Abstract
Since the discovery of μ-opioid receptor (MOR) gene two decades ago, various regulatory factors have been shown to interact with the MOR promoter and modulate transcript levels. However, the majority of early transcriptional studies on MOR gene have not addressed how intracellular signaling pathways mediate extracellular modulators. In this study, we demonstrate that MOR epigenetic regulation requires multiple coordinated signals converging at the MOR promoter, involving mitogen-activated protein kinase (MAPK) activation and mitogen- and stress-activated protein kinase 1 (MSK1)—ranges of intracellular signaling pathways similar to those activated by opioid agonists. Inhibiting p38 MAPK or extracellular signal-regulated kinase (ERK) 1/2 MAPK (upstream activators of MSK1) reduced MOR expression levels; accordingly, the functional role of MSK1, but not MSK2, was demonstrated using genetic approaches. However, for maximal MSK1 effect, an open chromatin configuration was required, because in vitro CpG methylation of the MOR promoter abolished MSK1 activity. Finally, endogenous MSK1 levels concomitantly increased to regulate MOR gene expression during neuronal differentiation of P19 cells, suggesting a conserved role of this kinase in the epigenic activation of MOR in neurons. Taken together, our findings indicate that the expression of MOR gene requires the activity of intracellular signaling pathways that have been implicated in the behavioral outcomes of opioid drugs, which suggests that an autoregulatory mechanism may function in opioid systems.
Introduction
Opioid drugs act through one of the four different G protein-coupled opioid receptor systems: μ, δ, κ, and opioid receptor-like 1 (ORL1) (Al-Hasani and Bruchas, 2011). These receptors are widely expressed in the central nervous system with varying densities among the nociceptive neural circuitry as well as critical brain regions involved in reward and emotion-related responses (Hwang et al., 2009). Since its discovery two decades ago, the genes encoding μ-opioid receptor (OPRM1 and Oprm1) have been extensively characterized at the cellular and molecular levels, and transgenic mouse models have been generated to demonstrate that the analgesic response to the prototypical opioid, morphine, depends on μ-opioid receptor (MOR) expression levels (Sora et al., 1997; Loh et al., 1998). Accordingly, transcriptional regulation of MOR gene in various cellular contexts has been reported, and several positive [e.g., interleukin-4 response element, sex determining region Y box (SOX), poly(rC)-binding protein, specificity protein 1 (SP1), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), cAMP response-element binding protein (CREB)] and negative regulatory factors [e.g., neuron-restrictive silencer factor, octamer-1, PU.1, SP3, poly-ADP ribose polymerase 1] that affect MOR transcription have been identified [for review see Wei and Loh (2011)]. However, the majority of these early studies on MOR gene transcription were performed in silico and applied genetic approaches to analyze whether various factors were bound to putative recognition site(s) within the MOR promoter to modulate expression. Nevertheless, a number of recent studies have shown that MOR gene transcription is activated in response to various extracellular stimuli and follows multiple coordinated intracellular signals that converge at the MOR promoter (Kim et al., 2011; Wagley et al., 2013). Quite interestingly, MOR gene expression follows a unique spatial and epigenetic regulation in different mouse brain regions where opioid drugs act (Hwang et al., 2009). As such, stimulation of P19 embryonic carcinoma cells with epigenetic modifiers such as a histone deacetylase inhibitor or a demethylating agent has been shown to increase de novo transcription of MOR gene in a tightly regulated event that repositions nucleosomes to mount the transcription machinery (Hwang et al., 2007, 2010). Yet, the precise role of signal transduction events during this critical chromatin-directed event remains largely elusive.
Histone deacetylase (HDAC) inhibitors result in the accumulation of hyperacetylated core histones in nucleosomes, leading to transcriptional reprogramming, which is considered to enhance the therapeutic benefits of HDAC inhibitors on several diseases, such as cancers, cardiovascular diseases, neurodegenerative disorders, and pulmonary diseases (Johnstone, 2002; Glaser et al., 2003; Delcuve et al., 2012; Chueh et al., 2015). Even so, only a small portion (2–10%) of expressed genes in transformed cells are truly modulated by HDAC inhibitors, which suggests that the net transcriptional outcome largely depends on functions of the regulatory proteins that can interact with the reconfigured nucleosomes (Dokmanovic et al., 2007). Indeed, phylogenetic analyses have suggested that HDACs preceded the evolution of histones (Dokmanovic et al., 2007) and can target the activity of nonhistone regulatory proteins to determine the transcriptional outcome of various genes, such as p21, γ-globin, and myostatin (Rivero and Adunyah, 1996; Zhong et al., 2003; Sangerman et al., 2006; Han et al., 2010; Simboeck et al., 2010). For example, treatment with the classic HDAC inhibitor trichostatin A (TSA) resulted in mitogen-activated protein kinase (MAPK) signaling pathway–mediated increase of histone H3 serine 10 phosphorylation, which was crucial for acetylation of lysine 14 and recruitment of activated RNA polymerase II at the p21 promoter (Simboeck et al., 2010). Additionally, sodium butyrate (NaB) has been shown to directly activate p38 MAPK and extracellular signal-regulated kinase 1 (ERK-1) in myocytes and lymphoid cells (Rivero and Adunyah, 1996; Zhong et al., 2003; Sangerman et al., 2006; Han et al., 2010), and in some cellular models, TSA stimulation has been shown to increase epidermal growth factor receptor tyrosine phosphorylation and phosphatidyl inositol-3-kinase /protein kinase B (AKT) activation to modulate survivin expression levels (Zhou et al., 2006).
In this study, we used classic HDAC inhibitors as a tool to investigate whether intracellular signaling cascades regulate the de novo transcriptional activation of MOR gene in P19 embryonic carcinoma cells. Interestingly, we observed that HDAC inhibition of P19 cells led to MAPK activation, and an increase in expression (and function) of downstream mitogen- and stress-activated kinase (MSK1) to affect MOR gene transcription. Analogous increase in MSK1 expression (and function) occurred alongside MOR gene transcription during retinoic acid-mediated neuronal differentiation of P19 cells, which indicates a central regulatory role of MAPK/MSK1 cascade in the epigenetic activation of MOR gene.
Materials and Methods
Materials.
Trichostatin A, valproic acid (VPA), sodium butyrate, retinoic acid, H89, and SP600125 (SP) were purchased from Sigma-Aldrich (St. Louis, MO). 4-(4-Flurophenyl)-2-(4-methylsulfinylphenyl)-5-(4-pyridyl) imidazole [SB203580 (SB)], and 1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio]butadiene (U0126) were purchased from Cell Signaling Technology (Danvers, MA). Anti-phospho-ERK 1/2, anti-phospho-p38 MAPK, anti-ERK 1/2, anti-p38 MAPK-α, anti-GAPDH, and anti-MSK1 antibodies were obtained from Cell Signaling Technology. Anti-phospho- histone H3 (Ser 28), anti-p65, anti-p50, anti-β-III tubulin, and anti-β-actin were obtained from Santa Cruz Biotechnology (Dallas, TX). Anti-CREB, anti-p38 MAPK-β-2, anti-acetyl histone H3, anti-phosphoacetyl histone H3 (phospho-Ser10-acetyl-Lys14) were obtained from EMD Millipore (Billerica, MA). Anti-phospho-histone H3 (Ser 10), anti-phospho-MSK1 (Ser 360), and anti-MSK2, were obtained from Abcam (Cambridge, MA). Alkaline phosphatase-conjugated goat anti-rabbit, goat anti-mouse, and rabbit anti-goat immunoglobulin G were supplied by BioRad (Hercules, CA). Restriction enzymes and buffers were obtained from New England Biolabs (Ipswich, MA). Supplies for cell cultures were from Gibco/Thermo Fisher Scientific (Sunnyvale, CA). Other reagents for molecular studies were supplied by Sigma-Aldrich.
Cells and Stimulations.
P19 cells were used throughout this study because epigenetic activation of MOR gene [upon stimulation with TSA or retinoic acid-mediated neuronal differentiation (see below)] has been demonstrated (Hwang et al., 2007), which makes this an excellent model to investigate the role of intracellular signaling cascades. For cell stimulations, the stock solutions of TSA (10 μM) and retinoic acid (1.0 mM) were prepared in ethanol. Stock solutions of NaB (0.5 M), VPA (1.0 M), and H89 (10 mM) were prepared in sterile distilled water and used within 2 months of preparation. Stock solutions of SB (25 mM), SP (25 mM), and U0126 (10 mM) were prepared in cell-culture grade dimethyl sulfoxide (Sigma-Aldrich). All stock solutions for cell stimulation were stored at –20°C in the dark. One day before treatment, 3 × 105 cells were seeded into each well of a six-well plate. On the treatment day, cells were washed twice with serum-free medium and cultured further for 4 hours before stimulating with TSA (25 nM), VPA (10 μM), or NaB (5 mM) for various lengths of time. The final concentration of the vehicles (ethanol or dimethyl sulfoxide) used for cell stimulation were between 0.1–0.2%, which do not affect MOR gene expression in P19 cells (Hwang et al., 2007). At the end of stimulation, cells were washed twice with ice-cold phosphate buffered saline (PBS), and total RNA or protein extract was prepared as required for analysis.
Neuronal Differentiation of P19 Cells.
P19 embryonic carcinoma cells were cultured and differentiated into neuronal cells as previously described (Hwang et al., 2007; Wagley et al., 2013) with minor modifications. Briefly, for induction of differentiation, 5 × 106 cells were cultured in differentiation medium [α-MEM (Life Technologies/Thermo Fisher Scientific) supplemented with 5% fetal bovine serum (HyClone Laboratories/GE Healthcare Life Sciences, Logan, UT)] in the presence of 0.5 μM retinoic acid in bacteriological-grade Petri dishes for 4 days to induce aggregation. On the fourth day, cell aggregates were extensively washed with serum-free α-MEM and dissociated using 0.1% trypsin-EDTA and 50 μg/ml DNase I. The dissociated cells were seeded on tissue culture dishes that were treated with 0.1 mg/ml poly-l-ornithine. Cell stimulations and transfection of the neuronally differentiating cells were carried out 4–8 hours after seeding and designated as day 1. The differentiating cells received fresh media with or without inhibitor every 48 hours after plating.
Preparation of Acid-Soluble Proteins.
Acid-soluble histone proteins were extracted as described previously (Zhong et al., 2001) with minor modifications. Briefly, 1 × 106 P19 cells stimulated with HDAC inhibitors for various lengths of time were collected and resuspended in 180 μl of cold PBS. Twenty microliters of 2 N HCl was added to lyse the cells. Acid-soluble supernatant fractions were collected after centrifugation at 12,000 rpm for 10 minutes and treated with a final concentration of 25% TCA on ice for 30 minutes to precipitate the proteins. The protein pellets were collected by centrifugation at 12,000 rpm for 10 minutes at 4°C, and washed twice with ice-cold acetone. Finally, the protein pellets were air-dried for 5 minutes at room temperature and solubilized using 1% SDS. The protein concentration was determined using a bicinchoninic acid (BCA) protein assay as described by the manufacturer (Thermo Fisher Scientific), and 500 ng to 1 μg of the preparation was used to perform immunoblotting with various anti-histone H3 antibodies.
RT-PCR and qRT-PCR
Total RNA was extracted using TRI Reagent (Molecular Research Center, Cincinnati, OH) and analyzed by reverse transcription–polymerase chain reaction (RT-PCR) using the MOR gene-specific primers 5′- CATCAAAGCACTGATCACGATTCC-3′ (mMOR-S) and 5′- TAGGGCAATGGAGCAGTTTCTGC-3′ (mMOR-AS) (Hwang et al., 2007; Wagley et al., 2013). Semiquantitative RT-PCR (qRT-PCR) was performed in 500 ng of total RNA using a Qiagen OneStep RT-PCR kit (Valencia, CA). Similar reactions were performed using β-actin as an internal control (Hwang et al., 2007). qRT-PCR was performed as previously described (Hwang et al., 2010) using the Quantitect SYBR Green RT-PCR kit (Valencia, CA). To calculate relative mRNA gene expression, amplification curves of test sample and standard samples that contained 101 to 106 molecules of the MOR gene (using pcDNA3.1-mMOR, constructed in our laboratory) were monitored, and the number of target molecules in the test sample was analyzed using qCalculator version 1.0 software (http://www.gene-quantification.de/download.html#qcalculator) on the basis of the mathematical model of Pfaffl (Pfaffl, 2001). The numbers of target molecules was normalized against that obtained for β-actin, which was used as an internal control (Hwang et al., 2007). The specificity of qRT-PCR reaction was determined using a melt curve after the amplification and the PCR products were also verified on an agarose gel. The RT-PCR and qRT-PCR experiments were performed in triplicate and each experiment was repeated with at least three different biologic replicates. Results from at least two separate experiments were combined and presented as mean ± S.EM.
Immunoblotting.
Immunoblotting was performed as described previously (Wagley et al., 2013). Briefly, 3 × 105 cells were seeded into each well of six-well dishes and treated as required. After washing the monolayer three times with ice-cold PBS, lysis buffer composed of 50 mM Tris-Cl, 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, protease inhibitor cocktail, and phosphatase inhibitors was added. Cell lysates were collected, vortexed vigorously, and clarified by centrifugation. The protein concentrations in the supernatant were determined using BCA protein assay. Thirty micrograms of each lysate was loaded into SDS-polyacrylamide gels and electrotransferred onto polyvinyl difluoride membranes. Membranes were blocked for 30 minutes in 3% bovine serum albumin solution in T-TBS (Tris-buffered saline containing 0.05% Tween-20), and then incubated overnight at 4°C using primary antibody dilutions as suggested by the manufacturers. Membranes were washed three times with T-TBS and incubated with alkaline-phosphatase–conjugated secondary antibodies for 1 hour at room temperature. Data were collected on a Storm 860 PhosphorImager Fluorescent Scanner (Molecular Dynamics, Chatsworth, CA) with appropriate settings for each antibody. Band densities were determined using the ImageQuant software (GE Healthcare Life Sciences, Marlborough, MA) and statistical analysis was performed using GraphPad Prism (version 5.04; La Jolla, CA).
Preparation of Cytosolic and Nuclear Protein Fractions.
Nuclear and cytosolic protein preparations were carried out as described previously (Wagley et al., 2013). At the end of treatment, cells were washed three times in ice-cold PBS. Cytosolic protein fraction was prepared by directly adding hypotonic lysis buffer (10 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol, 0.5 mM phenylmethyl sulfonylfluoride (PMSF), and protease inhibitor cocktail) to the monolayer and stored for 5 minutes on ice. Cell lysates were collected by scraping, and following centrifugation supernatants were collected as cytosolic protein fractions. Nuclear pellets were washed twice by centrifugation with hypotonic lysis buffer and resuspended in hypertonic buffer solution (20 mM HEPES, 400 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM PMSF, and protease inhibitor cocktail). Nuclear membrane was disrupted by vigorous vortexing and stored on ice for 30 minutes. Following the removal of insoluble particles by centrifugation, the supernatant was collected as nuclear fraction. A BCA protein assay was performed to determine the protein yield. Ten to twenty micrograms of protein from each fraction were used for immunoblot analysis as described above.
Transient Transfection Analysis.
Approximately 1.5 × 106 P19 cells were seeded in 60-mm dishes 1 day before transient transfection. Cells were transfected using a complex containing 10 μl of Lipofectamine 2000, 300 ng of MOR promoter construct (pGL450), 1.0 μg of pCMV5–constitutively active MSK1 (S376D, T581D, T700D) (CA-MSK1) (received from Dr. James C. Hastie, University of Dundee), and 50 ng of pCH110-β-galactosidase per transfection sample diluted in 500 μl of Opti-MEM (Life Technologies/Thermo Fisher Scientific). Forty eight hours after transfection, cells were washed with serum-free medium and treated with HDAC inhibitors for 8 hours, and cell lysates were analyzed for firefly luciferase activity and β-galactosidase activity as described by the manufacturer’s protocol [Promega (Madison, WI) and Tropix (Middlesex, MA), respectively]. Results were expressed as relative luciferase activity compared with the control cells (Hwang et al., 2003; Wagley et al., 2013).
RNA Interference.
The gene-specific effect on MOR transcription was analyzed by transfecting small-interfering RNA (siRNA) against p38 MAPK-α or p38 MAPK-β (Cell Signaling Technology), and MSK1 or MSK2 (Santa Cruz Biotechnology). For transfection, approximately 1 × 105 P19 cells/well in 1 ml of P19 growth medium (α-MEM supplemented with 7.5% newborn calf serum and 2.5% fetal bovine serum) were plated into 12-well plates. Cells were allowed to attach for 4–8 hours and transfected with 50 nM each of control siRNA (Cell Signaling Technology) or 50 nM gene-specific siRNAs using 3 μl of Lipofectamine 2000 in a transfection mixture prepared in 200 μl of Opti-MEM. Twelve hours after transfection, 1 ml of fresh growth medium was added to the cells and continued to culture for another 24 hours. At the end of transfection, cells were stimulated with HDAC inhibitors as required and harvested for qRT-PCR and immunoblot assays.
CpG Methylation of MOR Promoter.
CpG Methylation of MOR promoter construct pGL450 was carried out using SssI methylase as described by the manufacturer. Briefly, 2 μg of MOR promoter construct was added to a reaction mixture containing 640 μM S-adenosylmethionine buffer and 4 units of SssI methylase in a final reaction volume of 20 μl. The reaction was allowed to continue for 3 hours at 37°C. At the end of incubation period, the reaction was terminated by heating at 65°C for 30 minutes. Parallel reactions that lacked SssI enzyme were carried out as control. For analyses, one fourth (approximately 500 ng) of the methylated or sham-methylated MOR promoter was cotransfected with 1 μg of MSK1 expression construct and 50 ng of pCH110-β-galactosidase for 48 hours as described above. At the end of transfection, cells lysates were prepared and assayed for luciferase and β-galactosidase activity, and presented as relative activity compared with control.
Statistical Analysis.
All data presented herein were performed in at least three different biologic replicates. Numerical values were presented as the mean ± S.E.M. For comparison between two samples, t-test analysis was performed. For multiple comparisons, analysis of variance with Bonferroni’s post-hoc test was used. All statistical analyses were performed in GraphPad Prism 5 software. P values representing significances of <0.05, 0.01 are denoted with symbols *, **, whereas significances <0.05, 0.01 among various treatment groups are represented with †, ††, respectively.
Results
Time Delay between Histone Acetylation and Opioid Receptor Gene Expression.
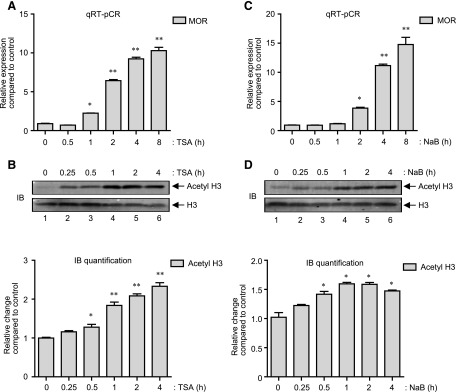
Although it is known that the de novo transcription of μ-opioid receptor (MOR) gene increases upon histone deacetylase (HDAC) inhibition and requires an extensive chromatin remodeling at the MOR promoter (Lin et al., 2008; Kraus, 2012), it is largely unknown how signal transduction cascades influence changes at the MOR promoter. A kinetic analysis of MOR gene expression induced by a classic HDAC inhibitor, Trichostatin A (TSA) in MOR-negative P19 embryonic carcinoma cells (P19 cells) was performed. As shown by quantitative real-time RT-PCR (qRT-PCR) analysis in Fig. 1A, TSA stimulations of P19 cells increased MOR gene expression in a time-dependent fashion, which appeared 1 hour after the treatment (∼2-fold), dramatically increased at 2 hours (**P < 0.01; ∼6-fold), and continued further (∼10-fold) by 8 hours of treatment. Quite interestingly, immunoblot analysis (Fig. 1B) of the acid-soluble histone proteins prepared from cells stimulated in a similar way (0–4 hours) showed that the increase in global histone H3 acetylation had already reached high significance (**P < 0.01 at t = 1 hour) (Fig. 1B, upper histogram) compared with the dramatic increase in MOR gene expression (Fig. 1 A, from ∼2-fold to ∼6-fold between 1 and 2 hours), indicating a time delay in between the two events. Further experiments were performed to investigate whether NaB, another HDAC inhibitor, also demonstrates a similar pattern of MOR gene transcription and histone acetylation kinetics. As shown in Fig. 1C, qRT-PCR analysis showed that NaB stimulation of P19 cells also significantly increases MOR gene expression by 2 hours (∼3.8-fold, **P < 0.01), which dramatically increases further to reach ∼12-fold by 4 hours. As observed with TSA stimulation (Fig. 1B), NaB stimulation of P19 cells also led to similar histone H3 acetylation kinetics (Fig. 1D) with maximum acetyl H3 achieved at 1 hour (**P < 0.01) (Fig. 1D, upper histogram). Taken together, there exists a similar time delay between the dramatic increase in MOR gene expression (starting 2 hours after stimulation) and significant histone H3 acetylation (starting 1 hour after stimulation) with two different HDAC inhibitors (TSA and NaB), which suggests that HDAC effects on MOR transcription are mediated by biochemical changes unique to histone acetylation.
Fig. 1.
HDAC inhibitors increase MOR transcription in a time-dependent fashion. (A) P19 cells were stimulated with TSA (25 ng/ml) for 0–8 hours as indicated, and total RNA was extracted. MOR expression levels were determined from total RNA samples by qRT-PCR analysis and presented as relative expression as described in Materials and Methods (*P < 0.05, **P < 0.01, n = 4). (B) Gel image: P19 cells were stimulated with TSA (0– 4 hours) as indicated, and acid-soluble protein fractions were prepared. A representative immunoblot that shows changes in the levels of acetylated histone H3 (Acetyl H3) is presented. The levels of total histone H3 were monitored as internal control (histogram). The pixel densities obtained for acetyl H3 and total H3 were measured for each time-point and presented as relative change compared with control (*P < 0.05, **P < 0.01, n = 3). (C) P19 cells were treated with NaB (5 mM) for (0–8 hours) as indicated and qRT-PCR analysis to determine relative MOR expression was performed as described in (A) (*P < 0.05, **P < 0.01, n = 5). (D) Gel image: P19 cells were stimulated with NaB (0–4 hours) as indicated, and the levels of acetyl H3 and H3 in the acid-soluble protein fractions were determined by immunoblot analysis. Histogram: The pixel densities for acetyl H3 and total H3 were measured for each sample and presented as relative change compared with control (*P < 0.05, n = 5).
p38 MAPK and ERK 1/2 Regulates HDAC Inhibition Mediated MOR Gene Expression.
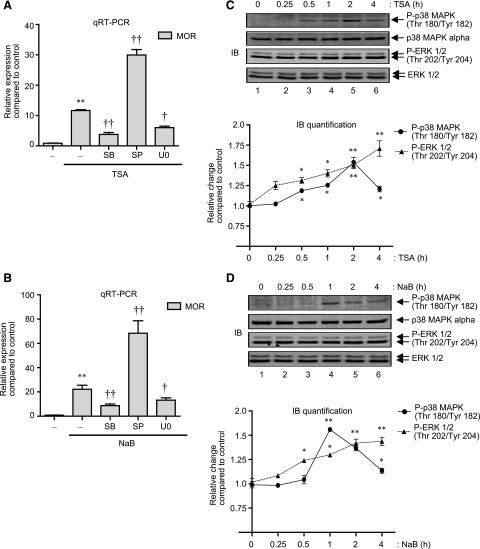
As mentioned previously, HDAC inhibitor-mediated transcription is a combinatorial outcome of histone modifications and functions of proteins of the signal-transduction cascade that directs sequence-specific transcription factors and components of the basal transcription machinery to the responsive promoter (Dokmanovic et al., 2007). Thus, we next examined if HDAC inhibitor-mediated increase of MOR gene expression is dependent on the activity of classic MAPK components such as p38 MAPK, c-Jun N-terminal kinase (JNK), or ERK. For this purpose, P19 cells were pretreated with pharmacological inhibitors of p38 MAPK [SB203580 (SB)], JNK [SP600125 (SP)], and MEK/ERK 1/2 (U0126, U0) for 1 hour and stimulated with TSA or NaB for a further 8 hours. Total RNA was extracted from stimulated cells, and analyzed for MOR expression. Figure 2, A and B, shows that SB and U0 each significantly blocked the MOR gene expression induced by TSA or NaB, suggesting the involvement of MAPK activities. Intriguingly, JNK inhibitor (SP) showed a synergistic effect and further potentiated MOR expression levels (∼3-fold increase in MOR expression levels compared with TSA or NaB stimulation alone) (Fig. 2, A and B). As a control, we have previously demonstrated that SP increases MOR gene expression via a p38 MAPK-dependent mechanism in P19 cells whereas SB or U0 treatment has minimal effects (Wagley et al., 2013).
Fig. 2.
HDAC inhibition activates p38 MAPK and ERK 1/2 to regulate MOR mRNA expression. (A) P19 cells were pretreated for 1 hour with 25 μM SB (p38 MAPK inhibitor), 25 μM SP (JNK inhibitor), or 10 μM U0 (MEK/ERK inhibitor) and then treated for 8 hours with 25 ng/ml of TSA. Total RNA was extracted, reverse transcribed, and analyzed by qRT-PCR to determine changes in MOR expression levels. Results were normalized using the levels of β-actin as internal control [F(4, 15) = 167.8, **P < 0.01 versus control; †P < 0.05, ††P < 0.01 versus TSA stimulation]. (B) P19 cells were pretreated for 1 hour with SB, SP, or U0 as in (A) and then treated for 8 hours with 5 mM NaB. Total RNA was extracted, and changes in MOR expression levels were determined as explained above [F(4, 14) = 74.07, *P < 0.01 versus control; †P < 0.05, ††P < 0.01 versus NaB stimulation]. (C) Gel image: P19 cells were treated with 25 ng/ml of TSA for various lengths of time as indicated (0–4 hours), and total cell lysates were analyzed by immunoblotting with anti-phospho-p38 MAPK, anti-p38 MAPK-α, anti-phospho-ERK 1/2, and anti-ERK 1/2 antibodies. Graph: The pixel densities obtained for phospho-p38 MAPK and phospho-ERK 1/2 were normalized against the pixel densities obtained for p38 MAPK and ERK 1/2, respectively. Data from three independent results were combined and presented as relative change compared with nontreated control (*P < 0.05, **P < 0.01). (D) Gel image: P19 cells were treated with 5 mM NaB (0–4 hours) as indicated, and the levels of phospho-p38 MAPK, p38 MAPK, phospho-ERK 1/2, and ERK 1/2 were determined in the total cell lysates as above. Graph: The pixel densities obtained for phospho-p38 MAPK and phospho-ERK 1/2 were plotted against the pixel densities obtained for p38 MAPK and ERK 1/2, respectively, as in (C) (*P < 0.05, **P < 0.01).
Since inhibition of either p38 MAPK or ERK 1/2 reduced MOR gene expression induced by TSA or NaB stimulation, we determined if p38 MAPK and ERK 1/2 phosphorylation levels are increased in the stimulated cells. As shown in Fig. 2C, the levels of phosphorylated p38 MAPK increased after 0.5 hours of TSA stimulation and continued further to reach maximum levels by 2 hours (**P < 0.01) (Fig. 2C, lanes 3–5). As a control, the levels of total p38 MAPK were analyzed, and found to be minimally changed. Interestingly, unlike p38 MAPK phosphorylation kinetics, which started to decline after reaching maximum levels at 2 hours, increases in the levels of phosphorylated ERK 1/2 (but not total ERK 1/2) were sustained, and continued throughout the 4-hour stimulation period (Fig. 2C, cf. lanes 1–6). Similar analyses in NaB-stimulated P19 cells (Fig. 2D) revealed analogous increase in ERK 1/2 phosphorylation that was sustained over the 4-hour stimulation period (Fig. 2D, cf. lanes 1 and 5), whereas p38 MAPK phosphorylation followed a slightly different course, and showed maximum levels at 1 hour after stimulation (compared with 2 hours for TSA stimulation). Collectively, these results demonstrate that HDAC inhibition increases p38 MAPK and ERK 1/2 phosphorylation in P19 cells, and that pharmacological inhibition of these kinases reduce MOR gene expression levels induced by the same HDAC inhibition.
MSK1 Expression Increases by HDAC Inhibition.
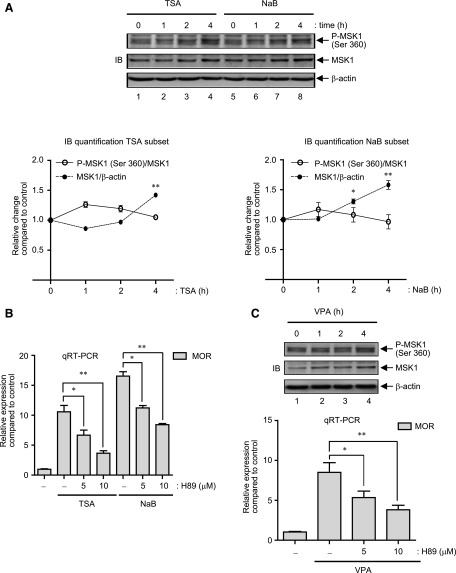
Depending on the stimulus, activation of MAPKs, specifically p38 MAPK and ERK 1/2, can lead to the phosphorylation and activation of the downstream kinase, mitogen- and stress-activated protein kinase 1 (MSK1), which affects transcription by increasing the phosphorylation of histone proteins to remodel transcriptionally inactive chromatin into a more active configuration, and/or by increasing the phosphorylation and downstream activity of transcription factors such as ER81, ATF1, NF-κB, CREB, HMG14 etc. (Zhong et al., 2003; Keum et al., 2013). Thus, MSK1 activation was examined by immunoblot analyses of total cell lysates prepared from TSA- and NaB-stimulated P19 cells (0–4 hours). As shown in Fig. 3A, MSK1 phosphorylation at serine 360 [a common substrate of p38 MAPK and ERK 1/2 within the linker domain of MSK1 (Arthur, 2008)] increased upon either TSA or NaB stimulation in a time-dependent fashion that started 1 hour after the stimulation and continued up to 4 hours (cf. lanes 1–4 and 5–8). Unexpectedly, analyses of total MSK1 also showed increased expression levels that were clearly evident by stimulations with TSA and NaB after 4 hours (Fig. 3A, cf. lanes 1, 4 and 5, 8). Meanwhile, densitometric analyses (Fig. 3A lower graphs) showed that the ratio of phosphorylated MSK1 to that of total MSK1 increased nominally between 1–2 hours of TSA stimulation (Fig. 3 A, lower panel, left graph) but remained fairly indistinguishable throughout NaB stimulation (Fig. 3A, lower panel, right graph). Further analyses at time-points earlier than 1 hour (0, 15 minutes, and 30 minutes) upon TSA or NaB stimulations were also carried out and showed minimal changes in both forms of MSK1 (data not shown). To rule out the possibility of a general increase of cellular protein levels upon stimulation, immunoblot analyses of β-actin levels were carried out, which showed minimal changes (Fig. 3A). Additionally, as already demonstrated, since the changes in the levels of total p38 MAPK and ERK 1/2 also remained minimal after TSA or NaB stimulation (Fig. 2, C and D), the increase of MSK1 levels appears to be a specific effect of HDAC inhibition.
Fig. 3.
HDAC inhibitors increase MSK1 protein levels in P19 cells. (A) Gel image: P19 cells were treated with 25 ng/ml TSA or 5 mM NaB for 0–4 hours as indicated and total cell lysates were prepared. Immunoblotting was performed to determine the expression levels of phospho-MSK1 and MSK1. Each blot was then reprobed with anti-β-actin as control. Graph: The pixel density values for phospho-MSK1, MSK1 and β-actin were measured for each time-point, and the relative changes in the ratio of phospho-MSK1/MSK1 and MSK1/β-actin was presented (TSA, left graph; NaB, right graph; *P < 0.05, **P < 0.01, n = 4). (B) P19 cells were pretreated for 1 hour with various concentrations of H89 (0, 5 or 10 μM) followed by TSA (25 ng/ml) or NaB (5 mM) stimulation for 6 hours. Total RNA was prepared and analyzed by qRT-PCR to determine changes in MOR expression levels as described previously (*P < 0.05, **P < 0.01, n = 4–8). (C) Gel image: P19 cells were treated with 10 μM VPA for 0–4 hours as indicated and total cell lysates were analyzed by immunoblotting with phospho-MSK1, MSK1 and β-actin antibodies. Histogram: P19 cells were pretreated for 1 hour with various concentrations of H89 (0–10 μM) followed by VPA treatment of 6 hours. MOR mRNA levels were determined by qRT-PCR as described in Fig. 1. Error bars represent the range of standard errors, and asterisks represent statistically significant findings (*P < 0.05; **P < 0.01, n = 3).
Since increased MSK1 expression and phosphorylation (Fig. 3A) occurred simultaneously with increases in MOR expression levels upon TSA or NaB stimulation (Fig. 1, A and C), we investigated whether MSK1 activity is involved by using a small molecule protein kinase A (PKA)/MSK1 inhibitor, H89. Therefore, P19 cells were stimulated with TSA or NaB in presence of a range of H89 (0–10 μM), and MOR expression levels were determined by qRT-PCR analyses. As presented in Fig. 3B, MOR gene expression induced by both HDAC inhibitors reduced in a dose-dependent fashion by H89 treatment (∼40% and ∼65% reduction compared with control for TSA, and ∼32% and ∼47% reduction compared with control for NaB stimulation by 5 μM or 10 μM H89, respectively), which suggests that MSK1 activity is involved. In further experiments, we determined if VPA (valproic acid, a different class of HDAC inhibitor used for the treatment of bipolar disorders), shares a common mechanism to regulate MOR gene transcription involving increased expression (and activity thereof) of MSK1 proteins. As suspected, and as shown in Fig. 3C (upper gel image), MSK1 expression (and phosphorylation) indeed increased in a time-dependent fashion (Fig. 3C, cf. 1, 3 and 4) upon VPA stimulation of P19 cells. Further qRT-PCR analyses showed that MOR gene expression too increased in a similar time-dependent fashion (data not shown), and that H89 treatment exerted a dose-dependent antagonistic effect on those MOR expression levels (Fig. 3C, lower histogram).
p38 MAPK-β Regulates MOR Gene Transcription but Minimally Affects MSK1 Expression.
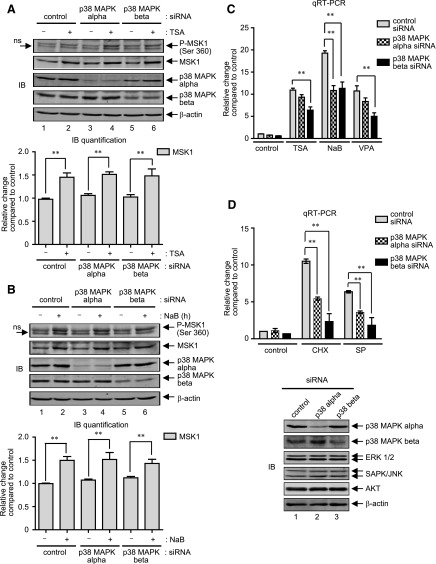
Although ∼60% homologous in their amino acid sequences, each of the four mammalian p38 MAPK isoforms (α, β, γ, and δ ) differ considerably in their tissue-specific expression patterns, substrate specificities, and susceptibility to chemical inhibitors (Cuenda and Rousseau, 2007; Coulthard et al., 2009). Of note, only the activities of p38 MAPK-α and p38 MAPK-β are inhibited by the chemical compound SB (Cuenda and Rousseau, 2007). As demonstrated earlier (Fig. 2, A and B), since SB inhibited MOR gene expression induced by TSA (∼65% reduction compared with control) and NaB (∼60% reduction compared with control), and given the fact that activation of p38 MAPK can lead to the phosphorylation and activation of downstream MSK1, we next determined which p38 MAPK isoform (α and/or β) regulates MSK1 phosphorylation in response to HDAC inhibition. P19 cells were transfected with siRNA against p38 MAPK-α or p38 MAPK-β for 36 hours, and were further stimulated with TSA or NaB for 4 hours to analyze changes in MSK1 expression and phosphorylation levels. The 4-hour time-point was chosen because both the expression and phosphorylation levels of MSK1 are clearly increased at this time-point (Fig. 3A). As shown in Fig. 4, A and B, immunoblot analyses, quite unexpectedly, revealed that although each p38-MAPK siRNA specifically and significantly decreased the expression level of its respective target, neither phosphorylation nor the expression of MSK1 levels decreased under stimulated conditions. In an attempt to determine whether both p38 MAPK activities simultaneously regulate MSK1 phosphorylation, further analyses using the pharmacological inhibitor SB were carried out, which also proved insignificant (data not shown). Taken together, these results demonstrate that HDAC inhibitors do not use p38 MAPK pathway to increase the phosphorylation and expression of MSK1 proteins in P19 cells.
Fig. 4.
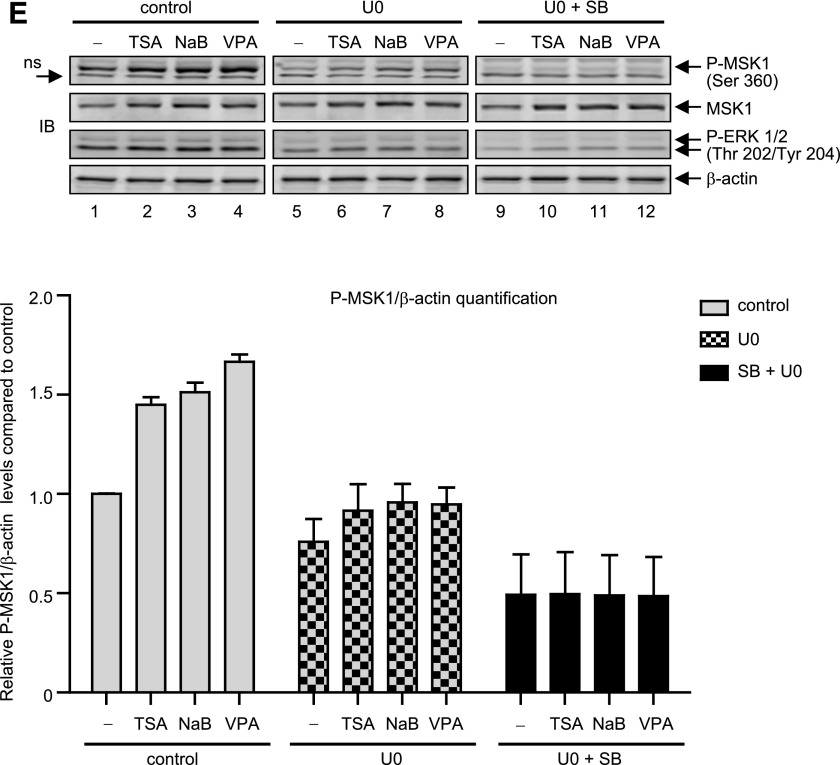
HDAC inhibition-mediated MAPK activity does not affect MSK1 expression. (A) Gel image: P19 cells were transfected with control siRNA, p38-MAPK-α siRNA or p38-MAPK-β siRNA for 36 hours. At the end of transfection, cells were left untreated or treated with TSA (25 ng/ml) for 4 hours, and changes in the expression levels of phospho-MSK1 and MSK1 were analyzed by immunoblotting. The levels of p38 MAPK-α and p38 MAPK-β were analyzed to show specificity of the siRNA transfection, and β-actin levels were analyzed as internal control. Histogram: The pixel densities obtained for MSK1 were normalized against the pixel densities obtained for β-actin values and presented as relative change compared with control (**P < 0.01, n = 4). (B) P19 cells transfected with control siRNA or siRNA specific for p38-MAPK-α and -β isoforms for 36 hours were treated with NaB (5 mM) for a further 4 hours, and immunoblot analysis (gel image) and MSK1 level quantification (histogram) were performed as described above (**P < 0.01, n = 4). (C) Total RNA extracted after 6-hour stimulation with TSA (25 ng/ml), NaB (5 mM), or VPA (10 μM) in P19 cells transfected as above were analyzed by qRT-PCR to determine changes in MOR mRNA levels. Results were normalized against the levels of β-actin and presented as relative change compared with control (**P < 0.01, n = 7–10). (D) Histogram: qRT-PCR analysis to determine changes in MOR mRNA levels were performed as above from cells transfected as in (B) but stimulated with CHX (10 μg/ml) or SP (25 μM) (**P < 0.01, n = 3). Gel image: Immunoblot analysis to detect changes in p38 MAPK-α, p38 MAPK-β, p42/p44MAPK, SAPK (stress-activated protein kinase)/JNK, AKT, and β-actin in the siRNA-transfected cells used for qRT-PCR are shown. (E) Gel image: P19 cells were pretreated for 1 hour with 10 mM U0 (lanes 5–8) or 10 mMU0 plus 25 mM SB (lanes 9–12), and then treated for 4 hours with TSA (25 ng/ml), NaB (5 mM), or VPA (10 mM). Total cell lysates were prepared and analyzed by immunoblotting to determine changes in the levels of phospho- MSK1 and MSK1. The same blots were reprobed with phospho-ERK 1/2 to show specificity of U0 treatment, and b-actin levels were analyzed as internal control. Histogram: The pixel densities obtained for phospho-MSK1 were measured and plotted against the values obtained for b-actin, which was used as internal control. Error bars represents the range of standard errors from three independent experiments.
In further experiments, we examined whether MOR expression levels are affected by each p38 MAPK isoform. Thus, P19 cells were transfected with p38 MAPK-α- or p38-MAPK-β–specific siRNAs for 36 hours and stimulated with HDAC inhibitors (TSA, NaB, and VPA) for a further 8 hours. As shown in Fig. 4C, qRT-PCR analyses showed that only p38-MAPK-β siRNA transfection significantly reduced MOR expression levels upon stimulation by TSA (∼35% reduction compared with control siRNA) or VPA (∼50% reduction compared with control siRNA); but both siRNAs effectively reduced MOR expression levels induced by NaB (∼50% reduction by each siRNA compared with control). Since the role of p38 MAPK-β appeared to be conserved equally across three different types of HDAC inhibitors, we further tested the effect of these siRNAs across two more stimuli that are known to increase MOR expression via a p38 MAPK-dependent mechanism: protein synthesis inhibitor cycloheximide (CHX) and JNK inhibitor (SP) (Kim et al., 2011) (Wagley et al., 2013). As shown in Fig. 4D, although both p38-MAPK siRNAs significantly reduced MOR gene expression induced by either stimulus, the effects were more pronounced with p38-MAPK-β siRNA (∼60–70% reduction in MOR expression levels compared with ∼50% reduction by p38-MAPK-α siRNA). As a control, immunoblot analyses were performed (Fig. 4D, lower panel) and revealed that each siRNA specifically and significantly decreased the expression levels of their respective p38 MAPK targets, without affecting ERK 1/2, JNK, or AKT levels. Collectively these results suggest that p38 MAPK-β has a variable MOR regulatory function, dependent largely on the type of stimulus and whether p38 MAPK-α is also involved.
We investigated next whether ERK 1/2 mediates MSK1 phosphorylation and expression because sustained and time-dependent increases were observed for both the kinases after TSA or NaB stimulation (Fig. 2, C and D, Fig. 3A). P19 cells were pretreated with U0 for 1 hour to inhibit MEK/ERK activities, and further stimulated with TSA, NaB, or VPA for 4 hours. As shown by immunoblot analysis (Fig. 4E), although U0 significantly blocked the phosphorylation of MSK1 (Fig. 4E, cf. lanes 1–4 and 5–8) induced by HDAC inhibition, the expression levels of MSK1 proteins remained unchanged. As a control, ERK 1/2 phosphorylation levels were analyzed and found to be reduced by U0 pretreatment. In further experiments, we used SB and U0 together to completely block upstream MAPK signals, and then exposed the cells to HDAC inhibitors to analyze the expression and phosphorylation levels of MSK1 proteins. As expected, basal as well as the HDAC inhibitor–mediated MSK1 phosphorylation levels were completely abolished by SB and U0 pretreatment (Fig. 4E, lanes 9–12), which indicates that complete inhibition of the MSK1 phosphorylation requires a simultaneous inhibition of both upstream kinase activities (p38 MAPK and ERK 1/2) under either stimulated or nonstimulated conditions. Nonetheless, despite complete lack of p38 MAPK and ERK 1/2 activities, MSK1 expression levels were still increased by HDAC inhibition. Collectively, these results indicate that HDAC inhibitor-mediated increase in MSK1 expression levels is independent of MAPK activities; yet, MAPK activities do act upon the MSK1 proteins to increase their phosphorylation status.
MSK1 Expression Increases and HDAC Inhibitors Potentiate MOR Promoter Activities.
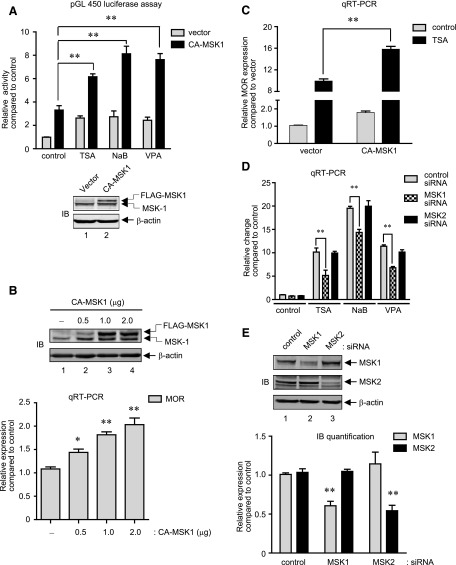
The core MOR promoter contains binding sites for two of the transcription factors known to be activated by MSK1—NF-κB p65 and CREB (Wei and Loh, 2011). We examined whether forced MSK1 expression potentiates MOR promoter activities. Thus, cell lysates were prepared from P19 cells cotransfected with MOR promoter reporter (pGL450) and CA-MSK1–expression constructs and further stimulated with HDAC inhibitors. As shown in Fig. 5A, upper histogram, CA-MSK1 expression increased the luciferase activity (∼3.3-fold compared with vector-transfected cells), and upon HDAC inhibition, it was further potentiated (∼2-fold for TSA, ∼3-fold each for NaB and VPA stimulations). As an internal control, CA-MSK1 expression was analyzed by immunoblotting, and showed correct expression (Fig. 5A, lower image). Since, the increase in MOR promoter activity by CA-MSK1 overexpression (∼3.3-fold) alone was comparable to that induced by HDAC inhibitors (TSA ∼2.6-fold, NaB ∼2.7-fold, VPA ∼2.4-fold); we determined whether MOR expression levels are also increased. Thus total RNA was prepared from P19 cells transfected with a varying amounts of CA-MSK1 for 48 hours, and MOR expression levels were determined. As shown in Fig. 5B, both the expression levels of CA-MSK1 (upper gel image) and MOR mRNA (lower histogram) increased in a dose-dependent fashion. In additional experiments, the effect of HDAC inhibition on MOR mRNA expression was analyzed by qRT-PCR in CA-MSK1 expressing cells. As shown in Fig. 5C, TSA stimulation of CA-MSK1 expressing cells showed a significant increase in MOR expression levels (∼10-fold in vector-transfected cells compared with ∼16-fold in CA-MSK1-transfected cells). Additional qRT-PCR analyses performed with NaB and VPA stimulations under conditions expressing CA-MSK1 also showed increased MOR expression levels (data not shown).
Fig. 5.
MSK1 regulates MOR gene expression induced by HDAC inhibitors. (A) Histogram: P19 cells were cotransfected with the MOR promoter construct (pGL 450, 300 ng) and expression plasmids for FLAG-tagged CA-MSK1 (1 μg) for 48 hours followed by stimulation with TSA, NaB, or VPA for a further 8 hours. Luciferase activities in the cell lysates were measured and expressed as relative activity compared with control. Gel image: A representative immunoblot analysis to demonstrate the expression of CA-MSK1 in the samples used for luciferase assay is presented (**P < 0.01, n = 4–6). (B) Gel image: Immunoblot analysis to detect MSK1 expression was performed with total cell lysates prepared from P19 cells transfected with increasing amounts of CA-MSK1 as indicated (0–2.0 μg) for 48 hours. The levels of β-actin were monitored as internal control. Histogram: qRT-PCR analyses were performed to determine changes in MOR mRNA levels from cells transfected as above (*P < 0.05, **P < 0.01, n = 5). (C) P19 cells were transfected with 1 μg of CA-MSK1 for 48 hours and further stimulated with TSA for 8 hours. Total RNA was extracted and qRT-PCR was performed to determine changes in MOR mRNA expression levels as described previously (**P < 0.01, n = 6). (D) P19 cells were transfected with control siRNA, MSK1 siRNA, or MSK2 siRNA for 36 hours. At the end of transfection, cells were treated with TSA (25 ng/ml), NaB (5 mM), or VPA (10 μM) for a further 8 hours and total RNA was analyzed by qRT-PCR to determine MOR mRNA levels. Results were normalized against the levels of β-actin as internal control and presented as relative change compared with control (**P < 0.01, n = 3–6). (E) Gel image: Immunoblot analysis to detect changes in MSK1, MSK2, and β-actin in the siRNA-transfected cells used for qRT-PCR in (D) is shown. Histogram: The pixel densities obtained for MSK1 and MSK2 were calculated, normalized against the values obtained for β-actin, and plotted as relative expression compared with control. Error bars represents the range of standard errors, and asterisks represent statistically significant findings (**P < 0.01, n = 3–4).
Since, MSK1 and MOR gene expression kinetics was similar upon HDAC inhibition, inhibition of PKA/MSK1 activity by a small molecule inhibitor, H89, abrogated the MOR gene expression, and MSK transfection increased and HDAC inhibitor-potentiated MOR promoter activities, we used siRNA strategy to directly confirm MSK1’s role in MOR gene expression. P19 cells were transfected with control siRNA or siRNA specific for MSK1 or MSK2 (a closely related protein to MSK1) for 36 hours, and stimulated for 8 hours with HDAC inhibitors (TSA, NaB, or VPA). As shown by qRT-PCR analysis in Fig. 5D, although MSK1 siRNA transfection did not completely abolish MOR expression induced by HDAC inhibitors, the levels of MOR expression were significantly decreased (∼50% for TSA, ∼25% for NaB, and ∼45% for VPA). On the other hand, MSK2 siRNA transfection showed minimal changes, and MOR expression levels remained comparable to those obtained with control siRNA transfection. To confirm the specificity of each siRNA, immunoblot analyses of the total cell lysates prepared at the end of transfection period showed that (Fig. 5E, upper image and quantification in lower histograms) each siRNA significantly reduced the expression levels of their respective target proteins, MSK1 and MSK2, but not vice versa. Collectively, these results demonstrate that HDAC inhibition-mediated MOR gene expression depends on the activity of MSK1.
CpG Methylation Abolishes MSK1-Mediated Increase in MOR Promoter Activity.
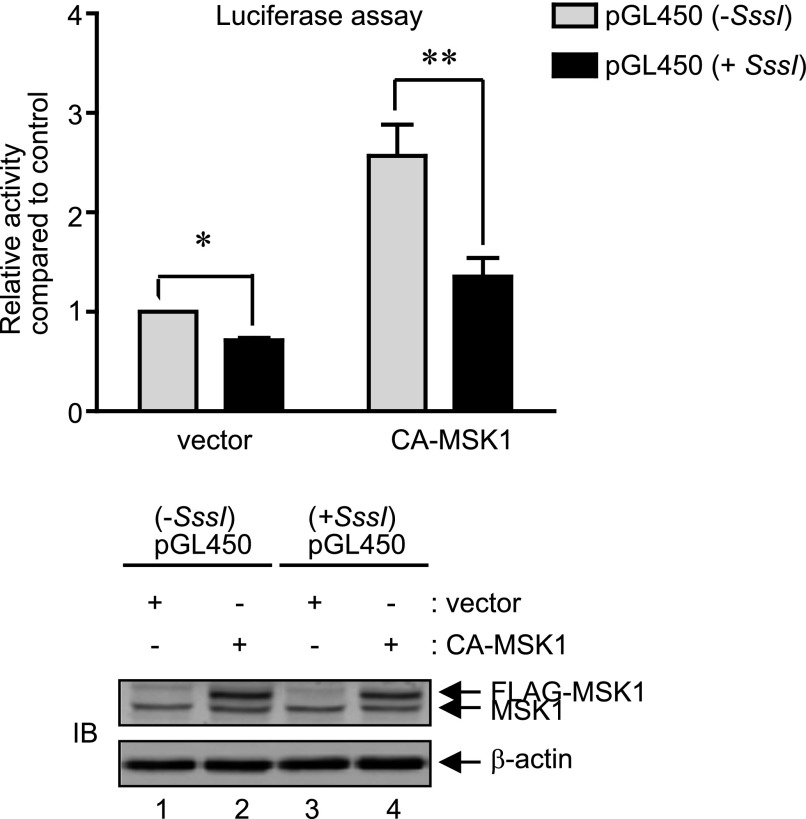
As reported earlier from our laboratory (Hwang et al., 2010), the CG residues of the endogenous MOR promoter in P19 cells are heavily methylated and are recognized by methyl-CpG-binding protein 2, which recruits repressive Brm proteins under unstimulated conditions. However, upon neuronal differentiation of P19 cells or stimulation with TSA, these repressive chromatin marks are removed, which allows the binding of activating transcription factors such as SP1. As demonstrated above, MSK1 strongly activated the MOR promoter (and activity was further potentiated by HDAC inhibition), thus we were interested to determine if in vitro methylation of the MOR promoter construct affects MSK1 activity. P19 cells were cotransfected with pGL450 promoter construct methylated in vitro, and the ability of CA-MSK1 to increase the MOR promoter activities were analyzed. As shown in Fig. 6 (upper histogram), CA-MSK1–mediated luciferase reporter activities from the methylated MOR promoter constructs were significantly reduced (∼47%) compared with the activities obtained from sham-methylated constructs. As an internal control, immunoblot analyses of the same cell lysates (Fig. 6, lower panel) showed that MSK1 expressions are comparable. Collectively, these results indicate that the repressive CpG methylation mark in the endogenous MOR promoter hinders efficient MSK1 action during the transcriptional activation of MOR gene.
Fig. 6.
CA-MSK1 fails to activate MOR promoter upon CpG methylation. Histogram: MOR promoter construct pGL450 was mock-methylated or SssI-methylated and cotransfected into P19 cells with 1 μg of FLAG-tagged CA-MSK1 for 48 hours. Luciferase activities were measured in the cell lysates and normalized against pCH110 β-galactosidase activity to calculate relative activity compared with control. The data represents the mean ± S.E.M of three independent experiments with at least two different plasmid preparations (*P < 0.05; **P < 0.01). Gel image: A representative immunoblot for CA-MSK1 expression in the cell lysates used for one of the luciferase assays is shown. The levels of β-actin were monitored as internal control.
MSK1 Expression Increases and Regulates MOR Gene Expression in Differentiating Neurons.
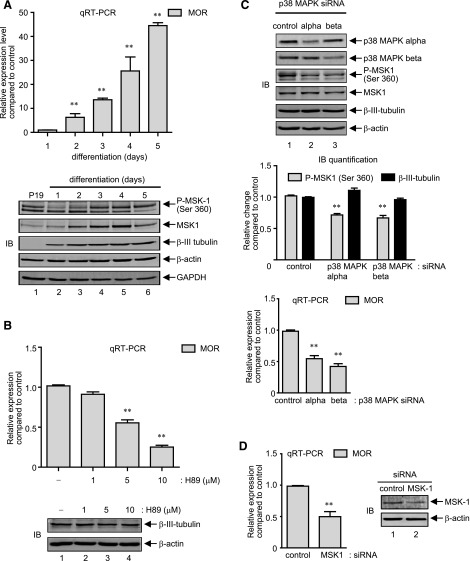
P19 cells can be induced to differentiate into neuronal cells by retinoic acid, a process that closely resembles mammalian neurogenesis in vivo (Monzo et al., 2012). As reported earlier, MOR gene expression starts to increase within 2 days after the induction of differentiation, and continues further as the cells fully differentiate into neuronal subtypes (Hwang et al., 2010). Interestingly, phosphorylation of all three major MAPKs (p38 MAPK, ERK 1/2, and JNK) also increases during neuronal differentiation of P19 cells, and follows MOR expression kinetics, among which pharmacological inhibition of p38 MAPK abrogates MOR gene expression (Wagley et al., 2013). Thus, to determine whether MSK1 is also activated alongside MOR expression, total cell lysates and total RNA samples were prepared in parallel from neuronally differentiating P19 cells each day for a total of 5 days, and analyzed by immunoblotting and qRT-PCR analyses. As shown in Fig. 7A, upper histogram, and as reported previously, qRT-PCR analysis showed that MOR expression levels start to increase 2 days after the induction of differentiation and dramatically increase up to the 5th day, when the cells completely differentiate into the neuronal types (the MOR levels on the 1st day of neuronal differentiation was indistinguishable with parent P19 cells and were not included in the analysis). Quite interestingly, as shown in Fig. 7A, lower panel gel images, biphasic MSK1 phosphorylation kinetics were observed: a first reduction phase which lasted until the cells started to differentiate into neuronal types (Fig. 7A, lanes 1, 2, and 3, and data not shown); however, the levels gradually increased and surpassed the levels seen with nondifferentiating cells within 3 days after the induction of neuronal differentiation (Fig. 7A, cf. lanes 1, 4, 5, 6). Corresponding analyses with total MSK1 also followed a biphasic course, but unlike the phosphorylated form, its levels followed a sustained increase (∼1.3- to ∼2.0-fold; Fig. 7A, lower) in their expression levels until the cells reached maximum neuronal differentiation at day 4, after which levels slightly declined (∼1.5-fold in day 5, Fig. 7A, cf. lanes 1–5 and 6). As an internal control for neuronal differentiation, immunoblot analyses of neuron-specific β-III tubulin levels were performed, which showed a continued increase and sustained its levels after 4 days. Further analyses to detect the expression changes in levels of β-actin and GAPDH (total p38 MAPK, ERK 1/2, and NF-κB p65 etc., data not shown) showed minimal changes, and thus, suggested that the changes in MSK1 levels are specific events observed during neuronal differentiation of P19 cells.
Fig. 7.
MSK1 level increases and regulates MOR expression in differentiating neurons. (A) Histogram: P19 cells were induced to differentiate in presence of 0.5 μM retinoic acid for 4 days in bacteriological Petri dishes. Cell aggregates were treated with trypsin-EDTA to obtain homogenous cell suspension and replated onto poly-l-ornithine–coated tissue culture plates. Total RNA was extracted each day for 5 days after plating, and qRT-PCR was performed to determine relative MOR mRNA expression levels compared with day 1 (**P < 0.05, n = 3). Gel image: Total cell lysates were collected each day for 5 days from P19 cells undergoing neuronal differentiation as above. Immunoblotting was performed with anti-phospho-MSK1 and anti-MSK1 as indicated. β-III tubulin (a neuron-specific protein) immunoblot was included to show correct neuronal differentiation of the P19 cells. The levels of β-actin and GAPDH were analyzed as internal control to show comparable protein loading across the samples. (B) Histogram: Neuronally differentiating P19 cells on day 1 were treated with a range of H89 (0–10 μM) for 2 days. RNA was extracted and MOR expression was determined by qRT-PCR as explained previously (**P < 0.01, n = 3). Gel image: Total cell lysates from neuronally differentiating P19 cells treated as above were analyzed by immunoblotting with neuron-specific anti-β-III tubulin antibodies. The levels of β-actin were monitored as internal control. (C) Gel image: Neuronally differentiating P19 cells on day 1 were transfected with control siRNA, p38-MAPK-α siRNA, or p38-MAPK-β siRNA for a further 2 days. At the end of transfection, total cell lysates were prepared and changes in the levels of p38 MAPK-α, p38 MAPK-β , phospho-MSK1, MSK1, β-III tubulin, and β-actin were determined by immunoblotting. Middle panel histogram: The pixel densities of phospho-MSK1 and β-III tubulin were measured from the siRNA-transfected cells and normalized against the values for β-actin levels to be plotted as relative change compared with control (**P < 0.01 versus control siRNA, n = 3). Lower histogram: The changes in the MOR mRNA expression levels were determined from the siRNA-transfected cells as described previously (**P < 0.01 versus control siRNA, n = 3). (D) Neuronally differentiating P19 cells (day 1) were transfected with MSK1 siRNA for 2 days. MOR mRNA expression (histogram, qRT-PCR) and MSK1 expression levels (gel image, immunoblotting) were determined as described previously. Results are representative of three separate experiments, and error bars represent the range of standard errors. Asterisks represent statistically significant findings compared with control siRNA (**P < 0.01).
We determined next whether increase in MSK1 expression (and phosphorylation) during neuronal differentiation of P19 cells is functionally relevant to increase MOR gene expression. Thus, qRT-PCR analysis was performed with total RNA extracted from neuronally differentiating P19 cells cultured in the presence of varying concentrations (0–10 μM) of PKA/MSK1 inhibitor, H89, for 3 days. Figure 7B, upper histogram, shows that H89 blocked the MOR expression in a dose-dependent fashion (∼40% to ∼70% reduction by 5–10 μM H89), which indicates that MSK1 activity is required for MOR expression in differentiating neurons. In a following set of experiments, we analyzed whether H89 decreased MOR expression levels owing to a general inhibition of neuronal differentiation. Thus, the levels of the neuron-specific β-III tubulin protein levels were immuno-detected in total cell lysates prepared from neuronally differentiating P19 cells cultured in presence of H89 as above. As shown in the lower gel image (Fig. 7B), β-III tubulin levels remained comparable amid H89, and suggested that PKA/MSK1 inhibition in general minimally affects the neuronal differentiation process.
As mentioned previously, depending on the stimulus, the phosphorylation and activation of MSK1 is regulated by either one or both of the upstream MAPKs—p38 MAPK or ERK 1/2. Also, in our previous report, we demonstrated that the pharmacological inhibition of p38 MAPK or NF-κB, but not ERK 1/2, abolishes MOR gene expression during neuronal differentiation of P19 cells (Wagley et al., 2013). Thus, to determine whether or which p38 MAPK isoform regulates MSK1 phosphorylation during neuronal differentiation of P19 cells, we transfected differentiating cells with p38 MAPK-α or p38 MAPK-β siRNA, and cell lysates were analyzed. As shown in Fig. 7C, top panel gel image, transfection with each p38-MAPK siRNA showed a specific and significant reduction of its respective target, and an ∼30% reduction in the levels of phosphorylated MSK1 (Ser 360) by each siRNA (Fig. 7C, gel image and quantification in middle panel histograms). However, the overall changes in neuronal differentiation were insignificant because the expression levels of the neuron-specific β-III tubulin protein changed minimally by p38 MAPK knockdown. In further experiments, MOR expression levels were determined by qRT-PCR analysis of cells likewise transfected, and demonstrated that each of these siRNAs, by themselves, significantly reduced MOR expression levels (∼40% and ∼60% reduction by p38 MAPK-α and p38-MAPK-β siRNA, respectively, compared with control siRNA), which suggest a coordinate p38MAPK/MSK1 pathway may be functioning to regulate MOR expression during neuronal differentiation of P19 cells. Additional qRT-PCR analysis using simultaneous transfections of both siRNAs did not produce additive inhibition of MSK1 phosphorylation levels or MOR expression levels, and was identical to the data obtained with p38-MAPK-β siRNA (data not shown).
To finally prove MSK1’s role in MOR gene regulation during neuronal differentiation of P19 cells, a specific siRNA targeting MSK1 was introduced into the differentiating cells, and 3 days later MOR expression level was determined by qRT-PCR analysis. As expected, MOR expression level was significantly reduced in MSK1 siRNA–transfected cells (∼50% reduction compared with control siRNA) (Fig. 7D, left panel), which was analogous to the inhibition levels obtained with p38-MAPK siRNA (Fig. 7C, qRT-PCR histograms). Corresponding immunoblot analysis (Fig. 7D, right panel) showed the specificity of the siRNA and showed that MSK1 expression was reduced in the transfected cells. Collectively, these results identify a regulatory role of MAPK/MSK1 cascade on MOR gene expression during neuronal differentiation of P19 cells.
Discussion
The transcriptional activation of μ-opioid receptor (MOR) gene is a complex process and requires extensive chromatin remodeling that allows the binding of various transcription factors to the MOR promoter [for review see Wei and Loh (2011)]. Here, we provide evidence that HDAC inhibition activates components of the MAPK cascade, thereby augmenting MOR transcription through regulatory signals that converge at the MOR promoter. Stimulation of P19 cells with classic HDAC inhibitors TSA or NaB increased MOR expression after p38 MAPK and ERK 1/2 were phosphorylated; and inhibiting the activities of either of these MAPKs decreased MOR expression levels (Figs. 1 and 2). The expression levels of their common downstream effector kinase, MSK1, also increased upon HDAC inhibition, and regulated MOR transcript levels (Fig. 3). In further analyses, although inhibition of p38 MAPK activities failed to modulate MSK1 levels (Fig. 4, A and B), MOR expression decreased in cells transfected with p38-MAPK-β siRNA or MSK1 siRNA (Figs. 4C and 5D). On the other hand, ERK 1/2 activities were found to regulate the phosphorylation but not the expression of MSK1 proteins (Fig. 4E), suggesting different modes of MSK1 expression and functional regulation. Likewise, the expression and function of MSK1 proteins increased to regulate MOR expression during neuronal differentiation of P19 cells (Fig. 7, A and B), suggesting a central functional role of this kinase in epigenetic regulation of MOR gene (Fig. 8).
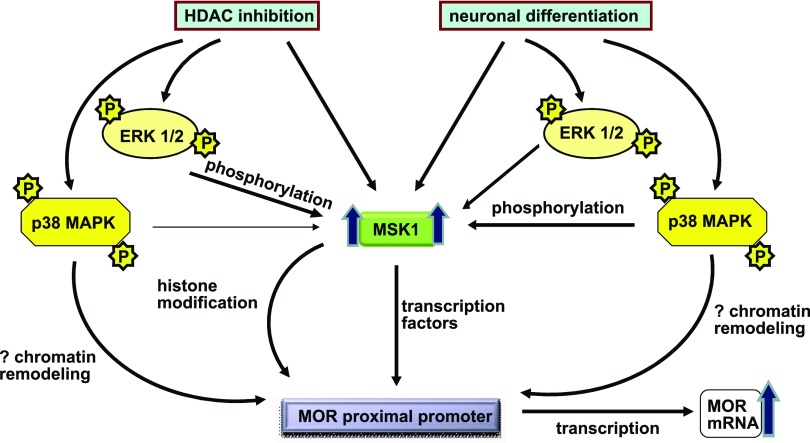
Fig. 8.
Schematic representation of events during MOR gene transcription after stimulation of P19 cells with HDAC inhibitors or retinoic acid-mediated neuronal differentiation. HDAC inhibitor stimulation or neuronal differentiation of P19 cells leads to a general increase in the expression of MSK1 proteins, which are concurrently activated by upstream ERK 1/2 or p38 MAPK. Upon activation, MSK1 increases the activity of transcription factors that bind to the remodeled MOR proximal promoter to increase transcription of MOR gene.
Although most of the transcriptional effects of such HDAC inhibitors as TSA or NaB have been attributed to direct chromatin remodeling by histone acetylation (Zhong et al., 2003), HDAC inhibitors only modulate a small proportion (2–10%) of expressed genes in transformed cells, suggesting that the net effect on gene transcription is a combinatorial outcome of histone modifications and functions of regulatory proteins, such as components of signal transduction cascade and basal transcriptional machinery, which specifically recognize the modified histone via protein domains specialized for such purpose (Dokmanovic et al., 2007). Indeed, the data presented in this study also supports the active role of such specialized proteins, specifically the components of the MAPK cascades, because p38 MAPK and ERK 1/2 were activated concurrently with a dramatic increase in MOR gene expression induced by TSA or NaB (Figs. 1 and 2). In agreement to this observation, a number of studies have shown that HDAC inhibitors can directly activate ERK 1/2 and p38 MAPK pathways in erythroid cells and lymphoid cells (Witt et al., 2002; Xie et al., 2010) and lead to increased transcription of genes such as p21, γ-globin, and myostatin (Rivero and Adunyah, 1996; Zhong et al., 2003; Sangerman et al., 2006; Han et al., 2010; Simboeck et al., 2010). Arguably, in some instances HDAC inhibitors have been shown to inhibit MAPK activity, possibly by increased activities of MAPK phosphatase owing to acetylation (Cao et al., 2008). Therefore, it appears that the net outcome of HDAC inhibition of MAPK depends on whether the MAPK kinase or MAPK phosphatase activities are affected. Nonetheless, p38 MAPK and ERK 1/2 activities were important for TSA- or NaB-mediated MOR transcription, because inhibition of either MAPK blunted MOR levels (Fig. 2, A and B). Further, HDAC inhibitor-induced MOR expression was potentiated in the presence of JNK inhibitor (SP) (Fig. 2, A and B). Since MOR gene expression upon JNK inhibition, protein synthesis inhibition (CHX, puromycin), or neuronal differentiation of P19 cells (see below) has already been shown to depend on p38 MAPK activities (Kim et al., 2011; Wagley et al., 2013); and that p38-MAPK-β siRNA transfection reduced MOR expression induced with all these stimuli (Fig. 4, C and D, and Fig. 7C), p38 MAPK-β appears to be essential for MOR gene regulation.
The regulation of MOR gene expression upon HDAC inhibition or neuronal differentiation of P19 was found to be a conserved mechanism, because MOR gene expression occurred concurrently with increases in MSK1 expression and activation (Figs. 3 and 7A). Although both p38 MAPK and ERK1/2 are known to effectively activate MSK1, different stimuli have been shown to require at least one or a combination of both MAPK modules (p38 and ERK1/2) to efficiently activate MSK1 (McCoy et al., 2005). As mentioned above, and as shown in this study, HDAC inhibitors used ERK 1/2 as the major upstream kinase (Fig. 4E), whereas p38 MAPK was equally functional during neuronal differentiation of P19 cells (Fig. 7C). Nevertheless, the definitive role of MSK1 in MOR gene expression is supported, because pharmacological inhibition of PKA/MSK1 with H89 (Fig. 5D) or MSK1 siRNA transfection abrogated the increases in MOR expression levels (Fig. 7D). Yet, MSK1 activity appeared to play a regulatory role rather than an absolute turn on/off switch because only a modest increase in the MOR mRNA levels was observed after the expression of constitutively active-MSK1 (CA-MSK1) construct (Fig. 5, B and C), and that CA-MSK1 failed to efficiently activate the in vitro methylated MOR promoter constructs (Fig. 6). These results indicate that MSK1 requires epigenetic modification at the MOR promoter and probably increases MOR transcription via activation of the transcription factors such as NF-κB or CREB, each of which contains binding sites in the functional MOR promoter (Wei and Loh, 2011). In support to this hypothesis, we have observed that the levels of MSK1 and its downstream transcription factor NF-κB p65 both increase in the nucleus of TSA-stimulated P19 cells or P19 cells undergoing neuronal differentiation, and pharmacological inhibition of NF-κB activity using QNZ reduces the MOR promoter activity in HDAC inhibitor-stimulated cells (unpublished observation). The role of ribosomal s6 kinase (RSK) 1/2 (another group of MAPK effector kinases ) is dubious in all these processes, since p38 MAPK pathway (involved in both models of MOR gene transcription presented here) does not activate RSK 1/2 (Healy et al., 2012), whereas the effect of ERK 1/2 was partial and only affected MOR expression levels induced by HDAC inhibitors. Regardless, it is clear that other intracellular pathways may also play a functional role in MOR expression mechanism, because the increase in total MSK1 levels was sustained even after simultaneous inhibition of p38 MAPK and ERK 1/2 pathways (Fig. 4E).
An earlier study from our laboratory has shown that TSA treatment shifts the border of the mononucleosome surrounding the MOR minimal promoter from –345 to –262, which suggests chromatin modification in this region (Hwang et al., 2010). With regard to this observation, the role of MAPK cascade in MOR gene expression involving the MOR minimal promoter region involves two steps, both chromatin modification and transcription factor activation. Quite obviously, although the data presented and discussed in this study supports MAPK/MSK1-dependent transcription factor activation, further studies on chromatin remodeling are also warranted because increasing evidence suggests MAPK activation regulates epigenetic changes in multiple ways (see below) and this was not fully addressed in this study. For example, during smooth muscle cell differentiation, p38 MAPK is directly recruited to the myogenic loci by SWItch/sucrose nonfermentable (SWI-SNF) chromatin remodeling complexes; and forced activation of p38 MAPK in myoblast by expression of a constitutively active dual specificity mitogen-activated protein kinase kinase 6 (MKK6) promotes unscheduled SWI-SNF recruitment to the myogenin promoter (Simone et al., 2004). In recent years, MSK-mediated histone phosphorylation is regarded as the critical event that directs extracellular cues to nucleosomal response, and transcriptional activation (Healy et al., 2012). According to this model, MSK1-mediated histone H3 phosphorylation allows recruitment of the 14-3-3 proteins and the Brm-related gene 1 (Brg1), component of the ATP-dependent chromatin remodeling SWI-SNF complex, to the promoter of the target genes by transcription factors such as Elk-1 or NF-κB (Drobic et al., 2010; Gehani et al., 2010). Indeed, since Brg1 has been shown to be recruited to the MOR promoter (Hwang et al., 2010) and that NF-κB activity directly affects MOR expression during the neuronal differentiation of P19 cells (Wagley et al., 2013), it would be interesting to determine how MAPK/MSK1 activity links the chromatin remodeling events with transcriptional initiation at the MOR promoter.
In recent years, accumulating evidence has pointed out that opioid agonists can stabilize the same receptor in multiple active confirmations to propagate differential intracellular signals (ERK 1/2, JNK, protein kinase A or C pathways), which determines the behavioral plasticity associated with these drugs (Hofford et al., 2009; Melief et al., 2010; Al-Hasani and Bruchas, 2011 ). Quite interestingly, HDAC inhibitors also modulate behavioral outcomes of opioid drugs such as acute morphine-induced hyperactivity, opioid dependence, the development of behavioral sensitization, and the precipitation of morphine withdrawal syndrome in vivo (Sanchis-Segura et al., 2009; Jing et al., 2011; Rehni et al., 2012). In addition, emerging studies suggest that HDAC inhibitors facilitate the extinction of rewarding memory of drug taking (Wang et al., 2012), and drug-induced conditioned place preference (Wang et al., 2010). In these contexts, it is valuable to understand that the same range of intracellular signaling cascades elicited by opioid drugs to modulate behavioral outcomes can also be modulated by HDAC inhibitors (which are increasingly used in clinical trials for the treatment of various disorders) to regulate MOR expression levels (Fig. 8), which suggests that the intracellular signaling cascades might play an autoregulatory role in pharmacological outcomes of opioid drugs.
In conclusion, we demonstrate that HDAC inhibitor stimulation or retinoic acid-mediated neuronal differentiation of P19 cells activate coordinated intracellular signals that converge at the MOR promoter via increased expression and MAPK-dependent function of MSK1 proteins to regulate MOR transcription (Fig. 8).
Acknowledgments
The authors thank Drs. Simon Arthur and James Hastie (University of Dundee, Scotland) for the MSK1 constructs used in this study and Dr. Kurt D. Hankenson for valuable discussion and feedback during final drafting of the manuscript.
Abbreviations
- AKT
protein kinase B
- BCA
bicinchoninic acid
- CHX
cycloheximide
- CREB
cAMP response-element binding protein
- ERK
extracellular signal-regulated kinase
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- H-89
N-[2-[[3-(4-bromophenyl)-2-propenyl]amino]ethyl]-5-isoquinolinesulfonamide
- HDAC
histone deacetylase
- JNK
c-Jun NH2-terminal kinase
- MAPK
mitogen-activated protein kinase
- MOR
μ-opioid receptor
- MSK
mitogen- and stress-activated protein kinase
- NaB
sodium butyrate
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- PBS
phosphate-buffered saline
- PKA
protein kinase A
- QNZ
6-amino-4-(4-phenoxyphenylethylamino)quinazoline
- qRT-PCR
quantitative reverse transcription–polymerase chain reaction
- RSK
ribosomal s6 kinase
- RT-PCR
reverse transcription–polymerase chain reaction
- SB
SB203580, [4-(4-flurophenyl)-2-(4-methylsulfinylphenyl)-5-(4-pyridyl)imidazole]
- si
small-interfering
- SP
SP600125 anthra(1,9-cd)pyrazol-6(2H)-one
- SWI-SNF
SWItch/sucrose nonfermentable
- TSA
trichostatin A
- U0126
1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio]butadiene
- VPA
valproic acid
Authorship Contributions
Participated in research design: Wagley, Law, Wei, Loh.
Conducted experiments: Wagley.Performed data analysis: Wagley, Law, Wei, Loh.
Wrote or contributed to the writing of the manuscript: Wagley, Law, Wei, Loh.
Footnotes
This work was supported by National Institutes of Health National Institute on Drug Abuse [Grant DA001583]. The authors declare no conflict of interest.
References
- Al-Hasani R, Bruchas MR. (2011) Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology 115:1363–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JS. (2008) MSK activation and physiological roles. Front Biosci 13:5866–5879. [DOI] [PubMed] [Google Scholar]
- Cao W, Bao C, Padalko E, Lowenstein CJ. (2008) Acetylation of mitogen-activated protein kinase phosphatase-1 inhibits Toll-like receptor signaling. J Exp Med 205:1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh AC, Tse JW, Togel L, Mariadason JM. (2015) Mechanisms of histone deacetylase inhibitor-regulated gene expression in cancer cells. Antioxid Redox Signal 23:66–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard LR, White DE, Jones DL, McDermott MF, Burchill SA. (2009) p38(MAPK): stress responses from molecular mechanisms to therapeutics. Trends Mol Med 15:369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S. (2007) p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta 1773:1358–1375. [DOI] [PubMed] [Google Scholar]
- Delcuve GP, Khan DH, Davie JR. (2012) Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokmanovic M, Clarke C, Marks PA. (2007) Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res 5:981–989. [DOI] [PubMed] [Google Scholar]
- Drobic B, Pérez-Cadahía B, Yu J, Kung SK, Davie JR. (2010) Promoter chromatin remodeling of immediate-early genes is mediated through H3 phosphorylation at either serine 28 or 10 by the MSK1 multi-protein complex. Nucleic Acids Res 38:3196–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. (2010) Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell 39:886–900. [DOI] [PubMed] [Google Scholar]
- Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. (2003) Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther 2:151–163. [PubMed] [Google Scholar]
- Han DS, Huang HP, Wang TG, Hung MY, Ke JY, Chang KT, Chang HY, Ho YP, Hsieh WY, Yang WS. (2010) Transcription activation of myostatin by trichostatin A in differentiated C2C12 myocytes via ASK1-MKK3/4/6-JNK and p38 mitogen-activated protein kinase pathways. J Cell Biochem 111:564–573. [DOI] [PubMed] [Google Scholar]
- Healy S, Khan P, He S, Davie JR. (2012) Histone H3 phosphorylation, immediate-early gene expression, and the nucleosomal response: a historical perspective. Biochem Cell Biol 90:39–54. [DOI] [PubMed] [Google Scholar]
- Hofford RS, Hodgson SR, Roberts KW, Bryant CD, Evans CJ, Eitan S. (2009) Extracellular signal-regulated kinase activation in the amygdala mediates elevated plus maze behavior during opioid withdrawal. Behav Pharmacol 20:576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, Kim CS, Kim DK, Law PY, Wei LN, Loh HH. (2010) Up-regulation of the mu-opioid receptor gene is mediated through chromatin remodeling and transcriptional factors in differentiated neuronal cells. Mol Pharmacol 78:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, Wei LN, Loh HH. (2007) Evidence of endogenous mu opioid receptor regulation by epigenetic control of the promoters. Mol Cell Biol 27:4720–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, Wei LN, Loh HH. (2009) Epigenetic programming of mu-opioid receptor gene in mouse brain is regulated by MeCP2 and Brg1 chromatin remodelling factor. J Cell Mol Med 13 (9B):3591–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, Wu X, Wang G, Kim CS, Loh HH. (2003) Mouse mu opioid receptor distal promoter transcriptional regulation by SOX proteins. J Biol Chem 278:3742–3750. [DOI] [PubMed] [Google Scholar]
- Jing L, Luo J, Zhang M, Qin WJ, Li YL, Liu Q, Wang YT, Lawrence AJ, Liang JH. (2011) Effect of the histone deacetylase inhibitors on behavioural sensitization to a single morphine exposure in mice. Neurosci Lett 494:169–173. [DOI] [PubMed] [Google Scholar]
- Johnstone RW. (2002) Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov 1:287–299. [DOI] [PubMed] [Google Scholar]
- Keum YS, Kim HG, Bode AM, Surh YJ, Dong Z. (2013) UVB-induced COX-2 expression requires histone H3 phosphorylation at Ser10 and Ser28. Oncogene 32:444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK , Hwang CK, Wagley Y, Law P-Y, Wei L-N and Loh HH (2011) p38 mitogen-activated protein kinase and PI3-kinase are involved in up-regulation of mu opioid receptor transcription induced by cycloheximide. J Neurochem 116:1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus J. (2012) Expression and functions of μ-opioid receptors and cannabinoid receptors type 1 in T lymphocytes. Ann N Y Acad Sci 1261:1–6. [DOI] [PubMed] [Google Scholar]
- Lin YC, Flock KE, Cook RJ, Hunkele AJ, Loh HH, Ko JL. (2008) Effects of trichostatin A on neuronal mu-opioid receptor gene expression. Brain Res 1246:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh HH, Liu HC, Cavalli A, Yang W, Chen YF, Wei LN. (1998) mu Opioid receptor knockout in mice: effects on ligand-induced analgesia and morphine lethality. Brain Res Mol Brain Res 54:321–326. [DOI] [PubMed] [Google Scholar]
- McCoy CE, Campbell DG, Deak M, Bloomberg GB, Arthur JS. (2005) MSK1 activity is controlled by multiple phosphorylation sites. Biochem J 387:507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melief EJ, Miyatake M, Bruchas MR, Chavkin C. (2010) Ligand-directed c-Jun N-terminal kinase activation disrupts opioid receptor signaling. Proc Natl Acad Sci USA 107:11608–11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzo HJ, Park TI, Montgomery JM, Faull RL, Dragunow M, Curtis MA. (2012) A method for generating high-yield enriched neuronal cultures from P19 embryonal carcinoma cells. J Neurosci Methods 204:87–103. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehni AK, Singh N, Rachamalla M, Tikoo K. (2012) Modulation of histone deacetylase attenuates naloxone-precipitated opioid withdrawal syndrome. Naunyn Schmiedebergs Arch Pharmacol 385:605–619. [DOI] [PubMed] [Google Scholar]
- Rivero JA, Adunyah SE. (1996) Sodium butyrate induces tyrosine phosphorylation and activation of MAP kinase (ERK-1) in human K562 cells. Biochem Biophys Res Commun 224:796–801. [DOI] [PubMed] [Google Scholar]
- Sanchis-Segura C, Lopez-Atalaya JP, Barco A. (2009) Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology 34:2642–2654. [DOI] [PubMed] [Google Scholar]
- Sangerman J, Lee MS, Yao X, Oteng E, Hsiao CH, Li W, Zein S, Ofori-Acquah SF, Pace BS. (2006) Mechanism for fetal hemoglobin induction by histone deacetylase inhibitors involves gamma-globin activation by CREB1 and ATF-2. Blood 108:3590–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simboeck E, Sawicka A, Zupkovitz G, Senese S, Winter S, Dequiedt F, Ogris E, Di Croce L, Chiocca S, Seiser C. (2010) A phosphorylation switch regulates the transcriptional activation of cell cycle regulator p21 by histone deacetylase inhibitors. J Biol Chem 285:41062–41073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. (2004) p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 36:738–743. [DOI] [PubMed] [Google Scholar]
- Sora I, Takahashi N, Funada M, Ujike H, Revay RS, Donovan DM, Miner LL, Uhl GR. (1997) Opiate receptor knockout mice define mu receptor roles in endogenous nociceptive responses and morphine-induced analgesia. Proc Natl Acad Sci USA 94:1544–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagley Y, Hwang CK, Lin HY, Kam AF, Law PY, Loh HH, Wei LN. (2013) Inhibition of c-Jun NH2-terminal kinase stimulates mu opioid receptor expression via p38 MAPK-mediated nuclear NF-κB activation in neuronal and non-neuronal cells. Biochim Biophys Acta 1833:1476–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhang Y, Qing H, Liu M, Yang P. (2010) The extinction of morphine-induced conditioned place preference by histone deacetylase inhibition. Neurosci Lett 483:137–142. [DOI] [PubMed] [Google Scholar]
- Wang WS, Kang S, Liu WT, Li M, Liu Y, Yu C, Chen J, Chi ZQ, He L, Liu JG. (2012) Extinction of aversive memories associated with morphine withdrawal requires ERK-mediated epigenetic regulation of brain-derived neurotrophic factor transcription in the rat ventromedial prefrontal cortex. J Neurosci 32:13763–13775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LN, Loh HH. (2011) Transcriptional and epigenetic regulation of opioid receptor genes: present and future. Annu Rev Pharmacol Toxicol 51:75–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt O, Mönkemeyer S, Kanbach K, Pekrun A. (2002) Induction of fetal hemoglobin synthesis by valproate: modulation of MAP kinase pathways. Am J Hematol 71:45–46. [DOI] [PubMed] [Google Scholar]
- Xie N, Wang C, Lin Y, Li H, Chen L, Zhang T, Sun Y, Zhang Y, Yin D, Chi Z. (2010) The role of p38 MAPK in valproic acid induced microglia apoptosis. Neurosci Lett 482:51–56. [DOI] [PubMed] [Google Scholar]
- Zhong S, Goto H, Inagaki M, Dong Z. (2003) Phosphorylation at serine 28 and acetylation at lysine 9 of histone H3 induced by trichostatin A. Oncogene 22:5291–5297. [DOI] [PubMed] [Google Scholar]
- Zhong S, Zhang Y, Jansen C, Goto H, Inagaki M, Dong Z. (2001) MAP kinases mediate UVB-induced phosphorylation of histone H3 at serine 28. J Biol Chem 276:12932–12937. [DOI] [PubMed] [Google Scholar]
- Zhou C, Qiu L, Sun Y, Healey S, Wanebo H, Kouttab N, Di W, Yan B, Wan Y. (2006) Inhibition of EGFR/PI3K/AKT cell survival pathway promotes TSA’s effect on cell death and migration in human ovarian cancer cells. Int J Oncol 29:269–278. [PubMed] [Google Scholar]