by qRT-PCR, and the results were normalized with the HPRT and the GAPDH mRNAs. The combinations of two or three gRNAs produced higher increased of Lama1 expression than a single gRNA (between 8,000 to 80,000 copies per microgram of total RNA) (Figures 4A–4D), demonstrating an important synergic effect when two or three gRNA are binding to the Lama1 promoter. These synergic effects were also clearly detected when the results were normalized relative to HPRT mRNAs (Figures 4A, 4C, and 4E) and relative to GAPDH mRNAs (Figures 4B, 4D, and 4F). The Lama1 mRNA was not detected in the negative control cells transfected with pAC154 plasmid containing the dCas9VP160 but not coding for a gRNA.
Figure 4.
Synergic Increased Expression of Lama1 by Two or Three gRNAs with dCas9-VP160
The C2C12 cells were transfected using Lipofectamine (A–D) or TransfeX (E and F) with two or three plasmids pAC154 each coding for dCas9-VP160 and for a gRNA. The control cells (C) were transfected with a pAC154 plasmid containing the dCas9VP160 but not coding for a gRNA. The Lama1 mRNA was quantified by qRT-PCR and expressed either as the number of copies per microgram of total RNA (left axis and white bars) or normalized with HPRT mRNA (A, C, and E, right axis, black bars) or Gaphd mRNA (B, D, and F, right axis, black bar). Each bar represents the result of an independent experiment. The combination of gRNA3 and gRNA4 produced the best induction of Lama1 expression. In the best situation, there were 300,000 copies of Lama1 mRNA per microgram of total RNA, i.e., 22% of the HPRT mRNA and 0.7% of the GAPDH mRNA. The Lama1 mRNA was not detected in the negative control cells.
The increase of Lama1 mRNA was further improved by transfecting 2 μg of each pAC154 plasmid with a new agent called TransfeX, instead of using Lipofectamine 2000. With the combination of gRNA3 and gRNA4 300,000 mRNA were detected per microgram of total RNA (Figures 4E and 4F). This is 22% of the HPRT mRNA and 0.7% of the GAPDH mRNA. The Lama1 mRNA was not detected in the negative control cells transfected with pAC154 plasmid containing the dCas9VP160 but not coding for a gRNA.
Combinations of Two gRNAs and dCas9-P300 Further Increased the Expression of the Lama1 mRNA
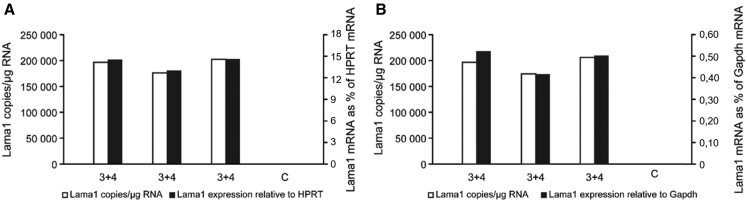
C2C12 cells were transfected with two pAC154 plasmids (2 μg of each plasmid) coding for two different gRNAs and dCas9-P300 using the optimized protocol with TransfeX. The Lama1 mRNA was quantified by qRT-PCR, and the results were normalized with the HPRT and the GAPDH mRNAs. As in the previous experiment the combination of gRNA3 and gRNA4 produced higher increased of Lama1 expression than a single gRNA, but the increased of Lama1 with P300 was less than the increased of Lama1 with VP160. dCas9 VP160 produced the highest increase of the Lama1 mRNA with 300,000 copies per microgram of total RNA, while 200,000 copies per microgram of total RNA when we used dCas9-P300 (Figure 5).
Figure 5.
Combinations of gRNA3 and gRNA4 with dCas9-P300 Further Increased the Expression of the Lama1 mRNA
The C2C12 cells were transfected with TransfeX with two plasmids coding for dCas9-P300 and for gRNA3 and gRNA4. The control cells (C) were transfected with a pAC154 plasmid containing the dCas9VP160 but not coding for a gRNA. The Lama1 mRNA was quantified by qRT-PCR and expressed either as the number of copies per microgram of total RNA (left axis and white bars) or normalized with HPRT mRNA (A, right axis, black bars) or GAPDH mRNA (B, right axis, black bar). The figure illustrates three independent replications of the experiment.
Detection of Lama1 Protein by Immunocytochemistry
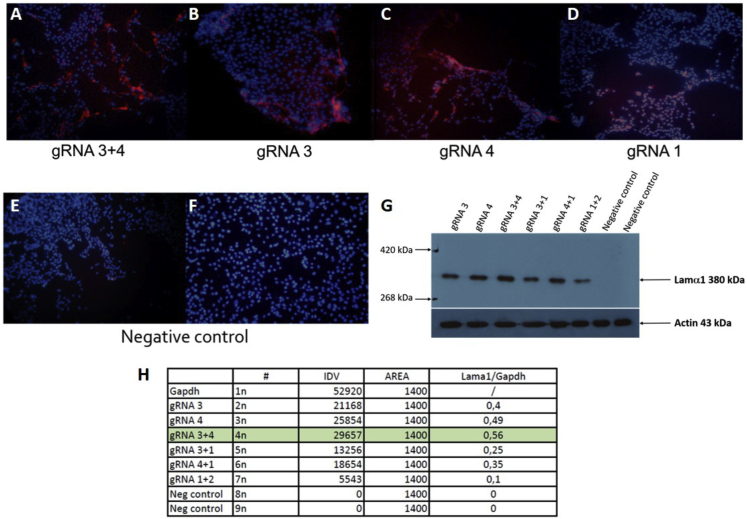
C2C12 cells were transfected with Lipofectamine 2000 with 2 μg of pAC154 plasmid coding either for gRNA1, gRNA3, and gRNA4 or for a combination of gRNA3 and gRNA4. The laminin subunit alpha 1 protein was detected by immunocytochemistry with all gRNAs but was higher with the combination of gRNA3 and gRNA4 (Figure 6). No fluorescence was observed in the control cells, which were transfected with the empty pAC154 plasmid.
Figure 6.
Detection of Laminin Subunit Alpha 1 Protein by Immunocytochemistry in C2C12 Cells
C2C12 cells were transfected with pAC154 coding for dCas9-VP160-2A-puro and a gRNA. The cells were selected with puromycin, expanded during 7 days and fixed with ethanol 95%. (A–F) The laminin subunit alpha 1 protein was detected by immunofluorescence (red label). The nuclei were stained with Hoechst 33258. (E and F) The negative control cells were transfected with a plasmid pAC154 not coding for a gRNA. (G) Proteins were extracted 18 days later, and laminin subunit alpha 1 protein was detected by western blot. (H) The best induction of the laminin subunit alpha 1 protein was observed in the cells transfected with gRNA3 and gRNA4. No laminin subunit alpha 1 protein was detected in the negative control cells transfected either with pAC154 not coding for a gRNA or a plasmid coding for eGFP.
Detection of Lama1 Protein by Western Blot
C2C12 cells were also transfected with 1, 2, or 3 plasmids coding for gRNAs. The total amount of plasmid was kept constant at 4 μg. The cells were selected with puromycin and proliferated during 3 weeks before protein extraction. The Lama1 protein was detected by western blot more strongly in the cells treated with the gRNA3 and gRNA4 (Figure 6). Since Lama1 is not expressed in the negative controls, it is impossible to indicate by how many folds its expression was increased. Quantification of the western blot and normalization relative to GAPDH protein indicated that the smallest increase was obtained with the combination of gRNA1 and gRNA2. gRNA3 or gRNA4 alone increased the Lama1 expression, respectively, 4 and 4.9 times more than the combination of gRNA1 and gRNA2. Finally, the best increased was obtained with the combination of gRNA3 and gRNA4, which increased Lama1 by 5.6-fold relative to the combination of gRNA1 and gRNA2.
Increase of Lama1 Expression in rag/mdx Muscles
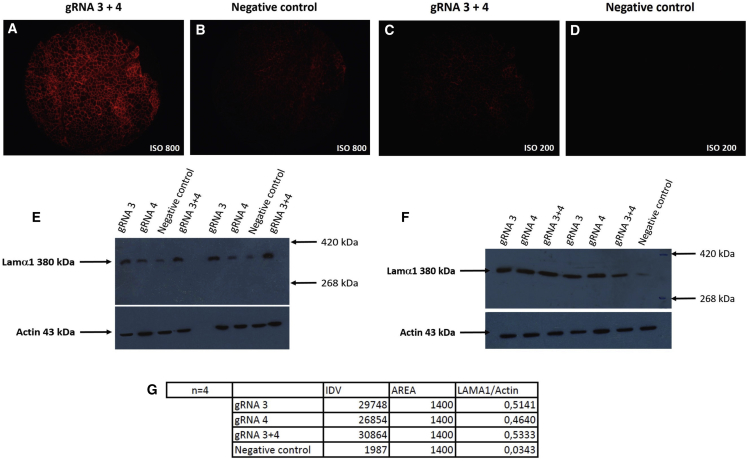
Tibialis anterior rag/mdx muscles of four mdx/rag mice (4 months old) were electroporated with a total of 40 μg of pAC154 plasmids coding for one or two gRNAs. The negative control muscles were subjected to the same treatment but with a pAC154 plasmid not containing a gRNA and the dCas9-VP160 gene. The muscles were removed 2 weeks later, and heterogeneous distributions of Lama1 protein were detected by immunohistochemistry in the muscles treated with a gRNA. The most intense immunolabeling was obtained with a combination of two plasmids, one coding for gRNA3 and the other for gRNA4 (Figure 7). Only background staining was detected in the control muscles. The immunohistochemistry results were also confirmed by western blots showing an increase of Lama1 by the gRNA and the dCas9-VP160 (Figure 7).
Figure 7.
Detection of Laminin Subunit Alpha 1 Protein In Vivo in Tibialis Anterior of rag/mdx Mouse by Immunohistochemistry and Western Blot
The muscles of Rag/mdx mice were injected with plasmids coding for dCas9- VP160 and for gRNA3 and gRNA4 and electroporated. The control muscles were injected with saline and electroporated with the same parameters. Muscles were collected 2 weeks later, and laminin subunit alpha 1 protein was detected by immunohistochemistry in cryostat sections of the muscles electroporated with dCas9-VP160 and both gRNAs (A and C) The staining appeared non-uniform at ISO 200 (C). In the control muscle (B and D), only background staining was detected at ISO 800 (B) and no staining at ISO 200 (D). Proteins were extracted from the muscle sections, and laminin subunit alpha 1 protein was detected by western blot. (E) Induction of the laminin subunit alpha 1 protein was observed in muscles transfected with gRNA3 and/or gRNA4. No laminin subunit alpha 1 protein was detected in the negative control cells transfected either with pAC154 not coding for a gRNA or a plasmid coding for eGFP. (F) Quantification of the laminin subunit alpha 1 bands did not indicate a synergic effect in vivo. (G) The western blot was quantified with the Alpha Imager. The integrated density value (IDV) was measured for a constant area, and the value obtained for the LAMA1 band was normalized with the value of the actin band in the same lane.
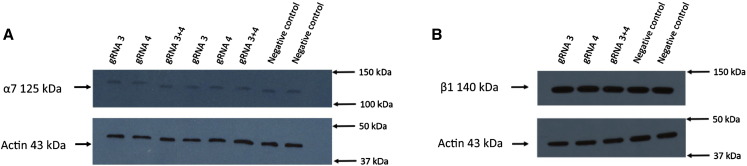
No Increase of α7 and β1 Integrins Expression in rag/mdx Muscles
The possible modification of α7 and β1 integrins expression induced by the expression of Lama1 was also investigated by western blots in the muscles of the rag/mdx mice electroporated with the pAC154 plasmids. No increase of these proteins was observed (Figure 8).
Figure 8.
Detection of α7 and β1 Integrins by Western Blots
The expression of the α7 (A) and β1 (B) integrins was also investigated by western blots (n = 3 for α7 integrin and n = 2 for α7 for β1 integrin) in the same muscles as for Figure 7. There was no significant change of the expression of these integrins following the increase of laminin subunit alpha 1 protein induced by the Cas9-VP160.
No Modification of the Expression of Potential Off-Target Genes by gRNAs 3 and 4
The potential off-target effects of the gRNAs 3 and 4 were identified with the Benchling software. None of these off-target sequences were located in promoters. The expression of the genes located close to these off-target binding sites (i.e., phosphatidylinositol transfer protein, membrane-associated (GenBank 18739), Dimethylglycine Dehydrogenase (GenBank 29958), Microtubule Associated Tumor Suppressor 1 (GenBank 57509), and predicted gene 3329 (GenBank 100041424) were nevertheless investigated by qRT-PCR (primers in Table 2). No modification of the expression of these genes was detected.
Table 2.
Sequence primers for PCR
| Gene Symbol | Description | GenBank | Primer Sequence 5'→3' (S/AS) | Denaturation | Annealing | Elongation |
|---|---|---|---|---|---|---|
| LAMA1 | Mus musculus laminin, alpha 1 (Lama1) | Gene ID: 16772 | TTCCCAGAGGTCTCCATCAATAAC/GCGCTTGCTTCCTTTACACTCAG | 95°C, 10s | 60°C, 10s | 72°C, 14s |
| Pitpnm1 | Mus musculus, phosphatidylinositol transfer protein, membrane-associated 1 | Gene ID: 18739 | AGGTCTCGATTAGCGATTTAACTG/ATTACCGTGGATCTAAGTTTCGT | 95°C, 10s | 58°C, 10s | 72°C, 14s |
| Dmgdh | Mus musculus, Dimethylglycine Dehydrogenase | Gene ID: 29958 | ACCGTGTATTAAAGCGCTGGCTGA/AGCGTGGTACTCCCCAGTCTTAAA | 95°C, 10s | 60°C, 10s | 72°C, 12s |
| Mtus1 | Mus musculus, microtubule-associated tumor suppressor 1 | Gene ID: 57509 | CCGGATTATGGCGCTTTAGAATGT/CGGATTAAGCCTCTATTGTAGAAC | 95°C, 10s | 61°C, 10s | 72°C, 16s |
| Gm3329 | Mus musculus, predicted gene 3329 | Gene ID: 100041424 | TTATCGGCGTGTTAGAAAGTTCGT/CACCTCCTGAAGTGCGTGGGGTCA | 95°C, 10s | 60°C, 10s | 72°C, 13s |
| Hprt1 | Mus musculus hypoxanthine guanine phosphoribosyl transferase 1 | NM_013556 | CAGGACTGAAAGACTTGCTCGAGAT/ CAGCAGGTCAGCAAAGAACTTATAGC | 95°C, 10s | 60°C, 10s | 72°C, 14s |
| GAPDH | Mus musculus glyceraldehyde-3-phosphate dehydrogenase | NM_008084 | GGCTGCCCAGAACATCATCCCT/ ATGCCTGCTTCACCACCTTCTTG | 95°C, 10s | 60°C, 10s | 72°C, 14s |
| ADNg Ctrl | Mus musculus chromosome 3 genomic contig, strain C57BL/6J (HSD3B1 intron) | NT_039239 | CACCCCTTAAGAGACCCATGTT/ CCCTGCAGAGACCTTAGAAAAC | 95°C, 10s | 60°C, 10s | 72°C, 14s |
Discussion
The CRISPR technology is revolutionizing molecular biology and permits to develop new therapies based on genome editing or modification the expression of a specific gene. In the present article, we have been able to strongly induce the expression of the silent Lama1 gene with one to three gRNAs and the dCas9 coupled with the VP160 Transcription Activation Domain (TAD), as previously reported for other genes.33, 34, 35
The gRNA3 produced the best induction of Lama1 expression. This may be because it targets a sequence ending with GG located just before the NGG PAM. Indeed, such a target sequence ending with GG has been previously reported to be more effective.36 The gRNA4 also produced a large increase of the Lama1 expression. This gRNA binds to a nucleotide sequence close to the binding site of Klf4. The Klf4 factor inhibits the expression of the Lama1 gene.37 The binding of the gRNA4 may thus inhibit the inhibitory action of Klf4. Thus, it is not surprising that gRNA3 and gRNA4 produced synergic effects because they act on different sites through different mechanisms.
It is possible that the gRNAs targeting the promoter of the Lama1 gene may also affect the expression of other genes. Possible off-target sequences of the best gRNAs (i.e., gRNA3 and gRNA4) were identified with the Benchling software (https://benchling.com/editor). None of the possible off-target sequences were located in promoters. We nevertheless verified by qRT-PCR whether the expression of the genes located closest to these off-target binding sites was modified. We did not observe any significant change in their expression.
For an eventual clinical application, the potential off-target binding sites in humans may be further reduced by using shorted gRNAs38 or the modified eSpCas9, containing four mutated amino acids to reduce non-specific binding.39 A non-functional (dead) version of eSpCas9 (deSpCas9) or eSaCas9 would have to be tested. Using this deSpCas9-VP160 or deSaCas9-VP160, potential modifications of the expression of all genes could be investigated using the RNA sequencing (RNA-seq) technique.40 For an eventual therapeutic application, the deSaCas9-VP160 and the gRNAs should be delivered systemically to all skeletal muscles using an AAV vector. To limit Lama1 expression only to skeletal muscles, the promoter of deSaCas9-VP160 should be muscle specific. Moreover, to prevent overexpression of Lama1, the Cas9-VP64 gene or the sequences coding for the sgRNAs can be placed under a promoter inducible by doxycycline and 4-hydroxytamoxifen.41, 42, 43, 44
Lama1 is not expressed in adult muscles, but laminin subunit alpha 2 (Lama2), laminin subunit beta 1 (Lamb1), and laminin subunit gamma 1 (Lamc1) are still expressed (http://www.genecards.org/) forming laminin-211. The induction of the Lama1 expression in adult muscles should also permit the formation of the laminin-111 complex. The expression of Lama1 will thus change the equilibrium between the different types of laminin chains. Maybe more laminin subunit beta 1 (Lamb1) and laminin subunit gamma 1 (Lamc1) will be produced to compensate. This will require further studies.
The laminin-111 complex is secreted and participates to the formation of the extracellular matrix. This complex binds with integrin α7β1 present in the muscle fiber membrane. This thus forms a link between the sarcolemma and the extracellular matrix. Dystrophin forms a complex in the sarcolemma with dystroglycans and sarcoglycans. This complex forms links with proteins of the extracellular matrix. This link between the sarcolemma and the extracellular matrix normally protects the muscle fiber membrane during contraction. This protection is missing in the DMD patients because the dystrophin complex does not form in the sarcolemma in the absence of dystrophin. The formation of interactions between laminin-111 and the integrin α7β1 would partially compensate for the absence of the dystrophin complex. It is important to note that the increased expression of Lama1 did not increase the expression of the α7 and of the β1 integrins. Thus, Lama1 would have to bind to α7β1 integrins already in the membrane.
Gawlik et al.45 have shown that transgenic overexpression of the mouse Lama1 does not prevent disease progression in mdx mice. The main difference between the between laminin-111 protein delivery versus Lama1 transcription in muscle is that the delivery of the protein only permits a transient effect whereas the CRISPR/dCas9-VP160 system permits a sustained Lama1 expression without treatment repetition. The results of Gawlik et al.45 are surprising in the light of articles by Burkin’s group28, 29 and our group31 that reported beneficial effects in mdx mice of delivering laminin-111. These discrepancies require further investigation. However, Lama1 increase also improved α7 integrin congenital myopathy and the viability of a mouse model of merosin-deficient congenital muscular dystrophy.11, 12 Thus, upregulation of Lama1 may have beneficial effects for other muscular diseases than DMD, and it is thus worth continuing investigation.
Our group also showed that laminin-111 significantly improved the success of myoblast transplantation by improving myoblast proliferation and migration in vitro.31 Similarly, Zou et al.46 demonstrated that laminin-111 increased muscle cell quantity and function following eccentric exercise. Thus, in follow-up studies, we will investigate whether the increase of Lama1 by the CRISPR/Cas9 technology also improves myoblast transplantation.
Different possible DMD treatments have been investigated. Most of them are aiming to restore the expression of the dystrophin gene. In the present article, we are presenting a different potential therapeutic strategy that could by itself reduce the damages induced to the muscle fibers during contractions and thus on the long-term prevent the progressive muscle weakness characteristic of this disease. Our proposed treatment has the advantage that it would be applicable to all DMD patients whatever their mutations, which is not the case for therapies based on exon skipping or gene editing with the CRISPR/Cas9 technology.47, 48, 49, 50, 51
In conclusion, the increased expression of Lama1 will permit to links this component of the extracellular matrix with the integrin α7β1 receptors already in the muscle fibers. This will replace to some extent the links between the matrix proteins and the dystrophin complex, which is absent in DMD patients. Thus, in future experiments, we will verify whether Lama1 induction with the CRISPR/Cas9 technology is therapeutic by itself and improve the success of myoblast transplantation.
Materials and Methods
Expression Vector
The plasmid px330-U6-Chimeric-HsPCas9 (Addgene plasmid #42230) was used for expression of the gRNA and of dCas9. It contains two BbsI restriction sites for insertion of an oligonucleotide, which selects the target sequence of the gRNA. This gRNA is under the control of the U6 promoter. The plasmid pAC154-dual-dCas9-VP160-sgExpression (Addgene plasmid #48240) was used for expression of the gRNA and dCas9-VP160. It contains two BbsI restriction sites for insertion of an oligonucleotide, which selects the target sequence of the gRNA. This gRNA is under the control of the U6 promoter. This plasmid was modified to include a gene encoding resistance to puromycin, the latter is separated from the gene encoding dCas9 via a sequence coding for the 2A peptide. There is a nuclear localization sequence (NLS) fused with the dCas9-VP160 gene.
Construction and Oligonucleotide Synthesis
The 20 nucleotide sequences, which will be targeted by the guide RNAs (gRNAs) in the Lama1 promoter, were identified through the Leiden Muscular Dystrophy Pages Web Site (http://www.dmd.nl/seqs/murefDMD.html) (GenBank NM004006.1). Sequence analysis of Lama1 promoter permitted to identify the Protospacer Adjacent Motif (PAM) in the sense and antisense strands. These PAMs are NGG for Cas9 of Streptococcus pyogenes. Oligonucleotides to target these sequences were synthesized by IDT (Integrated DNA Technologies) to produce seven gRNAs targeting the mouse Lama1 promoter.
Phosphorylation, Formation of Double-Stranded Oligonucleotides, and Ligation
Lyophilized oligonucleotides were obtained from IDT diluted in double distilled water to a final concentration of 1 μg/μL. The oligonucleotides were then phosphorylated using T4 PolyNucleotid Kinase (NEB). The double-stranded oligonucleotide formation was carried out by rising the temperature from 37°C to 95°C during 5 min, followed by a progressive temperature reduction of 5 degrees every 30 s until reaching 25°C. The PAC154-2A-puro plasmid was cut with BbsI (NEB) and purified by gel extraction (Thermo Scientific). A double-stranded oligonucleotide diluted 1–200 (5 ng of oligonucleotide) in double-distilled water was added to the purified pAC154 plasmid. The ligation was done with the Quickligase (NEB) during incubation at 25°C for 25 min.
Transformation of Competent Bacteria
For transformation of DH5α competent bacteria (Invitrogen), 50 ng of PAC154-2A-puro plasmid DNA was used, followed by a heat shock at 42°C for 45 s. The DH5α bacteria were then cultivated on lysogeny broth (LB) agar medium with ampicillin. The second day, colonies were picked and inoculated in LB broth containing ampicillin. Plasmid DNA was extracted on the third day with the Miniprep Kit (Thermo Scientific) and assayed using a Nanodrop (Thermo Scientific).
Transfection of Px330 Plasmids in C2C12 Myoblasts
To check the activity of each gRNA, Px330 plasmids encoding each gRNA were transfected in C2C12 murine myoblasts. The C2C12 cell line was grown in DMEM (Dulbecco’s modified Eagle medium) (Invitrogen) containing 10% fetal bovine serum (FBS) and antibiotics (penicillin/streptomycin). The 6-well plates containing 300,000 cells per well (about 70%–80% confluent) were transfected using 4 μg of plasmid DNA and 2 μL of Lipofectamine 2000 (Invitrogen) previously diluted in Opti-Mem (Invitrogen). The plates were incubated at 37°C in the presence of 5% CO2 for 48 hr. The successful transfection was evaluated by the cell survival after puromycin selection (2 μg/mL). 48 hr after transfection, the genomic DNA was extracted from the C2C12 cells with the phenol-chloroform method. Lama1 promoter was amplified by PCR with the thermal cycler C1000 Touch Bio-Rad. The amplification program was the following: 98°C for 1 min, 98°C for 10 s, 58°C for 20 s, 72°C for 1 min and 72°C for 5 min for a total of 35 cycles.
Transfection of Plasmid PAC154 VP160 in Murine C2C12 Myoblasts and Extraction of Cells for mRNA and Protein Analysis
Pac154-2A-puro plasmids encoding each gRNA were transfected in mouse C2C12 myoblasts with the same procedure as for the Px330 plasmids. The plates were incubated at 37°C in the presence of 5% CO2 for 48 hr. The transfection success was evaluated by cell survival after puromycin selection (2 μg/mL). Enrichment of transfected cells was performed during 15 days after transfection to have sufficient cell RNA to perform the RT-PCR. Cells were transfected with a plasmid PAC154-2A-puro not encoding any gRNA to obtain a negative control that has undergone the same stress as the treated cells. To extract the RNA, the cells were centrifuged to form a dry pellet, which was sent to the sequencing platform RT-PCR. For protein analysis, transfected cells were lysed directly in the well with 200 μL of lysis buffer (20 mM Tris-HCl [pH 7.5], 1 mM DTT, 1 mM PMSF, 1% SDS).
Surveyor Enzyme Test on PCR Products
DNA was extracted from cells 48 hr after transfection and the Lama1 promoter was PCR amplified. The amplicons were isolated, heated at 95°C to denature the DNA and slowly cooled at room temperature (RT) in the Bio-Rad thermocycler (Bio-Rad) to permit the re-annealing of the single stranded DNA chains. If the SpCas9/gRNA complex had induced INDELs, mismatched DNA double strands were formed. The re-annealed amplicons were then digested with the Surveyor/Cel II enzyme (Integrated DNA Technologies) in a water bath at 37°C for 1 hr. This enzyme cuts mismatched DNA strands. The digestion products were separated by electrophoresis on 1.5% agarose gel in 1 × TBE (Tris borate EDTA) at 100 V for 40 min.
Sequencing
To verify that the oligonucleotides have been properly inserted into the plasmid px330-U6-Chimeric-HsPCas9 (plasmid Addgene #42230) and plasmid PAC154-2A-PURO, these constructions were sent for Sanger sequencing to the Sequencing and Genotyping Platform Genomes CHUL/CHUQ using a common forward primer since both plasmids have the same backbone. The sequencing results were analyzed by the BLAST platform.
Plasmid Electroporation in Rag/mdx Mouse Muscles
The Rag/mdx mice were housed in the animal facility of the Centre de Recherche du Centre Hospitalier Universitaire de Québec (CRCHUQ), in accordance with the ethics committee of the institution. They originated from a cross between mdx mice and immunodeficient Rag mice from the Jackson Laboratory. The Tibialis anterior (TA) muscles of Rag/mdx mice were surgically exposed and injected with 40 μL of Tyrode saline containing 40 μg PAC154 plasmids coding for dCas9-VP160 and either for gRNA3 or gRNA4. One muscle was injected with 40 μg mixture of these two plasmids (20 μg of each plasmid), and one control muscle was injected only with saline. The muscle was then electroporated using the ECM 830 Electro Square Porator (BTX Harvard Apparatus) and the following parameters: 100 V/cm muscle, eight pulses of 20 ms, and interval 1 s. Two mice were used for each experimental condition. One mouse of each pair was sacrificed after 2 weeks and the other after 3 weeks.
qRT-PCR
Cells were homogenized in Qiazol buffer (QIAGEN), and total RNA was extracted using the RNeasy microkit on-column DNase (QIAGEN) treatment following the manufacturer’s instructions. Total RNA was measured using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies), and the RNA quality was analyzed on an Agilent BioAnalyzer 2100 (Agilent Technologies). First-strand cDNA was synthetized 2 μg of RNA in a reaction containing 200 U of Superscript III Rnase H-RT (Invitrogen Life Technologies), 300 ng of oligo-dT18, 50 ng of random hexamers, 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 500 μM deoxynucleotides triphosphate, 5 mM dithiothreitol, and 40 U of Protector RNase inhibitor (Roche Diagnostics) in a final volume of 50 μL. This mixture was incubated at 25°C for 10 min and at 50°C for 1 hr. A PCR purification kit (QIAGEN) was used to purify cDNA. The primer pairs were designed using the GeneTool 2.0 software (Biotools), and their specificity was verified with a blast in the GenBank database. The primers were synthetized by IDT (Integrated DNA Technology) (Table 1). cDNA corresponding to 20 ng of total RNA was used to perform fluorescent-based real-time PCR quantification using the LightCycler 480 (Roche Diagnostics). The LightCycler 480 SYBRGreen I Master mix (Roche Diagnostics) was used as described by the manufacturer. The conditions for PCRs were 45 cycles, DMSO 2% denaturation at 95°C for 10 s, annealing at 60°C for 10 s, and elongation and reading at 72°C for 14 s. A melting curve was performed to assess non-specific signal. Calculation of the number of copies of each mRNA was performed according to Luu-The et al.52 using second derivative method and a standard curve of a crossing point (Cp) value versus logarithm of the quantity. The standard curve was established using known amounts of purified PCR products (10, 102, 103, 104, 105, and 106 copies) and a LightCycler 480 v.1.5 program provided by the manufacturer (Roche Diagnostics). PCR amplification efficiency was verified. Normalization was performed using reference genes shown to have stable expression levels from embryonic life through adulthood in various tissues: hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Quantitative real-time PCR measurements were performed by the CHU de Québec Research Center (CHUL) Gene Expression Platform, Quebec, Canada and were compliant with minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines.
Table 1.
Sequence Primers for RT-PCR
| Gene Symbol | Description | GenBank | Size (pb) | Primer Sequence 5'→3' (S/AS) |
|---|---|---|---|---|
| Promotor Lamα1 | Mus musculus laminin, alpha 1 (Lama1) gRNA promotor | – | 1,261 | GTTAAGGACCTGTGCTTAGAGA/TCACCTGCTTTCTAGCTTAGTG |
| Promotor Lamα1 | Mus musculus laminin, alpha 1 (Lama1) gRNA promotor | – | 1,602 | GCATTACTGAGCCACAGTGGATT/ACCTTGAGATCCAGGAGTACAGT |
The four potential off-target genes (i.e., phosphatidylinositol transfer protein, membrane-associated [GenBank 18739], Dimethylglycine Dehydrogenase [GenBank 29958], Microtubule Associated Tumor Suppressor 1 [GenBank 57509], and predicted gene 3329 [GenBank 100041424]) were also quantified by RT-PCR (primers and parameters in Table 2). The other parameters were the same as for Lama1.
Immunocytochemistry on C2C12 Transfected or Not with Puro pAC154-dual-dCas9VP160-sg Expression
C2C12 cells were transfected in a 24-well plate (50,000 cells per well) with the pAC154-dual-dCas9VP160-sgExpression (Addgene plasmid #48240) coding for dCas9-VP160 and one gRNA. After puromycin selection and enrichment, cells were rinsed with Hank’s balanced salt solution (HBSS; Gibco). 500 μL of 95% ethanol was added during 10 min at RT to fix the cells. Ethanol was removed, and each well was washed with 1 mL PBS for 10 min at RT with stirring. The sites of non-specific interactions were then blocked with 1 mL of blocking solution (10% goat serum, Gibco) in PBS for 1 hr at RT with stirring. A mouse monoclonal antibody (mAb) against Lama1 (MAB4656, R&D Systems) was the added to each well at a final concentration of 15 μg/mL overnight at 4°C. The antibody was diluted in 1% goat serum, and 100 μL was added to each well. The next day, three 10 min washes with stirring using 1 mL of PBS were done to remove the excess antibody. Thereafter, 10 μg/mL of secondary antibody was used for one hour at RT, i.e., a goat antibody against rat immunoglobulin G (IgG) coupled with Alexa Fluor 456 (Molecular Probes) diluted 1:200 in PBS containing 1% goat serum. Two other 10-min PBS washes were then done with stirring at RT. During the third wash, Hoechst 33258 (Sigma Aldrich) was added to PBS (1:10,000) for nuclear staining.
Detection of Lama1 by Immunohistochemistry
2 or 3 weeks after electroporation, the mice were anesthetized with isoflurane and euthanized by cervical dislocation. The muscles were dissected out and embedded in Cryomatrix Shandon (Thermo Electron) and frozen in liquid nitrogen. 12-μm transverse cryo-sections were placed on gelatinized slides and stored at −20°C. The sections were fixed with 4% paraformaldehyde for 10 min, washed with 1 × PBS for 5 min, and permeabilized 10 min with PBS containing 0.15% Triton X-100. Non-specific interactions were blocked with PBS 10% fetal bovine serum (Gibco) for 1 hr at room temperature with slow stirring. The sections were incubated overnight at 4°C with a mouse mAb against Lama1 (MAB4656, R&D Systems) at a concentration of 20 μg/ml diluted in the blocking solution. The sections were washed, three times with 1 × PBS for 5 min. and incubated in the dark during 1 hr with secondary antibody coupled with Alexa Fluor 456 (Molecular Probes) at a concentration of 10 μg/ml. The sections were washed with PBS three times and covered with PBS/Glycerol 50/50 solution and a coverslip for fluorescence microscopy observation.
Western Blot
After 15–20 days under puromycin selection, the transfected cells were lysed with 200 μL of lysis buffer (20 mM Tris-HCl [pH 7.5], 1 mM DTT, 1 mM PMSF, 1% SDS). 600 μL of cold methanol was added to 200 μL of cell lysate. 200 μL of chloroform and 500 μL of distilled water were also added to the mixture. This solution was stirred with a vortex before centrifugation during 3 min at 13,000 rpm. The upper and lower phases were removed, and 800 μL of methanol was added to wash the proteins again. The mixture was stirred with a vortex and centrifuged again for 5 min. The supernatant was completely removed, and the protein pellet was concentrated for 10 min using the Savant Speed Vac Concentrator (Thermo Scientific). These concentrated proteins were then resuspended in 50 μL of loading buffer (0.06 M Tris-HCl [pH 6.8], 1% SDS, 10% glycerol). Proteins were subsequently dosed by the colorimetric method bicinchoninic acid (BCA; Thermo Scientific) according to the manufacturer’s protocol. Thereafter, 1% 2-mercaptoethanol and 0.025% bromophenol blue were added to the protein solution. The proteins were then boiled for 5 min and placed on ice for 2 min before centrifugation at 13,200 rpm for 3 min. 15 μg of each protein solution was placed on SDS-PAGE (gel concentration of 4% and 6% separation gel). Migration at 70 V for 15 min and 100 V for 150 min was carried out in running buffer (50 mM Tris, 0.2 M glycine, and 0.1% SDS). Proteins were transferred to a nitrocellulose membrane (Bio-Rad) at 20 V at 4°C overnight using a transfer buffer (50 mM Tris, 0.2 M glycine, 0.1% SDS, and 20% methanol). This was subsequently incubated 1 hr at RT in a blocking solution (0.1% PBS, 0.05% Tween 20 [Lab Mat], and 5% skimmed milk powder) to block non-specific interaction sites. Afterward, the membrane was incubated overnight at 4°C with a rat antibody (Mab4656, R&D Systems) directed against mouse Lama1 (15 μg/ml diluted 1:200 in blocking solution; Life Technologies). The membrane was washed three times for 10 min with agitation in the washing solution (0.1% PBS and 0.05% of Tween 20) to remove the excess or loosely bound antibody following nonspecific binding. Rabbit immunoglobulin anti-rat coupled with horseradish peroxidase (HRP; Dako), diluted 1:1,000 in blocking solution, was incubated on the membrane for 1 hr at RT with stirring. Another series of three washes was done before soaking the membrane for 4 min in a solution to induce chemiluminescence (ECL Western Clarity substrate, Bio-Rad). The result was visualized by exposing the membrane for 28 min to a autoradiographic HyBlot CL film (Denville Scientific).
Western blots were also done to verify whether the Lama1 protein increase modified the expression of the α7 and β1 integrins. For the α7 integrin, the antibody was obtained from Miltenyi Biotec and for the β1 integrin a rabbit mAb Abcam was used.
Author Contributions
A.P. conducted the experiments and wrote the first draft of the paper, J.R. participated in the construction of the plasmids, and J.P.T. designed the experiments and finalized the manuscript.
Conflicts of Interest
A patent application has been submitted by Laval University based on the results reported in the present article.
Acknowledgments
This work was supported by grant 257171 from the Canadian Institute of Health Research (CIHR).
References
- 1.Sussman M. Duchenne muscular dystrophy. J. Am. Acad. Orthop. Surg. 2002;10:138–151. doi: 10.5435/00124635-200203000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E.P., Brown R.H., Jr., Kunkel L.M. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Watkins S.C., Hoffman E.P., Slayter H.S., Kunkel L.M. Immunoelectron microscopic localization of dystrophin in myofibres. Nature. 1988;333:863–866. doi: 10.1038/333863a0. [DOI] [PubMed] [Google Scholar]
- 4.Ervasti J.M., Campbell K.P. Dystrophin-associated glycoproteins: Their possible roles in the pathogenesis of Duchenne muscular dystrophy. Mol. Cell Biol. Hum. Dis. Ser. 1993;3:139–166. doi: 10.1007/978-94-011-1528-5_6. [DOI] [PubMed] [Google Scholar]
- 5.Wagner K.R., Lechtzin N., Judge D.P. Current treatment of adult Duchenne muscular dystrophy. Biochim. Biophys. Acta. 2007;1772:229–237. doi: 10.1016/j.bbadis.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Cossu G., Sampaolesi M. New therapies for Duchenne muscular dystrophy: Challenges, prospects and clinical trials. Trends Mol. Med. 2007;13:520–526. doi: 10.1016/j.molmed.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Aurino S., Nigro V. Readthrough strategies for stop codons in Duchenne muscular dystrophy. Acta Myol. 2006;25:5–12. [PubMed] [Google Scholar]
- 8.Welch E.M., Barton E.R., Zhuo J., Tomizawa Y., Friesen W.J., Trifillis P., Paushkin S., Patel M., Trotta C.R., Hwang S. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 9.Wilton S. PTC124, nonsense mutations and Duchenne muscular dystrophy. Neuromuscul. Disord. 2007;17:719–720. doi: 10.1016/j.nmd.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Wilton S.D., Fletcher S. Exon skipping and Duchenne muscular dystrophy: Hope, hype and how feasible? Neurol. India. 2008;56:254–262. doi: 10.4103/0028-3886.43443. [DOI] [PubMed] [Google Scholar]
- 11.Rooney J.E., Knapp J.R., Hodges B.L., Wuebbles R.D., Burkin D.J. Laminin-111 protein therapy reduces muscle pathology and improves viability of a mouse model of merosin-deficient congenital muscular dystrophy. Am. J. Pathol. 2012;180:1593–1602. doi: 10.1016/j.ajpath.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Ry P.M., Minogue P., Hodges B.L., Burkin D.J. Laminin-111 improves muscle repair in a mouse model of merosin-deficient congenital muscular dystrophy. Hum. Mol. Genet. 2014;23:383–396. doi: 10.1093/hmg/ddt428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belkin A.M., Stepp M.A. Integrins as receptors for laminins. Microsc. Res. Tech. 2000;51:280–301. doi: 10.1002/1097-0029(20001101)51:3<280::AID-JEMT7>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 14.Durbeej M. Laminins. Cell Tissue Res. 2010;339:259–268. doi: 10.1007/s00441-009-0838-2. [DOI] [PubMed] [Google Scholar]
- 15.Hynes R.O., Lively J.C., McCarty J.H., Taverna D., Francis S.E., Hodivala-Dilke K., Xiao Q. The diverse roles of integrins and their ligands in angiogenesis. Cold Spring Harb. Symp. Quant. Biol. 2002;67:143–153. doi: 10.1101/sqb.2002.67.143. [DOI] [PubMed] [Google Scholar]
- 16.Hynes R.O. A reevaluation of integrins as regulators of angiogenesis. Nat. Med. 2002;8:918–921. doi: 10.1038/nm0902-918. [DOI] [PubMed] [Google Scholar]
- 17.Hynes R.O. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 18.Humphries J.D., Byron A., Humphries M.J. Integrin ligands at a glance. J. Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barczyk M., Carracedo S., Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M., O’Connor K.L. Integrin alpha6beta4 promotes expression of autotaxin/ENPP2 autocrine motility factor in breast carcinoma cells. Oncogene. 2005;24:5125–5130. doi: 10.1038/sj.onc.1208729. [DOI] [PubMed] [Google Scholar]
- 21.Linton J.M., Martin G.R., Reichardt L.F. The ECM protein nephronectin promotes kidney development via integrin alpha8beta1-mediated stimulation of Gdnf expression. Development. 2007;134:2501–2509. doi: 10.1242/dev.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzu J., Marinkovich M.P. Bridging structure with function: Structural, regulatory, and developmental role of laminins. Int. J. Biochem. Cell Biol. 2008;40:199–214. doi: 10.1016/j.biocel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yee K.L., Weaver V.M., Hammer D.A. Integrin-mediated signalling through the MAP-kinase pathway. IET Syst. Biol. 2008;2:8–15. doi: 10.1049/iet-syb:20060058. [DOI] [PubMed] [Google Scholar]
- 24.Legate K.R., Montañez E., Kudlacek O., Fässler R. ILK, PINCH and parvin: The tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 2006;7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- 25.Millward-Sadler S.J., Salter D.M. Integrin-dependent signal cascades in chondrocyte mechanotransduction. Ann. Biomed. Eng. 2004;32:435–446. doi: 10.1023/b:abme.0000017538.72511.48. [DOI] [PubMed] [Google Scholar]
- 26.Zhu C.Q., Popova S.N., Brown E.R., Barsyte-Lovejoy D., Navab R., Shih W., Li M., Lu M., Jurisica I., Penn L.Z. Integrin alpha 11 regulates IGF2 expression in fibroblasts to enhance tumorigenicity of human non-small-cell lung cancer cells. Proc. Natl. Acad. Sci. USA. 2007;104:11754–11759. doi: 10.1073/pnas.0703040104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishiuchi R., Takagi J., Hayashi M., Ido H., Yagi Y., Sanzen N., Tsuji T., Yamada M., Sekiguchi K. Ligand-binding specificities of laminin-binding integrins: A comprehensive survey of laminin-integrin interactions using recombinant alpha3beta1, alpha6beta1, alpha7beta1 and alpha6beta4 integrins. Matrix Biol. 2006;25:189–197. doi: 10.1016/j.matbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Rooney J.E., Gurpur P.B., Yablonka-Reuveni Z., Burkin D.J. Laminin-111 restores regenerative capacity in a mouse model for alpha7 integrin congenital myopathy. Am. J. Pathol. 2009;174:256–264. doi: 10.2353/ajpath.2009.080522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rooney J.E., Gurpur P.B., Burkin D.J. Laminin-111 protein therapy prevents muscle disease in the mdx mouse model for Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA. 2009;106:7991–7996. doi: 10.1073/pnas.0811599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rooney J.E., Welser J.V., Dechert M.A., Flintoff-Dye N.L., Kaufman S.J., Burkin D.J. Severe muscular dystrophy in mice that lack dystrophin and alpha7 integrin. J. Cell Sci. 2006;119:2185–2195. doi: 10.1242/jcs.02952. [DOI] [PubMed] [Google Scholar]
- 31.Goudenege S., Lamarre Y., Dumont N., Rousseau J., Frenette J., Skuk D., Tremblay J.P. Laminin-111: A potential therapeutic agent for Duchenne muscular dystrophy. Mol. Ther. 2010;18:2155–2163. doi: 10.1038/mt.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo C., Willem M., Werner A., Raivich G., Emerson M., Neyses L., Mayer U. Absence of alpha 7 integrin in dystrophin-deficient mice causes a myopathy similar to Duchenne muscular dystrophy. Hum. Mol. Genet. 2006;15:989–998. doi: 10.1093/hmg/ddl018. [DOI] [PubMed] [Google Scholar]
- 33.Wu J., Hunt S.D., Xue H., Liu Y., Darabi R. Generation and characterization of a MYF5 reporter Human iPS cell line using CRISPR/Cas9 mediated homologous recombination. Sci. Rep. 2016;6:18759. doi: 10.1038/srep18759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farboud B., Meyer B.J. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics. 2015;199:959–971. doi: 10.1534/genetics.115.175166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piccinni S.A., Bolcato-Bellemin A.L., Klein A., Yang V.W., Kedinger M., Simon-Assmann P., Lefebvre O. Kruppel-like factors regulate the Lama1 gene encoding the laminin alpha1 chain. J. Biol. Chem. 2004;279:9103–9114. doi: 10.1074/jbc.M305804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2015 doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pepke S., Wold B., Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6(11, Suppl):S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu K.I., Ramli M.N., Woo C.W., Wang Y., Zhao T., Zhang X., Yim G.R., Chong B.Y., Gowher A., Chua M.Z. A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing. Nat. Chem. Biol. 2016;12:980–987. doi: 10.1038/nchembio.2179. [DOI] [PubMed] [Google Scholar]
- 42.Cao J., Wu L., Zhang S.M., Lu M., Cheung W.K., Cai W., Gale M., Xu Q., Yan Q. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016;44:e149. doi: 10.1093/nar/gkw660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Solis C.A., Ho A., Holehonnur R., Ploski J.E. The Development of a Viral Mediated CRISPR/Cas9 System with Doxycycline Dependent gRNA Expression for Inducible In vitro and In vivo Genome Editing. Front. Mol. Neurosci. 2016;9:70. doi: 10.3389/fnmol.2016.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng J., Jing J., Sanchez-Lara P.A., Bootwalla M.S., Buckley J., Wu N., Yan Y., Chai Y. Generation and characterization of tamoxifen-inducible Pax9-CreER knock-in mice using CrispR/Cas9. Genesis. 2016;54:490–496. doi: 10.1002/dvg.22956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gawlik K.I., Oliveira B.M., Durbeej M. Transgenic expression of Laminin α1 chain does not prevent muscle disease in the mdx mouse model for Duchenne muscular dystrophy. Am. J. Pathol. 2011;178:1728–1737. doi: 10.1016/j.ajpath.2010.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou K., De Lisio M., Huntsman H.D., Pincu Y., Mahmassani Z., Miller M., Olatunbosun D., Jensen T., Boppart M.D. Laminin-111 improves skeletal muscle stem cell quantity and function following eccentric exercise. Stem Cells Transl. Med. 2014;3:1013–1022. doi: 10.5966/sctm.2014-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson-Hamm J.N., Gersbach C.A. Gene therapies that restore dystrophin expression for the treatment of Duchenne muscular dystrophy. Hum. Genet. 2016;135:1029–1040. doi: 10.1007/s00439-016-1725-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016 doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Rivera R.M. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016 doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tabebordbar M., Zhu K., Cheng J.K., Chew W.L., Widrick J.J., Yan W.X. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016 doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyombe-Engembe J.P., Ouellet D.L., Barbeau X., Rousseau J., Chapdelaine P., Lagüe P., Tremblay J.P. Efficient restoration of the dystrophin gene reading frame and protein structure in DMD myoblasts using the CinDel method. Mol. Ther. Nucleic Acids. 2016;5:e283. doi: 10.1038/mtna.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luu-The V., Paquet N., Calvo E., Cumps J. Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. Biotechniques. 2005;38:287–293. doi: 10.2144/05382RR05. [DOI] [PubMed] [Google Scholar]