Abstract
Neovascular age-related macular degeneration (nvAMD) is characterized by choroidal blood vessels growing into the subretinal space, leading to retinal pigment epithelial (RPE) cell degeneration and vision loss. Vessel growth results from an imbalance of pro-angiogenic (e.g., vascular endothelial growth factor [VEGF]) and anti-angiogenic factors (e.g., pigment epithelium-derived factor [PEDF]). Current treatment using intravitreal injections of anti-VEGF antibodies improves vision in about 30% of patients but may be accompanied by side effects and non-compliance. To avoid the difficulties posed by frequent intravitreal injections, we have proposed the transplantation of pigment epithelial cells modified to overexpress human PEDF. Stable transgene integration and expression is ensured by the hyperactive Sleeping Beauty transposon system delivered by pFAR4 miniplasmids, which have a backbone free of antibiotic resistance markers. We demonstrated efficient expression of the PEDF gene and an optimized PEDF cDNA sequence in as few as 5 × 103 primary cells. At 3 weeks post-transfection, PEDF secretion was significantly elevated and long-term follow-up indicated a more stable secretion by cells transfected with the optimized PEDF transgene. Analysis of transgene insertion sites in human RPE cells showed an almost random genomic distribution. The results represent an important contribution toward a clinical trial aiming at a non-viral gene therapy of nvAMD.
Keywords: neovascular age-related macular degeneration, non-viral ex vivo gene therapy, sleeping beauty transposon system, pFAR4 miniplasmids, primary pigment epithelial cells, pigment epithelium-derived factor
Introduction
Age-related macular degeneration (AMD) is the most common cause of severe vision loss in patients over the age of 60. There are two major forms of AMD, a non-neovascular or atrophic and a neovascular form. The neovascular form (nvAMD) is characterized by the formation of subretinal choroidal neovascularization (CNV) and is the major cause of severe vision loss in developed countries. It has been estimated that approximately 23.5 million people worldwide are affected with AMD.1 Globally, it is the fourth most common cause of blindness after cataracts, preterm birth, and glaucoma.2 AMD affects about 0.4% of people between the ages of 50 and 60 years and increases to about 12% of people over the age of 80 years.3
Evidence that in nvAMD overexpression of vascular endothelial growth factor (VEGF) and decrease in the expression of pigment epithelium-derived factor (PEDF) trigger the growth of choroidal blood vessels into the subretinal space4, 5 led to the development of anti-angiogenic therapies for nvAMD. With the introduction of anti-VEGF antibody therapy in 2006, and more recently the introduction of the recombinant fusion protein aflibercept,6 about 30%–40% of nvAMD patients regain three or more lines of visual acuity with stabilization occurring in 90% of the remaining patients. However, in addition to the fact that the majority of nvAMD patients do not regain vision with anti-VEGF therapy, the frequent, often monthly, intravitreal injections are a significant impediment to compliance, considering that these patients are old and visually impaired. Furthermore, these treatments represent a significant economic burden to healthcare systems.
A desirable alternative to overcome the difficulties associated with frequent, lifelong intravitreal injections would be a treatment modality that introduces an inhibitor of neovascularization to the retina, which would last for the life of the patient. To meet such treatment modality, in 2006, Campochiaro and colleagues delivered the gene encoding PEDF to the retina of nvAMD patients using adenoviral (Ad) delivery and reported significant improvement in 25% of patients after 12 weeks and no harmful side effects.7, 8 However, no follow-up to the trial has been reported. Currently, three additional gene therapy clinical trials are ongoing (https://clinicaltrials.gov/): trial NCT01494805 is based on the use of an adeno-associated viral (AAV) vector encoding sFlt-1, a splice variant of VEGF receptor 1; trials NCT01301443 and NCT01678872 are based on the use of a lentiviral vector expressing endostatin and angiostatin (RetinoStat). Aside from the results reported by Campochiaro et al.,8 the only other available data are from two Association for Research in Vision and Ophthalmology abstracts in 2014 and 2016 (M. Davies et al., 2014, Invest. Ophthalmol. Vis. Sci., abstract; A.K. Lauer et al., 2016, Invest. Ophthalmol. Vis. Sci., abstract) in which substantial transgene expression as well as a safe and well-tolerated treatment exhibiting signs of clinical benefit were reported for RetinoStat.
Even though efficient, the use of viral vectors to deliver genetic information is problematic. Transgene delivery via non-integrating viral vectors, such as Ad and AAV vectors, is episomal, often necessitating re-administration, which can elicit an immune response leading to a decline in transgene expression.9, 10 However, several clinical trials for Leber congenital amaurosis (LCA) showed that the AAV-based subretinal delivery of the human RPE65 gene is effective and safe.11 Lentiviral and retroviral vectors integrate the transgene into the host cell’s genome and could possibly express the transgene for the life of the host cells. However, the preference of lentiviral and retroviral vectors to integrate into transcriptionally active genomic regions is associated with a high risk of vector-associated insertional mutagenesis, which could be harmful to the host cell and to the patient.12, 13, 14, 15, 16, 17, 18, 19
In nvAMD, choroidal blood vessel growth into the subretinal space disrupts the normal architecture of the retina, leading to retinal pigment epithelial (RPE) cell degeneration and vision loss. The use of vectors to deliver an inhibitor of neovascularization into the subretinal space of nvAMD patients will benefit those patients, in which the RPE cells retain normal function. However, in nvAMD patients, RPE cells degenerate rapidly.20 Regain of vision in most nvAMD patients will require not only the expression of an inhibitor of neovascularization, but more importantly a functional retinal pigment epithelium. It has previously been shown that transplantation of RPE or iris pigment epithelial (IPE) cells, as a substitute for RPE cells, to the subretinal space did not result in vision improvement in nvAMD patients.21, 22, 23
We have postulated that the inhibition of CNV will require that the transplanted cells overexpress PEDF, which would not only inhibit neovascularization but also act as a neuroprotective agent for the retina. The complication associated with viral vectors can be avoided by the use of the non-viral Sleeping Beauty (SB) transposon system, which provides efficient and stable gene transfer and transgene expression, has a safer integration profile compared to other integrating vectors24 and offers ease of production. In fact, we have previously shown that the ARPE-19 cell line and primary bovine pigment epithelial cells transfected ex vivo by electroporation with a PEDF-encoding SB transposon delivered as plasmid DNA express elevated and stable levels of PEDF.25 Importantly, transplantation of these cells in a rabbit model of corneal neovascularization suppressed and reversed neovascularization.26 In these experiments, we used the pT2-CMV-PEDF/EGFP plasmid, which is 7,258 bp in size. The backbone contains an ampicillin resistance marker, and the expression cassette is composed of a His-tagged human PEDF transgene, an intervening sequence (IVS)/internal ribosomal entry site (IRES) element, and the EGFP gene,25 making the plasmid unsuitable for use in humans.
Although safer than viral vectors, the use of plasmids to deliver genes for therapeutic use also displays some drawbacks. Specifically, efficient production of plasmids in bacteria requires that the plasmid encodes a marker that favors the growth of the bacteria containing the plasmid, generally an antibiotic resistance gene. However, the presence of antibiotic resistance markers in gene therapy vectors is a matter of concern. Residues of antibiotics could contaminate the final product, placing at risk patients with severe hypersensitivity to antibiotics, which is relatively common for β-lactam antibiotics.27 Furthermore, the removal of antibiotic resistance genes allows for a reduction in the size of the plasmid vector, resulting in an increase in transfection efficiency.28
Finally, careful design of not only vector sequences, but of the therapeutic genes themselves can affect the outcome of somatic gene transfer. Namely, transgenes are usually encoded by intronless cDNA constructs, which, however, can still carry functionless exonic splice enhancer (ESE) sequences. In intron-containing genes, rates of evolution are lower near exon-intron boundaries than in exon cores.29, 30 Analysis of the rate of sequence evolution in retrogenes, which are derived from intron-containing genes by retroposition and may be considered as mimics of transgenes, led to the suggestion that intronless genes might be under selection to avoid some or all ESE motifs, as the genes might need to avoid attracting a splicing machinery. Based on this proposition, it was suggested that intronless transgenes could be improved by directed modification at synonymous sites of ESE motifs to degrade them while simultaneously improving RNA stability. Recent evidence has suggested that in intronless genes some ESE motifs remain under selection for splice-independent functions, while recent retrogenes—the best mimics of transgenes—evolved unusually fast at ESE motif sites,31 supporting the hypothesis that loss of some ESE motifs could be beneficial. Here, we used an optimization strategy to decrease ESE motifs in the PEDF coding sequence.
Since the ultimate goal of our research is to transplant RPE and/or IPE cells transfected with the PEDF gene into the subretinal space of nvAMD patients and to avoid the use of plasmids encoding antibiotic resistance genes, we have developed a protocol for the efficient delivery of the PEDF gene encoded in plasmids free of antibiotic resistance markers (pFAR) combined with the enhanced Sleeping Beauty (SB100X) transposon system. Here, we report that SB100X-mediated delivery of the PEDF gene, using pFAR4 miniplasmids to encode the PEDF gene and the SB100X transposase, is efficient and results in stable, long-term gene expression and protein secretion in as few as 5 × 103 primary bovine IPE and human RPE cells.
Results
Definition of the SB100X Transposase to PEDF Transposon Ratio
We previously showed that ratios ranging from 0.038 μg SB100X transposase expression plasmid DNA and 0.462 μg PEDF transposon plasmid DNA (1+12) to 0.017 μg SB100X transposase and 0.483 μg PEDF transposon (1+28) resulted in good transposition efficiencies, as determined by the secretion of recombinant PEDF.25 Since transfection experiments with varying amounts of pFAR4-CMV SB100X SV40 transposase and pFAR4-ITRs CMV PEDF BGH (pFAR-PEDF) transposon miniplasmid DNA exhibited similar results, with the highest PEDF secretion rates observed for the ratios 1+16 and 1+20 (Figure S1), all subsequent experiments were carried out using 0.03 μg SB100X transposase expression plasmid DNA and 0.47 μg PEDF transposon plasmid DNA.
Optimization of the PEDF DNA Sequence
Using the implemented algorithm, the number of ESE motifs in the PEDF coding sequence was decreased from 135 to 55, and GC nucleotide content increased from 63.3% to 71.8%. This optimized sequence is hereafter referred to as PEDFo.
SB100X-Mediated Transfection of Cultured Primary Bovine IPE Cells with the PEDF Gene Delivered by the pFAR4 Miniplasmid
Western blot analysis 21 days after transfection showed approximately an 18-fold increase in the secretion of PEDF by 1 × 104 bovine IPE cells transfected with the pFAR-PEDF transposon miniplasmid and approximately a 9-fold increase by 1 × 104 cells transfected with the pFAR-PEDFo transposon miniplasmid (Table 1; Figures S2A and S3A) compared to non-transfected cells. Transfection of 5 × 103 bovine IPE cells showed a 5-fold increase in PEDF secretion when transfected with the pFAR-PEDF transposon miniplasmid and approximately an 8-fold increase when transfected with the pFAR-PEDFo transposon miniplasmid (Table 1; Figures S2B and S3B) compared to non-transfected cells.
Table 1.
Western Blot-Based Quantification of PEDF Secretion by PEDF-Transfected Bovine IPE and Human RPE Cells
| Signal Intensities (Mean ± SD) | pFAR-PEDF versus pFAR-PEDFo | ||
|---|---|---|---|
| 1 × 104 bIPE cells | Control 1.00 ± 0.03 (n = 19) | Control 1.00 ± 0.00 (n = 9) | |
| pFAR-PEDF 17.96 ± 35.28 (n = 211) *p = 0.0376 | pFAR-PEDFo 8.84 ± 7.65 (n = 106) **p = 0.0028 | **p = 0.0090 | |
| 5 × 103 bIPE cells | Control 1.00 ± 0.11 (n = 18) | Control 1.00 ± 0.00 (n = 11) | |
| pFAR-PEDF 5.25 ± 5.20 (n = 200) ***p = 0.0007 | pFAR-PEDFo 8.42 ± 9.95 (n = 132) *p = 0.0149 | ***p = 0.0002 | |
| 1 × 104 hRPE cells | Control 1.00 ± 0.18 (n = 52) | Control 1.00 ± 0.17 (n = 26) | |
| pFAR-PEDF 2.54 ± 0.99 (n = 312) ****p < 0.0001 | pFAR-PEDFo 1.93 ± 0.76 (n = 156) ****p < 0.0001 | ****p < 0.0001 | |
| 5 × 103 hRPE cells | Control 1.00 ± 0.13 (n = 48) | Control 1.00 ± 0.19 (n = 26) | |
| pFAR-PEDF 2.02 ± 1.09 (n = 288) ****p < 0.0001 | pFAR-PEDFo 2.25 ± 1.14 (n = 156) ****p < 0.0001 | *p = 0.0378 | |
For each sample, one or two control transfections without the addition of miniplasmid DNA and ten to 12 transfections using 0.03 μg pFAR4-CMV SB100X SV40 transposase and 0.47 μg pFAR4-ITRs CMV PEDF BGH or pFAR4-ITRs CMV PEDFoptimized BGH miniplasmid DNA were carried out. Culture supernatants were analyzed for total PEDF secretion 21 days after transfection (Figures S2–S5). Signal intensities of PEDF-transfected and PEDFo-transfected cells were normalized to the signal intensities of control cells. Statistical analysis showed a significant difference in secretion between PEDF-transfected and PEDFo-transfected cells.
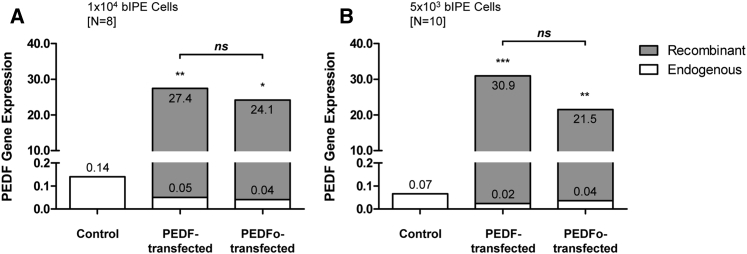
Endogenous and recombinant PEDF gene expression were analyzed by real-time qPCR in bovine IPE cells cultured for 21 days after transfection. PEDF gene expression was significantly increased after transfection with the pFAR-PEDF and the pFAR-PEDFo transposon miniplasmids compared to non-transfected cells (Figure 1). Endogenous PEDF gene expression in non-transfected and transfected cells did not show any significant differences. High gene expression levels of recombinant PEDF were detected in 1 × 104 and 5 × 103-transfected cells, but not in non-transfected cells. However, recombinant PEDF expression in pFAR-PEDF-transfected cells was not significantly different from recombinant PEDF expression in pFAR-PEDFo-transfected cells (Figure 1).
Figure 1.
PEDF Gene Expression in Primary Bovine IPE Cells after SB100X-Mediated Transfection
For each IPE sample, control transfections without the addition of miniplasmid DNA and transfections using 0.03 μg pFAR4-CMV SB100X SV40 transposase and 0.47 μg pFAR4-ITRs CMV PEDF BGH or pFAR4-ITRs CMV PEDFoptimized BGH miniplasmid DNA were performed. Total RNA was isolated 21 days after transfection. PEDF gene expression by cells transfected with pFAR-PEDF and pFAR-PEDFo was analyzed for (A) 1 × 104 cells for eight bovine eyes and (B) 5 × 103 cells for ten bovine eyes. Statistical analysis using a Kruskal-Wallis test showed a significant difference between transfected cells and non-transfected control cells. No significant difference was observed between cells transfected with pFAR-PEDF and pFAR-PEDFo.
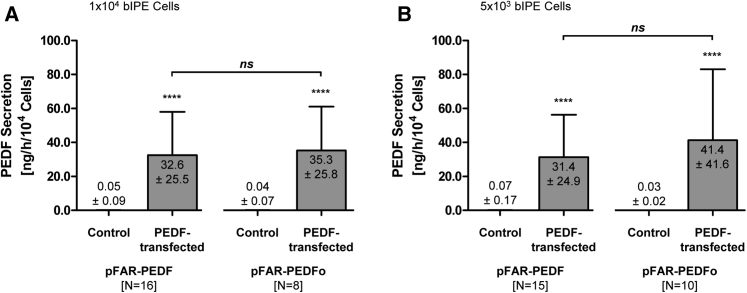
At about 4 weeks post-transfection, quantification by ELISA showed that 1 × 104 PEDF-transfected bovine IPE cells secreted into the media an average of 32.6 ± 25.5 versus 0.05 ± 0.09 ng for non-transfected cells. Similarly, 1 × 104 PEDFo-transfected cells secreted an average of 35.3 ± 25.8 versus 0.04 ± 0.07 ng for non-transfected cells (Figure 2A). When 5 × 103 cells were transfected, PEDF was secreted at a concentration of 31.4 ± 24.9 ng for PEDF-transfected cells and 41.4 ± 41.6 ng for PEDFo-transfected cells. PEDF secretion of cells transfected without miniplasmid DNA varied from 0.07 ± 0.17 to 0.03 ± 0.02 ng (Figure 2B). Levels of PEDF protein secreted by both 1 × 104 and 5 × 103 transfected cells were statistically different from non-transfected cells, regardless of the miniplasmid transposon used. However, the amount of PEDF secreted by cells transfected with the optimized PEDF sequence was not significantly different from the amount secreted by cells transfected with the native recombinant PEDF sequence.
Figure 2.
ELISA Quantification of Total PEDF Secreted by PEDF-Transfected Primary Bovine IPE Cells
For each IPE sample, two control transfections without the addition of miniplasmid DNA and two transfections using 0.03 μg pFAR4-CMV SB100X SV40 transposase and 0.47 μg pFAR4-ITRs CMV PEDF BGH or pFAR4-ITRs CMV PEDFoptimized BGH miniplasmid DNA were carried out. (A) PEDF secretion by 1 × 104 initially used cells. Mean ± SD of 16 bovine eyes transfected with pFAR-PEDF 27.5 ± 9.8 days after electroporation and mean ± SD of eight bovine eyes transfected with pFAR-PEDFo 24.0 ± 7.4 days after electroporation. (B) PEDF secretion by 5 × 103 initially used cells. Mean ± SD of 15 bovine eyes transfected with pFAR-PEDF 28.5 ± 13.8 days after electroporation and mean ± SD of ten bovine eyes transfected with pFAR-PEDFo 27.1 ± 16.3 days after electroporation. Total PEDF secretion of PEDF- and PEDFo-transfected cells was compared to the respective control cells (p < 0.0001, unpaired two-tailed t test) and total PEDF secretion of PEDF-transfected cells was compared to total PEDF secretion of PEDFo-transfected cells (not significant for 1 × 104 cells and 5 × 103 cells, unpaired two-tailed t test).
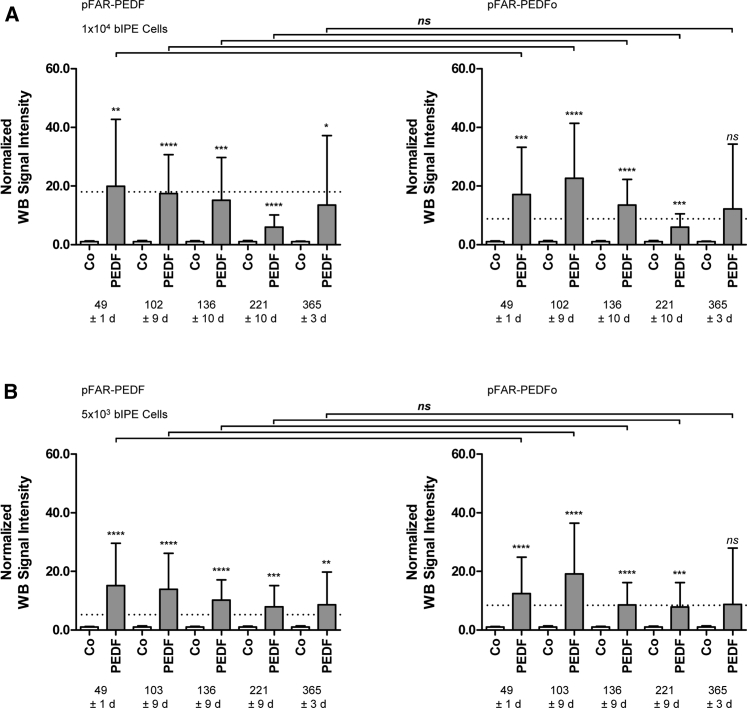
Long-term secretion of PEDF by cells transfected with the pFAR-PEDF and the pFAR-PEDFo transposon miniplasmids was examined by immunoblotting at regular intervals for 1 year. In almost all cases, normalized PEDF signal intensities were significantly enhanced compared to non-transfected cells, though they gradually decreased with time. During the 1 year, mean normalized signal intensities for 1 × 104 cells were reduced by 32.1% from 49 to 365 days post-transfection for pFAR-PEDF-transfected cells and by 28.7% for pFAR-PEDFo-transfected cells (Figure 3A; Table S1). For 5 × 103 cells, the mean band intensity decreased by 43.2% for PEDF-transfected cells, whereas for PEDFo-transfected cells the decrease was 29.3% (Figure 3B; Table S1). However, analysis at each time period revealed that the differences observed between PEDF secretion by PEDF-transfected and PEDFo-transfected cells were not statistically significant.
Figure 3.
Western-Blot-Based Quantification of Long-Term PEDF Secretion in Primary Bovine IPE Cells after SB100X-Mediated Transfection
For each IPE sample, two control transfections without the addition of miniplasmid DNA and two transfections using 0.03 μg pFAR4-CMV SB100X SV40 transposase and 0.47 μg pFAR4-ITRs CMV PEDF BGH or pFAR4-ITRs CMV PEDFoptimized BGH miniplasmid DNA were carried out. Culture supernatants were analyzed for total PEDF secretion 7, 15, 19, 32, and 52 weeks after transfection. Western blot signal intensities of PEDF-transfected and PEDFo-transfected cells were normalized to the signal intensities of the control cells. Data are presented as mean ± SD. Mean signal intensities of PEDF secretion 21 days after transfection are indicated by the dashed line. For 365 ± 3 days, levels of PEDF secreted by pFAR-PEDF- and pFAR-PEDFo-transfected cells were analyzed for (A) 1 × 104 cells and (B) 5 × 103 cells. Statistical analysis using an unpaired two-tailed t test showed a significant difference between transfected cells and non-transfected control cells. No significant difference was observed between cells transfected with pFAR-PEDF and pFAR-PEDFo at any time period.
SB100X-Mediated Transfection of Cultured Primary Human RPE Cells with the PEDF Gene Delivered by the pFAR4 Miniplasmid
Similar to the results of cultured bovine IPE cells, western blot analysis at 21 days after transfection showed a significant increase in the secretion of PEDF by human RPE cells transfected with the pFAR-PEDF transposon miniplasmid (Table 1; Figure S4) and the pFAR-PEDFo transposon miniplasmid (Table 1; Figure S5) compared to cells transfected without the addition of miniplasmid DNA. 1 × 104 cells transfected with the pFAR-PEDF transposon miniplasmid exhibited 31.6% higher normalized PEDF signal intensities than cells transfected with the pFAR-PEDFo transposon miniplasmid. For 5 × 103 cells, a 10.2% decrease in the mean band intensity for PEDF-transfected cells was observed compared to PEDFo-transfected cells (Table 1).
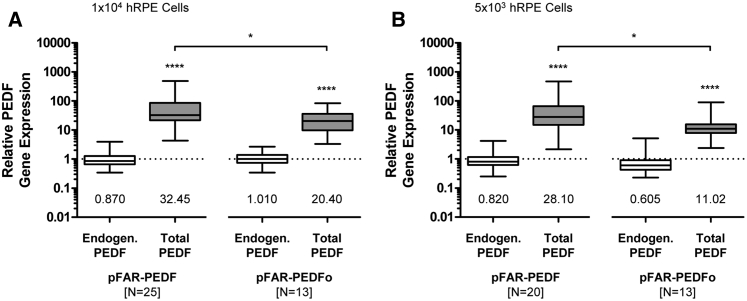
PEDF gene expression was analyzed by real-time qPCR in human RPE cells cultured for 21 days after transfection (Figure 4). In all cases, a significant increase in total PEDF gene expression was observed compared to endogenous PEDF gene expression. Transfection of 1 × 104 (Figure 4A) and 5 × 103 (Figure 4B) cells resulted in higher PEDF gene expression for pFAR-PEDF-transfected cells compared to pFAR-PEDFo-transfected cells.
Figure 4.
Relative PEDF Gene Expression in Primary Human RPE Cells after SB100X-Mediated Transfection
For each donor sample, four control transfections without the addition of miniplasmid DNA and four transfections using 0.03 μg pFAR4-CMV SB100X SV40 transposase and 0.47 μg pFAR4-ITRs CMV PEDF BGH or pFAR4-ITRs CMV PEDFoptimized BGH miniplasmid DNA were performed. Total RNA was isolated 21 days after transfection. Endogenous and total (endogenous + recombinant) PEDF gene expression in PEDF-transfected and PEDFo-transfected cells were related to the PEDF gene expression in control cells, whose expression was set to 1 (dashed line). (A) For 1 × 104 cells, PEDF expression by cells transfected with pFAR-PEDF was analyzed for 25 human donor eyes (age: 65.8 ± 14.2 years; gender: 12 males and 13 females; postmortem time: 32.4 ± 14.5 hr; cultivation time before transfection: 43.0 ± 15.7 days); PEDF expression by cells transfected with pFAR-PEDFo was analyzed for 13 human donor eyes (age: 61.1 ± 16.3 years; gender: seven males and six females; postmortem time: 33.5 ± 16.2 hr; cultivation time before transfection: 39.8 ± 17.1 days). (B) For 5 × 103 cells, PEDF expression by cells transfected with pFAR-PEDF was analyzed for 20 human donor eyes (age: 63.7 ± 14.5 years; gender: 13 males and seven females; postmortem time: 34.6 ± 14.9 hr; cultivation time before transfection: 45.3 ± 16.5 days); PEDF expression for cells transfected with pFAR-PEDFo was analyzed for 13 human donor eyes (age: 61.1 ± 16.3 years; gender: eight males and five females; postmortem time: 32.5 ± 16.1 hr; cultivation time before transfection: 42.6 ± 18.6 days). Data are presented as box-and-whisker plots (whiskers: min to max). Total PEDF gene expression was compared to the endogenous PEDF gene expression (p < 0.0001, unpaired two-tailed t test) and total PEDF gene expression of PEDF-transfected cells was compared to total PEDF gene expression of PEDFo-transfected cells (p = 0.0248 for 1 × 104 cells and p = 0.0161 for 5 × 103 cells, unpaired two-tailed t test).
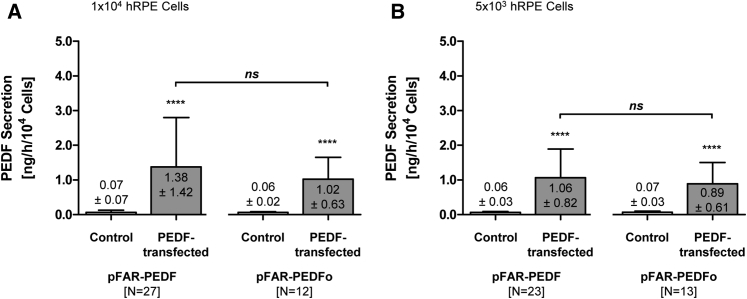
ELISA-based quantification of PEDF secretion corresponded to the differences observed for PEDF gene expression. At 3 weeks post-transfection, 1 × 104 cells PEDF-transfected cells secreted into the media an average of 1.38 ± 1.42 versus 0.07 ± 0.07 ng for non-transfected control cells, whereas PEDFo-transfected cells secreted 1.02 ± 0.63 versus 0.06 ± 0.02 ng for non-transfected control cells (Figure 5A). When transfection was done with 5 × 103 cells, PEDF was secreted at a concentration of 1.06 ± 0.82 ng for PEDF-transfected cells and 0.89 ± 0.61 ng for PEDFo-transfected cells; PEDF secretion by cells transfected without miniplasmid DNA varied from 0.06 ± 0.03 to 0.07 ± 0.03 ng (Figure 5B). Statistical analysis showed significant differences between PEDF secretion from transfected and non-transfected cells. However, the amount of PEDF protein secreted by cells transfected with the native PEDF sequence was not significantly different than the amount secreted by cells transfected with the optimized PEDF sequence.
Figure 5.
ELISA Quantification of Total PEDF Secretion by PEDF-Transfected Primary Human RPE Cells
For each donor sample, two control transfections without the addition of miniplasmid DNA and two transfections using 0.03 μg pFAR4-CMV SB100X SV40 transposase and 0.47 μg pFAR4-ITRs CMV PEDF BGH or pFAR4-ITRs CMV PEDFoptimized BGH miniplasmid DNA were carried out. Culture supernatants were analyzed 3 weeks after transfection. (A) PEDF secretion for 1 × 104 cells transfected with pFAR-PEDF was analyzed in 27 human donor eyes (age: 67.8 ± 11.9 years; gender: 13 males and 14 females; postmortem time: 31.5 ± 14.2 hr; cultivation time before transfection: 43.0 ± 15.1 days). For 1 × 104 cells transfected with pFAR-PEDFo, PEDF secretion was analyzed in 12 human donor eyes (age: 63.8 ± 13.5 years; gender: six males and six females; postmortem time: 33.0 ± 16.8 hr; cultivation time before transfection: 39.7 ± 17.8 days). (B) For 5 × 103 cells transfected with pFAR-PEDF, PEDF secretion was analyzed in 23 human donor eyes (age: 65.7 ± 14.7 years; gender: 13 males and ten females; postmortem time: 32.9 ± 14.8 hr; cultivation time before transfection: 44.4 ± 15.7 days); for cells transfected with pFAR-PEDFo, PEDF secretion was analyzed in 13 human donor eyes (age: 61.1 ± 16.3 years; gender: eight males and five females; postmortem time: 32.5 ± 16.1 hr; cultivation time before transfection: 42.6 ± 18.6 days). Data are presented as mean ± SD. Total PEDF secretion of PEDF-transfected and PEDFo-transfected cells was compared to the respective control cells (p < 0.0001, unpaired two-tailed t test) and total PEDF secretion of PEDF-transfected cells was compared to total PEDF secretion of PEDFo-transfected cells (not significant for 1 × 104 cells and 5 × 103 cells, unpaired two-tailed t test).
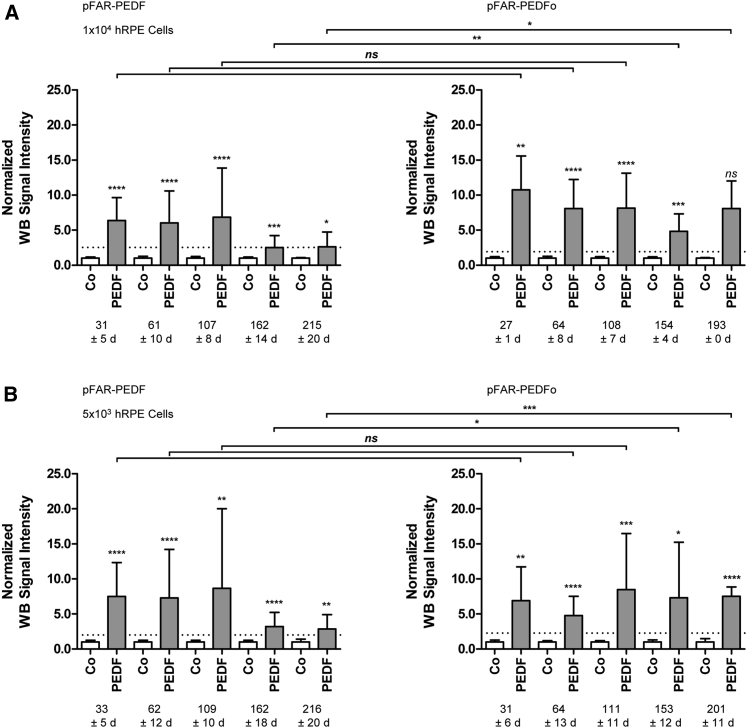
Long-term secretion of PEDF was analyzed by immunoblotting for a minimum of 6.5 months. The mean normalized signal intensity for 1 × 104 pFAR-PEDF-transfected cells was reduced by 59.0% from 31 to 215 days and by 24.6% for 1 × 104 pFAR-PEDFo-transfected cells from 27 to 193 days (Figure 6A; Table S2). The mean intensity decrease for 5 × 103 PEDF-transfected cells was 61.7% from day 33 to day 216. However, for 5 × 103 PEDFo-transfected cells, the mean normalized signal intensities increased by 9.5% from day 31 to day 201 (Figure 6B; Table S2). No statistical difference was observed between PEDF secretion by PEDF-transfected and PEDFo-transfected cells for the first 3.5 months. However, at later time periods, secretion by both 1 × 104 and 5 × 103 pFAR-PEDFo-transfected cells was significantly higher than secretion by cells transfected with pFAR-PEDF (Figure 6; Table S2).
Figure 6.
Western-Blot-Based Quantification of Long-Term PEDF Secretion in Primary Human RPE Cells after SB100X-Mediated Transfection
For each donor sample, two control transfections without the addition of miniplasmid DNA and two transfections using 0.03 μg pFAR4-CMV SB100X SV40 transposase and 0.47 μg pFAR4-ITRs CMV PEDF BGH or pFAR4-ITRs CMV PEDFoptimized BGH miniplasmid DNA were carried out. Culture supernatants were analyzed for total PEDF secretion every 4–8 weeks. Western blot signal intensities of PEDF-transfected and PEDFo-transfected cells were normalized to the signal intensities of the control cells. Data are presented as mean ± SD. Mean signal intensities of PEDF secretion 21 days after transfection are indicated by the dashed line. (A) For 1 × 104 cells, PEDF secretion was analyzed for 215 ± 20 days for pFAR-PEDF-transfected cells and for 193 days for pFAR-PEDFo-transfected cells. (B) For 5 × 103 cells, PEDF secretion was analyzed for 216 ± 20 days for pFAR-PEDF-transfected cells and for 201 ± 11 days for pFAR-PEDFo-transfected cells. Statistical analysis using an unpaired two-tailed t test showed a significant difference between transfected cells and non-transfected control cells. A comparison of PEDF-transfected cells to PEDFo-transfected cells showed no significant difference in PEDF secretion for 110 days, but by 160 days, a significant decrease in PEDF secretion by pFAR-PEDF-transfected cells compared to pFAR-PEDFo-transfected cells was observed.
Integration Profile of the pFAR4-ITRs CMV PEDF BGH Transposon Plasmid into Human RPE Cells
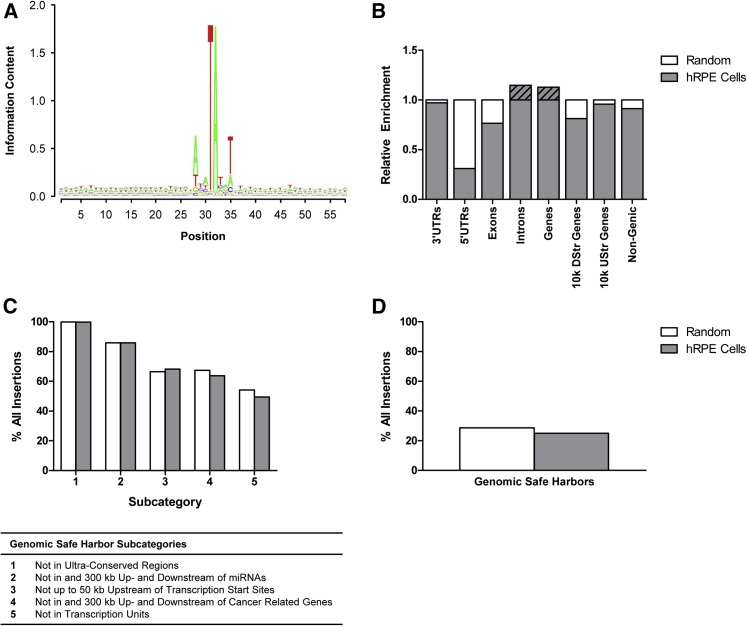
To investigate the integration profile of the PEDF transposon into human RPE cells, 1 × 104 cells were transfected with 0.03 μg pFAR4-CMV SB100X SV40 transposase and 0.47 μg pFAR4-ITRs CMV PEDF BGH transposon. After 21 days, cultures were terminated and genomic DNA extracted. Sequences neighboring the insertion sites were PCR-amplified using transposon inverted repeat (IR)-specific primers, and the resulting libraries were subjected to Illumina sequencing. Mapping the sequence reads on the human genome identified 3,114 unique transposon integration sites. As expected, the insertions occurred preferentially at the central dinucleotides of ATATATAT palindromic sequences (Figure 7A). To profile the SB insertions on a genomic scale, we next determined the frequencies of integrations into annotated genomic features including 3′ UTRs, 5′ UTRs, exons, introns, gene bodies, and regions around transcription start sites of RefSeq genes. When comparing the insertion profiles to a control set of 10,000 computationally generated random human genomic loci, no apparent bias was detected for any of the categories analyzed (Figure 7B). Furthermore, SB transposon integrations occur in genomic safe harbors, defined by five criteria32, 33 (Figure 7C), at about 25% (Figure 7D). These results are in good agreement with numerous previous studies, which established a close-to-random insertion profile for the SB system in human cells.34, 35, 36, 37
Figure 7.
Integration Profiling of the pFAR4-ITRs CMV PEDF BGH Transposon in Human RPE Cells
(A) Consensus sequence of the Sleeping Beauty insertion sites. The y axis in the Seqlogo analysis represents the strength of the information, with two bits being the maximum for a DNA sequence. (B) SB integration sites in human RPE cells were compared to a random control set of 1 × 104 computationally generated loci, determining the frequencies of integrations into annotated genomic features including 3′ UTRs, 5′ UTRs, exons, introns, gene bodies, and regions around transcription start sites. The integration profile was found to be close-to-random. (C) Relative frequencies of SB insertions into chromosomal sites defined as genomic safe harbors in comparison with the random dataset. (D) Relative frequency of SB integrations into genomic safe harbors, cumulative for all five subcategories shown in (C), in comparison with the random dataset.
Discussion
The development of the pFAR plasmid has enabled the maintenance and propagation of miniplasmids in the absence of antibiotics, eliminating a possible safety complication in the production of gene therapeutic vectors for use in human clinical trials. Instead of antibiotic selection, the propagation of the pFAR plasmid is based on the suppression of a chromosomal nonsense mutation in the thyA gene of the E. coli producer strain, ensuring efficient miniplasmid amplification using commercially available thymidine and animal components-free growth medium, which meets the requirements of regulatory authorities. In addition, the removal of the antibiotic resistance gene results in significant plasmid size reduction, which allows for a more efficient transgene38, 39 and SB transposon delivery (C. Marie and S.J., unpublished data).
The hyperactive form of the SB transposon system, SB100X,40 merges the advantages of viral vectors and non-viral plasmid DNA, since it possesses integrative properties and can be introduced into the cells by plasmid vectors, which reduces immunogenicity and simplifies its manufacturing. The SB100X transposase meditates the stable genomic integration of the expression cassette into the host cell’s genome, which allows for long-term transgene expression. Effective SB-mediated gene transfer has been reported for various in vitro and in vivo disease models,41, 42, 43 and it has been approved for clinical application of genetically modified T cells to treat B-lymphoid malignancies.44, 45 We have reported long-term and continuous PEDF secretion by RPE cells that were transfected with the PEDF transgene using the SB100X transposon system25 and demonstrated that the subretinal transplantation of PEDF-transfected cells in a rat model of CNV reduces neovascularization and inhibits new vessel formation.46
In animal studies, several gene therapeutic approaches for nvAMD have been described in which the transgenes have been delivered in vivo to resident cells by subretinal injection of viral vectors, and a clinical trial in patients with nvAMD has indicated that delivery of a gene to enhance the endogenous anti-angiogenic environment is a feasible approach (reviewed by Campochiaro7 in 2011). However, delivery of transgenes via viral vectors can distribute the transgene into tissues adjacent to the tissue of interest.47 We have proposed to treat nvAMD using a protocol comprising cell isolation from a patient, ex vivo transfection of the cells with the PEDF transgene using SB100X and subretinal transplantation of the transfected cells to the subretinal space of the same patient within a single surgical session lasting approximately 1 hr. Since the isolation of IPE or RPE cells from a patient’s biopsy yields a limited number of cells, i.e., 5 × 103 to 1 × 104 cells, classical methodologies for non-viral or virally mediated gene delivery are not suitable, since they require large numbers of cells and the necessity of selecting stably transfected cells in vitro, e.g., the generation of CD19-specific chimeric-antigen receptor (CAR) T cells.48
Here, we report the development of a protocol for the efficient transfection of as few as 5 × 103 primary pigment epithelial cells using pFAR miniplasmids carrying the SB100X transposase and the PEDF gene. IPE cells isolated from a number of bovine eyes and RPE cells isolated from a number of human donor eyes and co-transfected with both plasmids secrete high levels of PEDF continuously for the approximately 6 months that the cells have been followed in culture. The lower increase in PEDF secretion by PEDF-transfected primary human cells than the secretion by primary bovine cells may be due to the difference in the age of the human donors compared to the young bovine animals as well as to the fact that human cells were obtained from donor eyes 31.8 ± 13.7 hr postmortem whereas bovine cells were obtained from eyes within 3 hr postmortem. The increase in PEDF secretion by pFAR-PEDFo-transfected bovine IPE cells after 102 ± 9 (Figure 3A) and 103 ± 9 days (Figure 3B) suggests that the transfected cells were still able to proliferate.
Most importantly, we did not observe transgene silencing, which often occurs within a few days using the cytomegalovirus (CMV) promoter,49, 50 although the amount of PEDF secretion by the transfected cells decreased gradually over time, which was likely the result of the reduction of fetal bovine serum (FBS) in the cell-culture medium from 10% to 1%, once the primary cell cultures reached confluence.
To improve the stability of the human PEDF gene, the optimization strategy implemented here intended to mimic natural transgenes, removed potential ESE motifs by modulating synonymous sites and also included modulation of AT versus GC nucleotide content. However, removal of the ESE motifs did not affect PEDF gene expression significantly. In fact, analysis of the relative PEDF gene expression and quantification of total PEDF protein secretion in human RPE cells 21 days after transfection revealed higher expression and secretion levels by pFAR-PEDF-transfected cells than in pFAR-PEDFo-transfected cells. No statistical difference was observed between PEDF secretion by PEDF-transfected and PEDFo-transfected human RPE cells for the first 3.5 months. However, at 5 and 7 months after transfection, secretion by 1 × 104 and 5 × 103 pFAR-PEDFo-transfected human RPE cells was significantly higher than secretion by cells transfected with pFAR-PEDF, suggesting that the optimization protocol improved stability.
In contrast to the genomic integration profile of retroviral systems and other transposon systems (piggyBac and Tol2), which show integration preferences for actively transcribed genes,51, 52, 53 SB-based integration is random.37, 54 Compared to computationally generated control datasets, SB100X-mediated genomic integration of the PEDF gene delivered by the pFAR4 vector into primary human RPE cells showed a close-to-random profile. Noteworthy, the ratio of integrations of the PEDF gene into exons, representing 4% of the total integration events, was lower than the ratio expected by random distribution. In human T cells, SB transposon insertions were shown to have the lowest deviation from a random genome-wide distribution as well as the highest theoretical chance of targeting a safe site of the genome.35, 55 Although site-specific gene insertion into safe harbor sites by designer nucleases, such as zinc finger nucleases, TALENs, and the CRISPR/Cas system, is appealing from a safety point of view, the overall efficiency of transgene integration in polyclonal RPE cell populations by these technologies is expected to be far lower than with our transposon-based approach. In contrast, robust gene delivery in primary human cells by the hyperactive SB100X transposase is matched by an attractive safety profile in the context of currently available integrating gene vector systems.
Thus, using a transfection protocol that combines the use of the SB100X transposase and the pFAR4 miniplasmid, we have shown efficient transgene delivery to as few as 5 × 103 primary cells, sustained expression of the transgene, a close-to-random transgene integration profile, without the potential integration of plasmid-encoded antibiotic resistance genes into the genome of the host cell. These results are an important step of the TargetAMD project toward the approval of a clinical trial, where the PEDF transgene is introduced into autologous pigment epithelial cells ex vivo, followed by the transplantation to the subretinal space of nvAMD patients. Further deliverables that comprise, for example, different animal studies to evaluate efficacy and safety, the production of transfection-related agents under good manufacturing practice (GMP) conditions as well as administrative work are already finished or currently in process.
Materials and Methods
Miniplasmid Description
pFAR4 is a miniplasmid vector devoid of antibiotic resistance markers. Its propagation relies on the suppression by a suppressor t-RNA, encoded in the pFAR4 miniplasmid, of an amber mutation introduced into the thyA gene of Escherichia coli.56 The pFAR4-CMV SB100X SV40 (pFAR-SB100X) miniplasmid was obtained after extraction of the hyperactive SB100X transposase expression cassette from pCMV(CAT)T7-SB100X40 and introduction into the antibiotic-free pFAR4 vector.56 The PEDF-encoding transposon miniplasmid, pFAR4-ITRs CMV PEDF BGH (pFAR-PEDF), contains ITR sequences amplified from pT2/HB (a gift from Perry Hackett, Addgene plasmid #26556), a human PEDF cDNA that was generated from ARPE-19 cells (ATCC CRL-2302) and regulatory sequences amplified from pT2-CMV-PEDF/EGFP.25 The optimized PEDF DNA sequence was synthesized and used to generate pFAR4-ITRs CMV PEDFoptimized BGH (pFAR-PEDFo). The pFAR-PEDF and pFAR-PEDFo miniplasmids have exactly the same size.
All pFAR4 derivatives were constructed and propagated using the dedicated bacterial strain (TM #47-9a).56 Miniplasmids were purified using EndoFree plasmid preparation kits (Macherey-Nagel) and a bacterial growth medium that meets the requirements for use in clinical trials.
Optimization of the PEDF cDNA Sequence by Reduction of ESE Motifs
An algorithm was implemented, in which synonymous sites embedded in ESE motifs were identified and recoded to decrease purine content, potentially increasing RNA stability. A set of 238 human ESE motifs was employed from the RESCUE-ESE set.57 ESE motifs are very rich in the nucleotide A (∼50% of all sites in ESEs), followed by the nucleotide G with about 25% of all sites, whereas the nucleotides C and T each occupying about 12%–13% of all sites. ESE motifs were matched in all possible frames and synonymous sites identified in the human PEDF gene. All representations of codons within the ESE motifs for leucine were set to CTT and for arginine to CGT. At 4-fold degenerate sites embedded in ESE motifs, the C-ending codon was preferred. At 2-fold degenerate sites A was replaced with G, C with T, and T with C, whereas G remained unchanged. The C to T swap is necessary to break ESE motifs. Stop codons were avoided. The optimization program can be run iteratively until an equilibrium density of ESE motifs is reached or stopped prior to equilibrium to preserve a higher ESE density.
Isolation and Cultivation of Primary Human Retinal Pigment Epithelial Cells
Human eyes from 29 donors (age 66.1 ± 13.8 years; 16 males and 13 females) were obtained from the Aachen Cornea Bank of the Department of Ophthalmology, University Hospital RWTH Aachen. The eyes were removed 31.8 ± 13.7 hr postmortem after informed consent was obtained in accord with the Declaration of Helsinki protocols. Procedures for the collection and use of human samples were approved by the institutional ethics committee. For RPE cell isolation, the anterior segment was removed by a circumferential cut approximately 3 mm posterior to the limbus. After careful removal of the vitreous and the retina, the posterior eyecup was filled with 1 mL DMEM/Ham’s F-12 (Biochrom) supplemented with 10% FBS (PAA Laboratories), 80 U/mL penicillin and 80 μg/mL streptomycin (Lonza), and 2.5 μg/mL amphotericin B (Sigma-Aldrich). The cells were harvested by gently brushing the retinal pigment epithelium with a fire-polished glass spatula. The procedure was repeated once, and the cell mixture was centrifuged at 1,000 rpm for 10 min. The cell pellet from each eye was suspended in DMEM/Ham’s F-12 supplemented with 10% FBS, 80 U/mL penicillin, 80 μg/mL streptomycin, and 2.5 μg/mL amphotericin B and plated into three wells of a 24-well tissue culture plate. Cultures were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2 until confluence was reached. Cell-culture medium was changed twice a week.
Isolation and Cultivation of Primary Bovine Iris Pigment Epithelial Cells
Bovine eyes were obtained from a local abattoir and brought to the laboratory within 3 hr of sacrifice. The anterior segment was cut approximately 3.5 mm posterior to the limbus and the iris was dissected from the ciliary body. After incubation of the iris in 0.25% trypsin (GE Healthcare Europe) for 20 min at 37°C, IPE cells were isolated by gently brushing the posterior iris surface with a fire-polished glass spatula. The detached cells were centrifuged at 1,000 rpm for 10 min, and the cell pellet was suspended in DMEM/Ham’s F-12 supplemented with 10% FBS, 80 U/mL penicillin, 80 μg/mL streptomycin, and 2.5 μg/mL amphotericin B and plated at a density of 1 × 105 cells/cm2 in a 6-well tissue culture plate. The cultures were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2 until confluence was reached. Cell-culture medium was changed twice a week.
Electroporation of Primary Pigment Epithelial Cells and Cultivation of the Transfected Cells
Transfections were performed with the Neon Transfection System using the 10 μL Kit (Life Technologies) according to the manufacturer’s protocol. The electroporation parameters were as follows: two pulses, 1,100 V (pulse voltage), 20 ms (pulse width). 1 × 104 or 5 × 103 cells in 11 μL resuspension buffer R (Life Technologies) were combined with 2 μL of purified plasmid mixture containing 0.03 μg pFAR-SB100X transposase and 0.47 μg pFAR-PEDF or 0.47 μg pFAR-PEDFo transposon. Each experiment comprised eight transfection controls without electrical field application and without miniplasmid DNA, six transfection controls with electrical field application without miniplasmid DNA, and 12 transfections with electrical field application and miniplasmid DNA for each transposon miniplasmid. Transfected cells were transferred into 48-well tissue culture plates containing 0.5 mL of DMEM/Ham’s F-12 supplemented with 10% FBS, without antibiotics and antimycotics. Penicillin (80 U/mL), streptomycin (80 μg/mL), and amphotericin B (2.5 μg/mL) were added with the first medium exchange 3 days after electroporation. Cell cultures were either used after 3 weeks for further analyses or maintained for the analysis of long-term PEDF secretion.
SDS-PAGE and Western Blot Analysis
For SDS-PAGE, 15 μL of culture supernatant was mixed with an equal volume of 2 × SDS sample buffer58 and heated for 5 min at 95°C. Proteins were separated on a 10% SDS-PAGE gel and transferred onto a 0.45-μm pore-size nitrocellulose membrane (Whatman) using the semi-dry transfer system (Bio-Rad), followed by Ponceau S staining to confirm the transfer. For the detection of total (endogenous plus recombinant) PEDF, blots were blocked with 3% BSA/Tris-buffered saline (TBS) for 2 hr at room temperature and incubated for 1 hr at room temperature and overnight at 4°C with anti-PEDF antibodies (rabbit polyclonal; 1:4,000 diluted in 3% BSA/TBS; BioProducts MD), followed by incubation for 1 hr at room temperature with horseradish peroxidase-conjugated anti-rabbit antibodies (goat polyclonal; 1:2,000 diluted in 10% milk powder/TBS; Abcam). Proteins bands were visualized by chemiluminescence using the LAS-3000 imaging system (Fujifilm) or the Omega Lum G Imaging System (Aplegen) and evaluated by the open source image processing program ImageJ (Rasband, W.S., ImageJ, U.S. NIH, http://imagej.nih.gov/ij/, 1997–2014).
ELISA-Based Quantification of Total PEDF Secretion
Total PEDF secretion was analyzed in culture supernatants of control and PEDF-transfected cells 3–4 weeks after transfection. Cells were incubated in a defined volume of 0.5 mL cell-culture medium. After 24 hr, the culture media were analyzed by ELISA for human PEDF using the ELISAquant kit (BioProducts MD) according to the manufacturer’s protocol. Secreted PEDF was related to the cell number determined in each well. Cells were trypsinized with 0.05% trypsin-0.02% EDTA (PAA Laboratories) and counted using a Neubauer chamber or the CASY Cell Counter Model TT (Roche Diagnostics).
Isolation of Total RNA and Genomic DNA
Total RNA was isolated using the RNeasy Mini Kit together with the RNase-free DNase Set (QIAGEN) according to the manufacturer’s protocol. Reverse transcription was carried out on 0.1 μg total RNA using the Reverse Transcription System (Promega). Genomic DNA was isolated using the QIAamp DNA Mini Kit (QIAGEN) according to the manufacturer’s protocol for cultured cells.
Real-Time qPCR
For human RPE cells, real-time qPCRs were performed on a LightCycler 1.2 Instrument using the LightCycler FastStart DNA Master SYBR Green I kit (Roche) according to the manufacturer’s recommendations. The cDNA samples were run in duplicate using the following primers: GAPDH (GenBank: NM_002046.3) as the internal control gene (F: 5′-ATC CCA TCA CCA TCT TCC AG-3′ and R: 5′-ATG AGT CCT TCC ACG ATA CC-3′), endogenous PEDF (GenBank: NM_002615.4) (F: 5′-GCT GGC TTT GAG TGG AAC GA-3′ and R: 5′-GTG TCC TGT GGA ATC TGC TG-3′), and endogenous plus recombinant PEDF (F: 5′-CCT GCA GGA GAT GAA GCT GCA-3′ and R: 5′-TCC ACC TGA GTC AGC TTG ATG-3′), for the detection of the endogenous PEDF gene plus the PEDF and the optimized PEDF genes encoded in the pFAR4 miniplasmids. Reactions were performed with diluted cDNA corresponding to 2 ng of initially used total RNA and a primer concentration of 0.25 μM. Thermal cycler conditions were as follows: initial denaturation at 95°C for 10 min followed by 50 cycles with denaturation at 95°C for 10 s, annealing at 60°C for 8 s, and elongation at 72°C for 15 s. Melting curve analysis confirmed the application specificity of each primer pair. Data were processed by the LightCycler software 3.5.3 and evaluated using the comparative CT (2-ΔΔCT) method, which describes relative gene expression.59
For bovine IPE cells, real-time qPCRs were performed on a LightCycler 480 II instrument (Roche) using PerfeCta SYBR Green FastMix (Quanta Biosciences) according to manufacturer’s recommendations. The cDNA samples were run in duplicate in three independent experiments using the following primers: GAPDH (GenBank: NM_001034034.2) as the internal control gene (F: 5′-ATC ATC CCT GCT TCT ACT GG-3′ and R: 5′-CTC AGT GTA GCC TAG AAT GC-3′), endogenous PEDF (GenBank: AF017058.1) (F: 5′-TTC CCT CTG GAC TAT CAC CT-3′ and R: 5′-AAT CTG CCA TCC CTC TAG TG-3′), and recombinant PEDF (F: 5′-CCT GCA GGA GAT GAA GCT GCA-3′ and R: 5′-TCC ACC TGA GTC AGC TTG ATG-3′), for the detection of the PEDF transgene encoded in the pFAR-PEDF and pFAR-PEDFo miniplasmids. Reactions were performed with diluted cDNA corresponding to 1.2 ng of initially used total RNA, and primer concentrations of 3.5 μM for GAPDH and recombinant PEDF, and 1.75 μM for endogenous PEDF. Thermal cycler conditions were as follows: initial denaturation at 95°C for 10 min followed by 45 cycles with denaturation at 95°C for 5 s, annealing at 60°C for 10 s, and elongation at 72°C for 10 s. Melting curve analysis confirmed the application specificity of each primer pair; standard curves demonstrated high efficiency and good reproducibility of each experiment. Data were processed by the LightCycler 480 software 1.5.1.62 and evaluated using the standard curve-based absolute quantification method. The values were analyzed for statistical significance using a non-parametric Kruskal-Wallis test (GraphPad Prism, GraphPad).
Integration Site Library Construction
A computation-assisted hemi-specific PCR scheme was used to generate the insertion site libraries. The PCR assays were based on the use of computationally designed hemi-specific primers carrying four specific nucleotides (4-mers) at their 3′ ends followed by random sequences and a specific overhang.60 The computational primer design comprised: (1) choosing 4-mers that cannot give rise to unwanted amplicons on the transposon end or on the primer overhangs and (2) predicting the combination of those six 4-mers, which theoretically would result in the most comprehensive insertion site library on the human genome. Nested PCRs were performed to obtain indexed, Illumina-flow cell compatible insertion site libraries. Primer sequences are listed in Table S3. Following a short enrichment PCR using 300 ng of genomic DNA as template (primer SB_20_hmr, PCR conditions: 95°C for 2 min, 50 cycles of 94°C for 30 s, ramp to 63°C: 1°C/s, 63°C for 30 s, 72°C for 30 s), six parallel PCRs/sample were carried out, each containing 5 pmol of six different hemi-specific 4-mer primers using the following conditions: 95°C for 1 min, 40 cycles of 94°C for 30 s, ramp to 63°C: 1°C/s, 63°C for 30 s, 72°C for 30 s, two cycles of 94°C for 30 s, 25°C for 1 min and 30 s, ramp to 72°C: 0.2°C/s, 72°C for 1 min. Then each PCR was supplemented with 25 μL of PCR master mix containing 15 pmol of SB_7 primer. The PCR program for the nested PCR was: 15 super-cycles of three cycles of 94°C for 30 s, ramp to 64°C: 1°C/s, 64°C for 30 s, 72°C for 40 s, one cycle of 94°C for 30 s, ramp to 60°C: 0.5°C/s, 60°C for 30 s, 72°C for 40 s, final extension of 72°C for 5 min. The PCR products were column-purified, and 2 μL of the 30-μL elutes was used for the first exponential PCRs, with the primer PE_first and SB_PE_noTA_BC_n using the following cycling conditions: 95°C for 30 s, 20 cycles of 94°C for 30 s, ramp to 64°C: 1°C/s, 64°C for 30 s, 72°C for 1 min, final extension 72°C for 5 min. The first exponential PCR products diluted 10-fold were used to append the Illumina adaptors to the amplicons using Pfx polymerase (Life Technologies) with the following cycling conditions: 95°C for 30 s, 20 cycles of 94°C for 15 s, 68°C for 1 min, final extension 68°C for 5 min. The final PCR products were separated on agarose gels and amplicons between 200 and 500 bp were excised and column purified (Zymoclean Gel DNA Recovery Kit; Zymo Research). Sequencing of the resulting libraries was carried out on an Illumina HiSeq 2500 instrument with rapid run flow-cells at the Beckman Coulter Genomics sequencing facility.
Computational Analysis
The raw reads were processed for mapping using R (R: A language and environment for statistical computing; R Foundation for Statistical Computing, https://www.r-project.org/) as follows: primer-, transposon-, and right Illumina adaptor-related sequences were trimmed and the resulting reads were quality filtered by omitting reads containing “N” bases and by trimming reads as soon as two of five bases have quality encoding less than a phred score of 20. All trimmed reads shorter than 24 bases were not included in further analysis. The remaining sequences were mapped against the hg19 human genome assembly with Bowtie.61 We identified 3,114 independent integration events from three different human RPE samples for the pFAR4-ITRs CMV PEDF BGH transposon. Reads mapped to exactly the same strand and genomic positions were merged to a single site. To improve data quality, we discarded all sites supported by less than ten reads. The sequence logos for the nucleotide composition around the integration sites were generated with the seqLogo package in R. To compute the representation of the insertion sites in various annotated segments of the human genome, databases for various genomic features were downloaded from the UCSC database (http://genome.ucsc.edu), and the representation of the insertion sites were counted in the feature intervals using the BEDTools suite.62 A set of 10,000 computationally generated random genomic loci of the human genome was used as a reference.
Statistical Analysis
Unless otherwise stated, statistical analysis was performed using an unpaired two-tailed t test (GraphPad Prism, GraphPad).
Author Contributions
Conceptualization, G.T. and S.J.; Methodology, G.T., C. Marie, L.D.H., D.S., Z. Ivics, Z. Izsvák, and S.J.; Investigation, N.H., C.P.-S., M.P., A.S., C. Miskey, S.D., M.K., and S.J.; Resources, C. Marie and P.W.; Writing – Original Draft, G.T. and S.J.; Writing – Review & Editing, G.T., C. Marie, L.D.H., P.W., D.S., Z. Ivics, Z. Izsvák, and S.J.; Funding Acquisition, G.T.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the European Union’s Seventh Framework Programme for research, technological development and demonstration, grant agreement no. 305134. C. Miskey and Z. Ivics were supported by the Center for Cell and Gene Therapy of the LOEWE (Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz) program in Hessen, Germany. L.D.H. was supported by the European Research Council (advanced grant ERC-2014-ADG 669207) and the Medical Research Council (MR/L007215/1). The authors thank Anna Dobias, Antje Schiefer (Department of Ophthalmology, University Hospital RWTH Aachen), Gregg Sealy (Laboratory of Ophthalmology, University of Geneva) for excellent technical support, the Aachen Cornea Bank (Department of Ophthalmology, University Hospital RWTH Aachen) for providing the human donor eyes, and Perry B. Hackett (Center for Genome Engineering, The University of Minnesota) for the gift of pT2/BH.
Footnotes
Supplemental Information includes five figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.02.002.
Supplemental Information
References
- 1.Velez-Montoya R., Oliver S.C., Olson J.L., Fine S.L., Quiroz-Mercado H., Mandava N. Current knowledge and trends in age-related macular degeneration: Genetics, epidemiology, and prevention. Retina. 2014;34:423–441. doi: 10.1097/IAE.0000000000000036. [DOI] [PubMed] [Google Scholar]
- 2.Vos T., Barber R.M., Bell B., Bertozzi-Villa A., Biryukov S., Bolliger I., Global Burden of Disease Study 2013 Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386:743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta S. Age-related macular degeneration. Prim. Care. 2015;42:377–391. doi: 10.1016/j.pop.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Kwak N., Okamoto N., Wood J.M., Campochiaro P.A. VEGF is major stimulator in model of choroidal neovascularization. Invest. Ophthalmol. Vis. Sci. 2000;41:3158–3164. [PubMed] [Google Scholar]
- 5.Ohno-Matsui K. [Molecular mechanism for choroidal neovascularization in age-related macular degeneration] Nippon Ganka Gakkai Zasshi. 2003;107:657–673. [PubMed] [Google Scholar]
- 6.Hanout M., Ferraz D., Ansari M., Maqsood N., Kherani S., Sepah Y.J., Rajagopalan N., Ibrahim M., Do D.V., Nguyen Q.D. Therapies for neovascular age-related macular degeneration: Current approaches and pharmacologic agents in development. BioMed Res. Int. 2013;2013:830837. doi: 10.1155/2013/830837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campochiaro P.A. Gene transfer for neovascular age-related macular degeneration. Hum. Gene Ther. 2011;22:523–529. doi: 10.1089/hum.2011.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campochiaro P.A., Nguyen Q.D., Shah S.M., Klein M.L., Holz E., Frank R.N., Saperstein D.A., Gupta A., Stout J.T., Macko J. Adenoviral vector-delivered pigment epithelium-derived factor for neovascular age-related macular degeneration: Results of a phase I clinical trial. Hum. Gene Ther. 2006;17:167–176. doi: 10.1089/hum.2006.17.167. [DOI] [PubMed] [Google Scholar]
- 9.Dijkhuizen P.A., Pasterkamp R.J., Hermens W.T., de Winter F., Giger R.J., Verhaagen J. Adenoviral vector-mediated gene delivery to injured rat peripheral nerve. J. Neurotrauma. 1998;15:387–397. doi: 10.1089/neu.1998.15.387. [DOI] [PubMed] [Google Scholar]
- 10.Hermens W.T., Verhaagen J. Adenoviral vector-mediated gene expression in the nervous system of immunocompetent Wistar and T cell-deficient nude rats: Preferential survival of transduced astroglial cells in nude rats. Hum. Gene Ther. 1997;8:1049–1063. doi: 10.1089/hum.1997.8.9-1049. [DOI] [PubMed] [Google Scholar]
- 11.Pierce E.A., Bennett J. The status of RPE65 gene therapy trials: Safety and efficacy. Cold Spring Harb. Perspect. Med. 2015;5:a017285. doi: 10.1101/cshperspect.a017285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beard B.C., Dickerson D., Beebe K., Gooch C., Fletcher J., Okbinoglu T., Miller D.G., Jacobs M.A., Kaul R., Kiem H.P., Trobridge G.D. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol. Ther. 2007;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- 13.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cesana D., Sgualdino J., Rudilosso L., Merella S., Naldini L., Montini E. Whole transcriptome characterization of aberrant splicing events induced by lentiviral vector integrations. J. Clin. Invest. 2012;122:1667–1676. doi: 10.1172/JCI62189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hargrove P.W., Kepes S., Hanawa H., Obenauer J.C., Pei D., Cheng C., Gray J.T., Neale G., Persons D.A. Globin lentiviral vector insertions can perturb the expression of endogenous genes in beta-thalassemic hematopoietic cells. Mol. Ther. 2008;16:525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- 17.Montini E., Cesana D., Schmidt M., Sanvito F., Ponzoni M., Bartholomae C., Sergi Sergi L., Benedicenti F., Ambrosi A., Di Serio C. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 18.Stein S., Ott M.G., Schultze-Strasser S., Jauch A., Burwinkel B., Kinner A., Schmidt M., Krämer A., Schwäble J., Glimm H. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y., Keating K., Thorpe R. Comparison of toxicogenomic profiles of two murine strains treated with HIV-1-based vectors for gene therapy. Toxicol. Appl. Pharmacol. 2007;225:189–197. doi: 10.1016/j.taap.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Real J.P., Granero G.E., De Santis M.O., Juarez C.P., Palma S.D., Kelly S.P., Luna J.D. Rate of vision loss in neovascular age-related macular degeneration explored. Graefes Arch. Clin. Exp. Ophthalmol. 2015;253:1859–1865. doi: 10.1007/s00417-014-2885-y. [DOI] [PubMed] [Google Scholar]
- 21.Aisenbrey S., Lafaut B.A., Szurman P., Hilgers R.D., Esser P., Walter P., Bartz-Schmidt K.U., Thumann G. Iris pigment epithelial translocation in the treatment of exudative macular degeneration: A 3-year follow-up. Arch. Ophthalmol. 2006;124:183–188. doi: 10.1001/archopht.124.2.183. [DOI] [PubMed] [Google Scholar]
- 22.Falkner-Radler C.I., Krebs I., Glittenberg C., Povazay B., Drexler W., Graf A., Binder S. Human retinal pigment epithelium (RPE) transplantation: Outcome after autologous RPE-choroid sheet and RPE cell-suspension in a randomised clinical study. Br. J. Ophthalmol. 2011;95:370–375. doi: 10.1136/bjo.2009.176305. [DOI] [PubMed] [Google Scholar]
- 23.Lappas A., Foerster A.M., Weinberger A.W., Coburger S., Schrage N.F., Kirchhof B. Translocation of iris pigment epithelium in patients with exudative age-related macular degeneration: Long-term results. Graefes Arch. Clin. Exp. Ophthalmol. 2004;242:638–647. doi: 10.1007/s00417-003-0764-z. [DOI] [PubMed] [Google Scholar]
- 24.Izsvák Z., Hackett P.B., Cooper L.J., Ivics Z. Translating Sleeping Beauty transposition into cellular therapies: Victories and challenges. BioEssays. 2010;32:756–767. doi: 10.1002/bies.201000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnen S., Izsvák Z., Stöcker M., Harmening N., Salz A.K., Walter P., Thumann G. Sleeping Beauty transposon-mediated transfection of retinal and iris pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 2012;53:4787–4796. doi: 10.1167/iovs.12-9951. [DOI] [PubMed] [Google Scholar]
- 26.Kuerten D., Johnen S., Harmening N., Souteyrand G., Walter P., Thumann G. Transplantation of PEDF-transfected pigment epithelial cells inhibits corneal neovascularization in a rabbit model. Graefes Arch. Clin. Exp. Ophthalmol. 2015;253:1061–1069. doi: 10.1007/s00417-015-2954-x. [DOI] [PubMed] [Google Scholar]
- 27.Solensky R. Hypersensitivity reactions to beta-lactam antibiotics. Clin. Rev. Allergy Immunol. 2003;24:201–220. doi: 10.1385/CRIAI:24:3:201. [DOI] [PubMed] [Google Scholar]
- 28.Kreiss P., Cameron B., Rangara R., Mailhe P., Aguerre-Charriol O., Airiau M., Scherman D., Crouzet J., Pitard B. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res. 1999;27:3792–3798. doi: 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parmley J.L., Chamary J.V., Hurst L.D. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol. Biol. Evol. 2006;23:301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- 30.Parmley J.L., Urrutia A.O., Potrzebowski L., Kaessmann H., Hurst L.D. Splicing and the evolution of proteins in mammals. PLoS Biol. 2007;5:e14. doi: 10.1371/journal.pbio.0050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savisaar R., Hurst L.D. Purifying selection on exonic splice enhancers in intronless genes. Mol. Biol. Evol. 2016;33:1396–1418. doi: 10.1093/molbev/msw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papapetrou E.P., Lee G., Malani N., Setty M., Riviere I., Tirunagari L.M., Kadota K., Roth S.L., Giardina P., Viale A. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 34.Ammar I., Gogol-Döring A., Miskey C., Chen W., Cathomen T., Izsvák Z., Ivics Z. Retargeting transposon insertions by the adeno-associated virus Rep protein. Nucleic Acids Res. 2012;40:6693–6712. doi: 10.1093/nar/gks317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gogol-Döring A., Ammar I., Gupta S., Bunse M., Miskey C., Chen W. Genome-wide profiling reveals remarkable parallels between insertion site selection properties of the MLV retrovirus and the piggyBac transposon in primary human CD4(+) T cells. Mol. Ther. 2016;24:592–606. doi: 10.1038/mt.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voigt K., Gogol-Döring A., Miskey C., Chen W., Cathomen T., Izsvák Z., Ivics Z. Retargeting sleeping beauty transposon insertions by engineered zinc finger DNA-binding domains. Mol. Ther. 2012;20:1852–1862. doi: 10.1038/mt.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yant S.R., Wu X., Huang Y., Garrison B., Burgess S.M., Kay M.A. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chabot S., Orio J., Schmeer M., Schleef M., Golzio M., Teissié J. Minicircle DNA electrotransfer for efficient tissue-targeted gene delivery. Gene Ther. 2013;20:62–68. doi: 10.1038/gt.2011.215. [DOI] [PubMed] [Google Scholar]
- 39.Ribeiro S., Mairhofer J., Madeira C., Diogo M.M., Lobato da Silva C., Monteiro G., Grabherr R., Cabral J.M. Plasmid DNA size does affect nonviral gene delivery efficiency in stem cells. Cell. Reprogram. 2012;14:130–137. doi: 10.1089/cell.2011.0093. [DOI] [PubMed] [Google Scholar]
- 40.Mátés L., Chuah M.K., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 41.Hyland K.A., Olson E.R., Clark K.J., Aronovich E.L., Hackett P.B., Blazar B.R., Tolar J., Scott McIvor R. Sleeping Beauty-mediated correction of Fanconi anemia type C. J. Gene Med. 2011;13:462–469. doi: 10.1002/jgm.1589. [DOI] [PubMed] [Google Scholar]
- 42.Kren B.T., Unger G.M., Sjeklocha L., Trossen A.A., Korman V., Diethelm-Okita B.M., Reding M.T., Steer C.J. Nanocapsule-delivered Sleeping Beauty mediates therapeutic Factor VIII expression in liver sinusoidal endothelial cells of hemophilia A mice. J. Clin. Invest. 2009;119:2086–2099. doi: 10.1172/JCI34332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sjeklocha L.M., Wong P.Y., Belcher J.D., Vercellotti G.M., Steer C.J. β-Globin sleeping beauty transposon reduces red blood cell sickling in a patient-derived CD34(+)-based in vitro model. PLoS ONE. 2013;8:e80403. doi: 10.1371/journal.pone.0080403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kebriaei P., Huls H., Jena B., Munsell M., Jackson R., Lee D.A., Hackett P.B., Rondon G., Shpall E., Champlin R.E., Cooper L.J. Infusing CD19-directed T cells to augment disease control in patients undergoing autologous hematopoietic stem-cell transplantation for advanced B-lymphoid malignancies. Hum. Gene Ther. 2012;23:444–450. doi: 10.1089/hum.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh H., Figliola M.J., Dawson M.J., Olivares S., Zhang L., Yang G., Maiti S., Manuri P., Senyukov V., Jena B. Manufacture of clinical-grade CD19-specific T cells stably expressing chimeric antigen receptor using Sleeping Beauty system and artificial antigen presenting cells. PLoS ONE. 2013;8:e64138. doi: 10.1371/journal.pone.0064138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnen S., Djalali-Talab Y., Kazanskaya O., Möller T., Harmening N., Kropp M., Izsvák Z., Walter P., Thumann G. Antiangiogenic and neurogenic activities of Sleeping Beauty-mediated PEDF-transfected RPE cells in vitro and in vivo. BioMed Res. Int. 2015;2015:863845. doi: 10.1155/2015/863845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stieger K., Colle M.A., Dubreil L., Mendes-Madeira A., Weber M., Le Meur G., Deschamps J.Y., Provost N., Nivard D., Cherel Y. Subretinal delivery of recombinant AAV serotype 8 vector in dogs results in gene transfer to neurons in the brain. Mol. Ther. 2008;16:916–923. doi: 10.1038/mt.2008.41. [DOI] [PubMed] [Google Scholar]
- 48.Kebriaei P., Singh H., Huls M.H., Figliola M.J., Bassett R., Olivares S., Jena B., Dawson M.J., Kumaresan P.R., Su S. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J. Clin. Invest. 2016;126:3363–3376. doi: 10.1172/JCI86721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liew C.G., Draper J.S., Walsh J., Moore H., Andrews P.W. Transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1521–1528. doi: 10.1634/stemcells.2006-0634. [DOI] [PubMed] [Google Scholar]
- 50.Orbán T.I., Apáti A., Németh A., Varga N., Krizsik V., Schamberger A., Szebényi K., Erdei Z., Várady G., Karászi E. Applying a “double-feature” promoter to identify cardiomyocytes differentiated from human embryonic stem cells following transposon-based gene delivery. Stem Cells. 2009;27:1077–1087. doi: 10.1002/stem.45. [DOI] [PubMed] [Google Scholar]
- 51.Grabundzija I., Irgang M., Mátés L., Belay E., Matrai J., Gogol-Döring A., Kawakami K., Chen W., Ruiz P., Chuah M.K. Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson M.H., Coates C.J., George A.L., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 53.Wu X., Li Y., Crise B., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 54.Vigdal T.J., Kaufman C.D., Izsvák Z., Voytas D.F., Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 55.Monjezi R., Miskey C., Gogishvili T., Schleef M., Schmeer M., Einsele H., Ivics Z., Hudecek M. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia. 2017;31:186–194. doi: 10.1038/leu.2016.180. [DOI] [PubMed] [Google Scholar]
- 56.Marie C., Vandermeulen G., Quiviger M., Richard M., Préat V., Scherman D. pFARs, plasmids free of antibiotic resistance markers, display high-level transgene expression in muscle, skin and tumour cells. J. Gene Med. 2010;12:323–332. doi: 10.1002/jgm.1441. [DOI] [PubMed] [Google Scholar]
- 57.Fairbrother W.G., Yeo G.W., Yeh R., Goldstein P., Mawson M., Sharp P.A., Burge C.B. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh393. W187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 59.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 60.Ewing A.D., Kazazian H.H., Jr. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan A.R., Hall I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.