ABSTRACT
By attaching infected erythrocytes to the vascular lining, Plasmodium falciparum parasites leave blood circulation and avoid splenic clearance. This sequestration is central to pathogenesis. Severe malaria is associated with parasites expressing an antigenically distinct P. falciparum erythrocyte membrane protein 1 (PfEMP1) subset mediating binding to endothelial receptors. Previous studies indicate that PfEMP1 adhesins with so-called CIDRα1 domains capable of binding endothelial protein C receptor (EPCR) constitute the PfEMP1 subset associated with severe pediatric malaria. To analyze the relative importance of different subtypes of CIDRα1 domains, we compared Pfemp1 transcript levels in children with severe malaria (including 9 fatal and 114 surviving cases), children hospitalized with uncomplicated malaria (n = 42), children with mild malaria not requiring hospitalization (n = 10), and children with parasitemia and no ongoing fever (n = 12). High levels of transcripts encoding EPCR-binding PfEMP1 were found in patients with symptomatic infections, and the abundance of these transcripts increased with disease severity. The compositions of CIDRα1 subtype transcripts varied markedly between patients, and none of the subtypes were dominant. Transcript-level analyses targeting other domain types indicated that subtypes of DBLβ or DBLζ domains might mediate binding phenomena that, in conjunction with EPCR binding, could contribute to pathogenesis. These observations strengthen the rationale for targeting the PfEMP1-EPCR interaction by vaccines and adjunctive therapies. Interventions should target EPCR binding of all CIDRα1 subtypes.
KEYWORDS: Plasmodium falciparum, antigenic variation, gene expression, malaria, PfEMP1
INTRODUCTION
Based on simple clinical observations, malaria patients can be divided into a smaller group with severe manifestations and a much larger group with uncomplicated disease (1). In areas of Africa with high to moderate rates of malaria transmission, severe malaria is seen almost only in children below 5 years of age (2). At the first entry point for health care, the majority of African children diagnosed with Plasmodium falciparum malaria suffer from uncomplicated febrile disease and are treated as outpatients. However, based on the initial assessment, the examining physician hospitalizes some patients with more manifest symptoms. These children are not well, but based on simple triage, they can be further divided into a large group with uncomplicated disease, who, upon correct treatment, will be very likely to survive the disease, and a smaller group with severe disease, where a proportion will die despite the administration of what is currently considered optimal care (3). It is estimated that around 10 million children suffer from severe malaria every year and that 5 to 10% of these children die (4). Even though most children in areas where malaria is endemic are expected to experience several bouts of malaria during childhood, only one to three of these bouts are likely to cause severe illness, and they usually occur early in life (5). This epidemiological picture has spurred the hypothesis that parasites that cause severe malaria are phenotypically different from those that cause uncomplicated disease and that children acquire immunity to severe malaria by mounting an antibody response to the parasite proteins that convey the phenotype associated with severe outcomes (6–9).
P. falciparum parasites depend on evading splenic destruction by anchoring infected erythrocytes to endothelial cells. The sequestration of parasites in host capillaries drives malaria pathogenesis, and immunoepidemiological studies have indicated that an antigenically restricted subset of the polymorphic P. falciparum erythrocyte membrane protein 1 (PfEMP1) adhesins is associated with life-threatening infections experienced during childhood in regions where malaria is endemic (7, 8). PfEMP1 adhesins are expressed on the surface of erythrocytes infected with late-blood-stage parasites (trophozoites), where they mediate attachment to receptors on the vascular lining (10), allowing infected cells to avoid circulation and passage though the spleen, where they are destroyed.
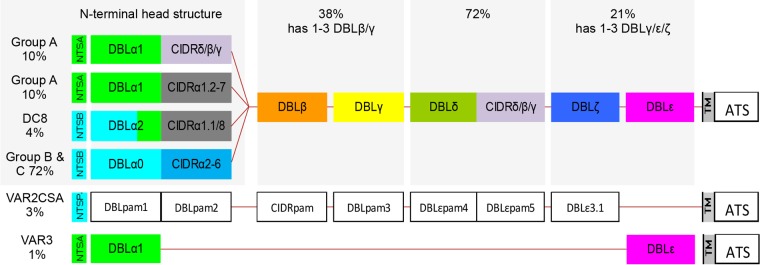
PfEMP1 adhesins are encoded by var genes. Each parasite genome harbors about 60 variants (11–13), but each parasite expresses only one var gene at a time (14). Although PfEMP1 sequences are extremely diverse, their domain architectures are highly organized, and all parasites carry similar PfEMP1 repertoires, which appear to bestow on all parasites the same fundamental repertoire of human receptor specificities (15). The large multidomain PfEMP1 adhesins consist of 2 to 9 Duffy-binding-like (DBL) and cysteine-rich interdomain region (CIDR) domains, which, based on sequence similarity, can be further subdivided into different groups (16, 17). A single distinct group of PfEMP1 proteins, VAR2CSA, binds parasites to receptors in the placenta and is a known virulence factor for pregnancy malaria (18, 19). It has proven more challenging to characterize the PfEMP1 types or traits linked to parasites causing severe pediatric malaria. Early studies implicated the so-called group A and B var gene variants (Fig. 1), which are separated from each other by chromosomal orientation and the encoding of distinct N-terminal domains (group A DBLα1 versus group B DBLα0/2 domains) (20–24). Subsequent studies specified that severe malaria is associated with parasites expressing the group B var gene subset, encoding domain cassette 8 (DC8) (has DBLα2 domains) and group A var genes, including those encoding DC13 (25, 26). These PfEMP1 types were found to share a binding phenotype (27), as both DC8 and DC13 bind endothelial protein C receptor (EPCR) via their CIDRα1.1 and CIDRα1.4 domains, respectively. Recently, the crystal structure of the CIDRα1-EPCR interaction was solved, and a more precise description of which CIDRα1 subclasses bind EPCR was obtained (28), showing that most CIDRα1 domain variants bind EPCR. Jespersen et al. (29), who reported the near-full-length sequence annotation of var transcripts in 44 patients, found that CIDRα1 was the only domain type that was common between different PfEMP1 variants expressed by severe malaria isolates. Parasites expressing DC13 PfEMP1 were recently shown to bind both EPCR and intercellular adhesion molecule 1 (ICAM-1) (30), suggesting that other PfEMP1 traits such as binding to ICAM-1 in combination with EPCR binding could increase the risk of developing specific syndromes. Similarly, PfEMP1 with C-terminal DC6 containing DBLγ14-DBLζ5-DBLε4 domains (31) was linked to the hospitalization of P. falciparum-infected Indian adults. Those studies included relatively few patients, and it is possible that var type quantification by sequence tags or by primers with limited sequence variant coverage led to the overestimation of these PfEMP1 traits or the underestimation of others.
FIG 1.
Schematic representation of typical PfEMP1 domain compositions. The N-terminal “head structure” confers mutually exclusive receptor-binding phenotypes: CSA (VAR2CSA), EPCR (CIDRα1), CD36 (CIDRα2-6), and as-yet-unknown phenotypes (CIDRδ/β/γ andVAR3). Group A PfEMP1 adhesins are encoded by subtelomeric genes transcribed toward the telomere. Group A PfEMP1 adhesins include both EPCR-binding and non-EPCR-binding PfEMP1 adhesins. Group B PfEMP1 adhesins are encoded by telomeric genes transcribed toward the centromere and include PfEMP1 adhesins that bind EPCR (DC8) and CD36. Group C PfEMP1 adhesins bind CD36 and are encoded by centromeric genes. PfEMP1 adhesins typically have two to six domains C terminal to the head structure. The subclass compositions of these domains vary but in general follow the depicted order. The C-terminal domain subclass composition is generally unrelated to the division of the N-terminal head structure, although some DBLβ sequences occur only in either group A or group B PfEMP1 adhesins. Most group A and DC8 group B PfEMP1 adhesins have four or more domains, whereas about two-thirds of the remaining group B and the group C PfEMP1 adhesins have only DBLδ-CIDR tandem domains. (See reference 17 for a detailed description of PfEMP1 diversity and domain architecture.) The estimated proportions of var genes of each group or with the indicated domain classes are given. TM, transmembrane domain; ATS, intracelluar acidic terminal segment.
Here, we show by quantitative PCR (qPCR) and by using a new set of var type-specific primers that is more comprehensive than those used previously (25, 26) that parasites from Tanzanian children hospitalized with malaria and diagnosed with severe malarial anemia, cerebral malaria, or no severe complications (uncomplicated malaria) were all characterized by high transcript levels of var genes encoding DC8 and the subset of group A var genes encoding EPCR-binding PfEMP1. Furthermore, in a few cases, severe disease was associated with increased levels of transcripts encoding specific subsets of DBLβ or DBLζ domains.
RESULTS
Clinical characteristics of patients.
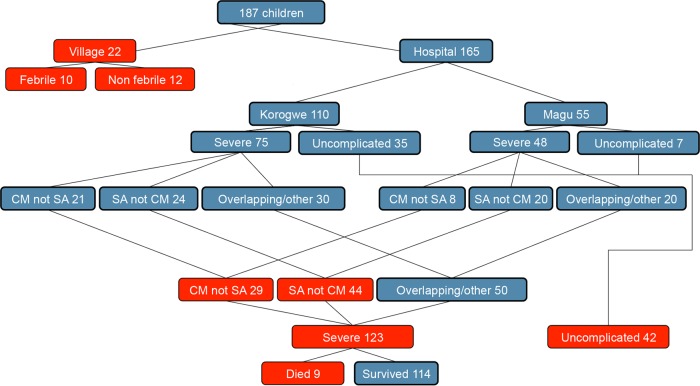
One hundred sixty-five Tanzanian children admitted to the hospital with P. falciparum malaria and 22 P. falciparum-positive children identified by a positive rapid diagnostic test during cross-sectional village surveys were included in this study. Children were classified as having severe malaria if the hemoglobin (Hb) level was <5 g/dl, the Blantyre coma score was <3, there were clinical signs of respiratory distress, or parasitemia was >200,000 parasites/μl (Table 1 and Fig. 2). Children admitted to the hospital with an Hb level of >8.0 g/dl, a Blantyre coma score of 5, and parasitemia of <200,000 parasites/μl were categorized as having uncomplicated malaria. Children with severe malaria were divided into nonoverlapping groups of those with a Blantyre coma score of <3 and a Hb level of >5 g/dl (cerebral malaria), those with a Hb level of <5 g/dl and a Blantyre coma score of 5 (severe anemia), and those with overlapping symptomatology or severity signs other than coma or low Hb levels (Fig. 2). Village children were divided into febrile (temperature of >37.5°C) and afebrile children. All children received prompt treatment and care, and as a result, the mortality rates were 0% among the children from the village and hospitalized children with uncomplicated malaria and 7.3% among those with severe disease (Table 1). The hospital studies were conducted at Magu District Hospital on the shore of Lake Victoria and Korogwe District Hospital, 100 km from the Indian Ocean coastline. These sites are in separate ecological zones 650 km apart.
TABLE 1.
Clinical characteristics of patients
| Location of sample collection | Malaria outcome | No. of patients | Mean age (yr) (SD) | % female patients | Median temp (°C) (p25–p75)a | Mean Hb level (g/dl) (SD) | Mean Blantyre score (SD) | Mean lactate level (mmol/liter) (min–max) | Mean glucose level (mmol/liter) (SD) | Mortality |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean (SD) | % | ||||||||||
| Village | No fever | 12 | 6.1 (3.0) | 50 | 36.8 (36.4–37.0) | 0.3 (0.2–1.6) | 9.8 (2.0) | 5.0 (5.0–5.0) | NDb | ND | 0.0 |
| Village | Fever | 10 | 6.1 (4.5) | 50 | 38.9 (38.5–39.0) | 1.0 (0.5–5.8) | 10.8 (2.3) | 5.0 (5.0–5.0) | ND | ND | 0.0 |
| Hospital | Uncomplicated | 42 | 3.1 (1.6) | 61 | 38.1 (37.5–39.5) | 10.0 (4.5–45.0) | 10.0 (2.0) | 5.0 (5.0–5.0) | 3.2 (1.7) | 5.8 (1.7) | 0.0 |
| Hospital | Severe | 123 | 2.8 (1.8) | 51 | 38.4 (37.6–39.4) | 272.0 (32.5–515.2) | 6.0 (2.3) | 3.6 (0.0–5.0) | 5.0 (3.4) | 6.3 (2.3) | 7.3 |
| Hospital | Severe anemia | 44 | 2.8 (1.9) | 59 | 38.0 (37.2–38.6) | 40.6 (9.2–330.0) | 4.0 (0.6) | 4.7 (3.0–5.0) | 4.9 (3.3) | 6.8 (2.1) | 4.5 |
| Hospital | Cerebral malaria | 29 | 3.1 (1.6) | 38 | 39.0 (38.0–39.8) | 210.0 (80.0–800.0) | 6.8 (2.5) | 1.1 (0.0–2.0) | 6.5 (4.0) | 5.5 (2.4) | 6.9 |
| Hospital | Died | 9 | 2.7 (1.4) | 33 | 39.0 (37.8–39.2) | 130.0 (31.3–174.4) | 5.7 (2.3) | 1.9 (0.0–4.0) | 9.0 (5.5) | 5.3 (2.9) | 100.0 |
p25–p75, 25th to 75th percentiles.
ND, not determined.
FIG 2.
Malaria patients enrolled in the study. Samples were collected in villages or at district hospitals in Korogwe or Magu. Children were categorized into nonoverlapping groups. At the hospital, this included patients with cerebral malaria (CM), those with severe anemia (SA), and those with overlapping syndromes and/or other signs linked to severity (respiratory distress and hyperparasitemia). Boxes in red are categories presented in Tables 1 and 2.
Malaria patients have high levels of transcripts encoding PfEMP1 adhesins predicted to bind EPCR, and increasing levels are associated with increased disease severity.
The median transcript level reported for each primer set and the summarized transcript levels for combinations of primer sets targeting same main domain class (e.g., EPCR-binding CIDRα1) were stratified according to clinical presentation (Table 2). The most striking differences were found for the levels of transcripts encoding CIDRα1 domains, which increased with disease severity (median levels of transcripts [transcript units, Tu] for CIDRα1 of all subtypes combined were 1, 12, 49, and 62 for village malaria without fever, village malaria with fever, uncomplicated hospital malaria, and severe hospital malaria, respectively; P = 0.0001 by a Kruskal-Wallis rank test). For all CIDRα1 primers but CIDRα1.5 and CIDRα1.6, there was a statistically significant association between disease severity and transcript level (P < 0.001 for all comparisons by a Kruskal-Wallis rank test). There was considerable heterogeneity within groups, and in comparisons between two disease outcomes (Table 2; see also Fig. S3 in the supplemental material), the difference in transcript levels reached statistical significance for only some of the CIDRα1 subclasses (e.g., the level of CIDRα1.1 was higher for village malaria with fever than for village malaria without fever [P = 0.021], the level of CIDRα1.1 was higher for severe hospital malaria than for uncomplicated hospital malaria [P = 0.004], and the level of CIDRα1.4/6 was higher for uncomplicated hospital malaria than for village malaria with fever [P = 0.012]).
TABLE 2.
Median var transcript levels (Tu and 10th and 90th percentiles) by patient groupa
| Primer(s)b | Target PfEMP1 |
Tu (10th–90th percentiles) for village |
P value for village with vs without feverc | Tu (10th–90th percentiles) for hospital, UM (n = 42) | P value for village fever vs UM | Tu (10th–90th percentiles) for hospital, SM (n = 123) | P value for UM vs SM | Tu (10th–90th percentiles) for hospital |
P value for SA vs CM | Tu (10th–90th percentiles) for hospital, died (n = 9) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group(s)d | Bindinge | No fever (n = 12) | With fever (n = 10) | SA (n = 44) | CM (n = 29) | ||||||||
| CIDRa1.all | A and BA | EPCR | 1.0 (1.0–29.2) | 11.8 (1.0–58.0) | 49.1 (7.1–134.0) | 0.0013 | 61.5 (7.6–288.1) | 57.5 (11.2–314.4) | 60.9 (3.4–210.2) | 120.5 (1.0–350.7) | |||
| CIDRa1.DC8 | BA | EPCR | 1.0 (1.0–7.2) | 9.9 (1.0–15.9) | 0.047 | 12.6 (1.0–90.8) | 21.7 (2.3–183.6) | 0.007 | 20.2 (2.3–184.2) | 21.1 (2.3–191.5) | 21.1 (1.0–194.3) | ||
| CIDRa1.1 | BA | EPCR | 1.0 (1.0–1.0) | 4.3 (1.0–14.2) | 0.021 | 1.3 (1.0–44.6) | 12.9 (1.0–99.2) | 0.004 | 11.8 (1.0–102.2) | 9.7 (1.0–121.9) | 12.9 (1.0–94.5) | ||
| CIDRa1.8 | BA | EPCR | 1.0 (1.0–1.0) | 1.0 (1.0–12.0) | 1.6 (1.0–30.4) | 5.6 (1.0–90.1) | 4.1 (1.0–82.4) | 6.7 (1.0–106.0) | 8.2 (1.0–124.1) | ||||
| CIDRa1.A | A | EPCR | 1.0 (1.0–7.5) | 2.4 (1.0–45.3) | 23.3 (1.0–91.8) | 0.037 | 25.5 (2.5–105.0) | 18.7 (3.1–89.0) | 18.8 (1.8–104.2) | 40.5 (1.0–246.2) | |||
| CIDRa1.4/6a | A | EPCR | 1.0 (1.0–1.3) | 1.0 (1.0–2.1) | 2.8 (1.0–22.7) | 0.012 | 4.5 (1.0–41.8) | 3.4 (1.0–35.5) | 4.1 (1.0–36.2) | 1.5 (1.0–143.0) | |||
| CIDRa1.5 | A | EPCR | 1.0 (1.0–1.0) | 1.0 (1.0–1.0) | 1.0 (1.0–32.8) | 0.03 | 1.3 (1.0–24.9) | 1.0 (1.0–17.6) | 2.3 (1.0–15.5) | 1.0 (1.0–62.1) | |||
| CIDRa1.6b | A | EPCR | 1.0 (1.0–1.0) | 1.0 (1.0–2.0) | 1.0 (1.0–8.9) | 1.0 (1.0–9.3) | 1.0 (1.0–10.2) | 1.0 (1.0–11.0) | 1.0 (1.0–17.9) | ||||
| CIDRa1.7 | A | EPCR | 1.0 (1.0–1.2) | 2.1 (1.0–43.6) | 9.4 (1.0–57.1) | 8.0 (1.0–51.3) | 8.9 (1.0–40.6) | 9.1 (1.0–51.3) | 10.1 (1.0–176.1) | ||||
| CIDRd | A | 1.0 (1.0–1.0) | 1.4 (1.0–4.4) | 0.0025 | 1.0 (1.0–30.5) | 1.5 (1.0–17.5) | 1.4 (1.0–18.6) | 1.6 (1.0–35.3) | 3.6 (1.0–6.8) | ||||
| CIDRg3.1 | A | 1.0 (1.0–1.0) | 1.0 (1.0–2.3) | 1.0 (1.0–2.7) | 1.0 (1.0–2.0) | 1.0 (1.0–1.5) | 1.0 (1.0–1.0) | 1.0 (1.0–4.0) | |||||
| CIDRa3.1/2 | A | CD36 | 1.0 (1.0–1.0) | 1.0 (1.0–1.6) | 0.046 | 1.0 (1.0–5.0) | 1.0 (1.0–5.1) | 1.1 (1.0–6.5) | 1.0 (1.0–3.5) | 1.3 (1.0–18.6) | |||
| DBLa1all | A | 1.0 (1.0–20.3) | 11.3 (1.0–120.1) | 44.6 (3.6–138.9) | 0.027 | 53.6 (4.7–155.4) | 39.5 (1.0–146.5) | 65.2 (10.2–194.3) | 120.3 (4.7–284.0) | ||||
| DBLa2/1.1/2/4/7 | A | 3.1 (2.4–31.8) | 24.5 (2.4–153.5) | 0.048 | 85.6 (6.2–169.5) | 0.037 | 78.4 (21.7–297.1) | 74.0 (21.7–303.4) | 65.3 (5.9–229.9) | 124.7 (8.5–410.1) | |||
| DBLa1.5/6/8 | A | 1.0 (1.0–3.1) | 12.3 (1.0–17.9) | 0.047 | 17.5 (1.0–112.6) | 27.6 (1.0–109.9) | 16.5 (1.0–216.4) | 20.4 (1.3–135.3) | 29.4 (1.0–166.9) | ||||
| DBLb1/3-1 | A | (ICAM1) | 1.0 (1.0–9.4) | 1.8 (1.1–27.4) | 1.0 (1.0–11.0) | 1.0 (1.0–11.7) | 1.0 (1.0–4.3) | 1.0 (1.0–39.8) | 0.039 | 1.0 (1.0–5.8) | |||
| DBLb1/3-2 | A | (ICAM1) | 1.0 (1.0–1.0) | 1.1 (1.0–45.8) | 0.0071 | 8.3 (1.0–71.5) | 6.4 (1.0–61.8) | 6.1 (1.0–70.3) | 3.4 (1.0–34.5) | 22.0 (1.0–83.9) | |||
| DBLb5 | B | ICAM1 | 1.0 (1.0–1.0) | 1.0 (1.0–1.0) | 1.0 (1.0–1.6) | 1.0 (1.0–8.3) | 1.0 (1.0–2.1) | 1.7 (1.0–18.1) | 1.0 (1.0–2.0) | ||||
| DBLz_all | 1.1 (1.0–18.0) | 8.3 (1.8–77.3) | 0.01 | 12.7 (2.3–64.5) | 20.4 (3.9–96.1) | 21.4 (3.6–133.4) | 22.1 (3.8–110.8) | 19.5 (1.0–84.9) | |||||
| DBLz2a | 1 (1–1.1) | 1 (1–1) | 1.0 (1.0–5.6) | 1.8 (1.0–14.4) | 0.0017 | 1.7 (1.0–20.2) | 2.4 (1.0–40.1) | 4.4 (1.0–6.1) | |||||
| DBLz3 | 2.5 (1–63.8) | 1 (1–1) | 0.0009 | 1.0 (1.0–4.7) | 0.0072 | 1.7 (1.0–9.3) | 0.0013 | 1.0 (1.0–9.7) | 2.0 (1.0–6.1) | 2.2 (1.0–37.8) | |||
| DBLe_all | 1.0 (1.0–6.7) | 2.0 (1.0–57.6) | 6.9 (1.0–79.7) | 0.027 | 7.7 (1.0–75.1) | 10.2 (1.0–128.3) | 4.2 (1.0–101.6) | 5.5 (1.0–75.1) | |||||
| DC5 | A | (PECAM1) | 1.0 (1.0–5.9) | 1.4 (1.0–10.5) | 3.6 (1.0–32.8) | 4.0 (1.0–37.4) | 2.9 (1.0–26.1) | 2.7 (1.0–42.6) | 2.7 (1.0–10.6) | ||||
| var2csa | E | CSA | 1.0 (1.0–23.4) | 12.4 (1.8–77.9) | 5.8 (1.0–40.2) | 3.4 (1.0–28.7) | 3.4 (1.0–23.8) | 3.3 (1.0–21.4) | 1.7 (1.0–24.6) | ||||
| var3 | A | 1.0 (1.0–1.0) | 1.0 (1.0–5.3) | 1.2 (1.0–14.6) | 1.1 (1.0–29.7) | 2.1 (1.0–61.9) | 1.0 (1.0–36.1) | 1.1 (1.0–78.2) | |||||
A Tu value of 32 corresponds to the mean transcript level of the two control genes. UM, uncomplicated malaria; SM, severe malaria; SA, severe anemia; CM, cerebral malaria.
Boldface type indicates that the Tu values are summarized levels reported by several primers. CIDRa1.DC8 summarizes CIDR1.1 and -1.8; CIDRa1.A summarizes CIDR1.4 to -7; CIDRa1.all summarizes CIDRa1.DC8 and CIDRa1.A. Italic type indicates that the primer detects transcripts predicted to encode EPCR-binding PfEMP1 domains.
P values were calculated by using the Wilcoxon rank sum test. Only P values of <0.05 are shown.
Indicates which (if any) PfEMP1 group is associated with the targeted sequence.
Indicates the predicted human receptor binding specificity (if any) of the domain type encoded by the targeted sequence. The receptors ICAM-1 and PECAM1 are indicated in parentheses, as evidence of the indicated domains conferring binding to these receptors is sporadic and thus not clearly linked to the domain type.
The assay allowed only crude comparisons between transcript levels within patients or patient groups. However, the data indicate that within hospitalized patient groups, the CIDRα1-encoding transcripts were generally found at higher levels than were transcripts for the other main group A var types characterized by encoding CIDRδ domains. In hospitalized patients, the abundance of transcripts encoding EPCR-binding domains of DC8 (i.e., CIDRα1.1 and CIDRα1.8 [“CIDRα1.DC8”]) was roughly similar to the abundance of transcripts encoding group A EPCR-binding domains (CIDRα1.4 to -7 [CIDRα1.A]) (Table 2). var2csa was found at high levels in 25.1% of the patients across all patient groups but with no statistically significant relation to severity.
The subclass of the N-terminal DBLα domain is predictive of the adjacent CIDR class. For this reason, primers targeting loci encoding the 3′ end of DBLα domains can to a certain degree be used to infer the expression level of PfEMP1 with specific receptor-binding phenotypes. The “DBLa2/1.1/2/4/7” primer set reports transcripts encoding CIDRα1 domains predicted to bind EPCR (both DC8 and group A genes), whereas the “DBLa1.5/6/8” primer set primarily detects transcripts of group A genes encoding N-terminal CIDRβ/γ/δ domains predicted not to bind EPCR. The DBLα1all primer set reports transcripts of most group A genes encoding N-terminal CIDRα1/β/γ/δ domains predicted not to bind CD36. Transcript levels reported by these DBLα domain primer sets confirmed the results obtained with the CIDR primers for hospitalized children, showing a higher abundance of transcripts encoding EPCR-binding PfEMP1 than of transcripts encoding other group A DBLα1.5/6/8 domains.
var expression patterns in patients with respect to mid- and C-terminal PfEMP1 domain classes.
The DBLβ, -γ, and -δ domain classes each cover broad sequence variation and include few well-defined subclasses, which makes it difficult to design subclass-specific primers that retain target coverage. However, a few distinct loci were identified in a subset of sequences encoding the DBLβ1/3 and DBLβ5 domains, including some sequence variants previously associated with ICAM-1 binding (30, 32, 33). Primers targeting these loci did not report significantly different levels between patients with severe and those with uncomplicated malaria.
The only C-terminal sequence trait unique to group A PfEMP1 is DC5. DC5 is found in about 15% of group A PfEMP1 adhesins and was previously associated with parasites binding to platelet endothelial cell adhesion molecule 1 (PECAM1) (34). The median transcript abundance was low in all patient groups.
Twelve primer sets targeting all different DBLζ subclasses and most DBLε subclasses not associated with VAR1, VAR2CSA, or VAR3 were applied (Table 2; see also Fig. S1 in the supplemental material). The DBLζ and DBLε domains are most often found together and form different domain cassettes (DC1 to -3, -6, -7, and -9 to -12). When transcript levels were summarized for all DBLζ primers, median transcript levels increased with increasing severity for the patient group. However, most DBLζ subclass primers reported low transcript levels, which did not differ significantly between patient categories (data not shown). Although the median levels were low, a small proportion of patients with severe outcomes had transcripts targeted by the DBLz2a or DBLz3 primers, resulting in a significant difference in the medial DBLz2a or DBLz3 levels between children with severe malaria and those with uncomplicated malaria (Table 2 and Fig. S3). The DBLz3 primers reported fewer transcripts in villagers with fever than in those without fever (P = 0.0009). When transcript levels were summarized for all DBLε primers, the median transcript level was higher in hospitalized children than in children from the village.
The var transcript profiles for patients with severe anemia or cerebral malaria and for those who died are largely similar.
Transcript profiles were compared between patients with severe anemia and those with cerebral malaria (Table 2). For nearly all primers, the median transcript levels were comparable between the two groups of patients; the only exception was a higher level of transcripts reported for cerebral malaria patients with the DBLb1/3-1 primer set (P = 0.039). The median transcript level was low in both patient groups, and the observed difference reflected that ∼25% of the patients with cerebral malaria had high levels of these transcripts (see Fig. S4 and Table S1 in the supplemental material).
There was no statistically significant difference between the transcript levels in those who died and those who survived a complicated malaria episode. Among those who died, the median transcript levels of genes encoding EPCR-binding PfEMP1 and particularly those belonging to group A were high, but there was considerable variation in transcript levels within this patient group (Table 2).
CIDRα1 transcript profiles differ considerably between patients but not according to patient group.
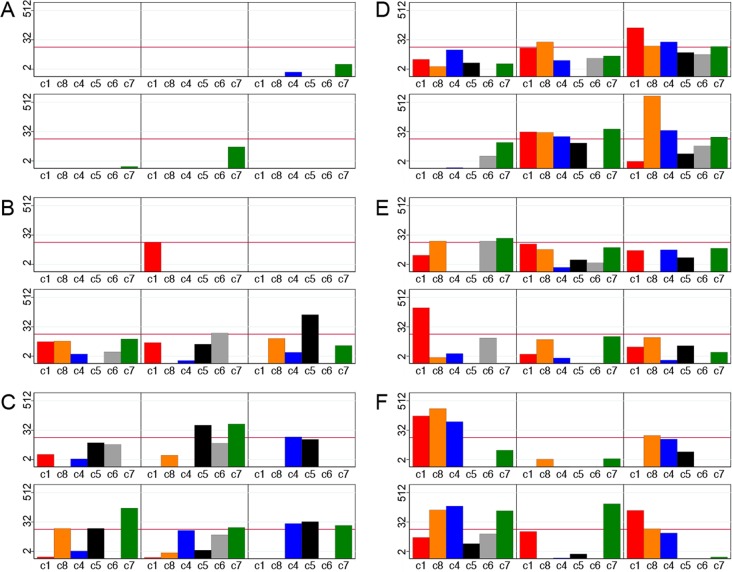
The primer sets used to measure transcripts encoding the six subclasses of EPCR-binding domains (CIDRα1.1 and CIDRα1.4-8) had similar good coverage and specificity (see Fig. S1 in the supplemental material); therefore, the transcript abundances reported with these primers within a patient should be compared with caution. The transcript patterns varied markedly between patients. In most patients, several different CIDRα1 types were expressed, but the dominant CIDRα1 subtypes varied from patient to patient. There was no association between disease outcome and the relative abundance of different CIDRα1 subtypes. The CIDRα1 subtype transcript patterns in 36 randomly selected patients are illustrated, with Fig. 3A to C showing 6 patients with parasitemia without fever, 6 with mild malaria, and 6 who were hospitalized with uncomplicated malaria, illustrating the increasing levels of CIDRα1 subtype transcripts with increasing disease severity, and Fig. 3D and E showing 6 patients with severe anemia, 6 with cerebral malaria, and 6 who died, illustrating that the majority of patients with severe disease had high transcript abundances of several CIDRα1 subtypes and that none of the subtypes were particularly dominant.
FIG 3.
Examples of levels (Tu) of var transcripts encoding different CIDRα1 subclasses in six children with parasitemia without ongoing fever (A), six children with mild malaria not requiring hospitalization (B), six children hospitalized with uncomplicated malaria (C), six children with severe malarial anemia (D), six children with cerebral malaria (E), and six children who succumbed to infection (F). Bars represent transcript levels reported by primer sets CIDRa1.1 (C1) (red), CIDRa1.8 (C8) (orange), CIDRa1.4/6a (c4) (blue), CIDRa1.5 (C5) (black), CIDRa1.6b (C6) (gray), and CIDRa1.7 (c7) (green). The red line indicates a Tu value of 16.
DISCUSSION
Here, we took advantage of the improved resolution of var sequence diversity gained through var genes extracted from over 200 recently sequenced P. falciparum genomes to design a new set of primers for quantitative PCR transcript analysis of var subclasses. These primers allowed unprecedented sensitivity and specificity in the detection and quantification of var transcripts encoding conserved sequence traits. In particular, var transcripts encoding group A PfEMP1 as well as transcripts encoding DBLε or DBLζ domains were well covered. Moreover, the coverage of specific DBLβ domains was improved. In general, the primers are predicted to underestimate the expression level of the targeted traits. This was particularly true for the primers targeting genes encoding CD36-binding CIDRα2-6, for which coverage was estimated to be 17%. In addition to the improved primer set, this study included a higher number of malaria patients and a broader spectrum of disease outcomes than in previous studies employing qPCR (25, 26, 31). Patient groups included children with parasitemia without fever, children with mild malaria who could be treated in the village, children who required hospitalization, children with severe disease manifestations, and children with a fatal infection outcome. Our study is based on the hypothesis that the PfEMP1 parasite phenotype is a determinant of disease outcome. Other factors relating to the patient (e.g., host genotype and health-seeking behavior) and parasite (e.g., drug resistance phenotype) will also contribute to the infection outcome. It should also be borne in mind that disease categorization was based on clinical presentation at diagnosis. Thus, parasites with a pathogenic phenotype may be detected in patients with mild symptoms who are diagnosed early. On the other hand, parasites with a nonpathogenic phenotype may be detected in patients for whom the symptoms are not caused by P. falciparum but by competing pathologies. A recent study from Malawi showed that more than 20% of patients classified as having cerebral malaria based on clinical criteria similar to those used in the present study were concluded to have died from causes other than malaria upon autopsy (35). Despite these inherent limitations, the median CIDRα1 transcript levels increased significantly with disease severity, being 50 to 100 times higher in those with severe disease than in those with parasitemia without fever. Unexpectedly, even patients with mild fever symptoms had increased expression levels of diverse, but in particular EPCR-binding, PfEMP1 compared to those in asymptomatic P. falciparum-infected individuals. This may imply that disease onset is associated with increased expression levels of, in particular, EPCR-binding PfEMP1; however, patient numbers were low, and more extensive studies are required to confirm this hypothesis. The transcript levels in children with severe malaria were similar to or exceeded the var2csa transcript level (encoding the PfEMP1-binding parasites in placenta) in parasites isolated from pregnant women (36, 37). The var2csa transcript levels were relatively high in one of four children regardless of disease severity. This could reflect that VAR2CSA was expressed on the infected erythrocytes of these children. However, the var2csa gene is unique among var genes in containing an upstream open reading frame, which can repress the translation of transcripts (38).
In line with previously reported observations (25, 26, 39), DC8 and group A PfEMP1 were highly expressed in most hospitalized children, and the abundance of transcripts encoding DC8 PfEMP1 was statistically significantly higher in patients with severe malaria than in those with uncomplicated disease. In contrast to data from previous reports, the difference in the abundances of transcripts encoding DC13 (containing CIDRα1.4) in patients with severe malaria or uncomplicated disease did not reach statistical significance. However, EPCR-binding group B (i.e., DC8) and group A PfEMP1 adhesins were estimated to be expressed at similar levels in patients with severe malaria. Moreover, transcripts for all EPCR-binding CIDRα1 subclasses were found to be highly expressed in individual patients, and in many patients, there were high levels of transcripts encoding several CIDRα1 subclasses. Thus, the development of severe malaria symptoms appears to be associated with EPCR-binding CIDRα1 in general. This is in line with the observation that most CIDRα1 variants bind EPCR with high affinity (28) and implies that a vaccine to protect children against severe malaria should target all or most CIDRα1 subtypes predicted to bind EPCR.
Due to the poor sensitivity of the detection of genes encoding CIDRα2-6 or CIDRβ/γ, the abundance of these transcripts might have been underestimated. However, a recent study by Jespersen et al. (29), which did not rely on specific var gene primers, showed that genes encoding CIDRα2-6- or CIDRβ/γ-containing PfEMP1 did not dominate var transcript profiles in patients with severe malaria. In the present study, the coverage of primers targeting genes encoding group A N-terminal CIDRδ domains was high, and in agreement with the results reported by Jespersen et al., the data showed that these transcripts were highly expressed in only a few patients with severe malaria. Together with data from early studies (20–24) linking group A PfEMP1 with severe disease, these studies suggest that parasites that bind to CD36, or unknown receptors through other group A-linked N-terminal CIDRβ/γ/δ subclasses, do not commonly precipitate severe malaria symptoms in young children.
In support of the suggested additive pathogenic effect of ICAM-1 binding (30), primers targeting group A DBLβ domains, some of which have been shown to bind ICAM-1, reported elevated transcript levels in severe malaria patients, in particular among those with cerebral complications. The median transcript levels were low, and a minority of patients with cerebral complications (24%) exhibited high expression levels of genes encoding PfEMP1 predicted to bind EPCR and ICAM-1. Both CD36- and EPCR-binding PfEMP1 adhesins can bind ICAM-1 (30, 32, 33, 40), but ICAM-1 and CD36 binding does not appear to be a common phenotype for parasites that cause severe malaria (29). Thus, EPCR binding by CIDRα1 domains appears to be required for the development of severe symptoms, but ICAM-1 binding and other host receptor interactions may, in some individuals (41), act in concert to strengthen cytoadhesion and aggravate disease. It is conceivable that this notion also explains the inconsistent association of the expression of diverse DBLζ variants with both uncomplicated (DBLζ4) (25) and severe (DBLζ2a and DBLζ3) (this study) pediatric malaria and severe malaria in adults (DBLζ5) (31).
It was previously reported that var expression levels differed among cerebral malaria patients with differing histopathologies (42); however, the present study could not address this. Future studies with a better resolution of cerebral manifestations, e.g., qualified by examination for retinopathy (43), are required to elucidate the suggested increased risk of development of cerebral complications when parasites can bind both EPCR and ICAM-1. However, our results suggest that circumstances relating to the patient, such as the expression of EPCR in different parts of the vasculature, regulation of the local inflammatory response, or prior immune priming, may be more important determinants of disease manifestation than the subtype of the EPCR-binding PfEMP1 gene that is being expressed.
In around one of five of the severe malaria patients in the present study, none of the primers used reported high levels of var transcripts (here defined as a transcript unit [Tu] value of >16). Although most primers used have target coverage below 100%, this could reflect that these patients suffered from infection with parasites expressing CD36-binding PfEMP1 or that the symptoms of some of these patients were not caused by malaria parasites.
Rosetting is a PfEMP1-mediated parasite phenotype previously associated with severe malaria (44–46). Rosetting appears to be mediated by some group A DBLα1 domains (47–49) but has also been suggested to be mediated by RIFIN (50) and STEVOR (51) proteins. As the PfEMP1 trait involved in the rosetting phenotype is yet to be resolved and cannot be predicted from domain subgrouping or amino acid sequence, this study cannot predict whether or to what degree, for example, DBLα1 domains adjacent to the EPCR-binding CIDRα1 domains contributed to pathology by mediating rosetting.
Data from this study in combination with previously reported observations from other studies of var transcription (25, 26, 29, 31) suggest that the expression of EPCR-binding PfEMP1 is the overarching parasite phenotype associated with the development of symptomatic and severe malaria and that its clinical relevance is stable across various confounding host or environmental factors. In line with this are the observations that parasites from children with severe disease and in vitro-adapted parasites expressing CIDRα1 PfEMP1 bind endothelial cells via EPCR (22, 52–54) and that CIDRα1 domains bind EPCR with high affinity, and binding inhibits the ability of EPCR to bind protein C, thereby potentially driving pathogenic endothelial inflammation (27, 28, 55–57). Finally, in regions where malaria is endemic, IgG to EPCR-binding CIDRα1 domains is acquired early in life and before antibodies to other classes of CIDR domains are acquired (54). Altogether, these findings argue that a vaccine that induces IgG to inhibit the PfEMP1-EPCR interaction could reduce severe disease and death due to P. falciparum and even reduce the number of malaria infections causing hospitalization. In addition, an adjunctive therapy aimed at alleviating the potential damaging consequences directly associated with the PfEMP1-EPCR interaction (58) may reduce the fatality rate or degree of sequelae for malaria patients.
MATERIALS AND METHODS
Sample collection.
Samples were collected from 187 children who were blood smear positive for P. falciparum. The children were enrolled after informed consent was obtained from a parent or legally acceptable guardian. Of the 187 children, 165 were admitted to either Korogwe District Hospital in northeast Tanzania (n = 110) or Magu District Hospital in the northwest (n = 55). Children were clinically evaluated by study clinicians, and a blood sample was collected for diagnostic and research purposes, after which treatment was instigated according to national guidelines. Samples were collected in 2013 and 2014. Blood samples from 22 nonhospitalized children living in Korogwe district who had mild malaria or malaria not accompanied by fever were also included in the study. These children were recruited as part of cross-sectional surveys in a village in 2007 and 2008. This study received ethical clearance and approval from the National Health Research Ethics Committee in Tanzania (reference no. NIMR/HQ/R.8c/Vol.II/436).
Primer design.
The sequence diversity of the different PfEMP1 domain classes differs from the relatively clear division of CIDRα1 domains into a few distinct subclasses to no particular subgrouping of DBLδ domains (17). For this reason, the design of informative primers was possible only for the best-defined domain subclasses (see Fig. S1 in the supplemental material). To maximize coverage while maintaining specificity for regions encoding specific domain subclasses, primers were designed based on full-length DBL and CIDR domain-encoding sequences from 7 P. falciparum genomes (17) and 226 Illumina whole-genome-sequenced P. falciparum field isolates (28, 59) (Fig. S1).
A particular effort was made to secure good coverage of group A var genes, in order to resolve which group A and DC8 var gene subclasses are associated with severe malaria. Group A PfEMP1 adhesins are characterized by having NTSA-DBLα1 and CIDRα1, -β, -γ, or -δ domains, where the NTSA sequences exhibit very little subgrouping, and the subclass of DBLα1 variants is predicted largely by the subgrouping of the following CIDR domain (17, 25). Group A CIDRα1.4-7 domains bind EPCR, whereas group A CIDRα1.2/3 domains are found in var1 pseudogenes, which do not bind EPCR (28). The functions of CIDRδ and the more diverse CIDRβ and -γ domains are unknown but have been suggested to be associated with rosetting (15). Group B and C PfEMP1 proteins have DBLα0- and CD36-binding CIDRα2-6 domains, apart from the atypical DC8-type group B PfEMP1 adhesins, which carry the DBLα2-CIDRα1.1/8 domains (EPCR-binding CIDR). Good-coverage primers (see Fig. S1 and S2 in the supplemental material) were successfully designed for all CIDRα1 and N-terminal CIDRδ domain subclasses, although the diversity of CIDRα2-6 and N-terminal CIDRβ and -γ domains was difficult to capture.
The unusually conserved PfEMP1 variants VAR2CSA (binding placental chondroitin sulfate A [CSA]) and VAR3 (unknown binding specificity) were targeted by specific DBL primers. Good coverage was also achieved for primers targeting C-terminal DBLε and DBLζ domains. ICAM-1 binding has been mapped to group A DBLβ1/3 and group B and C DBLβ5 domains. Primer sets with limited coverage but good specificity for DBLβ5 and two subsets of DBLβ1/3 (DBLb1/3-1 and DBLb1/3-2) were designed. For all primer sets, amplification efficiencies of >94% were ascertained by qPCR measurements of serial 10-fold dilutions of 3D7, HB3, and IT4/FCR3 genomic DNAs (gDNAs), and the predicted size or absence of PCR amplicons was validated by gel electrophoresis. The specificity and coverage of each primer set were evaluated in silico by using USEARCH (“search_pcr”) (60) against full-length DBL and CIDR domains extracted from the 233 genome sequences, calling targets by allowing up to 2 mismatches in each primer. The result was manually parsed to remove hits with 3′-terminal mismatches to either primer and to remove all but one (best) hit for each domain/contig (multiple reports of an amplicon from the same locus may be generated when degenerate primers are employed). These criteria were previously found to give good estimates of target amplification (25).
In summary, good specificity and coverage were achieved for primer sets capturing transcripts encoding subclasses of CIDRα1 as well as CIDRδ, VAR2CSA, and VAR3. The coverage for var sequences encoding DBLζ and DBLε domains was also good, while the coverage for CIDRγ, CD36-binding CIDRα2-6, and DBLβ was low.
Parasite RNA and qPCR.
Erythrocytes (50 to 100 μl) pelleted by centrifugation from venous blood samples were completely dissolved in 1 ml TRIzol reagent (Invitrogen) and stored in liquid nitrogen, in dry ice, or at −80°C until RNA purification was performed. Total RNA was extracted, treated with DNase (DNase I; Sigma-Aldrich), and verified for lack of residual genomic DNA by qPCR using primers for the endogenous seryl-tRNA synthetase housekeeping gene before being reverse transcribed (Superscript II; Invitrogen) as described previously (25). qPCR analyses were performed by using QuantiTect SYBR green PCR master mix (Qiagen). Master mix was distributed into primer-loaded tubes for qPCR performed in 20-μl reaction mixtures by using the Rotorgene thermal cycler system (Corbett Research) and cycling conditions described previously (25).
var transcript abundances were determined in relation to the averaged transcript abundances of the endogenous seryl-tRNA synthetase and aldolase housekeeping genes (ΔCTvar_primer = CTvar_primer − CTaverage_control primers). ΔCTvar_primer values were translated into transcript units [Tu = 2(5−ΔCt)] (25), where low-abundance transcripts with threshold cycle (ΔCT) values of >5 were all assigned a ΔCT value of 5 (Tu = 1).
For some primer sets, the reported transcript abundance was summarized by adding the reported transcript units for each primer set and subtracting a value of 1 for each additional value added (subtracting was done to ensure that the summarized Tu value was equal to 1 if no signal was detected for any of the transcripts summarized).
Due to the differences in sequence-type coverage between primer sets, and the unknown sequence diversity of the targeted genes in each sample leading to variations in primer set sensitivities between samples, exact estimates of the relative expression levels of var gene types within individual samples cannot be made. However, reported transcript levels can be used to make rough assessments of the relative transcript abundances of different var types within a patient or in patient groups (25). A transcript abundance value of 16 was chosen across primers to reflect a high level of transcripts and to ease data interpretation. This threshold was chosen based on the var2csa transcript level measured in parasites from pregnant women and knowledge about transcript levels in cultured parasite lines selected to predominantly express a PfEMP1 adhesin.
Statistical analyses.
Quantitative comparisons of transcript levels were done separately for each primer set or groups of primers between patient groups by using a Kruskal-Wallis rank sum test or a Wilcoxon rank sum test using Stata statistical software. Fisher's exact test was used to test proportions of patients with high levels (Tu > 16) of selected transcripts.
There was no statistically significant difference in the transcript abundances measured in Korogwe and Magu (data not shown). Hence, data from the two sites were pooled.
Supplementary Material
ACKNOWLEDGMENTS
We are deeply grateful to the Tanzanian donors.
This work received financial support from the Augustinus Fonden, the Lundbeckfonden, the Axel Muusfeldts Fond, the Grosserer L. F. Foghts Fond, the Danish International Development Agency (DANIDA), and the Danish Council for Independent Research (grants T1333-00220 1331-00089B and Sapere Aude program DFF-4004-00624B). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00841-16.
REFERENCES
- 1.Marsh K, Forster D, Waruiru C, Mwangi I, Winstanley M, Marsh V, Newton C, Winstanley P, Warn P, Peshu N, Pasvol G, Snow R. 1995. Indicators of life-threatening malaria in African children. N Engl J Med 332:1399–1404. doi: 10.1056/NEJM199505253322102. [DOI] [PubMed] [Google Scholar]
- 2.Okiro EA, Al-Taiar A, Reyburn H, Idro R, Berkley JA, Snow RW. 2009. Age patterns of severe paediatric malaria and their relationship to Plasmodium falciparum transmission intensity. Malar J 8:4. doi: 10.1186/1475-2875-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Seidlein L, Olaosebikan R, Hendriksen IC, Lee SJ, Adedoyin OT, Agbenyega T, Nguah SB, Bojang K, Deen JL, Evans J, Fanello CI, Gomes E, Pedro AJ, Kahabuka C, Karema C, Kivaya E, Maitland K, Mokuolu OA, Mtove G, Mwanga-Amumpaire J, Nadjm B, Nansumba M, Ngum WP, Onyamboko MA, Reyburn H, Sakulthaew T, Silamut K, Tshefu AK, Umulisa N, Gesase S, Day NP, White NJ, Dondorp AM. 2012. Predicting the clinical outcome of severe falciparum malaria in African children: findings from a large randomized trial. Clin Infect Dis 54:1080–1090. doi: 10.1093/cid/cis034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GBD 2015 Mortality and Causes of Death Collaborators. 2016. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goncalves BP, Huang CY, Morrison R, Holte S, Kabyemela E, Prevots DR, Fried M, Duffy PE. 2014. Parasite burden and severity of malaria in Tanzanian children. N Engl J Med 370:1799–1808. doi: 10.1056/NEJMoa1303944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh K, Howard RJ. 1986. Antigens induced on erythrocytes by P. falciparum: expression of diverse and conserved determinants. Science 231:150–153. doi: 10.1126/science.2417315. [DOI] [PubMed] [Google Scholar]
- 7.Bull PC, Lowe BS, Kortok M, Molyneux CS, Newbold CI, Marsh K. 1998. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat Med 4:358–360. doi: 10.1038/nm0398-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen MA, Staalsoe T, Kurtzhals JA, Goka BQ, Dodoo D, Alifrangis M, Theander TG, Akanmori BD, Hviid L. 2002. Plasmodium falciparum variant surface antigen expression varies between isolates causing severe and nonsevere malaria and is modified by acquired immunity. J Immunol 168:3444–3450. doi: 10.4049/jimmunol.168.7.3444. [DOI] [PubMed] [Google Scholar]
- 9.Bull PC, Kortok M, Kai O, Ndungu F, Ross A, Lowe BS, Newbold CI, Marsh K. 2000. Plasmodium falciparum-infected erythrocytes: agglutination by diverse Kenyan plasma is associated with severe disease and young host age. J Infect Dis 182:252–259. doi: 10.1086/315652. [DOI] [PubMed] [Google Scholar]
- 10.Leech JH, Barnwell JW, Miller LH, Howard RJ. 1984. Identification of a strain-specific malarial antigen exposed on the surface of Plasmodium falciparum-infected erythrocytes. J Exp Med 159:1567–1575. doi: 10.1084/jem.159.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. 1995. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 12.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82:101–110. doi: 10.1016/0092-8674(95)90056-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 14.Scherf A, Lopez-Rubio JJ, Riviere L. 2008. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol 62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 15.Smith JD, Rowe JA, Higgins MK, Lavstsen T. 2013. Malaria's deadly grip: cytoadhesion of Plasmodium falciparum-infected erythrocytes. Cell Microbiol 15:1976–1983. doi: 10.1111/cmi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavstsen T, Salanti A, Jensen AT, Arnot DE, Theander TG. 2003. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar J 2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T. 2010. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes—divide and conquer. PLoS Comput Biol 6:e1000933. doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, Hviid L, Theander TG. 2003. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol 49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 19.Salanti A, Dahlback M, Turner L, Nielsen MA, Barfod L, Magistrado P, Jensen AT, Lavstsen T, Ofori MF, Marsh K, Hviid L, Theander TG. 2004. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J Exp Med 200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warimwe GM, Keane TM, Fegan G, Musyoki JN, Newton CR, Pain A, Berriman M, Marsh K, Bull PC. 2009. Plasmodium falciparum var gene expression is modified by host immunity. Proc Natl Acad Sci U S A 106:21801–21806. doi: 10.1073/pnas.0907590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Normark J, Nilsson D, Ribacke U, Winter G, Moll K, Wheelock CE, Bayarugaba J, Kironde F, Egwang TG, Chen Q, Andersson B, Wahlgren M. 2007. PfEMP1-DBL1alpha amino acid motifs in severe disease states of Plasmodium falciparum malaria. Proc Natl Acad Sci U S A 104:15835–15840. doi: 10.1073/pnas.0610485104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen AT, Magistrado P, Sharp S, Joergensen L, Lavstsen T, Chiucchiuini A, Salanti A, Vestergaard LS, Lusingu JP, Hermsen R, Sauerwein R, Christensen J, Nielsen MA, Hviid L, Sutherland C, Staalsoe T, Theander TG. 2004. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J Exp Med 199:1179–1190. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rottmann M, Lavstsen T, Mugasa JP, Kaestli M, Jensen AT, Muller D, Theander T, Beck HP. 2006. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect Immun 74:3904–3911. doi: 10.1128/IAI.02073-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyriacou HM, Stone GN, Challis RJ, Raza A, Lyke KE, Thera MA, Kone AK, Doumbo OK, Plowe CV, Rowe JA. 2006. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol 150:211–218. doi: 10.1016/j.molbiopara.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavstsen T, Turner L, Saguti F, Magistrado P, Rask TS, Jespersen JS, Wang CW, Berger SS, Baraka V, Marquard AM, Seguin-Orlando A, Willerslev E, Gilbert MT, Lusingu J, Theander TG. 2012. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci U S A 109:E1791–E1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertin GI, Lavstsen T, Guillonneau F, Doritchamou J, Wang CW, Jespersen JS, Ezimegnon S, Fievet N, Alao MJ, Lalya F, Massougbodji A, Ndam NT, Theander TG, Deloron P. 2013. Expression of the domain cassette 8 Plasmodium falciparum erythrocyte membrane protein 1 is associated with cerebral malaria in Benin. PLoS One 8:e68368. doi: 10.1371/journal.pone.0068368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JE, Avril M, Brazier AJ, Freeth J, Jespersen JS, Nielsen MA, Magistrado P, Lusingu J, Smith JD, Higgins MK, Theander TG. 2013. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature 498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau CK, Turner L, Jespersen JS, Lowe ED, Petersen B, Wang CW, Petersen JE, Lusingu J, Theander TG, Lavstsen T, Higgins MK. 2015. Structural conservation despite huge sequence diversity allows EPCR binding by the PfEMP1 family implicated in severe childhood malaria. Cell Host Microbe 17:118–129. doi: 10.1016/j.chom.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jespersen JS, Wang CW, Mkumbaye SI, Minja DT, Petersen B, Turner L, Petersen JE, Lusingu JP, Theander TG, Lavstsen T. 2016. Plasmodium falciparum var genes expressed in children with severe malaria encode CIDRalpha1 domains. EMBO Mol Med 8:839–850. doi: 10.15252/emmm.201606188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avril M, Bernabeu M, Benjamin M, Brazier AJ, Smith JD. 2016. Interaction between endothelial protein C receptor and intercellular adhesion molecule 1 to mediate binding of Plasmodium falciparum-infected erythrocytes to endothelial cells. mBio 7:e00615-16. doi: 10.1128/mBio.00615-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernabeu M, Danziger SA, Avril M, Vaz M, Babar PH, Brazier AJ, Herricks T, Maki JN, Pereira L, Mascarenhas A, Gomes E, Chery L, Aitchison JD, Rathod PK, Smith JD. 2016. Severe adult malaria is associated with specific PfEMP1 adhesion types and high parasite biomass. Proc Natl Acad Sci U S A 113:E3270–E3279. doi: 10.1073/pnas.1524294113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janes JH, Wang CP, Levin-Edens E, Vigan-Womas I, Guillotte M, Melcher M, Mercereau-Puijalon O, Smith JD. 2011. Investigating the host binding signature on the Plasmodium falciparum PfEMP1 protein family. PLoS Pathog 7:e1002032. doi: 10.1371/journal.ppat.1002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bengtsson A, Joergensen L, Rask TS, Olsen RW, Andersen MA, Turner L, Theander TG, Hviid L, Higgins MK, Craig A, Brown A, Jensen AT. 2013. A novel domain cassette identifies Plasmodium falciparum PfEMP1 proteins binding ICAM-1 and is a target of cross-reactive, adhesion-inhibitory antibodies. J Immunol 190:240–249. doi: 10.4049/jimmunol.1202578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berger SS, Turner L, Wang CW, Petersen JE, Kraft M, Lusingu JP, Mmbando B, Marquard AM, Bengtsson DB, Hviid L, Nielsen MA, Theander TG, Lavstsen T. 2013. Plasmodium falciparum expressing domain cassette 5 type PfEMP1 (DC5-PfEMP1) bind PECAM1. PLoS One 8:e69117. doi: 10.1371/journal.pone.0069117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milner DA Jr, Whitten RO, Kamiza S, Carr R, Liomba G, Dzamalala C, Seydel KB, Molyneux ME, Taylor TE. 2014. The systemic pathology of cerebral malaria in African children. Front Cell Infect Microbiol 4:104. doi: 10.3389/fcimb.2014.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuikue Ndam NG, Salanti A, Bertin G, Dahlback M, Fievet N, Turner L, Gaye A, Theander T, Deloron P. 2005. High level of var2csa transcription by Plasmodium falciparum isolated from the placenta. J Infect Dis 192:331–335. doi: 10.1086/430933. [DOI] [PubMed] [Google Scholar]
- 37.Tuikue Ndam N, Bischoff E, Proux C, Lavstsen T, Salanti A, Guitard J, Nielsen MA, Coppee JY, Gaye A, Theander T, David PH, Deloron P. 2008. Plasmodium falciparum transcriptome analysis reveals pregnancy malaria associated gene expression. PLoS One 3:e1855. doi: 10.1371/journal.pone.0001855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amulic B, Salanti A, Lavstsen T, Nielsen MA, Deitsch KW. 2009. An upstream open reading frame controls translation of var2csa, a gene implicated in placental malaria. PLoS Pathog 5:e1000256. doi: 10.1371/journal.ppat.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdi AI, Kariuki SM, Muthui MK, Kivisi CA, Fegan G, Gitau E, Newton CR, Bull PC. 2015. Differential Plasmodium falciparum surface antigen expression among children with malarial retinopathy. Sci Rep 5:18034. doi: 10.1038/srep18034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gullingsrud J, Saveria T, Amos E, Duffy PE, Oleinikov AV. 2013. Structure-function-immunogenicity studies of PfEMP1 domain DBL2betaPF11_0521, a malaria parasite ligand for ICAM-1. PLoS One 8:e61323. doi: 10.1371/journal.pone.0061323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magallon-Tejada A, Machevo S, Cistero P, Lavstsen T, Aide P, Rubio M, Jimenez A, Turner L, Valmaseda A, Gupta H, De Las Salas B, Mandomando I, Wang CW, Petersen JE, Munoz J, Gascon J, Macete E, Alonso PL, Chitnis CE, Bassat Q, Mayor A. 2016. Cytoadhesion to gC1qR through Plasmodium falciparum erythrocyte membrane protein 1 in severe malaria. PLoS Pathog 12:e1006011. doi: 10.1371/journal.ppat.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tembo DL, Nyoni B, Murikoli RV, Mukaka M, Milner DA, Berriman M, Rogerson SJ, Taylor TE, Molyneux ME, Mandala WL, Craig AG, Montgomery J. 2014. Differential PfEMP1 expression is associated with cerebral malaria pathology. PLoS Pathog 10:e1004537. doi: 10.1371/journal.ppat.1004537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrera V, Hiscott PS, Craig AG, White VA, Milner DA, Beare NA, MacCormick IJ, Kamiza S, Taylor TE, Molyneux ME, Harding SP. 2015. Severity of retinopathy parallels the degree of parasite sequestration in the eyes and brains of Malawian children with fatal cerebral malaria. J Infect Dis 211:1977–1986. doi: 10.1093/infdis/jiu592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlson J, Helmby H, Hill AV, Brewster D, Greenwood BM, Wahlgren M. 1990. Human cerebral malaria: association with erythrocyte rosetting and lack of anti-rosetting antibodies. Lancet 336:1457–1460. doi: 10.1016/0140-6736(90)93174-N. [DOI] [PubMed] [Google Scholar]
- 45.Rowe A, Obeiro J, Newbold CI, Marsh K. 1995. Plasmodium falciparum rosetting is associated with malaria severity in Kenya. Infect Immun 63:2323–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ockenhouse CF, Ho M, Tandon NN, Van Seventer GA, Shaw S, White NJ, Jamieson GA, Chulay JD, Webster HK. 1991. Molecular basis of sequestration in severe and uncomplicated Plasmodium falciparum malaria: differential adhesion of infected erythrocytes to CD36 and ICAM-1. J Infect Dis 164:163–169. doi: 10.1093/infdis/164.1.163. [DOI] [PubMed] [Google Scholar]
- 47.Rowe JA, Moulds JM, Newbold CI, Miller LH. 1997. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature 388:292–295. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- 48.Ghumra A, Semblat JP, Ataide R, Kifude C, Adams Y, Claessens A, Anong DN, Bull PC, Fennell C, Arman M, Amambua-Ngwa A, Walther M, Conway DJ, Kassambara L, Doumbo OK, Raza A, Rowe JA. 2012. Induction of strain-transcending antibodies against group A PfEMP1 surface antigens from virulent malaria parasites. PLoS Pathog 8:e1002665. doi: 10.1371/journal.ppat.1002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Angeletti D, Albrecht L, Blomqvist K, Quintana MDP, Akhter T, Bachle SM, Sawyer A, Sandalova T, Achour A, Wahlgren M, Moll K. 2012. Plasmodium falciparum rosetting epitopes converge in the SD3-loop of PfEMP1-DBL1alpha. PLoS One 7:e50758. doi: 10.1371/journal.pone.0050758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goel S, Palmkvist M, Moll K, Joannin N, Lara P, Akhouri RR, Moradi N, Ojemalm K, Westman M, Angeletti D, Kjellin H, Lehtio J, Blixt O, Idestrom L, Gahmberg CG, Storry JR, Hult AK, Olsson ML, von Heijne G, Nilsson I, Wahlgren M. 2015. RIFINs are adhesins implicated in severe Plasmodium falciparum malaria. Nat Med 21:314–317. doi: 10.1038/nm.3812. [DOI] [PubMed] [Google Scholar]
- 51.Niang M, Bei AK, Madnani KG, Pelly S, Dankwa S, Kanjee U, Gunalan K, Amaladoss A, Yeo KP, Bob NS, Malleret B, Duraisingh MT, Preiser PR. 2014. STEVOR is a Plasmodium falciparum erythrocyte binding protein that mediates merozoite invasion and rosetting. Cell Host Microbe 16:81–93. doi: 10.1016/j.chom.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avril M, Tripathi AK, Brazier AJ, Andisi C, Janes JH, Soma VL, Sullivan DJ Jr, Bull PC, Stins MF, Smith JD. 2012. A restricted subset of var genes mediates adherence of Plasmodium falciparum-infected erythrocytes to brain endothelial cells. Proc Natl Acad Sci U S A 109:E1782–E1790. doi: 10.1073/pnas.1120534109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Claessens A, Adams Y, Ghumra A, Lindergard G, Buchan CC, Andisi C, Bull PC, Mok S, Gupta AP, Wang CW, Turner L, Arman M, Raza A, Bozdech Z, Rowe JA. 2012. A subset of group A-like var genes encodes the malaria parasite ligands for binding to human brain endothelial cells. Proc Natl Acad Sci U S A 109:E1772–E1781. doi: 10.1073/pnas.1120461109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner L, Lavstsen T, Mmbando BP, Wang CW, Magistrado PA, Vestergaard LS, Ishengoma DS, Minja DT, Lusingu JP, Theander TG. 2015. IgG antibodies to endothelial protein C receptor-binding cysteine-rich interdomain region domains of Plasmodium falciparum erythrocyte membrane protein 1 are acquired early in life in individuals exposed to malaria. Infect Immun 83:3096–3103. doi: 10.1128/IAI.00271-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petersen JE, Bouwens EA, Tamayo I, Turner L, Wang CW, Stins M, Theander TG, Hermida J, Mosnier LO, Lavstsen T. 2015. Protein C system defects inflicted by the malaria parasite protein PfEMP1 can be overcome by a soluble EPCR variant. Thromb Haemost 114:1038–1048. doi: 10.1160/TH15-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gillrie MR, Avril M, Brazier AJ, Davis SP, Stins MF, Smith JD, Ho M. 2015. Diverse functional outcomes of Plasmodium falciparum ligation of EPCR: potential implications for malarial pathogenesis. Cell Microbiol 17:1883–1899. doi: 10.1111/cmi.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sampath S, Brazier AJ, Avril M, Bernabeu M, Vigdorovich V, Mascarenhas A, Gomes E, Sather DN, Esmon CT, Smith JD. 2015. Plasmodium falciparum adhesion domains linked to severe malaria differ in blockade of endothelial protein C receptor. Cell Microbiol 17:1868–1882. doi: 10.1111/cmi.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mosnier LO, Lavstsen T. 2016. The role of EPCR in the pathogenesis of severe malaria. Thromb Res 141(Suppl 2):S46–S49. doi: 10.1016/S0049-3848(16)30364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, Maslen G, O'Brien J, Djimde A, Doumbo O, Zongo I, Ouedraogo JB, Michon P, Mueller I, Siba P, Nzila A, Borrmann S, Kiara SM, Marsh K, Jiang H, Su XZ, Amaratunga C, Fairhurst R, Socheat D, Nosten F, Imwong M, White NJ, Sanders M, Anastasi E, Alcock D, Drury E, Oyola S, Quail MA, Turner DJ, Ruano-Rubio V, Jyothi D, Amenga-Etego L, Hubbart C, Jeffreys A, Rowlands K, Sutherland C, Roper C, Mangano V, Modiano D, Tan JC, Ferdig MT, Amambua-Ngwa A, Conway DJ, Takala-Harrison S, Plowe CV, Rayner JC, et al. 2012. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487:375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.