Abstract
Aptamer-drug conjugates (ApDCs) have the potential to improve the therapeutic index of traditional chemotherapeutic agents due to their ability to deliver cytotoxic drugs specifically to cancer cells while sparing normal cells. This study reports on the conjugation of cytotoxic drugs to an aptamer previously described by our group, the pancreatic cancer RNA aptamer P19. To this end, P19 was incorporated with gemcitabine and 5-fluorouracil (5-FU), or conjugated to monomethyl auristatin E (MMAE) and derivative of maytansine 1 (DM1). The ApDCs P19-dFdCMP and P19-5FdUMP were shown to induce the phosphorylation of histone H2AX on Ser139 (γ-H2AX) and significantly inhibited cell proliferation by 51%–53% in PANC-1 and by 54%–34% in the gemcitabine-resistant pancreatic cancer cell line AsPC-1 (p ≤ 0.0001). P19-MMAE and P19-DM1 caused mitotic G2/M phase arrest and inhibited cell proliferation by up to 56% in a dose-dependent manner when compared to the control group (p ≤ 0.001). In addition, the cytotoxicity of P19-MMAE and P19-DM1 in normal cells and the control human breast cancer cell line MCF7 was minimal. These results suggest that this approach may be useful in decreasing cytotoxic side effects in non-tumoral tissue.
Keywords: aptamer-drug conjugates, ApDCs, nucleoside analog, cytotoxic agents, pancreatic cancer
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the 12th most common cancer worldwide and the 7th most common cause of cancer-related death.1 In contrast to other cancer types, the mortality rate of PDAC is increasing, and it has been predicted that PDAC will become the second leading cause of cancer-related mortality by 2020.2 Despite efforts to improve the treatment and outcome of patients with PDAC, limited progress has been made.2, 3 The survival rate remains less than 5% at 5 years taking into account all stages of the disease.4 Pancreatic resection remains the only curative option for patients with localized tumors; however, only 15%–20% of patients have resectable disease without metastatic spread at the time of presentation.5
For patients with locally advanced or metastatic disease, gemcitabine, a nucleoside analog with a structure similar to cytarabine, has been the standard treatment for more than 10 years.6 Gemcitabine, however, only increases the 1-year survival rate from 16% to 19%. Despite the introduction of new chemotherapy regimens, the median survival of PDAC patients remains less than 12 months.6, 7 Nevertheless, as systemic chemotherapy provides significant survival benefits and improves the quality of life of patients in comparison with best supportive care alone, it is still recommended by current guidelines.8, 9, 10 One of the current limitations of aggressive chemotherapy in PDAC is treatment-related toxicity.
Although combination therapy has been shown to be associated with higher response rates compared to single-agent regimens,7 multi-agent regimens are associated with more severe cytotoxic side effects, such as neutropenia, sensory neuropathy, vomiting, and diarrhea, all which have a negative impact on quality of life.7 In view of these problems, improvements in selective drug delivery are imperative for newer cytotoxic drugs.
The relatively new agents monomethyl auristatin E (MMAE), a synthetic analog of the natural product dolastatin,11 and the maytansinoid DM1, a derivative of maytansine, have both been investigated in several preclinical studies.12, 13 Both MMAE and DM1 are highly potent antimitotic drugs that bind to microtubules.14 Due to their high toxicity, however, both MMAE and DM1 cannot be used as cytotoxic drugs in their own right. Instead, both MMAE and DM1 have been used in the construction of antibody-drug conjugates (ADCs). ADCs are composed of a cytotoxic drug linked to a monoclonal antibody (mAb), which targets a tumor-associated antigen, thus delivering the cytotoxic drug directly to the cancer cell and so reducing toxicity in non-tumoral cells.15 ADCs can, however, induce an immune response driven by different parts of the conjugates thus compromising their safety and efficacy.16
This study aimed to improve current systemic chemotherapy for PDAC by constructing aptamer-drug conjugates (ApDCs). Aptamers are small structured single-stranded RNA chains capable of recognizing cell-specific receptor targets.
Results
Aptamer-Drug Conjugation: Nucleoside-Analog-Linked ApDCs
The construction of ApDCs involved conjugating the pancreatic-cancer-specific RNA aptamer P1917 to well-known cancer drugs. These included gemcitabine triphosphate (dFdCTP) and 5-fluorouracil (5-FU) triphosphate (5FdUTP). The active metabolites of the drugs, dFdCMP and 5FdUMP (Figure 1A), were incorporated enzymatically into the P19 RNA aptamer. The structure of P19 conjugated with dFdCMP and 5FdUMP is depicted in Figure 1B, where the inclusion of these drugs only marginally increased the molecular weight of P19-dFdCMP (to 1,862 g/mol) relative to P19 due to excess fluorine residues, while P19-5FdUMP remained unchanged (Figure S1A). The sequences of P19-dFdCMP and P19-5FdUMP are shown in Table S1.
Figure 1.
Construction of the ApDC Nucleoside Analogs
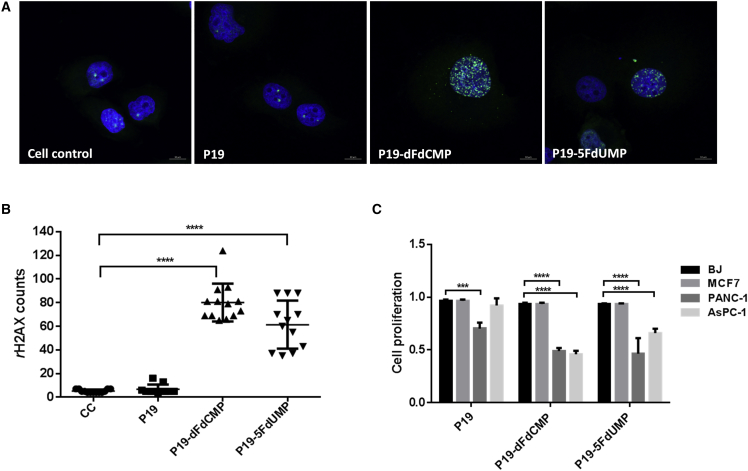
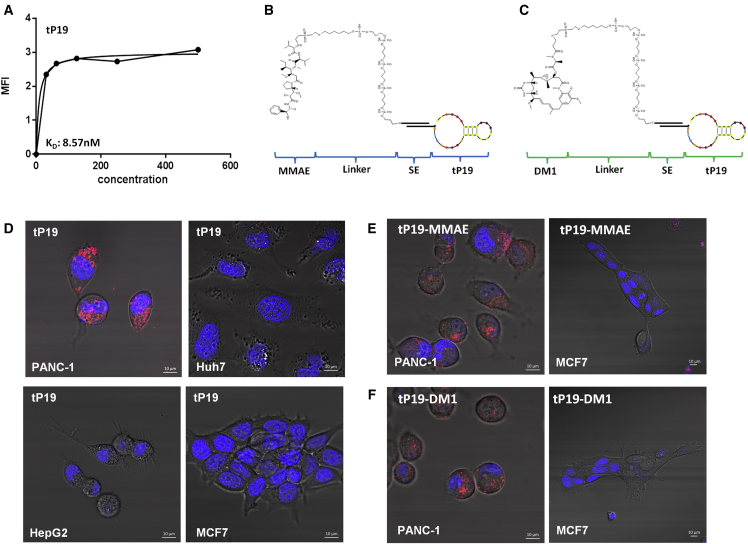
(A) The chemical structures of active metabolites of gemcitabine monophosphate (dFdCMP) and 5-F-2′-UTP monophosphate (5FdUMP) are shown. (B) The structure of P19 intrinsically conjugated with dFdCMP (P19-dFdCMP) and 5FdUMP (P19-5FdUM) is shown with red dots representing dFdCMPs and blue dots represent 5FdUMPs. (C) The dissociation constant (KD) of P19-dFdCMP and P19-5FdUMP was measured by flow cytometry using increasing concentrations of Cy3-labeled aptamers (from 15.6 to 500 nM). Mean fluorescence intensity (MFI) was measured and calculated using a one-site binding model for non-linear regression. (D) The pancreatic cancer cell line PANC-1 was treated with 200 nM of Cy3-labeled P19-dFdCMP and P19-5FdUMP for 4 hr and analyzed by confocal microscopy. Punctate regions of Cy3 labeling were observed in PANC-1 cells. Live-cell imaging. Red, Cy3; blue, Hoechst 33342. Scale bars, 10 μm.
To determine the binding affinity of the ApDCs, we performed fluorescent binding assays with PANC-1 human pancreatic cancer cells to calculate the apparent dissociation constant (KD). Treatment with P19 aptamer alone in PANC-1 was previously measured at a KD of 13.07 nM.17 Both P19-dFdCMP (KD = 7.76 nM) and P19-5FdUMP (KD = 6.9 nM) (Figure 1C, left and right panels) showed slightly increased binding affinity when compared to P19 alone. To determine whether the nucleoside analog ApDC was effectively internalized, we labeled P19-dFdCMP and P19-5FdUMP with Cy3 fluorescent dye and incubated them with our panel of cell types. P19-dFdCMP and P19-5FdUMP showed specific internalization into pancreatic cancer cell line PANC-l cells, while breast cancer cells (MCF7) did not show internalization (Figure 1D). From this, we concluded that both P19-dFdCMP and P19-5FdUMP maintained the same epitope specificity to PDAC cells as our original P19 aptamer.17
P19-dFdCMP and P19-5FdUMP Induce DNA Double-Strand Breaks and Inhibit Cell Proliferation in Pancreatic Cancer Cells
The most well-known mechanism of action for nucleoside analogs is inhibition of DNA synthesis and DNA damage.18 To characterize the level of DNA damage induced by P19-dFdCMP and P19-5FdUMP in PANC-1 cells, we used an indirect immunofluorescence assay (IFA) to measure the appearance of the phosphorylated form of histone H2AX Ser 139 (γ-H2AX), a specific biomarker of DNA double-strand breaks (DSBs).19 P19-dFdCMP and P19-5FdUMP significantly increased the incidence of nuclear DSBs, as indicated by increased γ-H2AX in ApDC-treated cells relative to negative control or P19-only-treated cells (Figure 2A). Quantification of γ-H2AX relative to control demonstrated a significant 16-fold increase in DSBs for P19-dFdCMP and a 12-fold increase for P19-5FdUMP (p ≤ 0.0001, one-way ANOVA) (Figure 2B; Table S2).
Figure 2.
The Effect of the ApDC Nucleoside Analogs on DNA Double-Strand Breaks and Cell Proliferation
(A) PANC-1 cells treated with 500 nM P19-dFdCMP and P19-5FdUMP ApDCs were stained with antibodies against γ-H2AX (indicative of DNA double-strand breaks). Fluorescent images were taken with confocal microscopy. Green, γ-H2AX; blue, Hoechst 33342. Scale bars, 10 μm. (B) Quantification of γ-H2AX was measured by Image-Pro 9.1 algorithm. One-way ANOVA test: ****p ≤ 0.0001. (C) After treatment of PANC-1, AsPC-1, BJ, and MCF7 cells with P19-dFdCMP and P19-5FdUMP for 72 hr, cell proliferation was measured by MTT assay. Cell proliferation was normalized with untreated control cells. One-way ANOVA test: ****p ≤ 0.0001, ***p ≤ 0.001.
To assess the effects of nucleoside analog ApDCs on cell proliferation, we used an MTT assay in PANC-1 and gemcitabine-resistant AsPC-1 cells and BJ and MCF7 cell lines. P19-dFdCMP and P19-5FdUMP (1 μM) significantly inhibited cell proliferation at 72 hr only in PANC-1 and AsPC-1 cells (Figure 2C). PANC-1 and AsPC-1 cells showed significant (and similar) rates of reduction in proliferation following treatment with P19-dFdCMP. In contrast, P19-5FdUMP was more potent in PANC-1 cells than in AsPC-1 cells. No reduction in proliferation was observed in MCF7 and BJ lines treated with P19-dFdCMP and P19-5FdUMP (Figure 2C).
Aptamer-Drug Conjugation: Cytotoxic-Drug-Linked ApDCs
We constructed antimitotic ApDCs by chemical conjugation of a truncated form of the P19 aptamer to MMAE and DM1. First, in order to facilitate increased binding affinity and allow large-scale chemical synthesis, the full length of P19 was truncated to a smaller 27-mer unit (tP19) (Table S1). The binding affinity of tP19 was confirmed at a KD of 8.7 nM, (Figure 3A), and this was also comparable to full-length P19. Next, to prevent structural hindrance of tP19, its 5′ end was attached to either MMAE (Figure 3B) or DM1 (Figure 3C) via a sticky sequence (SE). After chemical conjugation, each compound was high performance liquid chromatography (HPLC) purified. Then, to assemble the ApDCs, tP19-SE linked to MMAE-SE or DM1-SE was annealed in folding buffer to create functional tP19-MMAE and tP19-DM1 complexes (Figure S1B).
Figure 3.
Construction of Anti-mitotic ApDCs
(A) The dissociation constant (KD) of truncated P19 (tP19) was measured by flow cytometry using increasing concentrations of Cy3-labeled aptamers (from 15.6 to 500 nM). Mean fluorescence intensity (MFI) was measured and calculated using a one-site binding model for non-linear regression. (B and C) The schematic representation illustrates chemical conjugation of MMAE and DM1 to tP19. A carbon linker was used to conjugate the drugs to the 5′ end of a sticky sequence (SE). (D) Pancreatic cancer cell lines PANC-1 and non-pancreatic cell lines Huh7, HepG2, and MCF7 were treated with 200 nM Cy3-labeled tP19 for 4 hr and analyzed by confocal microscopy. Cy3 signal was only observed in the non-pancreatic cancer cell line. The staining represents live-cell imaging. Blue, Hoechst 33342. Scale bars, 10 μm. (E) The pancreatic cell line PANC-1 and non-pancreatic cancer cell line MCF7 were treated with 500 nM of the Cy3-labeled tP19-MMAE or (F) tP19-DM1 and analyzed by confocal microscopy. The staining represents live-cell imaging. Punctate regions of Cy3 labeling were observed only in PANC-1 cells. Red, Cy3; blue, Hoechst 33342. Scale bars, 10 μm.
To confirm whether this newly truncated tP19 complex maintained the same epitope specificity to PDAC cells as our previously reported full-length P19,17 a cell internalization assay was performed using PANC-1 and a panel of non-pancreatic cancer cells, including Huh7, HepG2, and MCF7. tP19 maintained binding specificity and internalization to PANC-1 cells only (Figure 3D). The ApDCs linked to the truncated aptamers were also screened for target cell specificity. Both tP19-MMAE and tP19-DM1 were tested against PANC-1 and MCF7 cells, but only PANC-1 cells showed internalization of tP19-MMAE (Figure 3E) and tP19-DM1 (Figure 3F).
tP19-MMAE and tP19-DM1 Induce Cell-Cycle Arrest and Inhibit Cell Proliferation in PANC-1 Cells
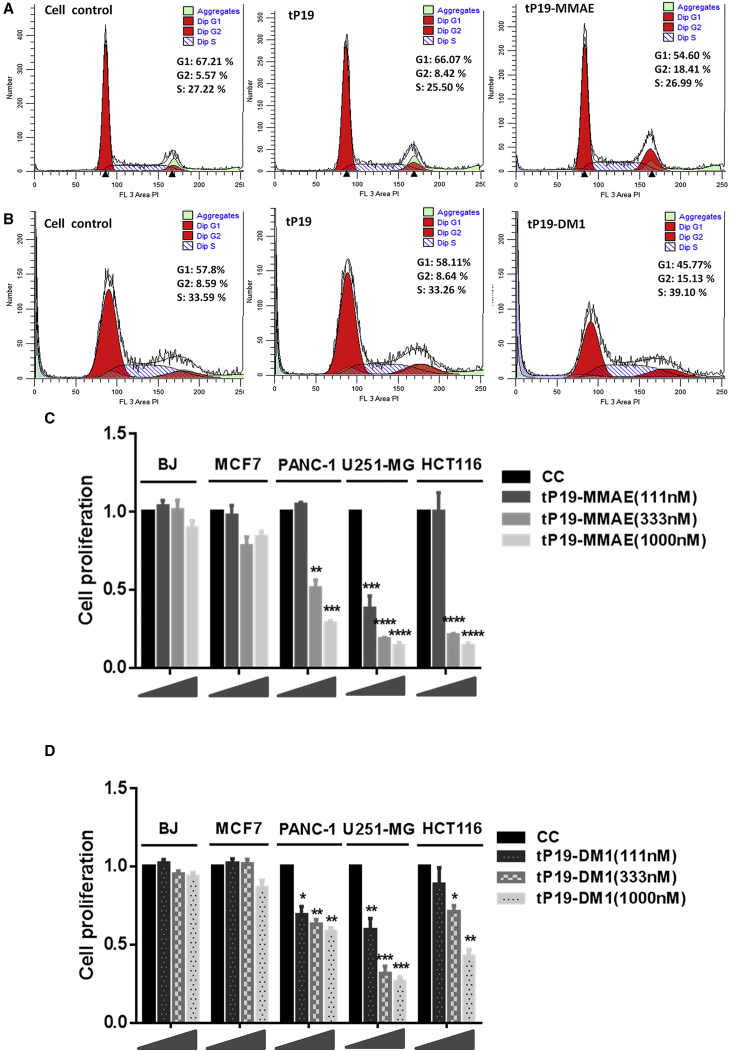
Since MMAE and DM1 are strong anti-mitotic drugs inhibiting microtubule polymerization, to characterize cell-cycle perturbation induced by tP19-MMAE and tP19-DM1, we used flow cytometry to measure the effect on cell-cycle progression. PANC-1 cells were treated with 500 nM tP19, tP19-MMAE, or tP19-DM1 for 72 hr. tP19 treatment did not disrupt any phase of the cell cycle (Figures 4A and 4B). tP19-MMAE and tp19-DM1 treatment caused a significant increase in cells entering the G2/M phase (Figures 4A and 4B). To confirm the antiproliferative effects of both ApDCs and their cell selectivity, an MTT cell proliferation assay was carried out in PANC-1, BJ, MCF7, U251-MG, and HCT116 cells treated with tP19-MMAE (Figure 4C) and tP19-DM1 (Figure 4D). tP19-MMAE and tP19-DM1, while showing no response in BJ and MCF7 cells, induced significant inhibition in proliferation of PANC-1 at 72 hr dose dependently. Since PANC-1 represents cells of an undifferentiated and aggressive metastatic phenotype, we tested both ApDCs on other cells bearing a similar phenotypic trait. U251-MG glioblastoma cells and HCT116 colorectal carcinoma cells were treated with tP19-MMAE and tP19-DM1. Both treatments cause significant inhibition in cell proliferation (Figures 4C and 4D). Interestingly, our original full-length P19 aptamer also showed suppression in proliferation of U251-MG and HCT116 similar to that observed with tP19 (Figure S2). When compared side by side, it was noted that both tP19-MMAE and tP19-DM1 caused stronger suppression in proliferation of U251-MG and HCT116 when compared to PANC-1 cells (Figures 4C and 4D). This observation suggests that P19/tP19 may also be a relevant vehicle for cells bearing an aggressive metastatic phenotype; however, this is an area that has yet to be investigated.
Figure 4.
Cell-Cycle Analysis and Cell Proliferation Assay of the Anti-mitotic ApDCs
(A and B) PANC-1 cells treated with tP19-MMAE (A) or tP19-DM1 (B) at 500 nM were stained with propidium iodide (PI). Fluorescence of the PI-stained cells was measured with flow cytometry and analyzed with ModFit deconvolution software. The percentage of cells in G1, S, and G2/M phases of the cycle is indicated (inset). (B and C) After treatment of PANC-1, U251, HCT116, BJ, and MCF-7 cells with various concentrations of tP19-MMAE (C) and tP19-DM1 (D) for 72 hr, cell proliferation was measured using an MTT assay. Cell proliferation was normalized with untreated control cells. One-way ANOVA test: ****p ≤ 0.0001, ***p ≤ 0.001 **p ≤ 0.01, *p ≤ 0.05.
Discussion
To improve drug-specific delivery to cancer cells, aptamers have been at the forefront of development in nanomedicine for targeted delivery of chemotherapeutic agents. These aptamers offer significant advantages over antibodies, including better stability, lower toxicity, lower immunogenicity, and a better safety profile.20, 21 RNA aptamers can be synthesized chemically or enzymatically, thus making it possible to incorporate nucleoside analogs for anti-cancer drugs into aptamers, such as gemcitabine and 5-FU.
In this study, ApDCs consisting of chimeric aptamers conjugated to the cytotoxic active metabolites of the nucleoside analogs gemcitabine and 5-FU and to the cytotoxins MMAE and DM1 were used for targeted delivery to PANC-1, a cell line representing PDAC (Figure 5). Successful construction and biological use of these ApDCs were demonstrated using a range of assays, including fluorescent internalization imaging and a MTT cell proliferation assay. In this setting, we demonstrated specific internalization of ApDCs into cancer cells bearing an aggressive metastatic phenotype, directly delivering active anti-metabolites and cytotoxins, and inducing selective and potent proliferative activity while sparing normal cells. To incorporate the nucleoside analogs gemcitabine and 5-FU in our previously reported PDAC-specific aptamer, P19, three-phosphate fluoropyrimidines of dFdCTP and 5FdUTP were used. During the conjugation process, two phosphates were removed and the final outputs were active monophosphate forms of the fluoropyrimidines (dFdCMP and 5FdUMP). Once internalized, dFdCMP and 5FdUMP would subsequently be phosphorylated to diphosphate or triphosphate fluorodeoxycytidine (dFdCDP and dFdCTP).18 Since fluorines are electronegativity charged, they strongly bind to other molecules and prevent DNA chain elongation and subsequently DNA synthesis.22 Their effects have been principally ascribed to misincorporation of fluoronucleotides into DNA, RNA and inhibition of thymidylate synthase (TS).23, 24, 25 From our study, we confirmed that ApDCs were internalized into cells (Figures 1D and 3D–3F) and induced DNA damage (Figures 2A and 2B). In vitro cell proliferation assays demonstrated significant cell growth inhibition in PANC-1 and even in the gemcitabine-resistant cell AsPC-1 (Figure 2C). Since low cell penetrance is the most likely paradigm for gemcitabine resistance, our data suggest that ApDCs may help ablate chemoresistance in pancreatic cancer by allowing internalization and delivery of the drug to otherwise inaccessible cells.26
Figure 5.
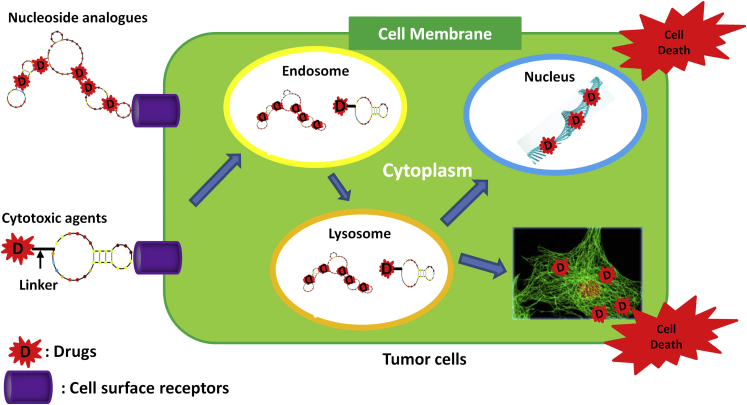
A Summary Schematic of ApDCs as Targeted Chemotherapeutics
To induce cytotoxic effects in cancer cells, intrinsically incorporated active metabolites of nucleoside analogs and anti-mitotic drugs conjugated at the 5′ end of aptamer were constructed. The ApDCs are internalized into the targeted cells by receptor mediation. The nucleoside analogs are disassociated from the aptamers and induce DNA damages to induce cell death. The cytotoxic agents are disassociated from the aptamer and inhibit the cell mitosis to induce cell death.
MMAE and DM1 are potent anti-mitotic agents. However, because of their high toxicity, they cannot be administered on their own. Most experimental evidence for the efficacy and safety of MMAE is derived from in vivo studies and xenograft models with ADCs.27 Up to now, there are few published studies regarding the use of MMAE with ADCs in pancreatic cancer. In a phase 1 study of patients with platinum-resistant ovarian cancer or unresectable PDAC, MMAE conjugated with a monoclonal antibody against mesothelin had antitumor activity in both types of cancer, with acceptable dose-limiting toxicity.28 In another phase 1 study of patients with gastric or pancreatic cancer, MMAE targeting SLC44A4 was generally well tolerated but had limited antitumor activity.29 Another promising cytotoxic drug is DM1, which is a potent microtubule-targeted compound, but in clinical trials, it showed lack of tumor specificity and unacceptable systemic toxicity. When combined with ADC, DM1 inhibited growth of grafted PDAC tumor nodules in preclinical models.13 However, ADCs have potential pitfalls that can affect their application in clinical practice, including instability, poor pharmacokinetic properties, and immunogenicity.30 In the field of targeted cancer therapy, ApDCs and ADCs are used similarly. However, ApDCs display different advantages over ADCs, as they do not aggregate, are more stable, have less toxicity, have greater specificity, and are easier and more economical to produce.31, 32, 33, 34
We previously demonstrated that aptamers could be used as potential drug-delivery vehicles with customized specificity to cell-surface receptors followed by efficient internalization to these cells.17 In this study, an ApDC was constructed to deliver chemotherapeutic agents specifically to PDAC cells. To construct ApDCs with MMAE and DM1, our previously reported P19 was truncated in order to increase binding affinity and allow large-scale chemical synthesis. This modified aptamer was termed tP19. An in vitro cell proliferation (MTT) assay demonstrated that tP19-MMAE and tP19-DM1 significantly inhibited growth in PANC-1 cells. We also observed cross anti-proliferative effects of tP19-MMAE and tP19-DM1 in the undifferentiated and highly proliferative U251-MG, glioblastoma (GBM) cell lines and HCT116, colorectal carcinoma cell lines. Interestingly, our original full-length P19 aptamer also showed cross-reactivity, with these two cells bearing an aggressive tumorigenic phenotype (Figure S2). This observation implies that the P19 aptamer or the truncated tP19 aptamers might be candidate vehicles to deliver chemotherapeutic agents to cells with an aggressive metastatic profile, such as glioblastomas and colon cancer cells.
As the technology to synthesize these oligonucleotides becomes more amenable to large-scale production under good manufacturing practice (GMP) conditions, ApDCs present as an attractive therapeutic moiety for safer cell-specific delivery of anti-cancer drugs.
Materials and Methods
Chemicals
Gemcitabine-5′-triphosphate (dFdCTP) was purchased from Sierra Bioresearch. 5-F-2′-UTP triphosphate (5FdUTP) was purchased from TriLink Biotechnologies. DuraScribe T7 transcription kit (Epicenter Biotechnologies) was used to incorporate dFdCTP and 5FdUTP into aptamers. Micro Bio-spin P30 columns (Bio-Rad) were used to remove unincorporated dFdCTP and 5FdUTP. Primary antibodies for γ-H2AX were purchased from Cell Signaling Technology. Secondary antibodies conjugated with Alexa 88 were purchased from Thermo Fisher Scientific.
Cell Lines
The following cell lines were purchased from the American Type Culture Collection (ATCC): PANC-1 (pancreatic epithelial carcinoma, CRL-1469), AsPC-1 (pancreatic adenocarcinoma, CRL-1682), MCF7 (breast adenocarcinoma, HTB-22), HCT116 (colorectal carcinoma, CCL-247), HepG2 (hepatocarcinoma, HB-8065), PC-3 (prostatic cancer, CRL-1435), and BJ (normal fibroblast, CRL-2522). Huh-7 cells were purchased from Japanese Collection of Research Bioresources (JCRB). U251-MG (glioblastoma, 09063001) cells were purchased from Sigma-Aldrich. The cells were cultured according to the suppliers’ instructions.
Aptamer Internalization Studies
1 × 105 PANC-1 cells were seeded in 35-mm glass-bottom dishes (MatTek) and grown in appropriate media for 24 hr. Aptamer RNA was labeled with Cy3 fluorescent dye using the Cy3 Silencer siRNA labeling kit (Thermo Fisher Scientific). Cy3-labeled aptamers were added to the cells at 200 nM or 500 nM and incubated for 2 hr. Live-cell confocal imaging was performed with a Zeiss LSM 510 Meta inverted two-photon confocal microscope system using a C-Apo 40×/1.2 numerical aperture (NA) water-immersion objective and AIM 4.2 software (Carl Zeiss).
Flow-Cytometry-Based Binding Assays
To determine the KD of truncated P19 interactions with PANC-1 cells, aptamer binding was assessed by flow cytometry. PANC-1 cells were detached using Accutase (Sigma-Aldrich), washed with PBS, and suspended in binding buffer (PBS solution [DPBS without Ca2+ and Mg2+, Corning] and 5 mM MgCl2). Next, Cy3-labeled aptamers were added and incubated with PANC-1 cells for 30 min at room temperature. Cells were washed with binding buffer and immediately analyzed by Fortessa flow cytometry (BD Biosciences). DAPI (1 μg/mL) was used to identify and exclude dead cells. Data were analyzed with FlowJo software. The mean fluorescence intensity (MFI) was calculated for each aptamer concentration and for the unselected library controls. Control values were considered to be background fluorescence and were subtracted from the aptamer values, as previously described by Sefah et al.35 KD was calculated using a one-site binding model. Non-linear regression was performed using GraphPad Prism (GraphPad Software).
MMAE and DM1 Aptamer Conjugation with a “Sticky Bridge Sequence”
MMAE and DM1 were chemically attached to the 5′ end of a sticky bridge sequence to maintain aptamer structure. The same concentration of tP19-SE and MMAE-SE or DM1-SE was annealed in binding buffer at 95°C for 5 min and slowly cooled down and incubated at 37°C for 20 min more to make the ApDCs.
γ-H2AX Evaluation
To investigate whether 5FdUMP and dFdCMP induced double-strand breaks in nuclear DNA, γ-H2AX evaluation was performed by IFA. 5 × 104 PANC-1 cells were seeded in four-chamber slides and grown in appropriate media for 24 hr. P19, P19-5FdUMP, and P19-dFdCMP were added to cells at 500 nM and incubated for 48 hr. The cells were fixed in 3% paraformaldehyde solution, permeabilized in 0.1% Triton X-100, and stained with anti-γ-H2AX (Cell Signaling, 1:2,000), followed by Alexa-488-labeled secondary antibody. Nuclear dots, indicative of nuclear DSBs, were counted using Image-Pro software (Media Cybernetics). The images were analyzed using Image Pro Premier, where the nuclear specks were counted using smart segmentation under count/size. The setting of smart segmentation was saved and then applied to subsequent images so that all images were analyzed in an identical manner. The representative images were depicted in Figure S3.
Cell-Cycle Analysis
For cell-cycle evaluation, 5 × 105 PANC-1 cells were treated with tP19, tP19-MMAE, or tP19-DM1 at 500 nM. After 4-hr incubation, cells were washed and new media was added. Cells were harvested after 72 hr, fixed in 70% ethanol overnight, and washed with PBS. Cells were centrifuged for 10 min at 300 × g and resuspended in 500 μL propidium iodide (PI)/Triton X-100 staining solution (0.40 mL of 500 μg/mL PI, 0.1% [v/v] Triton X-100, and 2 mg DNase-free RNase A, to 10 mL) for 15 min at 37°C. Fluorescence of the PI-stained cells was measured with flow cytometry and analyzed with ModFit deconvolution software (Verity Software House).
Cell Proliferation Assay
To determine the inhibition of cell proliferation, PANC-1 and AsPC-1 cells were seeded at 5 × 103 cells per well in 96-well plates and grown in appropriate media for 24 hr. P19, P19-5FdUMP, and P19-dFdCMP were added to cells at 1 μM and changed the media to remove the remaining ApDCs. Inhibition of cell proliferation was measured by MTT assay after 72-hr incubation.
PANC-1, MCF7, U251-MG, HCT116, and BJ cells were seeded at 5 × 103 cells per well in 96-well plates and grown in appropriate media for 24 hr. tP19-MMAE and tP19-DM1 were added to cells at various concentrations (from 1 μM to 0.45 μM, 3-fold dilutions) and incubated for 4 hr. Cells were washed and new media was added after 4-hr incubation to remove remaining ApDCs. After 72 hr, the inhibition of cell proliferation was measured by MTT assay (Promega).
Statistical Analysis
Statistically significant differences were determined by Student’s t test and ANOVA using GraphPad Prism software (GraphPad Software).
Author Contributions
S.Y., J.J.R., J.S., K.-W.H., and N.H. developed the concept, designed the experiments, and prepared the manuscript. V.R., D.S., I.R., and T.K. prepared the manuscript. S.Y. performed RNA aptamer truncation, measurements of cell binding affinity, the MTT assay, IFA, flow cytometry, data organization, and statistical analyses and wrote the manuscript. S.Y., D.Z., and K.D. made gemcitabine and 5-FU ApDCs. P.S. developed the concept and designed and synthesized aptamer-based conjugates, and L.L. conjugated chemically MMAE and DM1 to the aptamer. T.M.P. and Y.W. created an algorithm for the optimization of the sticky bridge sequences. B.A. performed the quantification of γ-H2AX using Image-Pro algorithm.
Conflicts of Interest
S.Y., J.J.R., P.S., and N.H. hold stock in Apterna Ltd. U.K.
Acknowledgments
We would like to thank the light microscopy digital imaging core facility of the Beckman Research Institute at the City of Hope for technical assistance. Research reported in this publication also included work performed in the Analytical Cytometry and Drug Discovery and Structural Biology Cores supported by the National Cancer Institute of the NIH under award number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also like to thank Sarah Wilkinson, scientific writer, for language editing. The authors would like to acknowledge support from Apterna Ltd. and NIAID R01 AI029329. T.M.P. and Y.W. are supported by the Intramural Research Program of the NIH, National Library of Medicine.
Footnotes
Supplemental Information includes three figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2016.11.008.
Supplemental Information
References
- 1.World Cancer Research Fund International. Pancreatic Cancer Statistics. http://www.wcrf.org/int/cancer-facts-figures/data-specific-cancers/pancreatic-cancer-statistics.
- 2.Stathis A., Moore M.J. Advanced pancreatic carcinoma: current treatment and future challenges. Nat. Rev. Clin. Oncol. 2010;7:163–172. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 3.(2011). Pancreatic cancer in the UK. Lancet 378, 1050. [DOI] [PubMed]
- 4.Jemal A., Siegel R., Ward E., Murray T., Xu J., Smigal C., Thun M.J. Cancer statistics, 2006. CA Cancer J. Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 5.Krempien R., Muenter M.W., Harms W., Debus J. Neoadjuvant chemoradiation in patients with pancreatic adenocarcinoma. HPB (Oxford) 2006;8:22–28. doi: 10.1080/13651820500468034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burris H.A., 3rd, Moore M.J., Andersen J., Green M.R., Rothenberg M.L., Modiano M.R., Cripps M.C., Portenoy R.K., Storniolo A.M., Tarassoff P. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 7.Conroy T., Desseigne F., Ychou M., Bouché O., Guimbaud R., Bécouarn Y., Adenis A., Raoul J.L., Gourgou-Bourgade S., de la Fouchardière C., Groupe Tumeurs Digestives of Unicancer. PRODIGE Intergroup FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 8.Yip D., Karapetis C., Strickland A., Steer C.B., Goldstein D. Chemotherapy and radiotherapy for inoperable advanced pancreatic cancer. Cochrane Database Syst. Rev. 2006;(3):CD002093. doi: 10.1002/14651858.CD002093.pub2. [DOI] [PubMed] [Google Scholar]
- 9.Sultana A., Smith C.T., Cunningham D., Starling N., Neoptolemos J.P., Ghaneh P. Meta-analyses of chemotherapy for locally advanced and metastatic pancreatic cancer. J. Clin. Oncol. 2007;25:2607–2615. doi: 10.1200/JCO.2006.09.2551. [DOI] [PubMed] [Google Scholar]
- 10.Glimelius B., Hoffman K., Sjoden P.O., Jacobsson G., Sellstrom H., Enander L.K., Linné T., Svensson C. Chemotherapy improves survival and quality of life in advanced pancreatic and biliary cancer. Ann. Oncol. 1996;7:593–600. doi: 10.1093/oxfordjournals.annonc.a010676. [DOI] [PubMed] [Google Scholar]
- 11.Pettit G.R. The dolastatins. Fortschr. Chem. Org. Naturst. 1997;70:1–79. doi: 10.1007/978-3-7091-6551-5_1. [DOI] [PubMed] [Google Scholar]
- 12.Koga Y., Manabe S., Aihara Y., Sato R., Tsumura R., Iwafuji H., Furuya F., Fuchigami H., Fujiwara Y., Hisada Y. Antitumor effect of antitissue factor antibody-MMAE conjugate in human pancreatic tumor xenografts. Int. J. Cancer. 2015;137:1457–1466. doi: 10.1002/ijc.29492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao H.P., Feng L., Zhou J.W., Zhang R.W., Wang M.H. Therapeutic evaluation of monoclonal antibody-maytansinoid conjugate as a model of RON-targeted drug delivery for pancreatic cancer treatment. Am. J. Cancer Res. 2016;6:937–956. [PMC free article] [PubMed] [Google Scholar]
- 14.Remillard S., Rebhun L.I., Howie G.A., Kupchan S.M. Antimitotic activity of the potent tumor inhibitor maytansine. Science. 1975;189:1002–1005. doi: 10.1126/science.1241159. [DOI] [PubMed] [Google Scholar]
- 15.Feld J., Barta S.K., Schinke C., Braunschweig I., Zhou Y., Verma A.K. Linked-in: design and efficacy of antibody drug conjugates in oncology. Oncotarget. 2013;4:397–412. doi: 10.18632/oncotarget.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauerborn M., van Dongen W. Practical considerations for the pharmacokinetic and immunogenic assessment of antibody-drug conjugates. BioDrugs. 2014;28:383–391. doi: 10.1007/s40259-014-0096-z. [DOI] [PubMed] [Google Scholar]
- 17.Yoon S., Huang K.W., Reebye V., Mintz P., Tien Y.W., Lai H.S., Saetrom P., Reccia I., Swiderski P., Armstrong B. Targeted delivery of C/EBPalpha -saRNA by pancreatic ductal adenocarcinoma (PDAC)-specific RNA aptamers inhibits tumor growth in vivo. Mol. Ther. 2016;24:1106–1116. doi: 10.1038/mt.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang P., Chubb S., Hertel L.W., Grindey G.B., Plunkett W. Action of 2′,2′-difluorodeoxycytidine on DNA synthesis. Cancer Res. 1991;51:6110–6117. [PubMed] [Google Scholar]
- 19.Soares D.G., Escargueil A.E., Poindessous V., Sarasin A., de Gramont A., Bonatto D., Henriques J.A., Larsen A.K. Replication and homologous recombination repair regulate DNA double-strand break formation by the antitumor alkylator ecteinascidin 743. Proc. Natl. Acad. Sci. USA. 2007;104:13062–13067. doi: 10.1073/pnas.0609877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 21.Tuerk C. Using the SELEX combinatorial chemistry process to find high affinity nucleic acid ligands to target molecules. Methods Mol. Biol. 1997;67:219–230. doi: 10.1385/0-89603-483-6:219. [DOI] [PubMed] [Google Scholar]
- 22.Pauling L. Cornell University Press; 1945. The Nature of the Chemical Bond. [Google Scholar]
- 23.Matuo R., Sousa F.G., Escargueil A.E., Grivicich I., Garcia-Santos D., Chies J.A., Saffi J., Larsen A.K., Henriques J.A. 5-Fluorouracil and its active metabolite FdUMP cause DNA damage in human SW620 colon adenocarcinoma cell line. J. Appl. Toxicol. 2009;29:308–316. doi: 10.1002/jat.1411. [DOI] [PubMed] [Google Scholar]
- 24.Ruiz van Haperen V.W., Veerman G., Vermorken J.B., Peters G.J. 2′,2′-Difluoro-deoxycytidine (gemcitabine) incorporation into RNA and DNA of tumour cell lines. Biochem. Pharmacol. 1993;46:762–766. doi: 10.1016/0006-2952(93)90566-f. [DOI] [PubMed] [Google Scholar]
- 25.Pourquier P., Gioffre C., Kohlhagen G., Urasaki Y., Goldwasser F., Hertel L.W., Yu S., Pon R.T., Gmeiner W.H., Pommier Y. Gemcitabine (2′,2′-difluoro-2′-deoxycytidine), an antimetabolite that poisons topoisomerase I. Clin. Cancer Res. 2002;8:2499–2504. [PubMed] [Google Scholar]
- 26.Papa A.L., Basu S., Sengupta P., Banerjee D., Sengupta S., Harfouche R. Mechanistic studies of Gemcitabine-loaded nanoplatforms in resistant pancreatic cancer cells. BMC Cancer. 2012;12:419. doi: 10.1186/1471-2407-12-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breij E.C., de Goeij B.E., Verploegen S., Schuurhuis D.H., Amirkhosravi A., Francis J., Miller V.B., Houtkamp M., Bleeker W.K., Satijn D., Parren P.W. An antibody-drug conjugate that targets tissue factor exhibits potent therapeutic activity against a broad range of solid tumors. Cancer Res. 2014;74:1214–1226. doi: 10.1158/0008-5472.CAN-13-2440. [DOI] [PubMed] [Google Scholar]
- 28.Weekes C.D., Lamberts L.E., Borad M.J., Voortman J., McWilliams R.R., Diamond J.R., de Vries E.G., Verheul H.M., Lieu C.H., Kim G.P. Phase I Study of DMOT4039A, an antibody-drug conjugate targeting mesothelin, in patients with unresectable pancreatic or platinum-resistant ovarian cancer. Mol. Cancer Ther. 2016;15:439–447. doi: 10.1158/1535-7163.MCT-15-0693. [DOI] [PubMed] [Google Scholar]
- 29.Coveler A.L., Ko A.H., Catenacci D.V., Von Hoff D., Becerra C., Whiting N.C., Yang J., Wolpin B. A phase 1 clinical trial of ASG-5ME, a novel drug-antibody conjugate targeting SLC44A4, in patients with advanced pancreatic and gastric cancers. Invest. New Drugs. 2016;34:319–328. doi: 10.1007/s10637-016-0343-x. [DOI] [PubMed] [Google Scholar]
- 30.Lee J.W., Kim H.J., Heo K. Therapeutic aptamers: developmental potential as anticancer drugs. BMB Rep. 2015;48:234–237. doi: 10.5483/BMBRep.2015.48.4.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoltenburg R., Reinemann C., Strehlitz B. SELEX—a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Jayasena S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 33.Ferreira C.S., Cheung M.C., Missailidis S., Bisland S., Gariépy J. Phototoxic aptamers selectively enter and kill epithelial cancer cells. Nucleic Acids Res. 2009;37:866–876. doi: 10.1093/nar/gkn967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan A.C., Levy M. Aptamers and aptamer targeted delivery. RNA Biol. 2009;6:316–320. doi: 10.4161/rna.6.3.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sefah K., Shangguan D., Xiong X., O’Donoghue M.B., Tan W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.