Summary
The budding yeast, Saccharomyces cerevisiae, harbors several prions that are transmitted as altered, heritable protein conformations. [SWI+] is one such prion whose determinant is Swi1, a subunit of the evolutionarily conserved chromatin-remodeling complex SWI/SNF. Despite the importance of Swi1, the molecular events that lead to [SWI+] prionogenesis remain poorly understood. In this study, we have constructed floccullin-promoter-based URA3 reporters for [SWI+] identification. Using these reporters, we show that the spontaneous formation frequency of [SWI+] is significantly higher than that of [PSI+] (prion form of Sup35). We also show that preexisting [PSI+] or [PIN+] (prion form of Rnq1), or overproduction of Swi1 prion-domain (PrD) can considerably promote Swi1 prionogenesis. Moreover, our data suggest a strain-specific effect of overproduction of Sse1 – a nucleotide exchange factor of the molecular chaperone Hsp70, and its interaction with another molecular chaperone Hsp104 on [SWI+] maintenance. Additionally, we show that Swi1 aggregates are initially ring/ribbon-like then become dot-like in mature [SWI+] cells. In the presence of [PSI+] or [PIN+], Swi1 ring/ribbon-like aggregates predominantly colocalize with the Sup35 or Rnq1 aggregates; without a preexisting prion, however, such colocalizations are rarely seen during Swi1-PrD overproduction-promoted Swi1 prionogenesis. We have thus demonstrated a complex interacting mechanism of yeast prionogenesis.
Keywords: prion, Swi1, SWI/SNF, aggregation, prion-interactions
Graphical abstract
We have dissected the yeast FLO11 promoter and constructed several truncated FLO11-promoter-based reporters for studying the events of [SWI+] de novo formation and propagation. We show that in the presence of [PSI+] or [PIN+], cross-seeding with Sup35 or Rnq1 is a prevalent mechanism promoting Swi1 aggregation and prionogenesis, but not when [SWI+] formation is merely promoted by Swi1 prion-domain overproduction. We also show a strain-specific sensitivity of [SWI+] to overproduction of the molecular chaperone Sse1.
Introduction
Prions, proteinaceous infectious particles, are composed of host proteins with altered and transmissible conformations (Wickner et al., 2015, Colby & Prusiner, 2011). Although the term prion was first used to describe the causative agent of the neurodegenerative diseases known as transmissible spongiform encephalopathies (TSEs) (Prusiner, 1982, Prusiner, 1998), this protein-only prion concept has been expanded to include a large number of fungal and mammalian proteins that can also undergo conformational changes and be transmitted as highly-ordered aggregates termed amyloids (Crow & Li, 2011, Munch et al., 2011, Sanders et al., 2014, Luk et al., 2012). In the budding yeast Saccharomyces cerevisiae, at least nine prion proteins have been identified and additional potential prions may exist (Crow & Li, 2011, Suzuki et al., 2012, Halfmann et al., 2012, Chakrabortee et al., 2016). When these prion proteins adopt prion conformation(s), the resulting prions usually manifest as dominant and heritable features, mostly through modulation of important cellular processes such as transcription and translation (Alberti et al., 2009, Du et al., 2015, Serio & Lindquist, 1999, Stein & True, 2014). In addition, it has been shown that some yeast prions can exist in wild strains and their prion conformational switches can be environmentally responsive, suggesting a possible role of prionogenesis in yeast adaptation to environmental fluctuations (Holmes et al., 2013, Suzuki et al., 2012, Jarosz et al., 2014, Tyedmers et al., 2008). Interestingly, protein misfolding, aggregation, and subsequent amyloid deposition have been proposed to be crucial etiological events associated with more than 40 incurable mammalian disorders, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS) and recently, cancers (Ano Bom et al., 2012, Luk et al., 2012, Munch et al., 2011, Sanders et al., 2014).
Amyloid yeast prions have become valuable systems for studying prion biology and other proteinopathies due to sharing similar structural features and transmission mechanisms as PrPSc, the causative agent of TSEs, and several other prion-like mammalian pathogenic proteins, such as tau (a microtubule-associated protein implicated in tauopathies when misfolded (Alonso et al., 2001)), β-amyloid (peptides of 36–43 amino acids derived from a protein known as amyloid protein precursor and implicated in Alzheimer’s disease (Vassar et al., 1999), and α-synuclein (an abundant protein in the brain implicated in a group of disorders known as synucleinopathies when it becomes insoluble (Spillantini et al., 1997)) . Utilizing the yeast prion systems, important knowledge has been gained regarding molecular mechanisms of prion formation and transmission and important cellular factors required for prionogenesis, such as molecular chaperones – Hsp104, Hsp40, and Hsp70 and its co-chaperones – have been identified (see reviews of (Guinan & Jones, 2009, Romanova & Chernoff, 2009, Winkler et al., 2012, Sporn & Hines, 2015)). In addition, valuable information regarding how heterologous prion proteins interact with each other and how such interactions influence their de novo prion formation and maintenance has also been reported (Derkatch et al., 2001, Derkatch et al., 1997, Du & Li, 2014, Osherovich & Weissman, 2001, Schwimmer & Masison, 2002).
Perhaps the most studied yeast prion is [PSI+], whose protein determinant is Sup35, a translational termination factor (Stansfield et al., 1995). When Sup35 enters a prion conformation, it results in a compromised translation termination function (Patino et al., 1996). Previously, it was reported that overproduction of Swi1, a subunit of the SWI/SNF chromatin-remodeling complex, can facilitate de novo formation of [PSI+] (Derkatch et al., 2001). We later reported that Swi1 can exist as a prion, [SWI+], and cells harboring [SWI+] exhibit a partial loss-of-function phenotype of SWI/SNF (Du et al., 2008). We also showed that preexisting [SWI+] can promote the de novo formation of [PSI+] and another prion known as [PIN+] or [RNQ+], whose protein determinant is Rnq1 (Du & Li, 2014, Sondheimer & Lindquist, 2000, Derkatch et al., 1997). Such [PSI+] or [PIN+] promotion by [SWI+] is likely realized mainly through a cross-seeding mechanism. The cross-seeding model proposes that a preexisting prion can provide a conformational template on which the first seeds of a different prion can form (Derkatch et al., 2001). In this case, the Swi1 prion aggregates could recruit Rnq1 or Sup35 to form ring/ribbon/rod-like (ribbon-like hereafter) heterogeneous aggregates in [SWI+] cells that are prionogenic, i.e. capable of leading to the formation of [RNQ+] or [PSI+] (Du & Li, 2014, Du et al., 2014). In addition, we also reported an antagonizing effect between [SWI+] and [PIN+] that [SWI+] can reduce [PSI+] promoting capacity of [PIN+] (Du & Li, 2014). These results demonstrate that the [SWI+] prion is an excellent system to study prion-prion interactions. However, the lack of a reliable reporter system that can distinguish [SWI+] from [swi−] has hindered our understanding of [SWI+] de novo formation and propagation. For instance, what is the spontaneous frequency of [SWI+] conversion and will [SWI+] conversion be affected by preexisting prions or overproduction of Swi1 PrD? Swi1 is an essential transcriptional activator for the expression of flocculin (FLO) genes (Mao et al., 2008). Flocculins or adhesins are a group of lectin-like cell wall proteins that confer a variety of multicellular features in several Candida albicans and S. cerevisiae strains (Barrales et al., 2008, Barrales et al., 2012, Li & Palecek, 2005). We have recently demonstrated that FLO gene expression is absent in [SWI+] cells so that [SWI+] cells cannot exhibit the multicellular features of flocculation, invasive growth or pseudohyphal formation (Du et al., 2015). In this study, by taking advantage of the tight regulation of [SWI+] on FLO gene expression, we explored the possibility of developing useful [SWI+] reporters that can report the prion status of Swi1 and be easily assayed. We report here that we have established several FLO promoter-based URA3 systems (FLOpr-URA3) that can be useful for [SWI+] research. With these reporters, we have characterized the [SWI+] propagation features with a focus on a strain-specific response of [SWI+] to functional alterations of Hsp104 and Hsp70-Ssa nucleotide-exchange-factor (NEF) co-chaperone Sse1. These reporter systems have also allowed us to monitor the events of [SWI+] de novo formation. We have determined and compared [SWI+] de novo formation frequencies in strains of S228C and 74D-694, and examined the influence of heterologous prions and overproduction of Swi1-PrD on such [SWI+] de novo formation and the underlying interacting mechanisms driving yeast prion de novo formation.
Results
Constructing URA3-based [SWI+] reporters using FLO gene promoters
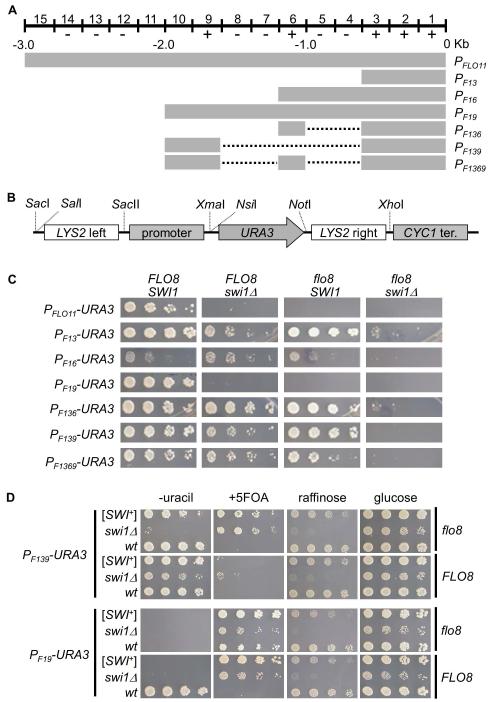
The yeast S288C strain has two active flocculins, Flo1 and Flo11, which are encoded by two FLO genes, FLO1 and FLO11, respectively, and whose functions are essential for exhibiting the multicellular phenotypes of this strain (Kobayashi et al., 1999). Due to containing an internal in-frame stop at codon 142 in the FLO8 gene, S288C-derived strains do not express FLO1 or FLO11 and thus cannot exhibit certain multicellular phenotypes (Liu et al., 1996). Flo8 is a key transcriptional activator in the cyclic adenosine monophosphate-dependent protein kinase A (cAMP-PKA) pathway and it acts in concert with other activators such as Mga1 and Mss11 in activating FLO promoters (Bester et al., 2006, Kim et al., 2014, Mayhew & Mitra, 2014). Upon repair of FLO8, the expression of FLO1 and FLO11 can be activated and multicellular phenotypes can be restored, such as flocculation and adhesive growth of haploid cells, as well as pseudohyphal growth of diploid cells (Fichtner et al., 2007). Our recent observations that in FLO8-repaired S288C strains that Swi1 is required for FLO gene expression and [SWI+] cells completely lack multicellularity encouraged us to test the idea of developing simple reporter assays of [SWI+] using the FLO promoters. Since the FLO1 promoter contains multiple reported interaction sites for SWI/SNF which can be > 5 kb upstream of the FLO1 start codon (Fleming & Pennings, 2001), it is difficult to clone or engineer the FLO1 promoter for this purpose. We thus focused on the FLO11 promoter in this study. As shown in Figure 1A, the FLO11 promoter contains multiple defined upstream activation sequences (UASs) and upstream repression sequences (URSs), which span a region of about 2.8 kb before the translation start codon (Rupp et al., 1999). Although published data showed that these UASs within the FLO11 promoter could respond independently to diverse external and internal signals, determination of the SWI/SNF-acting UASs was reported to be difficult (Rupp et al., 1999, Braus et al., 2003, Basu et al., 2004). Since Swi1 is required to activate FLO11 gene expression (Basu et al., 2004, Du et al., 2015), we assumed that the SWI/SNF (thus, Swi1)-acting sites are within the previously identified UASs but not URSs. To identify Swi1-acting sites, we generated six truncated FLO11 upstream regulatory region containing various numbers of UASs (+) and/or URSs (−) by PCR (Figure 1A and S1), and cloned them into the upstream of URA3 ORF in a p415-based plasmid harboring a CYC1 terminator. In these transcriptional fusion constructs, 50-bp of left and right sequences corresponding to the upstream and downstream regions of the LYS2 ORF are included to flank the expression cassette, which can be used for integrating the reporter into the LYS2 locus (Figure 1B).
Figure 1. Dissection of FLO11 promoter and construction of plasmid-based [SWI+] reporters for S288C strains.
(A) A diagrammatic illustration of the wild-type and truncation mutant FLO11 promoters. The 3-kb FLO11 promoter (PFLO11) spans 15 0.2-kb segments carrying 7 upstream repression sequences (URSs, −) and 5 upstream activation sequences (UASs, +). The dotted lines stand for regions that are deleted. Construction strategies are shown in Figure S1. (B) A diagram showing the structure of constructed reporter plasmids. URA3 gene serves as a reporter gene driven the wild type or a truncated FLO11 promoter shown in A. CYC1 terminator was included for all the constructs. 50-bp upstream and downstream extensions of LYS2 flank the reporter to provide an option to integrate a reporter into chromosome. (C) BY4741 non-prion strains with the indicated genotypes were transformed with individual p415-based URA3 reporter plasmids. Cells were spotted onto synthetic complete (SC) selective plates without uracil. Images were taken 3 days post spotting. Shown is a representative result of at least three independent experiments. (D) The indicated FLO8 or flo8 strains with distinct Swi1 status were transformed with either PF139-URA3 or PF19-URA3 plasmid and assayed for growth on glucose-containing SC medium (glucose) without uracil (−uracil), or with 5-FOA (+5FOA). Raffinose plate (glucose-free) was also included to verify the Swi1 status. Shown are representative results (3 days post-spotting) of at least three repeated tests.
To test if these reporter plasmids can be used for [SWI+] study, we first compared their URA3 expression in wild-type, single, and double mutant strains of flo8 and swi1Δ (Figure 1C). Except for PF16, a 1.2kb fragment from the 5′- site of the FLO11 start codon containing 4 UASs and 2 URSs, all tested truncations supported cell growth on media without uracil in the wild-type strain (FLO8/SWI1), suggesting that the two URSs at the region of −600 to −1000bp confer a strong repression. Considering PF19 behaved similarly to the wild-type FLO11 promoter (PFLO11), the 2000-bp upstream sequence of FLO11 seems to contain most of the critical regulatory elements in S288C strains. The fact that PF19 and the wild-type FLO11 were the only two inactive promoters in flo8 or swi1Δ mutant strain suggests that their de-repression requires both functions of Flo8 and Swi1. The activity of PF13 was slightly reduced in swi1Δ but not in the flo8 strain, suggesting that Swi1 but not Flo8 plays a major activator role in interacting with the UASs between −1 and −600bp. It was previously reported that that PF13 may serve as a core promoter containing the basal regulatory sites (Braus et al., 2003). Indeed, the activities of PF136, PF139, and PF1369, which contain one or two more activator binding sites beyond PF13, exhibited no significant enhancement of activity comparing to that of PF13 in the wild-type strain (Figure 1C). Based on the activities of PF16 and PF19, one may speculate that the region of −1600 and −2000 must contain UASs that interact with both Swi1 and Flo8 to de-repress the URSs between −600 and −1000 as PF16 grew poorly in SC-ura in all strain backgrounds, however, PF19 grew well in the wild-type strain, but not in any of the mutant strains (Figure 1C). Importantly, promoters other than PF19 and PFLO11 showed significant activities in single mutant strains (Figure 1C), suggesting that Flo8 and Swi1 likely have redundant functions in activating some of the truncated FLO11 promoters. Taken together, these results demonstrate a complex regulation of FLO11 promoter by SWI/SNF and Flo8, particularly of the Swi1 responding cis-elements of the promoter.
We next examined if plasmids built with the aforementioned truncated FLO11 promoters and three additional SWI/SNF-regulated promoters, SL (a chimeric SUC2-Leu2 promoter) (Neigeborn & Carlson, 1987), Alcohol DeHydrogenase II (ADH2) (Biddick et al., 2008), and HOmothallic switching endonuclease (HO) (Stern et al., 1984) (PLS, PADH2, and PHO), can be used to distinguish [SWI+] from [swi−] in S288C cells using the uracil/5-FOA growth assay. We also included a growth assay in raffinose media as an additional criterion to monitor the [SWI+] status as we previously showed that [SWI+] cells grow poorly in media using raffinose as the sole carbon source (Du et al., 2008). Isogenic [SWI+], [swi−], and swi1Δ strains with or without FLO8 repair were used in these assays. As shown in Figure S2, only two constructs, PF19-URA3 and PF139-URA3, were potentially able to serve as [SWI+] reporters. In the presence of Flo8, PF19-URA3 allowed non-prion ([swi−]) cells to grow in −uracil media but not +5-FOA media, whereas [SWI+] and swi1Δ cells could only grow in +5-FOA media but not in −uracil media (Figure 1D and S2). Differently, PF139-URA3 only allowed [SWI+]/swi1Δ - dependent growth in +5-FOA assay and such a growth phenotype was not FLO8-dependent. However, the −uracil growth assay of PF139-URA3 could not differentiate [swi−] cells from cells harboring [SWI+] either in the presence or absence of Flo8 (Figure 1D and S2). These results confirmed that both PF19-URA3 and PF139-URA3 could be potentially useful for [SWI+] study under selective conditions. We next investigated if we could establish a stable, chromosomal reporter of PF139-URA3 by replacing the endogenous FLO11 gene (including both the promoter region and ORF, see Experimental Procedures for details) at the FLO11 locus. Unexpectedly, the chromosomal PF139-URA3 (Chr::PF139-URA3) behaved differently from its plasmid version (compare Figure 1D and S3) – it could only discriminate [SWI+] and [swi−] in the −uracil assay in the presence of FLO8 but not in the +5-FOA assay in either flo8 or FLO8 background (Figure S3). As we showed previously (Du et al., 2015), Chr::PFLO1-URA3 can clearly distinguish [SWI+] and [swi−] cells in both −uracil and +5-FOA assays (Figure S3) and thus it is, applicable as a useful [SWI+] reporter. The Chr::PFLO11-URA3 behaves similarly to its plasmid version, which could only be used in the −uracil assay. After all, in above uracil/5 FOA assays, swi1Δ and [SWI+] strains could be noticeably distinguished by using some but not all of the reporter constructs, the [SWI+] status should be further verified by other reversible features such as aggregation and raffinose utilization.
Suitability of PF139- and PF19-URA3 reporters in [SWI+] research for the 74D-694 strain derivatives
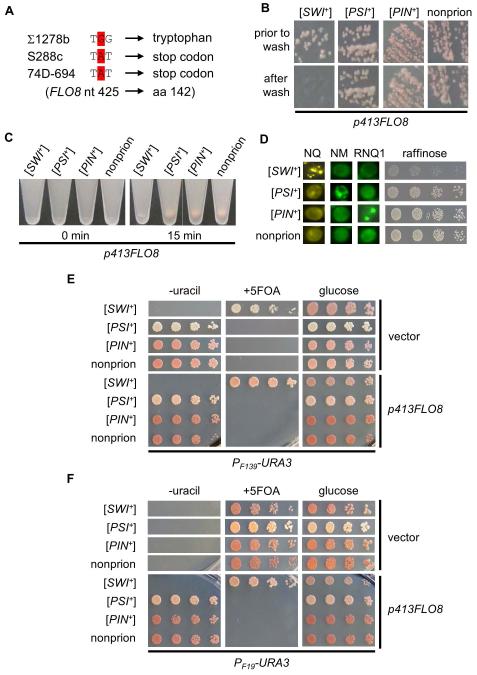
Next, we investigated if PF19-URA3 and PF139-URA3 could be used as [SWI+] reporters in 74D-694 strain and derivatives, which are commonly used for yeast prion studies. Upon examination, we found that the 74D-694 strain does not display multicellular phenotypes (data not shown), suggesting that the FLO genes are not expressed. DNA sequencing data demonstrated that the FLO8 gene contains a point mutation in the code of the 142th amino acid (tryptophan) to a stop code, which is identical to that in S288C strain (Figure 2A). Thus, the lack of multicellularity of 74D-694 is due to the same flo8 mutation of S288C. We transformed a plasmid expressing FLO8 into a wild-type 74D-694 strain and tested if FLO8 expression could restore the multicellular features, and if so, would these phenotypes be regulated by [SWI+]. We found that ectopic expression of FLO8 driven by its own promoter from a CEN-plasmid successfully restored the multicellular phenotypes of adhesive growth (Figure 2B) and flocculation (Figure 2C). Both phenotypes were abolished by [SWI+] but not by [PSI+] or [PIN+]. Taken together, we concluded that similar to S288C, the 74D-694 strain contains a mutation in FLO8 and thus lacks multicellularity; repairing Flo8 restores FLO gene expression and multicellularity; and [SWI+] abolishes FLO gene expression and multicellularity.
Figure 2. Performance of PF139-URA3 and PF19-URA3 plasmids as [SWI+] reporters for 74D-694 derivatives.
(A) DNA sequencing data demonstrate that the 74D-694 strain contains the same nonsense mutation in the FLO8 ORF as S288C-derivated strains. (B) Plasmid p413FLO8 was introduced into the four indicated strains to ectopically express Flo8 under its own promoter. Transformants were then tested for adhesion on SC selective plates using a wash assay (see Experimental Procedures). The remaining cells were imaged prior to and after washing (a representative result of at least three repeated assays). (C) The same strains used in panel B were assayed for flocculation (cell-cell adhesion, or cell aggregation, see details in Experimental Procedures). 0 min, immediately after vortexing the cell cultures; 15 min, 15 min after keeping cultures still on bench post-vortex. Shown is a typical result of at least three repeated tests. (D) Indicated isogenic 74D-694 strains were examined for raffinose phenotype (right) and aggregation patterns of Swi1-NQ-YFP (NQ), Sup35-NM-GFP (NM) and Rnq1-GFP (RNQ1) after transforming them with p416TEF-NQYFP, pCUP1-NMGFP, or pCUP1-RNQ1GFP. (E) Indicated strains were co-transformed with p415F139-URA3 and p413FLO8 or p413TEF (vector), and assayed for their growth on glucose-containing SC medium (glucose) without uracil (−uracil), or with 5-FOA (+5FOA). Shown are representative results (3 days post-spotting) of at least three repeated assays. (F) Experiment was carried out similar to that in panel E except that p415F19-URA3 was used instead of p415F139-URA3.
To test if PF19-URA3 and PF139-URA3 could be used to report the [SWI+] state in 74D-694 strains, we tested four isogenic 74D-694 strains harboring different prions (Figure 2D). As shown in Figure 2E, PF139-URA3 allowed [SWI+] cells to grow on 5-FOA-containing media but not on −uracil media, whereas [PSI+], [PIN+], and non-prion ([psi−][pin−][swi−]) strains grew well in −uracil media but had no growth in +5-FOA media. Interestingly, these phenotypes are independent of FLO8 (Figure 2E), indicating that PF139-URA3 can serve as a [SWI+]-specific reporter for 74D-694 strains in the presence or absence of FLO8. For PF19-URA3, all tested strains did not grow on media lacking uracil without ectopic FLO8 expression, whereas FLO8 expression allowed growth on −uracil media for [PSI+], [PIN+], and [swi−] but not for [SWI+] cells that were the only ones growing on +5-FOA plates (Figure 2F). These results demonstrate that PF19-URA3 can be a faithful [SWI+] reporter for 74D-694 strains in the presence of functional Flo8.
Although the PF139-URA3 reporter can be a decent [SWI+] reporter only in +5-FOA assay for S288C strains without Flo8 ectopic expression, it can be used in both −uracil and 5-FOA assays for 74D-694 strains independent of Flo8 expression (compare Figure 1D and 2E). However, PF19-URA3 can be used in either −uracil or +5-FOA assay for both S288C and 74D-694 strains only when Flo8 is ectopically expressed. These results demonstrate strain-specific performance of the two reporters.
Influence of Sse1-overproduction and its interaction with Hsp104 on [SWI+] propagation
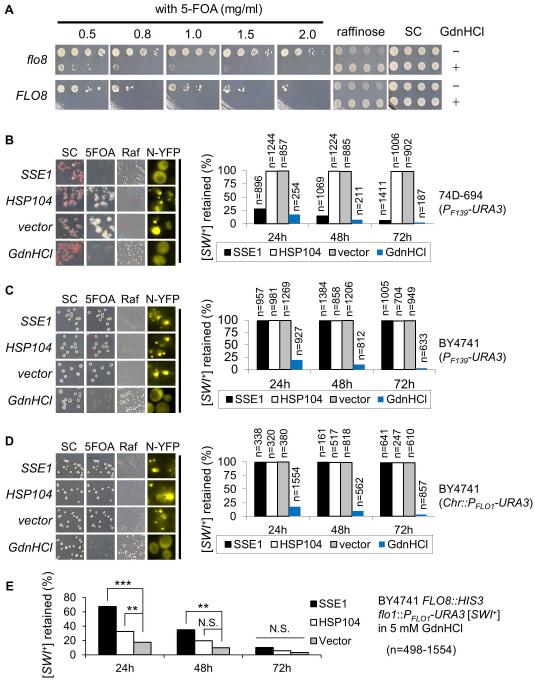
The maintenance of a yeast prion requires a concerted action from multiple molecular chaperones and other cellular factors (Winkler et al., 2012, Masison & Reidy, 2015). It was reported earlier that Sse1 overproduction can eliminate [SWI+] in 74D-694 strain whereas Hsp104 overproduction cannot (Du et al., 2008, Hines et al., 2011). To validate the suitability of the plasmid-based PF139-URA3 reporter in [SWI+] study, we examined if this reporter plasmid could be used to study the curing of [SWI+] by 5-mM guanidine hydrochloride (GdnHCl), a treatment known to eliminate [SWI+] through inactivating Hsp104 (Du et al., 2008). When [SWI+] S288C strains containing the PF139-URA3 plasmid were treated with 5-mM GdnHCl, the reporter plasmid was effective in detecting the [SWI+] curing as demonstrated by a lack of growth in +5-FOA media for both flo8 and FLO8 strains (Figure 3A). The 1.0 mg/mL 5-FOA concentration showed clear delineation between the prion and cured conditions and was used in the rest of the experiments described in this article.
Figure 3. The effect of altering Hsp104 and Sse1 activity on [SWI+] propagation.
(A) FLO8-repaired (FLO8) and unrepaired (flo8) BY4741 [SWI+] strains were transformed with p415F139-URA3, and streaked on SC selective plates (SC) with (+) or without (−) 5 mM GdnHCl and then cultured in SC-leu which was followed by growth on the indicated SC selective plates. Shown was a result (3 days post-spotting) of at least 3 independent experiments. (B, C, and D) As described in the Experimental Procedures, the influences of Hsp104 overproduction (HSP104), Hsp104 inactivation (GdnHCl), and Sse1 overproduction (SSE1) on [SWI+] propagation in 74D-694 (B) and BY4741 (C and D) backgrounds were investigated. vector, an empty vector control; SC, SC selective plate; 5FOA, SC selective plate with 5-FOA; Raf, raffinose plate; n, total colonies assayed. Note: the plasmid reporter PF139-URA3 was used for panel B and C, but chromosomal reporter of PFLO1-URA3 for panel D. Prion curing was summarized in the plot (right panel) based on the growth phenotypes (left). (E) Effect of Sse1 or Hsp104 overproduction on [SWI+] curing by GdnHCl was assayed as described in the Experimental Procedures. The remaining [SWI+] was assayed after growing cells in SC+GdnHCl medium for the indicated time while overproducing Hsp104 or Sse1. T-test was used to estimate the significance of the differences (N.S., not significant; **, P<0.01; ***, P<0.001). n, colonies assayed.
Experiments were conducted using the PF139-URA3 reporter plasmid in both 74D-694 and BY4741 (S288C derivative) strains. As expected, [SWI+] was eliminated by GdnHCl treatment but not by Hsp104 overproduction in both strains; however, unexpectedly, Sse1 overproduction only cured [SWI+] in 74D-694 but not the same [SWI+] variant in BY4741 (Figure 3B and 3C). To verify these results, experiments were repeated with a FLO8-repaired S288C [SWI+] strain with an integrated copy of PFLO1-URA3 at the FLO1 locus (Du et al., 2015). As shown in Figure 3D, similar results were obtained. These results indicate that [SWI+] is sensitive to Sse1 overproduction only in 74D-694 but not in S288C.
The insensitivity of [SWI+] to Sse1 overproduction in S288C background suggests a possibility that Sse1 does not participate in [SWI+] propagation in S288C since additional nucleotide exchange factors for Hsp70 do exist, such as Sse2 and Fes1 (Kabani et al., 2002, Mukai et al., 1993). To test this likelihood, we examined if Hsp104 or Sse1 overproduction would affect the [SWI+] curing capacity of GdnHCl using the chromosomal PFLO1-URA3 reporter in a S288C [SWI+] strain. Hsp104 and Sse1 were individually overproduced under a GPD promoter in media containing 5 mM GdnHCl. After expression and curing, [SWI+] loss was estimated by phenotypic assays on −uracil, +5-FOA, and raffinose plates. As shown in Figure 3E, overproduction of Hsp104 or Sse1 significantly delayed the prion curing process by GdnHCl. For instance, after 24 h of growth, there are larger fractions of cells overexpressing SSE1 or HSP104 retaining [SWI+] compared to that of the vector control (Figure 3E). As expected, excess amount of Hsp104 can antagonize the inactivating effect of GdnHCl. It is noteworthy that Sse1 showed a greater antagonizing effect against the prion curing by GdnHCl than Hsp104 overproduction. Although it is unclear why Sse1 overproduction decreases [SWI+]-curing capacity of GdnHCl, our results suggest that Sse1 does participate in [SWI+] maintenance and propagation via direct or indirect interaction with Hsp104, and a higher expression level of Sse1 favors [SWI+] propagation in the S288C strain derivatives (compare Figure 3B-3E).
The effects of strain, heterologous prions, and Swi1 PrD overproduction on [SWI+] formation
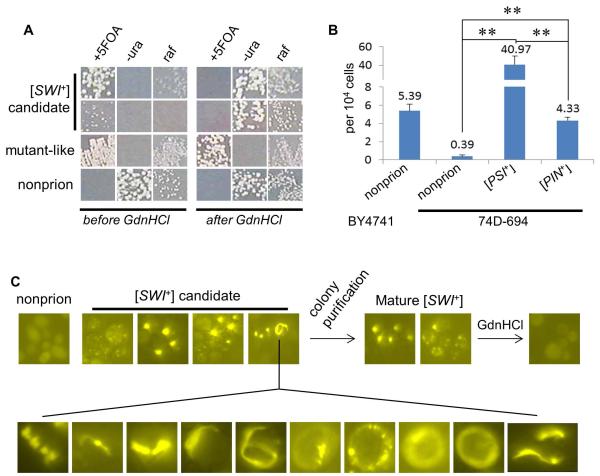
As the [SWI+] formation frequency remained elusive, next, we investigated the spontaneous rate of [SWI+] conversion in S288C and 74D-694 strains. For S288C, the FLO8-repaired BY4741FLO8::HIS3 flo1::PFLO1-URA3 non-prion ([psi−][pin−][swi−]) strain was used because the integrated PFLO1-URA3 reporter is a better [SWI+] reporter compared to PF139-URA3 for this strain background (Figure S3 and 1D). The 5-FOA resistant (5-FOA+) colonies were screened and then examined for their curability by 5 mM GdnHCl (Figure 4A and data not shown). Candidates that were able to convert from Ura− 5-FOA+ Raf± to Ura+ 5-FOA− Raf+ upon a treatment via growth on 5 mM GdnHCl plates were assayed for aggregation after being transformed with p415TEF-NQYFP, a plasmid expressing a -YFP fusion of N-terminal region of Swi1 containing the first 524 amino acid residues of Swi1 that is asparagine (N) and glutamine (Q)-rich. Only those 5-FOA+ isolates that displayed NQ-YFP aggregation and were GdnHCl-curable were scored as [SWI+]. As shown in Figure 4B, the average spontaneous [SWI+] formation frequency was approximately 5 × 10−4 for S288C under the examined experimental conditions.
Figure 4. [SWI+] spontaneous de novo formation promoted by pre-existing [PSI+] or [PIN+].
(A) Representative growth phenotypes of the BY4741 strain-derived [SWI+] candidate isolates before and after 5 mM GdnHCl treatment. The [SWI+] converting frequency is summarized in panel B. +5FOA (SC-his+5-FOA); −ura (SC-his-ura); raf (raffinose). (B) As described in the Experimental Procedures, [SWI+] spontaneous conversion frequencies in the indicated strain backgrounds were summarized based on at least three independent experiments. (C) Appearance, maturation and curability of Swi1-NQ-YFP aggregation during Swi1 prionogenesis in 74D-694 strains. Upon colony purification, aggregation appeared in 5-FOA+ cells could be stabilized, morphologically remodeled, and curable. Experiments were done for the [PSI+], [PIN+] and nonprion strains, and [SWI+] converting frequencies are summarized in panel B. Shown here are representative data (upper, from non-prion strain; lower, ring/ribbon-like aggregates from [PSI+] and [PIN+] strains). Statistical analysis was performed by T test (**, P<0.01).
For 74D-694, we showed that the PF139-URA3 reporter allows 74D-694 [SWI+] cells to grow in 5-FOA independent of Flo8 (Figure 2E). To monitor [SWI+] spontaneous formation rate in 74D-694, isogenic [PSI+], [PIN+], and non-prion ([psi−][pin−][swi−]) strains of 74D-694 were co-transformed with p415F139-URA3 and p413GAL1-NQYFP, and screened for growth on +5-FOA plates. Using the aforementioned criteria, 5-FOA+ colonies that exhibited NQ-YFP aggregation upon galactose induction and were curable by GdnHCl were scored as [SWI+] (Figure 4C). Regarding this, addition of 0.01 - 0.1% of galactose in sucrose-based media was sufficient to give rise to an aggregation frequency of about 30-80% for N-YFP (a YFP fusion containing the first 323 amino acids of Swi1 that are asparagine-rich and responsible for [SWI+] prion phenotypes) and NQ-YFP when expressed from a CEN-plasmid in [SWI+] cells, and no aggregation was detectable in [swi−] cells (Figure S4 and data not shown). Thus, 0.02% galactose serves as minimal concentrations to show [SWI+]-specific Swi1 N or NQ-YFP aggregation (Figure S4). Isolates with NQ-YFP aggregation were further colony-purified and re-assayed for aggregation stability, growth phenotypes on −uracil, +5-FOA, and raffinose plates, and curability by 5 mM GdnHCl (Figure 4C and data not shown).
As shown in Figure 4B, the spontaneous [SWI+] conversion frequency of 74D-694 is about 4 × 10−5 – approximately 10x lower than that of S288C under identical experimental conditions. Moreover, the tested [PSI+] and [PIN+] 74D-694 strains gave [SWI+] spontaneous formation frequency of about 4 × 10−3 and 4 × 10−4. Even though such an estimation of [SWI+] frequencies in [PSI+] and [PIN+] cells can be rough considering the possibility that the combination of [SWI+] with [PSI+] or [PIN+] may affect the fitness of cells carrying certain [SWI+] variants, our results suggest the [SWI+] conversion frequencies are approximately 100x and 10x greater in the [PSI+] and [PIN+] strains than that of the non-prion strain, respectively (Figure 4B). These data indicate that [PSI+] and [PIN+] can significantly promote [SWI+] de novo conversion. The unexpected observation that [PSI+] has a significantly higher inducibility of [SWI+] than [PIN+] suggests that the two prions might be structurally different and thus have different capability of cross-seeding Swi1 toward [SWI+] formation. Since [SWI+] could also promote [PSI+] and [PIN+] de novo formation (Du & Li, 2014), [SWI+], [PSI+], and [PIN+] seem to facilitate each another’s conversions. Ribbon-like NQ-YFP aggregation patterns were frequently observed in the newly formed [SWI+] cells and gradually lost and turned to dots upon a colony-purification process (Figure 4C). This is quite similar to the aggregation pattern changes of Sup35 and Rnq1 in the prionogenesis of [PSI+] and [PIN+] (Du & Li, 2014, Mathur et al., 2010, Zhou et al., 2001).
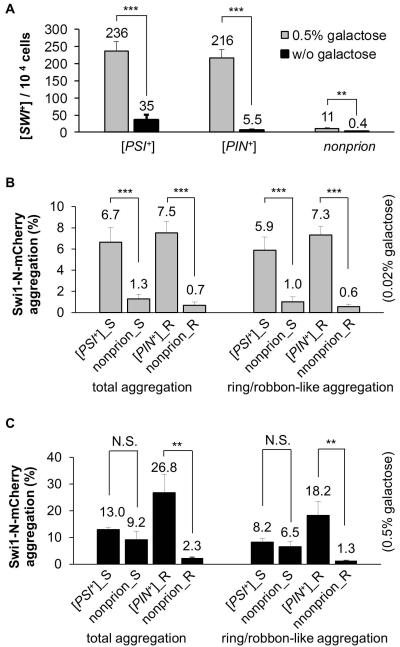
Next, we asked if overproduction of the Swi1 PrD would promote [SWI+] de novo formation. We showed previously that the first 323-amino acid of Swi1 (Swi1-N) contains the Swi1 PrD [57]. When Swi1-N was over-expressed under the TEF1 promoter from a CEN-plasmid in 74D-694 strains carrying the PF139-URA plasmid, we obtained significantly more 5-FOA+ colonies when compared to that of the vector control (data not shown). To better investigate the interplay between Swi1-N overproduction and heterologous prion interactions in regard to [SWI+] formation, we utilized N-YFP expression under the GAL1 promoter with supplementation of 0.5% galactose. The obtained 5-FOA+ colonies that exhibited GdnHCl-curable N-YFP-aggregation and growth phenotypes of Raf±, Ura−, and 5-FOA+ were scored as [SWI+]. As shown in Figure 5A, without N-YFP overproduction, the [SWI+] de novo forming frequencies were comparable to that shown in Figure 4B, however, the [SWI+] conversion rates were significantly increased by N-YFP overproduction. Interestingly, although [PSI+] has a considerably higher promoting ability for spontaneous [SWI+] formation than [PIN+], its capability of promoting [SWI+] formation under the N-YFP overproduction condition is similar to that of [PIN+] (comparing Figure 4B to 5A). In this case, N-YFP overproduction gave rise to [SWI+] forming frequencies of about 6.7x, 39.3x and 27.5x higher than the spontaneous frequencies for [PSI+], [PIN+], and non-prion strains, respectively (Figure 5A).
Figure 5. Aggregation and [SWI+] de novo formation are promoted by overproduction of Swi1 PrD.
(A) Overproduction of Swi1 PrD promotes [SWI+] conversion. As described in the Experimental Procedures, with Swi1-NYFP overproduction (0.5% gal) or without (w/o gal), [SWI+] frequencies were plotted for the indicated 74D-694 strains carrying the PF139-URA3 reporter plasmid. (B and C) The indicated three 74D-694 strains, [PSI+], [PIN+], and non-prion, were co-transformed with p423GAL1-NmCherry and pCUP1-NMGFP (for [PSI+], [PSI+]_S; and non-prion, nonprion_S), or with p423GAL1-NmCherry and pCUP1-RNQ1GFP (for [PIN+], [PIN+]_R and non-prion_R). SC-leu-ura sucrose cultures of the transformants were supplemented with 0.02% (B) or 0.5% (C) galactose and 10 μM CuSO4. After incubation for 48 h, percentages of cells with all types of aggregation (total aggregation) or ring/ribbon-like only aggregation of Swi1-N-mCherry are shown. Statistical analysis for panel A-C was performed by T test (N.S, not significant; **, P<0.01; ***, P<0.001).
Interacting mechanisms in spontaneous and pre-existing prion-induced [SWI+] initiation
Protein misfolding and aggregation are critical events that lead to prionogenesis (Landreh et al., 2016). Upon overproduction prionogenic aggregations of Sup35 and Rnq1 often occur and appear to be ring/ribbon-shaped (Zhou et al., 2001, Mathur et al., 2010, Du et al., 2014). Ring/ribbon-like aggregation was also seen for other prion and prion-like proteins [18]. As shown in Figure 4C, similar to the de novo formation of [PSI+] and [PIN+], Swi1 also appears primarily as ring/ribbon-like aggregates in premature [SWI+] cells and became dot-like in mature [SWI+] cells. To understand how [PSI+], [PIN+], and Swi1 PrD overproduction promotes [SWI+] de novo formation, we further investigated aggregation and interactions of Swi1 with Sup35 and Rnq1 in the process of [SWI+] initiation and maturation. To do so, Swi1-N-mCherry and Sup35 NM-GFP were co-expressed from the GAL1 and CUP1 promoters, respectively, in a [PSI+] or non-prion 74D-694 strain; GAL1-Swi1-N-mCherry and CUP1-Rnq1-GFP were similarly co-expressed in a [PIN+] or non-prion 74D-694 strain. Galactose was added to a final concentration of 0.02% as a minimal amount to visualize N-mCherry aggregation in [SWI+] cells, and this very low galactose induction condition will be referred to as non-overproduction (or low N-mCh) hereafter. In parallel, conditions using 0.5% galactose are referred to as overproduction (or high N-mCh). Similarly, we observed that 10 μM CuSO4 is the lowest concentration necessary to see Sup35 NM-GFP aggregation in [PSI+] cells (Figure S5). Therefore, such a minimal CuSO4 concentration was used to visualize Sup35 NM-GFP and Rnq1-GFP aggregation in [PSI+] and [PIN+] cells without unnecessary overproduction.
In the non-overproduction condition (0.02% galactose), the presence of [PSI+] or [PIN+] significantly increased both total and ring/ribbon-like aggregation of N-mCherry (Figure 5B). When compared to the non-prion cells, [PSI+] and [PIN+] cells had 5.2x and 10.7x more total aggregation of N-mCherry, respectively (Figure 5B). These results are consistent with the result of a higher [SWI+] conversion rate in [PSI+] or [PIN+] cells than that of non-prion cells (Figure 4B and 5A). The ring/ribbon-like aggregates of N-Cherry accounted for ~88%, 77%, 97%, and 86% of the total N-mCherry aggregates in [PSI+] cells expressing Sup35 NMGFP, non-prion expressing Sup35 NMGFP, [PIN+] cells expressing Rnq1GFP, and non-prion cells expressing Rnq1GFP, respectively. These results show that Swi1 aggregates are predominantly ring/ribbon-like at the [SWI+] initiation stage. Remarkably, only a small fraction of the N-mCherry aggregates are heritable to become mature [SWI+] whereas > 90% of them were ultimately lost (calculated by dividing the percent frequency of [SWI+] isolates (Figure 5A) by the percent frequency of total aggregates (Figure 5B and 5C). This result suggests that even though the ring/ribbon-like Swi1 aggregation was attributable to [SWI+] formation, most of the initially formed ribbon-like Swi1 aggregates could not lead to [SWI+] formation but are rather off-pathway products. Interestingly, although [PSI+] cells had a nearly 10x higher spontaneous [SWI+] formation rate than that of [PIN+] cells, [PSI+] cells overall had lower ring/ribbon-like aggregation of N-mCherry (compare Figure 5A and 5B). These results suggest that the [PSI+]-induced Swi1 aggregation is more prionogenic than that induced by [PIN+].
As expected, Swi1 PrD overproduction (0.5% galactose) promoted the total and ring/ribbon-like aggregation of N-mCherry (compare Figure 5B and 5C). Importantly, Swi1 PrD overproduction-induced N-mCherry aggregation is predominantly ring/ribbon-like (Figure 5C), which was consistent with the capacity of Swi1 PrD in promoting [SWI+] formation (Figure 5A). Although the dominance of ring/ribbon-like aggregation is relatively low under overproduction conditions compared to that under non-overproduction (compare Figure 5B and 5C), there were about 28.7%, 11.9%, and 0.17-8.5% of [PSI+], [PIN+], and non-prion cells, respectively, containing N-mCherry ring/ribbon-like aggregates that eventually became [SWI+] isolates under Swi1 PrD overproduction, which is much greater than their non-overproduction controls ([PSI+], 5.9%; [PIN+], 0.75%; non-prion, 0.4-0.67%). The aforementioned values were calculated by dividing the percent frequency of [SWI+] isolates (Figure 5A) by the percent frequency of ribbon-like aggregates (Figure 5B and 5C). Noticeably, [PSI+] cells generated approximately 21x more [SWI+] than the non-prion control with Swi1 PrD overproduction though the two strains showed similar amount of ring/ribbon-like aggregation of N-mCherry (Figure 5A and 5C). This again suggests that the Swi1 ring/ribbon-like aggregates formed in [PSI+] cells were more prionogenic. In other words, the Swi1 ring/ribbon-like aggregates formed in the [PIN+] and nonprion strains were less prionogenic or a larger fraction of such aggregates were off-pathway products compared to those formed in the [PSI+] strain. Taken together, these data suggest that the promotion of [SWI+] conversion by a preexisting prion, such as [PSI+] or [PIN+], or an event of Swi1-PrD overproduction, is achieved by facilitating Swi1 aggregation, including the ring/ribbon-like aggregation.
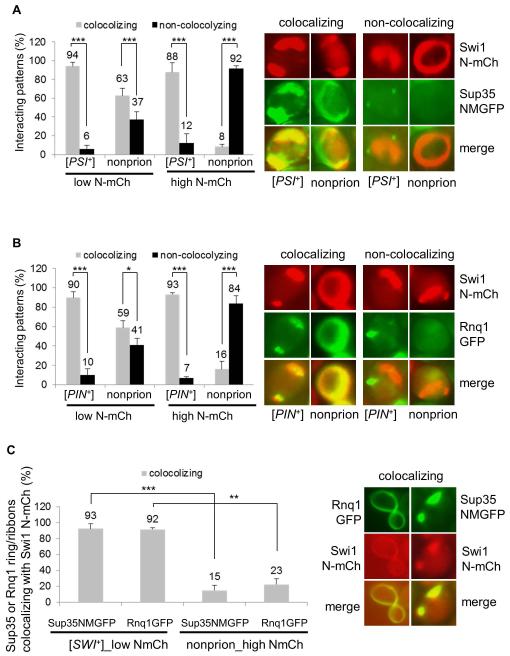
In the same experiments, we further assayed the interaction of Swi1 N-mCherry ring/ribbon-like aggregates with Sup35 NM-GFP and Rnq1-GFP under non-overproduction conditions and observed both colocalization (cross-seeding) and non-colocalization patterns for all tested strains (Figure 6A, 6B and data not shown). For instance, after 48 h of induction, about 94% and 90% of N-mCherry ribbon-like aggregates co-localized with Sup35 NM-GFP and Rnq1-GFP in [PSI+] and [PIN+] cells, respectively, in which the two preexisting prion aggregates were morphologically remodeled from dot-shaped to ring/ribbon/rod-like. Similarly, about 63% and 59% of Swi1 N-mCherry ring/ribbon-like aggregates co-localize with the signals of Sup35 NM-GFP and Rnq1-GFP in non-prion cells, respectively (Figure 6AC and 6B). Though colocalization signals of N-mCherry to Sup35 NM-GFP and Rnq1-GFP were prevalent in non-prion cells, the extremely low spontaneous aggregation frequency (Figure 5B) explains the low rate of [SWI+] conversion in this strain. These data suggest that under a Swi1 PrD non-overproduction condition, cross-seeding by Sup35 or Rnq1 is a major mechanism in promoting Swi1 aggregation and [SWI+] formation in all strains examined. However, a pre-existing [PSI+] or [PIN+] might provide a larger amount of ready-to-go templates for efficient cross-seeding of Swi1 to produce more on-pathway products to promote [SWI+] formation.
Figure 6. Interacting mechanisms of heterologous prion proteins, Sup35, Rnq1 and Swi1, in their prionogenesis.
(A) The indicated 74D-694 strains were co-transformed with p423GAL1-NmCherry and pCUP1-NMGFP. The ring/ribbon-like Swi1 N-mCherry aggregates (NmCh) that are supposed to be prionogenic in Swi1 prionogenesis were quantified for their interaction with Sup35-NMGFP (NMGFP) at low (low N-mCh, 0.02% galactose) or high (high N-mCh, 0.5% galactose) production of Swi1 PrD. Notably, 10 μM CuSO4 was supplemented, and both co-localizing and non-co-localizing patterns were observed after 48 h induction. The images shown on the right are representatives and plots are a summary of data from at least three independent experiments. (B) Similar to panel A, but the 74D-694 strains were co-transformed with p423GAL1-NmCherry and pCUP1-NMGFP, and the interaction is for Swi1 N-mCh and Rnq1-GFP. (C) A [SWI+] or a non-prion 74D-694 strain was co-transformed with p423GAL1-NmCherry and pCUP1-NMGFP, or with p423GAL1-NmCherry pCUP1-RNQ1GFP. Subsequently, the newly formed Sup35 NM-GFP (NMGFP) and Rnq1-GFP ring/ribbon-like aggregates were assayed for colocalization with Swi1 N-mCherry after 48 h of incubation. Low NmCh, 0.02% galactose; high NmCh, 0.5% galactose. In the experiment, 10 μM CuSO4 was supplied as minimum to visualize Sup35 NMGFP and Rnq1-GFP aggregation. Here shows a summary of the colocalizing frequencies (left) and representative images of the co-localization (right). Statistical analysis in the figure was performed by T test (*, P<0.05; **, P<0.01; ***, P<0.001).
Under the overproduction condition of Swi1 PrD, there were approximately 88% and 93% N-mCherry ring/ribbon-like aggregates co-localized with the morphologically remodeled Sup35 NM-GFP and Rnq1-GFP in [PSI+] and [PIN+] cells, respectively (Figure 6A and 6B), whereas there were only about 8% and 16% of the total N-mCherry ring/ribbon-like aggregates co-localized with Sup35 NM-GFP and Rnq1-GFP in non-prion cells, respectively (Figure 6A and 6B). These results suggest that in the event of Swi1 PrD-overproduction-promoted [SWI+] formation, cross-seeding is a major interacting mechanism between Swi1 and Sup35 or Rnq1 in [PSI+] and [PIN+] cells, but not in the examined non-prion cells under identical experimental conditions.
Sup35 and Rnq1 share similar mechanistic features in interacting with Swi1 in their prionogenesis processes
We showed previously that [SWI+] can promote [PSI+] and [PIN+] de novo formation (Du & Li, 2014). To examine the prionogenesis process of [PSI+] and [PIN+] by a preexisting [SWI+], we transformed a [SWI+] 74D-694 strain with p423GAL1-N-mCherry and pCUP1NMGFP or pCUP1RNQ1GFP. Upon addition of 10 μM CuSO4 and 0.02% galactose, an induction condition sufficient for visualizing prion protein aggregation without a significant overproduction, we examined the interactions of Swi1-N-mCherry with Sup35 NM-GFP and Rnq1-GFP. As shown in Figure 6C, under this induction condition, > 90% of Sup35 NM-GFP and Rnq1-GFP aggregates had ring/ribbon-like pattern and were co-localized with Swi1 N-mCherry aggregates, which were apparently remodeled from dot-shaped to ring/ribbon-like. These data indicate that cross-seeding is also a dominant interacting mechanism for [SWI+] to facilitate the conversion of [PSI+] and [PIN+] under non-overproduction conditions. With overproduction of Swi1 PrD (0.5% galactose) in pre-existing [SWI+] cells, the initially formed Sup35 NM-GFP and Rnq1-GFP ring/ribbon-like aggregates were also mostly colocalizing with morphologically remodeled Swi1 aggregates as we previously reported (Du & Li, 2014). In non-prion cells, when Swi1 N-mCherry was overproduced in the presence of 0.5% galactose, we found that Sup35 NMGFP and Rnq1-GFP ribbon-like aggregates rarely co-localize with Swi1 N-mCherry. In this case, only about 15% and 23% of the total ribbon-like aggregates of Sup35 NM-GFP and Rnq1-GFP colocalized with Swi1 N-mCherry, respectively (Figure 6C), suggesting that cross-seeding of Swi1 with Sup35 and/or Rnq1 rarely occurs in prionogenesis promoted merely by overproduction without a preexisting prion. Taken together, Sup35 and Rnq1 share similar interacting mechanistic features with Swi1 at their prion initiation stage.
Discussion
Based on our previous report that [SWI+] tightly regulates the expression of FLO genes that are essential for yeast multicellularity (Du et al., 2015), we have dissected the FLO11 promoter through a serial truncation analysis with the goals of understanding the FLO11 regulation by SWI/SNF and establishing a simple, faithful reporter system for [SWI+] study. As shown in Figure 1-2, we identified several truncated variants of the FLO11 promoter that can report the prion status of Swi1. Interestingly, we also uncovered a strain-specific requirement of Flo8 on some of the truncated FLO11 promoter variants. For example, PF19-URA3 can select for [swi−] cells using SC-ura media for both strains of 74D-694 and S288C but only in the presence of FLO8, whereas PF139-URA3 can select for [SWI+] cells using media supplemented with 5-FOA in the presence or absence of FLO8 for the 74D-694 strain. To be able to select for [SWI+] cells in the absence of a functional Flo8 in 74D-694 enabled us to estimate the spontaneous rate of [SWI+] formation by simply analyzing de novo formed 5-FOA+ isolates in any 74D-694 strain-derivatives without FLO8 repair. We show here that the spontaneous formation rate of [SWI+] is about 10−5 to 10−4 (Figure 4B), which is significantly higher than that of [PSI+], ~ 5 × 10−7 (Lancaster et al., 2010), but comparable to the observed rates for [ISP+] (Rogoza et al., 2010, Volkov et al., 2002), [MOT3+] (Alberti et al., 2009), and [URE3] (Brachmann et al., 2006, Wickner, 1994, Aigle & Lacroute, 1975), which can also be as high as 10−4. Previously, the rate of spontaneous formation of [RNQ+] ([PIN+]) was reported as ~ 2.96 × 10−6 (Huang et al., 2013). Interestingly, the protein determinants of [SWI+], [URE3], [MOT3+], and [ISP+] prions are low abundant transcriptional regulators with an asparagine (N)-rich PrD whereas the protein determinants of [PSI+] and [PIN+] prions are abundant cytoplasmic proteins with a PrD rich in both glutamine and asparagine (Q/N). It seems our newly determined spontaneous formation rate of [SWI+] is in agreement with an earlier prediction that PrDs with higher Q/N ratios seem to give rise to higher numbers of seeds (i.e. aggregates that can act as a template for the prion form) but lower prion formation rates whereas PrDs with lower Q/N ratios may give lower seed numbers but higher prion formation rates (Hines & Craig, 2011, Hines et al., 2011). However, the rate of a prion may be variable under different conditions, such as, in the absence of [PIN+], the [URE3] rate can be 10 fold lower (Bradley et al., 2002). Further research is needed to verify above observations and test the accuracy of the prediction.
Molecular chaperones are usually proteins that assist folding of nascent proteins and/or unfolding of mis-folded proteins. It has been shown that different prions can interact differently with molecular chaperones which are necessary for prion formation and propagation. For example, Hsp104 is needed for fragmentation of larger amyloid fibrils to produce prion seeds essential for prion propagation. Actually, Hsp104 overproduction can eliminate [PSI+] but has no effect on other prions (Crow & Li, 2011, Chernoff et al., 1995), and some N-rich prions, such as [URE3] and [SWI+], are sensitive to functional alterations of Hsp70-Ssa and its co-chaperones. The N-rich prions can be cured by Hsp70-Ssa overproduction, Sse1 overproduction, SSE1 deletion, or overproduction of Ydj1 or J-domain containing Hsp40 chaperones, whereas the Q/N-rich prions of [PSI+] and [PIN+] are generally resistant to such manipulations except weak [PSI+] variants that can be cured by SSE1 deletion (Fan et al., 2007, Hines et al., 2011, Kryndushkin & Wickner, 2007). In addition, [SWI+] is uniquely sensitive to Sis1 overproduction and YDJ1 deletion (Hines et al., 2011). Intriguingly, variants of the same prion can also interact with molecular chaperones dramatically different. For instance, Sse1 is required for propagating a weak [PSI+] variant but not for the propagation of a stronger variant under identical experimental conditions (Kryndushkin & Wickner, 2007, Fan et al., 2007). In this study, we also demonstrate a dramatic strain-specific interaction between [SWI+] and Sse1, the NEF of Hsp70-Ssa. We show that [SWI+] was eliminated by Sse1 overproduction in a 74D-694 strain but the same [SWI+] variant in a S288C strain was not (Figure 3B and 3C). Although it is possible that a different NEF other than Sse1, such as Sse2 or Fes1 (Kabani et al., 2002, Mukai et al., 1993), is required for interacting with Hsp70-Ssa for [SWI+] maintenance in the S288C strain, our observation that Sse1 is actively engaged in [SWI+] seed production in both strains of 74D-694 and S288C (Figure 3B-3E) indicates that it is not the case. The mechanism that underlies this strain-specific interaction of [SWI+] and Sse1 remains to be investigated.
Besides PrPSc, amyloid architecture is shared by a large number of other pathogenic proteins, including α-synuclein, β-amyloid, tau, and TDP-43, each of which is tightly linked to a fatal, devastating neurodegenerative disease (Aguzzi & O’Connor, 2010, Soto, 2003, Eisenberg & Jucker, 2012). As yeast prions are naturally occurring heritable amyloids, the budding yeast becomes a powerful model organism to study the behavior of amyloids. Although it is known that amyloidogenesis generally initiates from aberrant protein folding and aggregation, molecular events leading to amyloidosis remain poorly understood. In this study, we observed that in both cases of spontaneous [SWI+] formation in the absence of a known prion and Swi1-overproduction-facilitated [SWI+] conversion, Swi1 aggregates are predominantly ring/ribbon-like at the initiation stage of [SWI+] formation, which are then processed to become dot-like in mature [SWI+] cells (Figure 4C, 5B and 5C). Similar ribbon-to-dot transition events also take place in the prionogenesis of [PSI+], [PIN+], and a group of potential Q/N-rich prion candidates when overproduced (Alberti et al., 2009, Zhou et al., 2001, Du & Li, 2014). In the case of [PSI+] de novo formation, although both ring/ribbon-like and dot-like aggregation may appear upon Sup35 overproduction, only the former is usually prionogenic (Alberti et al., 2009, Derkatch et al., 2001, Zhou et al., 2001). In this study, we also observed that higher [SWI+] formation rates are associated with higher rates of ring/ribbon-like Swi1 aggregation (Figure 4B, 5B and 5C). The fact that both ring/ribbon-like and dot-like aggregates of [PSI+] share the same bundled fibrillar amyloid structure (Kawai-Noma et al., 2010, Tyedmers et al., 2010) suggests that this morphologic change of aggregation from ribbon-like to dot-like is unlikely to involve any significant changes in prion amyloid structures rather being a result of changes in their association partners.
We show here that the presence of a preexisting prion, [PSI+] or [PIN+], can facilitate the de novo formation of [SWI+]. Under Swi1-PrD overproduction conditions, although [PSI+] and non-prion strains had similar Swi1 N-mCherry aggregation frequency, the [PSI+] strain gave rise to a [SWI+] formation rate 21.5x higher than that of the non-prion strain (Figure 5A and 5C). In addition, in a [PIN+] strain, Swi1 PrD overproduction-induced N-mCherry aggregation (including total and ring/ribbon-like) and the [SWI+] conversion were significantly greater than in an isogenic non-prion strain (Figure 5A and 5C) – confirming that a preexisting prion can promote [SWI+] de novo formation. Interestingly, without overproduction, the N-mCherry ring/ribbon-like aggregation frequency is lower in [PSI+] cells than that in [PIN+] cells; however, the [SWI+] formation rate is opposite - higher in [PSI+] cells but lower in [PIN+] cells (Figure 4B and 5B). These data suggest that [PSI+] is a better inducer of [SWI+] than [PIN+]. Although the underlying mechanism of such preferable induction of [SWI+] by [PSI+] remains to be elucidated, we can speculate that perhaps the [PSI+] and [SWI+] prions are more structurally compatible than the [PIN+] and [SWI+] prions are and thus [PSI+] is a better cross-seeding template for [SWI+]. We also demonstrate here that a larger fraction of Swi1 aggregates are ring/ribbon-like in the non-overproduction condition than observed in N-mCherry overproduction conditions (compare Figure 5B and 5C), indicating that Swi1 PrD overproduction resulted in producing more non-prionogenic, off-pathway aggregation products.
Previous studies suggest that interactions of heterologous prion proteins mainly happen during the early phase of the prionogenesis process of [PSI+] and [PIN+] (Bagriantsev & Liebman, 2004, Derkatch et al., 2004, Du & Li, 2014). This is further confirmed by our finding that co-localization of Swi1 with Rnq1 or Sup35 mainly takes place at the initial prionogenic stage of [SWI+] formation (Figure 4C, 6A and 6B). We also show that with or without overproduction, the initially formed ring/ribbon-like aggregates of Swi1, Sup35, and Rnq1 predominantly overlap with the pre-existing prion facilitator even though some non-colocalizing aggregation can also occur simultaneously. In a non-prion strain, such a cross-seeding mechanism can also be prevalent without an overproduction event, but it rarely occurs when a prion protein is overproduced. The co-localization predominance of Swi1 with Sup35 or Rnq1 in a [PSI+] or [PIN+] strain is in agreement with an increase in [SWI+] conversion by the preexisting prion. These results suggest that the cross-seeding mechanism of using a pre-existing prion(s) as a template is an efficient means to facilitate the formation of another prion. The mechanistic events of yeast prion formation and interaction described here may have implications on etiologies of mammalian amyloidosis, which is also caused by protein misfolding and aggregation, similar to that of yeast prions (Chiti & Dobson, 2006, Prusiner, 2012, Soto, 2012). Many of these pathogenic proteins contain a Q/N-rich prion-like region (King et al., 2012). Notably, some of these pathogenic proteins often can co-aggregate (Gotz et al., 2001, Guo et al., 2013, Ono et al., 2012). It remains of great interest to understand how cross-seeding and other interaction mechanisms function in the aggregation and co-aggregation events of these pathogenic proteins. Thus the study of yeast prions can provide insights into our understanding of the general mechanisms involved in protein misfolding, aggregation, and protein misfolding diseases.
Experimental Procedures
Oligonucleotides, plasmids, and yeast strains that were used in this study are shown in Table S1, S2, and S3, respectively
Plasmid construction
Plasmids of p413TEF-NYFP, p413TEF-NQYFP, p413GAL1-NYFP, p413GAL1-NQYFP, p423GAL1-NYFP, p423GAL1-NQYFP, p423GPD-NYFP, p425GPD-NQYFP, p425GPD-NYFP, and p425GPD-SWI1YFP were constructed by sub-cloning the corresponding N-YFP, NQ-YFP, and SWI1-YFP fragment from p416TEF-NYFP, p416TEF-NQYFP, and p416TEF-SWI1-YFP into empty vectors of p413TEF, p413GAL1, p423GAL1, p423GPD and p425GPD through sites of SpeI/XhoI, respectively. A 714-bp mCherry PCR product was amplified with primer pair of mCherry-F/mCherry-R using p413TEF-SWI1mCherry as a template, and the PCR product was used to replace the YFP fragment of p413TEF-NYFP and p413TEF-NQYFP after digestion with XmaI and XhoI, generating p413TEF-NmCherry and p413TEF-NQmCherry, respectively. Further, NmCherry and NQmCherry from p413TEF-NmCherry and p413TEF-NQmCherry were sub-cloned into p423GAL1 through SpeI/XhoI sites, resulting in p423GAL1-NmCherry and p423GAL1-NQmCherry, respectively.
FLO11 promoter (PFLO11) was PCR-amplified with primer pair of FLO11S-F/1-3R from template DNA extracted from LY746. To dissect the FLO11 promoter, PF13 and PF16 were also PCR-amplified from LY746 genomic DNA using primer pairs of 1-3F/1-3R and 6F/1-3R, respectively. PF136 was generated by an overlapping PCR (bridge-PCR) – the primary PCR was done with primer pair of 6F/6R (with a 30-bp 3′ extension homologous to the 5′ sequence of PF13, PCR 6) using the FLO11 promoter PCR product as a template; and the secondary PCR was conducted with primer pair of 6F/1-3R using a mixture of PCR 6 and PF13 as template. PF1369 PCR product was also acquired by bridge-PCR – PCR was first performed with primer pair of 9-10F/9-10R2 (with a 30-bp 3′ extension homologous to the 5′ sequence of PCR 6, PCR 9-10) using the FLO11 promoter PCR product as a template followed by another round PCR that was carried out with primer pair of 9-10F/1-3R using PCR 9-10 and PF136 as templates. Similarly, PCR was done with a pair of primers,9-10F/9-10R1 (with a 30-bp 3′ extension homologous to the 5′ sequence of PF13, PCR 9-10a) using the full-length FLO11 promoter PCR product as a template. Further, PCR using the primer pair of 9-10F/1-3R and PCR 9-10a and PF13 as templates created PF139. The obtained PCR products of PFLO11, PF13, PF16, PF136, PF139, PF1369 were then digested with XmaI and SacII, and used to replace the SUC2-LEU2 promoter in p415SL-URA3, leading to plasmids of p415FLO11-URA3, p415F13-URA3, p415F16-URA3, p415F136-URA3, p415F139-URA3, and p415F1369-URA3, respectively. Moreover, ADH2 and HO promoters were PCR-amplified with primer pairs of ADH2-F/ADH2-R and HO-F/HO-R using the chromosomal DNA of YJW 426 as PCR template followed by another round of PCR using the first PCR products as templates, and the primer pairs of ADH2S-F/ADH2X-R and HOS-F/HOX-R. The resulting PCR products were then digested with XmaI and SacII, and used to replace the SUC2-LEU2 promoter in p415SL-URA3 to generate p415ADH2-URA3 and p415HO-URA3, respectively. Plasmid p425GPD-SSE1 was constructed by sub-cloning SSE1 from p426GPD-SSE1 into p425GPD at the XhoI and SpeI sites. All the newly created plasmids were confirmed by restriction digestion and DNA sequencing.
Yeast Strain engineering and cultivation
DY902 was acquired by growing LY746 on media containing 5 mM GdnHCl to cure [PIN+]. A successful curing was verified by lack of Rnq1-GFP aggregation upon transformation of the plasmid pCUP1-RNQ1GFP. Plasmid pCUP1-RNQ1GFP was then removed by counter selection on media containing 5-FOA. To replace the endogenous PFLO11-FLO11 with PF139-URA3, PF139-URA3 fragment was firstly PCR-amplified with primers of Flo11ex-F and Flo11URA3-R using p415F139-URA3 as template. The acquired PCR product was used as template for a secondary PCR with primer pair of Flo11ex-F/Flo11ex-R. The generated PCR product was then used to transform DY902, a wild-type non-prion BY4741 strain. The obtained Ura+ isolates are potential gene replacement mutants which are further verified by PCR with primers of Flo11ex-F and Flo11ex-R, leading to BY4741 flo8 flo11Δ:: PF139-URA3 (DY767). DY767 was repaired for FLO8 gene by introducing SalI-digested pRS303-FLO8 into the strain to create the strain DY759. A successful FLO8 repair was indicated by a phenotypic switch from Adh− (adhesion negative) His− Ura− to Adh+ His+ Ura+. [SWI+] was introduced into DY767 and DY759 by protein extract transformation as described later, leading to strains flo8 flo11Δ:: PF139-URA3 [SWI+] (LY740) and flo8::FLO8::HIS3 flo11Δ:: PF139-URA3 [SWI+] (LY744).
General yeast cultivation was performed as described previously (Fan et al., 2007, Park et al., 2006). LB, YPD, and SC (complete synthetic) media with certain amino acids dropout were used in this study otherwise specified. SC+5-FOA is a SC medium containing 0.1% 5-FOA, pH4.0. SC+raffinose+antimycin A has been previously described (Du et al., 2008). In galactose induction experiments, 2% sucrose was used as the carbon source. To induce the expression of a CUP1 promoter-controlled Sup35 NMGFP or Rnq1-GFP for examining their prion states, 50 μM CuSO4 was usually supplemented into a medium otherwise 10 μM CuSO4 was used to study the interaction of Sup35, Rnq1 and Rnq1 at their prion initiation stage. For Escherichia coli cultures, 100 μg/ml ampicillin, or 30 mg/ml kanamycin was typically supplemented.
[SWI+] transfer and PCR experiments
A protein extract transformation-based protocol for [SWI+] transfer was described previously (Du et al., 2010) with minimal modifications (Tanaka & Weissman, 2006). In this study, DY902 (with a chromosomal TEF1-RNQ1-CFP fusion gene) was used as a [SWI+] donor because the Rnq1 prion status can be conveniently monitored. In the prion transfer experiment, plasmid p415TEF-NQYFP was also co-transformed into the recipient strain to facilitate a successful selection of [SWI+]. A successful transfer of [SWI+] was verified by Swi1 NQ-YFP aggregation and Raf± that were curable by 5 mM GdnHCl.
In this study, PrimeSTAR HS DNA Polymerase (TaKaRa) was used in PCR to amplify DNA for cloning purposes. To extract DNA from yeast cells for PCR, yeast cells from cultures or colonies were simply boiled for 10 min in 20 mM NaOH, briefly cleared by centrifugation at 600 g for 2 min, and the crude supernatants used as PCR templates.
Adhesive growth and flocculation assays
Adhesive growth on SC plates was performed as described previously (Braus et al., 2003, Fichtner et al., 2007, Roberts & Fink, 1994) with minor modifications. In brief, fresh cells were streaked or spread on proper SC selective plates, incubated for 3 days at 30°C and shifted to room temperature for additional 3 days before performing the washing assay. The washing assay was performed in a water bowl by swirling the plate for different times before taking images. A vigorous wash was done using tap water with or without rubbing.
Flocculation assay was carried our similarly to that reported before (Kobayashi et al., 1996). Basically, after 2-3 days of growth in SC media, flocculating ability of cells was assessed based on appearance of visible cell aggregates. Saturated cell cultures were vigorously vortexed and images were taken as controls. Then images were taken in a time course to see appearance of cell pellets at the bottom of the tubes.
Fluorescence microscopy assays
In this study, GFP, YFP, CFP, and mCherry serve as fluorescence tags for various fusion proteins. These fluorescent proteins were either expressed ectopically from a plasmid or endogenously from a chromosomal tag. Methods for fluorescence microscopy were described previously [34]. For a constitutive promoter-driven expression, fresh colonies or log-phase cultures were used for microscopic observations. With an inducible promoter such as CUP1 or GAL1, CuSO4 or galactose was added in log-phase cultures to induce the expression of a fluorescent protein. In this study, a concentration gradient was tested in a time course for both kinds of inducers to determine their minimal concentrations in liquid media and/or on solid plates. For instance, 10 μM CuSO4 was used as a minimal concentration for Sup35 NM-GFP and Rnq1-GFP expression to ensure visibility of their aggregation in [PSI+] and [PIN+] cells but to avoid unnecessary overproduction of the protein, respectively. Similarly, 0.02% of galactose in a sucrose-based medium was used as minimum to detect Swi1 aggregation in cells without unnecessary overproduction for GAL1 promoter.
Assays for [SWI+] propagation and stability impacted by chaperone activities
A 74D-694 strain (LY722) or a BY4741 [SWI+] strain (LY742) was co-transformed with p415F139-URA3 and p423GPD-SSE1 or p2HGHSP104 to overproduce Sse1 or Hsp104, or p423GPD as an empty vector control. Transformants were grown for 24, 48 or 72 h in liquid culture then spread onto SC selective plates. Colonies were then replica-plated onto SC, SC+5-FOA and SC-uracil to test the status of [SWI+]; and their prion state was further verified by assaying their raffinose-ultilyzing ability and aggregation of Swi1-NQYFP. Curing frequencies were normalized by total colony numbers obtained on SC plates. To see the effect of Hsp104 functional knock down by GdnHCl, the vector-transformed prion strains (with p415F139-URA3) were cultured in SC+5 mM GdnHCl medium in parallel before spreading and replica-plating experiments as described above. Swi1 N-YFP aggregation was also assayed for randomly picked colonies from SC plates after the phenotypic curing tests upon removing the reporter plasmid and reintroducing p416TEF-NYFP. Similar assays were performed with another BY4741 strain (LY735) carrying chromosomal reporter of PFLO1-URA3. In addition, to see how overproduction of Sse1 or Hsp104 affects [SWI+]-curing by GdnHCl, LY735 carrying p425GPD-SSE1, p425GPD-HSP104, or p425GPD was cultured for 24, 48, and 72 h in 5 mM GdnHCl-containing SC selective medium, and assayed for stability of [SWI+] similarly.
[SWI+] de novo formation assays
To assay spontaneous [SWI+] conversion frequency in BY4741 cells, a non-prion flo8::FLO8::HIS3 flo1::PFLo1-URA3 strain (LY735) was cultured overnight in liquid YPD until reaching the stationary phase and spread onto SC-his and SC-his+5-FOA plates after proper dilution. Colonies that appeared on the 5-FOA plates were treated at least twice on SC-his+5 mM GdnHCl before replica-plating back to SC-his plates without uracil and SC-his+5-FOA plates to check their curability. Curable isolates were then transformed with p415TEF-NQYFP for aggregation assay to confirm the [SWI+] prion status. For 74D-694 strains, isogenic [PSI+], [PIN+], and non-prion strains carrying the reporter plasmid p415F139-URA3 and p413GAL1-NQYFP were cultured in SC-leu for 2 days until the stationary phase before spreading onto SC-leu and SC-leu + 5-FOA plates after proper dilution. Colonies that appeared on 5-FOA plates were replica-plated twice onto the same plates to stabilize the prion conformations before replica-plating onto SC-leu-his sucrose plates supplemented with 0.02% galactose to verify the presence of [SWI+] prion by NQ-YFP aggregation assay. Isolates with at least more than 30% cells containing NQ-YFP aggregates were further colony-purified and assayed again. Eventually, isolates with at least 60% of cells containing NQ-YFP aggregation were scored as [SWI+] isolates. To assay Swi1 PrD-promoted [SWI+] formation, 74D-694 strains with different prion backgrounds Swi1-NYFP were co-transformed with p423GAL1-NYFP and p415F139-URA3. Sucrose-based log-phase cultures were then supplemented with or without 0.5% galactose and induced for 24 h before spreading onto SC-leu-his and SC-leu-his+5-FOA plates. 5-FOA+ isolates were then tested for the aggregation of NYFP on 0.02% galactose sucrose plates to verify their prion status as aforementioned. Aggregation and prion phenotypes were then verified by their curability by GdnHCl. In all these experiments, [SWI+] rates were normalized with the total cells achieved on SC plates.
Supplementary Material
Acknowledgments
The authors thank Drs. S. Lindquist (Whitehead Institute for Biomedical Research, Department of Biology, Massachusetts Institute of Technology and Howard Hughes Medical Institute, and G. Braus (Georg August University Göttingen, Institute for Microbiology and Genetics) for providing us plasmids, strains, and other experimental materials. This work was supported by grants from the U.S. National Institutes of Health (R01GM110045) and U.S. National Science Foundation (MCB 1122135) to LL.
Footnotes
Three tables and five figures are included
Author Contributions
ZD, LL initiated the project and designed the experiments. ZD performed most of the experiments. DKG performed part of [SWI+] de novo formation experiments. XC conducted part of the [SWI+] propagation experiments. ZD and LL analyzed data. ZD, DKG, and LL wrote the manuscript.
References
- Aguzzi A, O’Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nature reviews. Drug discovery. 2010;9:237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- Aigle M, Lacroute F. Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Molecular & general genetics : MGG. 1975;136:327–335. doi: 10.1007/BF00341717. [DOI] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Zaidi T, Novak M, Grundke-Iqbal I, Iqbal K. Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6923–6928. doi: 10.1073/pnas.121119298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano Bom AP, Rangel LP, Costa DC, de Oliveira GA, Sanches D, Braga CA, Gava LM, Ramos CH, Cepeda AO, Stumbo AC, De Moura Gallo CV, Cordeiro Y, Silva JL. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: implications for cancer. The Journal of biological chemistry. 2012;287:28152–28162. doi: 10.1074/jbc.M112.340638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. The Journal of biological chemistry. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- Barrales RR, Jimenez J, Ibeas JI. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae. Genetics. 2008;178:145–156. doi: 10.1534/genetics.107.081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrales RR, Korber P, Jimenez J, Ibeas JI. Chromatin modulation at the FLO11 promoter of Saccharomyces cerevisiae by HDAC and Swi/Snf complexes. Genetics. 2012;191:791–803. doi: 10.1534/genetics.112.140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U, Southron JL, Stephens JL, Taylor GJ. Reverse genetic analysis of the glutathione metabolic pathway suggests a novel role of PHGPX and URE2 genes in aluminum resistance in Saccharomyces cerevisiae. Mol Genet Genomics. 2004;271:627–637. doi: 10.1007/s00438-004-1015-7. [DOI] [PubMed] [Google Scholar]
- Bester MC, Pretorius IS, Bauer FF. The regulation of Saccharomyces cerevisiae FLO gene expression and Ca2+ -dependent flocculation by Flo8p and Mss11p. Current genetics. 2006;49:375–383. doi: 10.1007/s00294-006-0068-z. [DOI] [PubMed] [Google Scholar]
- Biddick RK, Law GL, Chin KK, Young ET. The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes. The Journal of biological chemistry. 2008;283:33101–33109. doi: 10.1074/jbc.M805258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A, Toombs JA, Ross ED. Reporter assay systems for [URE3] detection and analysis. Methods. 2006 doi: 10.1016/j.ymeth.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion "strains" in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(Suppl 4):16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braus GH, Grundmann O, Bruckner S, Mosch HU. Amino acid starvation and Gcn4p regulate adhesive growth and FLO11 gene expression in Saccharomyces cerevisiae. Molecular biology of the cell. 2003;14:4272–4284. doi: 10.1091/mbc.E03-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, Byers JS, Jones S, Garcia DM, Bhullar B, Chang A, She R, Lee L, Fremin B, Lindquist S, Jarosz DF. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell. 2016;167:369–381. e312. doi: 10.1016/j.cell.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annual review of biochemistry. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Colby DW, Prusiner SB. Prions. Cold Spring Harb Perspect Biol. 2011;3:a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow ET, Li L. Newly identified prions in budding yeast, and their possible functions. Seminars in cell & developmental biology. 2011;22:452–459. doi: 10.1016/j.semcdb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Crow ET, Kang HS, Li L. Distinct subregions of Swi1 manifest striking differences in prion transmission and SWI/SNF function. Molecular and cellular biology. 2010;30:4644–4655. doi: 10.1128/MCB.00225-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Li L. Investigating the interactions of yeast prions: [SWI+], [PSI+], and [PIN+] Genetics. 2014;197:685–700. doi: 10.1534/genetics.114.163402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Valtierra S, Li L. An insight into the complex prion-prion interaction network in the budding yeast Saccharomyces cerevisiae. Prion. 2014;8:387–392. doi: 10.4161/19336896.2014.992274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhang Y, Li L. The Yeast Prion [SWI(+)] Abolishes Multicellular Growth by Triggering Conformational Changes of Multiple Regulators Required for Flocculin Gene Expression. Cell reports. 2015;13:2865–2878. doi: 10.1016/j.celrep.2015.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Park KW, Du Z, Morano KA, Li L. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics. 2007;177:1583–1593. doi: 10.1534/genetics.107.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner L, Schulze F, Braus GH. Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell-cell and cell-substrate adherence of S. cerevisiae S288c. Molecular microbiology. 2007;66:1276–1289. doi: 10.1111/j.1365-2958.2007.06014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Pennings S. Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. The EMBO journal. 2001;20:5219–5231. doi: 10.1093/emboj/20.18.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz J, Chen F, van Dorpe J, Nitsch RM. Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science. 2001;293:1491–1495. doi: 10.1126/science.1062097. [DOI] [PubMed] [Google Scholar]
- Guinan E, Jones GW. Influence of Hsp70 chaperone machinery on yeast prion propagation. Protein Pept Lett. 2009;16:583–586. doi: 10.2174/092986609788490168. [DOI] [PubMed] [Google Scholar]
- Guo JL, Covell DJ, Daniels JP, Iba M, Stieber A, Zhang B, Riddle DM, Kwong LK, Xu Y, Trojanowski JQ, Lee VM. Distinct alpha-synuclein strains differentially promote tau inclusions in neurons. Cell. 2013;154:103–117. doi: 10.1016/j.cell.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Wright JR, Alberti S, Lindquist S, Rexach M. Prion formation by a yeast GLFG nucleoporin. Prion. 2012;6:391–399. doi: 10.4161/pri.20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines JK, Craig EA. The sensitive [SWI (+)] prion: new perspectives on yeast prion diversity. Prion. 2011;5:164–168. doi: 10.4161/pri.5.3.16895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines JK, Li X, Du Z, Higurashi T, Li L, Craig EA. [SWI], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in Hsp70 chaperone system activity. PLoS genetics. 2011;7:e1001309. doi: 10.1371/journal.pgen.1001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DL, Lancaster AK, Lindquist S, Halfmann R. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell. 2013;153:153–165. doi: 10.1016/j.cell.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang VJ, Stein KC, True HL. Spontaneous variants of the [RNQ+] prion in yeast demonstrate the extensive conformational diversity possible with prion proteins. PloS one. 2013;8:e79582. doi: 10.1371/journal.pone.0079582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Lancaster AK, Brown JC, Lindquist S. An evolutionarily conserved prion-like element converts wild fungi from metabolic specialists to generalists. Cell. 2014;158:1072–1082. doi: 10.1016/j.cell.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Beckerich JM, Brodsky JL. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Molecular and cellular biology. 2002;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai-Noma S, Pack CG, Kojidani T, Asakawa H, Hiraoka Y, Kinjo M, Haraguchi T, Taguchi H, Hirata A. In vivo evidence for the fibrillar structures of Sup35 prions in yeast cells. The Journal of cell biology. 2010;190:223–231. doi: 10.1083/jcb.201002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Lee SB, Kang HS, Oh GT, Kim T. Two distinct domains of Flo8 activator mediates its role in transcriptional activation and the physical interaction with Mss11. Biochemical and biophysical research communications. 2014;449:202–207. doi: 10.1016/j.bbrc.2014.04.161. [DOI] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain research. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi O, Suda H, Ohtani T, Sone H. Molecular cloning and analysis of the dominant flocculation gene FLO8 from Saccharomyces cerevisiae. Molecular & general genetics : MGG. 1996;251:707–715. doi: 10.1007/BF02174120. [DOI] [PubMed] [Google Scholar]
- Kobayashi O, Yoshimoto H, Sone H. Analysis of the genes activated by the FLO8 gene in Saccharomyces cerevisiae. Current genetics. 1999;36:256–261. doi: 10.1007/s002940050498. [DOI] [PubMed] [Google Scholar]
- Kryndushkin D, Wickner RB. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Molecular biology of the cell. 2007;18:2149–2154. doi: 10.1091/mbc.E07-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184:393–400. doi: 10.1534/genetics.109.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landreh M, Sawaya MR, Hipp MS, Eisenberg DS, Wuthrich K, Hartl FU. The formation, function and regulation of amyloids: insights from structural biology. J Intern Med. 2016 doi: 10.1111/joim.12500. [DOI] [PubMed] [Google Scholar]
- Li F, Palecek SP. Identification of Candida albicans genes that induce Saccharomyces cerevisiae cell adhesion and morphogenesis. Biotechnology progress. 2005;21:1601–1609. doi: 10.1021/bp050236c. [DOI] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Kehm V, Carroll J, Zhang B, O’Brien P, Trojanowski JQ, Lee VM. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Li Y, Wang H, Cao F, Chen J. Antagonistic interplay of Swi1 and Tup1 on filamentous growth of Candida albicans. FEMS microbiology letters. 2008;285:233–241. doi: 10.1111/j.1574-6968.2008.01236.x. [DOI] [PubMed] [Google Scholar]
- Masison DC, Reidy M. Yeast prions are useful for studying protein chaperones and protein quality control. Prion. 2015;9:174–183. doi: 10.1080/19336896.2015.1027856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur V, Taneja V, Sun Y, Liebman SW. Analyzing the birth and propagation of two distinct prions, [PSI+] and [Het-s](y), in yeast. Molecular biology of the cell. 2010;21:1449–1461. doi: 10.1091/mbc.E09-11-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew D, Mitra RD. Transcription factor regulation and chromosome dynamics during pseudohyphal growth. Molecular biology of the cell. 2014;25:2669–2676. doi: 10.1091/mbc.E14-04-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai H, Kuno T, Tanaka H, Hirata D, Miyakawa T, Tanaka C. Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene. 1993;132:57–66. doi: 10.1016/0378-1119(93)90514-4. [DOI] [PubMed] [Google Scholar]
- Munch C, O’Brien J, Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3548–3553. doi: 10.1073/pnas.1017275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L, Carlson M. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics. 1987;115:247–253. doi: 10.1093/genetics/115.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Takahashi R, Ikeda T, Yamada M. Cross-seeding effects of amyloid beta-protein and alpha-synuclein. Journal of neurochemistry. 2012;122:883–890. doi: 10.1111/j.1471-4159.2012.07847.x. [DOI] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Park KW, Hahn JS, Fan Q, Thiele DJ, Li L. De Novo Appearance and "Strain" Formation of Yeast Prion [PSI+] Are Regulated by the Heat-Shock Transcription Factor. Genetics. 2006;173:35–47. doi: 10.1534/genetics.105.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Cell biology. A unifying role for prions in neurodegenerative diseases. Science. 2012;336:1511–1513. doi: 10.1126/science.1222951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes & development. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, Volkov K, Mironova L. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10573–10577. doi: 10.1073/pnas.1005949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova NV, Chernoff YO. Hsp104 and prion propagation. Protein Pept Lett. 2009;16:598–605. doi: 10.2174/092986609788490078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp S, Summers E, Lo HJ, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. The EMBO journal. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, Serpell LC, Miller TM, Grinberg LT, Seeley WW, Diamond MI. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron. 2014;82:1271–1288. doi: 10.1016/j.neuron.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Molecular and cellular biology. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Lindquist SL. [PSI+]: an epigenetic modulator of translation termination efficiency. Annual review of cell and developmental biology. 1999;15:661–703. doi: 10.1146/annurev.cellbio.15.1.661. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Molecular cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- Soto C. Transmissible proteins: expanding the prion heresy. Cell. 2012;149:968–977. doi: 10.1016/j.cell.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Sporn ZA, Hines JK. Hsp40 function in yeast prion propagation: Amyloid diversity necessitates chaperone functional complexity. Prion. 2015;9:80–89. doi: 10.1080/19336896.2015.1020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. The EMBO journal. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KC, True HL. Prion strains and amyloid polymorphism influence phenotypic variation. PLoS pathogens. 2014;10:e1004328. doi: 10.1371/journal.ppat.1004328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. Journal of molecular biology. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Weissman JS. An efficient protein transformation protocol for introducing prions into yeast. Methods Enzymol. 2006;412:185–200. doi: 10.1016/S0076-6879(06)12012-1. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS biology. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Volkov KV, Aksenova AY, Soom MJ, Osipov KV, Svitin AV, Kurischko C, Shkundina IS, Ter-Avanesyan MD, Inge-Vechtomov SG, Mironova LN. Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics. 2002;160:25–36. doi: 10.1093/genetics/160.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Bateman DA, Gorkovskiy A, Dayani Y, Bezsonov EE, Mukhamedova M. Yeast prions: proteins templating conformation and an anti-prion system. PLoS pathogens. 2015;11:e1004584. doi: 10.1371/journal.ppat.1004584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J, Tyedmers J, Bukau B, Mogk A. Chaperone networks in protein disaggregation and prion propagation. Journal of structural biology. 2012;179:152–160. doi: 10.1016/j.jsb.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI+] and [PIN+] Molecular microbiology. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.