Abstract
Well-differentiated human cancers share transcriptional programs with the normal tissue counterparts from which they arise. These programs broadly influence cell behavior and function and are integral modulators of malignancy. Here, we show that the master regulator of motile ciliogenesis, FOXJ1, is highly expressed in cells along the ventricular surface of the human brain. Strong expression is present in cells of the ependyma and the choroid plexus as well as in a subset of cells residing in the subventricular zone. Expression of FOXJ1 and its transcriptional program is maintained in many well-differentiated human tumours that arise along the ventricle, including low-grade ependymal tumours and choroid plexus papilloma. Anaplastic ependymoma as well as choroid plexus carcinoma show decreased FOXJ1 expression and its associated ciliogenesis program genes. In ependymoma and choroid plexus tumours, reduced expression of FOXJ1 and its ciliogenesis program are markers of poor outcome and are therefore useful biomarkers for assessing these tumours. Transitions in ciliogenesis define distinct differentiation states in ependymal and choroid plexus tumours with important implications for patient care.
Keywords: FOXJ1, ependymoma, choroid plexus, dedifferentiation, stem cells, subventricular zone, cilia, ciliogenesis
Introduction
The differentiation state of tumour cells is a critical determinant of their biological behavior [1,2]. From the inception of oncogenesis, a cell’s lineage identity influences the effects of potentially oncogenic lesions [3]. Once tumours begin to develop, cell lineage information and epigenomic patterns are often faithfully maintained in tumour cells [4].
Lineage identity is principally conferred by transcriptional regulators [5–7]. In some cases, lineage transcription factors such as Caudal Type Homeobox 2 (CDX2) [8], MITF Microphthalmia-Associated Transcription Factor (MITF) [7] and SRY Sex Determining Region Y-Box 2 (SOX2) [9] can be amplified and can promote survival programs and oncogenesis. In other cases, transcription factors can be suppressed and can generate cellular states that are intrinsically resistant to targeted therapeutic agents [10,11]. Moreover, as tumours recur and progress to more malignant states, they can undergo marked dedifferentiation, losing features of their cell lineage identity and entering more primitive differentiation states [12,13]. Understanding the components that define a tumour’s differentiation state is of utmost importance for understanding the pathogenesis of each tumour type and for developing rational therapeutic strategies.
The ventricular spaces of the brain are lined with ciliated epithelial cells called ependymocytes. These ciliated ependymocytes foster the flow of cerebrospinal fluid, which provides cues for migratory subventricular zone (SVZ) cells such as neuroblasts [14,15]. Closely related to these ependymocytes, are highly specialized ependymal cells of the circumventricular organs and choroid plexus [16].
Ependymomas are brain tumours that arise throughout the central nervous system, within the supratentorial areas, the posterior fossa and the spinal cord. These two tumours form in close association with the cavities of the ventricular system and the central canal of the spinal cord which are all lined by ependymal cells [17]. Ependymomas are thought to arise from radial glial cells located just beneath the ventricular surface [18], the same progenitor cells that develop into the mature ependymocytes that line the ventricles [19]. Histologic low-grade variants of ependymoma such as subependymoma and myxopapillary ependymoma are generally indolent and are classified as World Health Organization (WHO) grade I neoplasms. Other ependymomas are designated as WHO grade II or WHO grade III tumours, the latter displaying anaplastic features such as hypercellularity, elevated mitotic activity, endothelial proliferation and necrosis [20]. Recent studies have revealed that oncogenic fusion genes involving V-Rel Avian Reticuloendotheliosis Viral Oncogene Homolog A (RELA) and Yes-Associated Protein 1 (YAP1) can drive the formation of a subset of ependymomas and have led to a proposed molecular classification system [21–24].
Choroid plexus tumours arise from the choroid plexus and are usually located within ventricles. These tumours are classified as either well-differentiated low-grade papilloma (grade I), atypical papilloma (grade II) or high-grade carcinoma (grade III). Papilloma and carcinoma have different copy number profiles, gene expression patterns and DNA methylation signatures [25]. Like anaplastic ependymomas, choroid plexus carcinomas have a strong tendency for recurrence and for metastatic dissemination [17].
Forkhead Box J1 (FOXJ1) is the master regulator of the transcriptional program that drives assembly of motile cilia [26–28]. In model systems, ectopic expression of FOXJ1 alone is able to promote cilia production [26]. In rodents, FOXJ1 expression in postnatal radial glial progenitor cells is necessary for differentiation into ciliated mature ependymocytes [29]. In addition, FOXJ1 expression is needed for the development of a subset of astrocytes that reside adjacent to the lateral ventricles [29]. Notably, some FOXJ1 positive cells have a role in self-renewal and neurogenesis both in the developing [30,31] as well as the adult brain [32] of mice supporting that FOXJ1 plays an important role in cell fate determination and in establishing progenitor states. In the mouse brain, FOXJ1 expression is maintained in ependymal cells as well as in cells of the choroid plexus [33] which are also ciliated [34]. Ultrastructural studies have shown that ependymomas produce structurally abnormal cilia that are improperly localized in extracellular microrosettes and in intracytoplasmic luminal areas [35]. The cilia of choroid plexus tumours are not well studied, but altered microtubule configuration has been noted in these tumours [36]. Evidence for molecular crosstalk between ciliary factors and regulators of the cell cycle raises the possibility dysregulation of ciliogenesis may contribute to tumourigenesis [37–40].
Here we show that the FOXJ1 transcriptional program is active in ependymoma and in choroid plexus tumours, particularly in well-differentiated low-grade tumours. In contrast, the ciliogenesis program is suppressed in both high-grade ependymoma and high-grade choroid plexus tumours. Reduced FOXJ1 expression is associated with tumour recurrence. Our work defines important differentiation states and supports the critical role of differentiation states in tumour pathogenesis.
Materials and Methods
Tissue Samples
We retrieved formalin-fixed, paraffin-embedded (FFPE) tumour (n = 127) and normal (n = 4) tissue samples from the Brigham and Women’s Hospital (BWH) and Boston Children’s Hospital Archives (Table S7). The study was reviewed and approved by the BWH and Dana-Farber Cancer Institute human subjects review boards. The histology of all samples was reviewed by two neuropathologists (SS, MAA.) using WHO classification/grading criteria.
Immunohistochemistry and scoring
We performed immunohistochemical staining for FOXJ1 on FFPE tissue sections using rabbit polyclonal antibody HPA005714 (Sigma-Aldrich, 1:75 dilution). Antigen retrieval was with a pressure cooker. Staining was attempted with anti-HFH4 antibody[3–19] (Abcam ab40869) but despite recognizing FOXJ1 in ciliated fallopian tube cells, staining was not obtained in ependymoma tumour cells. Negative control staining was performed using normal/nonimmune rabbit serum, immunoglobulin fraction (DakoCytomation X0936). Nuclear staining was scored as positive using a semi-quantitative approach, with intensity scored from 0 to 3 (0 = no staining, 1 = weak, 2 = moderate, or 3 = strong) and frequency scored from 0 to 4 (0 = no cell with nuclear positivity, 1 = 1–25%, 2 = 25–50%, 3 = 50–75%, 4 = 75–100%) [41]. The IHC score was generated by multiplying the intensity and frequency scores. FOXJ1-high was > 8. FOXJ1-low was < 8.
Expression profiling
We normalized expression profiling data as previously described [42] from publicly available datasets cataloged in the Gene Expression Omnibus that had been acquired on Affymetrix HG-U133_Plus_2 arrays. Heat maps were generated using Genepattern tools (http://www.broadinstitute.org/cancer/software/genepattern/). Publicly available expression profiling data from ependymoma and choroid plexus tumours was obtained [22–25,43] and plotted data is presented in supplementary tables. Gene set enrichment analysis was performed using GOrilla [44] (http://cbl-gorilla.cs.technion.ac.il/) and visualized using REVIGO [45] (http://revigo.irb.hr/).
We monitored FOXJ1 activity using a FOXJ1 ciliogenesis program signature. The human genes comprising this signature are orthologs of over 80 genes identified as FOXJ1 targets in xenopus [46] and zebrafish [26,46] by overexpression and deletion/silencing approaches. We filtered this list against a list of genes differentially expressed in mouse tracheal epithelial cells as they were differentiated in vitro into ciliated cells [47]. These tracheal cells expressed green fluorescent protein (GFP) under the control of a FOXJ1 cell-specific promoter. GFP-expressing ciliated cells were separated from GFP-negative non-ciliated cells and mRNA expression was performed on these populations. This comparative approach allowed us to identify a stringent set of endogenous genes associated with FOXJ1-mediated ciliogenesis in mammals. Genes present in all lists (derived from xenopus, from zebrafish, and from mouse early and late differentiation time points) were used as the human FOXJ1 ciliogenesis program (Table S4). The log2 mean-normalized data for each gene/probe (as available) across samples was averaged for the genes comprising the FOXJ1 ciliogenesis program signature in each public dataset and this value was plotted.
In situ hybridization
In situ detection of FOXJ1 transcripts in FFPE samples was performed using RNAscope® assay Probe-Hs-FOXJ1 (Cat# 430921, Advanced Cell Diagnostics) and RNAscope® 2.0HD Reagent-Kit (BROWN) (Cat# 310035, Advanced Cell Diagnostics) following manufacturer suggested protocols. Images from E13.5 and P14 mice are from the Allen Brain Atlas (http://developingmouse.brain-map.org/).
Outcome Analysis and Statistical Analysis
We used Graph Pad Prism to perform analysis of variance (ANOVA) when comparing differences between mRNA and protein expression between various tumour types. P-values listed. Pearson’s r was calculated to investigate correlation.
Cox proportional hazards modeling was used to assess the association between variables and the time to recurrence and/or outcome. Variables included FOXJ1 IHC score, FoxGT8 (IHC score >8), FoxLT8 (IHC score < 8), WHO grade, gender, radiation-treatment pre or post-operation, epithelial membrane antigen staining, primary or recurrence status, Ki67/MIB percent-positivity and patient age as well as mRNA levels of FOXJ1, RFX2 or ciliogenesis program (log 2 fold change > or < 0). Hazard ratios (HR) and 95% confidence intervals (CI) are reported. Statistical analyses were performed using SASv9.3 (SAS Institute, Cary, NC).
Results
FOXJ1 ciliogenesis program is activated in ependymoma
To identify the transcriptional programs that are enriched in ependymal tumours, we analyzed public mRNA expression profiling data from 356 samples spanning ten different types of brain tumours and normal brain (Table S1). We ranked the genes that are differentially expressed in ependymoma compared to all other samples and performed Gene Set Enrichment Analysis (GSEA) on that list (Table S2, S3). This analysis showed that ependymoma are very highly enriched for components of cilia (FDR q-value = 1.17e-24) and their assembly (FDR q-value = 5.67e-28) (Table S2, S3 and Figure S1). Consistent with this, transcripts for many axonemal components, as well as for the transcription factors that regulate their expression, were highly expressed in ependymoma (Figure 1A, S2).
Figure 1.
FOXJ1 and critical components of the ciliogenesis program are highly expressed in ependymoma. A. Heat map of normalized publicly-available expression profiling data in the Gene Expression Omnibus (GEO) of various brain tumours (n = 346) and normal brain tissues (n =10 samples). Columns represent the average log2 fold change from mean normalized data for each sample type (Table S1). High expression orange, low expression blue. Transcription factors that regulate ciliogenesis as well as genes that are major components of cilia are shown. Pilocytic astrocytoma (n = 41), ependymoma (n = 19), meningioma (n = 68), Central Nervous System (CNS) PNET (n = 22), high-grade astrocytoma (including glioblastoma; n = 84), medulloblastoma (n = 29), atypical teratoid/rhabdoid tumour (n = 21), primary CNS lymphoma (n = 34), pineoblastoma (n = 1), pediatric glioblastoma (n = 27), normal brain (n = 10). B. and C. Scatter plot of log2 fold change of the mean normalized data for the mRNA of the cilia transcriptional regulators FOXJ1 and RFX2. Each point represents the data from a single gene expression profile from the indicated tissue type (from Table S1).
In particular, FOXJ1, the master transcriptional regulator of genes involved in the production of motile cilia, was highly and selectively expressed in ependymoma (Figure 1A, 1B, S2). RFX2 (Regulatory Factor X, 2), another transcriptional regulator of cilia production was also highly and selectively expressed in ependymoma (Figure 1A,C, S2). Other transcriptional regulators of ciliogenesis such as RFX1, RFX3 and RFX4 to 7 lacked such selective expression in ependymoma (Table S1). Although expression of FOXJ1 mRNA was somewhat elevated in several cases of atypical teratoid/rhabdoid tumour (AT/RT) and primitive neuroectodermal tumours (PNET) (Figure 1B, C), FOXJ1 mRNA expression was significantly higher in ependymoma (4 to 6 fold differences, p < 0.0001).
FOXJ1 is expressed in the ependymal lining, in the choroid plexus and in cells in the subventricular zone of the human brain
Because of the selective expression of FOXJ1 and RFX2 mRNA in ependymoma, we investigated the expression of these two transcripts in normal brain tissues from developing mouse. In publicly available images of in situ hybridization experiments from the Allen Developing Mouse Brain Atlas, we found strong expression of FOXJ1 mRNA (Figure 2A, 2B, S3, S4) and RFX2 mRNA (Figure S5, S6) in both the ependymal lining and in the choroid plexus of mouse embryos and post-natal mice.
Figure 2.
FOXJ1 mRNA and protein are expressed in the ependymal lining and choroid plexus of the mouse and human brain. A. and B. FOXJ1 in situ hybridization in a P14 mouse brain (images from the Allen Developing Mouse Brain Atlas). Immunohistochemistry for FOXJ1 protein expression in ciliated cells lining C. the plicae of the human fallopian tube and D. the bronchial airways of the human lung. E. Immunohistochemistry for FOXJ1 protein expression in the ependymal line (strong) and the choroid plexus (weaker) of the mouse brain. F. H&E stained section and G. immunohistochemistry for FOXJ1 protein expression in the brain of a 76 year old woman. SVZ denotes the subventricular zone region. There are many scattered FOXJ1 immunoreactive cells in the subventricular zone of the adult human brain. H. RNAscope in situ hybridization for FOXJ1 mRNA in a serial section of the same brain. Scale bar sizes are indicated in each of the images.
We next assessed the expression of FOXJ1 protein levels using immunohistochemistry. To validate the antibody, we used sections of human fallopian tube and lung tissue. We found strong FOXJ1 protein expression in ciliated epithelial cells lining the oviduct and the bronchial airways (Figure 2C, 2D, S7A). In mouse brain, we detected strong FOXJ1 expression in ependymal cells and moderate expression in the choroid plexus (Figure 2E, S7B, S7C). In human brain, FOXJ1 protein was strongly expressed in ependymal cells (Figure 2F, 2G) and in the epithelial lining of the choroid plexus (Figure S7D). Occasional FOXJ1 expressing cells were present in the subventricular zone of the mouse brain (Figure S7E). In the human subventricular zone, however, we found numerous scattered FOXJ1 positive cells (Figure S7F–S7G). Using RNAscope mRNA in situ hybridization, we confirmed that cells in the human ependymal lining and in the subventricular zone express FOXJ1 (Figure 2F, 2H, S7H).
Expression of FOXJ1 and the ciliogenesis program is reduced in aggressive ependymoma
Having optimized conditions for detecting FOXJ1, we next characterized FOXJ1 expression in human ependymoma tumours. We found strong FOXJ1 expression across various subtypes of ependymoma including WHO grade II ependymoma (Figure 3A, 3B, S8A), myxopapillary ependymoma from the lumbar spinal cord (Figure 3C, 3D, S8B-C), cellular ependymoma (Figure 3E) and subependymoma (Figure 3F). In addition, a well-differentiated ependymoma that had metastasized to the lung also showed high FOXJ1 expression (Figure S9).
Figure 3.
FOXJ1 protein is expressed in various ependymoma subtypes. A. H&E stained section of a WHO grade II ependymoma and B. the corresponding FOXJ1 immunohistochemistry. C. H&E stained sections of a WHO grade I myxopapillary ependymoma and D. the corresponding FOXJ1 immunohistochemistry. E. FOXJ1 immunohistochemistry in a cellular ependymoma and F. FOXJ1 immunohistochemistry in a subependymoma. Scale bar sizes are indicated in each of the images.
We next examined public mRNA expression profiling data from a cohort containing 75 WHO grade II and III ependymoma [24]. We defined the genes that comprise the FOXJ1 ciliogenesis program using published experimental data from Xenopus, zebrafish and mouse cells (see Materials and Methods, Table S4). We confirmed a strong correlation of FOXJ1 expression and the expression of the FOXJ1 ciliogenesis program in normal mouse ependymocytes using large scale single-cell RNA-sequencing data from normal mouse cortex and hippocampus (Pearson r = 0.71, p < 0.0001) (Table S5) [48]. In ependymoma, we found that the mRNA expression of FOXJ1 and the genes that comprise the FOXJ1 ciliogenesis program that it regulates (Table S4, S6) were very strongly correlated (plot of the data shows a strong correlation with a Pearson r = 0.89, p < 0.0001; Figure 4A, 4B). The expression of FOXJ1 and the ciliogenesis program varied widely with some tumours expressing high levels of mRNA for FOXJ1, RFX2 and the ciliogenesis program while others had low levels of mRNA for these genes (Figure 4A, 4B). Tumours with low expression were not restricted to one compartment of the central nervous system but rather were found in the supratentorial region and in the posterior fossa (Figure 4A).
Figure 4.
Expression of FOXJ1 and the ciliogenesis program varies in ependymoma. A. Heat map of FOXJ1, RFX2 and the FOXJ1 ciliogenesis program (see Materials and Methods section for details of the program; Table S4) in ependymoma. The source data is from Witt et al. [24]. Locations of the tumours (supratentorial, posterior fossa and spinal) are listed (Table S6). B. Plot of FOXJ1 mRNA expression and the mean of the expression of FOXJ1 ciliogenesis program genes (average of mean fold change for the genes comprising this program). The source data is from Witt et al. [24]. Analysis of the correlation shows a Pearson r = 0.89. C. Plot of the FOXJ1 ciliogenesis program from data in Pajtler et al, [22] for posterior fossa ependymoma (PF-EPN-A or PF-EPN-B), spinal cord ependymoma (SP-EPN) or supratentorial ependymoma (ST) with RELA (ST-EPN-RELA) or YAP1 (ST-EPND-YAP) fusions (Table S7). D. H&E stained section and FOXJ1 immunohistochemistry from serial sections of a recurrent anaplastic ependymoma with dedifferentiated regions from the right parietal-occipital region of a 21 year old male. E. Plot of FOXJ1 immunohistochemistry score (intensity x frequency on a scale of 0–12) on a set of 97 ependymal tumours including 42 WHO grade II ependymoma, 30 WHO grade III anaplastic ependymoma, 20 WHO grade I myxopapillary ependymoma and five WHO grade I subependymoma. F. Kaplan Meier survival analysis. Plot of the cumulative proportion without recurrence among ependymoma WHO grades II and III according to the FOXJ1 level. The high group had an IHC score > 8 (red line) and the low group < 8 (blue line) (from Table S8). Only patients with available recurrence date and last follow up are included in this analysis.
A recent report has proposed a comprehensive molecular classification system for ependymal tumours based on DNA methylation patterns [22]. Nine-subgroups are proposed. Posterior fossa ependymomas are classified as either PF-EPN-A tumours which have a poor prognosis or PF-EPN-B tumours which have a good prognosis. Supratentorial tumours are classified as either ST-EPN-RELA which have a poor prognosis or ST-EPN-YAP1 which have a good prognosis. In this data set, the mRNA levels of FOXJ1 were highly correlated with those of the FOXJ1 ciliogenesis program (Pearson r = 0.71, p < 0.0001, Table S7, Figure S10).
We first examined the expression of the FOXJ1 ciliogenesis program in posterior fossa tumours annotated by their molecular subgroup (Table S7; Figure S11) from the expression profiling data in Pajtler et al. [22]. While PF-EPN-B tumours (n = 39) and spinal cord ependymomas (n = 11) had high levels of FOXJ1 mRNA and the FOXJ1 ciliogenesis program (Figure 4C), the PF-EPN-A tumours (n = 72) which have a poor prognosis had significantly reduced expression of the FOXJ1 ciliogenesis program (p < 0.05). Moreover, while ST-EPN-YAP1 tumours (n = 11) had relatively high levels of the ciliogenesis program, the ST-EPN-RELA tumours (n = 48), which have a poor prognosis, had significantly reduced expression of the ciliogenesis program (p < 0.05) (Figure 4C, Table S7). Thus, expression of the FOXJ1 ciliogenesis program is decreased in the PF-EPN-A and ST-EPN-RELA ependymoma classes, the ones that exhibit the most aggressive behavior.
Recently, a pathway that regulates the development of multiciliated cells has been described in Xenopus epidermis and in mouse airway epithelium [49,50]. Critical factors in this multiciliogenesis cycle are multicilin (MCIDAS, MCIN, IDAS) which functions upstream of v-Myb Avian Myeloblastosis Viral Oncogene Homolog (MYB) which in turn functions upstream of FOXJ1 [49]. We investigated the mRNA expression of multicilin and MYB in ependymoma. Because probes for multicilin were not included in the microarray chips used in the ependymoma profiling studies, we turned to RNA-sequencing data from St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project [23]. In this data from 77 ependymomas, multicilin mRNA levels were negligible. Nearly half had fragments per kilobase of transcript per million mapped reads (FPKM) values of zero and the median value was 0.02 FPKM. For comparison the same samples had a median FPKM value of 196 for FOXJ1 mRNA. Interestingly, expression of MYB mRNA was detected in these ependymoma and we identified a strong correlation between MYB mRNA and the FOXJ1 ciliogenesis program in the datasets from Witt et al. [24] and Pajtler et al. [22] (Pearson r = 0.56 and Pearson r = 0.64, p < 0.0001, Figure S12A and B, Tables S6 and S7) suggesting that MYB may be involved in positively regulating FOXJ1 expression in ependymoma. Analysis of large scale quantitative single-cell RNA-sequencing data from mouse cortex and hippocampus also reveals that FOXJ1 ependymocytes have low levels of multicilin but high levels of MYB, suggesting that the MYB may be an important regulator of FOXJ1 expression in both ependymocytes and ependymoma alike (Table S5, Figure S13).
Expression of FOXJ1 protein is reduced in aggressive ependymoma and is associated with poor outcome
To examine the relationship between the ciliogenesis program and the malignant potential of ependymoma, we characterized FOXJ1 protein expression by immunohistochemistry in a cohort of cases of WHO grade I, grade II and grade III ependymoma collected from our hospitals (Table S8, Figure 4D–F). Interestingly, while most grade I and grade II ependymoma had diffuse uniform expression of FOXJ1, many anaplastic ependymoma demonstrated heterogeneous expression of FOXJ1 with some cells expressing the protein while others lacked protein expression (Figure 4D). Consistent with the mRNA results, we found that FOXJ1 protein expression was also reduced in more aggressive tumours with FOXJ1 protein levels significantly lower in WHO grade III anaplastic ependymoma compared to WHO grade II ependymoma (p < 0.05, Figure 4E). Similarly, we found a negative correlation between the level of FOXJ1 expression and the Mib-1 proliferative index of ependymoma (Pearson r = −0.38; p = 0.001) with FOXJ1-low ependymoma (n = 21) having a significantly higher mean Mib-1 proliferative index compared to ependymoma that retained high FOXJ1 expression (n = 46) (19.77 ± 3.75 vs 7.95 ± 1.45).
We collected clinical outcome data on tumour recurrence from this cohort of 94 patients with ependymoma from our hospitals, which included patients with WHO grade I, II and III tumours to determine if there was an association between FOXJ1 protein level and clinical outcome. We found that ependymomas with high FOXJ1 expression were less likely to recur (HR = 0.82, 95% CI = 0.70 to 0.96; p = 0.012; Table S8, S9). We observed the same result even when we restricted the analysis to WHO grade II and grade III ependymoma only (n= 70; HR = 0.81, 95% CI= 0.67 to 0.97; p = 0.024; Table S8, S9). Furthermore, we next performed Kaplan-Meier analysis on cases that had a defined last follow up date or a defined date of recurrence. When we divide these WHO grade II and grade III ependymoma into high FOXJ1 (n = 37) and low FOXJ1 (n = 14) expressing groups, we found that the low expressing group was significantly associated with recurrence (HR = 5.80, 95% CI = 1.06 to 31.67; p = 0.043) (Figure 4F).
Expression of FOXJ1 and the ciliogenesis program is reduced in high-grade choroid plexus tumours in mice and humans
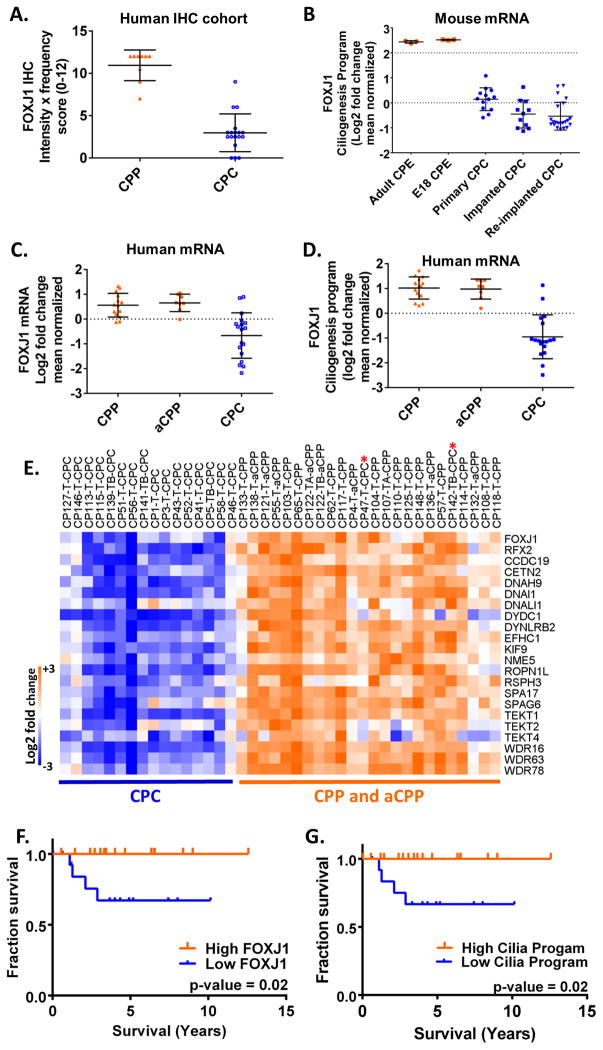
Because FOXJ1 is strongly expressed in the epithelial cells of the human and mouse choroid plexus, we next asked if choroid plexus papilloma also expressed FOXJ1. We found that the neoplastic epithelial cells of human choroid plexus papilloma uniformly expressed FOXJ1 protein by immunohistochemistry (Figure 5A–D). Analogous to the reduced expression of FOXJ1 in anaplastic ependymoma, we found that human choroid plexus carcinoma also had significantly lower expression of FOXJ1 protein than choroid plexus papilloma (Table S8, Figure 5E, 5F, 6A; p < 0.05). In these malignant tumours, FOXJ1 protein expression was heterogeneous and only found in a subpopulation of scattered carcinoma cells (Table S8, Figure 5F). While, outcome analysis was limited because of the relatively small size of the cohort (n = 23) and the lack of recurrences in the choroid plexus papilloma group, we did note that the three choroid plexus carcinomas that recurred each had low FOXJ1 expression (scores of 3, 3.5 and 5 out of 12).
Figure 5.
There is widespread expression of FOXJ1 protein in choroid plexus papilloma but only scattered cells express FOXJ1 in choroid plexus carcinoma. A. and C. H&E stained sections of choroid plexus papilloma and B. and D. FOXJ1 immunohistochemistry in choroid plexus papilloma. E. H&E stained section of choroid plexus carcinoma and F. FOXJ1 immunohistochemistry in a choroid plexus carcinoma. Scale bar sizes are indicated in the images.
Figure 6.
Expression of FOXJ1 protein and mRNA is reduced in human and mouse choroid plexus carcinoma. A. Plot of FOXJ1 immunohistochemistry score (intensity × frequency on a scale of 0–12) on a set of nine choroid plexus papillomas (CPP) formalin fixed paraffin embedded (FFPE) samples and 18 choroid plexus carcinomas (CPC) FFPE samples. B. Expression of FOXJ1 in mouse choroid plexus epithelium and choroid plexus carcinomas from Tong et al., [69]. Data was extracted from GEO data set GSE61659. Comparison of FOXJ1 mRNA expression levels from adult mouse choroid plexus epithelium (adult CPE), embryonic choroid plexus epithelium (E18 CPE), primary mouse choroid plexus carcinoma (CPC) removed following Cre-mediated recombinase driven inactivation of p53, Rb and Pten (primary CPC), or implanted and re-implanted mouse choroid plexus carcinoma (CPC) [69]. C. Plot of log2 fold change of FOXJ1 mRNA from public expression profiling data of human CPP, aCPP and CPC in Merino et al. [25] (Table S11). aCPP, atypical choroid plexus papilloma. D. Plot of log2 fold change of mean of the genes comprising the FOXJ1 ciliogenesis program (Table S11). E. Heat map of FOXJ1, RFX2 and the FOXJ1 ciliogenesis program from Merino et al. [25] (Table S11). CPC cases that cluster with the CPP and aCPP are marked with a red asterisk. F. and G. Kaplan-Meier Survival Analysis of cohort of patients with choroid plexus papilloma and choroid plexus carcinomas published in Merino et al., Clinical Cancer Research 2014 [25] and Tong et al., Cancer Cell 2015 [43] (Table S12). Kaplan-Meier survival curves were generated using GraphPad Prism version and p-values were generated by the Log-Rank (Mantel-Cox) test in GraphPad. FOXJ and ciliogenesis program expression profiles were binned into high and low expression by separating values above and below a log fold-change of 0.
We next turned to publicly available expression profiling data from murine and human choroid plexus tumours to characterize the mRNA levels of FOXJ1 and its ciliogenesis program. Recently, expression profiling data was published from choroid plexus carcinomas generated in a genetically-engineered murine model of choroid plexus carcinoma [43]. These tumours arise following Cre-mediated inactivation of p53, RB and PTEN. Strikingly, in this murine model, mRNA levels of FOXJ1 and its ciliogenesis program were also sharply decreased in all of the tumours that were generated (Figure 6B, S14; Table S10).
Using publicly available expression profiling data from humans with choroid plexus tumours [25], we also noted a strong correlation between the levels of FOXJ1 mRNA and the expression of the FOXJ1 ciliogenesis program (Figure S15A; Pearson r = 0.89, p < 0.0001; Table S11). Moreover, similar to our immunohistochemistry results, mRNA levels of FOXJ1, RFX2 and the ciliogenesis program were all abundant in choroid plexus papilloma but were significantly reduced in choroid plexus carcinoma (Figure 6C – E, S15B; p < 0.05; Table S11). Kaplan-Meier analysis of this public cohort [25,43] showed a very strong association between poor patient outcome and low levels of FOXJ1, RFX2 or the ciliogenesis program (Figure 6F, 6G, S16; Table S12).
Discussion
Our work shows that low-grade ependymal and choroid plexus tumours share a FOXJ1 ciliogenesis program with their normal tissue counterparts. This transcriptional program is decreased in high-grade ependymoma and choroid plexus tumours and in ependymomas that have poor outcome such as PF-EPN-A and ST-EPN-RELA ependymomas. Thus, shifts towards dedifferentiated states are associated with increased malignant potential in these tumours. From a practical perspective, our results show that FOXJ1 immunostaining is reliable as a diagnostic and prognostic marker in both ependymoma and choroid plexus tumours, thus providing an inexpensive, simple, and widely available tool for sample characterization. Few other markers of these tumours are definitive and reliable in clinical practice suggesting that FOXJ1 may be a clinically useful biomarker.
Although we show that transcriptional drivers of ciliogenesis such as FOXJ1 and RFX2 are highly expressed along with many critical components of the axoneme, ultrastructural studies of ependymoma demonstrate that many of the cilia structures that are produced have an aberrant arrangement and are improperly localized [37,51]. A recent analysis of serial sections of individual ependymoma cells by electron microscopy has shown that these cells have large basal bodies and up to three cilia per cell and that the cilia are mis-localized within regions that look like intracytoplasmic lumen [37]. Thus, despite the presence of high-level expression of the FOXJ1 master regulator of ciliogenesis, fateful recapitulation of normal ciliary structures and their appropriate localization is hindered. The reasons why these aberrations occur are unclear. It is also unclear whether the problem is linked to processes that dysregulate cell polarity in tumours. For example, gene set enrichment analysis (Table S3) identified a dysregulation of Wingless-Type (WNT) and Hedgehog signaling pathways in ependymoma, both pathways that are essential for establishing appropriate polarity [52]. However, the link between ependymoma tumour grade and the presence or absence of cilia is clearer. Consistent with our observation of reduced or absent expression of FOXJ1 in higher-grade and aggressive ependymoma, cells in such high grade tumours lack motile cilia when examined in serial sections by electron microscopy [37].
The presence of a FOXJ1 ciliogenesis program in many ependymomas may simply reflect a vestige from a prior cell lineage developmental program, one present in the cells of origin of ependymal and choroid plexus tumours. The emerging understanding of the complex link and crosstalk between ciliary factors and the regulators of entry into the cell cycle [38–40], however, raises the intriguing possibility that the ciliogenesis program and the abnormal structures that form may contribute to dysregulation of the cell cycle and may therefore promote cell growth and even oncogenesis [38–40].
In lower grade tumours such as grade II ependymoma, abnormal cilia might provide signals analogous to those that result during the disassembly and resorption of cilia. For example, Jouberin (Jbn) is localized to cilia in resting ciliated cells but could shift to act as a co-factor for beta-catenin during cilia disassembly, activating expression of cyclin D and facilitating G1/S transition [39]. Intraflagellar transport proteins such as IFT88, IFT27, and others, which are all present in cilia, have also been shown to influence cell cycle progression [39]. Interestingly a screen for chemical modulators using a genetically engineered murine model of ependymoma showed that these mouse tumour cells were particularly sensitive to compounds that interfere with centrosome duplication [53]. This finding suggests that certain ependymoma cells may be highly dependent on centrosome homeostasis and the ciliogenesis program very much in the same ways that tumours are addicted to oncogenic drivers [40,54,55]. It also suggests that the cell lineage identity of ependymal cells and choroid plexus cells which is enforced by FOXJ1 in tumours, might be an essential core component that fosters and supports aspects of tumourigenesis. Such a dependency creates vulnerabilities in the centrosome cycle that could be explored as a treatment strategy and further supports the dysregulation of centrosome biology as an important driver of malignancy in ependymoma.
In higher grade tumours such as anaplastic ependymoma, posterior fossa group A ependymoma, RELA fusion supratentorial ependymoma and choroid plexus carcinoma, the ciliogenesis program is further disrupted – with the FOXJ1 program significantly reduced or even extinguished. These aggressive tumours have likely accumulated additional aberrations that can facilitate highly malignant behavior and in that context lower FOXJ1 may be beneficial for tumour growth. For example, in certain contexts such as in ovarian tumours, a decrease in FOXJ1 can enhance cell migration and invasion [56] and is associated with poor outcome [57]. In other contexts, such as in hepatocellular carcinoma, however, overexpression of FOXJ1 is associated with worse outcome in gastric cancer [58].
Independent of the direct effects of decreased FOXJ1 in certain contexts, the molecular events that could lead to reduced FOXJ1 may be important for fostering malignancy. Suppression of lineage transcription factors by mutations directly in the transcription factor itself or by alterations in genes that regulate epigenetic processes can lead to lineage reversion and to dedifferentiation [13,59–67]. While mutations in FOXJ1 have been described infrequently in a few cancer types [68], various sequencing studies have failed to identify mutations in FOXJ1 in ependymoma, suggesting that the decrease in FOXJ1 that occurs in the more aggressive tumours likely results from alterations in epigenetic regulators and possibly from reduced levels of MYB. An important limitation of the present study is that mechanistic insights are not provided for how FOXJ1 is suppressed in high-grade ependymoma and choroid plexus tumours. Insights into the mechanisms that decrease the FOXJ1 program in these tumours may inform new therapeutic strategies.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Santagata lab and Adrian Dubuc for helpful discussions. The authors would like to thank Michelle Hadley and Lacie Cardillo for assistance with sample acquisition, Josh Francis for advice on computational analyses, Christopher Crum and Ronny Drapkin for advice on marker development, and Marian Slaney and Sebastian Valentin for technical assistance with tissue processing. We thank Vera Paulson for critically reading the manuscript. This study makes use of data generated by the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project. This study was supported by the Jared Branfman Sunflowers for Life Fund for Pediatric Brain and Spinal Cancer Research (S.S), the V Foundation (S.S), King Abdulaziz City for Science and Technology (KACST), Saudi Arabia (M.A.), and Sanad Children’s Cancer Support Association, Saudi Arabia (M.A).
Footnotes
Conflict of interest: No disclosures.
Statement of author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data, assisted in drafting the article or revising it critically for important intellectual content and made final approval of this version.
Conception and design (MAA, MPW, KLL, SS), Acquisition of data (MAA, MPW, PHM, ZD, TW, SH, SS), Analysis and interpretation of data (MAA, MPW, PHM, SH, KLL, SS), Contributed unpublished essential data (SHS).
Reference list
- 1.Santagata S, Thakkar A, Ergonul A, et al. Taxonomy of breast cancer based on normal cell phenotype predicts outcome. The Journal of clinical investigation. 2014;124:859–870. doi: 10.1172/JCI70941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ince TA, Richardson AL, Bell GW, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Blanpain C. Tracing the cellular origin of cancer. Nature cell biology. 2013;15:126–134. doi: 10.1038/ncb2657. [DOI] [PubMed] [Google Scholar]
- 4.Polak P, Karlic R, Koren A, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santagata S, Maire CL, Idbaih A, et al. CRX is a diagnostic marker of retinal and pineal lineage tumors. PloS one. 2009;4:e7932. doi: 10.1371/journal.pone.0007932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ligon KL, Alberta JA, Kho AT, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. Journal of neuropathology and experimental neurology. 2004;63:499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 7.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 8.Salari K, Spulak ME, Cuff J, et al. CDX2 is an amplified lineage-survival oncogene in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3196–3205. doi: 10.1073/pnas.1206004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature genetics. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller J, Krijgsman O, Tsoi J, et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nature communications. 2014;5:5712. doi: 10.1038/ncomms6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konieczkowski DJ, Johannessen CM, Abudayyeh O, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer discovery. 2014;4:816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abedalthagafi MS, Bi WL, Merrill PH, et al. ARID1A and TERT promoter mutations in dedifferentiated meningioma. Cancer genetics. 2015 doi: 10.1016/j.cancergen.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedmann-Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO reports. 2014;15:244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lun MP, Monuki ES, Lehtinen MK. Development and functions of the choroid plexus-cerebrospinal fluid system. Nature reviews Neuroscience. 2015;16:445–457. doi: 10.1038/nrn3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawamoto K, Wichterle H, Gonzalez-Perez O, et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science. 2006;311:629–632. doi: 10.1126/science.1119133. [DOI] [PubMed] [Google Scholar]
- 16.Lehman NL. Central nervous system tumors with ependymal features: a broadened spectrum of primarily ependymal differentiation? Journal of neuropathology and experimental neurology. 2008;67:177–188. doi: 10.1097/NEN.0b013e31816543a6. [DOI] [PubMed] [Google Scholar]
- 17.Louis DN, Ohgaki H, Wiestler OD, et al. WHO Classification of Tumours of the Central Nervous System. 4. International Agency for Research on Cancer; 2007. pp. 238–240. [Google Scholar]
- 18.Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Spassky N, Merkle FT, Flames N, et al. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghunathan A, Wani K, Armstrong TS, et al. Histological predictors of outcome in ependymoma are dependent on anatomic site within the central nervous system. Brain pathology. 2013;23:584–594. doi: 10.1111/bpa.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mack SC, Witt H, Piro RM, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pajtler KW, Witt H, Sill M, et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer cell. 2015;27:728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witt H, Mack SC, Ryzhova M, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer cell. 2011;20:143–157. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merino DM, Shlien A, Villani A, et al. Molecular characterization of choroid plexus tumors reveals novel clinically relevant subgroups. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:184–192. doi: 10.1158/1078-0432.CCR-14-1324. [DOI] [PubMed] [Google Scholar]
- 26.Yu X, Ng CP, Habacher H, et al. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nature genetics. 2008;40:1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]
- 27.Gomperts BN, Gong-Cooper X, Hackett BP. Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. Journal of cell science. 2004;117:1329–1337. doi: 10.1242/jcs.00978. [DOI] [PubMed] [Google Scholar]
- 28.Brody SL, Yan XH, Wuerffel MK, et al. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. American journal of respiratory cell and molecular biology. 2000;23:45–51. doi: 10.1165/ajrcmb.23.1.4070. [DOI] [PubMed] [Google Scholar]
- 29.Jacquet BV, Salinas-Mondragon R, Liang H, et al. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development. 2009;136:4021–4031. doi: 10.1242/dev.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devaraju K, Barnabe-Heider F, Kokaia Z, et al. FoxJ1-expressing cells contribute to neurogenesis in forebrain of adult rats: evidence from in vivo electroporation combined with piggyBac transposon. Experimental cell research. 2013;319:2790–2800. doi: 10.1016/j.yexcr.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 31.Jacquet BV, Muthusamy N, Sommerville LJ, et al. Specification of a Foxj1-dependent lineage in the forebrain is required for embryonic-to-postnatal transition of neurogenesis in the olfactory bulb. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:9368–9382. doi: 10.1523/JNEUROSCI.0171-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paez-Gonzalez P, Abdi K, Luciano D, et al. Ank3-dependent SVZ niche assembly is required for the continued production of new neurons. Neuron. 2011;71:61–75. doi: 10.1016/j.neuron.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim L, Zhou H, Costa RH. The winged helix transcription factor HFH-4 is expressed during choroid plexus epithelial development in the mouse embryo. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3094–3099. doi: 10.1073/pnas.94.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nonami Y, Narita K, Nakamura H, et al. Developmental changes in ciliary motility on choroid plexus epithelial cells during the perinatal period. Cytoskeleton. 2013;70:797–803. doi: 10.1002/cm.21132. [DOI] [PubMed] [Google Scholar]
- 35.Ho KL. Abnormal cilia in a fourth ventricular ependymoma. Acta neuropathologica. 1986;70:30–37. doi: 10.1007/BF00689511. [DOI] [PubMed] [Google Scholar]
- 36.McComb RD, Burger PC. Choroid plexus carcinoma. Report of a case with immunohistochemical and ultrastructural observations. Cancer. 1983;51:470–475. doi: 10.1002/1097-0142(19830201)51:3<470::aid-cncr2820510319>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 37.Alfaro-Cervello C, Soriano-Navarro M, Ramirez M, et al. Ultrastructural pathology of anaplastic and grade II ependymomas reveals distinctive ciliary structures--electron microscopy redux. Ultrastructural pathology. 2015;39:23–29. doi: 10.3109/01913123.2014.906526. [DOI] [PubMed] [Google Scholar]
- 38.Jackson PK. Do cilia put brakes on the cell cycle? Nature cell biology. 2011;13:340–342. doi: 10.1038/ncb0411-340. [DOI] [PubMed] [Google Scholar]
- 39.Kim S, Tsiokas L. Cilia and cell cycle re-entry: more than a coincidence. Cell Cycle. 2011;10:2683–2690. doi: 10.4161/cc.10.16.17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albee AJ, Kwan AL, Lin H, et al. Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3. 2013;3:979–991. doi: 10.1534/g3.113.006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santagata S, Hornick JL, Ligon KL. Comparative analysis of germ cell transcription factors in CNS germinoma reveals diagnostic utility of NANOG. The American journal of surgical pathology. 2006;30:1613–1618. doi: 10.1097/01.pas.0000213320.04919.1a. [DOI] [PubMed] [Google Scholar]
- 42.Jones RT, Abedalthagafi MS, Brahmandam M, et al. Cross-reactivity of the BRAF VE1 antibody with epitopes in axonemal dyneins leads to staining of cilia. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2015;28:596–606. doi: 10.1038/modpathol.2014.150. [DOI] [PubMed] [Google Scholar]
- 43.Tong Y, Merino D, Nimmervoll B, et al. Cross-Species Genomics Identifies TAF12, NFYC, and RAD54L as Choroid Plexus Carcinoma Oncogenes. Cancer cell. 2015;27:712–727. doi: 10.1016/j.ccell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eden E, Navon R, Steinfeld I, et al. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Supek F, Bosnjak M, Skunca N, et al. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS one. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stubbs JL, Oishi I, Izpisua Belmonte JC, et al. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nature genetics. 2008;40:1454–1460. doi: 10.1038/ng.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoh RA, Stowe TR, Turk E, et al. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PloS one. 2012;7:e52166. doi: 10.1371/journal.pone.0052166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeisel A, Munoz-Manchado AB, Codeluppi S, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 49.Tan FE, Vladar EK, Ma L, et al. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development. 2013;140:4277–4286. doi: 10.1242/dev.094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan JH, Adair-Kirk TL, Patel AC, et al. Myb permits multilineage airway epithelial cell differentiation. Stem cells. 2014;32:3245–3256. doi: 10.1002/stem.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kubota T, Ishise J, Yamashima T, et al. Abnormal cilia in a malignant ependymoma. Acta neuropathologica. 1986;71:100–105. doi: 10.1007/BF00687969. [DOI] [PubMed] [Google Scholar]
- 52.Ohata S, Nakatani J, Herranz-Perez V, et al. Loss of Dishevelleds disrupts planar polarity in ependymal motile cilia and results in hydrocephalus. Neuron. 2014;83:558–571. doi: 10.1016/j.neuron.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Atkinson JM, Shelat AA, Carcaboso AM, et al. An integrated in vitro and in vivo high-throughput screen identifies treatment leads for ependymoma. Cancer cell. 2011;20:384–399. doi: 10.1016/j.ccr.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pagliarini R, Shao W, Sellers WR. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO reports. 2015;16:280–296. doi: 10.15252/embr.201439949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siu MK, Wong ES, Kong DS, et al. Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene. 2013;32:3500–3509. doi: 10.1038/onc.2012.363. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Cai X, Xia L, et al. Decreased expression of FOXJ1 is a potential prognostic predictor for progression and poor survival of gastric cancer. Annals of surgical oncology. 2015;22:685–692. doi: 10.1245/s10434-014-3742-2. [DOI] [PubMed] [Google Scholar]
- 58.Chen HW, Huang XD, Li HC, et al. Expression of FOXJ1 in hepatocellular carcinoma: correlation with patients’ prognosis and tumor cell proliferation. Molecular carcinogenesis. 2013;52:647–659. doi: 10.1002/mc.21904. [DOI] [PubMed] [Google Scholar]
- 59.Eroglu E, Burkard TR, Jiang Y, et al. SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell. 2014;156:1259–1273. doi: 10.1016/j.cell.2014.01.053. [DOI] [PubMed] [Google Scholar]
- 60.Reiff T, Tsarovina K, Majdazari A, et al. Neuroblastoma phox2b variants stimulate proliferation and dedifferentiation of immature sympathetic neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:905–915. doi: 10.1523/JNEUROSCI.5368-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saha SK, Parachoniak CA, Ghanta KS, et al. Mutant IDH inhibits HNF-4alpha to block hepatocyte differentiation and promote biliary cancer. Nature. 2014;513:110–114. doi: 10.1038/nature13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tata PR, Mou H, Pardo-Saganta A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pirozzi CJ, Reitman ZJ, Yan H. Releasing the block: setting differentiation free with mutant IDH inhibitors. Cancer cell. 2013;23:570–572. doi: 10.1016/j.ccr.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sasaki M, Knobbe CB, Munger JC, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu J, Yu J, Mani RS, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller ML, Reznik E, Gauthier NP, et al. Pan-Cancer Analysis of Mutation Hotspots in Protein Domains. Cell Systems. 2015;1:197–209. doi: 10.1016/j.cels.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tong CK, Han YG, Shah JK, et al. Primary cilia are required in a unique subpopulation of neural progenitors. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12438–12443. doi: 10.1073/pnas.1321425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.